Abstract
Proteins containing [Fe–S] clusters perform essential functions in all domains of life. Previously, we identified the sufABCDSE operon as being necessary for virulence of the plant pathogen Erwinia chrysanthemi. In addition, we collected preliminary evidence that the sufABCDSE operon might be involved in the assembly of [Fe–S] clusters. Of particular interest are the sufB, sufC and sufD genes, which are conserved among Eubacteria, Archaea, plants and parasites. The present study establishes SufC as an unorthodox ATPase of the ABC superfamily that is located in the cytosol, wherein it interacts with both SufB and SufD. Moreover, under oxidative stress conditions, SufC was found to be necessary for the activity of enzymes containing oxygen-labile [Fe–S] clusters, but dispensable for glutamate synthase, which contains an oxidatively stable [Fe–S] cluster. Lastly, we have shown SufBCD to be essential for iron acquisition via chrysobactin, a siderophore of major importance in virulence. We discuss a model wherein the SufBCD proteins contribute to bacterial pathogenicity via their role in the assembly of [Fe–S] clusters under oxidative stress and iron limitation.
Keywords: ABC ATPase/bacterial pathogenicity/iron–sulfur proteins/oxidative stress
Introduction
Proteins that contain [Fe–S] clusters fulfil enzymatic or regulatory functions in various cellular processes ranging from respiration to gene expression (Johnson, 1998). It has been demonstrated that [Fe–S] clusters can form by mixing Fe3+/2+ and S2– under controlled conditions and that, in vitro, [Fe–S] clusters can be inserted into proteins in the presence of cysteine desulfurases, referred to as NifS-like enzymes (Zheng and Dean, 1994; Flint, 1996; Green et al., 1996; Hidalgo and Demple, 1996; Beinert and Kiley, 1999; Tse Sum Bui et al., 2000). Yet, the in vivo situation is expected to be more complex in order to meet constraints of efficiency and specificity. Therefore, the issue of in vivo assembly of [Fe–S] clusters into proteins has received increasing attention in recent years.
Genomic and biochemical investigations have identified determinants for cysteine desulfurase activity in most living organisms (Ellis et al., 2001; Tachezy et al., 2001). Frequently, there is more than one cysteine desulfurase encoding gene per organism, indicating how crucial this activity is in supporting cellular life.
In Escherichia coli, there are three cysteine desulfurase encoding genes, namely iscS, sufS and csdA (Mihara et al., 2000). The iscS gene belongs to the iscSUA hscBA fdx gene cluster. Recent investigations have provided evidence for the involvement of these six highly conserved genes in [Fe–S] cluster biogenesis in vivo in bacteria and yeast (Garland et al., 1999; Schilke et al., 1999; Takahashi and Nakamura, 1999; Kaut et al., 2000; Lange et al., 2000; Pelzer et al., 2000; Voisine et al., 2000; Kim et al., 2001; Tokumoto and Takahashi, 2001). Fdx is a [2Fe–2S] ferredoxin. IcsA and IscU interact with Fdx and IscS, respectively, and are thought to act as scaffolds for [Fe–S] cluster assembly (Ollagnier-de-Choudens et al., 2001; Urbina et al., 2001; Mansy et al., 2002). HscA and HscB are molecular chaperones that belong to the DnaK and DnaJ proteins families, respectively, but their roles in [Fe–S] cluster biogenesis remain unclear (Hoff et al., 2000).
The sufS gene belongs to the sufABCDSE operon, identified as being necessary for the stability of FhuF, a [2Fe–2S] cluster protein, potentially involved in iron assimilation (Patzer and Hantke, 1999). Recently, we identified the sufABCDSE operon of Erwinia chrysanthemi as being involved in iron metabolism as well as necessary for full virulence of this plant pathogen (Nachin et al., 2001). Erwinia chrysanthemi and E.coli suf operons are highly similar and the encoded proteins share 70–95% similarity. Non-polar insertions in each of the E.chrysanthemi suf genes led to increased free iron concentration. Recently, growth defects due to mutation in the isc genes were found to be phenotypically suppressed by modifying the expression of the suf genes (Takahashi and Tokumoto, 2002). Hence, the available results on the suf operon, although scarce, suggest that it encodes a pathway involved in [Fe–S] biosynthesis and/or repair.
The suf genes are present in numerous organisms but the suf operon is not (Ellis et al., 2001). In fact, the best conserved suf genes are sufB and sufC, which occur in Eubacteria, Archaea, plants and parasites. There are no genomes characterized thus far that lack sufC yet contain sufB (or sufD, as sufB and sufD genes are paralogous). This phylogenetic conservation supports the hypothesis that SufB, SufC and SufD functionally interact with each other. In Arabidopsis thaliana, mutation in sufB caused severe light signalling-related defects, and sufB and sufC genes were found to be essential in Synechocystis (Law et al., 2000; Moller et al., 2001). In E.chrysanthemi, a mutation in sufD caused increased sensitivity to oxidative stress and a mutation in sufC conferred increased intracellular accumulation of free iron, hypersensitivity to oxidative agents and reduced virulence (Nachin et al., 2001). In fact, phenotypic consequences of mutations in sufC were identical in their nature and extent to those inactivating the whole sufABCDSE operon, indicating that SufC is a key player in Suf functions (Nachin et al., 2001).
Primary sequence analysis of SufC revealed the presence of ABC ATPase signatures, i.e. both Walker A and B boxes as well as a C-motif. This class of ATPases is mostly found to be associated with membrane proteins, forming a complex that allows translocation across membranes of a wide range of allocrites (Fath and Kolter, 1993; Holland and Blight, 1999). Remarkably, phylogenetic analyses of ABC ATPases invariably put SufC as the sole member of its class, and failed to predict substrate specificity (Linton and Higgins, 1998; Quentin et al., 1999). Moreover, no potential transmembrane segments were found in any of the suf gene products. Taken together, these observations led us to two hypotheses: (i) SufC forms a typical ABC transporter with membrane partners encoded by genes outside of the suf operon and the resulting complex targets [Fe–S] clusters into extracytosolic proteins; or (ii) SufC forms, along with other Suf proteins, an atypical ABC-type cytoplasmic machinery that assembles and/or repairs [Fe–S].
The objective of the present study was to experimentally evaluate these two models. We report that SufC interacts with both SufB and SufD to form an unorthodox cytoplasmic ABC ATPase. We show that the SufBCD proteins function as a defence system for oxygen-labile [Fe–S] clusters present in proteins. Moreover, the present study helped us identify a link between SufBCD proteins and iron acquisition, thereby providing a rationale for the role of suf genes in the virulence of E.chrysanthemi.
Results
SufC is an ATPase
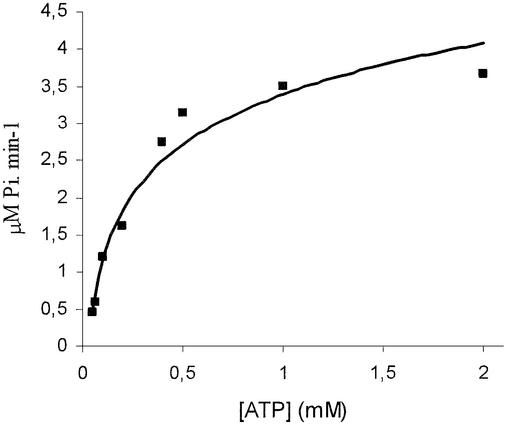
The three motifs of typical ATP-hydrolysing domains of ABC ATPases are found in SufC. These are Walker sites A and B as well as motif C (Linton and Higgins, 1998). We tested whether the E.chrysanthemi SufC is indeed a bona fide ABC ATPase. Preliminary attempts to purify the native SufC protein failed since it formed aggregates in the cell. Therefore, the sufC gene was fused at its 5′-end with the malE gene. The encoded MBP–SufC protein was soluble. Cleared lysate of cells overproducing MBP–SufC were loaded on a gel filtration column with an exclusion size of 600 kDa. The MBP–SufC-containing fractions were pooled, incubated with amylose beads and eluted with maltose. The purity of the resulting fraction was checked by mass spectrometry analysis (data not shown). The purified MBP–SufC was found to exhibit a specific ATPase activity value of 0.3 µmol of inorganic phosphate (Pi)/min/mg, at pH 7.5, in the presence of MnCl2 or MgCl2. No activity could be detected in the presence of CaCl2. Vmax and Km values obtained were 4.45 µM Pi/min and 0.29 mM, respectively (Figure 1). Note that the same specific activity was obtained with a SufC-His6 protein derivative (data not shown; see below). This study demonstrated that SufC possesses ATP-hydrolysing properties similar to other E.coli ABC ATPases such as HisP or MalK (Schneider and Hunke, 1998).
Fig. 1. Characterization of ATPase activity of the purified MBP–SufC protein. Activity was tested at pH 7.5 and 37°C, in the presence of 5 mM MgCl2, over a range of ATP concentrations (0.05–2 mM). Assays were performed in 25 mM Tris buffer with a MPB–SufC protein concentration of 9 µg/ml in a final volume of 200 µl. The amount of Pi released was measured at different time points over an 1 h period. The solid line is the theoretical curve for Km = 0.29 mM and Vmax = 4.45 µM/min, calculated using Graphpad Prism software.
