Abstract
The endocytic pathway in yeast leads to the vacuole, but resident proteins of the late Golgi, and some endocytosed proteins such as the exocytic SNARE Snc1p, are retrieved specifically to the Golgi. Retrieval can occur from both a late pre-vacuolar compartment and early or ‘post-Golgi’ endosomes. We show that the endosomal SNARE Pep12p, and a mutant version that reaches the cell surface and is endocytosed, are retrieved from pre-vacuolar endosomes. As with Golgi proteins, this requires the sorting nexin Grd19p and components of the retromer coat, supporting the view that endosomal and Golgi residents both cycle continuously between the exocytic and endocytic pathways. In contrast, retrieval of Snc1p from post-Golgi endosomes requires the sorting nexin Snx4p, to which Snc1p can be cross-linked. Snx4p binds to Snx41p/ydr425w and to Snx42p/ydl113c, both of which are also required for efficient Snc1p sorting. Our findings suggest a general role for yeast sorting nexins in protein retrieval, rather than degradation, and indicate that different sorting nexins operate in different classes of endosomes.
Keywords: endosome/grd19/pep12/snx4/sorting nexin
Introduction
Elucidating the ways in which proteins are localized to particular cellular compartments is a crucial step in understanding cellular organization. The organelles of the endomembrane system present a particular challenge. They are connected by a complex network of vesicular trafficking pathways, and the steady-state distribution of membrane proteins between them is determined by the kinetics with which these proteins move along the various pathways. The routes the proteins follow can be deduced from the effects of mutations, in particular those that disrupt sorting signals required for entry into a particular pathway. A major goal, therefore, is to define the organelles and pathways, the sorting signals on proteins and the machinery that recognizes them.
In yeast cells, the endocytic pathway leads to the vacuole, which shares the degradative properties of animal cell lysosomes. Once delivered to endosomes, from either the plasma membrane or the Golgi apparatus, integral membrane proteins require no particular cytoplasmic signals to proceed to the vacuole. Thus, removal of the cytoplasmic tails from proteins that reside in endosomes or the late Golgi results in their transfer to the vacuole. Such proteins are normally saved from this fate by a specific retention or retrieval process, and much evidence suggests that they are recycled actively from endosomes to the Golgi (reviewed by Conibear and Stevens, 1998). The retrieved proteins include receptors and enzymes that cycle between the Golgi and endosomes, and some endocytosed proteins such as the SNARE Snc1p and the chitin synthase Chs3p (Conibear and Stevens, 1998; Holthuis et al., 1998; Lewis et al., 2000; Valdivia et al., 2002).
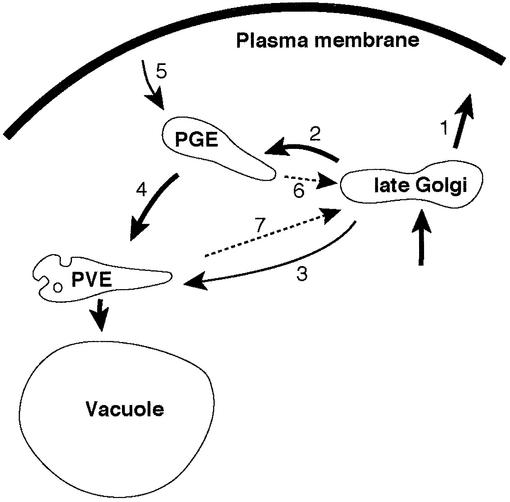
At least two distinct retrieval pathways have been identified. One originates in early or ‘post-Golgi’ endosomes (PGEs; Pelham, 2002), and is the route followed normally by Snc1p, Chs3p and probably by the late Golgi SNAREs Tlg1p and Tlg2p (step 6 in Figure 1) (Holthuis et al., 1998; Lewis et al., 2000; Valdivia et al., 2002). Another comes from late or ‘pre-vacuolar’ endosomes (PVEs; step 7), and is the pathway used by the carboxypeptidase Y sorting receptor Vps10p (Piper et al., 1995). These pathways can be distinguished in class E vacuolar protein sorting (vps) mutants, which accumulate multilamellar structures corresponding to abnormally enlarged PVEs (Rieder et al., 1996; Babst et al., 1997). In such mutants, retrieval from PVEs is inefficient, but recycling from PGEs continues (Lewis et al., 2000). However, many proteins, including Chs3p and the late Golgi proteins Kex2p and Ste13p, appear to carry multiple sorting signals that allow them to travel via either route, which complicates analysis (Brickner and Fuller, 1997; Bryant and Stevens, 1997; Valdivia et al., 2002).
Fig. 1. Trafficking pathways discussed herein. From the late Golgi, proteins can pass to the plasma membrane (1) or to post-Golgi endosomes (PGEs) (2) depending on the nature of their TMDs, or via the selective GGA protein-dependent pathway to pre-vacuolar endosomes (PVEs) (3). The FSD motif is required for Pep12p to follow this route. Proteins in PGEs can also pass, without special signals, to PVEs (4) and then to the vacuole. Snc1p is endocytosed to PGEs (5), then retrieved to the Golgi (6) by a mechanism that requires Snx4p. Proteins can also be retrieved from PVEs (7), a step that for Pep12p requires Grd19p. This is also the step mediated by retromer. Other pathways exist but have been omitted for clarity.
Signal-dependent sorting is usually the hallmark of protein coats, which gather particular subsets of proteins into transport carriers. There is evidence that two different kinds of clathrin coat can be found on endosomal membranes, at least in animal cells (Raiborg et al., 2002; Sachse et al., 2002), and also that COPI contributes to endosomal sorting (Daro et al., 1997; Piguet et al., 1999), but it remains unclear whether either clathrin or COPI is directly involved in retrieval in yeast. Both types of coat are also present on Golgi membranes, where they perform different sorting functions (Scales et al., 2000; Black and Pelham, 2001). Thus, even if the same coats are used, the cargo selection mechanism is likely to differ between Golgi and endosome.
One putative coat complex that seems to be involved specifically in retrieval from endosomes has been identified, namely the retromer. This consists of five proteins; Vps5p and Vps17p form a dimer, which associates with a complex of Vps26p, Vps29p and Vps35p (Seaman et al., 1998). All of these proteins are required for recycling of Vps10p from PVEs, but not for the retrieval of Snc1p from PGEs (Lewis et al., 2000). The retromer helps to retrieve a variety of late Golgi proteins, and there is good evidence that Vps35p recognizes sorting signals on the cytoplasmic tails of Ste13p and Vps10p (Nothwehr et al., 1999, 2000). Vps5p and Vps17p are examples of sorting nexins, peripheral membrane proteins originally identified in animal cells as proteins that bind to the cytoplasmic portions of receptor tyrosine kinases such as the epidermal growth factor receptor (Kurten et al., 1996). The functions of the sorting nexins have been somewhat unclear; they were postulated to mediate the down-regulation and degradation of receptors in animal cells, but the yeast data suggest instead that they may rescue proteins from degradation.
Sorting nexins can be defined as proteins that have a PX domain and a role in the sorting of membrane proteins. PX domains recently have been shown to bind lipids and to have specificity, in most cases, for the endosomal lipid phosphatidylinositol 3-phosphate (PI3P) (Bravo et al., 2001; Yu and Lemmon, 2001; for reviews, see Misra et al., 2001; Sato et al., 2001). Of the 15 PX domain proteins in yeast, five have known functions that do not involve cargo sorting in endosomes (Vam7p, Spo14p, Mdm1p, Bem1p and Bem3p). Besides the retromer components, two others have endosomal functions: Mvp1p is required for the efficient retrieval of Kex2p and Vps10p from endosomes (Chang and Fink, 1995; Ekena and Stevens, 1995), while Grd19p affects sorting of Kex2p and Ste13p, but not Vps10p (Voos and Stevens, 1998). The remaining proteins are largely uncharacterized, but all are candidates for additional sorting nexins.
In this study, we explore the roles of sorting nexins in the trafficking of two proteins that undergo endocytosis and then pass to the Golgi: the exocytic SNARE Snc1p and a mutated version of the endosomal SNARE Pep12p. We show that sorting of both the mutant Pep12p and of the endogenous wild-type protein is strongly dependent on Grd19p and retromer, which act in PVEs. In contrast, the sorting nexins Snx4p, Ydr425p (Snx41p) and Ydl113p (Snx42p) are required for recycling of Snc1p from PGEs. Thus, distinct sets of sorting nexins mediate retrieval from the two main types of yeast endosome. Snx4p and its companions are the clearest examples so far identified of proteins required for sorting in PGEs.
