Abstract
mRNAs that contain premature stop codons are degraded selectively and rapidly in eukaryotes, a phenomenon termed ‘nonsense-mediated mRNA decay’ (NMD). We report here molecular analysis of smg-5, which encodes a novel protein required for NMD in Caenorhabditis elegans. Using a combination of immunoprecipitation and yeast two-hybrid assays, we identified a series of protein–protein interactions involving SMG-5. SMG-5 interacts with at least four proteins: (i) SMG-7, a previously identified protein required for NMD; (ii) SMG-2, a phosphorylated protein required for NMD in worms, yeasts and mammals; (iii) PR65, the structural subunit of protein phosphatase 2A (PP2A); and (iv) PP2AC, the catalytic subunit of PP2A. Previous work demonstrated that both SMG-5 and SMG-7 are required for efficient dephosphorylation of SMG-2. Our results suggest that PP2A is the SMG-2 phosphatase, and the role of SMG-5 is to direct PP2A to its SMG-2 substrate. We discuss cycles of SMG-2 phosphorylation and their roles in NMD.
Keywords: mRNA decay/NMD/protein phosphatase 2A/SMG-2/SMG-5
Introduction
Eukaryotic mRNAs that contain premature translation stop codons are degraded more rapidly than their wild-type counterparts, a phenomenon termed ‘nonsense-mediated mRNA decay’ (NMD) or ‘mRNA surveillance’ (reviewed in Hentze and Kulozik, 1999; Hilleren and Parker, 1999; Wilusz et al., 2001). An important biological role of NMD may be to improve the fidelity of gene expression by eliminating mRNAs that encode truncated proteins, many of which are deleterious (Pulak and Anderson, 1993; Hall and Thein, 1994; Cali and Anderson, 1998). Aberrant mRNAs expressed from mutant genes or arising from various errors of gene expression are likely to be important substrates of NMD. NMD also degrades certain unspliced mRNAs (He et al., 1993; Li et al., 1995), mRNAs that have been spliced unproductively (Morrison et al., 1997; Mitrovich and Anderson, 2000) and, in the case of the vertebrate immune system, mRNAs emanating from out-of-frame gene rearrangements (Li and Wilkinson, 1998). NMD is a non-essential process in yeast and worms, but mouse mutants lacking NMD arrest development as early embryos (Medghalchi et al., 2001).
Translation termination at premature stop codons is distinguished from that occurring at normal stop codons by the manner in which the translational apparatus interacts with certain cis-acting elements of an mRNA. Nonsense codons are not sufficient to elicit NMD. Rather, specific cis-acting sequence elements must be located downstream of stop codons, and NMD ensues when translation terminates in that context (reviewed in Hilleren and Parker, 1999; Wilusz et al., 2001). The cis-acting downstream activators of NMD are short, discrete ‘downstream elements’ in yeast (Zhang et al., 1995), and a complex of proteins bound to exon–exon junctions in spliced mammalian mRNAs (Le Hir et al., 2000; Kim et al., 2001; Lykke-Andersen et al., 2001). Stop codons followed closely by downstream elements in yeast activate removal of the 5′ cap, after which decapped RNAs are degraded from their 5′ ends (Muhlrad and Parker, 1994). The decapping enzyme (Dcp1p) and 5′ exonuclease (Xrn1p) responsible for yeast NMD are the same as those involved in turnover of most wild-type mRNAs (Beelman et al., 1996). Nonsense mutant mRNAs are decapped, however, without prior deadenylation (Muhlrad and Parker, 1994), in contrast to wild-type mRNAs, which undergo deadenylation prior to decapping (Decker and Parker, 1993; Muhlrad et al., 1994). Thus, activation of decapping, which occurs during (or shortly after) translation termination in a manner that does not require deadenylation, is the key regulated event of NMD. The mechanism by which Dcp1p is activated to remove a cap and whether the yeast NMD pathway is typical of all eukaryotes presently are unknown.
Gene products required for NMD have been identified in yeast, Caenorhabdits elegans and mammals. Loss-of-function mutations affecting any of three yeast genes (UPF1, NMD2/UPF2 and UPF3) eliminate NMD (reviewed in Wilusz et al., 2001). Mammalian orthologs of the UPF genes have been identified (Perlick et al., 1996; Applequist et al., 1997; Lykke-Andersen et al., 2000; Mendell et al., 2000; Serin et al., 2001) and are known to be important for mammalian NMD (Sun et al., 1998; Lykke-Andersen et al., 2000; Le Hir et al., 2001; Wang et al., 2002). Upf1p, an ATPase with RNA helicase activity, appears in many ways to be the key component of NMD. It is the most highly conserved Upf protein, and its ATPase activity is required for NMD (Weng et al., 1996). Upf1p interacts directly with translation release factors and with the decapping complex (He and Jacobson, 1995; Czaplinski et al., 1998; Dunckley and Parker, 1999). Thus, Upf1p is present on the ribosome at the site of termination, and it interacts with many of the key components of termination and mRNA decay.
Upf1p, Upf2p and Upf3p form a complex that associates with polysomes (Atkin et al., 1997; Pal et al., 2001), with Upf2p bridging the interaction between Upf1p and Upf3p (He et al., 1997). Upf3p and its mammalian orthologs shuttle between the nucleus and cytoplasm and probably traffic with mRNA during nuclear export (Shirley et al., 1998; Lykke-Andersen et al., 2001; Kim et al., 2001). Upf1p, Upf2p and Upf3p may be components of a post-termination ‘surveillance complex’ that detects the presence of downstream elements (Ruiz-Echevarria et al., 1998). This model stipulates that, if such elements are detected, decapping ensues. An essential yeast protein, Hrp1p/Nab4p, binds to a downstream element and, in addition to other roles, is required for NMD in yeast (Gonzalez et al., 2000). Hrp1p/Nab4p interacts with Upf1p, possibly facilitating recognition of downstream elements by the surveillance complex. In a seemingly analogous manner, human Upf3 interacts with components of the exon–exon junction complex, certain of which appear to be involved in NMD (Kim et al., 2001; Le Hir et al., 2001). Thus, the detection of downstream signals may be similar in yeasts and mammals, even though the cis-acting RNA elements themselves are quite different.
The functions of seven genes, smg-1–smg-7, are required for NMD in C.elegans (Hodgkin et al., 1989; Pulak and Anderson, 1993; Cali et al., 1999). Caenorhabditis elegans smg-2, smg-3 and smg-4 are orthologs of yeast UPF1, UPF2 and UPF3, respectively (Page et al., 1999; Aronoff et al., 2001; S.Kuchma and P.Anderson, unpublished data). SMG-2 is a phosphorylated protein, and cycles of SMG-2 phosphorylation appear to be necessary for NMD (Page et al., 1999). SMG-2 detected in wild-type is not phosphorylated, but two circumstances cause phosphorylated SMG-2 to accumulate to abnormally high levels: (i) loss-of-function mutations affecting smg-5, smg-6 or smg-7; and (ii) certain single amino acid substitutions affecting the SMG-2 ATPase domain. In smg-1, smg-3 and smg-4 mutants, phosphorylation of SMG-2 is not detected, even in genetic backgrounds where it otherwise would occur. These observations suggest that SMG-1, SMG-3 and SMG-4 are needed to phosphorylate SMG-2, while SMG-5, SMG-6 and SMG-7 are needed to dephosphorylate SMG-2 efficiently. As all smg mutants have a similar phenotype concerning NMD, complete cycles of SMG-2 phosphorylation and dephosphorylation are required for NMD. Regulating the state of SMG-2 phosphorylation may be the key function of all smg genes. Human Upf1p, the ortholog of SMG-2, is phosphorylated in vivo (Pal et al., 2001) and in vitro by human SMG-1 (Denning et al., 2001; Yamashita et al., 2001). Important unanswered questions concerning SMG-2 phosphorylation are: what activities of SMG-2 are regulated by phosphorylation and what role does this play in NMD? What protein(s) is(are) the SMG-2 phosphatase? How are the activities of the SMG-2 kinase and phosphatase regulated?