Analysis of molecular interactions between SufB, SufC and SufD
Conservation of the sufC-sufB/D genetic organization in Eubacterial, Archaeal, plant and parasitic genomes suggested a functional link between the encoded products. Therefore, we tested whether SufB, SufC and SufD proteins interact physically by using the yeast two-hybrid system. The E.chrysanthemi sufB, sufC and sufD genes were cloned in-frame with either the LexA DNA-binding domain-encoding sequence, or with the B42 transcriptional activator encoding sequence. First, expression of the lacZ gene was used as a reporter. Diploid cells co-synthesizing pairs of potential interactants were scored on selective medium containing X-gal. Subsequently, β-gal<\\jy>actosidase activity tests were run on yeast cell extracts. The values obtained revealed an interaction between SufC and SufB, as well as between SufC and SufD (Table I). Also, the results suggested that SufD forms homodimers (Table I). Next, growth of diploid cells in liquid media lacking leucine was used as another reporter of efficient two-hybrid interaction. OD600 values obtained after overnight growth indicated interactions between SufB/SufC and SufD/SufC, but failed to confirm SufD/SufD interactions (Table I). Thus, the data provided the first evidence for a direct interaction of SufC with both SufB and SufD.
Table I. Interactions between SufB, SufC and SufD proteins in the yeast two-hybrid system.
| LexA | LexA–SufB | LexA–SufC | LexA–SufD | |
|---|---|---|---|---|
| β-galactosidase activity (U/mg protein) | ||||
| B42 | 28 ± 14 | 37 ± 23 | 29 ± 10 | 102 ± 3 |
| B42–SufB | 26 ± 14 | 37 ± 18 | 448 ± 50 | 72 ± 32 |
| B42–SufC | 27 ± 20 | 1334 ± 313 | 50 ± 17 | 1373 ± 398 |
| B42–SufD |
46 ± 33 |
23 ± 20 |
339 ± 95 |
1134 ± 33 |
| OD600 | ||||
| B42 | 0.017 ± 0.025 | 0.002 ± 0.004 | 0.007 ± 0.001 | 0.034 ± 0.011 |
| B42–SufB | 0.072 ± 0.082 | 0.009 ± 0.008 | 0.084 ± 0.004 | 0 |
| B42–SufC | 0.016 ± 0.021 | 0.150 ± 0.004 | 0.004 ± 0.002 | 0.514 ± 0.141 |
| B42–SufD | 0.037 ± 0.053 | 0.075 ± 0.001 | 0.449 ± 0.117 | 0.069 ± 0.001 |
Mating was carried out between appropriate yeast strains in order to produce the proteins indicated. Upper part: utilization of the LacZ reporter gene. The β-galactosidase activities were assayed from cultures grown overnight in inducible selective CM medium. Experiments were run at least in triplicate. Lower part: utilization of the Leu reporter gene. Diploid strains were grown in selective CM medium lacking leucine. Values indicated are differences in OD600 between cultures grown overnight in inducible (i.e. in the presence of galactose and raffinose) and non-inducible (i.e. in the presence of glucose) conditions. Experiments were run in duplicate.
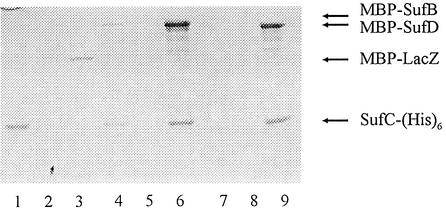
Interactions between the Suf proteins were further investigated using biochemical means. Both the sufB and sufD genes were fused at their 5′-end with the malE gene. The sufC gene was fused at its 3′-end with the His6 tag encoding DNA. The three chimeric proteins, MBP–SufB, MBP–SufD and SufC-His6, were purified. Equal amounts of SufC-His6 and MBP–SufB were mixed and loaded on an amylose column. Some residual amount of both proteins went with the flowthrough (Figure 2, lane 4). The column was then extensively washed without elution of either one of the two proteins (Figure 2, lane 5). In contrast, when a solution of maltose (2 mM) was applied, MBP–SufB and SufC-His6 were co-eluted (Figure 2, lane 6). In the same way, SufC-His6 was mixed with MBP–SufD. Here again, both proteins were retained on the column unless the maltose solution was applied (Figure 2, lanes 7–9). Bands observed right under those corresponding to MBP–SufB and MBP–SufD were identified by immunoblotting as corresponding to degradation products (data not shown). As a control, SufC-His6 was mixed with MBP–LacZ. In this case, SufC-His6 was not retained on the amylose column, while MBP–LacZ was eluted when the solution of maltose was applied (Figure 2, lanes 1–3). Thus, SufC-His6 was retained on amylose beads indirectly via its interactions with MBP–SufB in one case and MBP–SufD in the other. Thus, molecular and biochemical analyses allowed us to conclude that SufC interacts with both SufB and SufD.
Fig. 2. Interaction between SufB, SufC and SufD using affinity chromatography. Equal amounts of purified SufC-His6 and purified MBP–LacZ (lanes 1–3), MBP–SufB (lanes 4–6) or MBP–SufD (lanes 7–9) were mixed with 1 ml of amylose resin and loaded onto a column. Flowthrough (lanes 1, 4 and 7), washing (lanes 2, 5 and 8) and elution (lanes 3, 6 and 9) fractions were analysed by SDS–PAGE and Coomassie Blue staining.
Cytoplasmic location of the SufBCD proteins
In data banks, sufB, sufC and sufD genes are predicted to encode components of an ABC transporter. Yet, no potential transmembrane segment can be found in primary sequences of either SufB, SufC or SufD. Therefore, cell fractionation techniques were applied to investigate the location of the three proteins in vivo, using the E.chrysanthemi strain A3559. This strain possesses an in-frame deletion in the secretion gene outD, leading to periplasmic location of the Cel5 cellulase (Bortoli-German et al., 1994). The efficiency and reliability of fractionation procedures could therefore be ascertained by using Cel5, OutF, an inner membrane protein of the type II secretion machinery, and MsrA, a cytoplasmic methionine sulfoxide reductase (El Hassouni et al., 1999; Py et al., 2001). SufC protein was found to be exclusively cytoplasmic (Figure 3). Next, locations of SufB and SufD were studied. Erwinia chrysanthemi sufB, sufC or sufD genes were cloned into a pBAD-derived vector such that each Suf protein was fused to a haemagglutinin (HA) tag. HA-SufC and HA-SufB were exclusively found inside the cytosol (Figure 4). In the case of HA-SufD, cross-reacting material was also found in membrane fractions (Figure 4). However, inner membrane-bound forms of HA-SufD were shown to be aggregates that had co-sedimented with the membranes. Indeed, when the sedimented pellet was treated with NaCl, Triton X-100 or urea, only the latter chemical agent was able to solubilize HA-SufD (data not shown). This study showed that all three SufB, SufC and SufD proteins are localized in the cytosol.
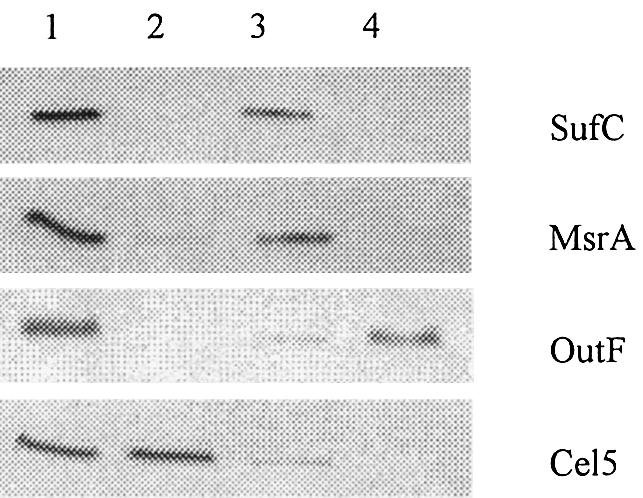
Fig. 3. Cytoplasmic location of SufC. Immunoblot analysis of crude extracts (lane 1), periplasmic (lane 2), cytoplasmic (lane 3) and membrane (lane 4) fractions prepared from E.chrysanthemi A3559 strain. On the right side of the panel are indicated the proteins under study. SufC protein was detected using anti-SufC polyclonal antibody. MsrA, OutF and Cel5 were used as markers for cytosolic, membrane and periplasmic compartments, respectively.