Results
Pep12p mutants that fail to be retrieved from endosomes
Pep12p normally travels from Golgi membranes to PVEs by a clathrin- and GGA protein-dependent pathway (step 3 in Figure 1). The wild-type protein is found in PVEs, but the localization mechanism is saturable: when overexpressed, a green fluorescent protein (GFP)-tagged version of Pep12p is found almost exclusively on the vacuolar membrane (Black and Pelham, 2000). By the introduction of appropriate mutations, Pep12p can be routed instead to PGEs either directly (step 2 in Figure 1) or via the plasma membrane (steps 1 and 5). In particular, the plasma membrane route is followed by a GFP-tagged derivative of Pep12p bearing the Sso1p transmembrane domain (TMD) and a mutation in the FSD motif that is required for sorting into the GGA pathway (Black and Pelham, 2000). In the steady state, this protein shows a punctate distribution that corresponds largely to PGEs, together with some plasma membrane fluorescence (Black and Pelham, 2000; see below, Figure 3A). We chose to study this version of Pep12p, since it successfully avoids accumulation in the vacuole and could thus yield information on how endocytosed proteins avoid this fate.
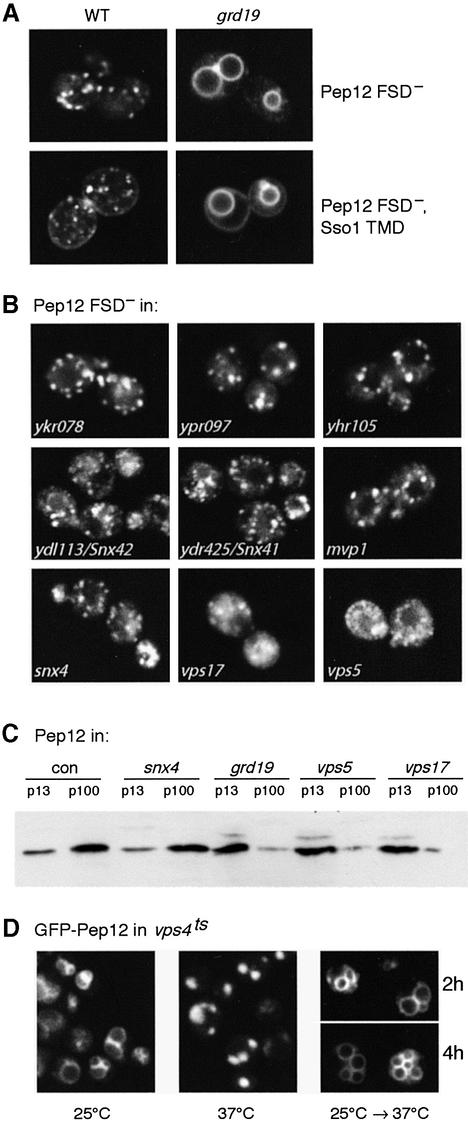
Fig. 3. Distribution of Pep12 derivatives in various mutants. (A) GFP–Pep12p is in punctate PGEs in wild-type cells (WT), whether it is delivered there directly from the Golgi (FSD–) or via the plasma membrane (FSD–, Sso1 TMD), but both constructs are on vacuolar membranes in grd19 cells. (B) Distribution of GFP-tagged FSD– Pep12p in deletion mutants of the nine sorting nexin mutants indicated. (C) Immunoblotting of endogenous Pep12p in medium-speed (p13) and high-speed pellet fractions (p100) of the indicated strains. (D) GFP–Pep12p in a vps4 temperature-sensitive mutant grown overnight at 25°C (only vacuoles are visible) or 37°C (pre-vacuolar structures). For the right hand panels, the protein was expressed from the GAL1 promoter for 5 h, expression stopped by shifting to glucose medium for 2 h, then the temperature shifted from 25 to 37°C and cells imaged 2 and 4 h later.
We randomly mutagenized the coding sequence of this Pep12p derivative by PCR, and selected variants that showed a vacuolar pattern by fluorescence microscopy. Two such mutants were analysed further. Both contained multiple mutations, but the phenotypic effect could be ascribed to those near the N-terminus. In one case, the effect was due to two simultaneous changes, F6L and I71T. Neither mutation alone was sufficient for mistargeting (Figure 2A), and indeed deletion of the first 20 residues of the protein also had no effect (data not shown). This suggests that either a patch on the surface of the protein is recognized, rather than a simple linear sequence, or the mutations have an indirect effect on the structure of the protein. The second mutant contained seven changes between positions 31 and 93, but we did not investigate these in detail.
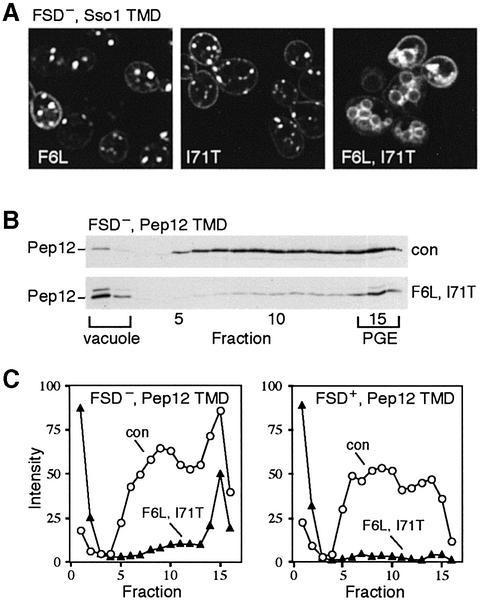
Fig. 2. Mutants that affect retrieval of Pep12p. (A) GFP–Pep12p bearing the Sso1p TMD and a mutant FSD signal (F20L) is in punctate PGE structures with either the F6L or I71T mutations, but with both it is on vacuolar membranes. (B) Sucrose gradient fractionation of Δpep12 cells expressing FSD– Pep12p, with its own TMD, with or without the F6L/I71T mutations. Fraction 1 is the top of the gradient. The positions of PGEs and vacuolar membranes, identified by the presence of Tlg1p and Vam3p, respectively (Black and Pelham, 2000), are indicated. PVEs, defined by Pep12p itself, run in the centre of the gradient. Note that the extracts were pre-cleared by medium-speed centrifugation, resulting in substantial under-representation of vacuoles on the gradients. (C) Quantitation of the indicated proteins in gradient fractions. The left hand panel corresponds to the gradients in (B).
We next examined the effects of the F6L/I71T mutations on Pep12p itself, expressed at wild-type levels in a pep12-null mutant. Initially, we continued to use a version of the protein mutated in the FSD motif, but with its own TMD. This protein is delivered directly from the Golgi to PGEs, without passing to the plasma membrane (step 2 in Figure 1; Black and Pelham, 2000). Even with the F6L/I71T changes, it complemented the growth and trafficking defects of the pep12 cells, indicating that it remained functional. Extracts from cells expressing Pep12p with or without the retrieval mutations were fractionated, and endosomal membranes remaining in the supernatant after centrifugation at 13 000 g were separated on sucrose density gradients (Figure 2B). On these gradients, PGE markers are found close to the bottom, while PVEs are spread through the centre; any vacuoles remaining after the 13 000 g centrifugation float to the top (Black and Pelham, 2000). The F6L/I71T mutations caused dramatic loss of Pep12p from PVEs, with a concomitant increase in the vacuolar fractions but, interestingly, loss from the PGE region was much less severe (Figure 2B and C). When the experiment was repeated using a version of Pep12p with the FSD motif intact, thus reducing delivery to PGEs and directing the protein instead to PVEs, the F6L/I71T mutations caused complete transfer to the vacuole (Figure 2C). Thus, although we had screened for loss from PGEs, the mutations we obtained affect sorting at the level of PVEs. Loss from PGEs may be secondary to this, reflecting a slow but steady delivery to PVEs.