We report here molecular analysis of smg-5 and its encoded protein. We cloned smg-5 and investigated the proteins with which SMG-5 interacts. Our results suggest that SMG-5 is a component of a SMG-2-specific protein phosphatase 2A (PP2A).
Results
Positional cloning of smg-5
smg-5 was mapped previously between dpy-5 and unc-13 on linkage group I (Hodgkin et al., 1989). We refined the position of smg-5 using three-factor mapping of nearby markers. Among 111 crossovers near unc-87, none separated smg-5 from unc-87. To identify the physical location of smg-5, we used cosmids from the unc-87 region (Goetinck and Waterston, 1994) as probes for Southern blots of wild-type and smg-5(–) DNAs. We anticipated that smg-5(r919) might exhibit Southern blot abnormalities, as this allele was isolated in a mut-2(r459) genetic background. mut-2(r459) activates several families of C.elegans transposons (Anderson, 1995), and smg-5(r919) spontaneously reverts to smg-5(+). Thus, r919 is likely to be a transposon insertion. Cosmid W02D3 identified an insertion in smg-5(r919) that is absent in wild-type and in three independent smg-5(+) revertants of r919. Subsequent analysis identified the inserted DNA as Tc3 (data not shown). Cosmid W02D3, therefore, probably contains all or part of smg-5.
To test whether cosmid W02D3 contains an intact smg-5(+) gene, we injected smg-5(r860) with W02D3 DNA and established that it fully rescues the Smg(–) phenotype of r860. Whereas unc-54(r293) smg-5(r860) mutants exhibit wild-type motility, several independent transformants derived from W02D3 were flaccidly paralyzed, the phenotype characteristic of unc-54(r293) smg-5(+) strains. The paralysis of transformants was heritable and depended both on the transgenic array and on unc-54(r293). We conclude that cosmid W02D3 contains all sequences sufficient for smg-5(+) function.
We identified a 2.5 kb subclone of W02D3 (plasmid TR 191; extending from nucleotides 14 196 to 16 745) that rescued smg-5(r860). We probed a mixed-stage cDNA library (Barstead and Waterston, 1989) with TR 191 and identified seven overlapping clones, five of which contain a poly(A) tract at their 3′ end, and four of which contain nine nucleotides of SL1 at their 5′ ends. Clones containing both SL1 and poly(A) are full length, and one of them, TR 162, was chosen for further study. TR 162 is 1.825 kb long and, when used as a probe on a northern blot, hybridizes to a single 2.0 kb species of poly(A)+ mRNA. This size is consistent with TR 162 being a full-length cDNA clone shortened only in the length of its 3′ poly(A) tail.
To confirm that the cDNA insert of TR 162 represents smg-5, we established that it provides smg-5(+) function when expressed as a transgene. We placed the cDNA insert of TR 162 into plasmid pPD30.38, an expression vector controlled by a body wall muscle-specific promoter (Mello and Fire, 1995). Using the transformation rescue assay described above, the resulting plasmid fully rescues the motility phenotype of smg-5(r860) unc-54(r293) mutants.
SMG-5 is a novel protein
We sequenced plasmids TR 191 (genomic DNA; DDBJ/EMBL/GenBank accession No. U64440) and TR 162 (full-length cDNA; GenBank accession No. U64441) and deduced that smg-5 contains four introns and a consensus 3′ splice acceptor at the site of SL1 trans-splicing. The single open reading frame (ORF) of TR 162 encodes a 549 amino acid (64 kDa) protein. Smg-5 corresponds to gene W02D3.8, an ORF predicted from the C.elegans genomic sequence, although Genefinder predictions of the W02D3.8 mRNA structure are incorrect at the 5′ end. SMG-5 is not strongly similar to other proteins of GenBank release 130.0, although we note weak similarity to an uncharacterized yeast protein, Tos5p. A 143 amino acid region of SMG-5 (amino acids 238–380) is 25% identical to a 144 amino acid region of Tos5p (amino acids 157–300). Weak homologs of SMG-5 have been detected in mammalian cells (S.Ohno and L.Maquat, personal communications), although such homologs are not evident following routine BLAST searches.
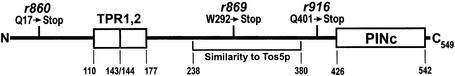
SMG-5 contains two conserved sequence motifs. Pattern and motif analyses using publicly available tools identify a PINc domain near the SMG-5 C-terminus (amino acids 426–542) and two TPR domains (amino acids 110–143 and 144–177). PINc domains are found in a diverse family of predicted nucleotide-binding proteins, members of which include several RNase components of the exosome. Interestingly, yeast Nmd4p, which was identified by its interaction with Upf1p in a two-hybrid screen, but has not been demonstrated to be involved in NMD (He and Jacobson, 1995), also contains a PINc domain (Clissold and Ponting, 2000). TPR (tetratricopeptide) repeats are 34 amino acid repeats found in many different proteins, wherein they facilitate specific interactions with partner proteins (Blatch and Lassle, 1999). SMG-7, with which SMG-5 interacts (see below), contains two TPR domains (Cali and Anderson, 1999).
Identifying smg-5-null alleles
In order to establish which, if any, smg-5 alleles are molecular nulls, we sequenced the coding region of three ethyl methanesulfonate (EMS)-induced smg-5 mutants. Smg-5(r860), smg-5(r869) and smg-5(r916) each contain a single G/C→A/T transition creating nonsense mutations at codons 17 (Q→UAA), 292 (W→UAG) and 401 (Q→UAA), respectively. The phenotypes of these nonsense alleles are indistinguishable from those of previously described smg-5 alleles (Hodgkin et al., 1989), demonstrating that this is the smg-5-null phenotype. The domain structure of SMG-5 and the location of smg-5 mutations are diagrammed in Figure 1.
Fig. 1. The domain structure of SMG-5. SMG-5 contains a PINc and two tetratricopeptide (TPR) repeats. smg-5(r860), smg-5(r869) and smg-5(r916) are nonsense alleles.
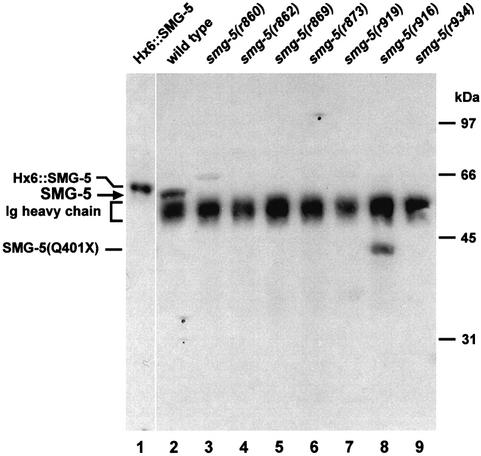
We prepared affinity-purified polyclonal antibodies against SMG-5 and identified SMG-5 on western blots. SMG-5 migrates as a single band with a relative mobility of 56 kDa (Figure 2, lane 2). Two observations demonstrate that the protein migrating at 56 kDa is SMG-5. First, a purified histidine-tagged SMG-5, which is 20 amino acids longer than SMG-5, migrates at 57 kDa (Figure 2, lane 1). Secondly, SMG-5 detected in wild type is absent from each of seven tested smg-5(–) mutants (Figure 2, lanes 3–9), including smg5(r860), an early nonsense mutation. Smg-5(r916) expresses a truncated protein whose relative mobility (42 kDa) is that predicted from its sequence, a nonsense mutation at codon 401.
