Abstract
cGMP-specific, cGMP-binding phosphodiesterase (PDE5) regulates such physiological processes as smooth muscle relaxation and neuronal survival. PDE5 contains two N-terminal domains (GAF A and GAF B), but the functional roles of these domains have not been determined. Here we show that recombinant PDE5 is activated directly upon cGMP binding to the GAF A domain, and this effect does not require PDE5 phosphorylation. PDE5 exhibited time- and concentration-dependent reversible activation in response to cGMP, with the highest activation (9- to 11-fold) observed at low substrate concentrations (0.1 µM cGMP). A monoclonal antibody directed against GAF A blocked cGMP binding, prevented PDE5 activation and decreased basal activity, revealing that PDE5 in its non-activated state has low intrinsic catalytic activity. Activated PDE5 showed higher sensitivity towards sildenafil than non-activated PDE5. The stimulatory effect of cGMP binding on the catalytic activity of PDE5 suggests that this mechanism of enzyme activation may be common among other GAF domain-containing proteins. The data also suggest that development of agonists and antagonists of PDE5 activity based on binding to this site might be possible.
Keywords: cyclic GMP/GAF domain/PDE5/sildenafil/smooth muscle
Introduction
A variety of physiological processes in the cardiovascular, nervous and immune systems are controlled by the nitric oxide (NO)/cGMP signaling pathway. In smooth muscle, NO and natriuretic peptides regulate vascular tone by inducing relaxation through stimulation of cGMP synthesis (Sausbier et al., 2000). Degradation of cGMP is controlled by cyclic nucleotide phosphodiesterases (PDEs), and PDE5 (also called cGMP-specific, cGMP-binding PDE) is the most highly expressed cGMP-hydrolyzing PDE in these cells. The physiological importance of PDE5 in regulation of smooth muscle tone has been demonstrated most clearly by clinical use of its specific inhibitor, sildenafil (Viagra®), in the treatment of erectile dysfunction (Ballard et al., 1998). Recently, development of other drugs targeting PDE5 in smooth muscle has also been reported. Both tadalafil (Cialis™) and vardenafil (Levitra™) can specifically inhibit PDE5 activity in the nanomolar concentration range, but differ somewhat in their inhibitory profiles towards PDEs from other families (Saenz de Tejada et al., 2001; Eardley and Cartledge, 2002).
However, the mechanisms by which PDE5 activity is regulated in the intact cell have not been resolved completely. PDE5 has two highly homologous domains in its N-terminal end, one of which is known to bind cGMP (McAllister-Lucas et al., 1993). These domains have been termed GAF A and GAF B based on their sequence homology to similar motifs in a wide group of proteins. The first members of this group included the cGMP-regulated phoshodiesterases, PDE2, PDE5 and PDE6, several adenylyl cyclases and the FhlA protein (a bacterial transcriptional factor). The first letters of these group names were used to derive the acronym, GAF. However, the GAF domain-containing proteins now include >1000 members (Aravind and Ponting, 1997; Anantharaman et al., 2001). It should be noted that the mere presence of this motif does not necessarily mean that a protein is able to bind cGMP. For example, the GAF domain of cyanobacterial adenylyl cyclase (cyaB1) specifically binds cAMP (Kanacher et al., 2002). The result of occupation of the GAF domain by cAMP, generated as a product of the adenylyl cyclase catalytic reaction, had a profound effect on the adenylyl cyclase catalytic activity, leading to a rapid exponential activation of this enzyme.
Among cGMP-binding proteins, PDE2, PDE5 and PDE6 have been reported to have high affinity binding sites for cGMP, and PDE2 is the only PDE with a well-determined role for cGMP-binding sites in regulation of its catalytic activity. cGMP binding to the GAF domain of PDE2 greatly stimulates its cAMP hydrolytic activity (Martins et al., 1982; Stroop and Beavo, 1992).
The cGMP-binding sites of PDE5 were found to be necessary for enhancement of Ser92 (bovine PDE5) phosphorylation by protein kinase A (PKA) or PKG in vitro (Thomas et al., 1990b). As a result of PDE5 phosphorylation, the Kd for cGMP binding shifted from 0.13 to 0.03 µM (Corbin et al., 2000). Recently, it has been suggested that PDE5, partially purified from rat cerebellum, was allosterically activated by cGMP when a fluorescent analog of cGMP was employed as a substrate (Okada and Asakawa, 2002). However, direct activation of catalytic activity by cGMP binding to the regulatory GAF domain was not demonstrated.
Previously, we showed PDE5 phosphorylation and activation by PKG in intact cells and tissues using a phospho-specific PDE5 antibody (Rybalkin et al., 2002). In vitro, we were also able to activate and phosphorylate PDE5 from human smooth muscle cells in the presence of different concentrations of cGMP and PKG. In order to answer the question of whether cGMP binding to the PDE5 GAF domain has any effect on its catalytic activity without phosphorylation, we expressed recombinant mouse PDE5 and examined its kinetic properties using conditions where cGMP-binding sites were partially or completely occupied.
Here, we report that recombinant PDE5 is directly activated upon cGMP binding when assayed at low substrate concentration (0.1 or 1.0 µM cGMP). Moreover, preventing cGMP binding to the GAF A domain by a specific monoclonal antibody (mAb) completely blocked PDE5 activation, and also significantly decreased PDE5 activity in the control samples, suggesting that in the absence of this GAF domain regulatory effect, PDE5 has a relatively low intrinsic hydrolytic activity.
Results
Pre-incubation of recombinant PDE5 with cGMP results in strong reversible activation of PDE5 catalytic activity at low substrate concentration
Recombinant mouse PDE5 was expressed in human embryonic kidney (HEK) 293 cells. The expression level of PDE5 in these cells was 200 times higher than in non-transfected cells in all experiments. The cells were lysed in a small volume of homogenization buffer in order to obtain a concentrated extract, and then the cell extract was pre-incubated with 50 µM cGMP on ice. At different time points, aliquots were diluted 1/400–1/500, and different amounts of diluted extract (additional dilution 1/5–1/10) were used for PDE5 activity assayed at 0.1 µM cGMP. After 5 min of pre-incubation with cGMP, <0.01 µM cGMP was carried over to the PDE5 assay mixture after all dilutions, and longer pre-incubations resulted in even less carry over of cGMP.
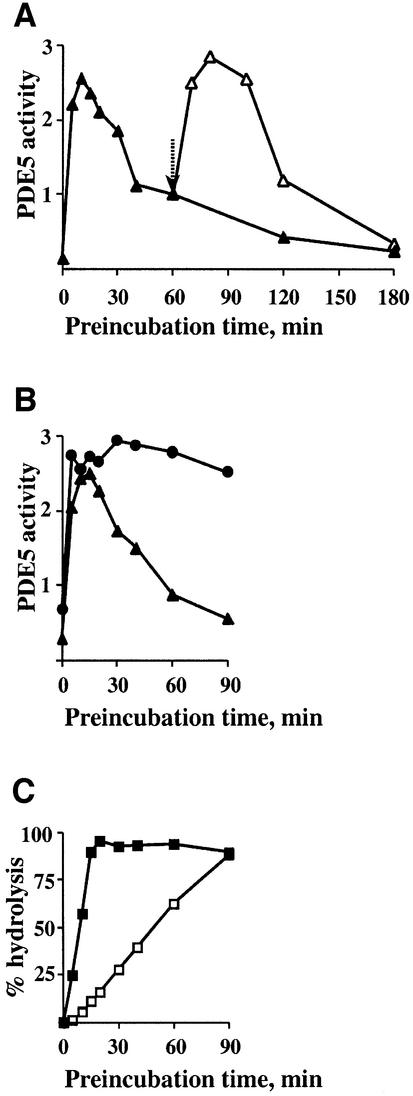
It was found that pre-incubation with cGMP led to a significant time-dependent reversible activation of PDE5 (Figure 1A). Maximum activation of PDE5 (10 times) was observed after 15 min of pre-incubation. Longer pre-incubation led to a gradual decline of PDE5 activity. By 2 h of pre-incubation, PDE5 activity was similar to the control samples. However, PDE5 activity could be reactivated after addition of another portion of cGMP to the pre-incubation mixture (Figure 1A). Again, cGMP-induced activation was time dependent, with maximum activation comparable with the first activation step.
Fig. 1. Time-dependent activation and reactivation of recombinant mouse PDE5 during pre-incubation with 50 µM cGMP on ice. (A and B) Lysate containing mouse recombinant PDE5 expressed in HEK 293 cells was pre-incubated with 50 µM cGMP on ice without PDE inhibitors (filled triangles, A and B) or with 0.2 µM sildenafil (filled circles, B). At different time points during pre-incubation, aliquots were taken and, after appropriate dilutions, PDE5 activity was assayed with 0.1 µM cGMP for 5 min at 30°C. An additional portion of 50 µM cGMP was added to the pre-incubation mixture at 60 min after the start of pre-incubation as indicated by an arrow (open triangles, A). PDE5 activity was expressed as pmol/min/µg of protein. (C) [3H]cGMP (total 100 000 c.p.m.) was added to the pre-incubation mixture containing 50 µM cGMP without (filled squares) or with 0.2 µM sildenafil (open squares). The percentage 50 µM cGMP hydrolysis at different time points was measured using the PDE5 activity assay.