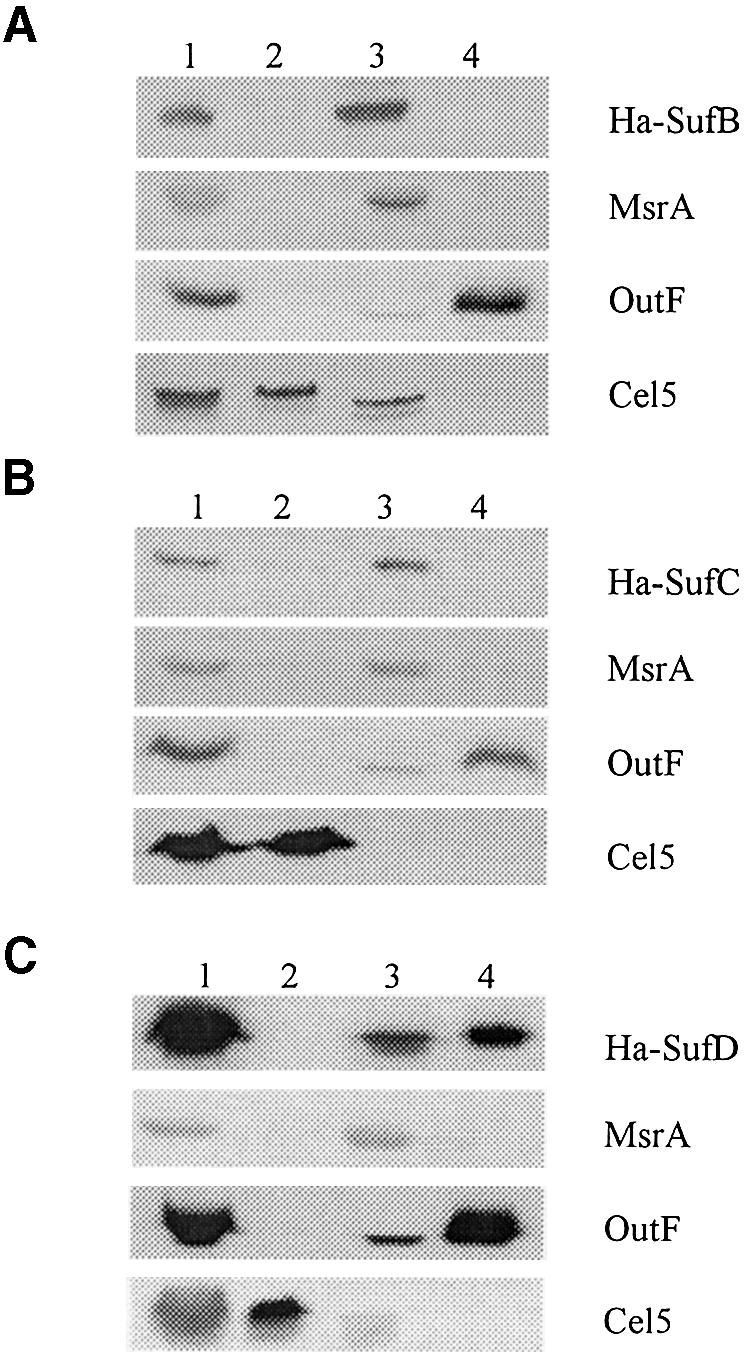
Fig. 4. SufB, SufC and SufD are located in the cytosol. Immunoblot analysis of crude extracts (lanes 1), periplasmic (lanes 2), cytoplasmic (lanes 3) and membrane (lanes 4) fractions prepared from E.chrysanthemi A3559 strain carrying pA-B (encoding HA-SufB) (A), pA-C (encoding HA-SufC) (B) and pA-D (encoding HA-SufD) (C), respectively. HA-SufB, HA-SufC and HA-SufD proteins were detected using anti-HA antibodies. On the right side of the panel are indicated the proteins under study. MsrA, OutF and Cel5 were used as markers for cytosolic, membrane and periplasmic compartments, respectively.
SufC is important for the protection of labile [Fe–S] clusters under oxidative stress conditions
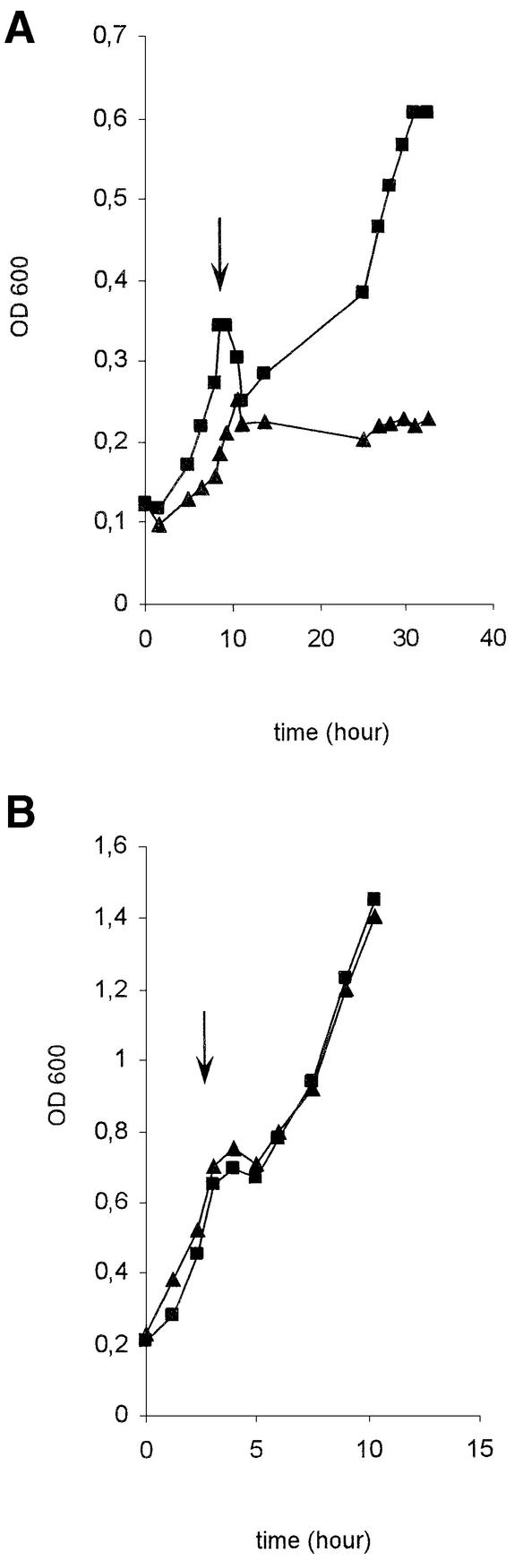
In order to test the involvement of sufC in the activity of [Fe–S] cluster-containing enzymes, we used E.coli as a model since many [Fe–S]-containing enzymes have been well studied in this species. Three enzymes were chosen: two containing oxygen-labile [Fe–S] clusters, e.g. 6-phosphogluconate dehydratase (referred to as 6-PGDH) and fumarase A, and one containing an oxidatively stable [Fe–S] cluster, e.g. glutamate synthase. In each case, we assayed the enzyme activity of exponentially growing wild-type and sufC strains exposed, or not, to the oxidative agent phenazine methosulfate (PMS). The values obtained showed that upon exposure to PMS, the wild-type strain retained 18% activity of 6-PGDH while the sufC mutant lost 97% activity (Table II). This loss in activity was further confirmed by comparing growth of the wild-type MG1655 and the sufC mutant on gluconate as a sole carbon source (Figure 5). A similar method was used by Gardner and Fridovich (1991) to evaluate phenotypic consequences of a lack of superoxide dismutase activity. In the absence of oxidative stress, both strains grew equally well (data not shown). However, when cultures were exposed to PMS, a dramatic difference was observed between the wild type and the sufC mutant. The wild-type strain stopped growing upon exposure to PMS, cell density declined slightly, but eventually growth resumed after ∼2 h (Figure 5A). Growth of the sufC mutant stopped immediately after exposure to PMS, but, in contrast to the wild-type strain, growth never resumed (Figure 5A). Control experiments showed no differences between wild type and sufC mutant when the carbon source was glycerol (Figure 5B). Use of paraquat as an oxidative agent had the same effect on wild type and sufC mutant as PMS (data not shown).
Table II. SufC protein is required for activity of oxidatively labile [Fe–S] cluster-containing enzymes.
| MG1655 |
Ratio | MG1655sufC |
Ratio | |||
|---|---|---|---|---|---|---|
| –PMS | +PMS | (%) | –PMS | +PMS | (%) | |
| 6-PGDH | 56 ± 9 | 10 ± 5 | 18 | 123 ± 36 | 4 ± 3 | 3 |
| Fumarase A | 126 ± 16 | 16 ± 2 | 13 | 138 ± 13 | 9 ± 2 | 6 |
| Glutamate synthase | 128 ± 2 | 313 ± 1 | 244 | 124 ± 4 | 283 ± 3 | 228 |
| PGM | 37 ± 6 | 36 ± 9 | 97 | 35 ± 10 | 39 ± 6 | 111 |
Escherichia coli wt and MG1655 sufC strains were grown in minimal A salts medium supplemented with gluconate and casamino acids for 6-PGDH and PGM assays, fumarate for fumarase assays, or glucose for glutamate synthase assays. During exponential growth phase, cultures were divided into two equal samples, one of which was treated with 10 M or 15 µM PMS (see Material and methods). After 30 min incubation at 37°C, 6-PGDH, fumarase A, glutamate synthase and PGM activities were assayed on clarified cell extracts. Activities are expressed as nmol of product/min/mg and are the average of at least two experiments. The percentage values indicate the ratio between the enzymatic activities measured in the presence and absence of PMS, the oxidative agent.
Fig. 5. SufC is required for gluconate utilization under oxidative stress. Escherichia coli strains were grown in minimal A medium supplemented with 0.2% gluconate (A) or glycerol (B) and 1 µg/ml thiamine. PMS (15 µM) was added to the cultures during the exponential growth phase. Squares, MG1655; triangles, MG1655 sufC. Arrows indicate the time of PMS addition.
To assay for fumarase A activity, both E.coli wild-type and sufC mutant strains were grown in minimal medium containing fumarate as a carbon source. At mid-exponential phase, without addition of PMS, the amount of fumarase activity was found to be similar between the two strains (Table II). In contrast, upon exposure to PMS, the decrease in activity was much more pronounced in the sufC strain (17-fold) as compared with the wild type (8-fold) (Table II). The levels of glutamate synthase activity were the same in wild-type and sufC strains in the absence of PMS. In the presence of PMS, the amount of glutamate synthase activity recovered was increased in both strains. Although we have no explanation for this increase, the results clearly showed that, in contrast to the two other enzymes, lack of the sufC gene had no effect on glutamate synthase activity under oxidative stress conditions (Table II). Lastly, we checked that the activity of phosphoglucomutase (PGM), a non-[Fe–S] cluster-containing enzyme, was identical in both wild-type and sufC strains, under both oxidative and non-oxidative conditions (Table II). These results demonstrated that a functional sufC gene is required under oxidative stress conditions for the activity of enzymes containing labile [Fe–S] clusters, but is dispensable for the activity of an oxidatively stable [Fe–S] cluster-containing enzyme.