Fig. 2. Expression of SMG-5 in wild-type and smg-5 mutants. SMG-5 was concentrated from wild-type and mutant extracts by immunoprecipitation and then analyzed by western blotting. Lane 1, purified recombinant His6-tagged SMG-5; lane 2, wild-type C.elegans; lanes 3–9, seven independent smg-5 alleles. Six tested alleles express no detectable SMG-5 even after prolonged exposure. Smg-5(r916) expresses an ∼42 kDa truncated protein [SMG-5(Q401X)]. Rabbit immunoglobulin (Ig) is present from the immunoprecipitations and detected in these experiments.
SMG-5 interacts with SMG-7
In both smg-5(–) and smg-7(–) mutants, a phosphorylated isoform of SMG-2 accumulates to abnormally high levels (Page et al., 1999), suggesting that both SMG-5 and SMG-7 are involved in SMG-2 dephosphorylation. We tested whether SMG-5 and SMG-7 physically interact by immunoprecipitating SMG-5 (or SMG-7) from wild-type and mutant crude extracts and then testing by western blots whether SMG-7 (or SMG-5) was present in the immune complexes. We used anti-SMG-5 antibodies described above and an anti-SMG-7 antibody described previously (Cali et al., 1999) for this analysis.
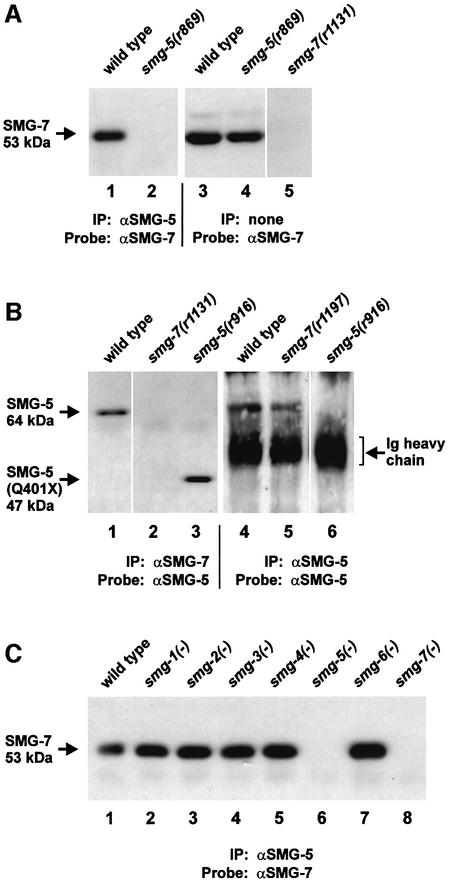
As shown in Figure 3A, SMG-7 co-purifies with immunoprecipitated SMG-5 from wild-type extracts (lane 1). SMG-7 does not co-immunoprecipitate with SMG-5 from smg-5(r869) extracts (a null allele; lane 2), demonstrating that SMG-7 is not trapped non-specifically in the immune complexes. Additional controls demonstrate that SMG-7 is expressed normally in smg-5(r869) (lane 4), and that the anti-SMG-7 antibody is specific for SMG-7 (lanes 3 and 5).
Fig. 3. Co-immunoprecipitation of SMG-5 and SMG-2. (A) Total soluble proteins (lanes 3–5) or proteins found in the pellet following immunoprecipitation of soluble extracts with anti-SMG-5 antibody (lanes 1 and 2) were electrophoresed and probed in western blots with anti-SMG-2 antibody. (B) Proteins immunoprecipitated by anti-SMG-7 or anti-SMG-5 antibodies were electrophoresed and probed in western blots with anti-SMG-5 antibodies. (C) Immunoprecipitations and western blotting performed as in (A). Mutants used for this analysis were smg-1(r861), smg-2(r908), smg-3(r867), smg-4(ma116), smg-5(r869), smg-6(r896) and smg-7(r1131).
The reciprocal immunoprecipitation experiment is shown in Figure 3B. SMG-5 (64 kDa) co-purifies with immunoprecipitated SMG-7 from wild-type extracts (lane 1), but not from smg-7(r1131) extracts (lane 2), an allele that does not express detectable SMG-7 (Cali et al., 1999). Additional controls demonstrate that SMG-5 is expressed normally in smg-7(r1137) (lane 5), a null allele of SMG-7 that is phenotypically indistinguishable from r1131 (Cali et al., 1999). SMG-5(Q401X), the 42 kDa truncated protein expressed in smg-5(r916), co-immunoprecipitates with SMG-7 (lane 3), demonstrating that the C-terminal ∼30% of SMG-5 is not required for the SMG-5–SMG-7 interaction. We conclude from these experiments that SMG-5 and SMG-7 physically interact, either directly or indirectly, as part of a larger protein complex.
SMG-5–SMG-7 interaction does not require activities of other smg genes
If SMG-5 and SMG-7 interact as part of a multiprotein complex, eliminating other components of the complex might disrupt the SMG-5–SMG-7 interaction. Products of the five remaining smg genes are good candidates for being part of an SMG-5–SMG-7 protein complex. We therefore tested whether the SMG-5–SMG-7 interaction detected above is maintained in representative alleles of all smg genes. As shown in Figure 3C, SMG-5 and SMG-7 co-immunoprecipitate in extracts of all tested smg mutants other than smg-5(–) and smg-7(–). Smg-2(r908) (lane 3) is a deletion of smg-2 (Page et al., 1999). The alleles of smg-1, -3, -4 and -6 used in this analysis have not been sequenced. Thus, we are unsure if stable mutant proteins are expressed in these alleles. We conclude that neither SMG-2 nor the activities of smg-1, -3, -4 and -6 are required for the SMG-5–SMG-7 interaction.
SMG-5 and SMG-7 probably interact directly
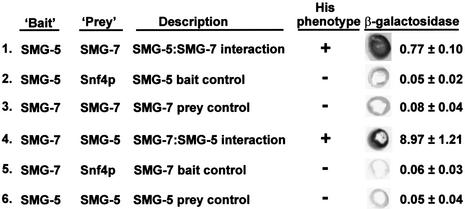
The experiments described above do not address whether SMG-5 interacts directly or indirectly with SMG-7. We therefore tested whether the SMG-5–SMG-7 interaction could be recapitulated in yeast. As neither protein is similar to any yeast proteins (see above; Cali et al., 1999), a positive interaction in a yeast two-hybrid assay would probably indicate that the proteins interact directly. We constructed ‘bait’ and ‘prey’ plasmids in which full- or nearly full-length SMG-5, SMG-7 or control proteins are fused to either the LexA DNA-binding domain (‘bait’) or the Gal4p activation domain (‘prey’). We assayed potential interactions both by the ability of strains to grow in media lacking histidine and by qualitative and quantitative β-galactosidase assays, using appropriate two-hybrid-responsive reporter genes. As shown in Figure 4, we detected positive SMG-5–SMG-7 interactions using either protein as bait and the other as prey (lines 1 and 4). Tests for SMG-5–SMG-5 and SMG-7–SMG-7 interactions were negative (lines 3 and 6). We conclude that SMG-5 and SMG-7 probably interact directly, but we cannot exclude the possibility that an unidentified yeast protein bridges their interaction in two-hybrid tests.
Fig. 4. Two-hybrid interactions between SMG-5 and SMG-7. ‘His phenotype’ (column 4) refers to the ability (‘+’) or inability (‘–’) of strains to grow on medium lacking histidine. Columns 5 and 6 show β-galactosidase colorimetric plate tests and enzymatic assays, respectively. Standard deviations were calculated from at least three independent assays. LexA DNA-binding domain fusions (‘bait’) and Gal4 activation domain fusions (‘prey’) contained SMG-5 amino acids 1–549 (full length) and 1–546, respectively. All SMG-7 fusion proteins contained SMG-7 amino acids 7–454 (out of 458).
