Abstract
DNA mismatch repair is an essential safeguard of genomic integrity by removing base mispairings that may arise from DNA polymerase errors or from homologous recombination between DNA strands. In Escherichia coli, the MutS enzyme recognizes mismatches and initiates repair. MutS has an intrinsic ATPase activity crucial for its function, but which is poorly understood. We show here that within the MutS homodimer, the two chemically identical ATPase sites have different affinities for ADP, and the two sites alternate in ATP hydrolysis. A single residue, Arg697, located at the interface of the two ATPase domains, controls the asymmetry. When mutated, the asymmetry is lost and mismatch repair in vivo is impaired. We propose that asymmetry of the ATPase domains is an essential feature of mismatch repair that controls the timing of the different steps in the repair cascade.
Keywords: ABC ATPase/alternating ATPase/asymmetry/DNA mismatch repair/MutS
Introduction
During replication of the genome, the DNA mismatch repair (MMR) system screens the genome for errors produced by the DNA polymerase. Additionally, MMR will remove base mispairs that may arise as a consequence of homologous recombination as well as some chemical errors (Aquilina and Bignami, 2001; Bellacosa, 2001; Marti et al., 2002). In Escherichia coli, three proteins initiate MMR. The first, MutS, recognizes mispaired and unpaired bases (1–3 bases), collectively called mismatches (Su and Modrich, 1986; Parker and Marinus, 1992). After binding to a mismatch, MutS binds the second protein MutL which, in turn, activates the endonuclease MutH. MutH then nicks the transiently unmethylated daughter strand. At the site of the single-stranded nick, exonucleases and helicases are loaded on the DNA and directed to remove the daughter strand containing the mispaired base. In eukaryotes, the system is conserved, albeit more complex, with several MutS homologues (MSHx) that form heterodimers (MSH2–MSH6, called MutSα; and MSH2–MSH3, called MutSβ) with different mismatch specificities (Bellacosa, 2001; Marti et al., 2002). Likewise, the MutL homologues (MLHx and PMSx) also form heterodimers called MutLα (composed of MLH1–PMS1 in yeast and MLH1–PMS2 in humans) and MutLβ (composed of MLH1–MLH3 in yeast and MLH1–PMS1 in humans). In humans, mutations in the mismatch repair genes hMSH2, hMSH6 and hMLH1 cause predisposition to a frequent form of hereditary cancer called hereditary non-polyposis colorectal cancer (HNPCC) (Lynch and de la Chapelle, 1999).
In order to bind MutL, MutS needs to both recognize a mismatch and bind ATP. The role of ATP binding and hydrolysis has been studied intensively, but has led to different interpretations. In one model, it is proposed that MutS is a motor protein that uses ATP turnover to propel itself along the DNA (Allen et al., 1997; Blackwell et al., 1998). A second model predicts that MutS is a molecular switch that uses ATP binding and ATP hydrolysis to toggle between an active and inactive state (Gradia et al., 1997). The molecular switch model also predicts that after MutS has bound a mismatch, addition of ATP will cause MutS to form a so-called ‘sliding clamp’ that will diffuse away from the mismatch along the DNA until it reaches an open end, where it falls off (Gradia et al., 1999; Iaccarino et al., 2000). A third model proposes that MutS uses ATP to help it discriminate between heteroduplex and homoduplex DNA, and to ‘authorize’ initiation of repair after mismatch binding. In contrast to the previous two models, it proposes that movement away from the mismatch is not required for initiation of repair (Junop et al., 2001).
Although it is apparent that mismatch recognition and ATP binding are both required for formation of the MutS–MutL complex (Galio et al., 1999; Blackwell et al., 2001; Bowers et al., 2001; Schofield et al., 2001), it remains unclear how the communication between ATP and mismatch binding is orchestrated. Also, the regulation of the subsequent steps in the repair cascade is still largely unsolved, although an important role for MutL is evident as it not only forms a complex with MutS, but also stimulates DNA nicking by MutH (Hall and Matson, 1999), and DNA unwinding by UvrD (helicase II) (Yamaguchi et al., 1998).