If the activation of PDE5 depends on the continued presence of a certain level of cGMP during the pre-incubation step, it should be possible to prolong the activation time period by adding a PDE5 inhibitor to the pre-incubation mixture. Indeed, in the presence of increasing concentrations of sildenafil, PDE5 was active for a longer period of time. At 0.2 µM sildenafil, PDE5 activation was sustained for at least 90 min (Figure 1B). At this concentration of sildenafil, the cGMP level declined much more slowly than without any PDE5 inhibitors (Figure 1C). The decline in PDE5 activation coincided with complete hydrolysis of cGMP during the pre-incubation step. In all further experiments, recombinant PDE5 was pre-incubated for 15 min with 50 µM cGMP in order to achieve maximum PDE5 activation.
Thus, a certain balance between the concentrations of PDE5 and cGMP and duration of pre-incubation needed to be maintained in order to achieve saturation of the cGMP-binding sites. For example, more concentrated PDE5 samples undoubtedly would require a higher amount of cGMP and different pre-incubation time to achieve the highest level of activation. PDE5 activation was found to be the highest when PDE5 activity was assayed at 0.1 µM cGMP (10 times) as shown above. Activation was less at 1.0 µM cGMP (2–3 times) and non-detectable when PDE5 activity was analyzed at 10 µM cGMP (Figure 2A). Similar data were obtained when PDE5 was partially purified by quick HPLC separation on a Mono Q column and immediately assayed.
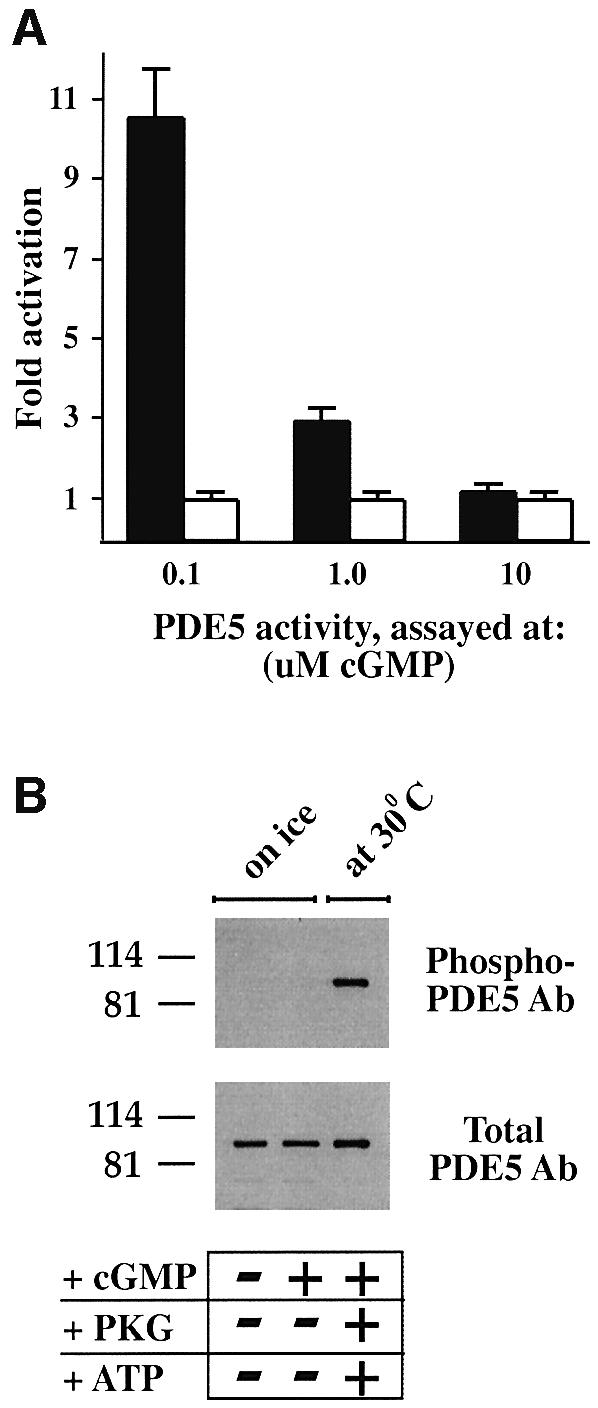
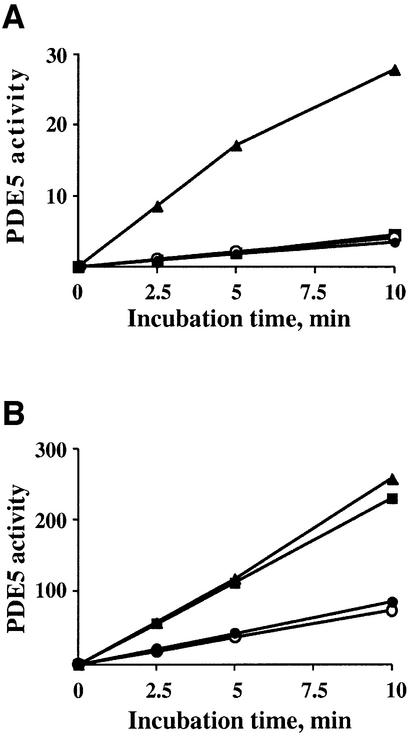
Fig. 2. Recombinant PDE5 is activated after pre-incubation with cGMP only at low substrate concentration, and is not a result of phosphorylation. (A) After pre-incubation of PDE5 with 50 µM cGMP on ice, an aliquot was diluted and assayed with either 0.1, 1.0 or 10 µM cGMP for 5 min at 30°C. PDE5 activity for each substrate concentration was expressed as fold activation of PDE5 (after pre-incubation with cGMP on ice) (black bars) relative to the control samples (without cGMP pre-incubation) (white bars), which were defined as 1. The data represent three or more different experiments with the mean fold activation ± SD. (B) Samples were prepared in SDS sample buffer before and after pre-incubation with 50 µM cGMP on ice and analyzed by western blot analysis with phospho-PDE5 and total PDE5 antibodies. As a control for phospho-PDE5, the same amount of recombinant PDE5 was phosphorylated in the presence of ATP, PKG and cGMP for 30 min at 30°C.
The observed activation of PDE5 is not a result of PDE5 phosphorylation
To explore the possibility that endogenous protein kinases from HEK 293 cells could phosphorylate and activate PDE5 during the pre-incubation with cGMP on ice, samples were prepared in SDS sample buffer before and after pre-incubation and analyzed by western blotting with a phospho-specific PDE5 antibody. However, no PDE5 phosphorylation was detected after pre-incubation with 50 µM cGMP for 15 min on ice (Figure 2B). Phosphorylation of PDE5 could be achieved only after addition of ATP and PKG, and incubation at 30°C.
In order to test if PKG or PKA could phosphorylate another site, not detected by application of phospho-specific PDE5 antibody (raised against phospho-Ser92), site-directed mutagenesis was applied. Ser92 was mutated to alanine, and the phospho-site mutant was subjected to in vitro phosphorylation by PKG or PKA (Figure 3A). However, no 32P incorporation was detected in the mutant PDE5 band, indicating that there is no other phosphorylation site besides Ser92. Moreover, when the phospho-site mutant PDE5 was expressed in HEK 293 cells, a similar pattern of PDE5 activation after pre-incubation with cGMP was found (Figure 3B). The highest level of activation in this mutant enzyme was also observed when PDE5 activity was measured at low substrate concentration (0.1 µM cGMP).