We delimited the regions of SMG-5 and SMG-7 responsible for their two-hybrid interaction by constructing a series of bait plasmids in which 12 different overlapping segments of SMG-5 were tested for two-hybrid interactions with nearly full-length SMG-7 (amino acids 7–454) as described above (see Supplementary table 1, available at The EMBO Journal Online). Similarly, 10 different overlapping segments of SMG-7 were tested for two-hybrid interactions with full-length SMG-5 (see Supplementary table 2). All constructs, including those proving to be negative in tests of interaction, were confirmed by western blotting using an anti-LexA antibody to express stable hybrid proteins (data not shown). The minimal SMG-5 fusion protein that retained interaction with SMG-7 contains SMG-5 amino acids 65–378 (out of 549). The minimal SMG-7 fusion protein that retained interaction with SMG-5 contains SMG-7 amino acids 7–393 (out of 458).
Proteins that interact with SMG-7
To identify possible additional proteins with which SMG-7 interacts, we transformed a library of C.elegans cDNAs fused to the Gal4p activation domain into a yeast strain containing SMG-7 amino acids 7–454 fused to LexA. Seven independent clones satisfied our tests as being positive for two separate two-hybrid-responsive reporter genes and specific for the SMG-7 hybrid partner. Three of these clones contained inserts of smg-5. Four additional clones encoded overlapping portions of gene Y37A1B.15. We investigated whether inhibiting expression of Y37A1B.15 by RNA interference (RNAi) yields a Smg(–) phenotype. Under conditions where microinjection of a control smg-2 double-stranded (ds) RNA yielded offspring showing a strong Smg(–) phenotype [as judged by suppression of unc-54(r293)], microinjection of Y37A1B.15 dsRNA yielded no detectable phenotype. Thus, the significance of possible SMG-7–Y37A1B.15 interactions and roles for Y37A1B.15 in NMD are unclear.
SMG-5 interacts with a subunit of protein phosphatase 2A
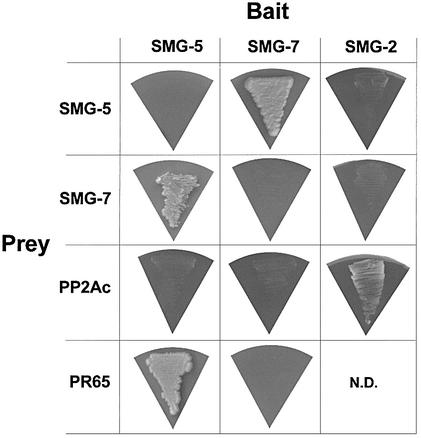
To identify possible additional proteins with which SMG-5 interacts, we transformed the two-hybrid library described above into a strain containing full-length SMG-5 fused to the Gal4p DNA-binding domain. Twenty-five independent clones satisfied our tests as being positive for two different two-hybrid-responsive reporter genes and specific for the SMG-5 hybrid partner. All 25 of these clones represent overlapping portions of ORF F48E8.5, which encodes PR65, the 65 kDa, A-type, structural subunit of PP2A. As shown in Figure 5, SMG-5 interacts with PR65 in directed two-hybrid tests, while SMG-7 does not.
Fig. 5. Two-hybrid interactions tested among SMG proteins and PP2A subunits. Growth tests were performed on histidine-free medium containing 1 mM 3-amino-1,2,4-triazole. ‘Bait’ and ‘prey’ proteins were fused to LexA DNA-binding domains and Gal4 activation domains, respectively.
Eukaryotic PP2As are a family of ubiquitous, abundant, Ser/Thr protein phosphatases (reviewed in Janssens and Goris, 2001). Active enzymes are usually heterotrimeric complexes containing: (i) a 36 kDa catalytic subunit (PP2AC); (ii) a 65 kDa structural subunit that scaffolds assembly of active enzyme (PR65); and (iii) one of several B-type regulatory subunits whose identity varies among different trimeric PP2A complexes. Identified B subunits are thought to regulate activity, subcellular localization and substrate specificity of the catalytic complex.
SMG-2 interacts with PP2AC
To identify possible additional proteins with which SMG-2 interacts, we transformed the two-hybrid library described above into a strain containing the Gal4p DNA-binding domain fused to SMG-2 amino acids 1–904 (out of 1069). Among 265 candidate positive clones, only a portion of which have been analyzed, one full-length clone of C.elegans gene F38H4.9 (318 amino acids) satisfied our tests as being positive for two different two-hybrid-responsive reporter genes and specific for the SMG-2 hybrid partner. F38H4.9 encodes C.elegans PP2AC, the catalytic subunit of PP2A. As shown in Figure 5, SMG-2 interacts with PP2AC in directed two-hybrid tests, while SMG-5 and SMG-7 do not.
SMG-5 and SMG-2 interact with PP2A in co-immunoprecipitation assays
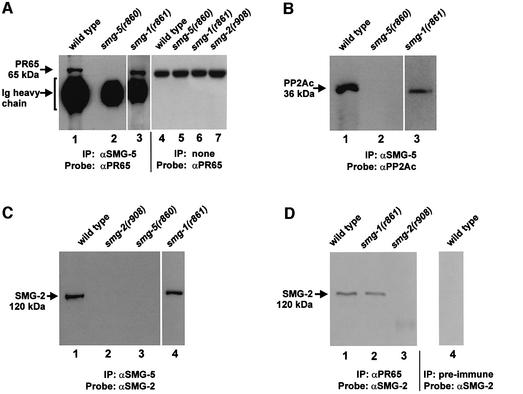
To investigate whether SMG-5 and SMG-2 interact with PP2A, we tested whether SMG-5 and SMG-2 co-immunoprecipitate with PR65 and PP2AC from C.elegans crude extracts. Our general strategy was to immunoprecipitate SMG-5 (or SMG-2) from wild-type and mutant extracts and then to test by western blots whether PR65, PP2AC or SMG-2 were present in the immune complexes.
As shown in Figure 6A and B, PR65 and PP2AC co-purify with immunoprecipitated SMG-5 from wild-type extracts (lanes 1), but not from smg-5(r860) (a null allele) mutant extracts (Figure 6A and B, lanes 2). An additional control demonstrates that PR65 is expressed at normal abundance in smg-5(r860) (Figure 6A, lane 5). Both PR65 and PP2AC co-immunoprecipitate with SMG-5 from smg-1(r861) mutant extracts (Figure 6A and B, lanes 3). Thus, failure to precipitate PR65 or PP2AC with SMG-5 in smg-5(r860) mutants is due to the absence of SMG-5, rather than because the extracts are smg(–) per se. We conclude that at least a fraction of SMG-5 interacts with both PP2AC and PR65 in crude extracts.
Fig. 6. SMG-5 and SMG-2 interact with PR65 and PP2AC. (A–C) Proteins immunoprecipitated by anti-SMG-5 antibody were electrophoresed and probed in western blots with antibodies to PR65, PP2AC and SMG-2, respectively. (D) Proteins immunoprecipitated by anti-PR65 antibody were probed with anti-SMG-2 antibody. Smg-1(r861), smg-2(r908) and smg-5(r860) are null alleles of the affected genes.
If SMG-5 functions as part of a complex that includes PP2A, an attractive function would be to target PP2AC to its phosphorylated SMG-2 substrate. If true, then SMG-5 and SMG-2 should interact. We tested this using co-immunoprecipitation assays similar to those described above. As shown in Figure 6C, SMG-2 co-purifies with immunoprecipitated SMG-5 from a wild-type extract (lane 1) and from a smg-1(r861) extract (lane 4), but not from a control smg-5(r860) extract (lane 3). Previous experiments demonstrate that SMG-2 is expressed normally in smg-5(r860) (Page et al., 1999). We conclude that at least a fraction of SMG-5, which was shown above to interact with PR65 and PP2AC, also interacts with SMG-2.
