Abstract
The budding yeast spindle aligns along the mother– bud axis through interactions between cytoplasmic microtubules (CMs) and the cell cortex. Kar9, in complex with the EB1-related protein Bim1, mediates contacts of CMs with the cortex of the daughter cell, the bud. Here we established a novel series of events that target Kar9 to the bud cortex. First, Kar9 binds to spindle pole bodies (SPBs) in G1 of the cell cycle. Secondly, in G1/S the yeast Cdk1, Cdc28, associates with SPBs and phosphorylates Kar9. Thirdly, Kar9 and Cdc28 then move from the SPB to the plus end of CMs directed towards the bud. This movement is dependent upon the microtubule motor protein Kip2. Cdc28 activity is required to concentrate Kar9 at the plus end of CMs and hence to establish contacts with the bud cortex. The Cdc28-regulated localization of Kar9 is therefore an integral part of the program that aligns spindles.
Keywords: APC/Bim1/Cdk1/Kar9/spindle alignment
Introduction
In mammalian cells, the targeting and capture of microtubule plus ends at special sites of the cell cortex are essential for directed cell migration, the alignment of mitotic spindles, axon outgrowth and polarization of T cells towards antigens (Goode et al., 2000). In budding yeast, cytoplasmic microtubules (CMs) interact with both the cortex of the mother cell and the bud to align the mitotic spindle with the mother–bud axis. Spindle alignment is essential to ensure that the elongation of the anaphase spindle will result in the correct distribution of the duplicated sister chromatids between the two cells. Two pathways establish spindle orientation in yeast. An early pathway involves the protein Kar9, the EB1 homologue Bim1, and a myosin V called Myo2. This pathway orients the spindle with respect to the mother–bud axis and positions the spindle close to the bud neck. Later in the cell cycle, a dynein-dependent pathway translocates the growing spindle through the mother–bud junction (bud neck) into the bud. Each pathway can partially compensate for a failure in the other; however, simultaneous loss of both pathways is lethal (Miller and Rose, 1998; Miller et al., 1998).
The capture of CMs by the bud cortex occurs during the G1/S interval of the cell cycle. Kar9 concentrates at the plus ends of CMs that contact the bud neck (Kusch et al., 2002). Kar9 is linked to microtubules by its association with Bim1 (Korinek et al., 2000; Lee et al., 2000), a microtubule-binding protein (Schwartz et al., 1997). Eventually, Myo2 binds to Kar9 and translocates Kar9 with the associated CMs along polarized actin cables into the bud (Beach et al., 2000; Yin et al., 2000). Later in the cell cycle, Kar9 associates with the bud tip in an actin-dependent manner. Significantly, this later association is not dependent upon microtubules (Miller et al., 1999).
The spindle pole body (SPB) is the functional equivalent of the mammalian centrosome (Byers and Goetsch, 1975). As such, it is responsible for nucleating and organizing all of the microtubules. The SPB duplicates by a conservative mechanism such that the newly formed SPB assembles next to the pre-existing old SPB from new components in G1/S of the cell cycle. The new and old SPBs do not behave in the same way. The old SPB is orientated towards the growing bud. It then translocates into the bud while the newly formed SPB is left behind in the mother cell (Pereira et al., 2002). The selective orientation of the old SPB towards the bud is dependent upon the presence of CMs and Kar9 (Pereira et al., 2002) and is regulated by Cdc28–Clb5 [cyclin-dependent kinase 1 (Cdk1) with the S phase cyclin Clb5] (Segal et al., 2000).
Here we address the molecular basis for the regulation of spindle orientation in budding yeast. We show that Kar9 associates with only the bud-ward-directed SPB. The microtubule motor protein Kip2 is used to translocate this population of Kar9 molecules from the SPB to the plus end of CMs that contact the bud neck in G1/S. The Kar9 cargo then associates with the bud neck. This association of Kar9 is regulated by Cdc28–Clb5. Cdc28 co-localizes with Kar9 both at SPBs and at the bud cortex.
Results
Cdc28 is associated with SPBs, CMs and the bud cortex
In budding yeast, coordinated spindle assembly and orientation requires Cdc28–Clb5 activity (Segal et al., 2000). To understand the molecular role of Cdc28 in spindle orientation, we fused the chromosomal CDC28 to four copies of the green fluorescent protein (CDC28–4GFP). CDC28–4GFP was the only source of Cdc28 activity in these cells. CDC28–4GFP cells divided with the same doubling time as isogenic CDC28 cells. This suggested that Cdc28–4GFP was fully functional.
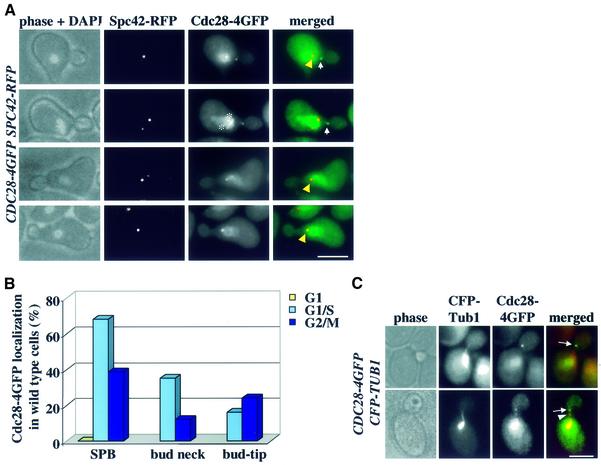
We investigated Cdc28–4GFP localization in cells in which cell cycle progression had been synchronized by α-factor arrest and release. Cdc28–4GFP was enriched in the nucleus throughout the cell cycle. A spindle-like staining of Cdc28–4GFP was observed in some cells (Figure 1A, asterisks). As cells developed a small bud in G1/S of the cell cycle, a strong dot of Cdc28–4GFP was seen at the nuclear periphery (Figure 1A, yellow arrowhead). This Cdc28–4GFP dot overlapped with the SPB marker Spc42 (Donaldson and Kilmartin, 1996) fused to a fast-folding version of the red fluorescent protein Spc42–RFP. Localization of a protein close to SPBs indicates that it is either associated with clustered kinetochores or with SPBs (Jin et al., 1998). Using ndc10-1 cells, which fail to assemble kinetochores at the restrictive temperature (Goh and Kilmartin, 1993), we excluded the possibility that the Cdc28–4GFP dot at the nuclear periphery was generated by association of this kinase with clustered kinetochores (data not shown). We concluded that Cdc28 associated with SPBs.
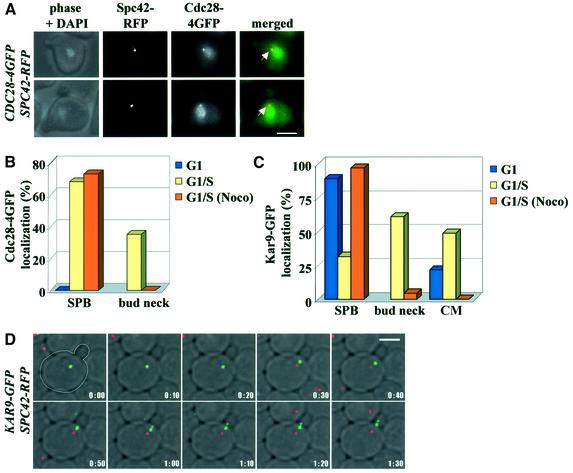
Fig. 1. Cdc28 binds to the SPB and bud cortex. (A) Localization of Cdc28–4GFP in G1/S cells. The yellow arrowheads point toward SPBs, the white arrows highlight Cdc28–4GFP at the bud cortex and the asterisks mark Cdc28–4GFP along a short nuclear spindle. (B) Quantification of Cdc28–4GFP localization. n > 100. (C) Combined images of CDC28–4GFP and CFP–TUB1 as indicated. The arrows point towards Cdc28–4GFP at CM plus ends. The arrowhead indicates Cdc28–4GFP along CMs. Bars: 5 µm.
A Cdc28–4GFP spot was also detected at the bud neck (Figure 1A, arrows). When CMs were labelled by fusing the α-tubulin gene, TUB1, with the cyan fluorescence protein (CFP-TUB1), a spot of Cdc28–4GFP co-localized with the plus end of CMs that contact the bud neck in G1/S (Kusch et al., 2002; Figure 1C, arrows). In addition, Cdc28–4GFP dots were observed along CMs (Figure 1C, arrowhead). Thus, Cdc28 is also found in association with sites of CM–bud neck interactions.