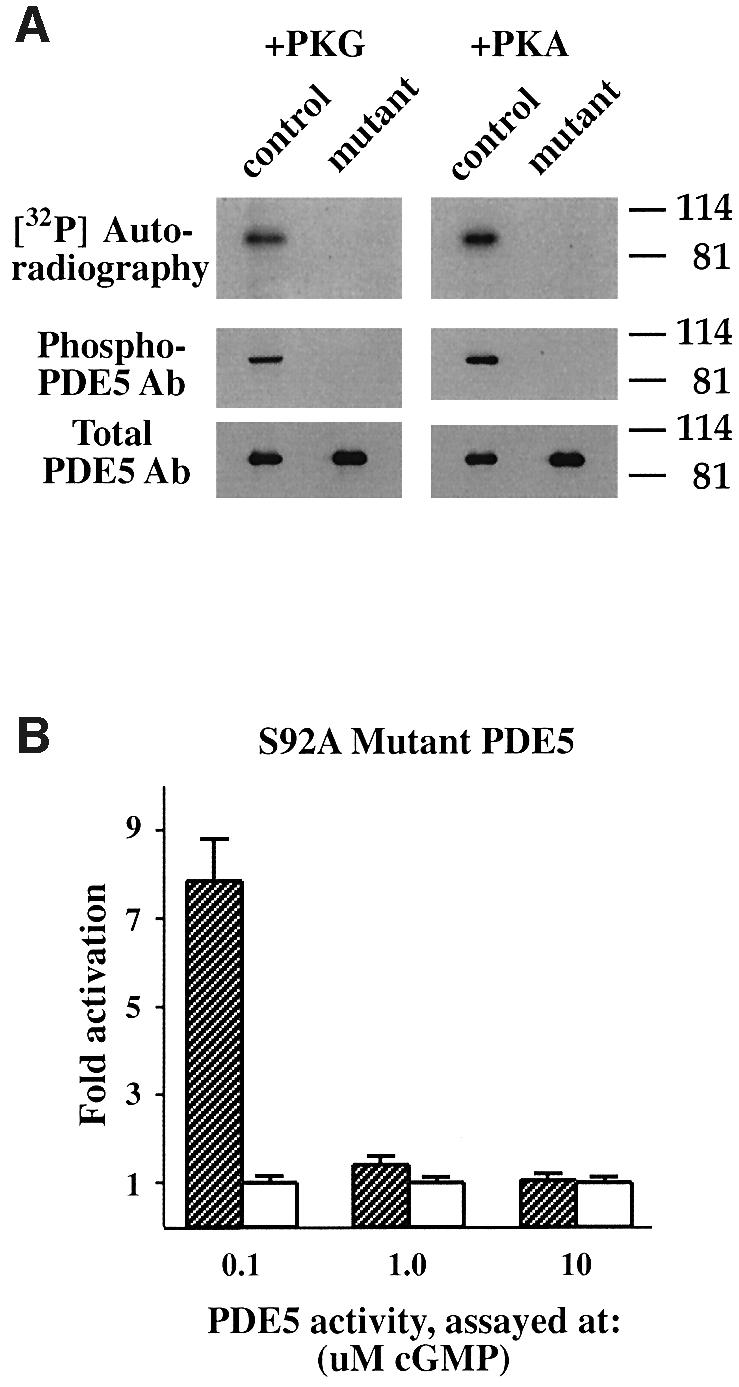
Fig. 3. PDE5 has only one phosphorylation site: phospho-site mutant PDE5 cannot be phosphorylated by either PKG or PKA in vitro. (A) Recombinant PDE5 (control) and phospho-site mutant PDE5 (mutant) were incubated with either PKG or the catalytic subunit of PKA in the phosphorylation buffer with [32P]ATP for 60 min at 30°C. After phosphorylation, PDE5 was immunoprecipitated, and the immunoprecipitates were analyzed by SDS–PAGE and then subjected to autoradiography to reveal 32P incorporation. For western blot analysis, samples were prepared directly after the phosphorylation step. The immunoblots were probed with phospho-PDE5 and total PDE5 antibodies. (B) Phospho-site mutant PDE5 expressed in HEK 293 cells was pre-incubated with 50 µM cGMP on ice and, after appropriate dilutions, assayed with either 0.1, 1.0 or 10 µM cGMP for 5 min at 30°C. Data are expressed as fold activation of PDE5 activity after pre-incubation with cGMP (hatched bars) relative to the control samples (white bars) and defined as in Figure 2A.
A mouse monoclonal antibody specifically blocks cGMP binding to the GAF A domain of PDE5
The previous studies showed that a short pre-incubation of PDE5 with cGMP on ice did not cause PDE5 phosphorylation, but was sufficient to induce PDE5 activation. To prove that the effect of PDE5 activation is due to the direct effect of cGMP occupancy of the cGMP-binding sites on the PDE5 catalytic activity, we developed mouse mAbs, generated against the cGMP-binding domain of PDE5, and screened them for their ability to affect cGMP binding.
The cGMP saturation binding assay was employed to determine the Kd for cGMP binding. Recombinant PDE5 was found to bind cGMP with high affinity (Kd ∼0.12 µM cGMP) (Figure 4A). All mAbs were pre-incubated with PDE5 for 20 min followed by incubation with different concentrations of [3H]cGMP (0.01–20 µM cGMP). One mAb (P3B2) was able to block cGMP binding to PDE5 substantially (Figure 4A). The apparent Kd for cGMP binding increased >100 times, but precise determination of Kd values at these high concentrations of cGMP (20–100 µM) was not possible using a Millipore filtration assay. Another mAb (P4D8) used as a control had no effect on cGMP binding.
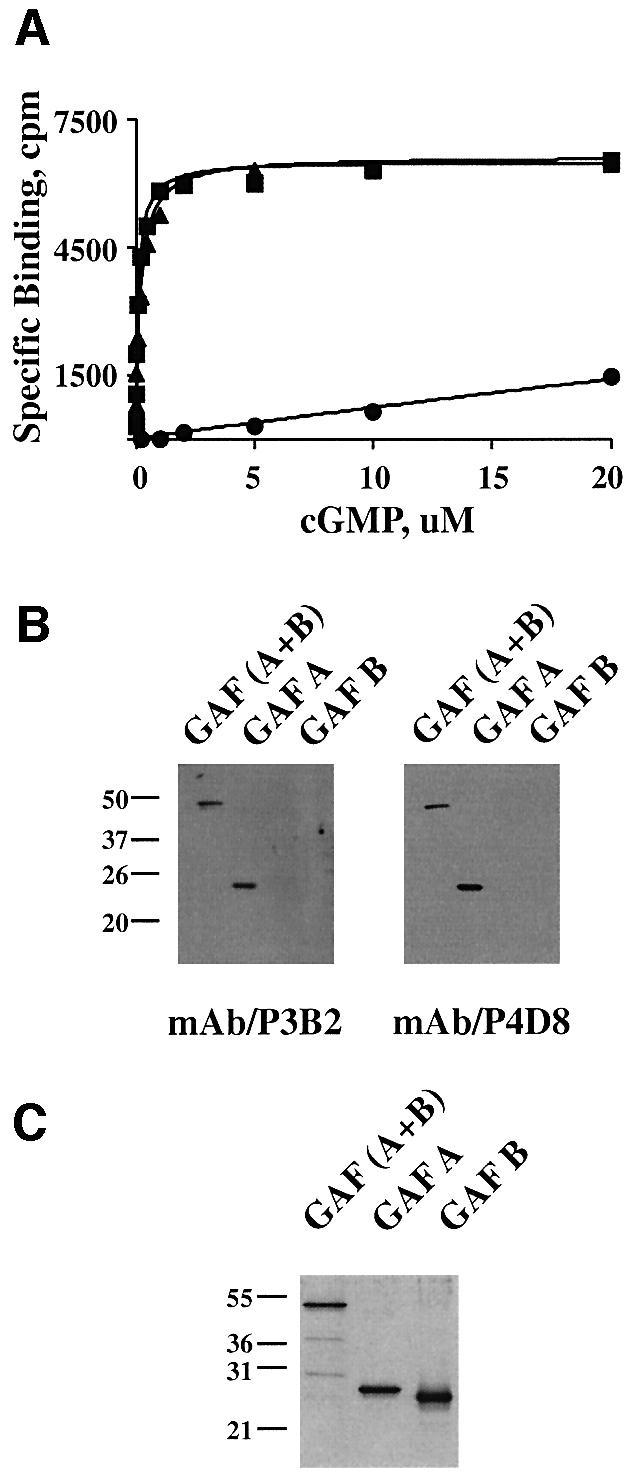
Fig. 4. Pre-treatment of PDE5 with mAb P3B2 blocks cGMP binding to the GAF A domain of PDE5. (A) cGMP saturation binding curves of recombinant PDE5 were measured without (filled squares) or with pre-incubation with mAbs P3B2 (filled circles) or P4D8 (filled triangles) for 30 min on ice. The binding assays were performed with 0.01–20 µM cGMP in the presence of 0.1 mM IBMX for 30 min on ice. The specific binding (c.p.m.) is calculated by subtraction of non-specific binding from total binding. (B and C) The epitope for mAb P3B2 lies in the GAF A region of PDE5. Recombinant mouse GAF(A + B), GAF A and GAF B were resolved by 12% SDS–PAGE and then either processed for western blot analysis (B) with mAbs P3B2 and P4D8, or analyzed by silver staining (C).
To determine which regions of the cGMP-binding domain contain the epitopes for these mAbs, purified mouse GAF A, GAF B and the whole cGMP-binding domain (GAF A and GAF B) were subjected to SDS–PAGE [12% (w/v) gel] and immunoblotted with either P3B2 or P4D8 (Figure 4B). While mAb P3B2 was much more effective in immunoprecipitation of PDE5 (data not shown) than in its detection by western blotting, we could clearly conclude that P3B2 binds to GAF A, but not to GAF B (Figure 4B and C).