If SMG-5 targets PP2A to its phosphorylated SMG-2 substrate, then SMG-2 and PP2A should interact. We tested this by co-immunoprecipitation assays. As shown in Figure 6D, SMG-2 co-purifies with immunoprecipitated PR65 in wild-type extracts (lane 1), but not in smg-2(r908) mutant extracts (lane 3). Interaction of PR65 with SMG-2 is maintained in smg-1(r861) extracts (lane 2). We conclude that at least a fraction of SMG-2 exists in a complex that includes PR65 in crude extracts. Two-hybrid experiments described above (see Figure 5) suggest that SMG-2 interacts directly with PP2AC.
Discussion
Smg-5 encodes a novel protein necessary for NMD
The functions of seven smg genes are required for accelerated decay of nonsense-mutant mRNAs in C.elegans (Pulak and Anderson, 1993; Cali et al., 1999). As part of an effort to characterize the genes involved in NMD, we cloned smg-5. Two lines of evidence demonstrate that the gene we describe is smg-5. First, clones of smg-5 restore NMD when introduced into smg-5(–) mutants. Secondly, three independent smg-5(–) mutants contain nonsense mutations in the cloned gene.
SMG-5 is not strikingly similar to ORFs of yeast, although weak similarity to an uncharacterized gene, TOS5, is evident. While the mechanisms of NMD could be different in yeasts and nematodes, the homology of Upf1p, Upf2p and Upf3p to SMG-2, SMG-3 and SMG-4, respectively (Page et al., 1999; Aronoff et al., 2001; S.Kuchma and P.Anderson, unpublished data) suggests that there are important similarities. Perhaps SMG-5 and an unknown yeast gene have diverged to the point where their evolutionary relationship is no longer apparent. Perhaps Nmd4p, which like SMG-5 contains a PINc domain, or Tos5p perform functions in yeast analogous to those of SMG-5 in C.elegans. Alternatively, perhaps Upf1p in yeast is controlled by mechanisms not involving phosphorylation. Human UPF1 is phosphorylated by human SMG1 (Denning et al., 2001; Pal et al., 2001; Yamashita et al., 2001), suggesting that the mechanisms of NMD are similar in nematodes and humans. Mammalian homologs of smg-5 have, furthermore, been identified (S.Ohno and L.Maquat, personal communication). We suggest that mammalian homologs of SMG-5, SMG-6 and SMG-7 will be involved similarly in dephosphorylation of human SMG2.
Complexes of SMG and related proteins
We report here that SMG-5 interacts with SMG-7, SMG-2 and the catalytic and structural subunits of PP2A (PP2Ac and PR65, respectively), as judged by yeast two-hybrid assays, co-immunoprecipitation assays or both. Whether these data reflect the existence of a single complex or multiple subcomplexes currently is unknown. What other proteins are components of these complexes? Upf1p of yeast, the ortholog of SMG-2, interacts directly or indirectly with several proteins known to be important for NMD, including Upf2p, Upf3p, translation release factors eRF1 and eRF3, and Dcp2p (He and Jacobson, 1995; He et al., 1997; Czaplinski et al., 1998; Dunckley and Parker, 1999). Mammalian PP2A interacts with translation release factor (Andjelkovic et al., 1996). Most of these proteins are associated with polysomes (Andjelkovic et al., 1996; Atkin et al., 1997; Pal et al., 2001), presumably reflecting the fact that the surveillance complex operates during translation termination to mediate alternative outcomes with regard to mRNA degradation.
SMG-5 is required for efficient dephosphorylation of SMG-2 (Page et al., 1999). We demonstrated using both co-immunoprecipitation and two-hybrid assays that SMG-5 interacts, directly or indirectly, with the structural (PR65) and catalytic (PP2AC) subunits of PP2A. We propose that interactions between SMG-5 and PP2A confer upon PP2A specificity for its SMG-2 substrate. SMG-5 might, therefore, be a B-type regulatory subunit of PP2A. While we have no direct biochemical evidence identifying a SMG-5-associated PP2A as the SMG-2 phosphatase, three observations suggest that this is the case. First, SMG-2 accumulates as a hyperphosphorylated isoform in smg-5(–) mutants. Secondly, SMG-2 interacts with SMG-5, PR65 and PP2AC (Figures 5 and 6). Thirdly, SMG-5 interacts with PR65 and SMG-2 (Figures 5 and 6). The most economical interpretation is that SMG-2 is the direct substrate of a SMG-5-associated PP2A. In smg-5(–) mutants, SMG-2 would be hyperphosphorylated due to its inability to interact with PP2A.
SMG-2, however, need not be the direct substrate of SMG-5-associated PP2A. In principle, any phosphorylated component of the NMD machinery could be the substrate of a SMG-5-associated PP2A. Dcp1p, for example, is a phosphoprotein, although a role for phosphorylation in decapping has not been established (LaGrandeur and Parker, 1998). Other known or unknown phosphoproteins of the NMD machinery could also be substrates of a SMG-5-associated PP2A. The simplest interpretation, however, is that SMG-2 is the substrate.
Role of SMG-2 phosphorylation in NMD
All known smg mutations affect either SMG-2 itself or the state of its phosphorylation (Page et al., 1999). Both mutations that block SMG-2 phosphorylation (smg-1, smg-3 and smg-4), and those that impair SMG-2 dephosphorylation (smg-5, smg-6 and smg-7) eliminate NMD (Pulak and Anderson, 1993; Cali et al., 1999). Thus, complete cycles of SMG-2 phosphorylation and dephosphorylation are required for NMD. In wild-type animals, the pool of SMG-2 is almost exclusively dephosphorylated (Page et al., 1999). Assuming that a substantial proportion of steady-state SMG-2 participates in NMD, phosphorylation of SMG-2, although essential for NMD, occurs for only a brief period of time and is followed quickly by its SMG-5-dependent dephosphorylation. When does this occur? The extent of phosphorylation of human UPF1 (the ortholog of SMG-2) is increased in polysome-associated UPF1 (Pal et al., 2001). Translation termination seems a logical step at which the phosphorylation cycle occurs. In support of this model, PP2A interacts with mammalian eRF1 (Andjelkovic et al., 1996), a protein that is required for and functions during translation termination.
What aspect of SMG-2 function is regulated by phosphorylation? Nucleotide exchange and/or hydrolysis is an attractive possibility. The ATPase/helicase activity of Upf1p is essential for NMD (Weng et al., 1996), and ATP is a cofactor of RNA binding by Upf1p in both yeast and humans (Weng et al., 1998; Bhattacharya et al., 2000). Two alleles of smg-2 are exceptional in that the mutant SMG-2 is dephosphorylated inefficiently (Page et al., 1999). With regard to SMG-2 phosphorylation, such alleles behave like smg-5(–) mutants. Both of these alleles, smg-2(r866) and smg-2(r895), are single amino substitutions of the P-loop of the SMG-2 nucleotide-binding site, and both are strongly predicted to eliminate ATP binding and/or hydrolysis. Thus, an ATPase-inactive SMG-2 adopts a conformation that promotes the phosphorylated isoform. In wild-type SMG-2, perhaps phosphorylated SMG-2 adopts a conformation that inhibits the ATPase. Activation of the SMG-2 ATPase via SMG-5-associated PP2A would be an essential step in NMD. If SMG-2 is not activated by dephosphorylation, or if it is not phosphorylated in the first place, NMD is eliminated. The activity of human p68 RNA helicase (an RNA-dependent ATPase and RNA helicase) is regulated similarly by phosphorylation (Buelt et al., 1994).
