Abstract
Small nucleolar RNAs (snoRNAs) guiding modifications of ribosomal RNAs and other RNAs display diverse modes of gene organization and expression depending on the eukaryotic system: in animals most are intron encoded, in yeast many are monocistronic genes and in plants most are polycistronic (independent or intronic) genes. Here we report an unprecedented organization: plant dicistronic tRNA–snoRNA genes. In Arabidopsis thaliana we identified a gene family encoding 12 novel box C/D snoRNAs (snoR43) located just downstream from tRNAGly genes. We confirmed that they are transcribed, probably from the tRNA gene promoter, producing dicistronic tRNAGly–snoR43 precursors. Using transgenic lines expressing a tagged tRNA–snoR43.1 gene we show that the dicistronic precursor is accurately processed to both snoR43.1 and tRNAGly. In addition, we show that a recombinant RNase Z, the plant tRNA 3′ processing enzyme, efficiently cleaves the dicistronic precursor in vitro releasing the snoR43.1 from the tRNAGly. Finally, we describe a similar case in rice implicating a tRNAMet-e expressed in fusion with a novel C/D snoRNA, showing that this mode of snoRNA expression is found in distant plant species.
Keywords: RNA processing/RNase Z/transcription/tRNA–snoRNA dicistronic genes
Introduction
In the last decade small nucleolar RNAs (snoRNAs) directing modification of ribosomal RNAs (rRNA), U RNAs and tRNAs have been identified in all eukaryotes and in Archaea. They fall into two major classes: the C/D snoRNAs guiding 2′-O-ribose methylation and the H/ACA snoRNA guiding pseudo-uridylation. The C/D snoRNAs have two phylogenetically conserved C (RUGAUGA) and D (CUGA) motifs flanked by short inverted repeats at the 5′ and 3′ termini of the snoRNA, respectively. These structural elements are essential for snoRNA stability and nucleolar accumulation. Adjacent to the terminal box D or to an additional internal box D′ there is an antisense sequence of 10–21 bases that forms an snoRNA/rRNA duplex selecting the targeted nucleotide. In vivo, all C/D snoRNAs are found within a snoRNP associated with fibrillarin, the methylase, and three other conserved core proteins identified in yeast (reviewed by Terns and Terns, 2002; Filipowicz and Pogacic, 2002).
The snoRNAs have diverse modes of expression depending on the organism. In vertebrates most snoRNAs are nested within introns of protein coding genes and are produced by exonucleolytic processing of the debranched intron (Kiss and Filipowicz, 1995). An alternate pathway implicates direct endonucleolytic cleavage of the unspliced mRNA to release the intronic snoRNA (Villa et al., 1998; Giorgi et al., 2001). Usually host genes encode abundant house-keeping proteins related to ribosome biogenesis but there are exceptional cases of non-coding genes expressing intronic snoRNAs (Tycowski et al., 1996; Pelczar and Filipowicz, 1998). In yeast, many snoRNAs are encoded by independent monocistronic genes, but there are also five polycistronic genes and a few intronic snoRNAs (Lowe and Eddy, 1999). In Archaea many C/D RNAs guiding rRNA methylation have been identified encoded by individual genes (Gaspin et al., 2000; Omer et al., 2000). Interestingly, one exceptional case is an archaeal C/D snRNA nested in an intron of pre-tRNATrp that guides methylation in cis of the same tRNATrp (Clouet d’Orval et al., 2001).
In plants, early biochemical data revealed that rRNAs are highly modified implicating overall 120 2′-O-ribose methylated residues (Lau et al., 1974; Cecchini and Miassod, 1979). The mechanisms guiding rRNA methylation depending on fibrillarin and C/D snoRNAs are conserved in plants (Barneche et al., 2000). Recently, three independent groups using computer-assisted approaches identified a total of 117 different C/D snoRNA genes expressed in Arabidopsis thaliana (Barneche et al., 2001; Brown et al., 2001; Qu et al., 2001). Moreover, nearly 50% of these snoRNAs display multiple isoforms resulting from gene and chromosomal duplication events which generated a massive expansion of snoRNA genes, a situation unique to plants (Barneche et al., 2001; Brown et al., 2001).
The plant snoRNAs are also distinguished by their modes of expression. Most Arabidopsis snoRNA genes are organized in independent clusters transcribed from a single promoter producing a polycistronic precursor, as first described for the C/D snoRNA U14 (Leader et al., 1997). The polycistronic precursor is then processed by endonucleolytic cleavages and exonucleolytic trimming to release the mature snoRNAs (Leader et al., 1999). Intronic snoRNAs also exist in Arabidopsis but are not frequent. Remarkably, three clusters were found within introns of protein coding genes (Barneche et al., 2001; Brown et al., 2001). These intronic clusters are unique to plants and occur frequently in rice, a monocotyledonous species (Liang et al., 2002). Finally, some snoRNAs have been annotated as monocistronic genes. Some of these could be intronic species disguised by the poor annotation of plant genes, but others probably represent independent genes expressed from their own promoters. This has been demonstrated for U3, a phylogenetically conserved C/D snoRNA implicated in pre-rRNA cleavage; however, in plants and the green alga Chlamydomonas reinhardii, U3 genes are transcribed by Pol III and not by Pol II as in the other eukaryotes (Kiss et al., 1991; Antal et al., 2000).
These observations show that plants have evolved specific modes of snoRNA gene expression. Here we report the existence of dicistronic tRNA–snoRNA genes in Arabidopsis and rice. We describe 12 novel C/D snoRNAs located at the 3′ end of the cognate tRNAGly gene isoforms in Arabidopsis and a similar organization implicating two novel C/D snoRNAs abutting tRNAMet-e genes in rice. The tight link between a tRNA and a snoRNA adds to the extraordinary diversity of snoRNA gene organization in plants and further illustrates how the expansion of snoRNAs implicated a colonization of all type of genes for expression.
Results
A family of C/D snoRNAs abuts tRNAGly genes in A.thaliana
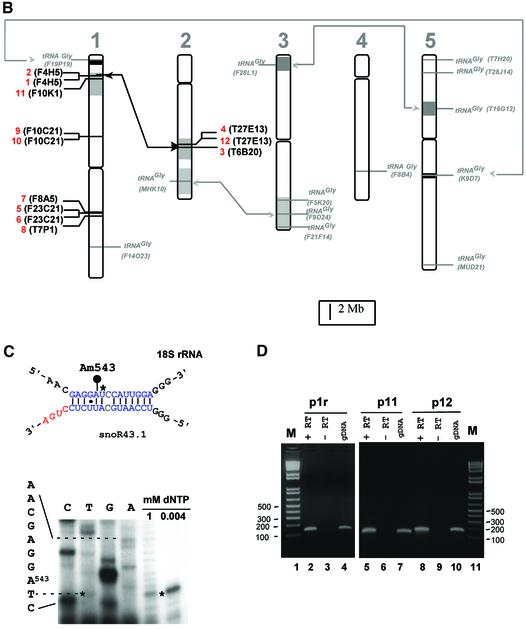
To unify the nomenclature of the Arabidopsis snoRNAs (Brown et al., 2001) the previously identified snoR34.1 (Barneche et al., 2001) was renamed snoR43.1 (standing for snoRNA 43 isoform 1). SnoR43.1 has all the functional elements of methylation guide C/D snoRNAs, i.e. inverted repeats flanking perfectly conserved C and D boxes, and a 14 nt antisense rRNA element associated with an internal D′ box (Figure 1A and C). The antisense element predicted to target 18S:A543 has one mismatch which should not impair its methylation guide function (Cavaillé and Bachellerie, 1998). Nucleotide A543 is conserved and methylated in both human and yeast 18S rRNAs, targeted by U62 and snR41, respectively (Bachellerie and Cavaillé, 1998; Lowe and Eddy, 1999). Using a dNTP-dependent reverse transcription assay (Maden, 2001) we confirmed that A543 is methylated in the 18S rRNA (Figure 1C), suggesting that snoR43.1 is functional in vivo. Nevertheless, in Arabidopsis, where snoRNA redundancy is common, 18S:A543 is also targeted by another C/D snoRNA, AtsnoR41Y (Barneche et al., 2001; Brown et al., 2001). Therefore direct evidence for the guide function of snoR43.1 in 18S:A543 methylation would require inactivation of its gene.