Blocking cGMP binding to the GAF A domain of PDE5 with a specific monoclonal antibody (P3B2) reveals a low intrinsic catalytic activity of PDE5
Treatment of PDE5 with a cGMP-blocking mAb (P3B2) before pre-incubation with cGMP not only completely blocked PDE5 activation but also lowered the PDE5 activity of control samples (Figure 5). In all experiments, conducted at different substrate concentrations (0.1–10 µM cGMP), both activated PDE5 (after pre-incubation with cGMP) and control PDE5 (without pre-incubation with cGMP) reached the same low level of enzymatic activity. These data indicate that without the activation effect of cGMP–GAF domain on the PDE5 catalytic domain, PDE5 has only a low intrinsic hydrolytic activity.
Fig. 5. PDE5 has a low intrinsic catalytic activity: blocking cGMP binding by mAb P3B2 results in a significant reduction of PDE5 activity. PDE5 activity was measured at 0.1 µM (A) or 10 µM cGMP (B). Samples were analyzed without any treatments (filled squares), after pre-incubation with 50 µM cGMP on ice (filled triangles) or after pre-incubation with mAb P3B2 for 30 min on ice (filled circles). Treatment of PDE5 with mAb P3B2 for 30 min on ice preceded pre-incubation with 50 µM cGMP (open circles). PDE5 activity was expressed as pmol/µg of protein.
Interestingly, we did not detect any changes in PDE5 activity after pre-incubation with cGMP when PDE5 activity was assayed at 10 µM cGMP (Figure 2A). Presumably this is because the enzyme is rapidly and fully activated by the high initial concentration of cGMP in the assay and pre-incubation is not necessary. However, by blocking cGMP binding with the specific mAb (P3B2), PDE5 activity dropped 60–70% (Figure 5B). This strongly suggests that under these conditions, PDE5 was already activated without the pre-incubation with cGMP.
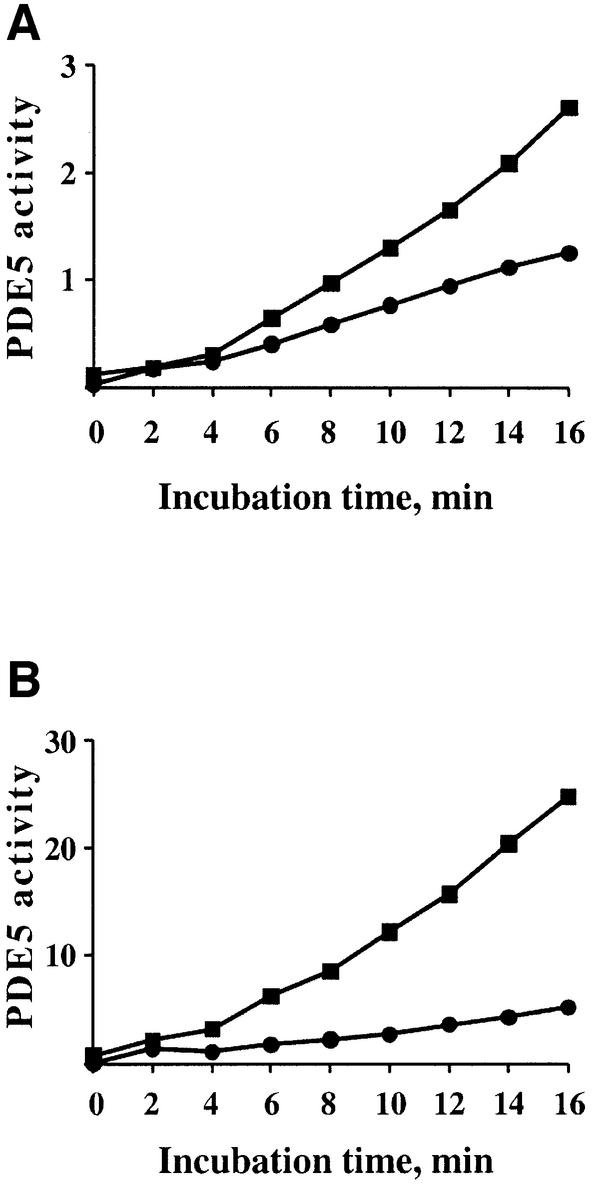
Activated PDE5 displayed linear kinetics of cGMP hydrolysis at least over 5 min of the incubation time at 0.1 µM cGMP and 10 min at 1.0 µM cGMP (data not shown). Meanwhile control (non-activated) PDE5 appeared to undergo activation at the end of 10 min of incubation at both substrate concentrations (Figure 5A).
To determine if PDE5 is activated during the assay, we measured cGMP hydrolysis every 2 min and increased the incubation time up to 16 min. A greater dilution of PDE5 was also necessary in order not to exceed 25% hydrolysis at the end of the activity assays. At both substrate concentrations (0.1 and 1.0 µM cGMP), after a short lag period (4–6 min), the activity of PDE5 gradually began to increase (Figure 6A and B). Once again, blocking cGMP binding by P3B2 linearized the kinetics of cGMP hydrolysis at the lower rate. Since 0.1 µM cGMP was near the Kd for cGMP binding of this protein, only a portion of the cGMP-binding sites were expected to bind cGMP. At 1.0 µM cGMP, more binding sites were occupied by cGMP, leading to the higher degree of PDE5 activation during the assay and subsequently a bigger reduction in total cGMP hydrolytic activity after treatment with the blocking mAb (Figure 6B).
Fig. 6. Time-dependent activation of PDE5 during an activity assay. PDE5 activity was measured at 0.1 µM (A) or 1.0 µM (B) cGMP at 30°C without any pre-incubation (filled squares) or after pre-incubation with mAb P3B2 for 20 min on ice (filled circles). PDE5 activity was expressed as pmol/µg of protein.
These experiments present another example of conversion of a low activity state enzyme to a high activity state upon cGMP binding. However, in contrast to PDE2, PDE5 needs a transitional period of several minutes to reach binding equilibrium at these low cGMP concentrations.
Kinetic and inhibitory characteristics of activated PDE5
At first, we detected changes in PDE5 activity after pre-incubation with cGMP only when PDE5 activity was assayed at 0.1 and 1.0 µm, but not at 10 µm (Figure 2A). These data suggested that activation affected the apparent value of Km, without any changes in the Vmax value for PDE5. However, the experiments with the P3B2 mAb revealed that a significant portion of PDE5 activity at 10 µM cGMP was already activated. Since activation of PDE5 was time and concentration dependent, in order to determine the kinetic characteristics of PDE5, we used enzyme treated with the mAb as a non-activated enzyme, and enzyme after pre-incubation with 50 µM cGMP on ice as fully activated. As revealed by a Lineweaver–Burk double-reciprocal plot (0.1–10 µM cGMP; data not shown), activation of PDE5 altered both the Km and the Vmax. Activated PDE5 had a higher apparent affinity for cGMP (the Km changed from 4.6 ± 0.7 to 0.96 ± 0.12 µM) and higher Vmax value (the Vmax changed from 8.9 ± 1.4 to 27 ± 3 pmol/min/µg of protein).
Activated PDE5 also showed a higher sensitivity towards the PDE5-specific inhibitor sildenafil. We found a shift in the IC50 for sildenafil inhibition from 2.1 ± 0.3 to 0.63 ± 0.10 nM when PDE5 activity was assayed at 0.1 µM cGMP (Figure 7A). At 1.0 µM cGMP, the IC50 decreased from 4.9 ± 0.7 to 1.2 ± 0.2 nM (data not shown). Since sildenafil is considered a competitive inhibitor, which binds at the catalytic site of PDE5 (Ballard et al., 1998), the different sildenafil inhibitory profile for activated PDE5 presumably reflects changes in kinetic characteristics caused by cGMP-induced PDE5 activation. These data are also one more demonstration that cGMP binding to the GAF domain affects the catalytic domain of PDE5.