Phosphorylation of SMG-2 might also (or alternatively) control assembly or dynamics of the surveillance complex. Conformational changes of SMG-2 caused by reversible phosphorylation might alter the affinity of SMG-2 for other components of the surveillance complex. Phosphorylation of eIF4E and eIF4E-binding proteins, for example, regulates assembly of eukaryotic translation initiation complexes (reviewed in Jackson and Wickens, 1997). Regulating the dynamics of SMG-2 protein–protein interactions in C.elegans might parallel those of Tap42p in yeast (Di Como and Arndt, 1996; Jiang and Broach, 1999). Tap42p is a downstream effector of signaling via the Tor protein kinase pathway. Tap42p is phosphorylated reversibly by the competing activities of a Tap42p kinase (probably Tor1p and Tor2p) and PP2A. Tap42p appears to compete with PP2A structural and regulatory subunits for binding to PP2AC, with formation of a novel Tap42p–PPA2C dimer. The binding of Tap42p to PP2AC is promoted by Tap42p phosphorylation. In NMD, SMG-2 is phosphorylated reversibly by the activities of a SMG-2 kinase (requiring SMG-1, SMG-3 and SMG-4) and a SMG-2 phosphatase (requiring SMG-5, SMG-6 and SMG-7). SMG-1 is a member of the Tor/phosphatidylinositol 3-kinase-related family of protein kinases (S.O’Connor and P.Anderson, unpublished data). As shown in Figure 5, SMG-2 interacts with PP2AC. This suggests the existence of an SMG-2–PP2AC dimer, analogous to the Tap42p–PP2AC dimer, with the remaining SMG proteins functioning to regulate this interaction.
Materials and methods
Strains and genetic methods
Genetic methods for C.elegans were described previously by Brenner (1974). Alleles used in this study were: dpy-5(e61), dpy-14(e188), him-5(e1467), him-5(e1490), mut-2(r459), unc-24(e138), unc-54(r293), unc-87(e843) (Hodgkin et al., 1988); smg-1(r861), smg-2(r863), smg-3(r867), smg-4(ma116), smg-5(r860), smg-5(r862), smg-5(r869), smg-5(r873), smg-6(r886), smg-6(r896) (Hodgkin et al., 1989); smg-1(r904), smg-2(r908), smg-5(r916), smg-5(r919::Tc3), smg-5(r931::Tc1), smg-5(r934) (R.Pulak and P.Anderson, unpublished data); smg-7(r1131) (Cali et al., 1999); and smg-5(r919r940), smg-5(r919r941) and smg-5(r919r942), isolated as smg(+) revertants of smg-5(r919::Tc3) (R.Pulak and P.Anderson, unpublished data).
Transformation rescue of smg-5
Motile smg-5(r860) or smg-5(r862) unc-54(r293) hermaphrodites were co-injected with DNA to be tested and marker plasmid pRF4[rol-6(su1006)] using established methods (Mello and Fire, 1995). Independent heritable transformants were established and subsequently scored for motility. Animals rescued for their smg-5 phenotype exhibited the flaccid paralysis characteristic of unc-54(r293) smg-5(+) strains. To confirm that the transforming DNA was supplying smg-5(+) activity and not directly causing a dominant Unc phenotype, rescued animals were crossed with wild-type males. Array-containing cross-progeny exhibited normal motility. cDNA expression construct TR 237 was constructed by inserting the cDNA fragment into the EcoRV site of expression vector pPD30.38 (Mello and Fire, 1995).
Analysis of smg-5 cDNA and genomic clones
A C.elegans mixed-stage cDNA library (Barstead and Waterston, 1989) was screened by filter hybridization using plasmid TR 191 as probe. Seven positive clones, including TR 162, were isolated. The complete coding regions of sequenced smg-5(–) alleles were amplified from genomic DNA by PCR, cloned and sequenced. Each identified mutation was confirmed by sequencing a second, independently cloned, PCR fragment.
Antibodies
SMG-5 antisera were generated by immunizing two rabbits with recombinant, His6-tagged, full-length SMG-5 (Hx6::SMG-5) purified from inclusion bodies (Williams et al., 1995) after expression in Escherichia coli (pET15 system; Novagen). Anti-SMG-5 antibodies were affinity purified by coupling Hx6::SMG-5 to Actigel ALD (Sterogene) and passing serum over the Hx6::SMG-5-conjugated resin. Anti-SMG-5 antibodies were eluted with ActiSep (Sterogene), desalted and concentrated. Anti-SMG-2 antibodies were described previously (Page et al., 1999). Anti-PR65 antisera were generated essentially as described above, except that the recombinant fusion protein used was tagged with GST and purified using glutathione–Sepharose 4B columns (pGEX4T system; Amersham Pharmacia). Anti-PP2AC was obtained commercially (Santa Cruz Biotechnology).
Two-hybrid analysis
Yeast strains L40 (Hollenberg et al., 1995) and PJ69-4A (James et al., 1996) were used to assay interacting proteins. Plasmid bait clones derived from pBTM116 were used with strain L40, and those derived from pGBD-C2 with strain PJ69-4A. Plasmid prey clones were derived from pACT2. All inserts used in two-hybrid analysis were generated by PCR with primers tailed with appropriate restriction sites; inserts and cloning junctions of completed clones were sequenced. Histidine growth tests contained 1 mM 3-amino-1,2,4-triazole. Segments of SMG-5 tested for interactions with SMG-2 (amino acids 7–454) were SMG-5 amino acids 1–549 (full length), 1–407, 1–346, 1–174, 65–549, 65–407, 65–378, 65–355, 65–329, 101–549, 101–407 and 203–512 (see Supplementary table 1). Segments of SMG-2 tested for interactions with full-length SMG-5 (amino acids 1–549) were SMG-2 amino acids 7–454, 7–393, 7–375, 7–362, 7–324, 7–255, 33–393, 33–362, 54–454 and 54–393 (see Supplementary table 2). The SMG-2 two-hybrid library screen was performed in yeast strain L40. The SMG-5 two-hybrid library screen was performed in yeast strain PJ69-4A. The two-hybrid library was a gift from the Barstead laboratory. Yeast plasmids were isolated using Zymoprep (Zymo Research). The cDNA inserts of candidate two-hybrid positive colonies were amplified by PCR and sequenced. Candidate clones were retransformed into yeast strains carrying different bait plasmids, in order to test for specificity.
RNAi
Templates for T3 RNA polymerase were generated by PCR. RNA was transcribed using MEGAscriptT3 (Ambion). Sense and antisense RNAs were mixed, hybridized and injected into unc-54(r293) animals using established methods (Mello and Fire, 1995).
Immunoprecipitation
Mixed-stage animals were suspended in 20 mM MOPS pH 7.2, homogenized by passing twice through a French pressure cell at 12 000 p.s.i. and centrifuged at 21 000 g for 20 min, retaining the supernatant fraction. For the experiments of Figure 3, affinity-purified anti-SMG-5 and anti-SMG-2 antibodies were coupled to Actigel ALD resin (Sterogene). Extracts were either incubated directly with Actigel-coupled antibodies or incubated with antibody followed by addition of protein A–Sepharose (Amersham-Pharmacia). Precipitated proteins were collected by centrifugation and washed 3–6 times with 20 mM MOPS pH 7.2.
Western blotting
Proteins were separated by SDS–PAGE and transferred to Immobilon-P membranes (Millipore) using a Trans-Blot semi-dry transfer cell (Bio-Rad). Western blotting was performed using standard procedures (Harlow and Lane, 1999) and the enhanced chemiluminescence (ECL) detection system (Amersham). To block immunoglobulin heavy chain, some blots were incubated for 1 h with goat anti-rabbit IgG (H + L) Fab fragments (Jackson ImmunoResearch Laboratories).
Supplementary data
Supplementary data are available at The EMBO Journal Online.
Acknowledgments
Acknowledgements
We thank Rock Pulak for providing smg-5(r919) and its revertants, Edward Maryon and Quinn Mitrovich for critical reading of the manuscript, Andrei Petcherski and the Kimble laboratory for gifts of yeast strains and plasmids, and the Barstead laboratory for a gift of the two-hybrid and cDNA libraries. This work was supported by the University of Wisconsin Training Grant in Genetics, by a Howard Hughes Predoctoral Fellowship (to A.G.) and by a research grant (GM50933) from the NIH.