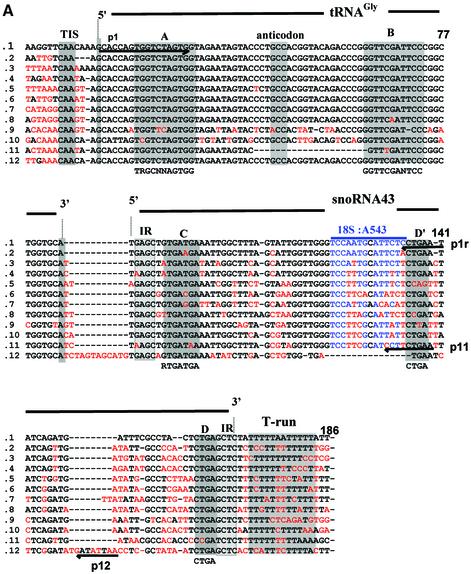
Fig. 1. Genomic organization of the tsnoR43 dicistronic genes. (A) Sequence alignment of the tsnoR43 genes. The tRNAGly sequences are in the Arabidopsis tRNA databanks (see Materials and methods for addresses). SnoRNAs were predicted by SnoScan (Lowe and Eddy, 1999). Divergent nucleotides are in red. Grey boxes indicate functional elements on the tRNA and snoRNA. Consensus sequences for tRNA A and B boxes (Geiduschek and Tocchini-Valentini, 1988) and snoRNA C and D boxes (Bachellerie and Cavaillé, 1998) are indicated below. The blue overline indicates the rRNA antisense element of snoR43.1 targeting 18S:A543 and blue nucleotides display rRNA/snoRNA pairing. IR indicates inverted repeats forming a terminal stem on C/D snoRNAs. The RT–PCR sense primer p1 annealing to the tRNA and the 5′ ends of the reverse primers p1r, p11 and p12 are underlined with arrows. (B) Chromosomal location of tsnoR43 genes. The 12 dicistronic tsnoR43 genes are shown in red. The 13 single tRNAGly genes are shown in grey. The name of the BAC containing these genes is given in parentheses in each case. Lines link isoforms located in large chromosomal duplications shown by the shaded regions (Blanc et al., 2000). (C) The 18S:A543 is 2′-O-methylated in the 18S rRNA. Methylation of 18S:A543 was detected by primer extension on total RNA at low dNTP concentration (Maden, 2001). Prematurely reverse transcriptase arrest one nucleotide before the methylated residue is shown by an asterisk. The 18S rRNA/snoR43.1 duplex is shown, and the target A543 is indicated by a black circle. (D) Expression of tsnoR43 genes detected by RT–PCR. The RT–PCR on total RNA from Arabidopsis seedlings was performed with p1 and each of the reverse primers as indicated: +RT and –RT refer to the presence and absence, respectively, of reverse transcriptase in the RT–PCR assay. The gDNA is a PCR reaction performed with genomic DNA as template for a size control to compare with the RT–PCR product. M is a DNA 100 bp ladder.
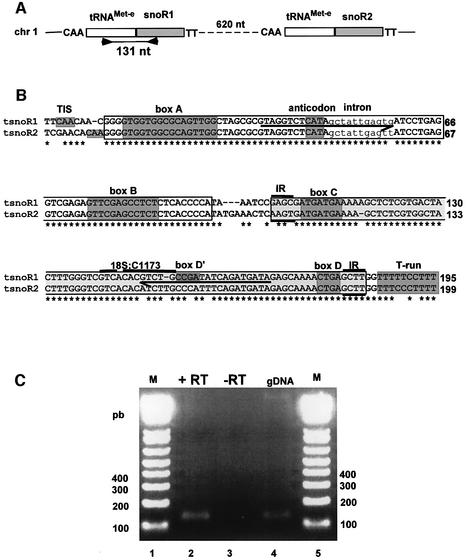
The sequence coding for snoR43.1 is located at the 3′ end of a tRNAGly gene in chromosome 1. The 5′ end of the C/D snoRNA predicted by the inverted repeat abuts the 3′ end of a tRNAGly (Figures 1A and 6). This tRNAGly is predicted to be a cytosolic isoacceptor with a GCC anticodon as reported in databanks (see Materials and methods for addresses). This tRNA–snoRNA locus has all the elements required for Pol III directed transcription of tRNA genes: the internal promoter A and B boxes, a TTA-rich instability region (see Figure 4A) upstream from a CAA element which is the transcription initiation site (TIS) in all plant tRNA genes and a T-run putative terminator of Pol III transcription found just downstream of the predicted 3′ end of the snoRNA (Geiduschek and Tocchini-Valentini, 1988; Yukawa et al., 2000). These structural data suggest that snoR43.1 is transcribed from the tRNA gene promoter and is synthesized in fusion with the tRNAGly. We called this dicistronic tRNA–snoRNA locus a tsnoRNA gene.
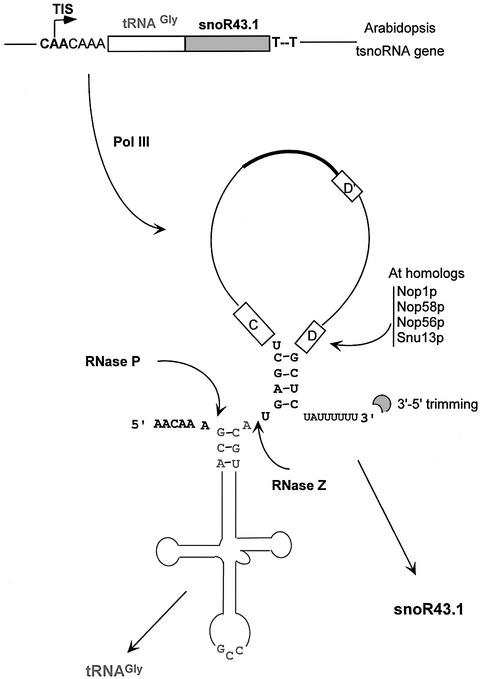
Fig. 6. Model for the expression of tRNA–snoRNA dicistronic genes in plants. The different activities predicted as participating in gene transcription and processing of the tRNAGly–snoR43.1 precursor are indicated. The assembly with the core conserved C/D snoRNP proteins may occur prior to 3′ end trimming to protect snoRNA from exonucleolytic degradation (Terns and Terns, 2002).