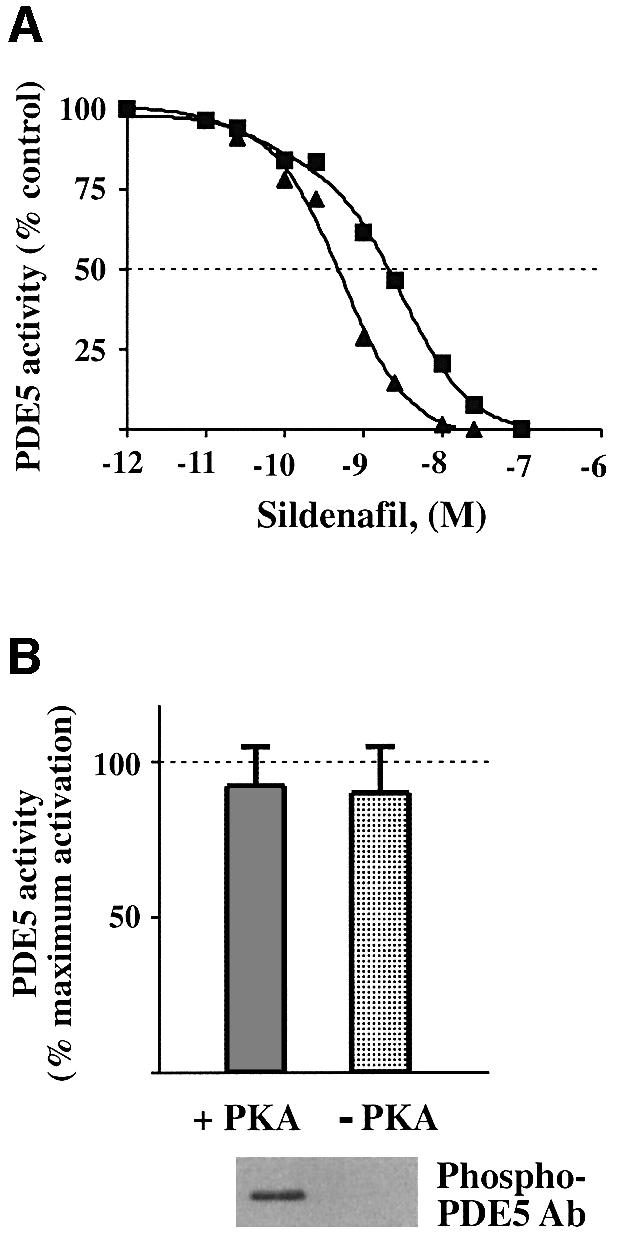
Fig. 7. PDE5 in an activated state is more sensitive to inhibition by sildenafil and is not affected by further phosphorylation. (A) Non- activated (without any pre-incubations) (filled squares) and activated (after pre-incubation with cGMP on ice) (filled triangles) samples of PDE5 were assayed for 5 min at 0.1 µM in the presence of different concentrations of sildenafil (0.01–250 µM). PDE5 activities, assayed without sildenafil, were expressed as 100%. (B) Activated PDE5 (after pre-incubation with 50 µM cGMP on ice for 15 min and subsequent dilution) was assayed with 0.1 µM cGMP before and after phosphorylation by the catalytic subunit of PKA at 30°C for 20 min. PDE5 activity was expressed as the percentage of the maximum PDE5 activation measured immediately after PDE5 pre-incubation with cGMP on ice and defined as 100%. Corresponding samples were prepared in SDS sample buffer and analyzed by western blot analysis with phospho-PDE5 antibody.
As shown in Figure 7B, in vitro phosphorylation of fully activated PDE5 by PKG or PKA does not have any additional effect on its activity. In this experiment, activated cGMP-bound PDE5 was phosphorylated by the catalytic subunit of PKA at 30°C. Since under these conditions, no extra cGMP was needed to perform the phosphorylation step, PDE5 activity was assayed at low substrate concentration. No significant changes of catalytic activity were detected upon phosphorylation (Figure 7B ). Still, blocking cGMP binding by pre-treatment of PDE5 with the mAb P3B2 substantially prevented PDE5 from phosphorylation by PKG or PKA, indicating that only when previously activated by cGMP could PDE5 undergo phosphorylation (data not shown).
PDE5 becomes activated and loses its ability for cGMP stimulation after 1–2 weeks of storage on ice
All previously described experiments were performed on recombinant PDE5 within a week of cell harvesting. Nevertheless, gradually, after a week of storage on ice, the basal PDE5 activity became higher and its response to pre-incubation with cGMP became weaker. By ∼2 weeks, the cGMP-hydrolyzing activity of PDE5 reached approximately the same level as cGMP-activated PDE5 from freshly transfected cells (Figure 5A), but completely lost its responsiveness to cGMP stimulation (Figure 8A). At the same time, the effect of the mAb P3B2 on PDE5 catalytic activity could no longer be observed, although this mAb was still able to immunoprecipitate PDE5 (Figure 8B). As with cGMP activation, storage caused the PDE5 to shift progressively to a form with a higher apparent affinity and higher Vmax value. The IC50 for sildenafil inhibition (2.5 ± 0.5, assayed at 0.1 µM cGMP) also shifted towards higher values. We do not know what type of physical change causes this slow activation. Since no changes in the amount of total PDE5 or its immunoprecipitation pattern with mAb P3B2 were found between freshly expressed PDE5 or PDE5 preparations stored on ice for 2 weeks, proteolytic degradation of PDE5 could be ruled out (Figure 8C). In addition, no change in PDE5 phosphorylation status was detected upon storage (data not shown). This slow progressive activation may explain, however, why PDE5 activation by cGMP has not been reported previously.
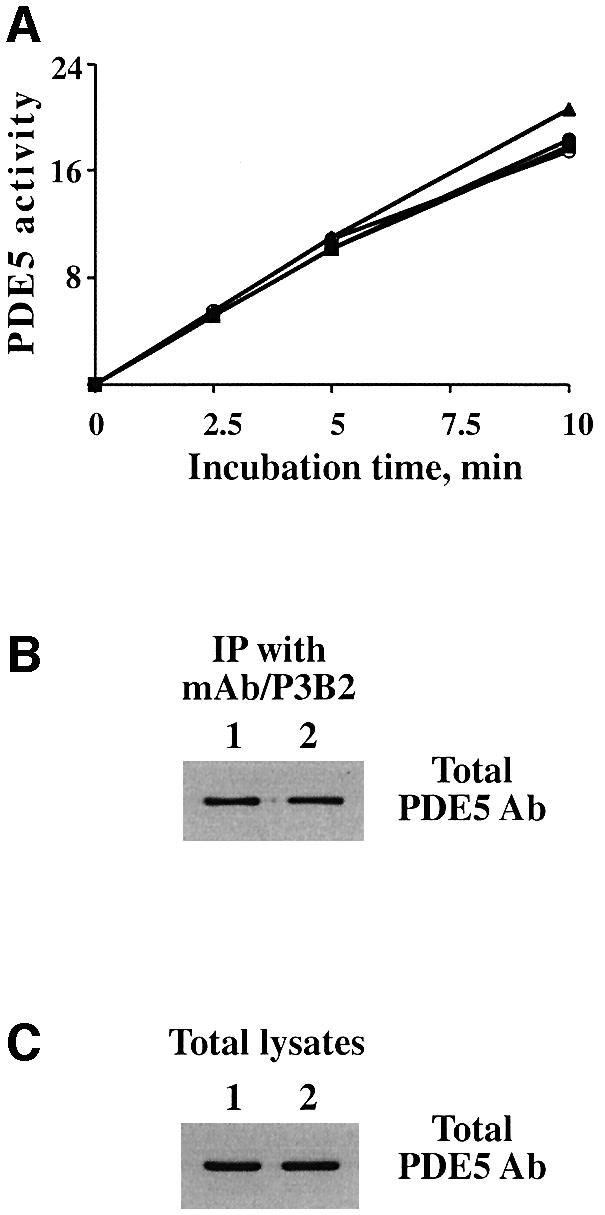
Fig. 8. PDE5 catalytic activity increases after 1–2 weeks of storage on ice accompanied by loss of its ability to be stimulated by cGMP. (A) PDE5 activity was measured at 0.1 µM after 2 weeks of storage on ice (filled squares). Samples were prepared under the same conditions as in Figure 5 and pre-incubated with cGMP (filled triangles), mAb P3B2 (filled circles) or both (open circles). PDE5 activity was expressed as pmol/µg of protein. (B and C) Samples of immunoprecipitates with mAb/P3B2 (B) and the total lysates (C) freshly expressed (1) or stored for 2 weeks on ice (2) were prepared in SDS sample buffer and analyzed by western blotting with total PDE5 antibody.
Discussion
Activation of PDE5 by cGMP binding to the GAF A domain is a fundamental feature of this enzyme
One of the most important findings of this study is that PDE5 can be activated directly by cGMP binding even at low, physiological substrate concentrations (0.1 and 1.0 µM cGMP). In fact, the data would suggest that occupancy of the GAF A site by cGMP is required for maximal activity of the holoenzyme and may be required for any appreciable activity. At high substrate levels, binding and activation occur very rapidly so the activation is not readily visualized. However, at lower concentrations, binding appears to take several minutes. Therefore, in order to detect full activation of PDE5 at low substrate concentrations, a pre-incubation step is required. Blocking cGMP binding by the application of the mouse mAb P3B2 allowed us to evaluate quantitatively the effect of occupancy of cGMP-binding sites on the total cGMP-hydrolyzing activity of PDE5. Pre-treatment of PDE5 with this blocking antibody linearized the kinetics of cGMP hydrolysis over time, substantially lowering PDE5 activity. If the cGMP-binding sites were saturated with cGMP, which was achieved after a short pre-incubation with 50 µM cGMP on ice, activated PDE5 showed a linear kinetic profile of cGMP hydrolysis, and blocking cGMP binding with the mAb P3B2 completely prevented activation. These data strongly indicate that PDE5 holoenzyme in a non-activated state possesses only a very low intrinsic catalytic activity if the stimulatory effect of the cGMP/GAF domain is blocked.
