Abstract
Prion diseases are infectious, sporadic and inherited fatal neurodegenerations that are propagated by an abnormal refolding of the cellular prion protein PrPC. Which chaperones assist the normal folding of PrPC is unknown. The linkage of familial Gerstmann– Sträussler–Scheinker (GSS) syndrome with proline substitutions in PrP raised the prospect that peptidylprolyl cis–trans isomerases (PPIases) may play a role in normal PrP metabolism. Here we used cyclo sporin A (CsA), an immunosuppressant, to inhibit the cyclophilin family of PPIases in cultured cells. CsA-treated cells accumulated proteasome-resistant, ‘prion-like’ PrP species, which deposited in long-lived aggresomes. PrP aggresomes also formed with disease-linked proline mutants when proteasomes were inhibited. These results suggest mechanisms whereby abnormally folded cytosolic PrP may in some cases participate in the development of spontaneous and inherited prion diseases.
Keywords: aggresome/cyclophilin/cyclosporin A/encephalopathy/prion protein
Introduction
Although protein folding and assembly are highly chaperoned processes, their success rate is astonishingly low. Overall, up to 30% of nascent proteins are defective and are readily destroyed (Schubert et al., 2000). For wild-type secretory and membrane proteins synthesized in the endoplasmic reticulum (ER), failure rates of 10–70% have been reported. Misfolded ER products are prevented from maturing in the Golgi apparatus and are instead dislocated into the cytosol and degraded by proteasomes through the ERAD pathway (reviewed in Ellgaard et al., 1999). Insufficient removal of misfolded polypeptides may lead to the intracellular deposition of potentially toxic aggregates (Kopito, 2000; Dobson, 2001). Indeed, the list of conditions characterized by insoluble protein deposits is growing rapidly. Among these ‘conformational diseases’ (Carrell and Lomas, 1997), the prion disorders such as Creutzfeldt–Jakob disease (CJD) and Gerstmann– Sträussler–Scheinker syndrome (GSS) of humans and scrapie of sheep occupy a unique place owing to their transmissibility and mechanism of propagation (for a review, see Prusiner, 1998).
Prion diseases are infectious, inherited and sporadic neurodegenerations that propagate by refolding the host protein, PrPC (Oesch et al., 1985), into a β-sheet (Pan et al., 1993) rich abnormal conformer called PrPSc. PrPSc is the only known component of prions. PrPC (Bolton et al., 1982; Basler et al., 1986) is a glycophosphatidylinositol (GPI)-anchored (Stahl et al., 1987), cell surface, copper-binding glycoprotein of unknown function (Prusiner, 1998). Because misfolded PrP can cause fatal neurodegenerations, it is crucial to understand its folding metabolism. Wild-type PrP molecules (wtPrP) are usually entirely translocated into the ER lumen where the N-terminal signal peptide and the C-terminal GPI attachment signal are cleaved and where the GPI anchor is added. They are then exported to the cell surface in association with cholesterol rafts (Naslavsky et al., 1997). In unstressed cells, ∼10% of nascent PrPC fails to fold properly and is degraded through ERAD, a process which includes the formation of polyubiquitylated PrP. When proteasomes are inhibited by drugs such as N-acetyl-leucinal-leucinal-norleucinal (ALLN), ERAD-bound PrPC accumulates throughout the cytoplasm (Yedidia et al., 2001).
Little is known about the chaperones and foldases that assist the folding of PrPC or that escort it through ERAD. Because there is an abundance of prolines in PrP, and since two proline substitutions (P102L and P105L) are linked with familial GSS (Hsiao et al., 1989; Yamazaki et al., 1999), we wondered whether cis–trans isomerization of peptidylprolyl bonds might participate either in the folding of PrPC or in its disposal. A rate-limiting step in the folding of many proteins, the isomerization of X-Pro bonds, is accelerated by ubiquitous peptidylprolyl isomerases (PPIases), which include the cyclophilins, the FKBPs and the parvulins (Hamilton and Steiner, 1998; Balbach and Schmid, 2000). Cyclophilins are found in most cellular compartments, including the cytosol (cypA) and the ER (cypB). Cyclosporin A (CsA), a fungal peptide used as an immunosuppressant, specifically inhibits the cyclophilin family.
Here we used CsA to study the importance of cyclophilins in the metabolism of an epitopically labeled wtPrP in Chinese hamster ovary (CHO-M) and mouse neuroblastoma (N2a-M) cells. Within 2 h from the addition of CsA, cells started accumulating misfolded 26 kDa PrP species that were Sarkosyl insoluble and resisted stringent proteolysis. This PrP was in part cytosolic. The CsA-induced PrP species were not detect ably ubiquitylated, they partially resisted proteasomal degradation, and they were metabolically very stable. In ∼10% of the CsA-treated cells, protease-resistant PrP deposited in a single juxtanuclear focus, a distribution strikingly different from the diffuse PrP deposits seen with proteasome inhibitors. Vimentin caging, co-localization with γ-tubulin, microtubule dependence and immuno-electron microscopy (EM) identified these deposits as aggresomes, which are microtubule-dependent inclusion bodies located at the centrosome (Johnston et al., 1998). To the best of our knowledge, this is the first instance of a wild-type cellular protein accumulating in stable aggresomes.
These results showed that cyclophilins play a role in the normal metabolism of PrP. To see whether this role includes a direct action on proline residues within PrP, we constructed three mutants on the mouse PrP platform: P101L and P104L, which are homologous to the disease-linked mutations in humans, and the combined mutant P101L,P104L-PrP (dmPrP). When proteasomes were inhibited with ALLN, both P101L and P104L formed diffuse perinuclear structures, whereas dmPrP produced bona fide vimentin-caged aggresomes. This indicates that cyclophilins intervene directly in the folding and/or processing of wtPrPC. Our observations raise the possibility that weakening of PPIase activity during aging may contribute to the development of sporadic prion diseases by inducing PrP deposits in the brain. Similarly, familial GSS might, in some cases, be initiated by proteasomal malfunction that enables the aggregation of proline-substituted mutant PrP to form ‘prion seeds’ within aggresomes.
Results
Protease-resistant PrP aggregates in CsA-treated cells
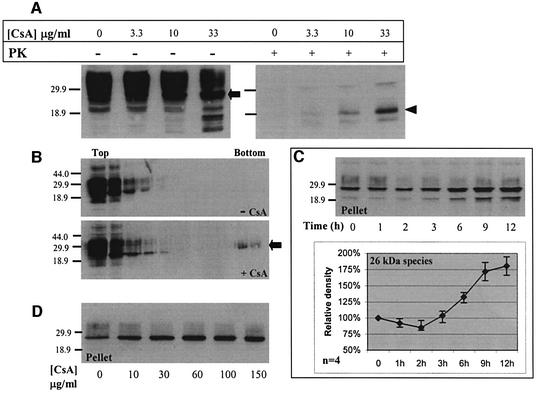
To see whether cyclophilins participate in the metabolism of PrPC, we used CsA. When N2a-M (Figure 1A) or CHO-M cells (data not shown) were treated for 24 h with CsA, there was a concentration-dependent increase in a 26 kDa PrP (Figure 1A, left panel, arrow). This species strictly co-migrated with the 26 kDa unglycosylated, full-length PrP found in untreated cells (Figure 1C and D; see also Figure 2). Since unglycosylated PrP is slightly ‘prion-like’ (Lehmann and Harris, 1996) we evaluated the protease resistance of this novel PrP species. In contrast to the endogenous unglycosylated 26 kDa band found in untreated cells, the CsA-induced PrP species resisted stringent proteolysis (proteinase K, 20 µg/ml, 37°C, 30 min) and produced a 19 kDa protease-resistant core (Figure 1A, right panel, arrowhead). Also contrasting with the endogenous unglycosylated species, the CsA-induced 26 kDa species formed large Sarkosyl-resistant aggregates that sedimented through 60% sucrose (Figure 1B, +CsA, arrow). Nineteen kilodalton protease-resistant cores and detergent insolubility are recurrent characteristics of pathologic or misfolded PrP (Meyer et al., 1986; Yedidia et al., 2001). These results thus indicated that CsA induced misfolded PrP.
Fig. 1. Protease-resistant and detergent-insoluble PrP species accumulate in CsA-treated cells (western blots). N2a-M cells were incubated with CsA as detailed below, and then lysed either in Triton-doc (A, C and D) or NOG (B) lysis buffers. Post nuclear supernatants (PNS) were then subjected to proteolysis or sedimentation as detailed below, and the PrP species were analyzed in western blots developed with the PrP mAb 3F4. (A) Cells were incubated with CsA at the indicated concentration for 24 h, and the lysates were analyzed with (+PK) or without (–PK) prior stringent proteolysis (20 µg/ml proteinase K, 30 min, 37°C). A major 26 kDa PrP species accumulated at high CsA concentrations (–PK, left panel, arrow). This band probably represents full-length, unglycosylated PrP. Proteolysis generated a major protease-resistant core of 19 kDa (+PK, right panel, arrowhead). (B) Cells were incubated for 36 h with or without 20 µg/ml CsA, as indicated. PNS were made 1% with Sarkosyl and centrifuged through 10–60% sucrose gradients containing 1% Sarkosyl. The CsA-induced 26 kDa species sedimented to the bottom of the gradient (arrow). (C) Cells were incubated with 15 µg/ml CsA for the indicated time. PNS were made 1% with Sarkosyl and separated into high-speed supernatants (not shown) and pellets. Densitometry of the 26 kDa band shows first a slight decrease and then a significant increase in the sedimenting PrP species (four independent experiments). (D) Cells were incubated for 6 h with the indicated CsA concentrations, and high-speed pellets were prepared as in (C). The insoluble PrP species increased until the CsA concentration reached 60 µg/ml, and then leveled off.