References
- Anderson P. (1995) Mutagenesis. Methods Cell Biol., 48, 31–58. [PubMed] [Google Scholar]
- Andjelkovic N., Zolnierowicz,S., Van Hoof,C., Goris,J. and Hemmings,B.A. (1996) The catalytic subunit of protein phosphatase 2A associates with the translation termination factor eRF1. EMBO J., 15, 7156–7167. [PMC free article] [PubMed] [Google Scholar]
- Applequist S.E., Selg,M., Raman,C. and Jack,H.M. (1997) Cloning and characterization of HUPF1, a human homolog of the Saccharomyces cerevisiae nonsense mRNA-reducing UPF1 protein. Nucleic Acids Res., 25, 814–824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aronoff R., Baran,R. and Hodgkin,J. (2001) Molecular identification of smg-4, required for mRNA surveillance in C.elegans. Gene, 268, 153–164. [DOI] [PubMed] [Google Scholar]
- Atkin A.L., Schenkman,L.R., Eastham,M., Dahlseid,J.N., Lelivelt,M.J. and Culbertson,M.R. (1997) Relationship between yeast polyribosomes and Upf proteins required for nonsense mRNA decay. J. Biol. Chem., 272, 22163–22172. [DOI] [PubMed] [Google Scholar]
- Barstead R.J. and Waterston,R.H. (1989) The basal component of the nematode dense body is vinculin. J. Biol. Chem., 264, 10177–10185. [PubMed] [Google Scholar]
- Beelman C.A., Stevens,A., Caponigro,G., LaGrandeur,T.E., Hatfield,L., Fortner,D.M. and Parker,R. (1996) An essential component of the decapping enzyme required for normal rates of mRNA turnover. Nature, 382, 642–646. [DOI] [PubMed] [Google Scholar]
- Bhattacharya A., Czaplinski,K., Triflissis,P., He,F., Jacobson,A. and Peltz,S.W. (2000) Characterization of the biochemical properties of the human Upf1 gene product that is involved in nonsense-mediated mRNA decay. RNA, 6, 1226–1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blatch G.L. and Lassle,M. (1999) The tetratricopeptide repeat: a structural motif mediating protein–protein interactions. BioEssays, 21, 932–939. [DOI] [PubMed] [Google Scholar]
- Brenner S. (1974) The genetics of Caenorhabditis elegans. Genetics, 77, 71–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buelt M.K., Glidden,B.J. and Storm,D.R. (1994) Regulation of p68 RNA helicase by calmodulin and protein kinase C. J. Biol. Chem., 269, 29367–29370. [PubMed] [Google Scholar]
- Cali B.M. and Anderson,P. (1998) mRNA surveillance mitigates genetic dominance in Caenorhabditis elegans. Mol. Gen. Genet., 260, 176–184. [DOI] [PubMed] [Google Scholar]
- Cali B.M., Kuchma,S.L., Latham,J. and Anderson,P. (1999) smg-7 is required for mRNA surveillance in Caenorhabditis elegans. Genetics, 151, 605–616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clissold P.M. and Ponting,C.P. (2000) PIN domains in nonsense-mediated mRNA decay and RNAi. Curr. Biol., 10, R888–R890. [DOI] [PubMed] [Google Scholar]
- Czaplinski K., Ruiz-Echevarria,M.J., Paushkin,S.V., Han,X., Weng,Y., Perlick,H.A., Dietz,H.C., Ter-Avanesyan,M.D. and Peltz,S.W. (1998) The surveillance complex interacts with the translation release factors to enhance termination and degrade aberrant mRNAs. Genes Dev., 12, 1665–1677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decker C.J. and Parker,R. (1993) A turnover pathway for both stable and unstable mRNAs in yeast: evidence for a requirement for deadenylation. Genes Dev., 7, 1632–1643. [DOI] [PubMed] [Google Scholar]
- Denning G., Jamieson,L., Maquat,L.E., Thompson,E.A. and Fields,A.P. (2001) Cloning of a novel phosphatidylinositol kinase-related kinase: characterization of the human SMG-1 RNA surveillance protein. J. Biol. Chem., 276, 22709–22714. [DOI] [PubMed] [Google Scholar]
- Di Como C.J. and Arndt,K.T. (1996) Nutrients, via the Tor proteins, stimulate the association of Tap42 with type 2A phosphatases. Genes Dev., 10, 1904–1916. [DOI] [PubMed] [Google Scholar]
- Dunckley T. and Parker,R. (1999) The DCP2 protein is required for mRNA decapping in Saccharomyces cerevisiae and contains a functional MutT motif. EMBO J., 18, 5411–5422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goetinck S. and Waterston,R.H. (1994) The Caenorhabditis elegans muscle-affecting gene unc-87 encodes a novel thin filament-associated protein. J. Cell Biol., 127, 79–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez C.I., Ruiz-Echevarria,M.J., Vasudevan,S., Henry,M.F. and Peltz,S.W. (2000) The yeast hnRNP-like protein Hrp1/Nab4 marks a transcript for nonsense-mediated mRNA decay. Mol. Cell, 5, 489–499. [DOI] [PubMed] [Google Scholar]
- Hall G.W. and Thein,S. (1994) Nonsense codon mutations in the terminal exon of the β-globin gene are not associated with a reduction in β-mRNA accumulation: a mechanism for the phenotype of dominant β-thalassemia. Blood, 83, 2031–2037. [PubMed] [Google Scholar]
- Harlow E. and Lane,D. (1999) Using Antibodies: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- He F. and Jacobson,A. (1995) Identification of a novel component of the nonsense-mediated mRNA decay pathway by use of an interacting protein screen. Genes Dev., 9, 437–454. [DOI] [PubMed] [Google Scholar]
- He F., Peltz,S.W., Donahue,J.L., Rosbash,M. and Jacobson,A. (1993) Stabilization and ribosome association of unspliced pre-mRNAs in a yeast upf1– mutant. Proc. Natl Acad. Sci. USA, 90, 7034–7038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He F., Brown,A.H. and Jacobson,A. (1997) Upf1p, Nmd2p and Upf3p are interacting components of the yeast nonsense-mediated mRNA decay pathway. Mol. Cell. Biol., 17, 1580–1594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hentze M.W. and Kulozik,A.E. (1999) A perfect message: RNA surveillance and nonsense-mediated decay. Cell, 96, 307–310. [DOI] [PubMed] [Google Scholar]
- Hilleren P. and Parker,R. (1999) Mechanisms of mRNA surveillance in eukaryotes. Annu. Rev. Genet., 33, 229–260. [DOI] [PubMed] [Google Scholar]
- Hodgkin J., Edgley,M., Riddle,D. and Albertson,D.G. (1988) The Nematode Caenorhabditis elegans. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Hodgkin J., Papp,A., Pulak,R., Ambros,V. and Anderson,P. (1989) A new kind of informational suppression in the nematode Caenorhabditis elegans. Genetics, 123, 301–313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollenberg S.M., Sternglanz,R., Cheng,P.F. and Weintraub,H. (1995) Identification of a new family of tissue-specific basic helix–loop–helix proteins with a two-hybrid system. Mol. Cell. Biol., 15, 3813–3822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson R.J. and Wickens,M. (1997) Translational controls impinging on the 5′-untranslated region and initiation factor proteins. Curr. Opin. Genet. Dev., 7, 233–241. [DOI] [PubMed] [Google Scholar]
- James P., Halladay,J. and Craig,E.A. (1996) Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics, 144, 1425–1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janssens V and Goris,J. (2001) Protein phosphatase 2A: a highly regulated family of serine/threonine phosphatases implicated in cell growth and signalling. Biochem. J., 353, 417–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Y. and Broach,J.R. (1999) Tor proteins and protein phosphatase 2A reciprocally regulate Tap42 in controlling cell growth in yeast. EMBO J., 18, 2782–2792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim V.N., Kataoka,N. and Dreyfuss,G. (2001) Role of the nonsense-mediated decay factor hUpf3 in the splicing-dependent exon–exon junction complex. Science, 7, 1832–1836. [DOI] [PubMed] [Google Scholar]
