Abstract
IQ motifs are widespread in nature. Mlc1p is a calmodulin-like myosin light chain that binds to IQ motifs of a class V myosin, Myo2p, and an IQGAP-related protein, Iqg1p, playing a role in polarized growth and cytokinesis in Saccharomyces cerevisiae. The crystal structures of Mlc1p bound to IQ2 and IQ4 of Myo2p differ dramatically. When bound to IQ2, Mlc1p adopts a compact conformation in which both the N- and C-lobes interact with the IQ motif. However, in the complex with IQ4, the N-lobe no longer interacts with the IQ motif, resulting in an extended conformation of Mlc1p. The two light chain structures relate to two distinct subfamilies of IQ motifs, one of which does not interact with the N-lobes of calmodulin-like light chains. The correlation between light chain structure and IQ sequence is demonstrated further by sedimentation velocity analysis of complexes of Mlc1p with IQ motifs from Myo2p and Iqg1p. The resulting ‘free’ N-lobes of myosin light chains in the extended conformation could mediate the formation of ternary complexes during protein localization and/or partner recruitment.
Keywords: analytical ultracentrifugation/IQ motif/myosin light chain/myosin V/X-ray crystallography
Introduction
Mlc1p is a calmodulin-like (CaM-like) protein from the budding yeast Saccharomyces cerevisiae. Mlc1p was first identified as a light chain of a class V myosin, Myo2p (Stevens and Davis, 1998). Myo2p localizes to the bud tip during bud formation and to the bud neck during cytokinesis (Brockerhoff et al., 1994; Lillie and Brown, 1994), and has been implicated in vesicle movement, polarized growth and mitotic spindle orientation (Johnston et al., 1991; Govindan et al., 1995; Pruyne et al., 1998; Yin et al., 2000). The domain structure of Myo2p is similar to that of other class V myosins (Cheney et al., 1993; Reck-Peterson et al., 2000). An N-terminal ‘head’ or motor domain, containing the actin-binding and ATP catalytic sites, is followed by an extended ‘neck’ domain formed by six in-tandem IQ motifs. Following the neck is the ‘tail’ domain, which contains a region predicted to form a coiled-coil and a C-terminal globular domain.
The IQ motifs constitute the binding sites for Ca2+-free (or apo) CaM and CaM-like light chains. IQ motifs are ∼25 amino acids long and conform to the consensus sequence IQxxxRGxxxR (Bahler and Rhoads, 2002). Although apo-CaM is the primary light chain of myosin V, occupying approximately four out of the six IQ sites, binding of light chains such as Mlc1p is essential (Cheney et al., 1993; Brockerhoff et al., 1994; Stevens and Davis, 1998; Reck-Peterson et al., 2000). For instance, overexpression of Myo2p in S.cerevisiae is toxic, causing a severe decrease in growth rate, which can be corrected by overexpression of Mlc1p but not CaM (Stevens and Davis, 1998).
Both CaM and the light chains are thought to play a role in myosin regulation and/or the stabilization of the neck, which is needed to produce steps of a definite size (Reck-Peterson et al., 2000). However, it remains unclear whether there are other functions associated with the light chains and why different classes of myosin carry varying numbers of IQ motifs. Also unknown is the distinction of roles between CaM and the light chains, and whether their ratio and IQ preferences can be regulated in response to cellular signals.
Myo2p is not the only target of Mlc1p. The finding that disruption of the MLC1 gene in S.cerevisiae causes a cytokinesis defect, which is independent of its Myo2p binding activity (Stevens and Davis, 1998), led to the search for different binding partners of Mlc1p. Thus, it was found that Mlc1p also binds to a class II myosin (Myo1p) in late mitosis and to Iqg1p, an IQGAP-like protein, during cytokinesis (Boyne et al., 2000; Shannon and Li, 2000). Mammalian IQGAP1 also binds a myosin light chain (Weissbach et al., 1998), suggesting that this function is conserved in eukaryotes. IQGAP proteins contain multiple domains including a calponin homology domain, an IQ repeat region and a Gap-related domain. They are thought to serve as effectors of Cdc42 and Rac, which are members of the Rho family of small GTPases, thus linking signaling pathways to the actin cytoskeleton (Machesky, 1998; Osman and Cerione, 1998). Binding of Mlc1p to Iqg1p, which is mediated by interactions with the IQ repeat region, recruits Iqg1p to the bud neck during cytokinesis (Boyne et al., 2000; Shannon and Li, 2000). The fact that the localization of Mlc1p occurs before and independently of Iqg1p, Myo2p, actin and Myo1p suggests that there is yet another target, possibly septin dependent, of Mlc1p (Boyne et al., 2000; Shannon and Li, 2000). An important implication of these findings is that the localization of Iqg1p to the bud neck would require Mlc1p to bind two different targets simultaneously. However, the existing structures of class II myosins that reveal the essential (ELC) and regulatory (RLC) light chains bound to IQ motifs (Rayment et al., 1993; Xie et al., 1994; Houdusse and Cohen, 1996; Dominguez et al., 1998) can not explain how a light chain could mediate the formation of such a ternary complex.
In this work, we describe the crystal structures of Mlc1p bound to IQ2 and IQ4 of Myo2p, which are representative members of two distinct subfamilies of IQ motifs found in a wide variety of proteins. Binding of Mlc1p to these two IQ motifs results in two markedly different light chain conformations: compact and extended. The correlation between light chain conformation and IQ motif subfamily is demonstrated further by sedimentation velocity studies of complexes of Mlc1p with IQ motifs from Myo2p and Iqg1p. The extended conformation frees the N-lobes of CaM-like light chains, providing a mechanism for how the binding of light chains to a subfamily of IQ motifs could mediate the formation of ternary complexes with multiple functional implications.
Results and discussion
IQ2 binds to both the N- and C-lobes of Mlc1p
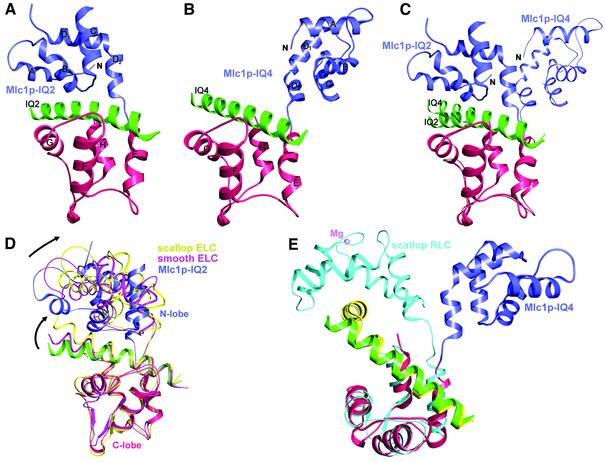
The structure of Mlc1p bound to IQ2 of Myo2p bears significant similarity to those of ELCs bound to IQ1 of the neck region of class II myosins (Rayment et al., 1993; Xie et al., 1994; Houdusse and Cohen, 1996; Dominguez et al., 1998) (Figure 1A and D). In all these structures, the light chains adopt a ‘compact’ conformation in which both the N- and C-lobes interact with the IQ motifs (Figure 1D). Like the ELCs and CaM (Babu et al., 1985), Mlc1p is a dumbbell-shaped molecule consisting of two homologous domains, the N- and C-lobes, connected by a flexible linker loop. Each lobe contains two helix–loop–helix (EF-hand) motifs joined by a loop. In contrast to CaM, the EF-hand motifs of Mlc1p do not bind Ca2+ despite the presence of 1 mM CaCl2 in the crystallization buffer, which is due to substitutions within several of the amino acids involved in the coordination of Ca2+ in CaM. Another noticeable difference is that the last α-helix of the N-lobe (helix-D) has an ∼70° bend in Mlc1p, resulting in two contiguous α-helices, a three-turn D1 helix and a two-turn D2 helix (Figure 1A). The splitting of helix D appears to be a genuine feature of Mlc1p since it is also observed in the structure of Mlc1p–IQ4 (see below).