It is known from the literature that cGMP binding to the GAF domain of PDE5 is a prerequisite for PDE5 phosphorylation (Thomas et al., 1990b; Rybalkin et al., 2002). Recently, it has been shown that PDE5 is a specific substrate for PKG in intact smooth muscle cells and tissues (Wyatt et al., 1998; Rybalkin et al., 2002). Thus, activation of both PDE5 and PKG by cGMP would depend on their cGMP-binding characteristics in vivo. We would speculate that activation of PDE5 by direct cGMP binding provides a rapid response to a burst of cGMP inside a cell, thereby limiting the amplitude and duration of cGMP at the submicromolar level. Further accumulation of cGMP eventually would activate PKG, leading to PDE5 phosphorylation and longer lasting activation, thus initiating a longer lasting cycle of negative feedback regulation of the cGMP signal.
PDE5 activity also appears to be a decisive factor in the control of cGMP signaling in cells other than smooth muscle. For example, in platelets, activation and phosphorylation of PDE5 were found to be responsible for a rapid decline in cGMP after NO stimulation, whereas neither guanylyl cyclase nor other PDEs were involved (Mullershausen et al., 2001).
Different conformational states of PDE5
Our data suggest that PDE5 can exist in at least three different conformational states: non-activated, activated by cGMP and activated after storage (Figure 9). The non-activated low intrinsic catalytic activity state and the cGMP-activated states are two reversible conformational states of PDE5 with different kinetic and inhibitory properties. It would seem likely that these are the ‘native’ states, which are present in vivo and respond attentively to fluctuations in cGMP levels via cGMP-induced allosteric transitions from the low catalytic activity state to the activated state.
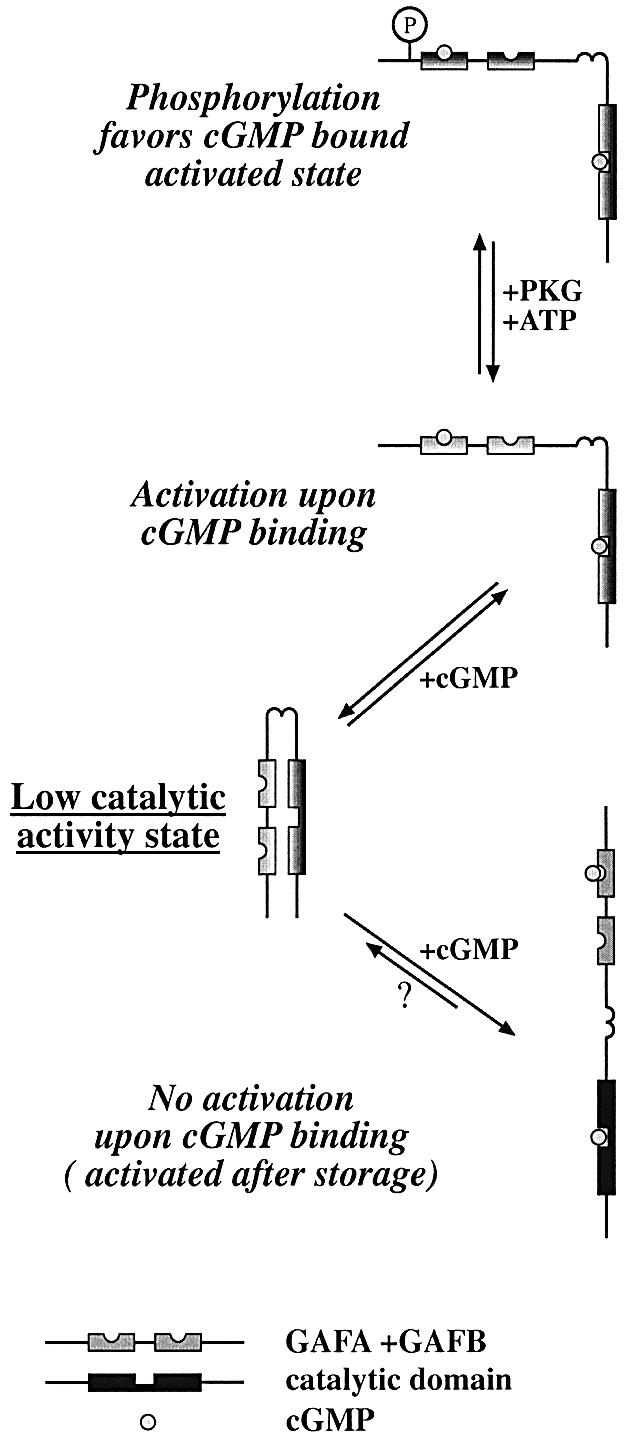
Fig. 9. PDE5 is activated directly upon cGMP binding to its GAF A domain. Without cGMP bound, PDE5 is in a non-activated state. When bound to cGMP, PDE5 is converted into an activated state. After storage, PDE5 converts to an activated state and loses its ability to be stimulated by cGMP. Although PDE5 is likely to exist as a dimer in all conformational states, in this cartoon, a monomeric structure of PDE5 is shown for reasons of simplification.
The ability of PDE5 to be activated directly by cGMP was limited to relatively fresh preparations (less than a week after harvesting transfected cells). Longer storage resulted in a complete loss of the cGMP–GAF domain stimulatory effect on the catalytic activity of PDE5. At the same time, this change might explain the absence of reported effects of cGMP on PDE5 catalytic activity in earlier studies. In particular, most classical purification procedures used to take many days (Thomas et al., 1990a). Apparently, PDE5 is more susceptible to storage or purification conditions than PDE2, which can also show less response to cGMP stimulation, but only after a much longer storage. What conditions are necessary to prolong the storage period during which PDE5 retains its ability for activation by cGMP are not clear at the present time. Thus, PDE5 activated after storage reveals an ‘artificial’, probably irreversible, conformational state of PDE5, suggesting that interaction between the cGMP–GAF domain and the catalytic domain has been interrupted (Figure 9).
Recently, it has been reported that the hydrolysis of a fluorescent analog of cGMP (Ant-cGMP-2′-O-anthranyloyl cGMP) by PDE5 could exhibit positive cooperativity at a few concentrations of cGMP, suggesting the possibility of a direct activation (Okada and Asakawa, 2002). However, under most conditions, only inhibition due to substrate competition was observed. It seems quite possible that this modest effect could be due to the aged nature of the enzyme preparation used by these investigators. Nevertheless, the earlier observation of stimulation of Ant-cGMP hydrolysis in intact cells (Hartell et al., 2001) is consistent with the direct activation caused by cGMP binding to the GAF domain reported herein. Our data also are consistent with the data from the laboratory of D.Koesling that indicate that cGMP by itself was sufficient to cause a sustained activation of PDE5 in intact platelets (D.Koesling, personal communication). They are also consistent with the recent data from the laboratory of J.Corbin that indicate that cGMP binding can increase [3H]sildenafil binding to the catalytic site (J.Corbin, personal communication).
PDE5 and other GAF domain-containing proteins
Recently, an electron microscopy study of PDE5 and PDE6, which were purified to homogeneity, showed a remarkable similarity in the molecular organization of these two PDEs (Kameni Tcheudji et al., 2001). Both enzymes had three electron-dense subdomains, including the catalytic domain and two GAF domains, which were organized linearly from the C-terminal to the N-terminal end. The only contact points were found between the corresponding GAF domains in the PDE5 dimeric structure. However, a linear organization of PDE5 does not easily explain the effect of cGMP binding on PDE5 hydrolytic activity since there is no interaction between the cGMP-binding GAF A domain and the catalytic domain. It is possible that the human platelet PDE5 used in that study (prepared using a long multistep purification process) could exist in a conformational state similar to that for the PDE5 observed in our study after 2 weeks of storage (Figure 8). If so, based on the similar electron microscopy data for PDE5 and PDE6, it is reasonable to suggest that PDE6 may also exist in the same ‘storage’-activated conformational state as PDE5. In this case, cGMP binding to the PDE6 GAF domain also may have regulatory effects on the catalytic domain of PDE6 in its ‘native’ state. So far, only the interaction between the γ-subunit and the catalytic domain of PDE6 has been shown to be affected by cGMP binding to its GAF domain (Yamazaki et al., 1980; Norton et al., 2000).
More generally, the mechanism of the GAF domain stimulatory effect on the catalytic domain might be extended not only to other cGMP-binding PDEs, but also to other GAF domain-containing proteins. Support for this idea is provided by the recent study of cAMP-activated adenylyl cyclase from cyanobacteria (Kanacher et al., 2002). In this study, replacement of the cAMP-binding GAF domain of the cyanobacteria adenylyl cyclase with the cGMP-binding GAF domain from rat cGMP-stimulated PDE (PDE2) caused the chimeric protein to become responsive to cGMP stimulation. Thus, despite the wide evolutionary gap, the basic mechanism of cyclic nucleotide/GAF domain stimulation of catalytic activity remained the same. These results, taken together with the well known activation of PDE2 and our new data on PDE5 reported here, suggest that the mechanism of GAF domain activation of adjacent catalytic domains may be generally applicable to many GAF domain-containing proteins. To date, stimulation has only been shown for proteins that contain two GAF domains, but the ability of single GAF domain-containing proteins to be regulated allosterically needs to be studied further. It is also possible that other unidentified small molecules could regulate activity in the same way as cGMP/or cAMP in this diverse family of GAF domain-containing proteins.