Role of SufBCD in iron assimilation in E.chrysanthemi
In the course of characterizing the E.chrysanthemi suf mutants, we made the serendipitous observation that the sufB, sufC or sufD mutants failed to grow on L-medium containing 2,2′-dipyridyl, a membrane-permeant ferrous iron chelator that causes intracellular iron deprivation. Therefore, we investigated the relationships between the suf genes and iron acquisition via ferric chrysobactin, a siderophore excreted by E.chrysanthemi that is of paramount importance in virulence (Masclaux and Expert, 1995). The two mutations cbsE-1 and acsA-37, which impair siderophore synthesis, were transduced into each of the three sufB, sufC and sufD mutants. This allowed us to determine the ability of each triple mutant to be cross-fed by exogenously added ferric chrysobactin. Growth of the cbs acs siderophore auxotroph strain was rescued by exogenous ferric chrysobactin (Table III). In contrast, growth of neither sufC cbs acs nor sufD cbs acs strains could be rescued by exogenous ferric chrysobactin, while growth of the sufB cbs acs strain was inefficiently rescued. We then carried out two additional controls. First, we showed that ferric chloride stimulated the growth of all suf mutants in a way similar to that of the parental strain, showing that iron was not toxic by itself (Table III). Secondly, use of [59Fe]chrysobactin allowed us to show that uptake of ferric chysobactin was not impaired by sufB or sufC mutation (data not shown). These experiments indicated that suf genes are required for utilization of ferric chrysobactin inside the bacterial cell.
Table III. SufB, SufC and SufD proteins are required for ferric chrysobactin utilization.
| Strain | Fe–chrysobactin | FeCl3 |
|---|---|---|
| cbs acs | 19 ± 1 | 20 ± 1 |
| sufB cbs acs | 12 ± 1 | 20 ± 1 |
| sufC cbs acs | No growth | 20 ± 1 |
| sufD cbs acs | No growth | 20 ± 1 |
The capacity of siderophore auxotroph strains to utilize exogenous ferric chrysobactin or FeCl3 as iron sources was assayed on L-agar plates containing 2,2′-dipyridyl (200 µM). The diameters of zones of growth of the tested strains were measured after 24 h and are expressed in millimetres. Experiments were carried out in duplicate.
Discussion
Biogenesis of [Fe–S] clusters in vivo has become an important area of study given the wide range of functions carried out by [Fe–S] proteins in all domains of life (Johnson, 1998; Beinert and Kiley, 1999). Preliminary evidence was obtained suggesting that the sufABCDSE operon has a role in [Fe–S] biosynthesis and/or repair (Patzer and Hantke, 1999; Nachin et al., 2001; Takahashi and Tokumoto, 2002). Here we have established the importance of the SufBCD proteins, and by inference of the sufABCDSE-encoded pathway, for the activity of two oxygen-labile [Fe–S] cluster-containing enzymes exposed to oxidative stress. Moreover, we have demonstrated that SufC is an ATPase from the ABC superfamily that is located in the cytoplasm. Lastly, we have identified a relationship between the Suf system and iron assimilation that provides a molecular basis for the role of suf genes in E.chrysanthemi virulence. A model is proposed wherein a [Fe–S]-containing reductase, dependent upon the Suf pathway, is necessary for both iron assimilation from incoming ferric siderophore and repair of oxidatively damaged [Fe–S] clusters.
In previous classifications of ABC ATPases, SufC was proposed to constitute a so-called orthologous class, i.e. no other ABC ATPase encoding gene clusters with sufC (Linton and Higgins, 1998; Quentin et al., 1999). It was, therefore, of great importance to assess experimentally whether SufC possesses ATPase activity and we demonstrate here that it is the case. Interestingly, a SufC-like enzyme from Thermotoga maritima was recently shown to have ATPase activity (Rangachari et al., 2002).
Genomes that contain a sufC ortholog also contain a proximal sufB-like gene (Ellis et al., 2001). This raised the possibility that SufC interacts with SufB and/or SufD since SufB and SufD are paralogous. Data presented here have shown that indeed SufC interacts with both SufB and SufD.
ABC ATPases are ubiquitous nucleotide hydrolysing domains that associate with membrane proteins (or domains) to build up an ABC transporter. ABC transporters allow export or import of a wide range of allocrites across biological membranes (Fath and Kolter, 1993; Holland and Blight, 1999). An intriguing feature of the SufBCD proteins is that none of the components is predicted to contain transmembrane segments. Here, we have shown that all three proteins are located in the cytosol. Other examples of cytoplasmic ABC ATPases have been reported, including Rad50, UvrA, MutS or SMC. All of these are related to DNA repair and serve as motors of molecular machineries instead of being true transporters (Linton and Higgins, 1998; Hopfner et al., 2000; Junop et al., 2001; Lowe et al., 2001). As discussed below, we propose that the SufBCD proteins provide energy to the SufABCDSE system to repair oxidatively damaged [Fe–S].
Previous studies showed that the Isc system has an important role in the biosynthesis of [Fe–S] clusters in vivo (Tokumoto and Takahashi, 2001). We have established here the importance of the Suf system in [Fe–S] biosynthesis as well. Interestingly, modified expression of suf genes was recently found to act as an extragenic suppressor of growth defects of isc mutation (Takahashi and Tokumoto, 2002). This raises the question of the specificities, if any, of each system. In E.coli cells growing aerobically, the Isc system was found to be important for 6-PGDH and fumarase A, the activities of which were reduced to 10 and 49%, respectively, in the iscS mutant (Schwartz et al., 2000). Using the same growth conditions, we observed no reduction in these two activities following inactivation of the sufC gene. In contrast, under oxidative stress conditions, the sufC gene was found to be of great importance, implying that the isc genes are of minor importance in these conditions. These results suggest that both pathways are used for different physiological processes. Moreover, a significant observation was made when analysing the activity of glutamate synthase. Contrary to 6-PGDH and fumarase A, which contain oxidatively labile [Fe–S] clusters, glutamate synthase contains an oxidatively stable [Fe–S] cluster. Inactivation of iscS was reported to cause a decrease in glutamate synthase activity (Schwartz et al., 2000). The importance of the Isc system for both types of enzyme argued for a role in de novo synthesis (Schwartz et al., 2000). In contrast, here, we observed no difference in glutamate synthase activity upon inactivation of sufC. Hence, a possibility is that the Suf system is involved in repair of oxidatively damaged [Fe–S] clusters. This proposal is consistent with our previous studies, which showed that lack of functional suf genes led to hypersensitivity to oxidative agents in E.chrysanthemi, and to modifications of the SoxR/S-dependent response to oxidative stress in E.coli (Nachin et al., 2001). This has now been further supported by the analysis of the E.coli transcriptome, which identified the suf operon as a member of the OxyR regulon (Zheng et al., 2001).
Our previous analysis of the suf genes in E.chrysanthemi revealed that they are important for full virulence of this plant pathogen. The present study provides a rationale for the precise role of the Suf system in virulence. Indeed, SufBCD proteins were found to be essential for use of ferric chrysobactin, a siderophore of primary importance in E.chrysanthemi virulence (Masclaux and Expert, 1995). At the molecular level, the simplest hypothesis is that a Suf-dependent [Fe–S] cluster-containing reductase is required for acting on incoming ferric chrysobactin. This would be reminiscent of the situation described in E.coli where FhuF, a [2Fe–2S] protein proposed to be necessary for use of ferrioxamine B, was affected by polar insertions in the sufD gene (Patzer and Hantke, 1999).
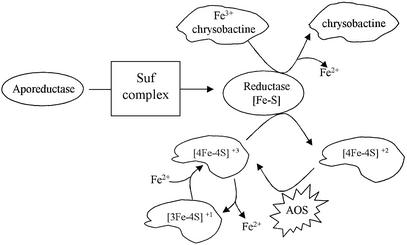
At the mechanistic level, one can only speculate on the processes involved in repair of oxidatively damaged [Fe–S]. 6-GPDH and fumarase A used in this study contain [4Fe–4S] clusters that actually consist of [(2Fe3+ 2Fe2+)–4S2–] species. Upon oxidation, this type of cluster is thought to be converted into [(3Fe3+ 1Fe2+)–4S2–] species, which readily lose one Fe2+ atom to form [3Fe3+–4S2–] clusters. Conversely, repair requires an Fe2+ atom, and reducing conditions to form [(2Fe3+ 2Fe2+)–4S2–] clusters (see Figure 6). Hence, the Suf system might activate a [Fe–S]-containing reductase that is required in the reducing step and/or help to recruit and integrate the Fe2+ atom. Interestingly, in vitro reactivation of oxidatively damaged [4Fe–4S] dehydratase was proposed to require energy since it could not proceed at 0°C (Flint et al., 1993). It is, therefore, tempting to speculate that the SufC ATPase provides the energy required for [Fe–S] cluster repair.