Grd19p and retromer are required for sorting of Pep12p
To search for proteins responsible for the sorting of Pep12p, we systematically examined sorting nexin mutants. This approach revealed that Grd19p was required for retrieval of the GFP-tagged FSD– variant of Pep12p; in a grd19 mutant, the GFP pattern changed from punctate to vacuolar (Figure 3A). Similar results were obtained whether the construct had the Pep12p TMD or the Sso1p TMD, i.e. whether it reached PGEs from the Golgi or from the plasma membrane. Mutations in the retromer components Vps5p and Vps17p also affected the distribution of Pep12p, but the results were less clear, in part because vps5 and vps17 mutants have fragmented vacuoles. Removal of any of the other seven putative sorting nexins had no obvious effect on the distribution of tagged Pep12p (Figure 3B).
To confirm these results, we fractionated cells by differential centrifugation and examined the distribution of endogenous Pep12p, which is normally distributed between the 13 000 g pellet (p13) and the 100 000 g pellet (p100). Figure 3C shows that in grd19, vps5 and vps17 cells, Pep12p was almost exclusively in p13, where the vacuoles are found. Similar results were obtained with another retromer mutant, vps35 (data not shown). In contrast, the Pep12p distribution was unaltered in another sorting nexin mutant, snx4 (Figure 3C). Surprisingly, despite their mistargeting of Pep12p, grd19 cells have a normal vacuolar morphology and sort vacuolar hydrolases efficiently. This is, however, consistent with previous observations that quite low levels of Pep12p are sufficient to maintain endosomal function (Reggiori et al., 2000).
Interestingly, in grd19 cells and retromer mutants, a minor form of Pep12p with an apparent size 2–3 kDa larger than normal was present (Figure 3C). A similar minor form could also be detected with the F6L/I71T mutant protein, in both the vacuolar and PGE fractions (Figure 2B). The nature of this apparent modification is unclear; preliminary experiments showed little effect of phosphatase treatment, suggesting that it does not represent phosphorylation. The modification correlates with mis-sorting, but whether it contributes to this, or is merely a reflection of the altered state of Pep12p, remains to be determined.
Previously, retromer and Grd19p have been implicated in the recycling of proteins from PVEs to the Golgi, and it seems likely that Pep12p follows this same route. However, it has been reported that an artificial construct bearing both a vacuole targeting signal and Golgi retrieval signal can reach the pre-vacuolar compartment of a class E vps mutant via the vacuole (Bryant et al., 1998). Such a vacuole–endosome pathway could, in principle, also help to maintain Pep12p in endosomes. To test this, we overexpressed GFP–Pep12p in a temperature-sensitive class E mutant, vps4 (Babst et al., 1997). As expected, cells grown at 25°C showed vacuolar membrane fluorescence after overnight growth at 37°C whereas the pattern was that of punctate pre-vacuolar compartments (Figure 3D). When GFP–Pep12p was transiently expressed from the GAL1 promoter, and then cells shifted from 25 to 37°C, the pre-existing protein remained on vacuolar membranes for at least 4 h (Figure 3D), eventually disappearing as a result of dilution and/or degradation without ever showing the typical pattern of pre-vacuolar accumulation. Thus, under conditions where delivery is blocked, Pep12p leaves the vacuole very inefficiently, if at all. We conclude that the endosomal location of Pep12p depends primarily on retrieval from PVEs, rather than the vacuole.
Snx4p, Snx41p and Snx42p are required for retrieval of Snc1p
We next sought to determine whether sorting nexins are involved in other pathways of retrieval from endosomes. We specifically investigated SNAREs that are candidates for direct retrieval from PGEs, namely Tlg1p, Tlg2p and Snc1p. Figure 4A shows that the distribution of all of these was normal in grd19 cells. We tested these proteins in strains lacking each of the other putative sorting nexins (as listed in Figure 3B). The only effects we observed were with Snc1p, which was significantly mislocalized to the vacuole in snx4, ydr425 and ydl113 cells. Because of the similarity of their phenotype to that of snx4, and other data discussed below, we refer henceforth to ydr425w as snx41, and to ydl113c as snx42. In these strains, Snc1p usually was found inside the vacuoles (Figure 4B), indicating entry into the internal membranes of multivesicular bodies, as was observed previously for mislocalized mutant versions of Snc1p (Lewis et al., 2000). Some Snc1p remained on the plasma membrane, indicating that mis-sorting was not complete. This did not seem to be due simply to redundancy of the nexins, because a triple mutant snx4 snx41 snx42 still showed some residual Snc1p at the cell surface (Figure 4B), which is not seen in mutants that completely block the recycling pathway such as ypt6 (Siniossoglou et al., 2000).
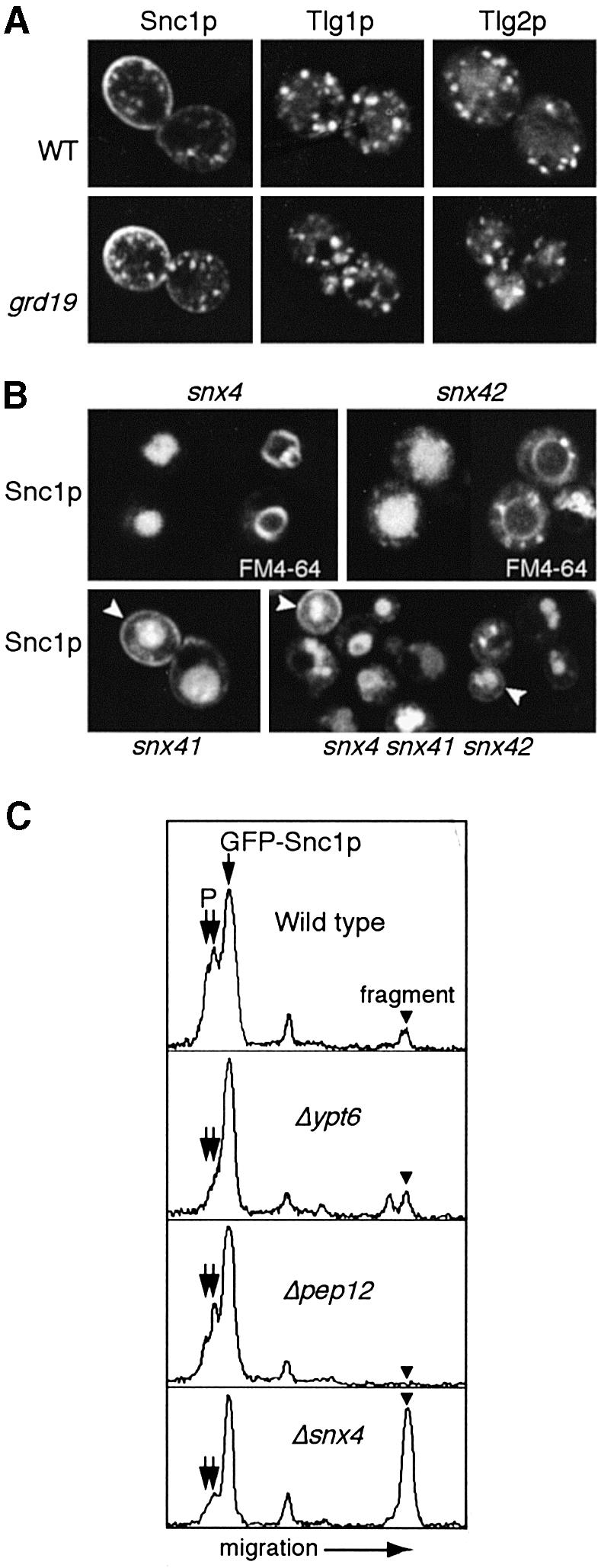
Fig. 4. Specific effects of different sorting nexin mutants. (A) The distribution of GFP-tagged Snc1p, Tlg1p and Tlg2p is the same in wild-type (WT) and grd19 cells. (B) GFP–Snc1p is mislocalized to the interior of the vacuole, identified in the upper panels by double labelling with FM4-64, in snx4, snx41 and snx42 cells. Some residual plasma membrane staining is apparent in some cells (arrowheads), and this is true even in an snx4 snx41 snx42 triple mutant (shown at lower magnification to illustrate the range of phenotypes). (C) Profiles of immunoblots of GFP–Snc1p in the indicated strains, detected with anti-GFP antibody. P indicates the phosphorylated forms, and the proteolytic fragment referred to in the text is also indicated. Note the reduction in phosphorylated forms and increased proteolysis of GFP–Snc1p in snx4 cells.