- LaGrandeur T.E. and Parker,R. (1998) Isolation and characterization of Dcp1p, the yeast mRNA decapping enzyme. EMBO J., 17, 1487–1496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Hir H., Izaurralde,E., Maquat,L.E. and Moore,M.J. (2000) The spliceosome deposits multiple proteins 20–24 nucleotides upstream of mRNA exon–exon junctions. EMBO J., 19, 6860–6869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Hir H., Gatfield,D., Izaurralde,E. and Moore,M.J. (2001) The exon–exon junction complex provides a binding platform for factors involved in mRNA export and nonsense-mediated mRNA decay. EMBO J., 20, 4987–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S. and Wilkinson,M.F. (1998) Nonsense surveillance in lymphocytes? Immunity, 8, 135–141. [DOI] [PubMed] [Google Scholar]
- Li Z., Paulovich,A.G. and Woolford,J.L. (1995) Feedback inhibition of the yeast ribosomal protein gene CRY2 is mediated by the nucleotide sequence and secondary structure of CRY2 pre-mRNA. Mol. Cell. Biol., 15, 6454–6464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lykke-Andersen J., Shu,M.D. and Steitz,J.A. (2000) Human Upf proteins target an mRNA for nonsense-mediated decay when bound downstream of a termination codon. Cell, 103, 1121–1131. [DOI] [PubMed] [Google Scholar]
- Lykke-Andersen J., Shu,M.D. and Steitz,J.A. (2001) Communication of the position of exon–exon junctions to the mRNA surveillance machinery by the protein RNPS1. Science, 293, 1836–1839. [DOI] [PubMed] [Google Scholar]
- Medghalchi S.M., Frischmeyer,P.A., Mendell,J.T., Kelly,A.G., Lawler,A.M. and Dietz,H.C. (2001) Rent1, a trans-effector of nonsense-mediated mRNA decay, is essential for mammalian embryonic viability. Hum. Mol. Genet., 10, 99–105. [DOI] [PubMed] [Google Scholar]
- Mello C. and Fire,A. (1995) DNA transformation. Methods Cell Biol., 48, 451–482. [PubMed] [Google Scholar]
- Mendell J.T., Medghalchi,S.M., Lake,R.G., Noensie,E.N. and Dietz,H.C. (2000) Novel Upf2p orthologues suggest a functional link between translation initiation and nonsense surveillance complexes. Mol. Cell. Biol., 20, 8944–8957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitrovich Q.M. and Anderson,P. (2000) Unproductively spliced ribosomal protein mRNAs are natural targets of mRNA surveillance in C.elegans. Genes Dev., 14, 2173–2184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison M., Harris,K.S. and Roth,M.B. (1997) smg mutants affect the expression of alternatively spliced SR protein mRNAs in Caenorhabditis elegans. Proc. Natl Acad. Sci. USA, 94, 9782–9785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muhlrad D. and Parker,R. (1994) Premature translational termination triggers mRNA decapping. Nature, 370, 578–581. [DOI] [PubMed] [Google Scholar]
- Muhlrad D., Decker,C.J. and Parker,R. (1994) Deadenylation of the unstable mRNA encoded by the yeast MFA2 gene leads to decapping followed by 5′ to 3′ digestion of the transcript. Genes Dev., 8, 855–866. [DOI] [PubMed] [Google Scholar]
- Page M.F., Carr,B., Anders,K.R., Grimson,A. and Anderson,P. (1999) SMG-2 is a phosphorylated protein required for mRNA surveillance in Caenorhabditis elegans and related to Upf1p of yeast. Mol. Cell. Biol. 19, 5943–5951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pal M., Ishigaki,Y., Nagy,E. and Maquat,L.E. (2001) Evidence that phosphorylation of human Upf1 protein varies with intracellular location and is mediated by a wortmannin-sensitive and rapamycin-sensitive PI 3-kinase-related kinase signaling pathway. RNA, 7, 5–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perlick H.A., Medghalchi,S.M., Spencer,F.A., Kendzior,R.J.,Jr and Dietz,H.C. (1996) Mammalian orthologues of a yeast regulator of nonsense transcript stability. Proc. Natl Acad. Sci. USA, 93, 10928–10932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pulak R. and Anderson,P. (1993) mRNA surveillance by the Caenorhabditis elegans smg genes. Genes Dev., 1885–1897. [DOI] [PubMed] [Google Scholar]
- Ruiz-Echevarria M.J., Gonzalez,C.I. and Peltz,S.W. (1998) Identifying the right stop: determining how the surveillance complex recognizes and degrades an aberrant mRNA. EMBO J., 17, 575–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serin G., Gersappe,A., Black,J.D., Aronoff,R. and Maquat,L.E. (2001) Identification and characterization of human orthologues to Saccharomyces cerevisiae Upf2 protein and Upf3 protein (Caenorhabditis elegans SMG-4). Mol. Cell. Biol., 21, 209–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirley R.L., Lelivelt,M.J., Schenkman,L.R., Dahlseid,J.N. and Culbertson,M.R. (1998) A factor required for nonsense-mediated mRNA decay in yeast is exported from the nucleus to the cytoplasm by a nuclear export signal sequence. J. Cell Sci., 111, 3129–3143. [DOI] [PubMed] [Google Scholar]
- Sun X., Perlick,H.A., Dietz,H.C. and Maquat,L.E. (1998) A mutated human homologue to yeast Upf1 protein has a dominant-negative effect on the decay of nonsense-containing mRNAs in mammalian cells. Proc. Natl Acad. Sci. USA, 95, 10009–10014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Vock,V.M., Li,S., Olivas,O.R. and Wilkinson,M.F. (2002) A quality control pathway that down-regulates aberrant T-cell receptor (TCR) transcripts by a mechanism requiring UPF2 and translation. J. Biol. Chem. 277, 18489–18493. [DOI] [PubMed] [Google Scholar]
- Weng Y., Czaplinski,K. and Peltz,S.W. (1996) Genetic and biochemical characterization of mutations in the ATPase and helicase regions of the Upf1 protein. Mol. Cell. Biol., 16, 5477–5490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weng Y., Czaplinski,K. and Peltz,S.W. (1998) ATP is a cofactor of the Upf1 protein that modulates its translation termination and RNA binding activities. RNA, 4, 205–214. [PMC free article] [PubMed] [Google Scholar]
- Williams J.A., Langeland,J.A., Thalley,B.S., Skeath,J.B. and Carroll,S.B. (1995) Expression of foreign proteins in E.coli using plasmid vectors and purification of specific polyclonal antibodies. In Glover,D.M. and Hames,B.D. (eds), DNA Cloning 2: Expression Systems: A Practical Approach. Oxford University Press, Oxford, pp. 15–58.
- Wilusz C.J., Wang,W. and Peltz,S.W. (2001) Curbing the nonsense: the activation and regulation of mRNA surveillance. Genes Dev., 15, 2781–2785. [DOI] [PubMed] [Google Scholar]
- Yamashita A., Ohnishi,T., Kashima,I., Taya,Y. and Ohno,S. (2001) Human SMG-1, a novel phosphatidylinositol 3-kinase related protein kinase, associates with components of the mRNA surveillance complex and is involved in the regulation of nonsense-mediated mRNA decay. Genes Dev., 15, 2215–2228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S., Ruiz-Echevarria,M.J., Quan,Y. and Peltz,S.W. (1995) Identification and characterization of a sequence motif involved in nonsense-mediated mRNA decay. Mol. Cell. Biol., 15, 2231–2244. [DOI] [PMC free article] [PubMed] [Google Scholar]