Fig. 1. Ribbon diagram of the structures of Mlc1p bound to IQ2 and IQ4 of Myo2p. (A) Compact structure of the Mlc1p–IQ2 complex. The N-lobe, the C-lobe and the IQ peptide are shown in blue, red and green, respectively (this color scheme is kept through all the figures). (B) Extended structure of the Mlc1p–IQ4 complex. (C) Superimposition of the structures of Mlc1p–IQ2 and Mlc1p–IQ4 (thinner trace). (D) Superimposition of the structures of Mlc1p–IQ2 with those of ELC–IQ complexes from scallop (yellow) and smooth muscle (pink) myosins (Xie et al., 1994; Houdusse and Cohen, 1996; Dominguez et al., 1998). Note how differences in the orientation of the N-lobes are mirrored by differences in the orientation of the C-terminal portions of the IQ motifs. (E) Superimposition of the extended structures of Mlc1p–IQ4 and scallop RLC–IQ (cyan) complexes. The myosin heavy chain fragment (yellow) bound to the scallop RLC is bent by ∼90°. The Mg2+-bound (and fully open) N-lobe of the RLC binds to a conserved sequence (WQWWKLYSKVKPLL) of the myosin heavy chain that follows immediately after the 90° turn. Note that this N-lobe-specific target sequence does not make up part of a canonical IQ motif.
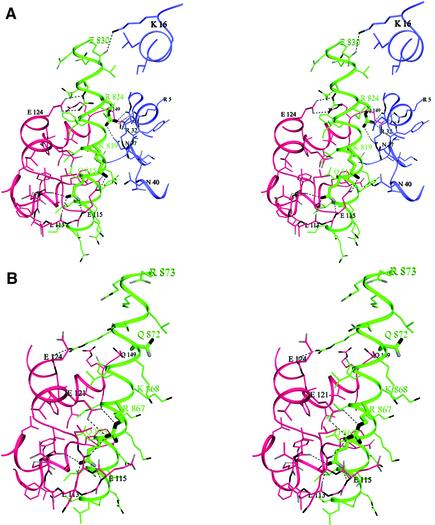
The C-lobe of Mlc1p interacts more extensively with IQ2 than the N-lobe (Figure 2A). The C-lobe adopts what is known as a semi-open conformation (Houdusse and Cohen, 1996), which is characterized by the partial exposure of the hydrophobic core of the domain. This lobe interacts primarily with the IQxxxR portion of the IQ motif (Figure 3). The N-lobe, on the other hand, is in a closed conformation and, as in the ELC–IQ complexes, interacts for the most part with the GxxxR part of the IQ motif (Figure 3). The 25 amino acids of IQ2 are folded as an uninterrupted seven-turn α-helix with a markedly amphiphilic character. The non-polar face of this α-helix flips sides after the fifth turn, so that the hydrophobic amino acids of the IQ motif face the hydrophobic core of the C-lobe during the first five turns and the N-lobe during the last two turns.
Fig. 2. Stereo diagrams showing details of the interactions of Mlc1p with IQ2 and IQ4. (A) Both the N-lobe (blue) and C-lobe (red) of Mlc1p interact with IQ2. A thicker trace is used to highlight the amino acids at positions 2, 6 and 11 of the IQ motif (Gln815, Lys819 and Arg824). Note that the main chain backbone at Gly820 (after Lys819) runs alongside Arg32 from the N-lobe. A side chain at this position would get in the way of the interactions of Arg824 with the N-lobe. (B) Interactions of IQ4 with the C-lobe of Mlc1p. The thicker trace identifies residues Gln863, Arg867 and Gln872 (Gln872 replaces the arginine residue found at this position in a canonical IQ motif). No interactions with the N-lobe were observed. In contrast, there is a striking similarity between the contacts of the C-lobe with IQ2 and IQ4. Note how the presence of the side chain of Lys868, which replaces a glycine residue at this position in a canonical IQ motif, would be inconsistent with a compact conformation in which the N-lobe would have to come close in order to interact with the IQ motif.
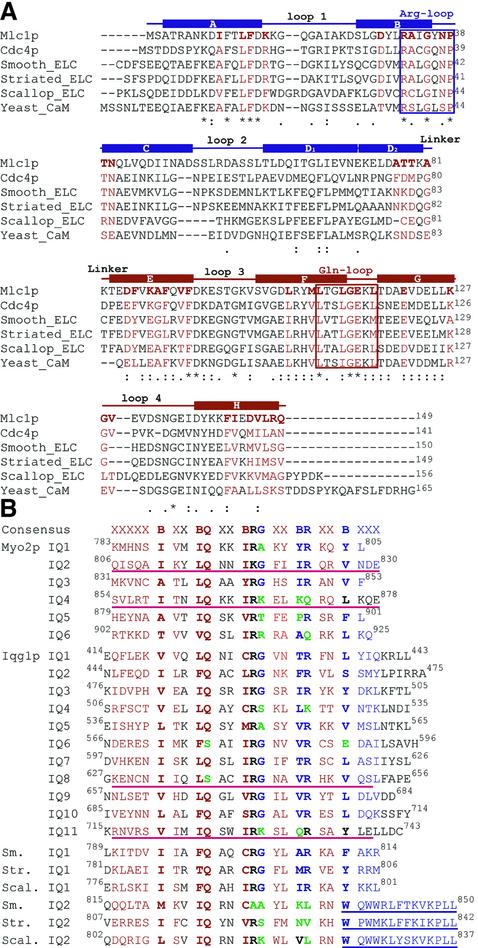
Fig. 3. Sequence alignment of light chains and IQ motifs. The alignments were done with the program Clustal W (Higgins et al., 1996). (A) Saccharomyces cerevisiae Mlc1p, Schizosaccharomyces pombe Cdc4p, smooth muscle myosin ELC, striated muscle myosin ELC, scallop myosin II ELC and S.cerevisiae CaM. The amino acids that form part of the eight α-helices of Mlc1p (helices A–H) are indicated by the secondary structure diagram shown on top of the sequences, where the two different colors (blue and red) represent the N- and C-lobes, respectively. The amino acids comprised within the ‘Arg-loop’ and ‘Gln-loop’ boxes are those that interact, respectively, with the glutamine at position 2 and arginine at postion 11 of IQ2. Loops 1–4 designate the four EF-hand Ca2+-binding loops. The amino acids of the light chains depicted in red are those that in the Mlc1p–IQ2 complex interact with the IQ motif (corresponding to the red surface in Figure 6). Note that the amino acids in the arginine and glutamine loop areas are more conserved than the four EF-hand loops, reflecting the fact that the light chains (except for CaM) bind few or no divalent cations but they all bind IQ motifs. CaM, on the other hand, binds IQ motifs in a Ca2+-free form (Jurado et al., 1999). (B) IQ motifs from Myo2p and Iqg1p. Also shown are the smooth, striated and scallop myosins IQ motifs 1 and 2 that bind, respectively, the ELC and RLC. The consensus IQ sequence is shown on top, where the letter B indicates positions generally occupied by hydrophobic amino acids. In red and blue are the amino acids that interact, respectively, with the C- and N-lobes of the light chain in a compact structure such as that of Mlc1p–IQ2. The amino acids in black are at the interface between the two lobes of the light chain. Amino acids depicted in green correspond to severe mutations (compared with the consensus sequence) that may be associated with a significant change in the conformation of the bound light chain. The four IQ peptides that were synthesized for this study are underlined (red). The three incomplete IQ2 motifs of class II myosins shown at the bottom of this figure bind the RLC. The RLC adopt an extended conformation in which the open N-lobes bind the sequence underlined (blue), corresponding to a separate α-helix after a sharp ∼90° bend of the myosin heavy chain centered at the tryptophan residue at position 14 of the motif.