Quantification of Cdc28 localization showed that bud neck and SPB association were cell cycle dependent. Cdc28–4GFP was not detected at SPBs of G1 cells [Figure 1B, α-factor cells or cycling cells (data not shown)]. On the other hand, over 60% of G1/S cells showed a Cdc28–4GFP SPB signal, indicating that Cdc28 bound to SPBs in G1/S. Cdc28–4GFP at SPBs decreased as cells entered mitosis (Figure 1B). Similarly, Cdc28–4GFP was not cell cortex associated in G1. Bud neck association was maximal in G1/S and decreased as cells entered mitosis. In summary, Cdc28 associated with SPBs, microtubules and at sites of CM bud neck interactions in a cell cycle-dependent manner.
Localization of Cdc28 at the bud cortex is dependent on Bim1 and Kar9
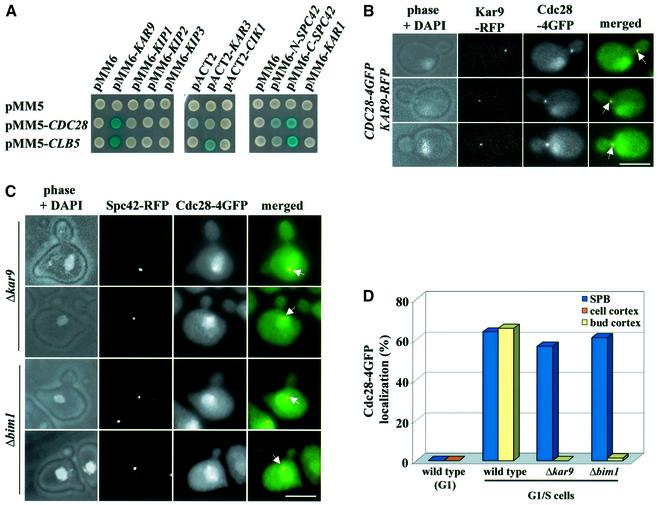
To identify the targets of Cdc28–Clb5 in spindle orientation, we screened for proteins that interacted with Cdc28 and Clb5. The motor protein Kar3 (Meluh and Rose, 1990), the SPB component Spc42 and Kar9 all showed two-hybrid interactions with Clb5 or Cdc28 (Figure 2A). The Kar9 interactor Bim1 showed a weak two-hybrid interaction with Cdc28 (data not shown). The interaction between Cdc28–Clb5 and Kar9 was of particular interest because of the role of Kar9 in spindle alignment and its localization to the bud neck (Miller and Rose, 1998; Korinek et al., 2000; Lee et al., 2000; Pereira et al., 2001). We therefore investigated cells, which contained both Kar9–RFP and Cdc28–4GFP, to determine whether the two proteins localized to the same structures. The signal of Cdc28–4GFP close to the bud neck of G1/S cells overlapped with the Kar9–RFP cortical signal (Figure 2B, arrows). In addition, Cdc28–4GFP and Kar9-RFP co-localized at SPBs (data not shown).
Fig. 2. Cdc28 interacts with Kar9. (A) Cdc28 and Kar9 interact in the yeast two-hybrid system. (B) Co-localization of Kar9 and Cdc28 at the bud cortex. Analysis of CDC28–4GFP KAR9–RFP cells by fluorescence microscopy. The arrows mark Cdc28–4GFP that co-localizes with Kar9–RFP. (C) Localization of Cdc28–4GFP in Δkar9 and Δbim1 cells. The arrows highlight SPB localization of Cdc28–4GFP. Note Cdc28–4GFP does not associate with the bud neck of Δkar9 and Δbim1 cells. (D) Quantification of Cdc28–4GFP localization in the indicated cell types. n > 100. Bars: 5 µm.
We next tested whether Cdc28 association with SPBs and the bud cortex was dependent upon KAR9. Since Kar9 interacts with Bim1, and Bim1 is required for the bud neck association of Kar9 (Korinek et al., 2000; Lee et al., 2000; Miller et al., 2000), we also investigated Cdc28 localization in Δbim1 cells. When synchronized cells were inspected 40 min after the release from the G1 block (>70% of cells were small budded), Cdc28–4GFP hardly associated with the bud cortex in Δbim1 and Δkar9 cells (Figure 2C and D), whereas 66% of wild-type cells displayed a Cdc28–4GFP bud cortex staining (Figure 2D). In addition, Cdc28–4GFP was not detected along CMs in Δbim1 and Δkar9 cells (data not shown). In contrast, SPB association of Cdc28–4GFP was not affected in Δbim1 and Δkar9 cells (Figure 2C, arrows). In conclusion, bud cortex localization, but not SPB binding of Cdc28, required Bim1 and Kar9.
Cdc28 phosphorylates Kar9
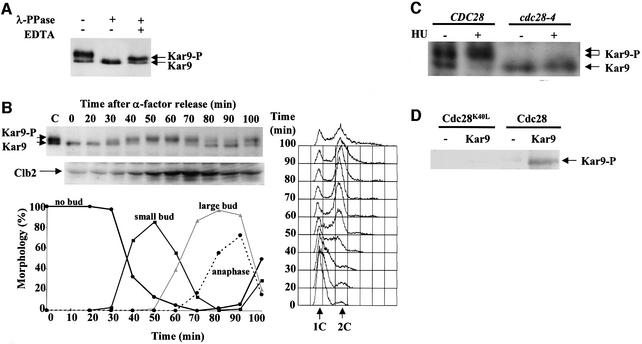
As is often the case for phosphoproteins, Kar9 migrated as a series of bands on SDS–PAGE (Miller et al., 2000). To test whether Kar9 is indeed subject to modification by phosphorylation, we immuno-precipitated Kar9 fused to three copies of the haemagglutinin epitope (Kar9-3HA) and treated it with λ phosphatase. If Kar9 is indeed a phosphoprotein, this treatment should convert the multiple forms into a single band. Kar9 collapsed into a single band upon incubation with phosphatase (Figure 3A). Phosphatase inhibitors delayed this conversion. Thus, Kar9 is a phosphoprotein.
Fig. 3. Phosphorylation of Kar9 is Cdc28 dependent. (A) Phosphorylated Kar9-3HA was incubated as indicated. Samples were analysed by immunoblotting with anti-HA antibodies (12CA5). (B) KAR9-3HA cells were synchronized by α-factor block and release and analysed for Kar9 phosphorylation, cell morphology and DNA content by flow cytometry. Clb2 content of cells was followed over time. (C) Wild-type and cdc28–4 cells with KAR9-3HA were arrested in S phase with HU. Kar9 phosphorylation was analysed by immunoblotting. (D) Buffer only (–) or GST–Kar9369–644 purified from E.coli were incubated with Cdc28-HA or the kinase-dead Cdc28K40L-HA from yeast cells in the presence of [32P]γ-ATP. Similar amounts of Cdc28-HA and Cdc28K40L-HA were used for the assay (data not shown).
Phosphorylation of Kar9 was studied in cells in which cell cycle progression had been synchronized. In G1 cells, Kar9 was not phosphorylated. Nearly all Kar9 became phosphorylated with the appearance of buds in G1/S phase and the beginning of DNA replication (Figure 3B; 40 min), which is well before the mitotic cyclin Clb2 accumulated in mitosis. Kar9 became dephosphorylated at the end of mitosis (Figure 3B; 80 min) with the degradation of Clb2. About 90% of Kar9 was phosphorylated when cell cycle progression of wild-type cells was arrested in S phase following treatment with hydroxyurea (HU) (Figure 3C). In contrast, Kar9 phosphorylation was not observed when cells carrying the conditional lethal cdc28-4 allele were similarly arrested in S phase with HU. Phosphorylation of Kar9 is therefore dependent upon Cdc28 and occurs at the same time as CMs establish interactions with the bud neck.