Fig. 6. Model of a link between iron assimilation and repair of [Fe–S] clusters under oxidative stress and iron limitation. This model postulates the existence of a reductase that depends upon the Suf system to acquire its [Fe–S] cluster (middle part). Under iron limitation and oxidative stress, this reductase allows, respectively, iron liberation from incoming iron (Fe3+)-loaded siderophore (chrysobactine) (upper part) and repair of the damaged [4Fe–4S] cluster (see text for details). AOS, activated oxygen species.
In this study, two processes, namely [Fe–S] biogenesis under oxidative stress and iron assimilation, were found to be dependent upon SufC. Our previous studies showed that the Suf system functions in low iron conditions, i.e. when iron might be limiting for [Fe–S] biogenesis. Therefore, we speculate that a Suf-dependent reductase acts on Fe3+-siderophore so as to liberate Fe2+ to be used for reactivation of the [Fe–S] cluster damaged by oxidative stress (Figure 6). Our next studies will aim at identifying such a reductase.
Materials and methods
Strains and culture conditions
All strains used are described in Table III. The E.coli TG1 strain was used for routine DNA manipulation. MG1655Δsuf strain was constructed in a one-step inactivation of suf genes as described by Datsenko and Wanner (2000). A DNA fragment containing cat gene flanked with a 5′ and 3′ region bordering the E.coli suf operon was amplified by PCR using pKD3 as a template and oligonucleotides 5Δsuf and 3Δsuf (Table IV). Strain BW25113, carrying the pKD46 plasmid, was transformed by electroporation with the amplified fragment and CmR colonies were selected. The replacement of chromosomal suf operon by cat gene was checked by PCR amplification in the CmR clones. Phage P1 was used to transduce the mutation into MG1655 strain, yielding MG1655suf strain. MG1655 sufC was described previously (Nachin et al., 2001).
Table IV. Characteristics of strains and plasmids used in this study.
| Strains and plasmids | Description | References |
|---|---|---|
|
Escherichia coli | ||
| TG1 | F′traD36 lacIq Δ(lacZ)M15 proA+B+/supE Δ(hsdM-mcrB)5 (rK–mK– McrB–) thi Δ(lac-proAB) | Sambrook et al. (1989) |
| BW25113 | lacIq rrnBT14 ΔlacZWJ16 hsdR514 ΔaraBADAH33 ΔrhaBADLD78 | Datsenko and Wanner (2000) |
| MG1655 | Wild type | Laboratory collection |
| MG1655suf | Δsuf, CamR | This study |
| MG1655sufC |
sufC::aphA-3, KanR |
Nachin et al. (2001) |
|
Erwinia chrysanthemi | ||
| A3559 | 3937 lacZ2 ΔoutD kdgR::Mu KanR | Bouley et al. (2001) |
| LNB-K3 | 3937 sufB ::aphA-3, KanR | Nachin et al. (2001) |
| LNC-K3 | 3937 sufC ::aphA-3, KanR | Nachin et al. (2001) |
| LND-K3 | 3937 sufD ::aphA-3, KanR | Nachin et al. (2001) |
| LNB-K3 cbs acs | 3937 sufB ::aphA-3 cbsE-1 acsA-37, KanR SpcR | This study |
| LNC-K3 cbs acs | 3937 sufC ::aphA-3 cbsE-1 acsA-37, KanR SpcR | This study |
| LND-K3 cbs acs |
3937 sufD ::aphA-3 cbsE-1 acsA-37, KanR SpcR |
This study |
|
Saccharomyces cerevisiae | ||
| EGY48 | MATα his3 trp1 ura3-52 leu2::3LexAop-LEU2 | Golemis et al. (1994) |
| RF206 | MATα his3Δ200 leu2-3 lys2Δ201 ura3-52 trp1Δ::hisG | Finley and Brent (1994) |
| Plasmids | ||
| pSH18–34 | Carries the GAL1-LexAop-lacZ reporter gene | Golemis et al. (1994) |
| pJG4-5 | Cloning vector for the construction of B42 hybrid proteins | Golemis et al. (1994) |
| pB42-B | pJG4-5 expressing B42–SufB hybrid protein | This study |
| pB42-C | pJG4-5 expressing B42–SufC hybrid protein | This study |
| pB42-D | pJG4-5 expressing B42–SufD hybrid protein | This study |
| pEG202 | Cloning vector for the construction of LexA hybrid proteins | Golemis et al. (1994) |
| pLexA-B | pEG202 expressing LexA–SufB hybrid protein | This study |
| pLexA-C | pEG202 expressing LexA–SufC hybrid protein | This study |
| pLexA-D | pEG202 expressing LexA–SufD hybrid protein | This study |
| pBAD24 | Cloning vector, AmpR | Guzman et al. (1995) |
| pA-B | pBAD24 expressing HA-tagged SufB | This study |
| pA-C | pBAD24 expressing HA-tagged SufC | This study |
| pA-D | pBAD24 expressing HA-tagged SufD | This study |
| pET22b+ | His6 tag fusion vector, AmpR | |
| pET-CHis | pET22 expressing SufC-His, AmpR | This study |
| pBADIK | Cloning vector, pBAD24 derivative, KmR | Py et al. (1999) |
| pK-CHis | pBADIK expressing SufC-His, KmR | This study |
| pSUF | pUC18 derivative containing sufA, sufB, sufC and 5′ part of sufD, AmpR | Nachin et al. (2001) |
| pMal-c2 | MBP fusion vector, AmpR | New England Biolabs |
| pMal-SufB | pMal-c2 expressing MBP–SufB hybrid protein, AmpR | This study |
| pMal-SufC | pMal-c2 expressing MBP–SufC hybrid protein, AmpR | This study |
| pMal-SufD | pMal-c2 expressing MBP–SufD hybrid protein, AmpR | This study |
| pKD3 | pANTSγ derivative containing an FRT-flanked chloramphenicol resistance gene, CmR | Datsenko and Wanner (2000) |
| pKD46 | Red helper plasmid, pINT-ts derivative containing araC-ParaB and γβ exo, AmpR | Datsenko and Wanner (2000) |
To construct E.chrysanthemi LNB-K3, LNC-K3 and LND-K3 strains, ΦEC2 phage was used to transduce both mutations cbsE-1 and acsA-37.
Escherichia coli strains were grown aerobically at 37°C in Luria–Bertani (LB) rich medium (Sambrook et al., 1989). For 6-PGDH and PGM assays, cells were grown in minimal A salts medium (Miller, 1972) containing 0.2% gluconate and 0.2% casamino acids. For the fumarase assay, cells were grown in minimal A salts containing 40 mM fumarate. For glutamate synthase, cells were grown in minimal A salts containing 0.2% glucose. When necessary, antibiotics were added at the following concentrations: 50 µg/ml ampicillin, 25 µg/ml kanamycin and 25 µg/ml chloramphenicol. Erwinia chrysanthemi strains were grown aerobically in LB medium at 30°C. Saccharomyces cerevisiae strains were grown in YPD or in appropriate minimal dropout media (CM) in which 2% (w/v) glucose or 2% (w/v) galactose and 2% (w/v) raffinose were added (Lundblad, 1993).
Plasmid construction
The pEG202 and pJG4-5 vectors were used to express Suf proteins fused to the DNA binding protein LexA and to the transcriptional activation motif B42, respectively. The sufB insert was obtained by PCR using plasmid pSUF as a template and primer 2s/2as (Table IV). The sufC and sufD inserts were amplified using E.chrysanthemi chromosomal DNA as a template and primer 3s/3as and 4s/4as, respectively (Table IV). The sufB and sufC inserts were digested by EcoRI–XhoI, and the sufD insert by MunI–XhoI. The restricted suf genes containing inserts were cloned into the EcoRI–XhoI-digested pEG202 and pJG4-5, yielding the pB42 and pLexA plasmid series, respectively.
The pBAD24 plasmid, containing the PBAD arabinose-inducible promoter, was used to express HA-tagged SufB, SufC and SufD proteins. The ha-sufB, ha-sufC and ha-sufD inserts were obtained by PCR amplification using pB42-B, pB42-C and pB42-D as templates, and oligonucleotides NcoHA/2as, NcoHA/3as and NcoHA/4as, respectively (Table V). The ha-sufC and ha-sufD PCR products were NcoI–XhoI digested and inserted into NcoI–SalI-digested pBAD24, yielding pA-C and pA-D, respectively. Insertion of ha-sufB into pBAD24 was realized in two steps. First, ha-sufB PCR product was digested by NcoI and XhoI, yielding a 5′-NcoI–NcoI fragment containing the HA encoding sequence and the 5′ part of sufB, and a 3′-NcoI–XhoI fragment containing the 3′ part of sufB. This latter fragment was inserted into NcoI–SalI-digested pBAD24, yielding pA-ΔB. The former fragment containing HA sequence was inserted into NcoI-digested pA-ΔB, yielding pA-B.