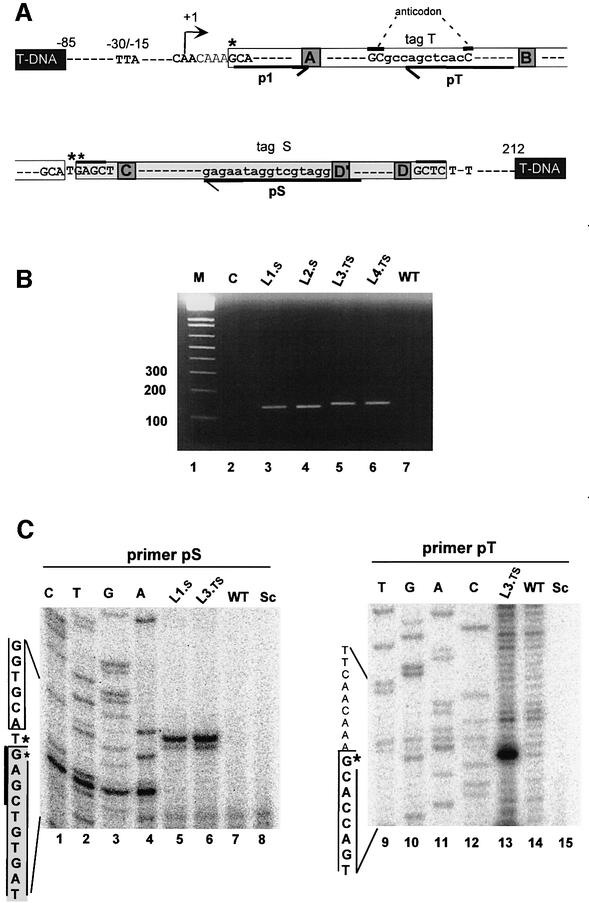
Fig. 4. Expression of tagged tRNAGly.t–snoR43.s in transgenic Arabidopsis lines. (A) Structure of the tagged tRNAGly.t–snoR43.s transgene. The TIS, nucleotide +1, is indicated by an arrow. The conserved elements controlling tRNA gene expression and snoRNA accumulation are indicated. White and grey boxes indicate the mature tRNA and snoRNA with their terminal sequences, respectively. The terminal inverted repeats of snoR43.1 are indicated by thick overlines. The tag t inserted in the GCC anticodon and the tag s replacing the antisense rRNA element adjacent to the D′ box are in lower-case letters. Black boxes indicate T-DNA flanking sequences. Four transgenic lines were produced: s-tagged L1.s and L2.s and ts-tagged L3.ts and L4.ts. Primers used for RT–PCR and primer extension analysis are indicated by p1, pt and ps. The asterisks indicate the positions of the primer extension signals detected in (C). (B) Detection of the dicistronic tagged precursors by RT–PCR. The assay was performed using primers p1/ps on total RNA extracted from transgenic lines L1.s, L2.s, L3.ts and L4.ts or wild-type (WT) non-transgenic plants as indicated. C is a control RT–PCR using total RNA from L1.s as substrate but omitting reverse transcriptase in the reaction. M indicates 100 bp DNA size markers. (C) Detection of tagged AtsnoR43.s and tRNAGly.ts by primer extension. Reverse transcription was performed with primers ps or pt and total RNA from transgenic lines L1.s and L3.ts and non-transgenic lines WT, as indicated. Sc is a control with yeast tRNA. CTAG is the sequence ladder of tRNAGlyt–snoR43.s gene. The tRNA and snoRNA sequences are indicated by white and grey boxes, respectively, on the left of the gels. The asterisks indicate the position of the extension signals.
In Arabidopsis 23 genes encode cytosolic tRNAGly isoaccceptors with the GCC anticodon (http://rna.wustl. edu/GtRDB/At/). Alignment of their flanking sequences revealed that nine of them were associated with a predicted C/D snoRNA at their 3′ end, very similar to AtsnoR43.1 (Figure 1A). Additional BLAST searches (Altschul et al., 1997) identified three other tsnoRNA genes. One is gene 9 which encodes the only tRNAGly with an ACC anticodon. The two other tsnoRNA genes encode non-functional tRNAs: gene 11 has a 20-nucleotide deletion in the tRNA anticodon region, and gene 10 is predicted to be a ψtRNA by the tRNAscan-SE algorithm (Lowe and Eddy, 1997). Remarkably, in contrast with the independent tRNAGly genes which are distributed randomly in the five chromosomes, the tsnoRNA genes are clustered in four loci in chromosomes 1 and 2 (Figure 1B). Two of these are within the large chromosomal duplications that characterize the Arabidopsis genome (Blanc et al., 2000).
All predicted snoR43 variants abut the 3′ end of the tRNA, except for AtsnoR43.12 which is separated from the tRNA by 12 nucleotides (Figure 1A). The 12 snoR43 isoforms, including snoR43.10 and snoR43.11 abutting the two ψtRNAs, have all structural elements required for their stability and nucleolar accumulation: conserved C and D boxes flanked by terminal inverted repeats. Remarkably, only snoR43.2 and snoR43.3 have antisense elements potentially targeting 18S:A543 as snoR43.1 (Figure 1A). The others have divergent antisense elements, some of which predict new target sites on the rRNAs (not shown). Interestingly, snoR43.6 and snoR43.11 contain no significant antisense rRNA element, suggesting that they are not involved in rRNA methylation and therefore may have another function.
The conserved arrangement of Pol III transcriptional cis elements in the 12 tsnoR43 genes suggests that they are all expressed from the tRNA gene promoter producing a tRNA–snoRNA precursor. To confirm this prediction we used RT–PCR to detect the dicistronic precursors of snoR43.1 that targets 18S:A543, snoR43.11 encoding a ψtRNA and snoR43.12 encoding the most divergent snoR43 variant. To ensure specific amplification the 3′ terminal nucleotide of the reverse primers was designed to anneal with a divergent residue not conserved in the other snoR43 variants (Figure 1A). We tested the specificity of p11 and p12 primers by PCR with tsnoR43.1 cloned gene as template. None of them gave a PCR product, showing that they discriminate snoR43 isoforms (result not shown). The RT–PCR performed on total seedling RNA specifically amplified one product of the expected size for the genes tsnoR43.1 (Figure 1D, lane 2), tsnoR43.11 (lane 5) and tsnoR43.12 (lane 8). A control PCR on genomic DNA with the same primers yielded similar products (lanes 4, 7 and 10). This shows that in vivo snoR43.1, snoR43.11 and snoR43.12 are expressed in fusion with the tRNAGly or ψtRNAs.
Identification of tsnoRNA genes in rice
To assess whether tsnoRNA dicistronic genes also occur in other plants we searched for analogous examples in rice, a monocotyledonous plant for which a first draft of the genome is available in databanks (Goff et al., 2002; Yu et al., 2002). In chromosome 1 we identified two tandemly duplicated genes separated by 620 nucleotides encoding two novel C/D snoRNAs, snoR1 and snoR2, abutting the 3′ end of a tRNAMet-e (Figure 2A and B). The tRNAMet-e encoded by the two genes are nearly identical and have a short intron conserved in all plant tRNAMet-e (Akama and Kashihara, 1996). The predicted C/D snoR1 and snoR2 have all the features of methylation guide snoRNAs, although they have divergent D′ box and rRNA antisense elements. Thus snoR1 would target methylation of 18S:C1173 residue, while snoR2 has no clear rRNA antisense element and could target an unidentified RNA substrate. Remarkably, both rice tsnoRNA genes have perfectly conserved tRNA transcriptional elements: a CAA putative TIS, the internal promoter A and B boxes and the T-run terminator downstream of the snoRNA. Expression of these genes was confirmed by RT–PCR with specific primers to detect the dicistronic precursor from the tRNAMet-e–snoR1 gene (Figure 2C). A single product of the expected size is specifically amplified on total RNA from rice seedlings (lane 2), similar to the product amplified in a PCR control on rice genomic DNA (lane 4). This indicates that the tRNAMet-e–snoR1 dicistronic gene is expressed in rice.
Fig. 2. Rice tRNAMet-e–snoRNA genes. (A) Chromosomal organization of tRNAMet-e–snoR1/snoR2 genes in chromosome 1. The putative TIS and terminator elements of Pol III are indicated by CAA and TT, respectively. Arrows indicate primers used for RT–PCR and the size of the expected product is given in nucleotides. The figure is not to scale. (B) Alignment of tsnoR1 and tsnoR2 gene sequences. Large white and grey boxes indicate the mature tRNAMet-e and snoRNAs, respectively. Conserved functional elements of the tRNA and snoRNA are depicted as in Figure 1A. The guide element of snoR1 with nucleotides pairing to rRNA is shown by thick overline. No rRNA antisense element can be found in snoR2. Primers used for RT–PCR are underlined by arrows. (C) Detection of the tRNAMet-e–snoR1 dicistronic precursor. RT–PCR with total RNA from rice seedlings was performed with the pair of primers indicated in (A); +RT and –RT refer to the presence and absence, respectively, of reverse transcriptase in the RT–PCR assay. gDNA indicates a PCR using genomic DNA from rice as template. M is the DNA 100 bp ladder.