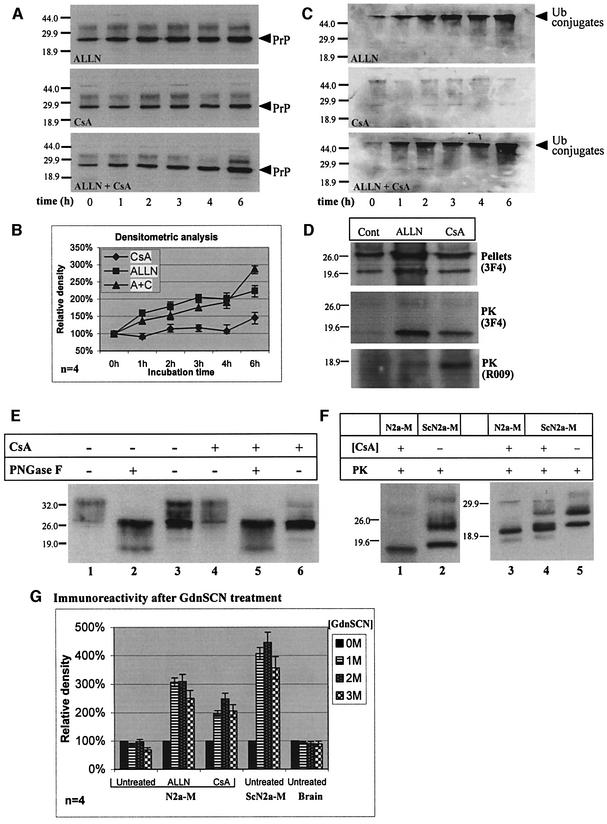
Fig. 2. CsA-induced PrP: kinetics of accumulation, proteasome resistance and comparison with PrPSc. (A–C) N2a-M cells were incubated with 150 µM ALLN, 25 µg/ml CsA, or both inhibitors (as indicated), for the indicated time. Sarkosyl-insoluble PrP species were then analyzed in western blots, which were first developed with 3F4 (A) and then reprobed with a ubiquitin (ub) mAb (C). Densitometry of the 26 kDa in (A) is shown in (B) (four independent experiments). (A and B) In contrast to CsA, ALLN caused an immediate increase in insoluble 26 kDa PrP. (C) ALLN, but not CsA, induced the accumulation of insoluble poly-ub conjugates (presumably proteasome bound). Since CsA did not inhibit the ubiquitylation machinery (ALLN + CsA), the lack of poly-ub conjugates with CsA alone indicates that proteasomes were not inhibited by this drug. (D) N2a-M cells were treated for 24 h with ALLN (75 µM), CsA (25 µg/ml) or left untreated (cont). PNS were separated into high-speed pellets (upper panel) and supernatants (data not shown) or subjected to stringent proteolysis (middle and bottom panels). The ALLN- and CsA-induced 26 and 22 kDa PrP bands co-migrated with the equivalent species in untreated cells. The 19 kDa protease-resistant cores elicited by CsA and ALLN also co-migrated, both when probed with 3F4 or with the C-terminal PrP antiserum R009. (E) N2a-M cells were either treated for 24 h with 25 µg/ml CsA or left untreated. Sarkosyl-insoluble fractions (lanes 3 and 6) were prepared by ultracentrifugation as described above. Lanes 1, 2, 4 and 5: total cell membranes were prepared from parallel cultures and extracted with sodium carbonate. Three-quarters of each membrane preparation was enzymatically deglycosylated with PNGase F (lanes 2 and 5), while the remaining membranes were analyzed without further treatment (lanes 1 and 4). That the Sarkosyl-insoluble 26 kDa bands strictly co-migrate with the deglycosylated membranal PrP confirms the ER origin of the former PrP species. (F) Comparison with PrPSc. The proteinase K-resistant core (20 µg/ml proteinase K, 30 min, 37°C) of CsA-treated (25 µg/ml CsA, 24 h) N2a-M cells (lanes 1 and 3) is smaller than that of the prion isoform PrPSc in untreated ScN2a-M cells (lanes 2 and 5). In ScN2a-M cells treated with CsA (lane 4), these two PK-resistant cores appeared as a doublet. (G) Histogram: denaturation-dependent immunoassay. Untreated, ALLN- (75 µM, 24 h) and CsA- (25 µg/ml, 24 h) treated N2a-M cells, untreated ScN2a-M cells and an uninfected hamster brain were lysed and spotted on nitrocellulose strips that were incubated with the indicated concentration of GdnSCN prior to development with 3F4. The ALLN and CsA samples, as well as the ScN2a-M lysates, were subjected to proteolysis (20 µg/ml, 37°C, 30 min) prior to blotting. The ALLN and CsA samples resembled the scrapie samples in that denaturation increased their immunoreactivity, in contrast to the PrPC present in the untreated N2a-M and the control brain.
To clarify the aggregation kinetics of PrP, N2a-M cells were exposed to 15 µg/ml CsA for 0–12 h, and their lysates were separated into Sarkosyl-soluble and -insoluble fractions. CsA affected insoluble PrP in two distinct phases (Figure 1C). (i) After a lag time during which insoluble 26 kDa slightly decreased, (ii) this insoluble PrP augmented continuously with time, especially after a longer (≥6 h) exposure. A still uncharacterized 22 kDa PrP band of variable intensity usually also appeared in the insoluble fraction, (see Figures 1C and 2D, pellets). In cells exposed to various CsA concentrations for 6 h, the levels of Sarkosyl-insoluble PrP increased until [CsA] reached 60 µg/ml, and thereafter leveled off (Figure 1D). Using densitometry (data not shown), we estimate that up to ∼10% of total PrP in the culture thus became insoluble. Incidentally, the absence of PrP molecular weight ladder in the CsA-treated samples suggests that insoluble PrP is not ubiquitylated in these cells.
Like most if not all GPI proteins, PrPC associates with cholesterol-rich ‘rafts’. To see whether CsA acts on this association, we used diagnostic Triton X-100 flotation gradients (Naslavsky et al., 1997). Our results (data not shown) indicate that: (i) CsA did not grossly alter the association of PrP with rafts; and (ii) the CsA-induced protease-resistant PrP is not associated with rafts.
CsA-induced PrP is not degraded by proteasomes
That CsA induced a 26 kDa PrP with a 19 kDa protease-resistant core resembles the effect of proteasome inhibitors, which accumulate PrP species of similar molecular weight by rescuing PrP from proteasomal degradation (Figure 2A, ‘ALLN’; Figure 7A, filled diamonds) (Ma and Lindquist, 2001; Yedidia et al., 2001). To understand the relationship between the effects of ALLN and CsA, we performed a combined kinetic experiment. N2a-M cells were exposed to 150 µM ALLN or 25 µg/ml CsA, or both, for 0–6 h (western blot in Figure 2A). Both drugs elicited Sarkosyl-insoluble 26 kDa PrP of indistinguishable electrophoretic mobility (see also Figure 2D), but with different kinetics (Figure 2A and B). The immediate effect of ALLN reflects the rescue of cytosolic, proteasome-bound PrP molecules (Yedidia et al., 2001) (Figure 7A, stage 5). In contrast, the more complex, non-monotonous kinetics observed with CsA suggests that this drug may exert multiple effects on PrP metabolism. One possibility is that CsA acts indirectly to slow the exit of misfolded PrP to the cytosol, where it aggregates more readily.