The effect of snx4 on Snc1p could also be observed by immunoblotting. GFP–Snc1p is phosphorylated on the plasma membrane, as revealed by the presence of more slowly migrating forms of the protein, and this has been used as a semi-quantitative assay for plasma membrane localization and hence recycling efficiency (Galan et al., 2001). Figure 4C shows that phosphorylated GFP–Snc1p could be detected in control cells, but not in a ypt6 mutant. It was still detectable in pep12 cells, consistent with continued recycling of Snc1p through PGEs even when delivery to PVEs is blocked. In snx4 cells, the phosphorylated species were reduced significantly, but the most striking effect was a large increase in a proteolytic fragment that is normally only present in minor amounts. This fragment evidently is generated in late endosomes or the vacuole, since it is absent when traffic to these is blocked by deletion of pep12 (Figure 4C). These data confirm that there is substantial mis-sorting of Snc1p in the snx4 mutant.
Despite the obvious effects on Snc1p, it was striking that recycling of Tlg1p and Tlg2p was not affected in snx4 cells. We also examined GFP–Chs3p, which is thought to recycle through PGEs, but this too was unaffected (data not shown). However, for none of these proteins has a specific retrieval signal yet been identified. A possible explanation is that there are multiple redundant signals, recognized by different receptors, and that removal of any one is not sufficient to perturb sorting.
Interactions between Snx41p, Snx42p and Snx4p
Snx41p and Snx42p are related throughout their sequence, and both show a more distant similarity to Snx4p. However, since all three are required for efficient sorting of Snc1p, and do not show an additive phenotype, they are not simply redundant. An alternative possibility is that they are components of a complex.
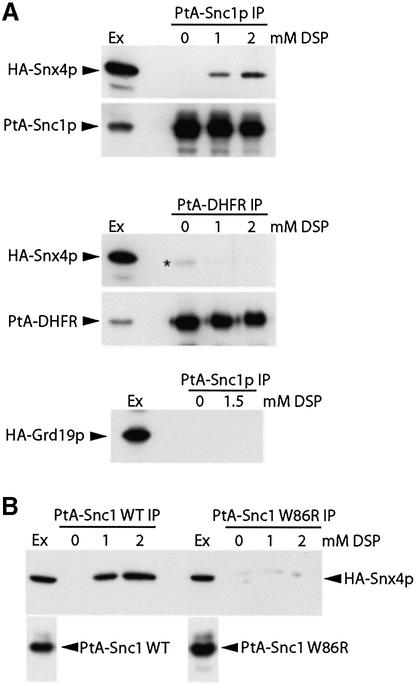
To test this, we co-expressed in yeast functional versions of the proteins tagged with either protein A (PtA) or a peptide epitope (haemagglutinin; HA), isolated the PtA chimeras using IgG–Sepharose beads, and tested for co-isolation of the HA-tagged partner. Using this approach, we were able to co-precipitate Snx41p with Snx4p, and vice versa, and also Snx42p with Snx4p, and vice versa, but could detect no interaction between Snx41p and Snx42p, or between any of these and Grd19p (Figure 5). The efficiency of precipitation of HA-Snx41p was comparable with that of PtA-Snx4p, implying that essentially all of the Snx41p was complexed with Snx4p; the recovery of HA-Snx42p was somewhat less efficient, but still substantial. In contrast, there was little interaction between differently tagged versions of the same protein, although a very weak signal was obtained for HA-Snx4p with PtA-Snx4p. Thus, under the conditions of the experiment, we can detect Snx4–Snx41 dimers and Snx4–Snx42 dimers, but not larger oligomers. From the mutant phenotypes, it appears that both dimers contribute to Snc1p sorting. In agreement with these results, high-throughput two-hybrid screens have also detected interaction of Snx41p and Snx42p with Snx4p, but not with each other (Uetz et al., 2000; Ito et al., 2001).

Fig. 5. Interactions between nexins. Yeast strains deleted for the appropriate gene or pair of genes were transformed with plasmids expressing the corresponding nexins, one protein A tagged and one HA tagged, as indicated. The protein A-tagged form was isolated, and co-purifying HA-tagged protein detected by immunoblotting (arrowheads). The immunoprecipitates (IP) contain more cell equivalents than the extracts (Ex), but the ratio is the same in each case. The band indicated with an asterisk represents binding of the anti-HA antibody to Snx42p– protein A.
Interaction of Snx4p with Snc1p
To explore the interactions between the Snx4p family and Snc1p, we co-expressed a PtA-tagged version of Snc1p with HA-tagged Snx4p, exposed isolated spheroplasts to the reversible cross-linker dithiobis(succinimidyl propionate) (DSP) and isolated the Snc1p on IgG–Sepharose. HA-Snx4p did not co-precipitate significantly with Snc1p in the absence of cross-linker, but was clearly detectable after cross-linking (Figure 6A). Controls showed that PtA fused to dihydrofolate reductase could not be cross-linked to Snx4p, and there was also no detectable cross-linking of PtA-Snc1p to HA-Grd19p (Figure 6A).
Fig. 6. Cross-linking of Snx4p to Snc1p. (A) Yeast strains deleted for snx4 or grd19 were transformed with plasmids expressing HA-tagged Snx4p or Grd19p, together with a plasmid expressing protein A–Snc1p or, as a control, protein A–dihydrofolate reductase. Spheroplasts were treated with the reversible cross-linker DSP prior to lysis, the protein A fusions immunoprecipitated with IgG–Sepharose beads, and protein A-tagged and associated HA-tagged proteins detected by immunoblotting. Results are shown for the cell extracts (Ex) and precipitates (IP). The band indicated by an asterisk corresponds not to HA-Snx4p, but to a small amount of IgG heavy chain eluted from the beads, which weakly cross-reacts with the secondary antibodies used in this experiment. (B) The effects of the W86R mutation in Snc1p on Snx4p binding were tested as in (A), except that in this experiment the bound protein was eluted from the IgG–Sepharose beads by specific proteolytic cleavage between the protein A and the Snc1p.
Previous studies identified one point mutation in the cytoplasmic domain of Snc1p that caused its mistargeting to the vacuole, namely W86R (Lewis et al., 2000). As shown in Figure 6B, this mutation also substantially reduced cross-linking of Snc1p to Snx4p, providing further evidence for the specificity and functional relevance of the interaction. We also failed to see significant cross-linking of Snx4p to the Pep12p derivative that is found in PGEs (data not shown). Thus, our data indicate a specific association of Snx4p with Snc1p, suggesting a direct role for this sorting nexin in the recycling of Snc1p.
Snx4 acts independently of retromer
The data suggest that Snx4p and its partners act in PGEs, and are quite independent of retromer, which is thought to act in PVEs. In agreement with this, we have shown previously that retromer mutants do not affect the cycling of Snc1p (Lewis et al., 2000). Furthermore, Figure 7A shows that the only sorting nexin mutants that substantially mis-sort carboxypeptidase Y are the retromer components, and to a lesser extent mvp1, whereas snx4 and the others do not.
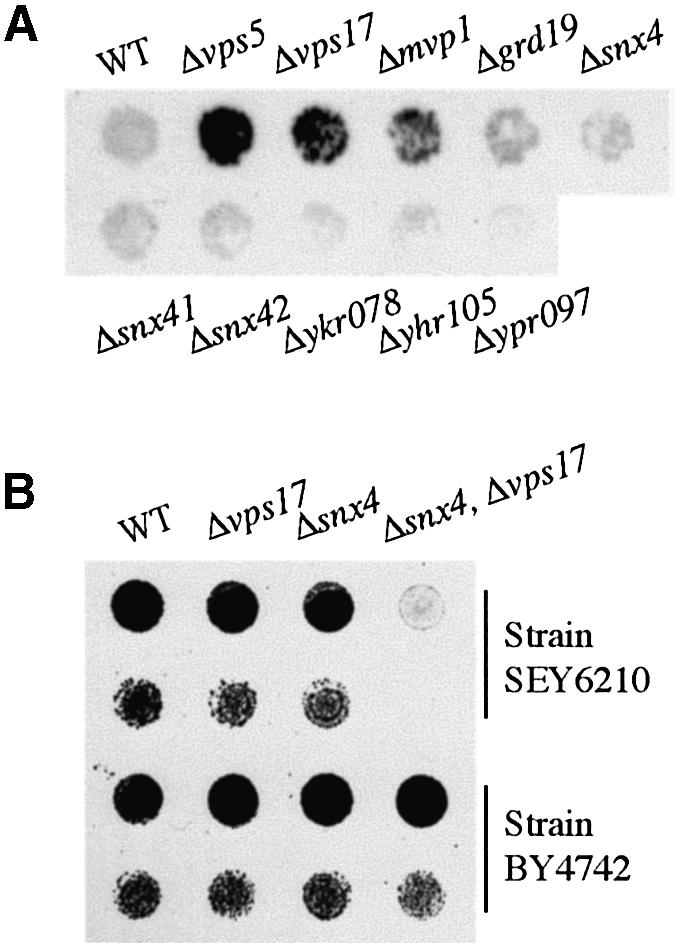
Fig. 7. The Snx4 family of nexins have functions distinct from that of retromer. (A) Detection of carboxypeptidase Y secreted by each sorting nexin mutant. Note that only vps5, vps17 (both encoding retromer subunits) and mvp1 show a detectable phenotype. (B) Synthetic growth defect. The indicated mutants, in two strain backgrounds, were grown to saturation in liquid culture and then spotted at high and low concentrations on plates.