Interestingly, PDE5 activity after 2 weeks of storage on ice could reach levels comparable with that of freshly expressed PDE5 after full activation by cGMP (compare Figure 5A with 8A). This suggests a possible mechanism for cGMP-induced allosteric transformation of PDE5. As shown in Figure 9, this model would include a possible physical interaction between the GAF domain and the catalytic domain. In the absence of cGMP when PDE5 is in a ‘native’ state, the GAF domain physically blocks the catalytic domain, resulting in a low intrinsic catalytic activity. cGMP binding relieves the inhibitory effect, and the catalytic domain becomes more open. After storage, all intramolecular domains may acquire a more linear organization in which the catalytic domain stays constitutively active. Certainly, additional experiments are needed to test this model of regulation of PDE5 activity. However, this type of activation has been reported for other proteins, including the Epac family, guanine nucleotide exchange factors that are directly activated by cAMP (de Rooij et al., 2000). In this case, the cAMP-binding domain suppresses the catalytic activities of Epac1 and Epac2, and cAMP binding results in a release of this inhibition of the catalytic domain.
The GAF A domain of PDE5 as a potential target for the development of new PDE5 inhibitors
All known PDE5 inhibitors were developed to compete with cGMP at the catalytic site. Since PDEs from different families share a substantial degree of sequence similarity in their catalytic domain, inhibitory profiles of specific PDE inhibitors towards different PDE families partially overlap. For example, PDE5 and PDE6 have a very high percentage identity in their catalytic domain, and sildenafil can also inhibit PDE6 in the nanomolar concentration range (Ballard et al., 1998). As a result, this drug may produce visual side effects in clinical applications due to its inhibitory effect on PDE6 in photoreceptor cells.
Therefore, the data in this report suggest that development of drugs that block cGMP binding to the GAF A domain of PDE5 (cGMP antagonists) could decrease PDE5 activity. Instead of targeting the catalytic domain, such agents may create a new class of PDE5-specific inhibitors with a greater isozyme selectivity. In addition, they might also prevent PDE5 from phosphorylation by PKG, which would otherwise lead to the continual activation of PDE5 and the potential development of tolerance. Perhaps more importantly, the fact that this enzyme can be activated by cGMP binding suggests that agonists interacting at the cGMP-binding sites could also be produced. Such agents would be expected to lower intracellular cGMP concentrations, and might be useful, for example, in treatment of conditions characterized by neuronal excitotoxicity or ischemia.
In conclusion, these data present evidence for direct activation of PDE5 upon cGMP binding to the GAF A domain as a new fundamental mechanism of regulation of PDE5 activity. This mechanism suggests that the cGMP-binding sites on the GAF domains could be good new targets for development of both agonists and antagonists of PDE activity. The data also suggest that the mechanism of enzyme alteration induced by ligand binding to the GAF domain may be common for other GAF domain-containing proteins.
Materials and methods
Cell culture
HEK 293 cells, obtained from the American Type Tissue Culture Collection, were grown in DMEM supplemented with 10% fetal bovine serum, 100 U/ml penicillin and 100 mg/ml streptomycin at 37°C in a humidified 5% CO2 atmosphere.
Transient transfection
Mouse PDE5A1 (DDBJ/EMBL/GenBank accession No. AF541937) was subcloned into pcDNA3 vector (Invitrogen). The plasmids for transfection were purified using a Qiagen plasmid maxi kit (Qiagen®). Cells plated on 100 mm plates were transiently transfected with DNA using the Lipofectamine 2000 (Invitrogen) method according to the manufacturer’s protocol. Protein expression was verified by western blot analysis of cell lysates with total PDE5 antibodies. At 2 days after transfection, cells were harvested for experiments.
After washing three times with cold PBS, cells were lysed in homogenization buffer (50 mM Tris–HCl pH 7.5, 2.0 mM EDTA, 1 mM DTT, 10 µg/ml aprotinin, 5 µg/ml pepstatin, 20 µg/ml leupeptin, 1 mM benzamidine, 0.2 mM sodium vanadate, 50 mM sodium β-glycerophosphate). Lysates were sonicated briefly (2–3 s) using a Virsonic 100 sonicator (Virtis Company, NY) and centrifuged at 16 000g for 15 min. The supernatants were used for activity assays. In some experiments, the cell extract was centrifuged at 100 000g for 1 h and loaded on a Mono Q anion exchange column HR 5/5 (Pharmacia). HPLC was conducted using conditions published previously (Rybalkin et al., 1997). Fractions containing PDE5 activity were used for experiments.
Site-directed mutagenesis of the PDE5 phospho-site
Site-directed mutagenesis was carried out using the QuickChange™ site-directed mutagenesis kit (Stratagene) according to the manufacturer’s protocol. The following oligonucleotide primers were used to substitute serine to alanine: 5′-GCCAGAAAAATAGCTGCCTCTGAATTTG-3′ and 5′-CAAATTCAGAGGCAGCTATTTTTCTGGC-3′. The mutated nucleotides are underlined. The mutation was confirmed by DNA sequencing.
In vitro phosphorylation and immunoprecipitation
Cell extracts were incubated for 60 min at 30°C in 30 µl of phosphorylation buffer containing homogenization buffer with 0.1 mM ATP, 1 µCi of [32P]ATP (sp. act. 30 Ci/mmol, Perkin-Elmer Life Sciences, Inc.), 10 mM MgCl2, 300 U of PKG (Promega) or 50 U of the catalytic subunit of PKA (Promega) and 50 µM cGMP. After phosphorylation, samples were diluted in immunoprecipitation buffer (homogenization buffer with 150 mM NaCl, 1% NP-40) and incubated with PDE5 mAb, followed by addition of protein G–agarose beads. After washing three times in immunoprecipitation buffer, the immunoprecipitates were prepared in SDS sample buffer, resolved by SDS–PAGE and analyzed by western blotting. Western blots were also subjected to autoradiography to detect radiolabeled phosphoproteins. In experiments without radiolabeled ATP, cGMP was added to the phosphorylation mixture where indicated, and samples were prepared in SDS sample buffer immediately after phosphorylation.
Antibody characterization
Production and purification of phospho-specific PDE5 rabbit polyclonal antibody raised against a phosphopeptide (amino acids 85–98 of bovine PDE5A), total PDE5 rabbit polyclonal antibody, raised against a peptide (amino acids 836–852, bovine PDE5A), and mouse mAbs, generated using recombinant protein (amino acids 125–539 of bovine PDE5A), were published previously (Rybalkin et al., 2002). mAbs were used as cell culture supernatants prepared from hybrydoma cells grown in culture. They were purified from culture media using an Ultrafree-4 centrifugal filter unit (Millipore Corp.).
Western blot analysis and autoradiography
Samples were prepared in 2× SDS sample buffer, heated at 95°C for 5 min and subjected to 8% SDS–PAGE. The separated proteins were transferred onto nitrocellulose membranes and immunostained with PDE5 antibodies. Immunoreactivity was detected by enhanced chemiluminescence (ECL) using horseradish peroxidase-conjugated goat anti-rabbit IgGs or goat anti-mouse IgGs and SuperSignal® West Pico Chemiluminescent Substrate (Pierce). BenchMark™ pre-stained protein standard was from Invitrogen. In experiments with radiolabeled ATP, the blots were also subjected to autoradiography using Kodak XAR-5 films with intensifying screens for 24–48 h at –80°C.
cGMP saturation binding assay
Binding studies were conducted using the Millipore filter binding assay in a total volume of 40 µl containing binding buffer (50 mM Tris–HCl pH 7.5, 2.0 mM EDTA, 1 mM DTT, 100 mM NaCI) and 0.1 mM IBMX in the presence of 0.01–20 µM [3H]cGMP. In some experiments, an additional 30 min pre-incubation of recombinant PDE5 with mAbs (P3B2 or P4D8) was performed before addition of cGMP. Following a 30 min incubation on ice, 1 ml of ice-cold binding buffer was added to the assays. The samples were filtered immediately through a Millipore HAWP nitrocellulose membrane (pore size 0.45 µM, 24 mm diameter) and rinsed three times with ice-cold binding buffer (10 ml each). The filters were dissolved in 5 ml of Filter Count™ (Packard Instrument) and counted in a liquid scintillation counter.