The most conserved amino acids of the IQ motif participate in numerous interactions with the light chain but do not appear to mediate light chain specificity. The unique exception appears to be the hydrophobic amino acid at position 1 of the IQ motif (Leu814), which is fully embedded within the hydrophobic core of the C-lobe (Figure 2A). This position, which is less conserved, can host a range of hydrophobic side chains including isoleucine, leucine, phenylalanine and valine (Figure 3). In contrast, the glutamine at position 2 (Gln815) is highly conserved. The side chain of this amino acid is involved in three hydrogen-bonding contacts with main chain atoms of the loop that connects the two EF-hands of the C-lobe (labeled glutamine loop in Figure 3A, see also Figure 2A). In a rather symmetric fashion, the conserved arginine at position 11 of the IQ motif (Arg824) makes two hydrogen-bonding contacts with backbone atoms of the corresponding loop in the N-lobe (labeled arginine loop in Figure 2A) and a third contact with the side chain Oδ1 atom of the conserved Asn37 from the same loop (Figures 2A and 3A). In IQ2, the conserved arginine at position 6 has been replaced by a lysine (Lys819). This substitution does not appear to affect the stability of the complex. The side chain of Lys819 is pointing toward the solvent and runs along the interface between the two lobes of the light chain. Such a location potentially may allow for interactions with residues from either lobe. In the current structure, Lys819 makes a salt bridge with Glu115 from the C-lobe. The next amino acid in the IQ motif typically is a glycine (Gly820 in IQ2). The absence of a side chain at this position is critical in order for the N-lobe to interact with the IQ motif in a compact light chain–IQ complex. Indeed, a side chain at this position (even a short one) would result in steric hindrance with the side chain of Arg32 of the N-lobe and would, in addition, prevent Arg824 of the IQ motif from forming the hydrogen-bonding contacts with the arginine loop characteristic of this compact structure. Note that Arg32 is one of 15 strictly conserved amino acids among a number of light chains and CaM (Figure 3A). Because the most conserved amino acids of the IQ motif (glutamine at position 2 and arginine at postion 11) interact mainly with main chain atoms of two of the most conserved regions of the light chains (the glutamine and arginine loop regions), these interactions probably are common to all light chain–IQ complexes that adopt a compact conformation (Figures 2A and 3). Other positions within the IQ motifs vary significantly but retain a hydrophobic character (indicated by the letter B in the consensus sequence, Figure 3B). Such positions may control light chain specificity.
IQ4 binds only to the C-lobe of Mlc1p, leaving the N-lobe free
In contrast to the structures of Mlc1p–IQ2 and ELC–IQ complexes (Rayment et al., 1993; Xie et al., 1994; Houdusse and Cohen, 1996; Dominguez et al., 1998), Mlc1p binds to IQ4 in an extended conformation in which the N-lobe no longer interacts with the IQ motif (Figures 1B and 2B). The C-lobe, however, interacts with the IQxxxR portion of IQ4 in a similar way as in the compact structure of Mlc1p–IQ2 (Figures 1C and 2B). One noticeable difference is that the arginine at position 6 of the IQ motif (which has been replaced by a lysine in IQ2) is preserved in IQ4 (Arg867) and forms a salt bridge with Glu121 from the C-lobe of the light chain.
Compared with the structure of Mlc1p–IQ2, the N-lobe is rotated away from the IQ peptide in the Mlc1p–IQ4 complex. However, it is important to note that the exact orientation and positioning of the N-lobe in the crystals are probably irrelevant since this lobe is connected to the rest of the structure by the flexible linker loop, which may allow it to move relatively freely when it is not interacting with the IQ motif. Thus, the most relevant feature characterizing the Mlc1p–IQ4 structure is that the N-lobe no longer interacts with the IQ motif. What prevents the N-lobe from interacting with the IQ motif? Comparison of the sequences of IQ2 and IQ4 reveals that the glycine residue at position 7 of the IQ motif has been replaced by a lysine (Lys868) in IQ4. Such a replacement is critical. As mentioned above, the presence of a side chain at this position prevents the arginine loop of the N-lobe from getting close enough to interact with the IQ motif (compare Figure 2A with B). In addition, a glutamine (Gln872) replaces the conserved arginine at position 11 of IQ4, making it even more unlikely for the N-lobe to interact with IQ4. Apart from Lys868, there are four additional positively charged amino acids of IQ4 that face the N-lobe (Lys864, Lys865, Lys871 and Arg857). If the N-lobe were to occupy a position similar to that in the compact structures, the conserved Arg32 of the light chain would be confronted against these positively charged amino acids. It is likely that such charge repulsion may repel the N-lobe farther away from IQ4 in the Mlc1p–IQ4 complex.
Is the extended light chain conformation unique to IQ4 or is it a more general feature? As shown above, mutations at positions 7 and 11, combined with the presence of numerous positively charged side chains, could explain the extended conformation of Mlc1p when bound to IQ4. We predict that other IQ motifs, containing mutations simultaneously at positions 7 and 11, will also be unable to interact with the N-lobe and potentially could induce an extended light chain conformation. However, other factors such as charge distribution may contribute to the actual positioning of the N-lobe when it is not interacting with the IQ motif. In some IQ motifs, only the arginine residue at position 11 (but not the glycine at position 7) is mutated. In such cases, it is possible that other interactions could compensate for the lost contacts with the arginine loop of the N-lobe while still allowing the N-lobe to interact with the IQ motif.