Table V. Sequence of oligonucleotides used for strain and plasmid constructions.
| Oligonucleotide | 5′–3′ sequence |
|---|---|
| 5Δsuf | ACATGCTGTTATACGCTGAAAGCGATGAAGTGAGGTAAATCGCATATGAATATCCTCCTTA |
| 3Δsuf | CCATCCGGCAATGTGAGCCAACCGGATGAAAGCTGTCCTTTTAGTGTGTAGGCTGGAGCTGCTTC |
| 2s | GCATCGAATTCATGGCGCGTAGCAATGTAGACG |
| 2as | GCCTTCTCGAGTCAGCCCACACTGTGTTCG |
| 3s | GCATCGAATTCATGTTAACGATTGAAAACTTG |
| 3as | GCCTTCTCGAGTTACTGTTGGTCGGTAAGCC |
| 4s | GCATCCAATTGATGGCTGGCTTACCGACCAAC |
| 4as | GCCTTCTCGAGTCACGCTGTTTCTCCTCCC |
| NdeI-3s | GGGAATTCCATATGTTAACGATTGAAAACTTG |
| XhoI-3as | GCCTTCTCGAGCTGTTGGTCGGTAAGC |
| NcoHA | CCCTACCATGGCTTACCCATATGATGTGCCAGATTATG |
In order to overproduce the SufC-His6 protein, the sufC gene was first cloned into pET22b+. The sufC insert was obtained by PCR amplification using oligonucleotides NdeI-3s/XhoI-3as and NdeI–XhoI digestion. The restricted fragment was inserted into pET22b+ digested by the same enzymes, yielding pET-CHis. In order to have sufChis expression under the control of the pBAD promoter, the insert, obtained by XbaI–XhoI digestion of pET-Chis, was inserted in the NheI–XhoI-digested pBADIK, yielding pK-CHis.
In order to overproduce MBP–SufB, MBP–SufC and MBP–SufD chimeric proteins, the sufB, sufC and sufD genes were cloned into the pMal-c2 vector downstream of the Ptac promoter. The sufB and sufC inserts were obtained by EcoRI–XhoI digestion of the pB42-B and pB42-C plasmid, respectively. The restricted fragments were inserted into EcoRI–SalI-digested pMal-c2, yielding pMal-SufB and pMal-SufC. The sufD insert was obtained by PCR amplification using pA-D as template and the oligonucleotides 4S and 4as. The sufD insert was MunI–XhoI digested and inserted into EcoRI–SalI-digested pMal-c2, yielding the pMal-SufC plasmid.
Purification of SufC-His6
TG1 cells harbouring pK-CHis were grown in LB medium up to an OD600 of 0.35. l-arabinose 0.02% (w/v) was added and cultures were incubated for 4 h at 30°C. Cells were harvested, then disrupted twice by French pressure treatment in 30 ml of buffer A (100 mM Tris pH 7.5, 50 mM NaCl). After centrifugation, most of the SufC-His6 protein was recovered in the insoluble fraction. The pellet was suspended in 50 ml of buffer A supplemented with 8 M urea, and stored on ice for 60 min with gentle shaking. The suspension was centrifuged for 60 min at 17 000 g. SufC-His6 was detected in the supernatant using Coomassie Blue staining of SDS–PAGE. The SufC-His6-containing supernatant was diluted in 200 ml of buffer A, then dialysed against buffer A and applied to a 1 ml Ni2+ affinity column (Pharmacia Biotech). After washing the column with 50 ml of buffer A containing 40 mM imidazole, SufC-His6 was eluted with a 40–500 mM imidazole gradient. Fractions containing SufC-His6 were pooled and dialysed against buffer A. Purity of SufC-His6 was estimated by Coomassie Blue staining of SDS–PAGE.
Immunization
Anti-SufC-His6 antibodies were raised in a New Zealand rabbit by an initial injection with 200 µg of affinity-purified SufC-His6, and followed by boosters of 100 µg at monthly intervals. Polyclonal anti-SufC-His6 antibodies were then purified against an E.coli MG1655sufC strain lysate as described previously (Sambrook et al., 1989).
Purification of MBP hybrid proteins
Escherichia coli TG1 cells transformed with pMal-SufC were grown at 37°C in 200 ml of LB medium to an OD600 of 1.0. Induction was performed with 0.1 mM IPTG, which was added for 2.5 h at 37°C. The bacterial pellet was resuspended in 2 ml of buffer B (20 mM Tris–HCl, 200 mM NaCl, 1 mM EDTA, 10 mM 2-mercaptoethanol pH 7.5), and disrupted twice by French pressure treatment. The cell lysate was centrifuged at high speed for 15 min at 4°C. Supernatant (200 µl) was loaded at a flow rate of 0.5 ml/min onto a Superdex 200 HR 10/30 column (Pharmacia Biotech). Fractions of 0.5 ml were analysed by immunoblotting using anti-SufC antibody and assayed for ATP hydrolysis.
ATPase activity and MBP–SufC protein were found within fractions corresponding to the void volume. These fractions were pooled and added to 1 ml of amylose resin (New England Biolabs). The mixture was stored for 18 h at 4°C. The resin was then washed with 10 resin vols of buffer B. The recombinant MBP–SufC protein was eluted with buffer B added with 2 mM maltose.
The MG1655Δsuf strains transformed with pMal-SufB, pMal-SufD or pMalc2 were grown at 30°C in 400 ml of LB medium to an OD600 of 1.0. Induction was performed with 0.1 mM IPTG for 5 h at 30°C. Then, bacteria were harvested by centrifugation. The bacterial pellet was resuspended in 15 ml of buffer A and disrupted twice by French pressure treatment. The cell lysate was centrifuged at high speed for 15 min at 4°C. Fifteen millilitres of supernatant were added to 1 ml of amylose resin (New England Biolabs). The mixture was stored for 18 h at 4°C. The resin was then washed with 30 resin vols of buffer A. The recombinant proteins, MBP–SufB, MBP–SufD and MBP–LacZ, were eluted with buffer A added with 2 mM maltose. Eluted solution (200 µl) was loaded at a flow rate of 0.5 ml/min onto a Superdex 200 HR 10/30 column (Pharmacia Biotech). Fractions of 0.5 ml were analysed by Coomassie Blue staining and immunoblotting using anti-MBP antibodies (New England Biolabs). Purity of proteins was checked by Coomassie Blue staining of SDS–PAGE.
Isolation of SufC-His6-associated proteins by affinity chromatography
Thirty micrograms of purified SufC-His6 were mixed with 30 µg of purified MBP–LacZα, MBP–SufB and MBP–SufD proteins in the presence of 1 ml of amylose resin. After 18 h at 4°C, the mixture was loaded onto a column (Bio-Spin Disposable Chromatography Columns; Bio-Rad) and washed with 8 resin vols of buffer A. Elution was performed with buffer A containing 2 mM maltose. Fractions corresponding to flowthrough, washing and elution were analysed by SDS– PAGE and visualized by Coomassie Blue staining and immunoblotting.
ATPase assay
ATPase activities were tested by measuring the amounts of Pi released by ATP hydrolysis with the use of Malachite Green reagent (Morbach et al., 1993). During the purification procedures, ATPase assays were performed in 25 mM Tris buffer pH 7.5 in the presence of 5 mM MgCl2 and 0.5 mM ATP at 37°C during 30 min. The reaction was stopped by the addition of 50 mM EDTA. For further characterization, purified MBP–SufC proteins were assayed for ATPase activity in the presence of 5 mM MgCl2, 5 mM MnCl2 or 5 mM CaCl2. The kinetic activities of purified MBP–SufC were tested at pH 7.5, in the presence of 5 mM MgCl2, over a range of ATP concentrations (0.05–2 mM), allowing Vm and Km values to be determined using Graphpad Prism software.
Cell fractionation procedures
Erwinia chrysanthemi A3559 strain was grown overnight in LB medium at 30°C. When plasmids pA-B, pA-C or pA-D were used, ampicillin was added to the medium. Cultures were used to inoculate fresh LB medium at an OD600 of 0.35. After growth for 1 h, 0.02% l-arabinose (w/v) was added and cultures were incubated for 4 h at 30°C. Cultures were then divided into two equal samples. On one hand, cells were pelleted and resuspended in 1.5 ml of Tris buffer (40 mM, pH 7.5), allowing for the estimation of the amount of Suf proteins present in the cells. On the other hand, spheroplasts were prepared (in 1.5 ml of Tris buffer) as described by Bortoli-German et al. (1994). After centrifugation (10 000 r.p.m. for 10 min at 4°C), periplasmic fractions containing supernatants were stored at 4°C. Spheroplasts were washed, resuspended in 1.5 ml of Tris buffer (40 mM, pH 7.5) and disrupted by French pressure treatment. After centrifugation (15 000 r.p.m. for 15 min at 4°C), supernatants were subjected to ultracentrifugation at 45 000 r.p.m. for 1.5 h at 4°C. The resulting supernatants corresponded to cytosol. The membranes were resuspended in 1.5 ml of Tris buffer (40 mM, pH 7.5). SufC protein was detected by immunoblotting using polyclonal antibody raised against SufC; HA-tagged Suf proteins were detected using antibody raised against the HA epitope. For each location experiment, the efficiency and reliability of the cell fractionation procedure were checked using antibody raised against Cel5, OutF and MsrA.
Yeast two-hybrid system
The yeast two-hybrid system assay was performed as described by Golemis et al. (1994). The β-galactosidase activity from diploid cells, obtained by mating of strains EGY48 and RF206 carrying the appropriate plasmid, was detected on plates containing X-gal (Golemis et al., 1994) and was quantified from liquid culture. The β-galactosidase activity was expressed as nanomoles of o-nitrophenyl-β-d-galactoside hydrolysed per minute per milligram of protein. The protein concentration in cell extracts was determined by Coomassie Blue colorimetric assay (Bio-Rad). The diploid cells were also assayed for the expression of the Leu reporter gene. Diploid strains were grown in liquid selective CM medium lacking leucine. The OD600 values were measured after overnight growth. Differences in OD600 values between cultures grown in inducible (galactose + raffinose) and non-inducible (glucose) conditions were calculated. The presence of hybrid proteins in cell extracts was checked by immunoblotting using monoclonal antibodies raised against either HA or LexA moieties.