That retromer and Snx4p have distinct functions is also indicated by a genetic interaction between them. We observed that in one genetic background (strain SEY6210), there was a synthetic growth defect between deletions of snx4 and the retromer component vps17: the double mutant grew significantly more slowly than either single mutant (Figure 7B), though in liquid culture the cells did eventually reach comparable densities. Growth could be restored to the double mutant by transformation with a plasmid encoding either of the genes (data not shown). This demonstrates formally that Snx4p and Vps17p can each function in the absence of the other, since each can improve growth of the double mutant.
PI3P-dependent binding of Snx4p, Snx41p and Snx42p to membranes
PX domains bind to PI3P, which is found in endosomes and the vacuole. To localize Snx4p and its partners, we tagged each with GFP and examined them in live cells. When expressed at endogenous levels, GFP–Snx4p and GFP–Snx42p were visible, though very faint. We were unable to detect GFP–Snx41p under these conditions, but clear localization was apparent when the protein was expressed from the TPI1 promoter. As shown in Figure 8, each protein had a punctate distribution consistent with a location on endosomes but not vacuoles, together with a slight diffuse haze corresponding to free cytosolic protein, or possibly to vesicles. The punctate localization was dependent on the presence of PI3P, since it was absent in vps34 cells, which lack the catalytic subunit of PI 3-kinase.
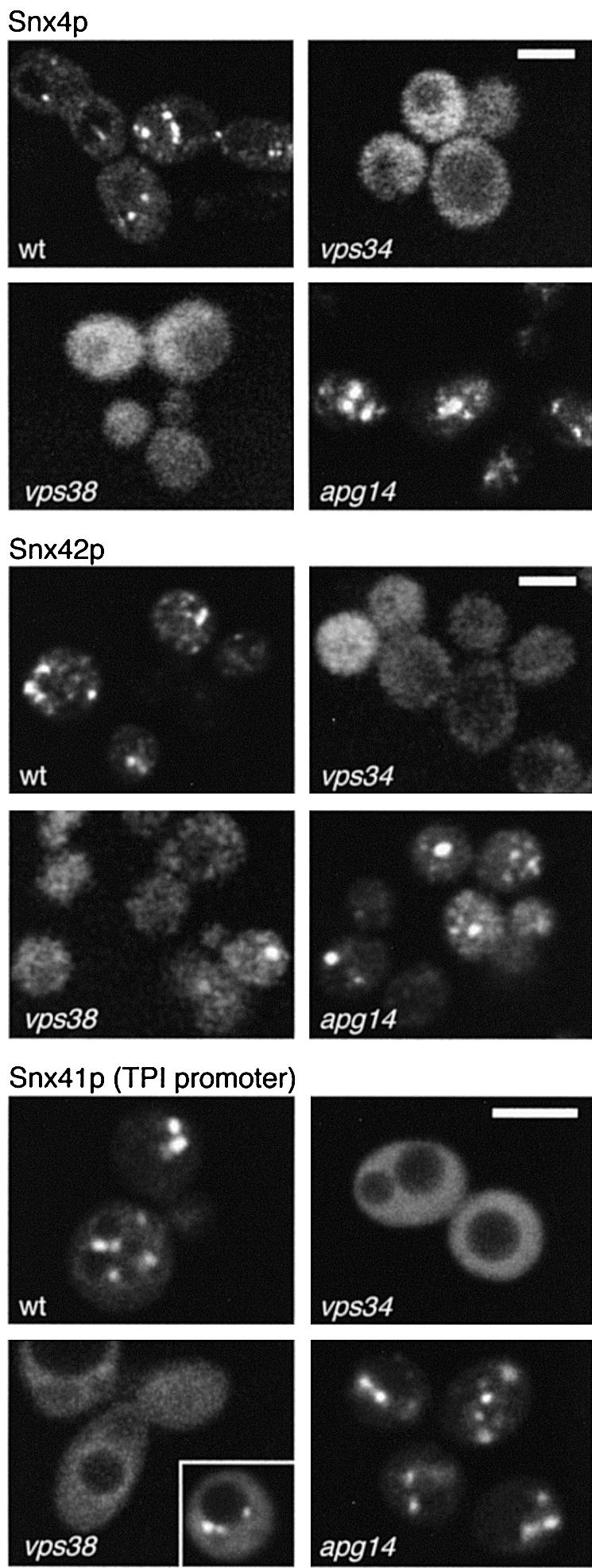
Fig. 8. Localization of Snx4p, Snx41p and Snx42p. GFP-tagged versions of the proteins were expressed in the indicated mutants and live cells imaged in a single focal plane. The ‘wt’ cells in each case lacked the endogenous copy of the corresponding gene, but the other strains did not. GFP–Snx4p and GFP–Snx42p were expressed from centromere vectors using the SNX4 promoter; GFP–Snx41p was expressed from the TPI1 promoter, which gives higher levels of fluorescence and hence better quality images. Bars are 3 µm.
While this manuscript was under review, it was reported that Snx4p (renamed Cvt13p) and Snx42p (named Cvt20p) are required for cytoplasm to vacuole transport (Cvt) and are present in specific perivacuolar structures called pre-autophagosomes (Nice et al., 2002). It was also reported that their presence in these structures depends both on their PX domains and on Apg14p, a protein that is thought to direct PI 3-kinase activity to the Cvt/autophagy pathway (Kihara et al., 2001). However, we found that the punctate distribution of Snx4p, Snx41p and Snx42p was largely unaffected in an apg14 mutant (Figure 8). In contrast, removal of Vps38p, a component of a distinct PI 3-kinase complex that is required for carboxypeptidase Y sorting but not for autophagy (Kihara et al., 2001), resulted in almost complete release of the sorting nexins from punctate structures. Interestingly, a few perivacuolar dots remained in vps38 cells, which reflect the presence of PI3P since they were completely absent in vps34 cells. These may well be pre-autophagosomes. Together, the data strongly suggest that all three of the sorting nexins are present both in endosomes, marked by PI3P in a Vps38p-dependent manner, and in pre-autophagosomes, whose PI3P content depends on Apg14p. These two locations appear to be matched by two distinct functions, in Cvt and endosomal sorting.
Discussion
We have identified sorting nexins required for the retrieval of two proteins from endosomes, namely Pep12p and Snc1p. One of our goals was to examine retrieval from PGEs, since this is largely uncharacterized. We have found that the sorting nexins Snx4p, Snx41p and Snx42p are required specifically for the retrieval of Snc1p from PGEs, and that this process is quite distinct from the retrieval of Pep12p, which requires Grd19p and retromer and appears to occur mainly, if not exclusively, from PVEs.