There must exist, however, significant evolutionary pressure for the conservation of the glycine residue at positions 7 of the IQ motif, which would explain the apparent paradox that a strong helix breaker amino acid (Chakrabartty et al., 1991) occupies a midpoint position within a motif that is folded as an uninterrupted α-helix. Analysis of the structures of Mlc1p–IQ2 and ELC–IQ complexes reveals that the glycine at position 7 is directly associated with the compact conformation of the light chain (see above). However, nearly 50% of IQ motifs from different protein families (Bahler and Rhoads, 2002) present substitutions of this glycine. To understand the potential effects of such substitutions, we modeled various types of side chains (alanine, serine, methionine, lysine and argine) into position 7 of IQ2 of the Mlc1p–IQ2 structure. Substitutions of this glycine by bulky side chains such as methionine, lysine and arginine would free the N-lobe as they force the arginine at position 11 of the IQ motif and the arginine loop of the N-lobe into positions where they can no longer interact. Smaller side chains (alanine, serine and threonine) are equally disruptive to these interactions, but we can not rule out the possibility of the N-lobe interacting with the IQ motif following changes elsewhere in the structure. In this respect, an analysis of the structures of RLCs bound to IQ2 of the heavy chains of class II myosins (Rayment et al., 1993; Houdusse and Cohen, 1996) could be revealing. Similarly to IQ4, the IQ motifs that bind the RLCs have fully functional IQxxxR portions but have lost the glycine and arginine residues at positions 7 and 11 (Figure 3B). Independently of the size of the amino acid replacing the glycine residue at position 7 (Figure 3B), the RLCs all adopt an extended conformation in which only the C-lobe interacts with the IQ motif (Figure 1E). However, in contrast to the complex of Mlc1p–IQ4, the N-lobes of the RLCs still interact with the myosin heavy chain (see below), albeit in a different way from that of the compact structures of Mlc1p–IQ2 and ELC–IQ complexes (compare Figure 1D and E).
Is the inability of the N-lobe to interact with certain IQ motifs unique to Mlc1p? As we have shown, the two conformations of Mlc1p are IQ dependent. The amino acids of the N-lobe that interact with the GxxxR portion of the IQ motifs are well conserved among the light chains (Figure 3A). These include the C-terminal end of helix A, the region around the ‘arginine loop’ and the less well conserved linker loop. We tested replacing the glycine residue at position 7 of the IQ motifs in the structures of ELC–IQ complexes by different types of side chains. As for Mlc1p–IQ4, such mutations would result in loss of interaction with the N-lobe. Moreover, CaM modeled into a compact conformation by analogy with the structure of Mlc1p–IQ2 appears equally sensitive to whether or not there is a glycine residue at position 7 of an IQ motif. As discussed above, the RLCs of class II myosins, which are more distantly related to Mlc1p than CaM and the ELCs, also adopt an extended conformation when bound to IQ motifs in which the glycine residue at position 7 has been replaced (Figures 1E and 3B). Taken together, these observations would indicate that if another light chain, such as CaM, were to bind IQ4 in vivo, it would most probably adopt a similar extended conformation to that of Mlc1p.
Sedimentation velocity analysis of Mlc1p bound to IQ motifs from Myo2p and Iqg1p
The structures of Mlc1p bound to IQ2 and IQ4 of Myo2p point toward a division of the IQ motif family into two categories: IQ motifs that interact with both the N- and C-lobes of the light chains and IQ motifs that interact with the C-lobe only. The most obvious distinction between these two subfamilies of IQ motifs is whether or not there is a glycine residue at position 7. To test this hypothesis further, we performed sedimentation velocity studies of Mlc1p alone and in complexes with IQ motifs from Myo2p and Iqg1p. In agreement with the crystal structures, the complex of Mlc1p with IQ2 is much more compact in solution than that of Mlc1p–IQ4 or Mlc1p alone, as indicated by a smaller value of the axial ratio (a/b) of the hydrodynamically equivalent prolate ellipsoid of revolution (Table II). Moreover, theoretically calculated sedimentation coefficients, derived from bead models based on the crystal structures, agree very well with the observed experimental values for both complexes (Table II). Such an agreement indicates that the observed sedimentation coefficients provide a good representation of the conformation of the complexes in solution and could, therefore, be used to assist homology-based modeling of other light chain–IQ complexes based on the crystal structures of Mlc1p–IQ2 and Mlc1p–IQ4 as starting models.
Table II. Sedimentation analysis and modeling.
| Protein | Ma | v2b (cm3/g) | δ1c (g/g) | s(20,w)d (S) | s(20,w)e calc. | f/fof | a/bg |
|---|---|---|---|---|---|---|---|
| Mlc1p–IQ2 | 19 303 | 0.739 | 0.413 | 1.95 | 1.94 | 1.105 | 2.90 |
| Mlc1p–IQ8gap | 18 995 | 0.738 | 0.412 | 1.93 | 1.110 | 2.96 | |
| Mlc1p–IQ4**h | 19 350 | 0.742 | 0.421 | 1.87 | 1.138 | 3.36 | |
| Mlc1p–IQ11gap | 19 248 | 0.740 | 0.410 | 1.87 | 1.147 | 3.50 | |
| Mlc1p | 16 313 | 0.740 | 0.444 | 1.64 | 1.160 | 3.67 | |
| Mlc1p–IQ4*i | 19 322 | 0.741 | 0.420 | 1.81 | 1.180 | 3.96 | |
| Mlc1p–IQ4 | 19 393 | 0.742 | 0.421 | 1.79 | 1.75 | 1.190 | 4.12 |
| Mlc1p–IQ2*j | 19 374 | 0.741 | 0.412 | 1.77 | 1.211 | 4.42 |
aMolar mass.
bPartial specific volume.
cHydration (grams of water per gram of protein).
dSedimentation coefficient in Svedbergs (1.e–13 s) obtained by fitting with the program SedAnal (see Materials and methods).
eSedimentation coefficient calculated from a bead model using the program HYDRO (Garcia de la Torre et al., 1994).
fFrictional ratio.
gAxial ratio of the hydrodynamically equivalent prolate ellipsoid of revolution. This value reflects the relative ‘compactness’ of each complex.
hMutant of IQ4 in which the lysine and glutamine at positions 7 and 11 were substituted by glycine and arginine, respectively.
iMutant of IQ4 in which the lysine at position 7 was substituted by glycine.
jMutant of IQ2 in which the glycine at position 7 was substituted by lysine.
In a separate series of experiments, we attempted to revert the conformation of Mlc1p from compact to extended and vice versa by introducing point mutations into IQ2 and IQ4. Thus, a single mutation of the glycine residue at position 7 of IQ2 (Gly820) to a lysine resulted in a very extended conformation of the complex Mlc1p–IQ2* (Table II). Conversely, mutation of the lysine residue at position 7 of IQ4 (Lys864) to a glycine resulted in a somewhat more compact complex Mlc1p–IQ4*. However, IQ4 also lacks the critical arginine at position 11, which in a compact conformation accounts for most of the interactions with the N-lobe of the light chain. To restore these interactions, we generated a double mutant peptide containing a glycine residue at position 7 and an arginine residue at position 11. The complex of this peptide with Mlc1p, Mlc1p–IQ4**, is much more compact than the original Mlc1p–IQ4 complex but not quite as compact as Mlc1p–IQ2 (compare the axial ratios a/b for these three complexes listed in Table II). From these experiments, we conclude that although replacement of the glycine residue at position 7 of an IQ motif generally would be sufficient to induce an extended light chain conformation, a compact conformation may require both a glycine at position 7 and an arginine at position 11.