Kar9 has 15 Cdc28 consensus phosphorylation sites (S/T P). To show direct phosphorylation of Kar9 by Cdc28, we expressed Kar9 as a glutathione S-transferase (GST) fusion in Escherichia coli. However, the GST–Kar9 fusion protein was unstable. We therefore expressed the C-terminal 275 amino acids of Kar9 fused to GST (GST–Kar9369–644), which contained 11 Cdc28 phosphorylation consensus sites, in E.coli and purified the GST fusion protein (GST–Kar9369–644). GST and GST–Kar9369–644 were then incubated with Cdc28-HA or the kinase-dead Cdc28K40L-HA enriched from yeast cells. Only Cdc28-HA, but not Cdc28K40L-HA, was able to phosphorylate GST–Kar9369–644 (Figure 3D). GST was not phosphorylated by Cdc28-HA (data not shown). Thus, Cdc28 phosphorylates Kar9 directly in vitro.
When taken together, the timing of Kar9 phosphorylation at the beginning of S phase, when the Cdc28–Clb5 complex provides most of the Cdc28 activity, is consistent with the notion that Cdc28 in association with Clb5 is mainly responsible for Kar9 phosphorylation in vivo. However, other Cdc28–Clb complexes may also be able to phosphorylate Kar9. In fact, we observed that Cdc28–Clb2-3HA was able to phosphorylate Kar9 in vitro (data not shown).
Cdc28 regulates Kar9 localization
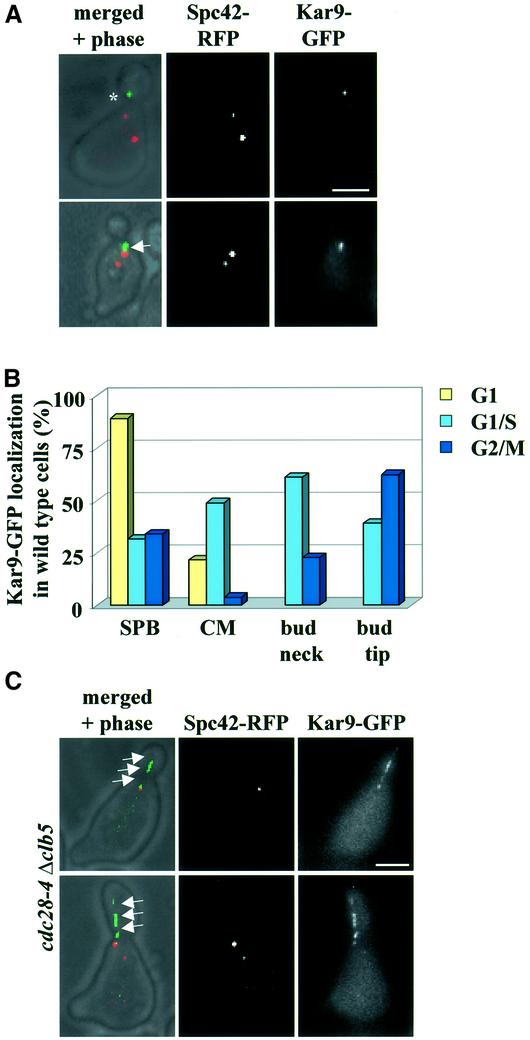
We assessed whether Cdc28 regulates localization of Kar9. First, localization of Kar9 was studied in synchronized wild-type cells. The chromosomal KAR9 was fused to GFP (KAR9–GFP) and KAR9–GFP was expressed from the endogenous promoter. This contrasts other studies that expressed GFP–KAR9 from the strong Gal1 promoter (Miller and Rose, 1998; Beach et al., 2000; Korinek et al., 2000; Lee et al., 2000). In most α-factor-arrested G1 wild-type cells, Kar9–GFP associated with SPBs (Figure 4B), a localization that was confirmed when cells were synchronized by CDC20 depletion in M phase and then upon re-induction of CDC20 progressed into G1 (data not shown). In G1/S cells, Kar9–GFP associated mainly as a dot with the bud cortex close to the bud neck (Kusch et al., 2002) and SPB localization was reduced from the G1 figure of 83 to 32% (Figure 4A and B). In G2/M, Kar9–GFP was predominantly associated with the bud tip (Miller et al., 1999; Figure 4B). It is important to note that in G1/S, more cells (>60%) had a Cdc28–4GFP signal at SPBs (Figure 1B) compared to Kar9–GFP (32%). This difference indicates that not all Cdc28 is in complex with Kar9. The additional Cdc28 at SPBs may be required for SPB separation (Lim et al., 1996). Thus, Kar9 binds to SPBs in G1 and associates with the bud neck in G1/S.
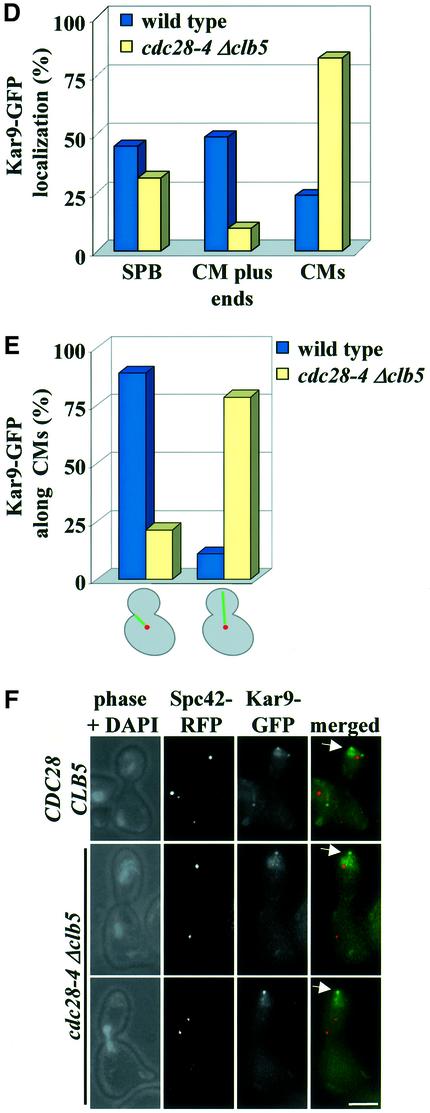
Fig. 4. Cdc28 regulates Kar9 localization. (A) Localization of Kar9–GFP in synchronized G1/S wild-type cells at 30°C. The asterisk indicates Kar9–GFP at the bud neck and the arrow Kar9–GFP along CMs. (B) Quantification of Kar9–GFP localization in wild-type cells. n > 100. (C) Kar9–GFP localization in G1/S of cdc28-4 Gal1-CLB5 cells. Cells were blocked in YPRG with α-factor and released in YPAD medium at 23°C. The arrows indicate Kar9–GFP along CMs. (D) Quantification of wild type and cdc28-4 Gal1-CLB5 cells in G1/S. Both strains were grown as described in (C). n > 100. (E) Direction of CMs (stained by Kar9–GFP) in wild-type and cdc28-4 Δclb5 cells. (F) Localization of Kar9–GFP with the bud tip (indicated by the arrows) in wild type and cdc28–4 Δclb5 cells in anaphase. Bars: 2.5 µm.
Next, we compared the distribution of Kar9–GFP in wild-type cells with that in cells in which Cdc28 activity was reduced. Cdc28 activity of cells carrying the cdc28-4 mutation becomes severely impaired at the permissive temperature if the B-type cyclin Clb5 is depleted (Δclb5) (Segal et al., 1998). In contrast to cells lacking all Cdc28 activity, cdc28-4 Δclb5 cells still progress through the cell cycle (Segal et al., 2000), allowing the analysis of the cell cycle-dependent distribution of Kar9. Wild-type and cdc28-4 Gal1-CLB5 cells arrested in G1 were released into glucose medium at 23°C, which caused rapid depletion of Clb5 from cdc28-4 Gal1-CLB5 cells. Most G1/S cdc28-4 Δclb5 cells did not show the enrichment of Kar9 at the plus end of CMs that was seen in wild-type cells (Figure 4C and D). Instead, Kar9 localization along CMs increased from 24% in wild-type cells to 82% in cdc28-4 Δclb5 cells. This indicated that Cdc28–Clb5 activity was important to concentrate Kar9 at the plus end of CMs. Because Kar9 at CMs plus ends is required for the interaction of CMs with the bud neck (Kusch et al., 2002), we would anticipate a reduction in CM bud neck interactions in cdc28-4 Δclb5 cells. Indeed, CMs of G1/S cdc28-4 Δclb5 cells were mostly orientated towards the bud tip. This contrasted with the predominant orientation towards the bud neck that was seen in wild-type cells, at the same stage of the cell cycle (Figure 4E). Together, these results indicate that Cdc28–Clb5 activity is required to concentrate Kar9 at the plus end of CMs and thereby to establish CM–bud neck interactions.