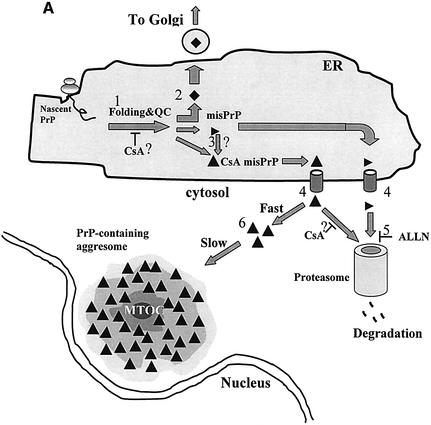
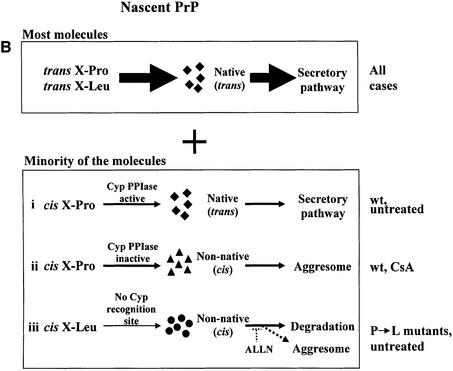
Fig. 7. Possible actions of CsA on PrP metabolism. (A) The influence of CsA on PrP metabolism. (i) In the absence of CsA, ∼90% of nascent PrP molecules in the ER gain their native conformation (stage 1) and are exported to the secretory pathway (stage 2, filled diamonds). ER cyclophilins (presumably cypB) may play a role in normal PrP folding, perhaps through their PPIase activity. PrP chains that fail to fold properly (horizontal triangles) are directed to degradation via the ERAD machinery. In the course of ERAD, these misfolded PrP molecules are dislocated into the cytosol (stage 4) and are degraded by proteasomes (stage 5). Degradation is inhibited by ALLN. (ii) The action of CsA is likely to be manifold and may target several of the normal metabolic steps outlined above. (a) By inhibiting the PPIase activity of cyclophilins, CsA may interfere with stage 1, perhaps by hampering the rescue of a minority of PrP molecules with cis X-Pro peptide bonds (see text and panel B). As a result, specific CsA-induced PrP molecules are formed (filled triangles). The CsA-induced PrP are likely to include non-native X-Pro isomers. (b) CsA could also act on some spontaneously misfolded PrP molecules (horizontal triangles) and convert them into CsA-induced species (stage 3, filled triangles). In either case, the CsA-induced species (filled triangles) are dislocated into the cytosol (stage 4). However, in contrast to the normally misfolded species, they escape proteasomal degradation and accumulate in aggresomes (stage 6). We envisage two mechanisms whereby CsA-induced PrP could avoid proteasomes. First, they might form tight aggregates at the exit of the sec61p dislocon, and these deposits may be unresolvable by the cytosolic chaperone system in the absence of cyclophilin activity. Another possibility is that these species cannot be unfolded to permit proteasomal degradation, perhaps because correcting their aberrant X-Pro bonds requires cytosolic cyclophilins, which are inhibited by CsA. (B) Model for the generation of misfolded PrP by wtPrP and by the P102L and P105L mutants. Upper panel: in all cases the majority of both wild-type and mutant (P102L and P105L) PrP is formed with predominantly trans X-Pro (or trans X-Leu) peptide bonds (which are energetically favored). Lower panel: (i) in untreated cells expressing wtPrP, the small minority of nascent PrP with cis X-Pro bonds is corrected to trans by ER cyclophilins and adopts a native conformation (filled diamonds); (ii) when cells expressing wtPrP are exposed to CsA, the cis-containing PrP molecules fail to be rescued and they misfold and adopt prion-like biochemical properties (panel A, filled triangle); (iii) a very small minority of mutant P→L-PrP molecules is formed with aberrant cis X-Leu bonds (at positions 102 or 105, depending on the mutant). Since cyclophilins fail to correct these molecules, they misfold into prion-like PrP. These molecules are normally degraded, hence ALLN is needed to enable their accumulation in aggresomes. Because spontaneous cis X-Leu bonds (in the mutants) are less probable than their cis X-Pro counterpart (in wtPrP), this model predicts that less prion-like PrP will be generated in (iii) than in (ii).
The observation that CsA-induced insoluble 26 kDa PrP accumulated in the absence of proteasome inhibitors (Figure 2A, CsA) suggested that this species is not degraded by proteasomes. To verify that CsA did not inhibit proteasomes, we reprobed the membranes shown in Figure 2A with a ubiquitin (ub) monoclonal antibody (mAb) (Figure 2C). Inhibiting proteasomes with 150 µM ALLN provoked the accumulation of insoluble poly-ub conjugates (Figure 2C, ALLN). That CsA did not prevent this effect of ALLN (ALLN + CsA) demonstrates that CsA does not interfere with the ubiquitylation machinery. Thus, the finding that CsA alone failed to induce such conjugates (CsA) indicates that proteasomes were active in the presence of CsA. Because CsA-induced PrP species did access the cytosol (Figures 3–5), their accumulation shows that they either resist proteasomes or are inaccessible to them. Proteasome sensitivity thus distinguishes the CsA-induced PrP (Figure 7A, filled triangles) from that revealed by ALLN (Figure 7A, filled diamonds). Whether this property is acquired in the ER or in the cytosol remains to be seen.
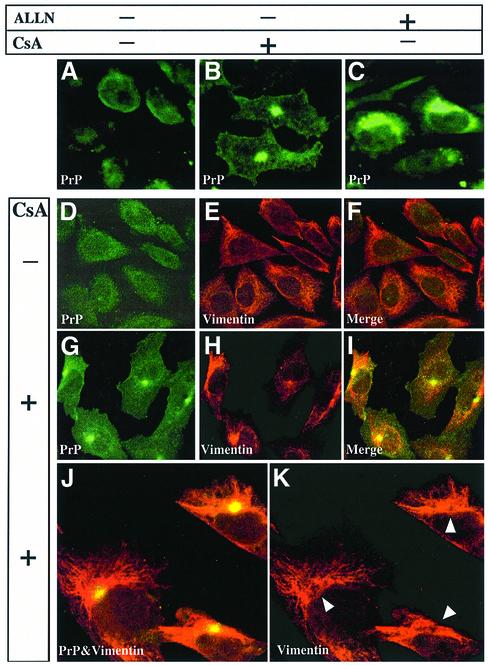
Fig. 3. PrP accumulates in vimentin-surrounded aggresomes in CHO-M cells treated with CsA but not with ALLN. (A–C) CHO-M cells were incubated for 24 h either with 20 µg/ml CsA (B) or with 75 µM ALLN (C), or left untreated (A). Focal PrP deposits were seen in ∼10% of the CsA-treated cells (B). In contrast, in ALLN-treated cells, PrP accumulated diffusely throughout the cytosol (C). (D–K) To further characterize the PrP-containing deposits, CHO-M cells were incubated for 48 h without (D–F) or with (G–K) 30 µg/ml CsA. The cells were labeled using the RO73 PrP antiserum (green) and with an anti-vimentin mAb (red). In the CsA-treated cells, the PrP foci were surrounded by collapsed vimentin fibers. A higher magnification (J and K) confirmed that vimentin cages enclose the PrP deposits (see arrowheads in K).
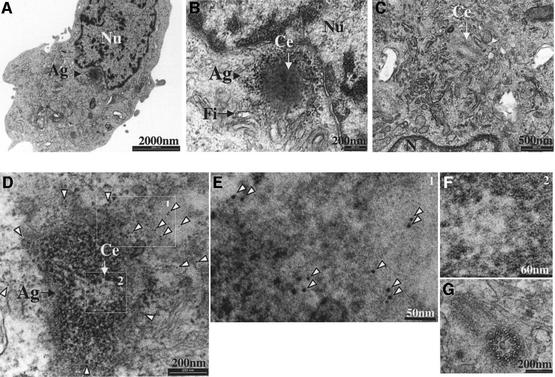
Fig. 5. PrP aggresomes characterized by thin-section EM. CHO-M cells were incubated with 15 µg/ml CsA for 48 h and examined by transmission EM. (A) Micrograph of a CHO-M cell containing a juxtanuclear aggresome. (B) A higher magnification of (A) shows that the structure impacts on the nucleus and is surrounded by filamentous material. A centriole (Ce) is clearly seen within the aggresome. (C) No similar structures were seen in the centriolar area of untreated cells (n = 100). (D–G) CHO-M cells were treated with 15 µg/ml CsA for 48 h and subjected to pre-embedding immunogold staining using RO73 (6 nm gold). Most gold particles were observed in the vicinity of the aggresome, indicating the presence of PrP (D; higher magnification in E). A centriole is clearly seen within the aggresome (D; higher magnification in F). No gold particles were seen around centrioles of untreated cells (G). Nu, nucleus; Ag, aggresome; Fi, filamentous material; Ce, centriole.
CsA-PrP and ALLN-PrP are biochemically similar, but differ from PrP27–30
We next sought to ascertain that the CsA-induced 26 kDa PrP band originated in the ER. Although PrP contains an ER signal peptide, one effect of CsA could have been to interfere with the targeting of signal peptide-containing proteins to the rough ER, or to otherwise prevent their translocations through the translocon. Although side-by-side electrophoresis confirmed that both the CsA- and the ALLN-induced 26 kDa bands co-migrated with the equivalent species found in the Sarkosyl-insoluble fraction of untreated N2a-M cells (Figure 2D, pellets), we reasoned that the identity of these detergent-insoluble PrP species has never been formally established, even for untreated cells.