Because Iqg1p is another target for Mlc1p in the cell, we used analytical ultracentrifugation to investigate the conformation of Mlc1p when bound to two distinctive IQ motifs from Iqg1p, IQ8 and IQ11. Based on our results, we predict that IQ11 of Iqg1p would not interact with the N-lobe of Mlc1p due to the presence of a lysine residue (Lys730) at position 7 (Figure 3B). Mlc1p–IQ11 is characterized by an intermediate axial ratio value of 3.5, compared with typical values of ∼3.0 and ∼4.0 for extended and compact complexes, respectively (Table II). It appears that specific features in the sequence of IQ11 of Iqg1p may account for a smaller a/b ratio of Mlc1p–IQ11 compared with that of Mlc1p–IQ4. Thus, IQ11 is unique in that it carries a tryptophan residue at position 4. Moreover, three out of five positively charged amino acids on the N-lobe side of IQ4 (lysine residues at positions 3, 4 and 10) have been substituted by non-charged amino acids in IQ11 (Figure 3B), resulting in different global charge distributions for these two IQ motifs.
Superimposition of the C-lobes and IQxxxR portions of the structures of Mlc1p–IQ2 and ELC–IQ complexes highlights a significant variability in the orientation that the N-lobes can adopt (Figure 1D). Interestingly, the different N-lobe orientations are mirrored by differences in the orientation of the GxxxR portions of the IQ motifs, indicating that the interaction between these two parts of the complexes is relatively strong. This is important since certain IQ motifs have fully functional GxxxR portions but have substitutions of key amino acids in their IQxxxR sides. For instance, a serine replaces the glutamine at position 2 of IQ6 and IQ8 of Iqg1p (Figure 3B), which so far has prevented these two sequences from being recognized as functional IQ motifs (Epp and Chant, 1997; Lippincott and Li, 1998). Can these two IQ motifs bind light chains? If so, does the binding involve interactions with both lobes of the light chains or with the N-lobe alone? The sedimentation velocity of Mlc1p with IQ8 of Iqg1p demonstrates that IQ8 does indeed bind Mlc1p and that the complex is as compact as that of Mlc1p–IQ2. This observation, together with conservation of the hydrophobic amino acids that interact with the C-lobe, would be consistent with IQ8 interacting with both the N- and C-lobes of Mlc1p in a manner analogous to that of the Mlc1p–IQ2 complex.
Implications of the presence of free N-lobes in light chain–IQ complexes
An analysis of IQ motifs from numerous proteins (Bahler and Rhoads, 2002), according to whether or not the glycine residue at position 7 is conserved, allows the prediction that a significant number of bound light chains will carry free N-lobes. Thus, IQ4 and IQ6 of Myo2p, and IQ4 and IQ11 of Iqg1p will not interact with the N-lobes of bound light chains. Most unconventional myosins have at least one IQ motif that is related to IQ4, and will therefore result in a bound light chain with a free N-lobe. Is there a functional role associated with these free N-lobes? It is striking that the localization of Mlc1p to the bud neck occurs before and independently of Iqg1p, suggesting that Mlc1p binds first to another (unknown) target and then recruits Iqg1p by interacting with its IQ motifs (Boyne et al., 2000; Shannon and Li, 2000). It is reasonable to suggest that the free N-lobes of Mlc1p could be involved in the formation of such a ternary complex. More generally, light chain–IQ-mediated ternary complexes may play a role in protein localization and/or the recruitment of partner molecules.
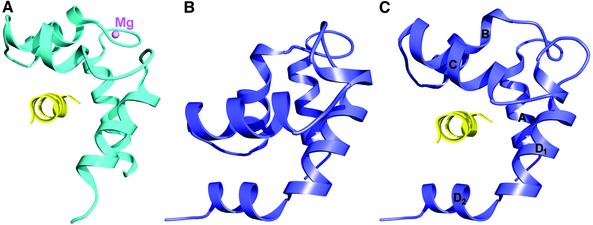
At least two models can be proposed for how the free N-lobes could mediate interactions with other proteins. The first model is by analogy with the RLCs of class II myosins (Rayment et al., 1993; Houdusse and Cohen, 1996). Indeed, there is a major difference between the IQ motifs that bind the RLCs and C-lobe-specific IQ motifs such as IQ4 (Figure 1E). While IQ4 is folded as an uninterrupted α-helix and is followed by other IQ motifs (Figure 3B), the IQxxxR portions of the IQ motifs that bind the RLCs are followed immediately by a highly conserved sequence, which is characterized by a sharp ∼90° bend of the myosin II heavy chain (Figures 1E and 3B) that precedes the coiled-coil region that forms the muscle thick filaments. After the bend, the heavy chain continues as an α-helix to which the N-lobes of the RLCs bind in an open conformation (Figure 4A). Opening of the N-lobe of the RLC is facilitated by the binding of Mg2+ to the first EF-hand and by the target sequence itself (Houdusse and Cohen, 1996). Figure 4 illustrates a model for how opening of the N-lobe of Mlc1p would be consistent with binding to a sequence related to that of the heavy chain of myosin II (Figure 3B). Such a sequence is absent in both Iqg1p and Myo2p, indicating that if this type of binding mechanism were to occur it would have to involve a third protein, as compared with the same target for the RLCs of class II myosins. Since Mlc1p does not appear to bind divalent cations, it remains unclear what factors, other than the target sequence itself, could promote the opening of the N-lobe. One possibility would be phosphorylation. Interestingly, Cdc4p, a closely related homologue of Mlc1p from Schizosaccharomyces pombe (Figure 3A), can be phosphorylated on either Ser2 or Ser6, though there is no known function of this modification (McCollum et al., 1999). A recent structure of CaM bound to the gating domain of a K+ channel (Schumacher et al., 2001) provides yet another example of how the N- and C-lobes of CaM-like light chains can bind separately to two different targets. In this structure, the Ca2+-loaded N-lobe of CaM binds to one α-helix and the Ca2+-free C-lobe binds to two α-helices of a dimer of the channel protein.
Fig. 4. Model of target binding to the open N-lobe of Mlc1p. This model is based on the structures of RLCs bound to the heavy chains of class II myosins (Rayment et al., 1993; Houdusse and Cohen, 1996). (A) The N-lobe of the scallop RLC (cyan) bound to the sequence WQWWKLYSKVKPLL of the myosin heavy chain (yellow). Opening of the N-lobe is brought about both by Mg2+ binding to the first EF-hand and by the target sequence itself. (B) The closed N-lobe of Mlc1p from the structure of Mlc1p–IQ4. The orientation of the molecule is the same as in (A). (C) Model, built by homology with the scallop RLC, showing the opening of the N-lobe of Mlc1p (blue) and binding of a sequence related to that of the myosin heavy chain (yellow). Only helices B and C move during opening of the N-lobe. Note that the splitting of helix D into helices D1 and D2 creates a deeper binding pocket in Mlc1p than in the RLC. It remains unclear, however, which factors would lead to the opening of the N-lobe of Mlc1p (see text).