Enzymatic assays
Cultures for assays of enzymatic activities were grown in media that would ensure consistent synthesis of the relevant enzymes. During exponential growth phase, cultures were divided into two fractions, one of which was treated with 15 µM PMS. Growth was continued for 30 min. Cells were harvested by centrifugation, resuspended in appropriate buffer and disrupted by French pressure treatment. The lysis buffer used for 6-PGDH and PGM assays contained 50 mM Tris buffer pH 7.5 and 10 mM MgCl2, while that for fumarase assays contained 50 mM potassium phosphate pH 7.5 and that for glutamate synthase assays contained 100 mM Tris–HCl pH 7.5. After centrifugation (15 000 r.p.m. for 15 min at 4°C), the enzymatic assays were performed on supernatant. Activity of 6-PGDH was assayed by the two-step procedure for the determination of pyruvate produced (Fraenkel and Horecker, 1964). PGM activity was determined by the reduction of NADP in the presence of glucose-1-phosphate and glucose-6-phosphate dehydrogenase (Bergmeyer, 1986). Total fumarase activity was assayed by the conversion of 50 mM malate to fumarate. Superoxide-resistant fumarase C activity was measured after fumarase A activity had been inactivated by incubation of the clarified extract with xanthine oxidase and xanthine (Gort and Imlay, 1998). Glutamate synthase activity was determined by the oxidation of NADPH in the presence of l-glutamine and α-ketoglutarate (Meister, 1985). The protein concentration in cell extracts was determined by Coomassie Blue colorimetric assay (Bio-Rad). Activity of enzymes was expressed as nanomoles of product per minute per milligram of protein.
Growth assays
Escherichia coli wt and sufC strains were grown overnight in LB medium at 37°C. Cultures were used to inoculate minimal A medium containing 0.2% gluconate or 0.2% glycerol, and 1 µg/ml thiamine. During exponential growth phase, PMS was added at a final concentration of 15 µM and growth was followed by measuring OD600 values of the culture.
Cross-feeding assay
Utilization of ferric chrysobactin by the tested strains was determined in a bioassay under low-iron conditions as described previously (Rauscher et al., 2002), with modifications. Plates were poured with 15 ml of L-agar containing the chelator 2,2′-dipyridyl (200 µM) and the strain to be tested at a final concentration of 104 c.f.u./ml. Sterile disks of 6 mm diameter were placed on the agar surface and 10 µl of chrysobactin, corresponding to 50 µM dihydroxybenzoic acid equivalents, were added. Controls were carried out with 10 µl of FeCl3 (10 mM). The assay was positive when bacterial growth was visible around the disk after 24 h of incubation. Diameters of growth zones were measured.
Acknowledgments
Acknowledgements
Thanks are due to all members of the F.B. group for fruitful discussions. Thanks are due to S.Ollagnier (CEA Grenoble), B.Holland (Orsay) and G.Storz (NIH Bethesda) for valuable discussions. We are grateful to V.Méjean (Marseille) and T.Nyström (Sweden) for their critical evaluation of these experiments and careful reading of the manuscript. Thanks to D.Moinier for mass spectrometry analysis. This work was funded by Centre National de la Recherche Scientifique and Université of Aix-Marseille II. L.N. was a recipient of a fellowship from Ministère de la Recherche.
References
- Beinert H. and Kiley,P.J. (1999) Fe–S proteins in sensing and regulatory functions. Curr. Opin. Chem. Biol., 3, 152–157. [DOI] [PubMed] [Google Scholar]
- Bergmeyer H.U. (1986) Methods in Enzymatics Analysis, 3rd edn. Verlag Chemie, Weinheim, Germany.
- Bortoli-German I., Brun,E., Py,B., Chippaux,M. and Barras,F. (1994) Periplasmic disulphide bond formation is essential for cellulase secretion by the plant pathogen Erwinia chrysanthemi. Mol. Microbiol., 11, 545–553. [DOI] [PubMed] [Google Scholar]
- Bouley J., Condemine,G. and Shevchik,V.E. (2001) The PDZ domain of OutC and the N-terminal region of OutD determine the secretion specificity of the type II out pathway of Erwinia chrysanthemi. J. Mol. Biol., 27, 205–219. [DOI] [PubMed] [Google Scholar]
- Datsenko K.A. and Wanner,B.L. (2000) One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl Acad. Sci. USA, 97, 6640–6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Hassouni M., Chambost,J.P., Expert,D., Van Gijsegem,F. and Barras,F. (1999) The minimal gene set member msrA, encoding peptide methionine sulfoxide reductase, is a virulence determinant of the plant pathogen Erwinia chrysanthemi. Proc. Natl Acad. Sci. USA, 96, 887–892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis K.E., Clough,B., Saldanha,J.W. and Wilson,R.J. (2001) Nifs and Sufs in malaria. Mol. Microbiol., 41, 973–981. [DOI] [PubMed] [Google Scholar]
- Fath M.J. and Kolter,R. (1993) ABC transporters: bacterial exporters. Microbiol. Rev., 57, 995–1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finley R.L. and Brent,R. (1994) Interaction mating reveals binary and ternary connections between Drosophila cycle regulators. Proc. Natl Acad. Sci. USA, 91, 12980–12984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flint D.H. (1996) Escherichia coli contains a protein that is homologous in function and N-terminal sequence to the protein encoded by the nifS gene of Azotobacter vinelandii and that can participate in the synthesis of the Fe–S cluster of dihydroxy-acid dehydratase. J. Biol. Chem., 271, 16068–16074. [PubMed] [Google Scholar]
- Flint D.H., Smyk-Randall,E., Tuminello,J.E., Draczynska-Lusiak,B. and Brown,O.R. (1993) The inactivation of dihydroxy-acid dehydratase in Escherichia coli treated with hyperbaric oxygen occurs because of the destruction of its Fe–S cluster, but the enzyme remains in the cell in a form that can be reactivated. J. Biol. Chem., 268, 25547–25552. [PubMed] [Google Scholar]
- Fraenkel D.G. and Horecker,B.L. (1964) Pathway of d-glucose metabolism in Salmonella typhimurium. A study of a mutant lacking phosphoglucose isomerase. J. Biol. Chem., 239, 2765–2771. [PubMed] [Google Scholar]
- Gardner P.R. and Fridovich,I. (1991) Superoxide sensitivity of the Escherichia coli 6-phosphogluconate dehydratase. J. Biol. Chem., 266, 1478–1483. [PubMed] [Google Scholar]
- Garland S.A., Hoff,K., Vickery,L.E. and Cizewski-Culotta,V. (1999) Saccharomyces cerevisiae ISU1 and ISU2: members of a well-conserved gene family for iron–sulfur cluster assembly. J. Mol. Biol., 294, 897–907. [DOI] [PubMed] [Google Scholar]
- Golemis E.A., Gyuris,J. and Brent,R. (1994) Interaction trap/two-hybrid system to identify interacting proteins. In Ausubel,F.M., Brent,R., Kingston,R.E., Moore,D.D., Seidman,J.G., Smith,J.A. and Struhl,K. (eds), Current Protocols in Molecular Biology, Unit 13.14. John Wiley and Sons, NY.
- Gort A.S. and Imlay,J.A. (1998) Balance between endogenous superoxide stress and antioxidant defenses. J. Bacteriol., 180, 1402–1410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green J., Bennett,B., Jordan,P., Ralph,E.T., Thomson,A.J. and Guest,J.R. (1996) Reconstitution of the [4Fe–4S] cluster in FNR and demonstration of the aerobic–anaerobic transcription switch in vitro. Biochem. J., 316, 887–892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guzman L.M., Belin,D., Carson,M.J. and Beckwith,J. (1995) Tight regulation, modulation and high-level expression by vectors containing the arabinose PBAD promoter. J. Bacteriol., 177, 4121–4130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hidalgo E. and Demple,B. (1996) Activation of SoxR-dependent transcription in vitro by noncatalytic or NifS-mediated assembly of [2Fe–2S] clusters into apo-SoxR. J. Biol. Chem., 271, 7269–7272. [DOI] [PubMed] [Google Scholar]
- Hoff K.G., Silberg,J.J. and Vickery,L.E. (2000) Interaction of the iron–sulfur cluster assembly protein IscU with the Hsc66/Hsc20 molecular chaperone system of Escherichia coli. Proc. Natl Acad. Sci. USA, 97, 7790–7795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holland I.B. and Blight,M. (1999) ABC-ATPases, adaptable energy generators fuelling transmembrane movement of a variety of molecules in organisms from bacteria to humans. J. Mol. Biol., 293, 381–399. [DOI] [PubMed] [Google Scholar]
- Hopfner K.P., Karcher,A., Shin,D.S., Craig,L., Arthur,L.M., Carney,J.P. and Tainer,J.A. (2000) Structural biology of Rad50 ATPase: ATP-driven conformational control in DNA double-strand break repair and the ABC-ATPase superfamily. Cell, 101, 789–800. [DOI] [PubMed] [Google Scholar]
- Johnson M.K. (1998) Iron–sulfur proteins: new roles for old clusters. Curr. Opin. Chem. Biol., 2, 173–181. [DOI] [PubMed] [Google Scholar]
- Junop M.S., Obmolova,G., Rausch,K., Hsieh,P. and Yang,W. (2001) Composite active site of an ABC ATPase: MutS uses ATP to verify mismatch recognition and authorize DNA repair. Mol. Cell, 7, 1–12. [DOI] [PubMed] [Google Scholar]
- Kaut A., Lange,H., Diekert,K., Kispal,G. and Lill,R. (2000) Isa1p is a component of the mitochondrial machinery for maturation of the cellular iron–sulfur proteins and requires conserved cysteine residues for function. J. Biol. Chem., 275, 15955–15961. [DOI] [PubMed] [Google Scholar]
- Kim R., Saxena,S., Gordon,D.M., Pain,D. and Dancis,A. (2001) J-domain protein, Jac1p, of yeast mitochondria required for iron homeostasis and activity of Fe–S cluster proteins. J. Biol. Chem., 276, 17524–17532. [DOI] [PubMed] [Google Scholar]
- Lange H., Kaut,A., Kispal,G. and Lill,R. (2000) A mitochondrial ferredoxin is essential for biogenesis of cellular iron–sulfur proteins. Proc. Natl Acad. Sci. USA, 97, 1050–1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law A.E., Mullineaux,C.W., Hirst,E.M.A., Saldanha,J. and Wilson,R.J.M. (2000) Bacterial orthologues indicate the malarial plastid gene ycf24 is essential. Protist, 151, 317–327. [DOI] [PubMed] [Google Scholar]
- Linton K.J. and Higgins,C.F. (1998) The Escherichia coli ATP-binding cassette (ABC) proteins. Mol. Microbiol., 28, 5–13. [DOI] [PubMed] [Google Scholar]
- Lowe J., Cordell,S.C. and Van den Ent,F. (2001) Crystal structure of the SMC head domain: an ABC ATPase with 900 residues antiparallel coiled-coil inserted. J. Mol. Biol., 306, 25–35. [DOI] [PubMed] [Google Scholar]
- Lundblad V. (1993) Saccharomyces cerevisiae. In Ausubel,F.M., Brent,R., Kingston,R.E., Moore,D.D., Seidman,J.G., Smith,J.A. and Struhl,K. (eds), Current Protocols in Molecular Biology, Unit 13. John Wiley and Sons, NY.