Characterization of the tsnoR43.1 gene transcripts expressed in vivo
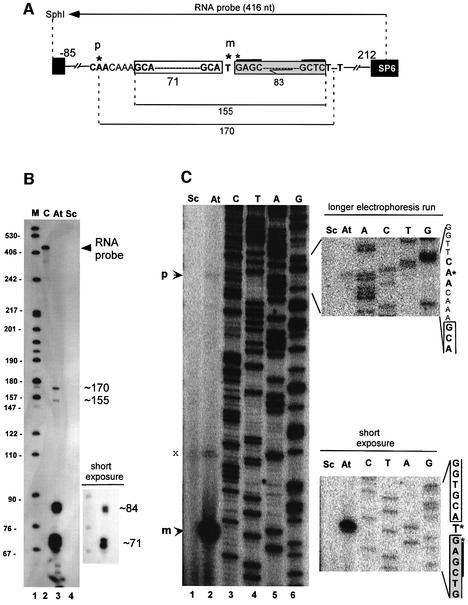
To characterize the different RNA products synthesized from the tsnoR43.1 gene we first performed an RNase protection assay with an RNA probe encompassing both tRNA and snoRNA (Figure 3A). Four protected fragments are detected in total RNA from Arabidopsis seedlings (Figure 3B, lane 3). The two major bands of ∼84 and ∼71 nucleotides have the expected size for mature snoR43.1 and tRNAGly, respectively (Figure 3A). In each case a doublet of fragments is distinguished. This is typical for C/D snoRNAs and arises from differences of one or two nucleotides produced by exonucleolytic trimming to the terminal stem (Kiss and Filipowicz, 1995). In the case of the tRNA the doublet could represent either two stages of tRNA maturation or two very similar tRNAGly isoforms protected by the RNA probe. In addition to these major products, two longer fragments of ∼170 and ∼155 nucleotides are protected (Figure 3B, lane 3). The 170-nucleotide fragment has the predicted size for a primary precursor extending from the putative CAA initiator to the T-run terminator (Figure 3A). The 155-nucleotide band could be a partially processed tRNA–snoRNA precursor extending from the 5′ end of the mature tRNA to the 3′ end of the snoRNA.
Fig. 3. Characterization of tsnoR43.1 gene transcripts produced in vivo. (A) Schematic structure of the cloned tsnoR43.1 gene. Genomic sequences from –85 to +212 relative to TIS were inserted in pGEM-T Easy vector. Black boxes indicate vector sequences. White and grey boxes indicate the tRNAGly and predicted snoR43.1 with their terminal sequences. CAA and T–T indicate the predicted TIS and T-run terminator. For RNase protection the RNA probe complementary to the genomic sequence was transcribed from the SP6 promoter to the SphI site, as indicated. The fragments protected from RNase digestion are shown by horizontal lines with their sizes given in nucleotides. The primer used for reverse transcription is shown by an underlining arrow annealing to AtsnoR43.1. The positions of the primer extension stops are indicated by asterisks. Letters p and m indicate the 5′ end of precursor and mature products, respectively. The figure is not to scale. (B) RNase A/T1 mapping. For RNase protection assay the antisense RNA probe was hybridized to total RNA from Arabidopsis seedlings (At) or yeast tRNA (Sc). Upon RNase treatment, products were separated on a sequencing gel. C is a control with an untreated full-length probe. M shows DNA size markers in nucleotides. A shorter time exposure at the bottom of the gel is shown. (C) Primer extension analysis. Reverse transcription was performed from a radiolabelled primer annealing to snoR43.1 in total RNA from Arabidopsis seedlings (At) or yeast tRNA (Sc). A shorter time exposure at the bottom of the gel is shown to improve visualization of the mature snoRNA ends. A gel with a longer electrophoresis run was done to map the precursor 5′ end better. Letters p and m indicate precursor and mature products, respectively. The position of the primer extension signal is shown by an asterisk. X is an unidentified signal. CTAG is the sequence of the tsnoR43.1 gene. The tRNA and snoRNA terminal sequences are indicated by white and grey boxes, respectively, on the right of the gels.
We mapped the 5′ end of the mature snoR43.1 and the discistronic precursor by primer extension using a radiolabelled primer annealing to snoR43.1 (Figure 3A). Reverse transcription of seedling total RNA produces a major signal mapping to the T residue at the 5′ end of the inverted repeat (Figure 3C, lane 2). A minor signal is detected on the neighbouring G residue in the inverted repeat, probably resulting from one- or two-nucleotide heterogeneity of the snoRNA termini due to trimming in vivo. In addition, an extended product is revealed by a weak signal that maps to the first A of the CAA element upstream from the 5′ end of the mature tRNA (Figure 3C). This signal was consistently observed in different experiments. Since tRNA processing is a rapid event in plants, pre-tRNA precursors do not accumulate and their 5′ ends are difficult to map (Yukawa et al., 2000).
Taken together, these results confirm that, in vivo, snoR43.1 is synthesized in fusion with the tRNAGly and show that transcription starts on the CAA and terminates in the T-run elements, the Pol III transcription initiation and terminator sites of tRNA genes.
Processing of tagged tRNA–snoRNA precursor expressed in transgenic plants
In Arabidopsis there are 13 independent genes encoding cytosolic tRNAGly with GCC anticodon. To establish whether a functional tRNAGly is produced from the dicistronic precursor, Arabidopsis transgenic lines expressing a double-tagged tRNAGly.t–snoR43.s gene were created. The transgenic sequence extends from the TTA-rich and CAA initiator elements to a few nucleotides downstream from the T-run terminator (Figure 4A) and thus its expression should depend on the tRNA gene promoter. The snoR43.1 was tagged by replacing the antisense element with a 16-nucleotide tag (s tag in Figure 4A) that should not impair its synthesis and its stability (Lange et al., 1998). The tRNAGly was tagged with a 10-nucleotide tag (t tag) placed into the anticodon between conserved boxes A and B (Figure 4A) to pre serve the internal promoter (Geiduschek and Tocchini-Valentini, 1988). Four different transgenic lines were selected: L1.s/L2.s producing tagged snoR43.s, and L3.ts/L4.ts producing a tagged tRNA.t in addition to the tagged snoR43.s.
Expression of the tagged dicistronic precursors in the transgenic lines was assessed by RT–PCR using primer p1 and the reverse primer ps with its 3′ ends annealing to the s tag (Figure 4A). This should amplify the 134 bp band from lines L1.s/L2.s and the 144 bp band from the double-tagged lines L3.ts/L4.ts. An RT–PCR specific product of the expected size is detected on RNA samples from lines L1.s and L2.s (Figure 4B, lanes 3 and 4) and, as predicted, a slightly larger product from lines L3.ts and L4.ts (lanes 5 and 6). These signals do not appear on RT–PCR using RNA samples from non-transgenic plants (lane 7).
Primer extension from primer ps revealed that snoR43.s is produced in lines L1.s and L3.ts (Figure 4C, lanes 5 and 6) and the 5′ ends of the major and minor bands precisely map to the T and G terminal nucleotides, respectively, as for the endogenous snoR43.1 (Figure 3C). These signals are not detected on RNA samples from non-transgenic plants (Figure 4C, lane 7) or in a control reaction using yeast tRNA (lane 8). Likewise, we detected expression of the tagged tRNAGly t in transgenic line L3.ts. Reverse transcription with primer pt (Figure 4A) revealed a specific product mapping precisely to the 5′ end of tRNAGly (Figure 4C, lane 13). We conclude that the tRNA.t accumulates in vivo and its 5′ end is correctly processed.
Taken together, these results demonstrate that the tagged tRNAGly.t and snoR43.s are co-transcribed producing a dicistronic precursor which is processed to the mature snoRNA and tRNA.