In a second binding model, opening of the N-lobe is not required. As discussed above, the interaction between the N-lobe of the light chains and the GxxxR portion of the IQ motifs appears strong enough to suggest that these two regions could interact independently of the interaction of the C-lobe with the IQxxxR portion of the IQ motif. Figure 5 shows how, upon binding of an IQ motif in an extended conformation, the N-lobe exposes a surface that normally would interact with the GxxxR portion of the IQ motif in a compact light chain–IQ complex. If a target protein were to contain a sequence related to that of the second half of an IQ motif, it could be expected to interact with this exposed surface of the N-lobe, without a need for the N-lobe to open. The affinity of such an interaction may be modulated additionally by cross-contacts between the targets bound to both lobes, as observed in the complex of CaM with the K+ channel protein (Schumacher et al., 2001).
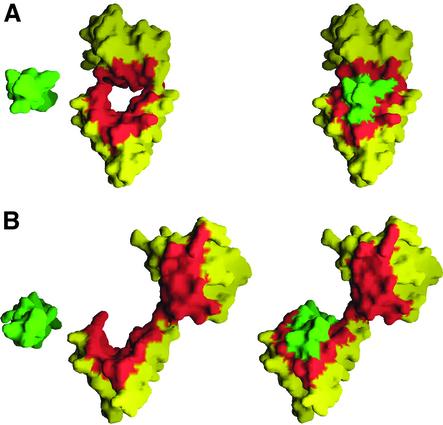
Fig. 5. IQ motif-binding interface of Mlc1p. (A) Compact conformation of the Mlc1p–IQ2 complex. The view is along the axis of the α-helices of IQ2 (shown in green). The amino acids of Mlc1p are colored yellow, except for those amino acids that make direct contacts with the IQ peptide, which are depicted in red (the same amino acids are also colored red in Figure 33A). Two views are shown: on the left side of the figure, the IQ peptide has been moved out of its binding cleft manually to illustrate better the binding interface, while the view on the right side corresponds to the actual position of the IQ peptide in the crystal structure. (B) Extended structure of Mlc1p–IQ4 (same color scheme and orientation as A). Note how all of the amino acids of the N-lobe that interact with IQ2 in the compact structure become solvent exposed in this complex. The resulting surface would be available for interactions with a GxxxR-containing target without a need for the N-lobe to open.
The NMR structure of a homologue of Mlc1p in S.pombe, Cdc4p, was determined in an uncomplexed form (Slupsky et al., 2001). Like Mlc1p, Cdc4p is essential for cytokinesis (McCollum et al., 1995; D’Souza et al., 2001). Both proteins bind similar targets: Cdc4p serves as a light chain for a class II myosin, Myo2p (Naqvi et al., 1999), and a class V myosin, Myo51p (D’Souza et al., 2001), and binds to Rng2p, which is an IQGAP-related protein from S.pombe (Eng et al., 1998; D’Souza et al., 2001). Taken together, these findings suggest that Mlc1p and Cdc4p fulfill similar roles in their respective hosts. By analogy with Mlc1p, we would like to propose that Cdc4p might bind via its N- and C-lobes to two separate targets, Rng2p (Eng et al., 1998; D’Souza et al., 2001) and an as yet unidentified target, to mediate cytokinesis in S.pombe.
Conclusions
Although IQ motifs are widely distributed and often occur in multiple copies, their function is poorly understood (Bahler and Rhoads, 2002). In myosins, they are thought to be involved in light chain-mediated regulation and/or to function as passive mechanical devices that determine the length of the myosin lever arm and hence the step size. This work presents evidence from both X-ray crystallography and analytical ultracentrifugation suggesting that the IQ motif family can be divided into IQ motifs that interact with both the N- and C-lobes of the light chains and those that interact with the C-lobe alone, leaving free N-lobes available for other interactions. Varying numbers of such free N-lobes, from either CaM or the light chains, are likely to be carried by most unconventional myosins, IQGAP, neuronal proteins and other IQ-containing proteins. The free N-lobes may mediate interactions with other proteins, which could, for instance, explain the mechanism of recruitment of Iqg1p to the bud neck by Mlc1p (Boyne et al., 2000; Shannon and Li, 2000). We have advanced two possible models for how CaM-like light chains could mediate the formation of such ternary complexes. Both models assume that the C-lobes of the light chains bind to the IQxxxR portion of a C-lobe-specific IQ motif of one target protein, while the N-lobes adopt either an open or a closed conformation to bind to a second target protein. Additional research is needed to validate these models and to identify the nature and functions of the N-lobe-specific targets. Analytical ultracentrifugation can be a powerful tool to extend these findings to other CaM– and light chain–IQ complexes.
Materials and methods
Details concerning the preparation of the proteins and IQ peptides, crystallization, data collection and structure determination have been published elsewhere (Terrak et al., 2002). Briefly, Escherichia coli-expressed Mlc1p complexed with IQ2 and IQ4 of Myo2p (Figure 3B) were dialyzed against 5 mM sodium acetate pH 4.0, 1 mM calcium chloride, 1 mM dithiothreitol (DTT), and concentrated to ∼10 mg/ml. Crystals of the Mlc1p–IQ2 complex were obtained in 20% (w/v) polyethylene glycol 3350 and 0.2 M sodium chloride. Crystals of both wild-type Mlc1p and an Se–Met derivative of a mutant, Ile64Met, of Mlc1p complexed with IQ4 were obtained in 2.2 M ammonium sulfate, 0.2 M potassium sodium tartrate tetrahydrate and 0.1 M tri-sodium citrate dihydrate pH 5.6.
Crystals of Mlc1p–IQ4 diffracted X-rays significantly better when collected at room temperature than under freezing conditions. In addition to a higher mosaicity and lower diffraction resolution, freezing caused a space group and unit cell change. However, in order to determine the structure of the Mlc1p–IQ4 complex, we performed an Se–Met multiple amorphous diffraction (MAD) experiment, which required freezing as a way to limit radiation damage of the Se–Met derivative Ile64Met Mlc1p–IQ4 crystals. A three-wavelength MAD experiment was collected at the BioCARS beamline 14-BM-D (APS, Argonne). The positions of the selenium atoms were found with the program SnB (Weeks and Miller, 1999) and refined to 3.0 Å resolution with MLPHARE (CCP4, 1994). The phases obtained from MLPHARE were refined further and extended to 2.4 Å resolution with the program DM (Cowtan and Main, 1998). The resulting electron density map was of excellent quality, allowing the construction of a rough model that subsequently was used to find a molecular replacement solution in the space group of the unfrozen crystals. The structure was then refined to 2.1 Å resolution with the program Refmac (Murshudov et al., 1999) (Table I).
Table I. Crystallographic refinement statistics.
| Mlc1p–IQ2 | Mlc1p–IQ4 | |
|---|---|---|
| Space group | P212121 | P21212 |
| Unit cell parameters | ||
| a, b, c (Å) | 43.6, 56.5, 56.9 | 48.5, 121.5, 29.1 |
| α, β, γ (°) | 90.0, 90.0, 90.0 | 90.0, 90.0, 90.0 |
| Resolution range (Å) | ||
| All data | 25.0–1.65 | 40.0–2.1 |
| Last shell | 1.69–1.65 | 2.17–2.1 |
| Completeness (%) | 98.0 (95.4)a | 99.2 (95.2) |
| σ cut-off | None | None |
| Rfactor (%)b | 19.1 (22.9) | 19.9 (25.8) |
| Rfree (%)c | 22.8 (27.5) | 23.9 (28.6) |
| Average B-factors (Å2) | ||
| Protein atoms | 22.0 | 36.4 |
| Water molecules | 36.5 | 43.9 |
| PDB accession code | 1M45 | 1M46 |
aValues in parentheses correspond to the last resolution shell.
bRfactor = Σ|Fo – Fc|/Σ|Fc|, where Fo and Fc are observed and calculated structure factors, respectively.
cRfree = Rfactor calculated for a subset of the reflections (5%), which were omitted during the refinement and used to monitor its convergence.