- Mansy S.S., Wu,G., Surerus,K.K. and Cowan,J.A. (2002) Iron–sulfur cluster biosynthesis. Thermatoga maritima IscU is a structured iron–sulfur cluster assembly protein. J. Biol. Chem., 277, 21397–21404. [DOI] [PubMed] [Google Scholar]
- Masclaux C. and Expert,D. (1995) Signalling potential of iron in plant–microbe interactions: the pathogenic switch of iron transport in Erwinia chrysanthemi. Plant J., 7, 121–128. [Google Scholar]
- Mihara H., Kurihara,T., Yoshimura,T. and Esaki,N. (2000) Kinetic and mutational studies of three NifS homologs from Escherichia coli: mechanistic difference between l-cysteine desulfurase and l-selenocysteine lyase reactions. J. Biochem., 127, 559–567. [DOI] [PubMed] [Google Scholar]
- Meister A. (1985) Glutamate synthase from Escherichia coli, Klebsiella aerogenes and Saccharomyces cerevisiae. Methods Enzymol., 113, 327–337. [DOI] [PubMed] [Google Scholar]
- Miller J.H. (1972) A Short Course in Bacterial Genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Moller S.G., Kunkel,T. and Chua,N.H. (2001) A plastidic ABC protein involved in intercompartmental communication of light signaling. Genes Dev., 15, 90–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morbach S., Tebbe,S. and Schneider,E. (1993) The ATP-binding cassette (ABC) transporter for maltose/maltodextrins of Salmonella typhimurium. Characterization of the ATPase activity associated with the purified MalK subunit. J. Biol. Chem., 268, 18617–18621. [PubMed] [Google Scholar]
- Nachin L., El Hassouni,M., Loiseau,L., Expert,D. and Barras,F. (2001) SoxR-dependent response to oxidative stress and virulence of Erwinia chrysanthemi: the key role of SufC, an orphan ABC ATPase. Mol. Microbiol., 39, 960–972. [DOI] [PubMed] [Google Scholar]
- Ollagnier-de-Choudens S., Mattioli,T., Takahashi,Y. and Fontecave,M. (2001) Iron–sulfur cluster assembly: characterization of IscA and evidence for a specific and functional complex with ferredoxin. J. Biol. Chem., 276, 22604–22607. [DOI] [PubMed] [Google Scholar]
- Patzer S.I. and Hantke,K. (1999) SufS is a NifS-like protein and is necessary for stability of the [2Fe–2S] FhuF protein in Escherichia coli. J. Bacteriol., 181, 3307–3309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelzer W., Mühlenhoff,U., Diekert,K., Siegmund,K., Kispal,G. and Lill,R. (2000) Mitochondrial Isa2p plays a crucial role in the maturation of cellular iron–sulfur proteins. FEBS Lett., 476, 134–139. [DOI] [PubMed] [Google Scholar]
- Py B., Loiseau,L. and Barras,F. (2001) An inner membrane platform in the type II secretion machinery of Gram-negative bacteria. EMBO rep., 2, 244–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quentin Y., Fichant,G. and Denizot,F. (1999) Inventory, assembly and analysis of Bacillus subtilis ABC transport systems. J. Mol. Biol., 287, 467–484. [DOI] [PubMed] [Google Scholar]
- Rangachari K., Davis,C.T., Eccleston,J.F., Hirst,E.M.A., Saldanha,J.W., Strath,M. and Wilson,R.J.M. (2002) SufC hydrolyzes ATP and interacts with SufB from Thermotoga maritima. FEBS Lett., 74, 225–228. [DOI] [PubMed] [Google Scholar]
- Rauscher L., Expert,D., Matzanke,B.F. and Trautwein,A.X. (2002) Chrysobactin-dependent iron acquisition in Erwinia chrysanthemi: functional study of a homolog of the Escherichia coli ferric enterobactin esterase. J. Biol. Chem., 277, 2385–2395. [DOI] [PubMed] [Google Scholar]
- Sambrook J., Fritsch,E.F. and Maniatis,T. (1989) Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Schilke B., Voisine,C., Beinert,H. and Craig,E. (1999) Evidence for a conserved system for iron metabolism in the mitochondria of Saccharomyces cerevisiae. Proc. Natl Acad. Sci. USA, 96, 10206–10211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider E. and Hunke,S. (1998) ATP-binding-cassette (ABC) transport systems: functional and structural aspects of the ATP-hydrolyzing subunits/domains. FEMS Microbiol. Rev., 22, 1–20. [DOI] [PubMed] [Google Scholar]
- Schwartz C.J., Djaman,O., Imlay,J.A. and Kiley,P.J. (2000) The cysteine desulfurase, IscS, has a major role in in vivo Fe–S cluster formation in Escherichia coli. Proc. Natl Acad. Sci. USA, 97, 9009–9014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tachezy J., Sanchez,L.B. and Müller,M. (2001) Mitochondrial type iron sulfur cluster assembly in the amitochondrial eucaryotes Trichomonas vaginalis and Giardia intestinalis, as indicated by the phylogeny of iscS. Mol. Biol. Evol., 18, 1919–1928. [DOI] [PubMed] [Google Scholar]
- Takahashi Y. and Nakamura,M. (1999) Functional assignment of the ORF2-iscS-iscU-iscA-hscB-hscA-fdx-ORF3 gene cluster involved in the assembly of Fe–S clusters in Escherichia coli. J. Biochem., 126, 917–926. [DOI] [PubMed] [Google Scholar]
- Takahashi Y. and Tokumoto,U. (2002) A third bacterial system for the assembly of iron–sulfur clusters with homologs in archaea and plastids. J. Biol. Chem., 277, 28380–28383. [DOI] [PubMed] [Google Scholar]
- Tokumoto U. and Takahashi,Y. (2001) Genetic analysis of the isc operon in Escherichia coli involved in the biogenesis of cellular iron–sulfur proteins. J. Biochem., 130, 63–71. [DOI] [PubMed] [Google Scholar]
- Tse Sum Bui B., Escalettes,F., Chottard,G., Florentin,D. and Marquet,A. (2000) Enzyme-mediated sulfide production for the reconstitution of [2Fe–2S] clusters into apo-biotin synthase of Escherichia coli. Sulfide transfer from cysteine to biotin. Eur. J. Biochem., 267, 2688–2694. [DOI] [PubMed] [Google Scholar]
- Urbina H.D., Silberg,J.J., Hoff,K.G. and Vickery,L.E. (2001) Transfer of sulfur from IscS to IscU during Fe/S cluster assembly. J. Biol. Chem., 276, 44521–44526. [DOI] [PubMed] [Google Scholar]
- Voisine C., Schilke,B., Ohlson,M., Beinert,H., Marszalek,J. and Craig,E.A. (2000) Role of the mitochondrial Hsp70s, Ssc1 and Ssq1, in the maturation of Yfh1. Mol. Cell. Biol., 20, 3677–3684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng L. and Dean,D.R. (1994) Catalytic formation of a nitrogenase iron–sulfur cluster. J. Biol. Chem., 269, 18723–18726. [PubMed] [Google Scholar]
- Zheng M., Wang,X., Templeton,L.J., Smulski,D.R., LaRossa,R.A. and Storz,G. (2001) DNA microarray-mediated transcriptional profiling of the Escherichia coli response to hydrogen peroxide. J. Bacteriol., 183, 4562–4570. [DOI] [PMC free article] [PubMed] [Google Scholar]