To verify the ER origin of the 26 kDa insoluble PrP species, we compared their Mr to that of mature PrPC before and after enzymatic deglycosylation (Figure 2E). A cytosolically synthesized PrP would contain both the ER signal peptide and the GPI attachment sequence, and would thus be heavier than ER species by an easily detectable 6 kDa. Carbonate-extracted microsomal preparations of untreated (lanes 1–3) or CsA-treated (lanes 4–6) N2a-M cells were either deglycosylated with PNGase F (lanes 2 and 5) or left untreated (lanes 1 and 4). They were then analyzed on western blots together with Sarkosyl-insoluble fractions extracted from cells in parallel dishes (lanes 3 and 6). Both in the presence of CsA and in its absence, the insoluble species strictly co-electrophoresed with the deglycosylated PrPC. This indicates that the Sarkosyl-insoluble 26 kDa PrP originates in the ER.
The 19 kDa protease-resistant cores elicited by CsA and ALLN also strictly co-migrated with each other, both when probed with 3F4 and with the C-terminal PrP antiserum (R009) (Figure 2D, PK). In contrast, the CsA-induced 19 kDa core was smaller than the unglycosylated, protease-resistant core of bona fide PrPSc found in scrapie-infected ScN2a-M cells (Figure 2F, lanes 1 and 2) (Butler et al., 1988). To confirm this Mr disparity, we extended our analysis to the effects of CsA on prion-infected cells. Treating ScN2a-M cells with CsA for 24 h resulted in the appearance of a protease-resistant doublet around 19 kDa (lane 4). The upper band in this doublet clearly represents the unglycosylated PrP27–30 endogenous to these cells (compare with untreated cells, lane 5), whereas the lower band was CsA specific (compare lane 3 with 4). This analysis suggests that the PrP conformation elicited by CsA resembles that provoked by ALLN, but that they both differ from that of the RML strain of PrPSc found in ScN2a-M cells (Collinge et al., 1996).
We further compared these various PrP species by using a denaturation-dependent immunoassay (Serban et al., 1990; Safar et al., 1999). Lysates of N2a-M, ScN2a-M and of a non-infected Syrian hamster brain were spotted on nitrocellulose strips that were subsequently incubated with the indicated concentration of guanidine thiocyanate (GdnSCN) prior to development with 3F4 (Figure 2G). The samples from ALLN- and CsA-treated cells, as well as the lysate of the scrapie-infected cells, were treated with proteinase K (20 µg/ml, 37°C, 30 min) prior to blotting. This assay showed that denaturation was needed to expose the 3F4 epitope in both ALLN- and CsA-induced protease-resistant PrP in that these species resembled the scrapie samples and differed from the normal PrPC found in the uninfected N2a-M and brain. The effects of CsA on PrPSc will be described further elsewhere (E.Cohen, manuscript in preparation).
CsA-induced PrP accumulates in aggresomes
The foregoing results showed that CsA and ALLN induce PrP species with similar biochemical properties but disparate cellular fates. Furthermore, these inhibitors also caused strikingly different PrP subcellular distribution. In ∼10% of N2a-M (data not shown) and CHO-M cells (Figure 3B) exposed to 20 µg/ml CsA for 24 h, PrP formed a single juxtanuclear focus. In comparison, there was a diffuse cytoplasmic PrP pattern in cells exposed to 75 µM ALLN for 24 h (Figure 3C). These results are consistent with previous studies (Ma and Lindquist, 2001; Yedidia et al., 2001). Several other ALLN regimes (from 150 µg/ml for 12 h to 15 µg/ml for 48 h) also failed to yield juxtanuclear foci (data not shown).
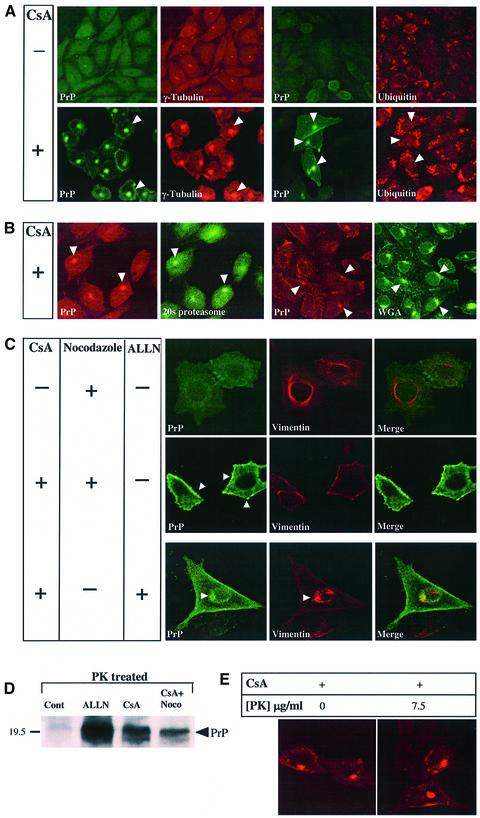
The PrP foci in CsA-treated cells strongly resembled the aggresomes previously observed with mutants of cystic fibrosis transmembrane regulator (CFTR), of presenilin 1 (PS1) (Johnston et al., 1998), and of other proteins (Garcia-Mata et al., 1999). The following results further strengthened this resemblance to aggresomes: (i) selective digitonin permeabilization, indicating that the PrP deposits were probably not enclosed by a membrane (data not shown); (ii) in CsA-treated CHO-M cells, the PrP foci (Figure 3G–K, green) were surrounded and caged by collapsed vimentin fibers (red); and (iii) the PrP foci co-localized with the microtubule-organizing center (MTOC) component γ-tubulin (Figure 4A) as well as with the 20S proteasome α-subunit (Figure 4B) (Wigley et al., 1999). In contrast, the PrP foci were not spots of general protein deposition, since they did not contain either PS1, APP, the raft resident endothelial surface antigen (ESA) (data not shown) or detectable ub (Figure 4A, ubiquitin). The CsA-induced PrP dots were found in the same general region as, but were more focused than, that stained with wheat germ agglutinin (Golgi and lysosomes; Figure 4B, WGA).
Fig. 4. The PrP aggresomes co-localize with γ-tubulin and 20S proteasomes but not with ubiquitin; they are inhibited by nocodazole and contain protease-resistant PrP. (A and B) CHO-M cells were incubated for 24 h either with or without 30 µg/ml CsA, as indicated, and examined by immunofluorescent confocal microscopy. PrP was detected using either RO73 (A, green channel) or 3F4 (B, red) and additional Abs as indicated. PrP aggresomes co-localized with the centrosomal marker γ-tubulin (A) and with 20S proteasomes (B) but not with ub (A), and they were found in the general region of the cells that stained with wheat germ agglutinin (WGA) (B). (C) CHO-M cells were incubated for 24 h either with 10 µg/ml nocodazole, with 30 µg/ml CsA and 10 µg/ml nocodazole or with 30 µg/ml CsA and 75 µM ALLN, and then examined by immunofluorescence for PrP (R073, green) and vimentin (red). Nocodazole prevented the formation of PrP aggresomes by CsA, but multiple PrP aggregates could be seen throughout the cell and especially underneath the cell surface (arrowheads). ALLN and CsA induced diffuse cytosolic accumulation as well as a single juxtanuclear, vimentin-caged deposit of PrP (arrowhead). (D) N2a-M cells treated for 24 h either with 25 µg/ml CsA, or with 25 µg/ml CsA and 10 µg/ml nocodazole, were subjected to proteolysis (20 µg/ml proteinase K, 30 min, 37°C) prior to western blotting with 3F4. Nocodazole did not prevent the formation of PK-resistant PrP, and the formation of aggresomes is thus not a prerequisite for the protease resistance of the CsA-induced PrP species. (E) CHO-M cells were treated with CsA (30 µM, 48 h), and then fixed with formalin and incubated with proteinase K (right panel; 7.5 µg/ml, 30 min, 37°C) prior to immunodetection of PrP with RO73 (red). The PrP immunoreactivity in aggresomes was protease resistant.
The PrP foci were further similar to aggresomes in that nocodazole (10 µg/ml), a microtubule-disrupting agent, prevented their formation (Figure 4C) (Johnston et al., 1998). However, nocodazole did not prevent the congregation of PrP into multiple aggregates that were spread throughout the cell (arrowheads), nor did it completely stop the formation of proteinase K-resistant PrP (Figure 4D). Thus, CsA induces the formation of small aggregates containing protease-resistant PrP even when aggresomes are prevented.
Combined treatment with both CsA (30 µg/ml) and ALLN (75 µM) (Figure 4C) resulted in juxtaposed PrP patterns of both drugs within the same cells, suggesting that these drugs acted largely independently on PrP accumulation (Figure 4C, bottom-most panel).
To verify that cyclosporin-induced aggresomes indeed contained protease-resistant PrP, we used an in situ proteolysis assay on fixed cells (Figure 4E) (Taraboulos et al., 1990). PrP immunoreactivity in the aggresomes resisted a stringent treatment with proteinase K (7.5 µg/ml, 30 min, 37°C). This finding thus correlates the microscopic observations with the biochemical data described in Figures 1 and 2.