Processing of the tRNAGly–snoR43.1 dicistronic precursor by RNase Z
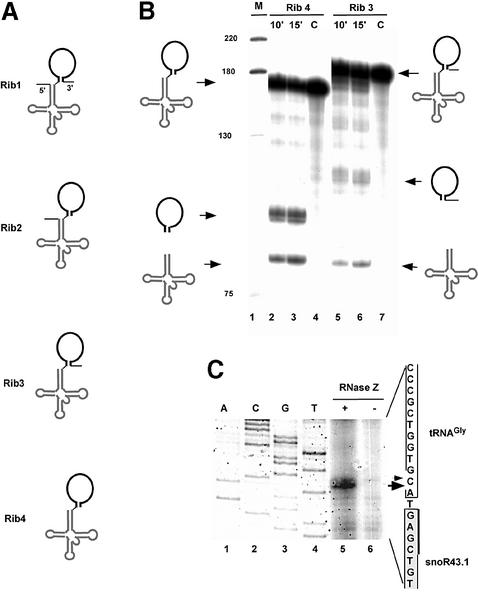
In vivo the tRNA–snoRNA precursor produces both snoRNA and tRNA and could involve activities processing the precursor tRNAs (pre-tRNA). Recently, the endonuclease RNase Z, which specifically cleaves the 3′ end of pre-tRNAs in vitro, has been identified in plants (Schiffer et al., 2002). Since the cleavage determinants of RNase Z reside solely in the tRNA (Schiffer et al., 2001), this enzyme is a good candidate for cleaving the dicistronic precursor separating snoRNA from tRNA. Recombinant RNase Z (rRNase Z) from Arabidopsis expressed in Escherichia coli specifically cleaves tRNA 3′ ends in vitro (Schiffer et al., 2002). Therefore we tested whether rRNase Z is able to cleave four different tRNAGly–snoR43.1 precursors to yield tRNA and snoRNA (Figure 5A). Only substrates Rib3 and Rib4 with mature 5′ tRNA were efficiently cleaved by rRNase Z (Figure 5B). Rib3 extends from tRNA 5′ end to the U tract predicted to be the unprocessed 3′ trailer sequence produced by Pol III transcription (see Figures 1A and 3A). Rib4 differs from Rib3 by the absence of the 3′ unprocessed snoRNA extension and extends from the 5′ tRNA to the 3′ snoRNA mature ends (Figure 5A); it may correspond to the 155-nucleotide intermediate precursor mapped by RNase protection (Figure 3B). The purified rRNase Z efficiently processed Rib4 yielding two products with the expected sizes for tRNA and snoRNA, respectively (Figure 5B, lanes 2 and 3). A doublet is observed for the cleaved snoRNA which probably reflects heterogeneity of one or two nucleotides at the 3′ end generated by in vitro transcription (Kholod et al., 1998). The rRNase Z also accurately processed substrate Rib3, producing the expected products (again a doublet in the case of snoRNA) but with reduced efficiency (Figure 5B, lanes 5 and 6).
Fig. 5. Cleavage of the tRNA–snoRNA precursor by recombinant RNase Z in vitro. (A) Structures of RNA substrates used for processing. All substrates contain tRNA (grey cloverleaf) and snoRNA (black stem loop). Substrates differ in the presence of the 5′ leader and 3′ trailer sequences shown by thin lines. (B) Processing of RNA substrates Rib3 and Rib4. Precursor RNAs Rib4 (5′ and 3′ end matured) and Rib3 (5′ matured) were incubated with rRNase Z for 10 min and 15 min as indicated. C are control reactions performed without addition of proteins. M is the DNA size marker given in nucleotides on the left. Precursor and products are shown schematically at the sides. (C) Primer extension analysis of the rRNase Z cleavage site on Rib4 substrate. Unlabelled Rib4 substrate was incubated in vitro with or without rRNase Z as indicated. After incubation, products of the reaction were extracted and analysed by primer extension with a radiolabelled primer snoR4 annealing to the snoRNA. ACGT refers to sequencing reactions started from primer sno4. The position of the cleavage site on the coding strand is indicated with an arrow. The small arrow indicates a minor cleavage site.
The RNase Z cleavage site on the Rib4 substrate was mapped by primer extension with a primer annealing to snoR43.1. A major signal mapping 5′ to the A residue on the tRNA 3′ end was detected (Figure 5C, lane 5) one nucleotide upstream from the tRNA–snoRNA cleavage site mapped in vivo (Figure 3C). This difference of a single nucleotide has been observed for other tRNA precursors processed by this enzyme in vitro (Mayer et al., 2000; Schiffer et al., 2001).
These results show that the plant RNase Z activity can cleave the 5′ mature tRNAGly–snoR43.1 precursor to separate the tRNA from the snoRNA in vitro.
Discussion
The plant tsnoRNA genes described here in Arabidopsis and rice represent a new mode of snoRNA gene organization in eukaryotes and raise the question of the elements controlling expression of these genes. In addition, they show that the link between small RNAs and tRNAs, first revealed in trypanosomes and more recently in Archaea, also occurs in plants.
Transcription of tsnoRNA genes
The plant tsnoRNA genes are reminiscent of the dicistronic tRNA genes found in yeast that are transcribed by Pol III from the upstream tRNA gene promoter (Mao et al., 1980; Willis et al., 1984; Reyes et al., 1986). The structural data and the mapping of the 5′ end of the dicistronic precursor (Figures 1 and 3) strongly suggest that the tsnoRNA are also transcribed by Pol III from the tRNA gene promoter.
Another precedent related to plant tsnoRNAs is found in trypanosomes in which the genes encoding U6, U3 and 7SL RNA are located ∼100 bp downstream of distinct tRNA genes oriented in the opposite direction (Nakaar et al., 1994). In this case the intragenic tRNA promoter functions as extragenic element to direct transcription of the small RNAs, but it is not clear how the divergently oriented promoter functions. Therefore, in contrast with plant tsnoRNAs, the U6, U3 and 7SL RNA precursors from trypanosomes are monocistronic and do not include the tRNA.
Interestingly, the U3 genes in plants are monocistronic and are also transcribed by Pol III (Kiss et al., 1991). Thus both snoRNA genes for which the transcription initiation sites have been determined in plants, U3 and the dicistronic tsnoR43.1, are transcribed by Pol III. It would be interesting to assess the proportion of non-polycistronic non-intronic snoRNAs that are transcribed by Pol III in plants. This would distinguish them from the plant polycistronic genes which lack conserved Pol III transcriptional elements and are probably transcribed by Pol II as the yeast (Qu et al., 1999) and the trypanosome clusters (Xu et al., 2001).
Processing of the tRNA–snoRNA dicistronic precursor
The plant tsnoRNA and polycistronic snoRNAs precursors have a distinct structural organization. The polycistronic snoRNA precursors contain multiple snoRNAs separated by spacers of 30 to 300 nucleotides (Barneche et al., 2001; Brown et al., 2001). The individual snoRNAs are released by endonucleolytic processing of the precursor and subsequent exonucleolytic trimming (Leader et al., 1999). A similar mechanism could release snoRNAs from intronic clusters, implying direct degradation of host mRNA precursor (Leader et al., 1999). Although the endonuclease has not been identified in plants, a candidate is an RNase III-like activity which catalyses the endonucleolytic cleavage of polycistronic snoRNAs in yeast (Chanfreau et al., 1998; Qu et al., 1999).
The tRNA–snoRNA dicistronic precursor has an adjacent tRNA and snoRNA which are both accurately processed; this probably implicates tRNA processing activities. The first event of tRNA processing in yeast is binding of Lhp1p protein (protein La in vertebrates) to the U tract at the 3′ extension of all pre-tRNAs, protecting them from 3′-5′exonucleases (Wolin and Matera, 1999). The Lhp1p-bound tRNA is then the substrate of the conserved RNase P that removes the 5′ leader by a single endonucleolytic cut. Subsequently, tRNA 3′ end processing occurs controlled by the Lhp1p protein and an as yet unidentified endonuclease (Kufel et al., 2002). In plants this enzyme could be RNase Z which generates the 3′ end of tRNAs in vitro (Schiffer et al., 2002). The recombinant RNase Z was shown to cleave the dicistronic precursor efficiently, releasing both tRNAGly and snoR43.1 (Figure 5) and may also cleave the rice tRNAMet-e –snoR1/2 precursor which contains a short intron. The pre-tRNA containing an intron folds into a typical tRNA three-dimensional structure that is accurately matured by tRNA processing activities, and RNase Z has been shown to process intron-containing pre-tRNAs (Mayer et al., 2000). Since the recombinant RNase Z was unable to cleave tRNAGly–snoR43.1 precursors with unprocessed 5′ extension (results not shown), we propose that the dicistronic precursor synthesized by Pol III is processed by RNase P and subsequently by RNase Z which releases the mature tRNA and the snoRNA (Figure 6). Whether RNase Z cleavage occurs before or after 3′ end maturation of the snoRNA is unknown. The 3′ extension of precursor C/D snoRNAs is removed in vivo by an endonucleolytic cut and subsequent exonucleolytic trimming to the terminal stem. Recently, a maturation pathway has been proposed for the yeast monocistronic U3 which implicates Lhp1p binding to the U3 to control the 3′ end trimming (Kufel et al., 2000, 2002). Whether the plant C/D snoRNA 3′ end processing follows a similar pathway is not known. Nevertheless, C/D snoRNP core proteins (Figure 6), which are highly conserved in plants (M.Echeverria, unpublished results), should assemble on the snoRNA before 3′ end trimming. These could assemble on the tRNA–snoRNA precursor, prior to RNase Z cleavage, or later on the released snoRNA. The detection of a partially processed precursor of ∼155 nucleotides probably lacking 5′ and 3′ trailer sequences (Figure 3) and the fact that rRNase Z is more efficient on 3′ processed dicistron in vitro (Figure 5B) suggest that 3′ end trimming could occur prior to RNase Z cleavage.