In contrast to the situation in G1/S, Kar9 associates with the bud tip of anaphase cells in an actin- but not tubulin- dependent manner (Miller et al., 1999). In both wild-type and cdc28-4 Δclb5 cells in anaphase, Kar9–GFP localized at the bud tip (Figure 4F, arrow). Cdc28–Clb5 is therefore only required for binding of Kar9 to the bud neck, but not for its association at later cell cycle stages with the tip of the bud.
Transport of Cdc28 and Kar9 from the SPB to the bud neck required CMs and the motor Kip2
Two models account for the binding of Cdc28 and Kar9 to the plus ends of CMs that contact the bud neck. First, Cdc28 and Kar9 bind to the CM plus ends when these ends contact the bud neck. Alternatively, both proteins bind first to SPBs and are then transported along CMs towards their plus end and so eventually Kar9 interacts with Myo2 at the bud neck. Both models predict that CMs are required for the bud neck localization of Cdc28 and Kar9. However, only when Kar9 binds first to SPBs and then moves along CMs would depolymerization of microtubules prevent the disappearance of Kar9 from the SPB. Determining the consequences of microtubule depolymerization upon Kar9 and Cdc28 distribution in an otherwise unperturbed cell cycle therefore offers a simple way to differentiate between these two possibilities.
To test both models, G1 cells were released from the cell cycle block in the presence of the microtubule depolymerizing drug nocodazole. Nocodazole treatment did not affect cell cycle progression through G1 and S phase as judged by flow cytometry and the formation of buds (data not shown). The binding of both Cdc28–4GFP (Figure 5A and B) and Kar9–GFP (Figure 5C) to the bud neck in G1/S was strongly reduced by the drug treatment, suggesting that microtubules were essential to target Cdc28 and Kar9 to the bud neck. Consistent with the second model, SPB dissociation of Kar9 was prevented when cells were treated with nocodazole. When untreated cells progressed from G1 into G1/S, SPB association of Kar9–GFP decreased from 83 to 32% (Figures 4B and 5C). However, in the presence of nocodazole, the number of G1/S cells with Kar9–GFP at SPBs did not drop, rather it increased to a level that was slightly higher than that seen in G1 cells prior to addition of the drug (Figure 5C). Cdc28–4GFP binding to SPBs in G1/S was not altered by nocodazole (Figure 5B). This result is consistent with a model in which Kar9 binds to SPBs in G1 and then moves from the SPB to the bud neck in G1/S. This movement requires the presence of microtubules. Moreover, Cdc28 binding to the bud neck, but not to SPBs in G1/S, is microtubule dependent.
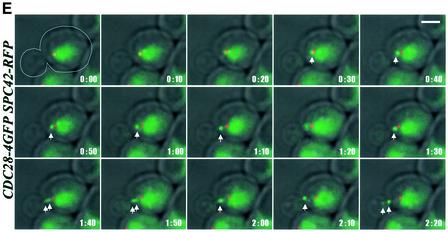
Fig. 5. Localization of Cdc28 and Kar9 with the bud cortex requires microtubules. (A) Cdc28–4GFP localization in nocodazole-treated G1/S cells. The arrows indicate Cdc28–4GFP co-localizing with the SPB marker Spc42–RFP. (B and C) Cells were synchronized by α-factor and released into medium with or without nocodazole. G1 and G1/S cells were quantified. (D) Time-lapse analysis of KAR9–GFP SPC42–RFP cells. Shown are the merged Kar9–GFP (shown in green) and Spc42–RFP signals (red) and phase contrast taken every 10 sec. (E) As (D) but with CDC28–4GFP SPC42–RFP cells. The arrows highlight the position of Cdc28–4GFP moving from the SPB to the bud cortex. Bars: 5 µm.
Time-lapse analysis confirmed that in G1/S cells the Kar9–GFP signal at SPBs (marked by red Spc42–RFP) migrated to the bud neck (Figure 5D). The linear nature of the migration suggests that this movement is most likely to be occurring along CM tracks. In some experiments, the Kar9–GFP signal, which originated at SPBs, even moved 1–2 µm backwards towards the SPB and then again reversed its movement to the bud neck (data not shown). It remains unclear whether the backward movement of Kar9 is due to a minus end-directed motor protein or to the Kar9–GFP holding on to the tip of a disassembling CM. Movements of Cdc28–4GFP from the SPB to the bud neck were also observed when CDC28–4GFP SPC42–RFP cells were used for the time-lapse experiment (Figure 5E, the green Cdc28–4GFP signal is marked by an arrow).
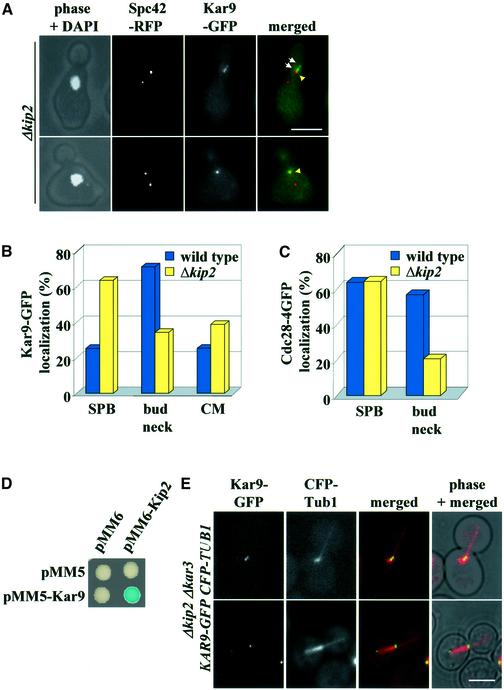
If Kar9 and Cdc28 were moving along CMs, we would expect to find that this movement was dependent upon the function of a microtubule-dependent motor protein. In budding yeast, the kinesin-like proteins Kar3, Kip2 and Kip3 and the dynein–dynactin complex all play an important role in spindle alignment (Hildebrandt and Hoyt, 2000). Smy1 shows homology to kinesin-like motor proteins; however, it functions together with the myosin motor Myo2 (Lillie and Brown, 1994). Kar9–GFP localization in G1/S was only affected in Δkip2 cells, but not drastically altered in Δkar3, Δkip3, Δsym1 or Δact5 cells [deletion of ACT5 impairs the function of the dynein–dynactin complex (Li et al., 1994)]. In contrast to wild-type cells, most Δkip2 cells in G1/S showed a Kar9–GFP signal close to the bud-directed SPB (Figure 6A and B). Moreover, when Kar9 was seen along CMs, the signal was much stronger nearer the SPB (Figure 6A, top) while in wild-type cells in G1/S, any association with the CMs was seen near the bud neck and not the SPB (Figure 4A, bottom). Localization of Cdc28 with the bud neck was also reduced in Δkip2 cells whereas the association of Cdc28 with the SPBs was not affected by KIP2 deletion (Figure 6C). This result suggests that Kip2 is important to concentrate Cdc28 and Kar9 at the bud neck, probably by interacting with Cdc28 or Kar9. Indeed, we observed a two-hybrid interaction of Kip2 and Kar9 (Figure 6D).
Fig. 6. Kip2 is required to concentrate Cdc28 and Kar9 at the bud neck. (A) Localization of Kar9–GFP in α-factor synchronized Δkip2 cells in G1/S. The arrows indicate Kar9–GFP close to the bud-ward-orientated SPB. (B) Quantification of (A). Cells (n > 100) were analysed 40 min after the release of the α-factor block. (C) Localization of Cdc28–4GFP in synchronized Δkip2 cells in G1/S. (D) Kip2 interacts with Kar9 in the yeast two-hybrid system. (E) Kar9–GFP of Δkip2 Δkar3 KAR9–GFP CFP-TUB1 cells associates with the minus end of CMs, but not with the site where CMs contact the bud cortex. Bars: 5 µm.