GAF domain protein analyses
Recombinant proteins of GAF(A + B) (amino acids 125–539 of mouse PDE5), GAF A (amino acids 125–320) and GAF B (amino acids 334–525) were expressed in Escherichia coli with polyhistidine tags and purified on a Talon Metal Affinity Resin (Clontech). Samples (0.1 µg) were resolved using a 12% SDS–polyacrylamide gel. Protein bands were either analyzed by western blotting or silver stained (SilverXpress, Invitrogen) according to the manufacturer’s protocol. Mark12™ unstained molecular weight standard was from Invitrogen.
PDE assays and protein determinations
Phosphodiesterase assays were carried out according to established procedures (Sonnenburg et al., 1998). All assays were performed for 5 min (unless otherwise indicated) at 30°C using different concentrations of cGMP (0.1–10 µM) as substrates. The total reaction volume was 125 µl containing 40 mM MOPS pH 7.5, 2.0 mM EGTA, 15 mM Mg acetate, 0.2 mg/ml bovine serum albumin and 50 000 c.p.m. [3H]cGMP. Recombinant PDE5 was diluted to produce <30% total cGMP hydrolysis at any given substrate concentration. Assays were stopped by placing the tubes in a boiling water bath for 1 min. After addition of 10 µl of snake venom (2.5 mg/ml) and incubation for 5 min at 30°C, samples were then applied to DEAE-Sephadex A-25 columns, and the radioactivity was counted in a liquid scintillation counter. Protein concentrations were determined by the Coomassie Plus protein assay (Pierce). Km, Vmax and IC50 values were calculated using GraphPad Prism 2.0C (GraphPad Software, San Diego, CA) and presented as an average of three experiments ± SD.
Acknowledgments
Acknowledgements
Much of the original cloning work for mouse PDE5A was carried out by Dr Fiona Burns while she was in this laboratory, and she supplied the original construct. This work was supported by grants DK 21723 and HL44948 from the National Institutes of Health.
References
- Anantharaman V., Koonin,E.V. and Aravind,L. (2001) Regulatory potential, phyletic distribution and evolution of ancient, intracellular small-molecule-binding domains. J. Mol. Biol., 307, 1271–1292. [DOI] [PubMed] [Google Scholar]
- Aravind L. and Ponting,C.P. (1997) The GAF domain: an evolutionary link between diverse phototransducing proteins. Trends Biochem. Sci., 22, 458–459. [DOI] [PubMed] [Google Scholar]
- Ballard S.A., Gingell,C.J., Tang,K., Turner,L.A., Price,M.E. and Naylor,A.M. (1998) Effects of sildenafil on the relaxation of human corpus cavernosum tissue in vitro and on the activities of cyclic nucleotide phosphodiesterase isozymes. J. Urol., 159, 2164–2171. [DOI] [PubMed] [Google Scholar]
- Corbin J.D., Turko,I.V., Beasley,A. and Francis,S.H. (2000) Phosphorylation of phosphodiesterase-5 by cyclic nucleotide-dependent protein kinase alters its catalytic and allosteric cGMP-binding activities. Eur. J. Biochem., 267, 2760–2767. [DOI] [PubMed] [Google Scholar]
- de Rooij J., Rehmann,H., van Triest,M., Cool,R.H., Wittinghofer,A. and Bos,J.L. (2000) Mechanism of regulation of the Epac family of cAMP-dependent RapGEFs. J. Biol. Chem., 275, 20829–20836. [DOI] [PubMed] [Google Scholar]
- Eardley I. and Cartledge,J. (2002) Tadalafil (Cialis) for men with erectile dysfunction. Int. J. Clin. Pract., 56, 300–304. [PubMed] [Google Scholar]
- Hartell N.A., Furuya,S., Jacoby,S. and Okada,D. (2001) Intercellular action of nitric oxide increases cGMP in cerebellar Purkinje cells. Neuroreport, 12, 25–28. [DOI] [PubMed] [Google Scholar]
- Kameni Tcheudji J.F., Lebeau,L., Virmaux,N., Maftei,C.G., Cote,R.H., Lugnier,C. and Schultz,P. (2001) Molecular organization of bovine rod cGMP-phosphodiesterase 6. J. Mol. Biol., 310, 781–791. [DOI] [PubMed] [Google Scholar]
- Kanacher T., Schultz,A., Linder,J.U. and Schultz,J.E. (2002) A GAF-domain-regulated adenylyl cyclase from Anabaena is a self-activating cAMP switch. EMBO J., 21, 3672–3680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martins T.J., Mumby,M.C. and Beavo,J.A. (1982) Purification and characterization of a cyclic GMP-stimulated cyclic nucleotide phosphodiesterase from bovine tissues. J. Biol. Chem., 257, 1973–1979. [PubMed] [Google Scholar]
- McAllister-Lucas L.M. et al. (1993) The structure of a bovine lung cGMP-binding, cGMP-specific phosphodiesterase deduced from a cDNA clone. J. Biol. Chem., 268, 22863–22873. [PubMed] [Google Scholar]
- Mullershausen F., Russwurm,M., Thompson,W.J., Liu,L., Koesling,D. and Friebe,A. (2001) Rapid nitric oxide-induced desensitization of the cGMP response is caused by increased activity of phosphodiesterase type 5 paralleled by phosphorylation of the enzyme. J. Cell Biol., 155, 271–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norton A.W., D’Amours,M.R., Grazio,H.J., Hebert,T.L. and Cote,R.H. (2000) Mechanism of transducin activation of frog rod photoreceptor phosphodiesterase. Allosteric interactions between the inhibitory γ subunit and the noncatalytic cGMP-binding sites. J. Biol. Chem., 275, 38611–38619. [DOI] [PubMed] [Google Scholar]
- Okada D. and Asakawa,S. (2002) Allosteric activation of cGMP-specific, cGMP-binding phosphodiesterase (PDE5) by cGMP. Biochemistry, 41, 9672–9679. [DOI] [PubMed] [Google Scholar]
- Rybalkin S.D., Bornfeldt,K.E., Sonnenburg,W.K., Rybalkina,I.G., Kwak,K.S., Hanson,K., Krebs,E.G. and Beavo,J.A. (1997) Calmodulin-stimulated cyclic nucleotide phosphodiesterase (PDE1C) is induced in human arterial smooth muscle cells of the synthetic, proliferative phenotype. J. Clin. Invest., 100, 2611–2621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rybalkin S.D., Rybalkina,I.G., Feil,R., Hofmann,F. and Beavo,J.A. (2002) Regulation of cGMP-specific phosphodiesterase (PDE5) phosphorylation in smooth muscle cells. J. Biol. Chem., 277, 3310–3317. [DOI] [PubMed] [Google Scholar]
- Saenz de Tejada I. et al. (2001) The phosphodiesterase inhibitory selectivity and the in vitro and in vivo potency of the new PDE5 inhibitor vardenafil. Int. J. Impot. Res., 13, 282–290. [DOI] [PubMed] [Google Scholar]
- Sausbier M., Schubert,R., Voigt,V., Hirneiss,C., Pfeifer,A., Korth,M., Kleppisch,T., Ruth,P. and Hofmann,F. (2000) Mechanisms of NO/cGMP-dependent vasorelaxation. Circ. Res., 87, 825–830. [DOI] [PubMed] [Google Scholar]
- Sonnenburg W.K., Rybalkin,S.D., Bornfeldt,K.E., Kwak,K.S., Rybalkina,I.G. and Beavo,J.A. (1998) Identification, quantitation and cellular localization of PDE1 calmodulin-stimulated cyclic nucleotide phosphodiesterases. Methods, 14, 3–19. [DOI] [PubMed] [Google Scholar]
- Stroop S.D. and Beavo,J.A. (1992) Sequence homology and structure–function studies of the bovine cyclic-GMP-stimulated and retinal phosphodiesterases. Adv. Second Messenger Phosphoprotein Res., 25, 55–71. [PubMed] [Google Scholar]
- Thomas M.K., Francis,S.H. and Corbin,J.D. (1990a) Characterization of a purified bovine lung cGMP-binding cGMP phosphodiesterase. J. Biol. Chem., 265, 14964–14970. [PubMed] [Google Scholar]
- Thomas M.K., Francis,S.H. and Corbin,J.D. (1990b) Substrate- and kinase-directed regulation of phosphorylation of a cGMP-binding phosphodiesterase by cGMP. J. Biol. Chem., 265, 14971–14978. [PubMed] [Google Scholar]
- Wyatt T.A., Naftilan,A.J., Francis,S.H. and Corbin,J.D. (1998) ANF elicits phosphorylation of the cGMP phosphodiesterase in vascular smooth muscle cells. Am. J. Physiol., 274, H448–H455. [DOI] [PubMed] [Google Scholar]
- Yamazaki A., Sen,I., Bitensky,M.W., Casnellie,J.E. and Greengard,P. (1980) Cyclic GMP-specific, high affinity, noncatalytic binding sites on light-activated phosphodiesterase. J. Biol. Chem., 255, 11619–11624. [PubMed] [Google Scholar]