PrP aggresomes observed by thin-section EM
We next turned to transmission EM. About 10% of CsA-treated CHO-M cells, but none of ∼100 untreated cells, displayed juxtanuclear foci in which centrioles (Figure 5, Ce) were seen within an electron-dense region (Figure 5B and D). These aggresomal structures impacted on the nucleus and were not enclosed by a membrane bilayer. That the electron-dense material contained PrP was confirmed in a pre-embedding immunogold experiment (Figure 5D and E). While this technique results in inferior ultrastructure, gold particles clearly stained the aggresome periphery (Figure 5D and E, arrowheads). Whether the distal immunogold distribution reflects the totality of PrP in these aggresomes, or whether it results from the inability of the gold particles to enter the dense core of aggresomes, is still obscure. In contrast, no gold particles were seen around the centrioles of untreated cells (Figure 5G, n = 100). Further studies will help characterize the molecular content of the CsA-induced aggresomes.
A double proline mutant accumulates in aggresomes when proteasomes are inhibited
The foregoing observations suggested that cyclophilins, which are the only known cellular CsA receptors, influence the metabolism of PrP. However, the results did not establish whether cyclophilins catalyze the isomerization of X-Pro bonds within PrP itself, or whether their action on PrP is indirect. To address this question, we turned to genetics. We reasoned that if cyclophilins act directly on PrP during normal folding, then replacing prolines in the PrP sequence might recapitulate some of the phenomena observed with CsA.
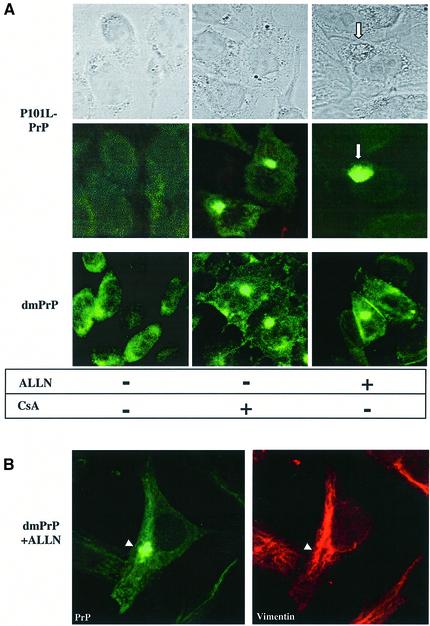
In a first step in this direction, we focused on residues P102 and P105. The substitutions P102L and P105L are linked, separately, to familial GSS (Hsiao et al., 1989; Yamazaki et al., 1999), but the etiology is poorly understood. The biochemical properties of MoP101L-PrP (the mouse homolog of HuP102L-PrP) have been characterized by Harris and colleagues, who have shown that this mutant is slightly protease resistant (Lehmann and Harris, 1995). We thus constructed plasmids encoding either one of these mutations, or both (dmPrP), in the mouse PrP–MHM2 background, to see whether they display CsA-like properties. In both CsA-treated and -untreated cells, the mutants had unremarkable, wild-type-like subcellular distributions (Figure 6A). Distinct properties of the mutants were revealed, however, when the cells were treated with ALLN (15 µM, 48 h). In a small number of cells, P101L-PrP (Figure 6A) and P104L-PrP (data not shown) deposited in large and diffuse perinuclear structures that we never observed in ALLN-treated cells (compare with Figure 3C). In contrast to this unfocused deposition, in ALLN-treated cells, dmPrP formed compact juxtanuclear foci (Figure 6A) that were caged by vimentin fibers (Figure 6B) and were thus identified as aggresomes. That proline mutants mimicked some of the effects of CsA suggests that P101 and P104 are normally targets of cyclophilins. This issue is discussed further below. The biochemical properties of P104L-PrP and dmPrP will be described elsewhere(E.Cohen, manuscript in preparation).
Fig. 6. The double proline PrP mutant (dmPrP) accumulates in aggresomes of ALLN-treated cells. (A) CHO cells expressing P101L-PrP or the double mutant (P101L,P104L) dmPrP, as indicated, were treated either with 30 µg/ml CsA or 15 µM ALLN for 48 h or left untreated and examined by confocal immunofluorescence microscopy using RO73 (green). In untreated cells as well as in cells treated with CsA, both mutants behaved similarly to the wtPrP. In contrast, in ALLN-treated cells, the distribution of the mutants differed significantly from that of wtPrP as: (i) P101L-PrP formed diffuse perinuclear deposits (lower panel), which could sometimes be seen by Nomarski optics as well (upper panel); (ii) in a few cells, dmPrP formed tight juxtanuclear foci, and because these deposits were surrounded by a vimentin cage (B), we identified them as aggresomes.
Discussion
We found that CsA, a widely used immunosuppressant, induces the formation of detergent-insoluble and protease-resistant aggregates of the wtPrP in cultured cells. These CsA-induced PrP species accumulate in aggresomes, a property that was somewhat mimicked by disease-linked proline mutants of PrP. Our results point to a novel possible link between neurodegeneration and aggresomes, and suggest mechanisms whereby sporadic and familial prion disease may in some cases be initiated by misfolded cytosolic PrP (see the model in Figure 7). Whether induction of aggresomes may contribute to the clinical side-effects of CsA remains to be determined.
How does CsA induce misfolded PrP?
CsA could act at one or more stages of the synthesis of PrP or of its degradation (Figure 7A). It is important to note that while CsA is an inhibitor of the PPIase activity of cyclophilins, it also interferes with non-enzymatic properties of these proteins, such as their participation in protein complexes (Uittenbogaard et al., 1998; Balbach and Schmid, 2000). We will, however, concentrate our discussion on the PPIase-inhibiting aspect of CsA. In relaxed peptides (and presumably in nascent polypeptides as well), the majority of X-Pro bonds are in the trans conformation, but there is a minority of bonds that are formed spontaneously in the cis conformation (Reimer et al., 1998). One way in which CsA could exert its influence on PrP is by preventing ER cyclophilins from isomerizing trans X-Pro peptide bonds within PrP to the cis conformation (assuming that the native conformation contains cis bonds; Figure7 A, stage 1). However, such an action would concern the majority of nascent PrP molecules. Thus, the findings that only a small portion of PrP molecules were misfolded by CsA (Figure 1B) and that the action of CsA was saturable (Figure 1D) militate against such a mechanism.
A more attractive possibility is that CsA acts on the small minority of PrP polypeptides that are synthesized with non-native cis X-Pro bonds. This model is depicted in Figure 7B (rows i and ii). In cells not exposed to CsA, ER cyclophilins (presumably cypB) could act by rescuing the minority of cis X-Pro-containing nascent PrP by isomerizing them to the presumably native trans conformation (Figure 7B, row i). By preventing such a rescue, CsA would cause the accumulation of a small number of cis X-Pro-containing, misfolded PrP molecules (Figure 7B and A, row ii and filled triangles, respectively). That the insoluble CsA-induced PrP originates in the ER is shown by the results of the deglycosylation experiment (Figure 2E). This scenario predicts that only a minority of PrP molecules are affected by CsA and is thus consistent with our quantitative data. The model could also provide a mechanism for the distinct properties of the CsA-induced PrP species. Since CsA inhibits cyclophilins in all subcellular compartments, cis X-Pro bonds would fail to be corrected by cyclophilins even after their dislocation into the cytosol and would thus retain aberrant properties, which would eventually target them to aggresomes.
Why do CsA-induced PrP species (Figure 7A, filled triangles) resist proteasomes? One possibility is that rapid aggregation of these species (Figure 7A, step 6) at the exit of the dislocon prevents their degradation. In this view, CsA-specific PrP would thus be more ‘aggregation-prone’ (Kopito and Ron, 2000; Kopito and Sitia, 2000) than the normally misfolded PrP that successfully undergoes ERAD in untreated cells (Figure 7A, horizontal triangles). Alternatively, the CsA-induced PrP could resist the unfolding required for degradation (Navon and Goldberg, 2001). Although this unfolding seems to take place on the surface of the proteasome, it is possible that it could require the assistance of cyclophilins and thus be inhibited by CsA (Navon and Goldberg, 2001). In addition, CsA could also act in the cytosol by protecting a subset of cytosolic PrP molecules from proteasomal-like activities such as the self-compartmentalizing protease (Geier et al., 1999). Whether such non-proteasomal degradation requires the assistance of cyclophilins is unknown.
Do cis X-Leu peptide bonds feature in the proline-linked GSS?
The model depicted in Figure 7B proposes that, by preventing cyclophilins from isomerizing X-Pro peptide bonds, CsA causes the formation of aberrant PrP molecules. Although this could apply to any of the X-Pro bonds in mature wild-type huPrP, it is tempting to focus on P102 and P105, the two proline residues that are linked to familial GSS [P102L (Hsiao et al., 1989); P105L (Yamazaki et al., 1999)]. If P102 and P105 are cyclophilin substrates, then the action of CsA on PrP could be exerted, at least in part, through these two residues. The model would then predict that cis bonds in positions 102 and/or 105 promote PrP with non-native prion-like properties.