CMs are shorter and less dynamic in Δkip2 than in wild-type cells. Deletion of the motor KAR3 reversed these defects of Δkip2 cells (Huyett et al., 1998). To exclude the possibility that the reduced binding of Kar9 with the bud neck in Δkip2 cells was due to a change in CM properties, rather than an active role for Kip2 in transporting Kar9, we investigated Kar9 localization in Δkip2 Δkar3 cells. In Δkip2 Δkar3 cells, Kar9–GFP still showed the same reduction in association with the bud neck that was seen in single Δkip2 mutant cells. In fact, most of the Kar9–GFP localized close to SPBs in Δkip2 Δkar3 KAR9–GFP SPC42–RFP cells (data not shown). To ensure that Δkip2 Δkar3 cells with relatively normal CMs fail to transport Kar9 from the SPB to the bud cortex, microtubules were stained with CFP–Tub1. The Kar9–GFP signal of G1/S and even S phase cells still associated with the minus end (the microtubule end associated with the SPB) of long CMs that were directed into the bud. No Kar9–GFP was detected at the site where CMs contacted the bud cortex (Figure 6E). This result indicates that even in Δkip2 cells with normal CMs, Kar9 fails to become transported from the SPB to the bud cortex. It is therefore likely that Kip2 is directly involved in the transport of Kar9.
Kar9 binds to the bud-ward-directed SPB
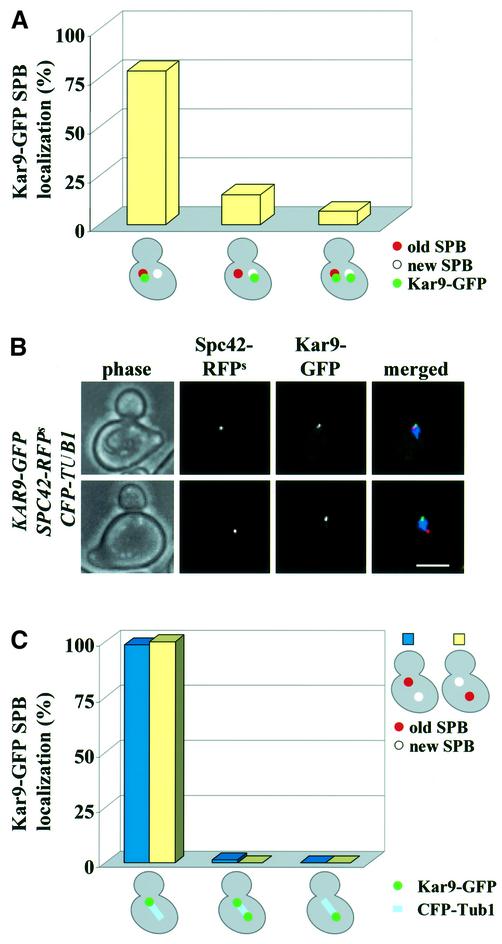
Kar9 predominately associates with the CMs that enter the bud and not the CMs that contact the cortex of the mother cell (Figure 4A, bottom; Beach et al., 2000). In most (90–95%), but not all, wild-type cells, the old SPB segregates into the bud while the new SPB stays in the mother cell (Pereira et al., 2001). This raises the possibility that Kar9 could bind exclusively to the old SPB, which has experienced at least one cell cycle and therefore may have been subjected to a modification that the new SPB is yet to experience. If this is the case, then Kar9 should only associate with the old SPB, even in the rare cases (5–10%) when the new SPB migrates into the bud. To test this possibility, the SPB protein Spc42 was fused with a slow folding version of RFPS (SPC42-RFPS) (Pereira et al., 2001). The slow folding property of this RFPS variant allows clear distinguishing of the old (fluorescent) from the new (non-fluorescent) SPB. To increase the percentage of cells with separated SPBs and Kar9 staining, SPC42-RFPS KAR9–GFP cells were treated with nocodazole. In ∼10% of cells, Kar9–GFP was only associated with the new SPB, which was not labelled by Spc42–RFPS (Figure 7A). This result suggested that Kar9 binding was not determined by the age of the SPB.
Fig. 7. Kar9 binds to the bud-ward-directed SPB. (A) SPB association of Kar9–GFP in KAR9–GFP CFP-TUB1 SPC42–RFPS cells was analysed after nocodazole treatment. (B) Kar9–GFP SPB association of synchronized KAR9–GFP CFP-TUB1 SPC42–RFPS Δkip2 cells. The CFP-Tub1 signal is shown in the merged figure in blue. Bar: 5 µm. (C) Quantification of (B). n > 200.
In Δkip2 cells, Kar9 remains associated with SPBs even when they separate in S phase (Figure 6A). In addition, due to the defect of CMs in interacting with the cell cortex (Huyett et al., 1998), SPB inheritance is disturbed in Δkip2 cells. In 19% of Δkip2 cells, the old SPB stayed in the mother cell and the new SPB segregated into the bud. This property of Δkip2 cells allowed us to study what determines SPB binding of Kar9. Kar9 localization of cells with CFP-TUB1 (to indicate the position of SPBs) and SPC42-RFPS (marks the old SPB) was followed through the cell cycle. In cells with a short spindle, Kar9–GFP always associated with the bud-ward-directed SPB independently, whether it was the old or the new (Figure 7B and C). This result established that it is not the age of the SPB that determines SPB binding of Kar9, rather the position of the SPB relative to the location of the bud determines SPB binding of Kar9.
Discussion
During asymmetric division of cells of Drosophila melanogaster and Caenorhabditis elegans and during spindle assembly in budding and fission yeast, the spindle becomes orientated relative to spatial cellular markers (Schaefer and Knoblich, 2001). Therefore, the two sets of CMs, each organized by one of the two SPBs or centrosomes, have to be captured by at least two distinct subdomains of the cell cortex. Cells must have developed mechanisms that ensure that the stable interaction of the two CM sets with the same cortical domain is rare since this would ultimately lead to a mispositioning of the spindle.
A yeast protein that is involved in the differential attachment of CMs to the cell cortex is Kar9. Using GFP–KAR9, mostly expressed from the Gal1 promoter, Kar9 was localized to the SPB, along CMs, the bud neck and the bud tip (Miller and Rose, 1998; Miller et al., 1999, 2000; Beach et al., 2000; Korinek et al., 2000; Lee et al., 2000; Kusch et al., 2002). However, the functional significance of Kar9 at SPBs and CMs, the interdependency of these localizations and their regulation remained unclear. Here we report the identification of a novel mechanism of how Cdc28 and Kar9 are transported from SPB to the bud neck in G1/S of the cell cycle. Thereby both proteins ensure the timely correct alignment of the mitotic spindle. Our data are consistent with a model in which Kar9 binds to SPBs in G1 of the cell cycle (Figure 8, step 1). Kar9 was always associated with the bud-ward-directed SPB upon spindle formation, suggesting a preferential binding to or stabilization of Kar9 at this SPB. How Kar9 is targeted to only one of the two SPBs is not understood. As one possibility, the CMs that contact the bud cortex first, which are in most cases organized by the old SPB (Segal et al., 2000), modify this SPB to allow the preferential binding of Kar9. Such a mechanism has been proposed for the checkpoint protein Bfa1 (Pereira et al., 2001). Pulling or pushing forces applied via CMs onto the SPB, or molecules, which move from the bud cortex along CMs to the SPB, may modify the SPB to allow Kar9 binding.
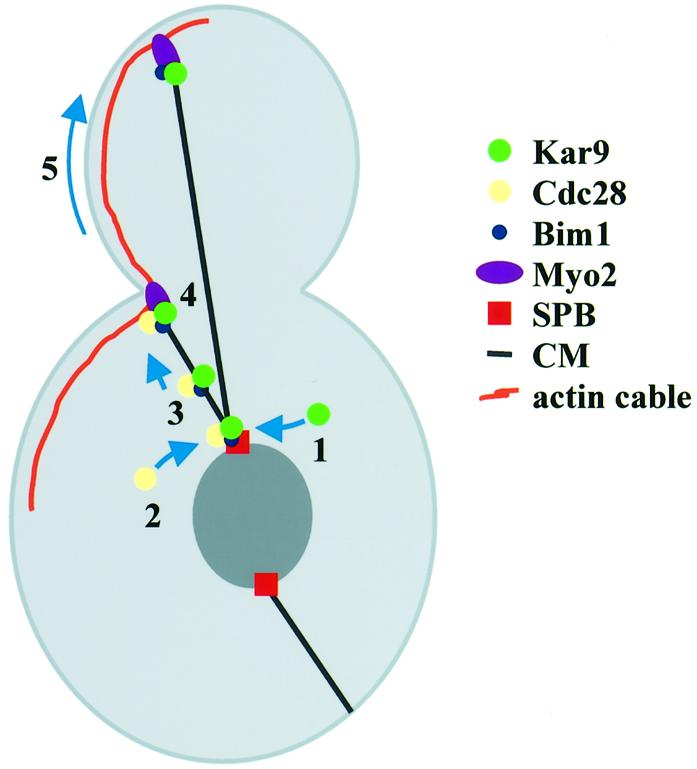
Fig. 8. Model for Cdc28–Kar9 translocation from the SPB to the bud neck. See text for details.