A data set to 1.65 Å resolution was collected at 100 K from a crystal of Mlc1p–IQ2 at the BioCARS beamline 14-BM-D (APS, Argonne). The differences between the Mlc1p–IQ2 and Mlc1p–IQ4 complexes were so pronounced that a molecular replacement solution of the Mlc1p–IQ2 structure could not be obtained using any part of the structure of Mlc1p–IQ4 as a search model. The structure of Mlc1p–IQ2 was determined subsequently by molecular replacement based on a complex of Mlc1p with IQ2,3, which was determined from a separate Se–Met MAD experiment collected at beamline F2, CHESS (Ithaca, NY) and is still under refinement (Terrak et al., 2002). The refinement of the structure of Mlc1p–IQ2 was then completed to 1.65 Å resolution with the program Refmac (Murshudov et al., 1999) (Table I).
Sedimentation analysis was carried out on a BeckmanCoulter Optima XL-I analytical ultracentrifuge. Sedimentation velocity runs were performed at 50 000 r.p.m. and 20°C, using sample concentrations of 0.3–0.5 mg/ml. Samples were prepared by mixing Mlc1p with a 1.2 molar excess of synthetic IQ peptides and dialyzing overnight against 100 mM NaCl, 10 mM HEPES pH 7.5 and 5 mM DDT using a 500 Da cut-off membrane. Sedimentation coefficients of the complexes were determined by curve fitting to the concentration profiles using the program SedAnal (Stafford, 1998). This program fits time difference data from the interference optical system using numerical solutions of the Lamm equation according to the method of Todd and Haschemeyer (1981). Since each sample contained an excess of IQ peptide over Mlc1p, the mixtures were fit using a two-species model that distinguished between Mlc1p–IQ complexes and free IQ peptides. Complex dissociation was not observed. The partial specific volume and hydration of each peptide and complex were computed from their amino acid composition (Stafford et al., 2001). Bead modeling was carried out using the program HYDRO (Garcia de la Torre et al., 1994). Bead models were generated from the crystal structures by replacing each amino acid with a bead of diameter ∼3.7 Å centered at the Cα position. The exact diameter of the beads was adjusted so that the volume of the model added up to the volume of the hydrated protein. The frictional ratio was calculated as f/fo = (so/s20,w)[v2/(v2 + δ1v1)]1/3, where so is the sedimentation coefficient calculated for an unhydrated sphere with mass equal to that of the Mlc1p–IQ complexes and is given by the relationship so = M(1 – v2ρ)/(6NπηRo), where Ro = [3Mv2/(4πN)]1/3 and N is Avogadro’s number. Axial ratios (a/b) of the equivalent ellipsoids of revolution were computed using Perrin’s equation (Table II).
Acknowledgments
Acknowledgements
We would like to thank the staff members at BioCARS and MacCHESS for assistance during data collection, and Gina Pagani and Ming-Jen Tsay for the synthesis of IQ peptides for this work. Supported by NIH grant R01 AR46524 to R.D. Use of the Advanced Photon Source was supported by the US Department of Energy, Basic Energy Sciences, Office of Science, under contract No. W-31-109-Eng-38. Use of the BioCARS facilities was supported by NIH grant RR07707. Use of the CHESS and MacCHESS facilities was supported by the NSF (award DMR 97-13424) and NIH (award RR-01646), respectively.
Note added in proof
A recent publication provides biochemical evidence for the binding under certain conditions of one molecule of calmodulin to up to two IQ-target sequences simultaneously.
Martin,S.R. and Bayley,P.M. (2002) Regulatory implications of a novel mode of interaction of calmodulin with a double IQ-motif target sequence from murine dilute myosin V. Protein Sci., 11, 2909–2923.
References
- Babu Y.S., Sack,J.S., Greenhough,T.J., Bugg,C.E., Means,A.R. and Cook,W.J. (1985) Three-dimensional structure of calmodulin. Nature, 315, 37–40. [DOI] [PubMed] [Google Scholar]
- Bahler M. and Rhoads,A. (2002) Calmodulin signaling via the IQ motif. FEBS Lett., 513, 107–113. [DOI] [PubMed] [Google Scholar]
- Boyne J.R., Yosuf,H.M., Bieganowski,P., Brenner,C. and Price,C. (2000) Yeast myosin light chain, Mlc1p, interacts with both IQGAP and class II myosin to effect cytokinesis. J. Cell Sci., 113, 4533–4543. [DOI] [PubMed] [Google Scholar]
- Brockerhoff S.E., Stevens,R.C. and Davis,T.N. (1994) The unconventional myosin, Myo2p, is a calmodulin target at sites of cell growth in Saccharomyces cerevisiae. J. Cell Biol., 124, 315–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CCP4 (1994) The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D, 50, 760–763. [DOI] [PubMed] [Google Scholar]
- Chakrabartty A., Schellman,J.A. and Baldwin,R.L. (1991) Large differences in the helix propensities of alanine and glycine. Nature, 351, 586–588. [DOI] [PubMed] [Google Scholar]
- Cheney R.E., O’Shea,M.K., Heuser,J.E., Coelho,M.V., Wolenski,J.S., Espreafico,E.M., Forscher,P., Larson,R.E. and Mooseker,M.S. (1993) Brain myosin-V is a two-headed unconventional myosin with motor activity. Cell, 75, 13–23. [DOI] [PubMed] [Google Scholar]
- Cowtan K. and Main,P. (1998) Miscellaneous algorithms for density modification. Acta Crystallogr. D, 54, 487–493. [DOI] [PubMed] [Google Scholar]
- Dominguez R., Freyzon,Y., Trybus,K.M. and Cohen,C. (1998) Crystal structure of a vertebrate smooth muscle myosin motor domain and its complex with the essential light chain: visualization of the pre-power stroke state. Cell, 94, 559–571. [DOI] [PubMed] [Google Scholar]
- D’Souza V.M., Naqvi,N.I., Wang,H. and Balasubramanian,M.K. (2001) Interactions of Cdc4p, a myosin light chain, with IQ-domain containing proteins in Schizosaccharomyces pombe. Cell Struct. Funct., 26, 555–565. [DOI] [PubMed] [Google Scholar]
- Eng K., Naqvi,N.I., Wong,K.C. and Balasubramanian,M.K. (1998) Rng2p, a protein required for cytokinesis in fission yeast, is a component of the actomyosin ring and the spindle pole body. Curr. Biol., 8, 611–621. [DOI] [PubMed] [Google Scholar]
- Epp J.A. and Chant,J. (1997) An IQGAP-related protein controls actin-ring formation and cytokinesis in yeast. Curr. Biol., 7, 921–929. [DOI] [PubMed] [Google Scholar]