How does our model accommodate the proline to leucine mutations? While proline is especially prone to form spontaneous cis peptide bonds, other amino acids can also form bonds in cis, albeit with considerably smaller probabilities (0.1–0.5%; see Balbach and Schmid, 2000). It is thus plausible that P102L- and P105L-PrP are sometimes (even if rarely) synthesized with cis X-Leu bonds at positions 102 or 105. Because these bonds are not efficient substrates for cyclophilins, they would remain uncorrected and result in the formation of prion-like PrP that is similar to the CsA-induced PrP (Figure 7B, row iii). This model has some verifiable predictions. (i) It forecasts that while a small number of P102L or P105L PrP molecules may acquire prion-like properties, the vast majority of these mutant molecules will form in the native conformation. This prediction is consistent with the observations that both P101L-PrP (Lehmann and Harris, 1995) and P104L-PrP as well as the double mutant PrP (dmPrP) (E.Cohen, manuscript in preparation) are only slightly prion like. (ii) The model also forecasts that much less prion-like PrP is formed by these mutants in untreated cells than by wtPrP in cells treated with CsA. (iii) Finally, the model depicted in Figure 7B suggests that the proline mutants might be similar to the CsA-induced PrP species in their tendency to form aggresomes. The results shown in Figure 6 confirm these predictions. dmPrP was directed by the cellular machinery to aggresomes. That ALLN was required to enable the detection of dmPrP in aggresomes may be explained by the small amounts of misfolded PrP formed by these mutants.
Aggresomes and prion diseases
How do these observations relate to prion diseases? The CsA-induced PrP displayed striking prion-like characteristics. Of particular interest is the induction by CsA of a 19 kDa, protease-resistant core. Such cores are found almost invariably in prion disease and their size has been used to type the strains of prion diseases (Collinge et al., 1996). Nineteen kilodalton cores have also been observed in the P101L mutant (Lehmann and Harris, 1995).
There seems to be a consensus in the prion literature that prion-like properties do not unequivocally correlate with infectivity. Many authors have reported that prion-like PrP can be induced by metabolic disturbances or PrP overexpression. For instance, tunicamycin (Lehmann and Harris, 1997), DTT and a combination of both inhibitors (Ma and Lindquist, 1999), as well as ALLN (Yedidia et al., 2001), cause the accumulation of insoluble, protease-resistant PrP in treated cells. Thus, we do not suggest that PPIases malfunction or their inhibition by drugs directly produces infectious prions. However, it is conceivable that a self-propagating ‘prion order’ could be spontaneously generated in long-lived aggresomes. Such an event could provide the ‘prion seed’ required for igniting spontaneous (familial or sporadic) (Figure 7B, lanes iii and ii, respectively) prion diseases. That these diseases appear very late in life could be explained in part by the paucity of misfolded PrP molecules produced by the cis bonds model (Figures 6 and 7B). The possible weakening of cyclophilin activity in old age could also contribute to the initiation of spontaneous prion diseases.
Because of their cytosolic location, putative prion seeds generated in aggresomes would initially be unable to interact with wtPrPC (which is found primarily in the luminal side of the endomembrane system). However, autophagocytic mechanisms could eventually bring these putative prion seeds to lysosomes, which are one natural location of the pathogenic PrPSc in prion-infected cells (McKinley et al., 1991). This scenario is supported by reports that autophagocytosis is increased in cells with aggresomes (Kopito, 2000). In any case, bioassays are needed to see whether prion infectivity is generated in CsA-treated cells.
Do aggresomes contribute to CsA cytotoxicity?
To the best of our knowledge, this is the first report of the induction of aggresomes by a therapeutic drug. The CsA-induced aggresomes might be more specific than their ALLN-induced counterparts because they are likely to contain only the proteins that need cyclophilins for proper handling. Although we detected no ubiquitin immuno reactivity in the CsA-induced aggresomes, proteasomes did concentrate in their vicinity (Figure 4). More studies are required to characterize the content of the CsA-induced aggresomes.
Can the phenomena described here contribute to the clinical side-effects of CsA? Although the CsA concentrations used in our experiments (15–30 µg/ml) are considerably higher then the serum levels obtained in patients (∼250 ng/ml; Warrens et al., 1999), chronic administration of the drug could result in cumulative effects. Side-effects of CsA, including neurological signs such as ataxia (Atkinson et al., 1984; Thompson et al., 1984), remain unexplained, and it will be pertinent to verify whether protein misfolding and aberrant aggregation and deposition could contribute to these episodes. In addition, neuroprotective protocols, which involve direct intraventricular administration of CsA, attain cerebrospinal fluid concentrations that are similar to the ones used here (∼20 µg/ml; Scheff and Sullivan, 1999).
When studying the possible contribution of aggregates to CsA toxicity, it is meaningful to recall that far from being innocuous, aggresomes impair the ubiquitin– proteasome pathway (Bence et al., 2001). These properties of aggresomes could well contribute to CsA-related cytopathological effects. It has also been suggested that aggresomes might contribute to runaway reactions whereby more misfolded proteins fail to be digested and join aggresomes (Kopito, 2000; Kopito and Ron, 2000).
Materials and methods
Materials
Cell culture reagents were purchased from Biological Industries (Beit Haemek, Israel). Tissue culture plates were purchased from Miniplast (Ein Shemer, Israel) or Nunc (Roskilde, Denmark). G418 and N-glycosidase F (PNGase F) (362185) were purchased from Calbiochem (San Diego, CA). Protein concentration was determined using a BCA kit (Pierce 23223). Proteinase K was from Roche (161519). Reagents for EM were purchased from Agar Scientific (Stansted, UK) or Sigma (St Louis, MO). CsA (C1832) and ALLN (A6185) were purchased from Sigma. All other chemicals were from Sigma.
Cell cultures and transfections
N2a-M and CHO-M cells stably express moderate levels of the MHM2–PrP chimeric protein (Scott et al., 1992), which carries the 3F4 epitope. Cells were grown at 37°C in DMEM16 (low glucose) supplemented with 10% FCS. Stable transfections of the PrP constructs were achieved with the non-liposomal reagent FuGENE 6 (Roche).
Construction of plasmids
In situ mutagenesis was achieved by the method of Kunkel, using the following primers: TGGAACAAGCTCAGTAAGCCAAAAACC for P101L-PrP, TGGAACAAGCCCAGTAAGCTAAAAACC for P104L-PrP and TGGAACAAGCTCAGTAAGCTAAAAACC for the double mutant P101L,P104L (dmPrP). The presence of the mutation was confirmed by DNA sequencing. The mutated gene was excised from pBluescript (EcoRI and NheI) and cloned into the vector pCI-neo (Promega, Madison, WI). Plasmid DNA for transfections was prepared with the WizardPlus maxiprep DNA purification system (Promega).
Antibodies
The rabbit antiserum R073 binds both mouse PrP and MHM2–PrP (Serban et al., 1990). The mAb 3F4 (Kascsak et al., 1987) binds to residues Met109 and Met112 in the chimeric MHM2–PrP but does not recognize the wild-type mouse PrP endogenous to N2a cells (Rogers et al., 1991). These antibodies were used at a dilution of 1:5000 for western blots or 1:2000 for immunofluorescence (of the serum or the ascitic fluid, as appropriate). The ubiquitin mAb (MMS-258R) was from Babco (Richmond, CA) and was used at a 1:10 000 dilution in western blots or 1:3000 in immunofluorescence. Vimentin mAb (clone 9; V-6630) and γ-tubulin mAb (clone GTU-88; T-6557) were from Sigma. Rabbit anti-20S proteasome α-subunit (539153) was purchased from Calbiochem. Secondary antibodies conjugated to FITC, Texas Red, horseradish peroxidase or gold were purchased from Jackson ImmunoResearch (West Grove, PA)
PrP analysis
SDS–PAGE and western immunoblotting of PrP were carried out as described previously (Taraboulos et al., 1995). Cells were lysed either in ice-cold Triton-doc lysis buffer (0.5% Triton X-100, 0.25% Na deoxycholate, 150 mM NaCl, 10 mM Tris–HCl pH 7.5, 10 mM EDTA) or in NOG lysis buffer (2% n-octyl-β-d-glucopyranoside in PBS), as indicated. NOG lysates were incubated for 30 min on ice prior to centrifugation, while Triton-doc lysates were immediately centrifuged. The lysates were spun for 1 min at 6000 r.p.m. in an Eppendorf microfuge. Biochemical analyses were performed on the post-nuclear supernatant (PNS). Aggregated PrP was separated from soluble fractions by one of two procedures. (i) Sedimentation: PNS were brought to 1% Sarkosyl, incubated on ice for 30 min and then spun at 45 000 r.p.m. for 1 h at 4°C in a TL45 rotor (109 000 g). The pellets were then resuspended in lysis buffer. (ii) Velocity sedimentation through sucrose gradient: 10–60% sucrose step gradient in TNS (10 mM Tris–HCl pH 7.5, 150 mM NaCl, 1% Sarkosyl) were prepared in 2 ml TLS-55 tubes (300 µl each of 60, 30, 25, 20, 15 and 10% sucrose). The PNS was laid on top of the gradient and the tubes were then spun at 55 000 r.p.m. (200 000 g) for 1 h at 4°C in a TLS-55 rotor (Beckman). Eleven fractions of 180 µl each were collected from the top of the tube. Western blots were developed using an ECL system. For reprobing, PVDF membranes were stripped by incubation in 300 mM NaOH [5 min, room temperature (RT)], followed by neutralization by several rinses in TBST (10 mM Tris–HCl pH 7.5, 150 mM NaCl, 0.3% Tween-20).