In G1/S, Cdc28 associates with SPBs (step 2) and Kar9 becomes phosphorylated. Four observations suggest that Cdc28 directly phosphorylates Kar9. First, Cdc28 and Clb5 interacted with Kar9 in the yeast two-hybrid system. Secondly, the in vivo phosphorylation of Kar9 coincides with Cdc28 binding to SPBs. Thirdly, phosphorylation of Kar9 is dependent on Cdc28 activity in vivo. Fourthly, only Cdc28, and not the kinase-dead Cdc28K40L, was able to phosphorylate recombinant Kar9 in vitro.
Kar9 and Cdc28 then move from the SPB to the plus end of CMs (step 3). The Bim1 and Kar9 dependency of the Cdc28 translocation, complex formation of Bim1 and Kar9 (Korinek et al., 2000; Lee et al., 2000; Miller et al., 2000) and the similar cell cycle-dependent movement of Cdc28 and Kar9 from the SPB to the bud neck (Figure 5D and E) are consistent with the notion that a complex of Cdc28 and Kar9 and possibly Bim1 translocates from the SPB to the bud cortex. The movement of Cdc28 and Kar9 was dependent upon the microtubule motor protein Kip2. A role for Kip2 in moving Kar9 to the bud cortex is further supported by the phenotype of Δkip2 cells. Δkip2 cells have a similar spindle orientation defect as Δkar9 cells (Miller et al., 1998). A likely interpretation of these results is that the microtubule-dependent motor Kip2 interacts with a component of the Kar9 complex and directly transports Cdc28 and Kar9 to the plus end of CMs. In this respect, it is interesting that Kar9 and Kip2 interact in the yeast two-hybrid system. Alternatively, Kar9, as is probably the case for the fission yeast cell end marker Tea1 (Mata and Nurse, 1997; Behrens and Nurse, 2002), binds to CM plus ends and becomes transported by microtubule growth to the cell cortex. In this case, the defect of Kar9 translocation would be the result of the shorter and less dynamic CMs of Δkip2 cells (Huyett et al., 1998). We consider this latter possibility less likely since the deletion of KAR3, which reverses the CM defect of Δkip2 cells (Huyett et al., 1998), did not restore the transport of Kar9 from the SPB to the bud cortex. In any case, additional studies are required to fully understand the molecular function of Kip2 in Kar9 translocation.
Based on the lethality of Δkip2 Δkar9 double mutants, it has been suggested that KIP2 functions together with the dynein-dependent pathway in spindle assembly (Miller et al., 1998). Our data now suggest that Kip2 functions also in the early Kar9-dependent spindle orientation pathway. The role of Kip2 in the early pathway was probably not detected by the genetic assay (viability of Δkip2 Δdyn1 double mutant) because of the redundancy of motor protein function in localizing Kar9 to the bud neck. Moreover, not all interactions of Kar9 with the bud cortex are affected in Δkip2 cells. Binding of Kar9 to the bud tip in mitosis occurs in Δkip2 cells with equal efficiency to that in wild-type cells (Miller et al., 1998).
Kar9 at CMs plus ends then facilitates the interaction of CM ends with the bud cortex probably through its binding to the actin motor Myo2 (step 4). Myo2 transports the CMs along actin cables towards the bud tip (step 5) (Yin et al., 2000) where Kar9 becomes anchored to the bud tip (Miller et al., 1999). Kar9 also binds to the bud tip in the absence of CMs, and this binding is not affected when Cdc28 activity is impaired (Figure 4F). Thus, the movement of Kar9 from the SPB to the bud cortex is not an absolute necessity for the bud tip localization.
What is the role of Cdc28 at CMs? As discussed above, one target of Cdc28 is likely to be Kar9 itself. Cdc28 may also regulate other proteins involved in Kar9 translocation. For example, Kip2 is separated into multiple bands by SDS–PAGE, which may represent phospho-forms of Kip2 (T.Usui, unpublished data). Cdc28 could regulate the cell cycle-dependent translocation of Kar9 from SPBs to the plus end of CMs. This model is consistent with the observation that Kar9 translocation coincides with Cdc28 binding to SPBs. Moreover, when Cdc28 activity is impaired, Kar9 transport to CMs plus ends is inefficient and hence CMs fail to establish interactions with the bud neck. In addition, Cdc28 could modulate the interaction of Kar9 with Myo2 at the bud cortex (Yin et al., 2000).
The selective binding of Kar9 to one SPB and its Cdc28-regulated translocation to CM plus ends ensure that only one of the two CM sets is able to establish Kar9–Myo2 contacts with the bud cortex. The other CMs, which are organized by the SPB that stays in the mother cell, do not carry Kar9 and therefore fail to establish stable contacts even if they accidentally touch the bud cortex. Due to the failure to concentrate Kar9 at CM plus ends, the differential binding mechanism is impaired in cdc28-4 Δclb5 cells. This deficiency of cdc28-4 Δclb5 cells may contribute to the frequent mis-segregation of the entire nucleus into the bud (Segal et al., 1998, 2000). Thus, the Cdc28-regulated localization of Kar9 at CMs plus ends is an important part of the program that aligns spindles and prevents chromosome mis-segregation in budding yeast.
Adherens junctions of Drosophila epithelial cells provide a planar cue to orient the mitotic spindle during symmetrical division. The adenomatous polyposis coli (APC) tumour suppressor, which shows limited homology to Kar9, and EB1, the homologue of Bim1, are involved in this orientation (Bienz, 2001). It is therefore likely that, in other organisms also, the capture of Kar9–Bim1-related complexes by the cell cortex contributes to spindle orientation. Moreover, the regulation of APC–EB1 complexes could be similar to budding yeast. In Schizosaccharomyces pombe and mammalian cells, Cdk1 and cyclin B1 also transiently associate with the SPB or centrosome (Bailly et al., 1989; Alfa et al., 1990). It will be important to investigate whether in higher eukaryotes APC–EB1 complexes are transported from the centrosome to the cell cortex in a Cdk-regulated manner.
Materials and methods
Plasmids and yeast strains
Strains and plasmids are listed in Table I. Yeast strains were derivatives of YPH499 (Sikorski and Hieter, 1989). Yeast strains were constructed by PCR-based methods (Knop et al., 1999). Plasmid pSM1023 (4GFP–KanMX6) was used for the construction of CDC28–4GFP, pSM822 for the slow folding SPC42–RFPS (Pereira et al., 2001) and pYM12 for KAR9–GFP (Knop et al., 1999). The RFP–natNT2 cassette, which encodes a fast folding RFP with nourseothricin resistance as selectable marker, was constructed by combining described mutations (Bevis and Glick, 2002; Knop et al., 2002). Two-hybrid interactions were determined as described previously (Janke et al., 2001).