Transcription of tRNA genes occurs in the nucleoplasm, and the first processing event is the cleavage of the 5′ end of the pre-tRNA by RNase P. In yeast the RNase P is also a nucleolar protein, and pre-tRNAs as well as yeast dicistronic tRNAs are processed in the nucleolus (Bertrand et al., 1998; Wolin and Matera, 1999). Additionally, processing of the yeast U3 snoRNA, which requires Lhp1p protein, also occur in the nucleolus (Verheggen et al., 2002). Thus, in yeast, the nucleolus is both a snoRNA and a tRNA processing compartment. In animals, processing of intronic snoRNAs probably begins in the nucleoplasm but snoRNP assembly and some steps of snoRNA processing occur in the Cajal bodies prior to their nucleolar localization (Verheggen et al., 2002; Filipowicz and Pogacic, 2002). In plants, polycistronic snoRNA precursors have been detected in Cajal bodies and in the nucleolus, suggesting that snoRNP assembly and processing probably occurs in these two compartments (Shaw et al., 1998). Whether plant dicistronic tRNA–snoRNA processing occurs in the nucleoplasm and Cajal bodies and/or in the nucleolus is an open question that requires localization of the dicistronic tRNA–snoRNA and the processing activities.
Plant tsnoRNA genes and evolution
A systematic analysis of all tRNA gene loci associated with C/D snoRNAs predicted by the Snoscan algorithm (Lowe and Eddy, 1999) conducted in Arabidopsis did not detect any other dicistronic gene similar to tsnoR43. However, many predicted C/D snoRNAs were located overlapping or very close to tRNA genes, either in the same or in the complementary strand (data not shown). Although we do not know whether all the predicted snoRNAs associated with tRNA genes are expressed, this confirms a genetic link between snoRNAs and tRNAs in eukaryotes. The recent identification of a C/D RNA nested in an intron of the pre-tRNATrp in Archaea reveals that this link occurred very early in evolution (Clouet d’Orval et al., 2001). These examples reinforce the hypothesis that snoRNAs derived from reverse transcription of abundant small RNAs (Lafontaine and Tollervey, 1998; Brosius, 1999) and targeted tRNAs for insertion and expression. The plant tsnoRNAs may represent traces of these ancient events in the evolution of eukaryotic snoRNAs.
Materials and methods
Plant material
Arabidopsis thaliana (col 0) and Oryza sativa (nipponbare) seedlings were cultivated in a growth chamber under normal daylight conditions.
Identification of tsnoRNA genes and sequence analysis
The Arabidopsis tRNA sequences were recovered from two distinct databanks (http://www.inra.fr/Internet/Produits/TAARSAT/; http://rna. wustl.edu/GtRDB/At/). BLAST searches (Altschul et al., 1997) were performed with each tRNA sequence against Arabidopsis and rice genomic sequences available in public databases. All tRNA sequences were recovered with 200 bp upstream and downstream flanking genomic sequences and processed successively by tRNA-scan SE (Lowe and Eddy, 1997; http://www.genetics.wustl.edu/eddy/tRNA-scan-SE/) and Snoscan (Lowe and Eddy, 1999; http://rna.wustl.edu/snoRNAdb/code/). Sequence alignments were performed with CLUSTAL W (Thompson et al., 1994).
The names and DDBL/EMBL/GenBank accession Numbers of the BACs containing the Arabidopsis tRNAGly given in Figure 1 are as follows. Chromosome 1: F19P19 (AC000104); F4H5 (AC011001); F10C21 (AC051630); F23C21 (AC079675); F8A5 (AC002292); F10K1 (AC067971); T7P1 (AC018908); F14O23 (AC012654). Chromosome 2: T6B20 (U93215); T27E13 (AC004165); MH10K (AC005956). Chromosome 3: F21F14 (AL138642); F5K20 (AL132960); F28L1 (AC018907); F9024 (AL137081). Chromosome 4: F8B4 (AL034567). Chromosome 5: T16G12 (AC068809); T28J14 (AL163652); T7H20 (AL162508); MUD21 (AB010700); K9O7 (AB016875). Rice snoR1 and snoR2 sequences in chromosome 1: PAC clone P0665A11 (AP003106).
The tsnoRNA gene sequences submitted to the EMBL database have the following accession Numbers: tsnoR43.1, AJ506163; tsnoR43.2, AJ506164; tsnoR43.3, AJ506165; tsnoR43.4, AJ506166; tsnoR43.5, AJ506167; tsnoR43.6, AJ506168; tsnoR43.7, AJ506169; tsnoR43.8, AJ506170; tsnoR43.9, AJ506171; tsnoR43.10, AJ506172; tsnoR43.11, AJ506173; tsnoR43.12, AJ506174; rice tsnoR1, AJ506175; rice tsnoR2, AJ506176.
Genomic DNA cloning and mutagenesis
The Arabidopsis tsnoR43.1 gene, extending from –85 to +212 relative to TIS, was amplified by PCR from genomic DNA of Arabidopsis using specific primers. The amplified product was then cloned in the pGEM-T Easy vector (Promega) producing plasmid pGtsnoR43 (see Figure 3).
Tagged dicistronic genes were obtained using the ExSite PCR-based Site-Directed kit (Stratagene). A 16 -nucleotide tag (tag s in Figure 4A) was inserted, replacing 15 nucleotides encompassing the antisense element of the snoR43.1, to produce plasmid pGtsnoR43. This produced plasmid pGtsnoR43.s. Subsequently, an additional tag was introduced into the anticodon of the tRNA (tag t in Figure 4A) producing pGtsnoR43.ts.
Arabidopsis tsnoR43 transgenic lines
For transfer into Arabidopsis the tagged dicistronic genes in pGtsnoR43.s and pGtsnoR43.ts were amplified by PCR and cloned in the binary vector pBin19, conferring resistance to kanamycin. Arabidopsis plants were transformed via Agrobacterium by the vacuum infiltration method (Bechtold et al., 1993) and kanamycin-resistant plants were selected. Stable insertion of the transgene into the genome in kanamycin-resistant plants was confirmed by PCR analysis. Four lines were selected: L1.s and L2.s transformed with tRNAGly–snoR43.s, and L3.ts and L4.ts transformed with tRNAGly.t–snoR43.s gene.
RNA extraction and analysis
Total RNA was extracted from 10-day-old A.thaliana or 15-day-old rice seedlings using a standard method (Chomczynski and Sacchi, 1987). RNA was subjected to DNase treatment for 30 min at 37°C in 100 µl reactions with 5 units of RQ1-DNase (Promega) and then extracted with phenol–chloroform before ethanol precipitation.
RNA samples were then used for primer extension and RNase A/T1 mapping performed as previously described (Barneche et al., 2000). For primer extension specific primers were designed as indicated in the text. For the RNase mapping the riboprobe was synthesized in vitro using the pGtsnoR43 plasmid digested by Sph1 (Figure 3) and SP6 RNA polymerase. The riboprobe was then purified on an acrylamide gel and used for RNase protection assay as described (Barneche et al., 2000).