Sorting of Pep12p
Wild-type Pep12p is considered a marker for PVEs, but appropriate mutations can direct it to the cell surface, from where it is endocytosed. In the steady state, this mutant form is found in PGEs, and thus we anticipated that it would be a suitable substrate with which to analyse retrieval from these earlier endosomes. However, mutagenesis of Pep12p and of the sorting nexins revealed instead signals and components involved in retrieval from PVEs. Our data most readily fit a model in which both Pep12p and its PGE-localized derivative avoid passage to the vacuole by moving on the well-established pathway from PVEs back to the Golgi. In principle, recycling could also occur from PVE to PGE, but we cannot easily distinguish such a pathway. It seems much less likely that retrieval occurs from the vacuole, in part because Pep12p and sorting components such as retromer are not normally found there, but also because we failed to see removal of Pep12p from this organelle when a temperature-sensitive vps4 mutant was used to block further delivery. Electron microscopy studies have also shown that Pep12p is normally removed from endosomes before they fuse with the vacuole (Prescianotto-Baschong and Riezman, 2002).
We were surprised to find that both forms of Pep12p were retrieved by the same machinery, because retrieval of GFP–Pep12p itself is readily saturable, resulting in leakage to the vacuole, whereas expression of the PGE form at the same level does not result in such leakage. A likely explanation is that default progression from PGE to PVE is intrinsically slow compared with the GGA-mediated transport of wild-type Pep12p from Golgi to PVE, and this prevents saturation of the retrieval machinery when Pep12p is directed via PGEs. Slow exit from the PGE/Golgi system coupled to rapid retrieval from PVEs would result in a steady-state distribution mainly in PGEs and Golgi, as is observed for this construct.
Strikingly, this is precisely the strategy used by proteins such as Kex2p and Ste13p which, despite being found in Golgi/PGE membranes at steady state, require constant retrieval from PVEs to avoid passage to the vacuole, and indeed use the same machinery in the form of retromer and Grd19p (Conibear and Stevens, 1998). The Golgi proteins do contain additional weak signals that help to retain them but, as with the Pep12p construct, PVE retrieval seems to be more important (Brickner and Fuller, 1997; Bryant and Stevens, 1997; Conibear and Stevens, 1998). We therefore argue that there is little intrinsic difference between the sorting of endosomal and Golgi proteins: endosomal residents cycle through the Golgi, just as Golgi residents cycle through endosomes. It is only the FSD signal that gives wild-type Pep12p a steady-state distribution different from that of Golgi proteins, by directing it into the efficient GGA-mediated pathway from Golgi to PVEs.
A remaining uncertainty is the precise mechanistic role of Grd19p. This sorting nexin is required for the retrieval of Kex2p, Ste13p and Pep12p, all of which also require retromer. In contrast, retrieval of Vps10p requires retromer but not Grd19p. This suggests that Grd19p may associate with retromer, improving the recruitment of some but not all cargo molecules. Such an association has not been demonstrated, and whether Grd19p directly recognizes cargo is also unclear; we have not observed binding of Grd19p to Pep12p, either by cross-linking or by affinity chromatography, and, although Grd19p has been reported to bind the cytoplasmic tail of Ste13p (Voos and Stevens, 1998), there is also evidence that recognition of Ste13p is mediated by the retromer subunit Vps35p (Nothwehr et al., 1999, 2000). Grd19p may therefore act indirectly, by modulating retromer binding.
Sorting of Snc1p
In contrast to Pep12p, Snc1p seems to be retrieved almost exclusively from PGEs, being sorted correctly even in a pep12 mutant (Lewis et al., 2000). It has proven hard to define a cytoplasmic sorting signal on Snc1p, but our finding that its retrieval requires Snx4p and its partners, and that specific interaction between Snx4p and Snc1p can be detected in cells, provides strong evidence that retrieval from PGEs to the Golgi is an active process. This in turn emphasizes the distinct functional identity of PGEs, which contain many of the same integral membrane proteins as late Golgi compartments and are difficult to distinguish from them. Moreover, since the nexins bind PI3P, a marker for the endocytic pathway, their involvement argues that PGEs are true endosomes. Golgi proteins such as Ypt6p are also required for recycling of Snc1p, but their effects are quite different; they are needed for vesicle fusion rather than budding. Thus, in ypt6 cells, Snc1p does not pass to the vacuole but instead accumulates in small vesicles (Siniossoglou et al., 2000). The only other protein that has been suggested to affect PGEs specifically is the F-box protein Rcy1p, whose removal causes an enlargement of presumptive endosomal structures in which Snc1p is trapped, though the mechanistic basis of this is unknown (Galan et al., 2001).
How might the nexins promote Snc1p sorting? Snx4p can be cross-linked to Snc1p and, moreover, this interaction is blocked by a cytoplasmic point mutation in Snc1p that inhibits sorting. However, efficient sorting of Snc1p also requires particular features of its TMD (Lewis et al., 2000). Thus, either there is a further transmembrane component that aids the interaction, or there is an additional process, such as partitioning between lipid domains, that is also important for sorting. Snx4p can bind either Snx41p or Snx42p, but whether it forms, or is part of, a conventional coat is unclear. A variety of other putative partners for these proteins have been detected in two-hybrid screens, including Apg17p, Ygl161c, Ygl198w, Ydr084c, Yjr110w and Yhr022c (Uetz et al., 2000; Ito et al., 2001), but none of these is essential for Snc1p sorting (our unpublished observations). Further more, Snx4p acts independently of retromer. This follows both from the formal genetic observation that Vsp17p and Snx4p can each improve cell growth in the absence of the other, and also from the fact that Snx4p-dependent sorting of Snc1p is unaffected by removal of retromer components (Lewis et al., 2000).
If the nexins do not themselves form or interact with a rigid coat, they may nevertheless help to partition Snc1p into a membrane domain that is destined to return to the Golgi, perhaps acting synergistically with partitioning events in the lipid bilayer. The final budding of such a membrane domain may then be facilitated by other components. This view of sorting nexins, as components that aid the partitioning of cargo proteins between membrane domains whose fates differ, explains why nexin mutants do not completely prevent recycling of Snc1p; some of it will reach the correct domain even if its distribution is random. In practice, much of the membrane in PGEs may actually return to the Golgi, which would explain the ease with which Pep12p remains in the PGE/Golgi cycle and progresses only slowly to PVEs, as well as the observed recycling of a substantial proportion of the endocytic tracer dye FM4-64 to the cell surface (Wiederkehr et al., 2000). Endocytosed membrane proteins that pass rapidly to PVEs and the vacuole may either be partitioned efficiently into the regions of PGEs that have this destination, or be sorted at the plasma membrane into carriers that bypass PGEs altogether.
Role of nexins in retrieval from endosomes
Sorting nexins are defined largely by the presence of a PX domain, whose main function seems to be to direct them to PI3P-containing endosomal membranes. Since PI3P is found throughout the endocytic pathway, including the vacuole (Gillooly et al., 2000), other components, and/or sequences outside of the PX domain, must determine the precise site of action of individual sorting nexins. Note, however, that although our data indicate that Snx4p must act at PGEs, and Grd19p and retromer at PVEs, it does not exclude the possibility that each is active on both types of endosome.
A surprising finding arising from this work and the independent study of Nice et al. (2002) is that Snx4p and Snx42p, and probably Snx41p as well, are present not only on endosomes but also on pre-autophagosomes, where they play a role in the Cvt pathway. These organelles contain PI3P, provided by a specific PI 3-kinase complex that contains Apg14p. They are clearly distinct from endosomes, though they may be derived from them since the Cvt pathway is stimulated by the PGE/Golgi SNARE Tlg2p (Abeliovich et al., 1999). Presumably, the ability of the sorting nexins to organize membranes has been harnessed for two distinct purposes in yeast.
In animal cells, sorting nexins have been shown to bind to the cytoplasmic tails of plasma membrane receptors, and they have been suggested variously to promote or hinder the transport of these proteins to lysosomes (Kurten et al., 1996; for a review, see Pelham, 2002). In yeast, the picture so far has been more consistent: sorting nexins in general help to prevent particular membrane proteins from reaching the vacuole. This has been shown previously for Vps5p, Vps17p, Mvp1p and Grd19p acting in PVEs, and our demonstration that Snx4p, Snx41p and Snx42p help to retrieve proteins from earlier endosomes extends these findings, though the role of Snx4p and Snx42p in the Cvt pathway may indicate greater complexity. It will be interesting to see whether the apparent differences between sorting nexin function in animal and yeast cells can be accounted for by differences in experimental approach, or whether these proteins have generic sorting properties which have been used for different purposes in different organisms.