Table I. Yeast strains and plasmids.
| Name | Genotype construction | Source or reference |
|---|---|---|
| Yeast strains | ||
| ESM356-1 | MATa ura3-52 trp1Δ63 his3 Δ200 leu2Δ1 | Pereira et al. (2001) |
| SHM285 | MATa ura3-52 trp1Δ63 his3Δ200 leu2Δ1 CDC28-4GFP-KanMX6 SPC42-RFP-NatNT2a | This study |
| SHM288 | MATa ura3-52 trp1Δ63 his3Δ200 leu2Δ1 CDC28-4GFP-KanMX6 KAR9-RFP-NatNT2 | This study |
| SHM289 | MATa ura3-52 trp1Δ63 his3Δ200 leu2Δ1 CDC28-4GFP-KanMX6 SPC42-RFP-NatNT2 Δkar9::klTRP1b | This study |
| SHM292 | MATa ura3-52 trp1Δ63 his3Δ200 leu2Δ1 KAR9-GFP-klTRP1 KanMX6-Gal1-HA-CLB5 cdc28-4 | This study |
| SHM296 | MATa ura3-52 trp1Δ63 his3Δ200 leu2Δ1 KAR9-3HA-klTRP1 KanMX6-Gal1-HA-CLB5 cdc28-4 | This study |
| SHM315 | MATa ura3-52 trp1Δ63 his3Δ200 leu2Δ1 KAR9-GFP-His3MX6 SPC42-RFP-NatNT2 | This study |
| SHM323 | MATa ura3-52 lys2-801 ade2-101 trp1Δ63 his3Δ200 leu2Δ1 KAR9-3HA-His3MX6 bar1 | This study |
| SHM326 | MATa ura3-52 trp1Δ63 his3Δ200 leu2Δ1 CDC28-4GFP-KanMX6 SPC42-RFP-NatNT2 Δbim1::hphMX4c | This study |
| SHM327 | MATa ura3-52 trp1Δ63 his3Δ200 leu2Δ1 CDC28-4GFP-KanMX6 SPC42-RFP-NatNT2 Δkip2::klTRP1 | This study |
| SHM342 | MATa ura3-52 trp1Δ63 his3Δ200 leu2Δ1 KAR9-GFP-His3MX6 SPC42-RFP-NatNT2 Δkip2::klTRP1 | This study |
| SHM357 | MATa ura3-52 trp1Δ63 his3Δ200 leu2Δ1 CDC28-4GFP-KanMX6 CFP-TUB1 | This study |
| SHM370 | MATa ura3-52 trp1Δ63 his3Δ200 leu2Δ1 SPC42-RFPs-KanMX6 KAR9-GFP-klTRP1 CFP-TUB1 | This study |
| SHM382 | MATa ura3-52 trp1Δ63 his3Δ200 leu2Δ1 SPC42-RFPs-KanMX6 KAR9-GFP-klTRP1 Δkip2::hphMX4 CFP-TUB1 | This study |
| SHM466 | MATa ura3-52 trp1Δ63 his3Δ200 leu2Δ1 SPC42-RFP-NatNT2 KAR9-GFP-His3MX6 Δkip2::klTRP1 Δkar3::hphMX4 CFP-TUB1 | This study |
| YAS8 |
MATa ura3-52 lys2-801 ade2-101 trp1Δ63 his3Δ200 leu2Δ1 bar1 |
Spang et al. (1996) |
| Plasmids | ||
| pACT2 | 2 µm, LEU2-based vector carrying the GAL4 activator domain | Durfee et al. (1993) |
| pCFP-TUB1 | pRS306 carrying CFP-TUB1 | Jensen et al. (2001) |
| pHM19 | pMM5 carrying CLB5 | This study |
| pHM21 | pMM5 carrying CDC28 | This study |
| pHM49 | pGEX-4T-1 carrying KAR9 (codons 369–644) | This study |
| pKB174 | CEN CDC28-HA TRP1 | Agarwal and Cohen-Fix (2002) |
| pKB173 | CEN cdc28K40L-HA TRP1 | Agarwal and Cohen-Fix (2002) |
| pMM5 | p423-pGal1-lexA-Myc | M.Knop |
| pMM6 | p425-pGal1-lexA-Myc | M.Knop |
| pSE72 | pMM6 carrying KAR1 (codons 1–410) | This study |
| pSE74 | pMM6 carrying SPC42 (codons 1–138) | This study |
| pSE76 | pMM6 carrying SPC42 (codons 96–363) | This study |
| pSM833 | pMM5 carrying KAR9 | This study |
| pSM834 | pMM6 carrying KAR9 | This study |
| pSM855 | pMM6 carrying KIP1 | This study |
| pSM856 | pMM6 carrying KIP2 | This study |
| pSM859 | pMM6 carrying KIP3 | This study |
| pUG41 | pACT2 carrying KAR3 | This study |
| pUG15 | pACT2 carrying CIK1 | This study |
aNatNT2 encodes the Streptomyces noursei nat1 gene.
bklTRP1 encodes the Kluyveromyces lactis TRP1 gene.
chphMX4 encodes the E.coli hph gene.
Cell cycle analysis and growth conditions
Yeast strains were grown in yeast extract, peptone, dextrose medium containing 100 mg/l adenine (YPAD medium) (Sherman, 1991). Cells were blocked in G1 by the addition of 10 µg/ml mating pheromone α-factor. After 2.5 h at 23°C or 2 h at 30°C, cells were released from the block by washing with YPAD medium. The old SPB was selectively labelled following a standard protocol (Pereira et al., 2001). cdc28-4 Gal1-CLB5 cells were grown in YP medium with 3% raffinose and 2% galactose (YPRG). Gal1-CLB5 expression was repressed by washing the cells with YPAD. Microtubules were depolymerized with 15 µg/ml nocodazole. DNA content was analysed by flow cytometry (Hutter and Eipel, 1979).
Phopshorylation of Kar9
Kar9-3HA was immunoprecipitated from logarithmically growing cells and incubated with buffer alone or 40 U λ phosphatase either with or without the inhibitor EDTA (5 mM) for 1 h at 30°C. For the Cdc28 dependence of Kar9 phosphorylation, wild-type and cdc28-4 Gal1-CLB5 cells were incubated with 200 mM HU at 23°C in YPRG until >90% of the cells had large buds. For the in vitro phosphorylation assay, an EcoRI–SalI fragment encoding amino acids 369–644 of Kar9 was subcloned into pGEX-4T-1 (Pharmacia). Cdc28-HA or the kinase-dead Cdc28K40L-HA were purified from Δkar9 yeast cells carrying plasmid pKB174 (CDC28-HA) or pKB173 (cdc28K40L-HA) (Agarwal and Cohen-Fix, 2002). The HA-tagged proteins were enriched using anti-HA beads (12CA5). For the in vitro phosphorylation, purified GST–Kar9369–644 and GST were incubated with beads carrying Cdc28-HA or Cdc28K40L-HA in kinase buffer (50 mM Tris–HCl, 20 mM MgCl2, 1 mM dithiothreitol and 0.5 mM sodium orthovanadate pH 7.5) containing [32P]γ-ATP for 45 min.
Image analysis
GFP- or RFP-labelled cells were analysed by fluorescence microscopy after fixing the cells with 3.7% formaldehyde. DNA of cells was stained with 4′,6-diamidine-2-phenylindole (DAPI) (Pereira et al., 2000). Single plane time-lapse analysis of living cells was performed as described previously (Pereira et al., 2001).
Acknowledgments
Acknowledgements
We are grateful to Dr I.Hagan for comments to the manuscript and Drs O.Cohen-Fix and S.Jensen for materials. E.S. is supported by Cancer Research UK. T.U. is financed by a grant of the Bioarchitect program of the RIKEN Institute.