- Garcia de la Torre J., Navarro,S., Lopez Martinez,M.C., Diaz,F.G. and Lopez Cascales,J.J. (1994) HYDRO: a computer program for the prediction of hydrodynamic properties of macromolecules. Biophys. J., 67, 530–531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Govindan B., Bowser,R. and Novick,P. (1995) The role of Myo2, a yeast class V myosin, in vesicular transport. J. Cell Biol., 128, 1055–1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins D.G., Thompson,J.D. and Gibson,T.J. (1996) Using CLUSTAL for multiple sequence alignments. Methods Enzymol., 266, 383–402. [DOI] [PubMed] [Google Scholar]
- Houdusse A. and Cohen,C. (1996) Structure of the regulatory domain of scallop myosin at 2 Å resolution: implications for regulation. Structure, 4, 21–32. [DOI] [PubMed] [Google Scholar]
- Johnston G.C., Prendergast,J.A. and Singer,R.A. (1991) The Saccharomyces cerevisiae MYO2 gene encodes an essential myosin for vectorial transport of vesicles. J. Cell Biol., 113, 539–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurado L.A., Chockalingam,P.S. and Jarrett,H.W. (1999) Apocalmodulin. Physiol. Rev., 79, 661–682. [DOI] [PubMed] [Google Scholar]
- Lillie S.H. and Brown,S.S. (1994) Immunofluorescence localization of the unconventional myosin, Myo2p and the putative kinesin-related protein, Smy1p, to the same regions of polarized growth in Saccharomyces cerevisiae. J. Cell Biol., 125, 825–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lippincott J. and Li,R. (1998) Sequential assembly of myosin II, an IQGAP-like protein and filamentous actin to a ring structure involved in budding yeast cytokinesis. J. Cell Biol., 140, 355–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machesky L.M. (1998) Cytokinesis: IQGAPs find a function. Curr. Biol., 8, R202–R205. [DOI] [PubMed] [Google Scholar]
- McCollum D., Balasubramanian,M.K., Pelcher,L.E., Hemmingsen,S.M. and Gould,K.L. (1995) Schizosaccharomyces pombe cdc4+ gene encodes a novel EF-hand protein essential for cytokinesis. J. Cell Biol., 130, 651–660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCollum D., Feoktistova,A. and Gould,K.L. (1999) Phosphorylation of the myosin-II light chain does not regulate the timing of cytokinesis in fission yeast. J. Biol. Chem., 274, 17691–17695. [DOI] [PubMed] [Google Scholar]
- Murshudov G.N., Lebedev,A., Vagin,A.A., Wilson,K.S. and Dodson,E.J. (1999) Efficient anisotropic refinement of macromolecular structures using FFT. Acta Crystallogr. D, 55, 247–255. [DOI] [PubMed] [Google Scholar]
- Naqvi N.I., Eng,K., Gould,K.L. and Balasubramanian,M.K. (1999) Evidence for F-actin-dependent and -independent mechanisms involved in assembly and stability of the medial actomyosin ring in fission yeast. EMBO J., 18, 854–862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osman M.A. and Cerione,R.A. (1998) Iqg1p, a yeast homologue of the mammalian IQGAPs, mediates cdc42p effects on the actin cytoskeleton. J. Cell Biol., 142, 443–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruyne D.W., Schott,D.H. and Bretscher,A. (1998) Tropomyosin-containing actin cables direct the Myo2p-dependent polarized delivery of secretory vesicles in budding yeast. J. Cell Biol., 143, 1931–1945. [DOI] [PubMed] [Google Scholar]
- Rayment I., Rypniewski,W.R., Schmidt-Base,K., Smith,R., Tomchick,D.R., Benning,M.M., Winkelmann,D.A., Wesenberg,G. and Holden,H.M. (1993) Three-dimensional structure of myosin subfragment-1: a molecular motor. Science, 261, 50–58. [DOI] [PubMed] [Google Scholar]
- Reck-Peterson S.L., Provance,D.W.,Jr, Mooseker,M.S. and Mercer,J.A. (2000) Class V myosins. Biochim. Biophys. Acta, 1496, 36–51. [DOI] [PubMed] [Google Scholar]
- Schumacher M.A., Rivard,A.F., Bachinger,H.P. and Adelman,J.P. (2001) Structure of the gating domain of a Ca2+-activated K+ channel complexed with Ca2+/calmodulin. Nature, 410, 1120–1124. [DOI] [PubMed] [Google Scholar]
- Shannon K.B. and Li,R. (2000) A myosin light chain mediates the localization of the budding yeast IQGAP-like protein during contractile ring formation. Curr. Biol., 10, 727–730. [DOI] [PubMed] [Google Scholar]
- Slupsky C.M., Desautels,M., Huebert,T., Zhao,R., Hemmingsen,S.M. and McIntosh,L.P. (2001) Structure of Cdc4p, a contractile ring protein essential for cytokinesis in Schizosaccharomyces pombe. J. Biol. Chem., 276, 5943–5951. [DOI] [PubMed] [Google Scholar]
- Stafford W.F. (1998) Time difference sedimentation velocity analysis of rapidly reversible interacting systems: determination of equilibrium constants by non-linear curve fitting procedures. Biophys. J., 74, A301. [Google Scholar]
- Stafford W.F., Jacobsen,M.P., Woodhead,J., Craig,R., O’Neall-Hennessey,E. and Szent-Gyorgyi,A.G. (2001) Calcium-dependent structural changes in scallop heavy meromyosin. J. Mol. Biol., 307, 137–147. [DOI] [PubMed] [Google Scholar]
- Stevens R.C. and Davis,T.N. (1998) Mlc1p is a light chain for the unconventional myosin Myo2p in Saccharomyces cerevisiae. J. Cell Biol., 142, 711–722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terrak M., Otterbein,L.R., Wu,G., Palecanda,L.A., Lu,R.C. and Dominguez,R. (2002) Crystallization, X-ray characterization and selenomethionine phasing of Mlc1p bound to IQ motifs from myosin V. Acta Crystallogr. D, 58, 1882–1885. [DOI] [PubMed] [Google Scholar]
- Todd G.P. and Haschemeyer,R.H. (1981) General solution to the inverse problem of the differential equation of the ultracentrifuge. Proc. Natl Acad. Sci. USA, 78, 6739–6743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weeks C.M. and Miller,R. (1999) The design and implementation of SnB v2.0. J. Appl. Crystallogr., 32, 120–124. [Google Scholar]
- Weissbach L., Bernards,A. and Herion,D.W. (1998) Binding of myosin essential light chain to the cytoskeleton-associated protein IQGAP1. Biochem. Biophys. Res. Commun., 251, 269–276. [DOI] [PubMed] [Google Scholar]
- Xie X., Harrison,D.H., Schlichting,I., Sweet,R.M., Kalabokis,V.N., Szent-Gyorgyi,A.G. and Cohen,C. (1994) Structure of the regulatory domain of scallop myosin at 2.8 Å resolution. Nature, 368, 306–312. [DOI] [PubMed] [Google Scholar]
- Yin H., Pruyne,D., Huffaker,T.C. and Bretscher,A. (2000) Myosin V orientates the mitotic spindle in yeast. Nature, 406, 1013–1015. [DOI] [PubMed] [Google Scholar]