Materials and methods
Yeast strains
The majority of strains used herein were from the EUROSCARF consortium (www.uni-frankfurt.de/fb15/mikro/euroscarf) and were complete deletions in strain B4742 (snx4, snx41, snx42, grd19, mvp1, vps5, vps17, vps27, ypt6, pep12, ykr078, ypr097, yhr105). Double mutants were made by replacement of the complete reading frame of the second gene with Schizosaccharomyces pombe HIS5, starting with the appropriate single mutant (snx4 snx41, snx4 snx42, snx41 snx42, snx4 vps27, snx4 vps17). For the triple mutant snx4 snx41 snx42, the first copy of HIS5 was removed using the cre-lox system (Sauer, 1994). The experiment in Figure 2A used strain SEY6210; Figure 2B and C used a pep12 deletion in this strain, with plasmids expressing the indicated Pep12p derivatives (Black and Pelham, 2000). Figure 7B (where indicated) also used SEY6210 or mutants in this background. We have observed minor differences in the behaviour of GFP–Snc1p in the SEY6210 and B4742 backgrounds, notably increased plasma membrane fluorescence in the latter. However, GFP–Snc1p showed vacuolar fluorescence in the snx4 derivative of SEY6210, just as in the snx4 EUROSCARF strain.
The experiment shown in Figure 3D used a derivative of SEY6210 with a temperature-sensitive allele of vps4 (Babst et al., 1997). GFP–Pep12p was expressed from the TPI1 promoter (left hand two panels) or the GAL1 promoter (right hand panels) on a centromere plasmid. This strain does not take up galactose efficiently, and hence galactose induction is weak; the induction conditions used (5 h in 2% galactose, 2% raffinose) produced about five times as much GFP–Pep12p as endogenous Pep12p, as estimated by immunoblotting.
Plasmids
Plasmids expressing N-terminally tagged sorting nexins were all based on Ycplac33 and Ycplac111 (Gietz and Sugino, 1988) containing the SNX4 promoter (EcoRI–SacI) and PGK1 terminator (just 3′ of the multiple cloning site) (Harmsen et al., 1993). Both the triple HA tag (in Ycplac33-based plasmids) and the PtA tag (in Ycplac111-based plasmids) were inserted as SacI–BamHI fragments, and the SNX4, SNX41, SNX42 and GRD19 open reading frames were amplified by PCR as BamHI–XhoI, BamHI–PstI, XbaI–PstI and BamHI–SalI fragments, respectively, and cloned into the cassettes. A similar strategy was used to express GFP fusions of the sorting nexins. Plasmids expressing GFP-tagged SNARE constructs were all based on pRS416 vectors (Sikorski and Hieter, 1989) and have been described previously (Black and Pelham, 2000; Lewis et al., 2000); the PtA-tagged Snc1 construct was made in the same way. Random PCR mutagenesis of the plasmid encoding Pep12p with the Sso1p TMD has been described (Black and Pelham, 2000). The mutant analysed in detail was M18, which contained the F20L mutation that inactivates the FSD motif as well as the F6L and I71T changes. These mutations subsequently were introduced one at a time using synthetic oligonucleotides for PCR mutagenesis.
Affinity purification of PtA fusions and in vivo cross-linking
Yeast deletion mutants were transformed with plasmids encoding either PtA-Snx4p, PtA-Snx41p, PtA-Snx42p or PtA-Grd19p (LEU2 marker) and plasmids encoding either HA-Snx4p, HA-Snx41p, HA-Snx42p or HA-Grd10p (URA3 marker). Both the PtA- and HA-tagged sorting nexins replaced the corresponding endogenous protein. Affinity purification of the PtA fusions was performed essentially as described (Siniossoglou et al., 2000). For in vivo cross-linking, 300 mg of freshly prepared spheroplasts from cells expressing either PtA-Snc1p, PtA-Snc1(W86R), PtA-DHFR or PtA-Pep12(FSD–) and either HA-tagged Snx4p, Snx41p, Snx42p or Grd19p were resuspended in 0.6 ml of 25 mM potassium phosphate pH 7.4, 200 mM sorbitol containing protease inhibitors and 0–2 mM DSP (Pierce), and incubated for 30 min at 4°C. The cross-linker was then quenched for 5 min by the addition of 150 mM Tris–HCl pH 7.4, cell suspensions were diluted with 5 ml of 150 mM KCl, 5 mM MgCl2, and Triton X-100 was added to a final concentration of 1%. PtA fusions were purified by IgG–Sepharose chromatography and eluted by low pH (Siniossoglou et al., 2000). Cross-links were cleaved using 20 mM dithiothreitol (DTT) in SDS sample buffer and samples were analysed by SDS–PAGE and western blotting. In some cases, PtA fusions were eluted from IgG–Sepharose with tobacco etch virus protease, which cleaves at a specific site introduced between the PtA and the fusion partner (Siniossoglou et al., 2000).
Other methods
Confocal microscopy of live cells in early log phase growth, FM4-64 staining, immunoblotting, quantitation and analysis of carboxypeptidase Y secretion were all performed as before (Black and Pelham, 2000). Averaging of multiple confocal scans was avoided because movement of punctate structures caused blurring. The anti-Pep12p was monoclonal 2C3 from Molecular Probes (Eugene, Oregon); anti-HA was monoclonal 12CA5 (a gift from Sean Munro), and PtA was detected with rabbit peroxidase–anti-peroxidase complex (Dako). Sucrose gradient fractionation of cell extracts (13 000 g supernatants) was performed exactly as described previously (Black and Pelham, 2000). P100 fractions were prepared by centrifugation of the 13 000 g supernatants at 55 000 r.p.m. for 30 min in a TLA100.2 rotor (Beckman).
Acknowledgments
Acknowledgements
We thank Symeon Siniossoglou for reagents and helpful advice, Matthew Seaman for a vps17 strain and plasmid, Greg Odorizzi for the vps4 temperature-sensitive mutant and Sean Munro for comments on the manuscript. M.W.B. was the recipient of a Burroughs Wellcome Fund Hitchings-Elion fellowship; E.H.H. was supported by a fellowship from the Human Frontiers Science Program Organisation.