Deglycosylation of PrP
PNGase F experiments were performed on purified membranes. Cells were rinsed, scraped and resuspended in ice-cold HEPES buffer (20 mM HEPES, 150 mM NaCl, 1 mM MgCl2, 0.1 mM CaCl2), and then homogenized by sonication in a bath sonicator (3 × 10 s) in glass tubes. The homogenate was spun in a microfuge (5000 r.p.m., 2 min). The supernatant was then brought to 100 mM Na2CO3 pH 11.5, incubated for 20 min on ice and then spun (1 h, 200 000 g) through a 300 µl sucrose cushion (20% sucrose in 100 mM Na2CO3). Membrane pellets were washed in 50 mM phosphate buffer (PB) pH 7.5 and boiled for 1 h in 0.2% SDS in PNGase F buffer (50 mM PB pH 7.5, 0.05 M βME). After cooling, TX-100 was added to a final concentration of 0.75%. Deglycosylation was performed with PNGase F (1 U/100 µl, 37°C, 24 h).
Denaturation-dependent immunoassays
Eighty microliter lysates of cell or brain were loaded on nitrocellulose membranes (Schleicher and Schuell, Dassel, Germany) using a suction apparatus. The membranes were rinsed for 10 min in TBST, and then strips incubated for 5 min at RT in the indicated concentration of guanidine thiocyanate (GdnSCN) prepared in 50 mM Tris–HCl pH 7.5. After extensive rinses in TBST, the strips were blocked with homogenized 1% fat milk and then processed for immune detection as described previously (Taraboulos et al., 1995).
Immunofluorescence microscopy
To detect total PrP, cells were grown on 8-well plastic slides (Nunc), fixed (8% formalin in PBS, 30 min, RT), quenched with cold 1% NH4Cl in PBS and then permeabilized (0.1% TX-100 in PBS, 2 min, RT) and blocked with 2% BSA in PBS (30 min, RT). The cells were then incubated with the relevant primary Ab (in 1% BSA in PBS, overnight, 4°C), rinsed, and the secondary Ab conjugated either to FITC or to Texas Red (diluted 1:200 in 1% BSA in PBS) was added for 30 min (RT). The labeled cells were mounted in an anti-fading preparation (5% n-propyl gallate, 100 mM Tris–HCl pH 9, 70% glycerol) (Giloh and Sedat, 1982) and viewed with a Zeiss axiovert 135 confocal microscope. For the in situ protease resistance experiment (Figure 4E), the cells were fixed and then treated with proteinase K (7.5 µg/ml, 30 min, 37°C) prior to the blocking as described previously (Taraboulos et al., 1990).
Transmission EM
To detect aggresomal structures, CHO-M cells were grown to 75% confluence in 100 mm dishes and then incubated with or without 20 µg/ml CsA for 48 h. The cells were then resuspended with 1 ml of trypsin–EDTA for 3 min (RT) and the trypsin then inhibited with 3 ml of complete medium. The cells were spun for 5 min at 1200 r.p.m. in a Beckman TJ-6 centrifuge, and the pellets rinsed in cold PBS and then fixed in 2% paraformaldehyde, 1.5% glutaraldehyde in 100 mM PB (22 mM NaH2PO4, 78 mM Na2HPO4 pH 7.3) for 3 h at RT. Pellets were washed three times in PB and collected by centrifugation for 5 min at 2500 r.p.m. in an Eppendorf microfuge. The fixed pellets were then embedded in 1.7% low-melting agarose and post-fixed for 1 h (RT) in reduced OsO4 (1% OsO4, 100 mM cacodylate pH 7.4, 1.5% potassium ferricyanide). The pellets were rinsed extensively in 100 mM cacodylate pH 7.4, followed by dehydration in a series of graded ethanol (30, 50, 70, 80, 90, 95 and 100%) and a 1 h incubation in 100% ethanol at RT (the ethanol was replaced every 20 min). Dehydrated pellets were then incubated for 10 min in 100% propylene oxide (RT). The pellets were embedded in Agar 100 resin and the blocks were polymerized for 48 h at 60°C. Thin sections were cut, collected onto copper grids and stained with 1% uranyl acetate for 3 min followed by 5 min in 1% lead citrate before viewing in a Tecnai 12 TEM electron microscope (Philips).
For pre-embedding immunogold staining, CHO-M cells were grown on 8-well plastic slides (Nunc) and either left untreated or treated with 20 µg/ml CsA for 48 h. They were then rinsed in cold PB and fixed in 2.5% paraformaldehyde in 100 mM PB for 1 h (RT). The cells were then washed in cold PB supplemented with 1% NH4Cl and permeabilized with 0.05% TX-100 for 3 min (RT) followed by blocking with 4% BSA in PBS for 30 min (RT) and incubation with the RO73 PrP antiserum (1:500 in 1% BSA in PBS) overnight at 4°C. The cells were then rinsed extensively with cold PBS, blocked again with 1% BSA in PBS for 1 h (RT) and incubated for 3 h (RT) with goat anti-rabbit IgG conjugated to 6 nm gold particles (1:50 in 1% BSA in PBS), followed by an extensive wash with cold PBS. The cell monolayer was then fixed, dehydrated, embedded and cut into thin sections as described above.
Acknowledgments
Acknowledgements
We thank Mrs Naomi Feinstein for expert assistance with the electron microscope. This work was supported by a generous grant from the Israel Center for the Study of Emerging Diseases.
References
- Atkinson K., Biggs,J., Darveniza,P., Boland,J., Concannon,A. and Dodds,A. (1984) Cyclosporin-associated central nervous system toxicity after allogeneic bone marrow transplantation. Transplanta tion, 38, 34–37. [DOI] [PubMed] [Google Scholar]
- Balbach J. and Schmid,F.X. (2000) Proline isomerization and its catalysis in protein folding. In Pain,R.H (ed.), Mechanisms of Protein Folding. Oxford University Press, Oxford, UK, pp. 212–249.