References
- Agarwal R. and Cohen-Fix,O. (2002) Phosphorylation of the mitotic regulator Pds1/securin by Cdc28 is required for efficient nuclear localization of Esp1/separase. Genes Dev., 16, 1371–1382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alfa C.E., Ducommun,B., Beach,D. and Hyams,J.S. (1990) Distinct nuclear and spindle pole body populations of cyclin-cdc2 in fission yeast. Nature, 347, 680–682. [DOI] [PubMed] [Google Scholar]
- Bailly E., Doree,M., Nurse,P. and Bornens,M. (1989) p34cdc2 is located in both nucleus and cytoplasm: part is centrosomally associated at G2/M and enters vesicles at anaphase. EMBO J., 8, 3985–3995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beach D.L., Thibodeaux,J., Maddox,P., Yeh,E. and Bloom,K. (2000) The role of the proteins Kar9 and Myo2 in orienting the mitotic spindle of budding yeast. Curr. Biol., 10, 1497–1506. [DOI] [PubMed] [Google Scholar]
- Behrens R. and Nurse,P. (2002) Roles of fission yeast tea1p in the localization of polarity factors and in organizing the microtubular cytoskeleton. J. Cell Biol., 157, 783–793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bevis B.J. and Glick,B.S. (2002) Rapidly maturing variants of the Discosoma red fluorescent protein (DsRed). Nat. Biotechnol., 20, 83–87. [DOI] [PubMed] [Google Scholar]
- Bienz M. (2001) Spindles cotton on to junctions, APC and EB1. Nat. Cell Biol., 3, E1–E3. [DOI] [PubMed] [Google Scholar]
- Byers B. and Goetsch,L. (1975) Behavior of spindles and spindle plaques in the cell cycle and conjugation of Saccharomyces cerevisiae. J. Bacteriol., 124, 511–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donaldson D. and Kilmartin,V. (1996) Spc42p: a phosphorylated component of the S. cerevisiae spindle pole body (SPB) with an essential function during SPB duplication. J. Cell Biol., 132, 887–901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durfee T., Becherer,K., Chen,P.L., Yeh,S.H., Yang,Y., Kilburn,A.E., Lee,W.H. and Elledge,S.J. (1993) The retinoblastoma protein associates with the protein phosphatase type 1 catalytic subunit. Genes Dev., 7, 555–569. [DOI] [PubMed] [Google Scholar]
- Goh P.Y. and Kilmartin,J.V. (1993) NDC10: a gene involved in chromosome segregation in Saccharomyces cerevisiae. J. Cell Biol., 121, 503–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goode B.L., Drubin,D.G. and Barnes,G. (2000) Functional cooperation between the microtubule and actin cytoskeletons. Curr. Opin. Cell Biol., 12, 63–71. [DOI] [PubMed] [Google Scholar]
- Hildebrandt E.R. and Hoyt,M.A. (2000) Mitotic motors in Saccharomyces cerevisiae. Biochim. Biophys. Acta, 1496, 99–116. [DOI] [PubMed] [Google Scholar]
- Hutter K.J. and Eipel,H.E. (1979) Microbial determination by flow cytometry. J. Gen. Microbiol., 113, 369–375. [DOI] [PubMed] [Google Scholar]
- Huyett A., Kahana,J., Silver,P., Zeng,X. and Saunders,W.S. (1998) The Kar3p and Kip2p motors function antagonistically at the spindle poles to influence cytoplasmic microtubule numbers. J. Cell Sci., 111, 295–301. [DOI] [PubMed] [Google Scholar]
- Janke C., Ortíz,J., Lechner,J., Shevchenko,A., Shevchenko,A., Magiera,M.M., Schramm,C. and Schiebel,E. (2001) The budding yeast proteins Spc24p and Spc25p interact with Ndc80p and Nuf2p at the kinetochore and are important for kinetochore clustering and checkpoint control. EMBO J., 20, 777–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen S., Segal,M., Clarke,D.J. and Reed,S.I. (2001) A novel role of the budding yeast separin Esp1 in anaphase spindle elongation: evidence that proper spindle association of Esp1p is regulated by Pds1p. J. Cell Biol., 152, 27–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin Q.-W., Trelles-Sticken,E., Scherthan,H. and Loidl,J. (1998) Yeast nuclei display prominent centromere clustering that is reduced in nondividing cells and in meiotic prophase. J. Cell Biol., 141, 21–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knop M., Siegers,K., Pereira,G., Zachariae,W., Winsor,B., Nasmyth,K. and Schiebel,E. (1999) Epitope tagging of yeast genes using a PCR-based strategy: more tags and improved practical routines. Yeast, 15, 963–972. [DOI] [PubMed] [Google Scholar]
- Knop M., Barr,F., Riedel,C.G., Heckel,T. and Reichel,C. (2002) Improved version of the red fluorescent protein (drFP583/DsRed/RFP). BioTechniques, 33, 592–602. [DOI] [PubMed] [Google Scholar]
- Korinek W.S., Copeland,M.J., Chaudhuri,A. and Chant,J. (2000) Molecular linkage underlying microtubule orientation toward cortical sites in yeast. Science, 287, 2257–2259. [DOI] [PubMed] [Google Scholar]
- Kusch J., Meyer,A., Snyder,M.P. and Barral,Y. (2002) Microtubule capture by the cleavage apparatus is required for proper spindle positioning in yeast. Genes Dev., 16, 1627–1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee L., Tirnauer,J.S., Li,J., Schuyler,S.C., Liu,J.Y. and Pellman,D. (2000) Positioning of the mitotic spindle by a cortical–microtubule capture mechanism. Science, 287, 2260–2262. [DOI] [PubMed] [Google Scholar]
- Li M.H., Karpova,T.S. and Cooper,J.A. (1994) A yeast actin-related protein homologous to that in vertebrate dynactin complex is important for spindle orientation and nuclear migration. Cell, 78, 669–679. [DOI] [PubMed] [Google Scholar]
- Lillie S.H. and Brown,S.S. (1994) Immunofluorescence localization of the unconventional myosin, Myo2p, and the putative kinesin-related protein, Smy1p, to the same regions of polarized growth in Saccharomyces cerevisiae. J. Cell Biol., 125, 825–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim H.H., Goh,P.Y. and Surana,U. (1996) Spindle pole body separation in Saccharomyces cerevisiae requires dephosphorylation of the tyrosine-19 residue of Cdc28. Mol. Cell. Biol., 16, 6385–6397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mata J. and Nurse,P. (1997) tea1 and the microtubular cytoskeleton are important for generating global spatial order within the fission yeast cell. Cell, 89, 939–949. [DOI] [PubMed] [Google Scholar]
- Meluh P.B. and Rose,M.D. (1990) KAR3, a kinesin-related gene required for yeast nuclear fusion. Cell, 60, 1029–1041. [DOI] [PubMed] [Google Scholar]
- Miller R.K. and Rose,M.D. (1998) Kar9p is a novel cortical protein required for cytoplasmic microtubule orientation in yeast. J. Cell Biol., 140, 377–390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller R.K., Heller,K.K. and Rose,M.D. (1998) The kinesin-related proteins, Kip2p and Kip3p, function differently in nuclear migration in yeast. Mol. Biol. Cell, 9, 2051–2068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller R.K., Matheos,D. and Rose,M.D. (1999) The cortical localization of the microtubule orientation protein, Kar9p, is dependent upon actin and proteins required for polarization. J. Cell Biol., 144, 963–975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller R.K., Cheng,S.-C. and Rose,M.D. (2000) Bim1p/Yeb1p mediates the Kar9p-dependent cortical attachment of cytoplasmic microtubules. Mol. Biol. Cell, 11, 2949–2959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira G., Höfken,T., Grindlay,J., Manson,C. and Schiebel,E. (2000) The Bub2p spindle checkpoint links nuclear migration with mitotic exit. Mol. Cell, 6, 1–10. [PubMed] [Google Scholar]
- Pereira G., Tanaka,T.U., Nasmyth,K. and Schiebel,E. (2001) Modes of spindle pole body inheritance and segregation of the Bfa1p/Bub2p checkpoint protein complex. EMBO J., 20, 6359–6370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira G., Manson,C., Grindlay,J. and Schiebel,E. (2002) Regulation of the Bfa1p–Bub2p complex at spindle pole bodies by the cell cycle phosphatase Cdc14p. J. Cell Biol., 157, 367–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaefer M. and Knoblich,A.J. (2001) Protein localization during asymmetric cell division. Exp. Cell Res., 271, 66–74. [DOI] [PubMed] [Google Scholar]
- Schwartz K., Richards,K. and Botstein,D. (1997) BIM1 encodes a microtubule-binding protein in yeast. Mol. Biol. Cell, 8, 2677–2691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segal M., Clarke,D.J. and Reed,S.I. (1998) Clb5-associated kinase activity is required early in the spindle pathway for correct preanaphase nuclear positioning in Saccharomyces cerevisiae. J. Cell Biol., 143, 135–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segal M., Clarke,D.J., Maddox,P., Salmon,E.D., Bloom,K. and Reed,S.I. (2000) Coordinated spindle assembly and orientation requires Clb5-dependent kinase in budding yeast. J. Cell Biol., 148, 441–451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman F. (1991) Getting started with yeast. Methods Enzymol., 194, 3–21. [DOI] [PubMed] [Google Scholar]
- Sikorski R.S. and Hieter,P. (1989) A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics, 122, 19–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spang A., Geissler,S., Grein,K. and Schiebel,E. (1996) γ-tubulin-like Tub4p of Saccharomyces cerevisiae is associated with the spindle pole body substructures that organize microtubules and is required for mitotic spindle formation. J. Cell Biol., 134, 429–441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin H., Pruyne,D., Huffaker,T.C. and Bretscher,A. (2000) Myosin V orientates the mitotic spindle in yeast. Nature, 406, 1013–1015. [DOI] [PubMed] [Google Scholar]