References
- Abeliovich H., Darsow,T. and Emr,S.D. (1999) Cytoplasm to vacuole trafficking of aminopeptidase I requires a t-SNARE–Sec1p complex composed of Tlg2p and Vps45p. EMBO J., 18, 6005–6016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babst M., Sato,T.K., Banta,L.M. and Emr,S.D. (1997) Endosomal transport function in yeast requires a novel AAA-type ATPase, Vps4p. EMBO J., 16, 1820–1831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black M.W. and Pelham,H.R. (2000) A selective transport route from Golgi to late endosomes that requires the yeast GGA proteins. J. Cell Biol., 151, 587–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black M.W. and Pelham,H.R. (2001) Membrane traffic: how do GGAs fit in with the adaptors? Curr. Biol., 11, R460–R462. [DOI] [PubMed] [Google Scholar]
- Bravo J. et al. (2001) The crystal structure of the PX domain from p40(phox) bound to phosphatidylinositol 3-phosphate. Mol. Cell, 8, 829–839. [DOI] [PubMed] [Google Scholar]
- Brickner J.H. and Fuller,R.S. (1997) SOI1 encodes a novel, conserved protein that promotes TGN–endosomal cycling of Kex2p and other membrane proteins by modulating the function of two TGN localization signals. J. Cell Biol., 139, 23–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryant N.J. and Stevens,T.H. (1997) Two separate signals act independently to localize a yeast late Golgi membrane protein through a combination of retrieval and retention. J. Cell Biol., 136, 287–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryant N.J., Piper,R.C., Weisman,L.S. and Stevens,T.H. (1998) Retrograde traffic out of the yeast vacuole to the TGN occurs via the prevacuolar/endosomal compartment. J. Cell Biol., 142, 651–663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang A. and Fink,G.R. (1995) Targeting of the yeast plasma membrane [H+]ATPase: a novel gene AST1 prevents mislocalization of mutant ATPase to the vacuole. J. Cell Biol., 128, 39–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conibear E. and Stevens,T.H. (1998) Multiple sorting pathways between the late Golgi and the vacuole in yeast. Biochim. Biophys. Acta, 1404, 211–230. [DOI] [PubMed] [Google Scholar]
- Daro E., Sheff,D., Gomez,M., Kreis,T. and Mellman,I. (1997) Inhibition of endosome function in CHO cells bearing a temperature-sensitive defect in the coatomer (COPI) component ε-COP. J. Cell Biol., 139, 1747–1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekena K. and Stevens,T.H. (1995) The Saccharomyces cerevisiae MVP1 gene interacts with VPS1 and is required for vacuolar protein sorting. Mol. Cell. Biol., 15, 1671–1678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galan J.M., Wiederkehr,A., Seol,J.H., Haguenauer-Tsapis,R., Deshaies,R.J., Riezman,H. and Peter,M. (2001) Skp1p and the F-box protein Rcy1p form a non-SCF complex involved in recycling of the SNARE Snc1p in yeast. Mol. Cell. Biol., 21, 3105–3117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gietz R.D. and Sugino,A. (1988) New yeast–Escherichia coli shuttle vectors constructed with in vitro mutagenized yeast genes lacking six-base pair restriction sites. Gene, 74, 527–534. [DOI] [PubMed] [Google Scholar]
- Gillooly D.J., Morrow,I.C., Lindsay,M., Gould,R., Bryant,N.J., Gaullier,J.M., Parton,R.G. and Stenmark,H. (2000) Localization of phosphatidylinositol 3-phosphate in yeast and mammalian cells. EMBO J., 19, 4577–4588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harmsen M.M., Langedijk,A.C., van Tuinen,E., Geerse,R.H., Raue,H.A. and Maat,J. (1993) Effect of a pmr 1 disruption and different signal sequences on the intracellular processing and secretion of Cyamopsis tetragonoloba α-galactosidase by Saccharomyces cerevisiae. Gene, 125, 115–123. [DOI] [PubMed] [Google Scholar]
- Holthuis J.C., Nichols,B.J. and Pelham,H.R. (1998) The syntaxin Tlg1p mediates trafficking of chitin synthase III to polarized growth sites in yeast. Mol. Biol. Cell, 9, 3383–3397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito T., Chiba,T., Ozawa,R., Yoshida,M., Hattori,M. and Sakaki,Y. (2001) A comprehensive two-hybrid analysis to explore the yeast protein interactome. Proc. Natl Acad. Sci. USA, 98, 4569–4574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kihara A., Noda,T., Ishihara,N. and Ohsumi,Y. (2001) Two distinct Vps34 phosphatidylinositol 3-kinase complexes function in autophagy and carboxypeptidase Y sorting in Saccharomyces cerevisiae. J. Cell Biol., 152, 519–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurten R.C., Cadena,D.L. and Gill,G.N. (1996) Enhanced degradation of EGF receptors by a sorting nexin, SNX1. Science, 272, 1008–1010. [DOI] [PubMed] [Google Scholar]
- Lewis M.J., Nichols,B.J., Prescianotto-Baschong,C., Riezman,H. and Pelham,H.R. (2000) Specific retrieval of the exocytic SNARE Snc1p from early yeast endosomes. Mol. Biol. Cell., 11, 23–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misra S., Miller,G.J. and Hurley,J.H. (2001) Recognizing phosphatidylinositol 3-phosphate. Cell, 107, 559–562. [DOI] [PubMed] [Google Scholar]
- Nice D.C., Sato,T.K., Stromhaug,P.E., Emr,S.D. and Klionsky,D.J. (2002) Cooperative binding of the cytoplasm to vacuole targeting pathway proteins, Cvt13 and Cvt20, to phosphatidylinositol 3-phosphate at the pre-autophagosomal structure is required for selective autophagy. J. Biol. Chem., 277, 30198–30207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nothwehr S.F., Bruinsma,P. and Strawn,L.A. (1999) Distinct domains within Vps35p mediate the retrieval of two different cargo proteins from the yeast prevacuolar/endosomal compartment. Mol. Biol. Cell., 10, 875–890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nothwehr S.F., Ha,S.A. and Bruinsma,P. (2000) Sorting of yeast membrane proteins into an endosome-to-Golgi pathway involves direct interaction of their cytosolic domains with Vps35p. J. Cell Biol., 151, 297–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelham H.R. (2002) Insights from yeast endosomes. Curr. Opin. Cell Biol., 14, 454–462. [DOI] [PubMed] [Google Scholar]
- Piguet V., Gu,F., Foti,M., Demaurex,N., Gruenberg,J., Carpentier,J.L. and Trono,D. (1999) Nef-induced CD4 degradation: a diacidic-based motif in Nef functions as a lysosomal targeting signal through the binding of β-COP in endosomes. Cell, 97, 63–73. [DOI] [PubMed] [Google Scholar]
- Piper R.C., Cooper,A.A., Yang,H. and Stevens,T.H. (1995) VPS27 controls vacuolar and endocytic traffic through a prevacuolar compartment in Saccharomyces cerevisiae. J. Cell Biol., 131, 603–617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prescianotto-Baschong C. and Riezman,H. (2002) Ordering of compartments in the yeast endocytic pathway. Traffic, 3, 37–49. [DOI] [PubMed] [Google Scholar]
- Raiborg C., Bache,K.G., Gillooly,D.J., Madshus,I.H., Stang,E. and Stenmark,H. (2002) Hrs sorts ubiquitinated proteins into clathrin-coated microdomains of early endosomes. Nat. Cell Biol., 4, 394–398. [DOI] [PubMed] [Google Scholar]
- Reggiori F., Black,M.W. and Pelham,H.R. (2000) Polar transmembrane domains target proteins to the interior of the yeast vacuole. Mol. Biol. Cell, 11, 3737–3749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieder S.E., Banta,L.M., Kohrer,K., McCaffery,J.M. and Emr,S.D. (1996) Multilamellar endosome-like compartment accumulates in the yeast vps28 vacuolar protein sorting mutant. Mol. Biol. Cell, 7, 985–999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sachse M., Urbe,S., Oorschot,V., Strous,G.J. and Klumperman,J. (2002) Bilayered clathrin coats on endosomal vacuoles are involved in protein sorting toward lysosomes. Mol. Biol. Cell, 13, 1313–1328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato T.K., Overduin,M. and Emr,S.D. (2001) Location, location, location: membrane targeting directed by PX domains. Science, 294, 1881–1885. [DOI] [PubMed] [Google Scholar]
- Sauer B. (1994) Recycling selectable markers in yeast. Biotechniques, 16, 1086–1088. [PubMed] [Google Scholar]
- Scales S.J., Gomez,M. and Kreis,T.E. (2000) Coat proteins regulating membrane traffic. Int. Rev. Cytol., 195, 67–144. [DOI] [PubMed] [Google Scholar]
- Seaman M.N., McCaffery,J.M. and Emr,S.D. (1998) A membrane coat complex essential for endosome-to-Golgi retrograde transport in yeast. J. Cell Biol., 142, 665–681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikorski R.S. and Hieter,P. (1989) A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics, 122, 19–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siniossoglou S., Peak-Chew,S.Y. and Pelham,H.R. (2000) Ric1p and Rgp1p form a complex that catalyses nucleotide exchange on Ypt6p. EMBO J., 19, 4885–4894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uetz P. et al. (2000) A comprehensive analysis of protein–protein interactions in Saccharomyces cerevisiae. Nature, 403, 623–627. [DOI] [PubMed] [Google Scholar]
- Valdivia R.H., Baggott,D., Chuang,J.S. and Schekman,R.W. (2002) The yeast clathrin adaptor protein complex 1 is required for the efficient retention of a subset of late Golgi membrane proteins. Dev. Cell, 2, 283–294. [DOI] [PubMed] [Google Scholar]
- Voos W. and Stevens,T.H. (1998) Retrieval of resident late-Golgi membrane proteins from the prevacuolar compartment of Saccharomyces cerevisiae is dependent on the function of Grd19p. J. Cell Biol., 140, 577–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiederkehr A., Avaro,S., Prescianotto-Baschong,C., Haguenauer-Tsapis,R. and Riezman,H. (2000) The F-box protein Rcy1p is involved in endocytic membrane traffic and recycling out of an early endosome in Saccharomyces cerevisiae. J. Cell Biol., 149, 397–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J.W. and Lemmon,M.A. (2001) All phox homology (PX) domains from Saccharomyces cerevisiae specifically recognize phosphatidylinositol 3-phosphate. J. Biol. Chem., 276, 44179–44184. [DOI] [PubMed] [Google Scholar]