- Basler K., Oesch,B., Scott,M., Westaway,D., Walchli,M., Groth,D.F., McKinley,M.P., Prusiner,S.B. and Weissmann,C. (1986) Scrapie and cellular PrP isoforms are encoded by the same chromosomal gene. Cell, 46, 417–428. [DOI] [PubMed] [Google Scholar]
- Bence N.F., Sampat,R.M. and Kopito,R.R. (2001) Impairment of the ubiquitin–proteasome system by protein aggregation. Science, 292, 1552–1555. [DOI] [PubMed] [Google Scholar]
- Bolton D.C., McKinley,M.P. and Prusiner,S.B. (1982) Identification of a protein that purifies with the scrapie prion. Science, 218, 1309–1311. [DOI] [PubMed] [Google Scholar]
- Butler D.A., Scott,M.R., Bockman,J.M., Borchelt,D.R., Taraboulos,A., Hsiao,K.K., Kingsbury,D.T. and Prusiner,S.B. (1988) Scrapie-infected murine neuroblastoma cells produce protease-resistant prion proteins. J. Virol., 62, 1558–1564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrell R.W. and Lomas,D.A. (1997) Conformational disease. Lancet, 350, 134–138. [DOI] [PubMed] [Google Scholar]
- Collinge J., Sidle,K.C., Meads,J., Ironside,J. and Hill,A.F. (1996) Molecular analysis of prion strain variation and the aetiology of ‘new variant’ CJD. Nature, 383, 685–690. [DOI] [PubMed] [Google Scholar]
- Dobson C.M. (2001) The structural basis of protein folding and its links with human disease. Philos. Trans. R. Soc. Lond. B. Biol. Sci., 356, 133–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellgaard L., Molinari,M. and Helenius,A. (1999) Setting the standards: quality control in the secretory pathway. Science, 286, 1882–1888. [DOI] [PubMed] [Google Scholar]
- Garcia-Mata R., Bebok,Z., Sorscher,E.J. and Sztul,E.S. (1999) Characterization and dynamics of aggresome formation by a cytosolic GFP-chimera. J. Cell Biol., 146, 1239–1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geier E., Pfeifer,G., Wilm,M., Lucchiari-Hartz,M., Baumeister,W., Eichmann,K. and Niedermann,G. (1999) A giant protease with potential to substitute for some functions of the proteasome. Science, 283, 978–981. [DOI] [PubMed] [Google Scholar]
- Giloh H. and Sedat,J.W. (1982) Fluorescence microscopy: reduced photobleaching of rhodamine and fluorescein protein conjugates by n-propyl gallate. Science, 217, 1252–1255. [DOI] [PubMed] [Google Scholar]
- Hamilton G.S. and Steiner,J.P. (1998) Immunophilins: beyond immunosuppression. J. Med. Chem., 41, 5119–5143. [DOI] [PubMed] [Google Scholar]
- Hsiao K., Baker,H.F., Crow,T.J., Poulter,M., Owen,F., Terwilliger,J.D., Westaway,D., Ott,J. and Prusiner,S.B. (1989) Linkage of a prion protein missense variant to Gerstmann–Straussler syndrome. Nature, 338, 342–345. [DOI] [PubMed] [Google Scholar]
- Johnston J.A., Ward,C.L. and Kopito,R.R. (1998) Aggresomes: a cellular response to misfolded proteins. J. Cell Biol., 143, 1883–1898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kascsak R.J., Rubenstein,R., Merz,P.A., Tonna-DeMasi,M., Fersko,R., Carp,R.I., Wisniewski,H.M. and Diringer,H. (1987) Mouse polyclonal and monoclonal antibody to scrapie-associated fibril proteins. J. Virol., 61, 3688–3693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopito R.R. (2000) Aggresomes, inclusion bodies and protein aggregation. Trends Cell Biol., 10, 524–530. [DOI] [PubMed] [Google Scholar]
- Kopito R.R. and Ron,D. (2000) Conformational disease. Nat. Cell Biol., 2, E207–E209. [DOI] [PubMed] [Google Scholar]
- Kopito R.R. and Sitia,R. (2000) Aggresomes and Russell bodies. Symptoms of cellular indigestion? EMBO rep., 1, 225–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehmann S. and Harris,D.A. (1995) A mutant prion protein displays an aberrant membrane association when expressed in cultured cells. J. Biol. Chem., 270, 24589–24597. [DOI] [PubMed] [Google Scholar]
- Lehmann S. and Harris,D.A. (1996) Mutant and infectious prion proteins display common biochemical properties in cultured cells. J. Biol. Chem., 271, 1633–1637. [DOI] [PubMed] [Google Scholar]
- Lehmann S. and Harris,D.A. (1997) Blockade of glycosylation promotes acquisition of scrapie-like properties by the prion protein in cultured cells. J. Biol. Chem., 272, 21479–21487. [DOI] [PubMed] [Google Scholar]
- Ma J. and Lindquist,S. (1999) De novo generation of a PrPSc-like conformation in living cells. Nat. Cell Biol., 1, 358–361. [DOI] [PubMed] [Google Scholar]
- Ma J. and Lindquist,S. (2001) Wild-type PrP and a mutant associated with prion disease are subject to retrograde transport and proteasome degradation. Proc. Natl Acad. Sci. USA, 98, 14955–14960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKinley M.P., Taraboulos,A., Kenaga,L., Serban,D., Stieber,A., DeArmond,S.J., Prusiner,S.B. and Gonatas,N. (1991) Ultrastructural localization of scrapie prion proteins in cytoplasmic vesicles of infected cultured cells. Lab. Invest., 65, 622–630. [PubMed] [Google Scholar]
- Meyer R.K., McKinley,M.P., Bowman,K.A., Braunfeld,M.B., Barry,R.A. and Prusiner,S.B. (1986) Separation and properties of cellular and scrapie prion proteins. Proc. Natl Acad. Sci. USA, 83, 2310–2314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naslavsky N., Stein,R., Yanai,A., Friedlander,G. and Taraboulos,A. (1997) Characterization of detergent-insoluble complexes containing the cellular prion protein and its scrapie isoform. J. Biol. Chem., 272, 6324–6331. [DOI] [PubMed] [Google Scholar]
- Navon A. and Goldberg,A.L. (2001) Proteins are unfolded on the surface of the ATPase ring before transport into the proteasome. Mol. Cell, 8, 1339–1349. [DOI] [PubMed] [Google Scholar]
- Oesch B. et al. (1985) A cellular gene encodes scrapie PrP 27–30 protein. Cell, 40, 735–746. [DOI] [PubMed] [Google Scholar]
- Pan K.M. et al. (1993) Conversion of α-helices into β-sheets features in the formation of the scrapie prion proteins. Proc. Natl Acad. Sci. USA, 90, 10962–10966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prusiner S.B. (1998) Prions. Proc. Natl Acad. Sci. USA, 95, 13363–13383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reimer U., Scherer,G., Drewello,M., Kruber,S., Schutkowski,M. and Fischer,G. (1998) Side-chain effects on peptidyl-prolyl cis/trans isomerisation. J. Mol. Biol., 279, 449–460. [DOI] [PubMed] [Google Scholar]
- Rogers M., Serban,D., Gyuris,T., Scott,M., Torchia,T. and Prusiner,S.B. (1991) Epitope mapping of the Syrian hamster prion protein utilizing chimeric and mutant genes in a vaccinia virus expression system. J. Immunol., 147, 3568–3574. [PubMed] [Google Scholar]
- Safar J., Wille,H., Itri,V., Groth,D., Serban,H., Torchia,M., Cohen,F.E. and Prusiner,S.B. (1998) Eight prion strains have PrPSc molecules with different conformations. Nat. Med., 4, 1157–1165. [DOI] [PubMed] [Google Scholar]
- Scheff S.W. and Sullivan,P.G. (1999) Cyclosporin A significantly ameliorates cortical damage following experimental traumatic brain injury in rodents. J. Neurotrauma, 16, 783–792. [DOI] [PubMed] [Google Scholar]
- Schubert U., Anton,L.C., Gibbs,J., Norbury,C.C., Yewdell,J.W. and Bennink,J.R. (2000) Rapid degradation of a large fraction of newly synthesized proteins by proteasomes. Nature, 404, 770–774. [DOI] [PubMed] [Google Scholar]
- Scott M.R., Kohler,R., Foster,D. and Prusiner,S.B. (1992) Chimeric prion protein expression in cultured cells and transgenic mice. Protein Sci., 1, 986–997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serban D., Taraboulos,A., DeArmond,S.J. and Prusiner,S.B. (1990) Rapid detection of Creutzfeldt–Jakob disease and scrapie prion proteins. Neurology, 40, 110–117. [DOI] [PubMed] [Google Scholar]
- Stahl N., Borchelt,D.R., Hsiao,K. and Prusiner,S.B. (1987) Scrapie prion protein contains a phosphatidylinositol glycolipid. Cell, 51, 229–240. [DOI] [PubMed] [Google Scholar]
- Taraboulos A., Serban,D. and Prusiner,S.B. (1990) Scrapie prion proteins accumulate in the cytoplasm of persistently infected cultured cells. J. Cell Biol., 110, 2117–2132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taraboulos A., Scott,M., Semenov,A., Avrahami,D., Laszlo,L., Prusiner,S.B. and Avraham,D. (1995) Cholesterol depletion and modification of COOH-terminal targeting sequence of the prion protein inhibit formation of the scrapie isoform. J. Cell Biol., 129, 121–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson C.B., June,C.H., Sullivan,K.M. and Thomas,E.D. (1984) Association between cyclosporin neurotoxicity and hypomagnesaemia. Lancet, 2, 1116–1120. [DOI] [PubMed] [Google Scholar]
- Uittenbogaard A., Ying,Y. and Smart,E.J. (1998) Characterization of a cytosolic heat-shock protein–caveolin chaperone complex. Involvement in cholesterol trafficking. J. Biol. Chem., 273, 6525–6532. [DOI] [PubMed] [Google Scholar]
- Warrens A.N., Waters,J.B., Salama,A.D. and Lechler,R.I. (1999) Improving the therapeutic monitoring of cyclosporin A. Clin. Transplant., 13, 193–200. [DOI] [PubMed] [Google Scholar]
- Wigley W.C., Fabunmi,R.P., Lee,M.G., Marino,C.R., Muallem,S., DeMartino,G.N. and Thomas,P.J. (1999) Dynamic association of proteasomal machinery with the centrosome. J. Cell Biol., 145, 481–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamazaki M., Oyanagi,K., Mori,O., Kitamura,S., Ohyama,M., Terashi,A., Kitamoto,T. and Katayama,Y. (1999) Variant Gerstmann–Straussler syndrome with the P105L prion gene mutation: an unusual case with nigral degeneration and widespread neurofibrillary tangles. Acta Neuropathol. (Berl.), 98, 506–511. [DOI] [PubMed] [Google Scholar]
- Yedidia Y., Horonchik,L., Tzaban,S., Yanai,A. and Taraboulos,A. (2001) Proteasomes and ubiquitin are involved in the turnover of the wild-type prion protein. EMBO J., 20, 5383–5391. [DOI] [PMC free article] [PubMed] [Google Scholar]