To detect methylation of 18S:A543 primer extension was performed in similar conditions, except that two different reactions were done with normal (1 mM) and limiting (0.004 mM) dNTP concentration, respectively, as previously reported (Barneche et al., 2000).
Reverse transcription polymerase chain reaction
RT–PCR was performed using the Superscript One-Step RT–PCR with Platinum Taq (Gibco-BRL). Briefly, reactions were carried out according to the supplier’s conditions in 50 µl reactions containing 0.5 µg of total RNA. For controls, two other reactions were carried out in parallel without Superscript RT: 2 units of Platinum Taq DNA Polymerase and either 0.5 µg of RNA (negative control) or 0.5 µg of genomic DNA (positive control). Amplification products were then analysed on 2% agarose gels.
In vitro processing assay with recombinant RNase Z
Templates for in vitro transcription of substrates Rib1–Rib4 were generated using primers sno1 and sno5 (Rib1), sno1 and sno4 (Rib2), sno3 and sno5 (Rib3), and sno3 and sno4 (Rib4). All templates contained the T7 promotor at the 5′ end. In vitro transcription by T7 polymerase and purification of transcripts was performed as described (Marchfelder and Brennicke, 1994)
The recombinant RNase Z was isolated as described (Schiffer et al., 2002). In vitro processing assays were performed as described (Mayer et al., 2000) with the following modifications: 30 ng of rRNase Z protein were used and all reactions were incubated for 10 or 15 min at 37°C.
For primer extension analysis of the RNase Z cleavage site an in vitro processing assay using unlabelled Rib4 precursor was performed. As a control reaction the precursor was incubated in the absence of protein. After extraction with phenol–chloroform, RNAs were precipitated and subsequently used in a reverse transcription reaction using primer sno4 annealing to the AtsnoR43.1 sequence. The same primer was used for the sequencing reaction.
Primers
All primer sequences used for RT–PCR, primer extension and cloning are available upon request.
Acknowledgments
Acknowledgements
We are grateful to Drs M.Delseny (CNRS, Perpignan), A.Brennicke (University of Ulm) and J.Saez Vasquez (CNRS, Perpignan) for their valuable comments which improved the manuscript. We are also grateful to E.Bruckbauer (University of Ulm) for technical assistance, B.Piegu for advice on the usage of computational tools and Dr T.Roscoe (CNRS, Perpignan) for correcting the English. F.B. was supported by an MENESR fellowship and K.K. was supported by a Bourse d’Accueil de Jeunes Chercheurs Etrangers from Ministère de la Recherche, France, and a Poste Chercheur Etranger Associée au CNRS.
References
- Akama K. and Kashihara,M. (1996) Plant nuclear tRNA(Met) genes are ubiquitously interrupted by introns. Plant Mol. Biol., 32, 427–434. [DOI] [PubMed] [Google Scholar]
- Altschul S.F., Madden,T.L., Schaffer,A.A., Zhang,J., Zhang,Z., Miller,W. and Lipman,D.J. (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search Programs. Nucleic Acids Res., 25, 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antal M., Mougin,A., Kis,M., Boros,E., Steger,G., Jakab,G., Solymosy,F. and Branlant,C. (2000) Molecular characterization at the RNA and gene levels of U3 snoRNA from a unicellular green alga, Chlamydomonas reinhardtii. Nucleic Acids Res., 28, 2959–2968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachellerie J.P. and Cavaillé,J. (1998) Small nucleolar RNAs guide the ribose methylation of eukaryotic rRNAs. In Grosjean,H. and Benne,R. (eds), Modification and Editing of RNA. ASM Press, Washington, DC, pp. 255–272.
- Barneche F., Steinmetz,F. and Echeverria,M. (2000) Fibrillarin genes encode both a conserved nucleolar protein and a novel small nucleolar RNA involved in ribosomal RNA methylation in Arabidopsis thaliana. J. Biol. Chem., 275, 27212–27220. [DOI] [PubMed] [Google Scholar]
- Barneche F., Gaspin,C., Guyot,R. and Echeverria,M., (2001) Identification of 66 box C/D snoRNAs in Arabidopsis thaliana: extensive gene duplications generated multiple isoforms predicting new ribosomal RNA 2′-O-methylation sites. J. Mol. Biol., 311, 57–73. [DOI] [PubMed] [Google Scholar]
- Bechtold N., Ellis,J. and Pelletier,G. (1993) In planta Agrobacterium mediated gene transfer by infiltration of adult Arabidopsis thaliana plants. C.R. Acad. Sci. Paris, 316, 1194–1199. [Google Scholar]
- Bertrand E., Houser-Scott,F., Kendall,A., Singer,R.H. and Engelke,D.R. (1998) Nucleolar localization of early tRNA processing. Genes Dev., 12, 2463–2468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanc G., Barakat,A., Guyot,R., Cooke,R. and Delseny,M. (2000) Extensive duplication and reshuffling in the Arabidopsis genome. Plant Cell, 12, 1093–1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brosius J. (1999) RNAs from all categories generate retrosequences that may be exapted as novel genes or regulatory elements. Gene, 238, 115–134. [DOI] [PubMed] [Google Scholar]
- Brown J.W, Clark,G.P., Leader,D.J., Simpson,C.G. and Lowe,T. (2001) Multiple snoRNA gene clusters from Arabidopsis. RNA, 7, 1817–1832. [PMC free article] [PubMed] [Google Scholar]
- Cavaillé J. and Bachellerie,J.P. (1998) SnoRNA guided ribose methylation of rRNA: structural features of the guide RNA duplex influencing the extent of the reaction. Nucleic Acids Res., 26, 1576–1587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cecchini J.P. and Miassod,R. (1979) Studies on the methylation of cytoplasm RNA from cultured higher plant cells. Eur. J. Biochem., 98, 203–214. [DOI] [PubMed] [Google Scholar]
- Chanfreau G. Legrain,P. and Jacquier,A. (1998) Yeast RNase III as a key processing enzyme in small nucleolar RNAs metabolism. J. Mol. Biol., 284, 975–988. [DOI] [PubMed] [Google Scholar]
- Chomczynski P. and Sacchi,N. (1987) Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal. Biochem., 162, 156–159. [DOI] [PubMed] [Google Scholar]
- Clouet d’Orval B., Bortolin,M., Gaspin,C. and Bachellerie,J.P. (2001) Box C/D RNA guides for the ribose methylation of archaeal tRNAs. The tRNATrp intron guides the formation of two ribose-methylated nucleosides in the mature tRNATrp. Nucleic Acids Res., 29, 4518–4529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filipowicz W. and Pogacic,V. (2002) Biogenesis of small nucleolar ribonucleoproteins. Curr. Opin. Cell Biol., 14, 319–327. [DOI] [PubMed] [Google Scholar]
- Gaspin C., Cavaillé,J., Erauso,G. and Bachellerie,J.P. (2000) Archaeal homologs of eukaryotic methylation guide small nucleolar RNAs: lessons from the Pyrococcus genomes. J. Mol. Biol., 297, 895–906. [DOI] [PubMed] [Google Scholar]
- Geiduschek E.P. and Tocchini-Valentini,G.P. (1988) Transcription by RNA pol III. Annu. Rev. Biochem., 57, 873–914. [DOI] [PubMed] [Google Scholar]
- Giorgi C., Fatica,A., Nagel,R. and Bozzoni,I. (2001) Release of U18 snoRNA from its host intron requires interaction of Nop1p with the Rnt1p endonuclease. EMBO J., 20, 6856–6865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goff S.A. et al. (2002) A draft sequence of the rice genome (Oryza sativa L. ssp. japonica). Science, 296, 92–100. [DOI] [PubMed] [Google Scholar]
- Kholod N., Vassilenko,K., Shlyapnikov,M, Ksenzenko,V. and Kisselev,L. (1998) Preparation of active tRNA gene transcripts devoid of 3′-extended products and dimers. Nucleic Acids Res., 26, 2500–2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiss T. and Filipowicz,W. (1995) Exonucleolytic processing of small nucleolar RNAs from pre-mRNA introns. Genes Dev., 9, 1411–1424. [DOI] [PubMed] [Google Scholar]
- Kiss T., Marshallsay,C. and Filipowicz,W. (1991) Alteration of the RNA polymerase specificity of U3 snRNA genes during evolution and in vitro. Cell, 65, 517–526. [DOI] [PubMed] [Google Scholar]
- Kufel J., Allmang,C., Chanfreau,G., Petfalski,E., Lafontaine,D.L. and Tollervey,D. (2000) Precursors to the U3 small nucleolar RNA lack small nucleolar RNP proteins but are stabilized by La binding. Mol. Cell Biol., 20, 5415–5424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kufel J., Allmang,C., Verdone,L., Beggs,J.D. and Tollervey,D. (2002) Lsm proteins are required for normal processing of pre-tRNAs and their efficient association with La-homologous protein Lhp1p. Mol. Cell Biol., 22, 5248–5256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lafontaine D.J. and Tollervey,D. (1998) Birth of the snoRNPs: the evolution of the modification guide snoRNAs. Trends Biochem. Sci., 23, 383–388. [DOI] [PubMed] [Google Scholar]
- Lange T.S., Borovjagin,A., Maxwell,E.S. and Gerbi,S.A. (1998) Conserved boxes C and D are essential nucleolar localization elements of U14 and U8 snoRNAs. EMBO J., 17, 3176–3187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau R.Y., Kennedy,T.D. and Lane,B.G. (1974) Wheat embryo ribonucleates. III: Modified nucleotide constituents in each of the 5.8S, 18S and 26S ribonucleates. Can. J. Biochem., 52, 1110–1123. [DOI] [PubMed] [Google Scholar]
- Leader D.J., ClarkG.P., Watters J, Beven,A.F., Shaw,P.J. and Brown,J.W. (1997) Clusters of multiple different small nucleolar RNA genes in plants are expressed as and processed from polycistronic pre-snoRNAs. EMBO J., 16, 5742–5751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leader D.J., Clark,G.P., Watters,J., Beven,A.F., Shaw,P.J. and Brown,J.W. (1999) Splicing-independent processing of plant box C/D and box H/ACA small nucleolar RNAs. Plant Mol. Biol., 39, 1091–1100. [DOI] [PubMed] [Google Scholar]
- Liang D., Zhou,H., Zhang,P., Chen,Y.Q., Chen,X., Chen,C.L. and Qu,L.H. (2002) A novel gene organization: intronic snoRNA gene clusters from Oryza sativa. Nucleic Acids Res., 30, 3262–3272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe T.M. and Eddy,S.R. (1997) tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res., 25, 955–964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe T.M. and Eddy,S.R. (1999) A computational screen for methylation guide snoRNAs in yeast. Science, 283, 1168–1171. [DOI] [PubMed] [Google Scholar]
- Maden B.E. (2001) Mapping 2′-O-methyl groups in ribosomal RNA. Methods, 25, 374–382. [DOI] [PubMed] [Google Scholar]
- Mao J.I., Schmidt,O. and Söll,D. (1980) Dimeric transfer RNA precursors in S.pombe. Cell, 21, 509–516. [DOI] [PubMed] [Google Scholar]
- Marchfelder A. and Brennicke,A. (1994) Characterization and partial purification of tRNA processing activities from potato mitochondria. Plant Physiol., 105, 1247–1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer M., Schiffer,S. and Marchfelder,A. (2000) tRNA 3′ processing in plants: nuclear and mitochondrial activities differ. Biochemistry, 39, 2096–2105. [DOI] [PubMed] [Google Scholar]
- Nakaar V., Dare,A.O., Hong,D., Ullu,E. and Tschudi,C. (1994) Upstream tRNA genes are essential for expression of small nuclear and cytoplasmic RNA genes in trypanosomes. Mol. Cell. Biol., 14, 6736–6742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omer A.D., Lowe,T.M., Russell,A.G., Ebhardt,H., Eddy,S.R. and Dennis,P.P. (2000) Homologs of small nucleolar RNAs in Archaea. Science, 288, 517–522. [DOI] [PubMed] [Google Scholar]
- Pelczar P. and Filipowicz,W. (1998) The host gene for intronic U17 small nucleolar RNAs in mammals has no protein-coding potential and is a member of the 5′-terminal oligopyrimidine gene family. Mol. Cell. Biol., 18, 4509–4518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu L.H. et al. (1999) Seven novel methylation guide small nucleolar RNAs are processed from a common polycistronic transcript by Rat1p and RNase III in yeast. Mol. Cell. Biol., 19, 1144–1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu L.H., Meng,Q., Zhou,H. and Chen,Y.Q., (2001) Identification of 10 novel snoRNA gene clusters from Arabidopsis thaliana. Nucleic Acids Res., 29, 1623–1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes V.M., Newman,A. and Abelson,J. (1986) Mutational analysis of the coordinate expression of the yeast tRNAArg–tRNAAsp gene in tandem. Mol. Cell. Biol., 6, 2436–2442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiffer S., Helm,M., Théobald-Dietrich,A., Giegé,R. and Marchfelder,A. (2001) The plant tRNA 3′ processing enzyme has a broad substrate spectrum. Biochemistry, 40, 8264–8772. [DOI] [PubMed] [Google Scholar]
- Schiffer S., Rösch,S. and Marchfelder,A. (2002) Assigning a function to a conserved group of proteins: the tRNA 3′-processing enzymes. EMBO J., 21, 2769–2777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw P.J., Beven,A.F., Leader,D.J. and Brown,J.W. (1998) Localisation and processing from a polycistronic precursor of novel snoRNAs in maize. J. Cell Sci., 111, 2121–2128. [DOI] [PubMed] [Google Scholar]
- Terns M.P. and Terns,R.M. (2002) Small nucleolar RNAs: versatile trans-acting molecules of ancient evolutionary origin. Gene Expr., 10, 17–39. [PMC free article] [PubMed] [Google Scholar]
- Thompson J.D., Higgins,D.G. and Gibson,T.J. (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, positions-specific gap penalties and weight matrix choice. Nucleic Acids Res., 22, 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tycowski K.T., Shu,M.D. and Steitz,J.A. (1996) A mammalian gene with introns instead of exons generating stable RNA products. Nature, 379, 464–466. [DOI] [PubMed] [Google Scholar]
- Verheggen C., Lafontaine,D.L., Samarsky,D., Mouaikel,J., Blanchard,J.M., Bordonne,R. and Bertrand,E. (2002) Mammalian and yeast U3 snoRNPs are matured in specific and related nuclear compartments. EMBO J., 21, 2736–2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villa T., Ceradini,F., Presutti,C. and Bozzoni,I. (1998) Processing of the intron-encoded U18 small nucleolar RNA in the yeast Saccharomyces cerevisiae relies on both exo- and endonucleolytic activities. Mol. Cell. Biol., 18, 3376–3383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willis I., Hottinger,H., Pearson,D., Chisholm,V., Leupold,U. and Söll,D. (1984) Mutations affecting excision of the intron from a eukaryotic dimeric tRNA precursor. EMBO J., 3, 1573–1580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolin S.L. and Matera,G. (1999). The trials and travels of tRNA. Genes Dev.,13, 1–10. [DOI] [PubMed] [Google Scholar]
- Xu Y., Liu,L., Lopez-Estrano,C. and Michaeli,S. (2001) Expression studies on clustered trypanosomatid box C/D small nucleolar RNAs. J. Biol. Chem., 276, 14289–14298. [DOI] [PubMed] [Google Scholar]
- Yu J. et al. (2002). A draft sequence of the rice genome (Oryza sativa L. ssp. indica). Science, 296, 79–92. [DOI] [PubMed] [Google Scholar]
- Yukawa Y., Sugita,M., Choisne,N., Small,I. and Sugiura,M. (2000). The TATA motif, the CAA motif and the poly(T) transcription termination motif are all important for transcription re-initiation on plant tRNA genes. Plant J., 22, 439–447. [DOI] [PubMed] [Google Scholar]