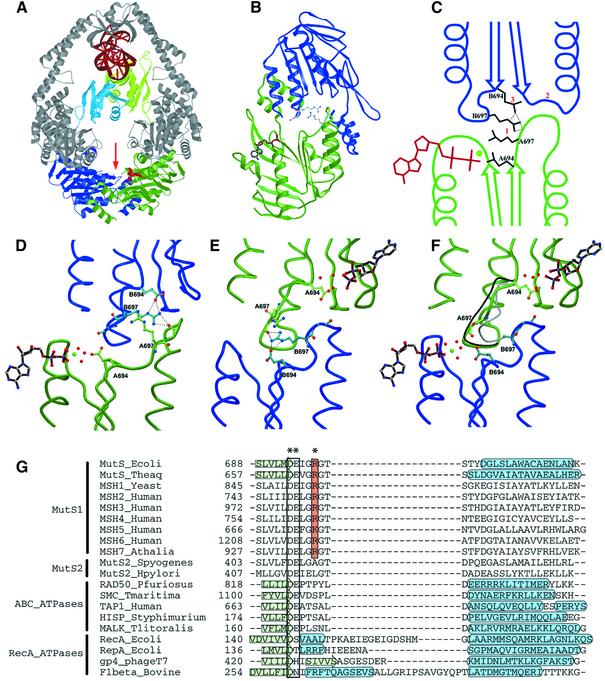
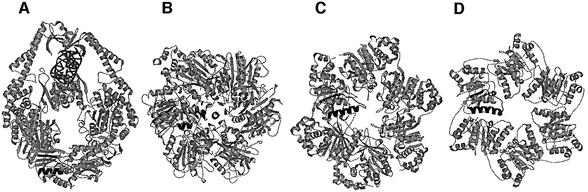
Previously, we have determined the crystal structure of E.coli MutS (ΔC800) binding to a DNA oligomer containing a single G:T mismatch (Lamers et al., 2000). The protein used for this structure lacks the C-terminal non-conserved 53 amino acids, but is proficient in mismatch recognition, ATPase activity and ATP-induced DNA release, and is able to complement mismatch repair in a MutS-deficient strain. In the crystal structure, two MutS molecules form a clamp that embraces the DNA (Figure 6A). The DNA-binding domains and ATPase domains are located at opposite ends of the complex, >90 Å apart. At the DNA-binding end, the two monomers encircle the DNA, but only one monomer contacts the mismatch directly. At the other end of the MutS complex, the two ATPase domains form the major interface of the MutS dimer. The overall features of this structure are very similar to those of a Taq MutS structure reported at the same time (Obmolova et al., 2000). Previously, the ATPase domain of MutS has been classified as belonging to the superfamily of ABC ATPases (Gorbalenya and Koonin, 1990) which includes the ABC membrane transporters such as cystic fibrosis transmembrane conductance regulator (CFTR), histidine permease and P-glycoprotein, and several DNA maintenance/repair enzymes including UvrA, SMC and RAD50. In the different crystal structures of ABC ATPases, several conformations of the ATPase dimer have been observed [HisP (Hung et al., 1998), MalK (Diederichs et al., 2000), RAD50 (Hopfner et al., 2000), MutS (Lamers et al., 2000; Obmolova et al., 2000), MsbA (Chang and Roth, 2001) and BtuCD (Locher et al., 2002)]. However, among these, only one dimer type has been observed multiple times: in the two MutS structures, in RAD50 and recently in the structure of BtuCD. It is also this dimer conformation that fits best with biochemical data (Jones and George, 1999; Hopfner et al., 2000; Junop et al., 2001; Mannering et al., 2001; Qu and Sharom, 2001).
Fig. 6. Structural analysis of asymmetry. (A) Structure of the MutS dimer in complex with mismatched DNA. Protein is coloured in grey, with ATPase domains of monomer A and B coloured in green and blue, respectively, and mismatch binding domains of monomer A and B in light green and light blue. DNA and ADP are in red. (B) The two ATPase domains viewed along the arrow in (A). The ADP molecule in monomer A is coloured in brown; the side chains of the two Arg697 (coloured in grey) are located at the centre of the interface of the two ATPase domains. (C) Schematic representation of the asymmetric interactions of the ATPase domains of monomer A (green) and B (blue). (1) ArgB697 hydrogen-bonds to the backbone of GlyA698 in the DE-loop of monomer A, while the reverse contact does not take place. (2) As a result, the DE-loop of monomer A clashes with the P-loop of monomer B, which is thus inhibited from nucleotide binding. (3) Simultaneously, ArgB697 also hydrogen-bonds to and displaces GluB694. (D) Close-up of the nucleotide-binding site of monomer A in green, with the opposing monomer B in blue. (E) Same view as (D), but now viewed from monomer B. (F) Same view as (E), but of R697A MutS. The position of the DE-loop in monomer A and B of wild-type MutS is indicated in grey and black, respectively. In the absence of Arg697, no contacts are made between the two DE-loops, and both monomer A and B now bind ADP-Mg2+. (G) Structure-based sequence alignment of MutS homologues and paralogues, and related ATPases. The conserved Arg697 is coloured in orange, and marked with an asterisk (*). The Walker B motif is boxed, and indicated by (**). Secondary structural elements are indicated by a green arrow (β-strand) or a blue tube (α-helix). PDB accession codes E.coli MutS, 1E3M; Taq MutS, 1EWQ; RAD50, 1F2U; SMC, 1E69; TAP1, 1JJ7; HisP, 1B0U; MalK, 1G29; RecA, 2REB; RepA, 1G8Y; gp4, 1E0J; F1, 1BMF.
The structure of E.coli MutS is not only asymmetric in mismatch binding but the chemically identical ATPase domains are also asymmetric in their nucleotide-binding mode. The monomer recognizing the mismatch binds an ADP molecule while the other monomer is empty. In contrast, the Taq MutS structures show no asymmetry in nucleotide binding (Obmolova et al., 2000; Junop et al., 2001). Nevertheless, it was shown that the two Taq MutS ATPase sites interact closely, suggesting that they could act sequentially (Junop et al., 2001). Asymmetry of the ATPase domains has also been observed in the differential effect of ATPase mutants for the human and yeast MSH2 and MSH6 proteins (Iaccarino et al., 1998; Studamire et al., 1998; Bowers et al., 2000), but it remains unclear if the asymmetry in the ATPase domains plays a role in the DNA mismatch repair. Here we show that the two ATPase domains of the E.coli MutS dimer are asymmetric not only in the crystal structure but also in solution. The two chemically identical ATPase domains bind ADP with different affinities and alternate during ATP hydrolysis even in the absence of DNA binding. Loss of asymmetry decreases the rate of hydrolysis, prevents ATP-induced release of DNA and ultimately affects mismatch repair in vivo.
Results
Nucleotide binding properties of MutS
To investigate the structurally observed asymmetry of the ATPase domains (Lamers et al., 2000), we asked if this asymmetry is present in solution and whether it affects the ATPase reaction. In this analysis, we make use of comparisons between wild-type MutS and an Arg697 to alanine mutant (R697A), that we created because analysis of the structure predicted that it could be important for the asymmetry of the ATPase domains.
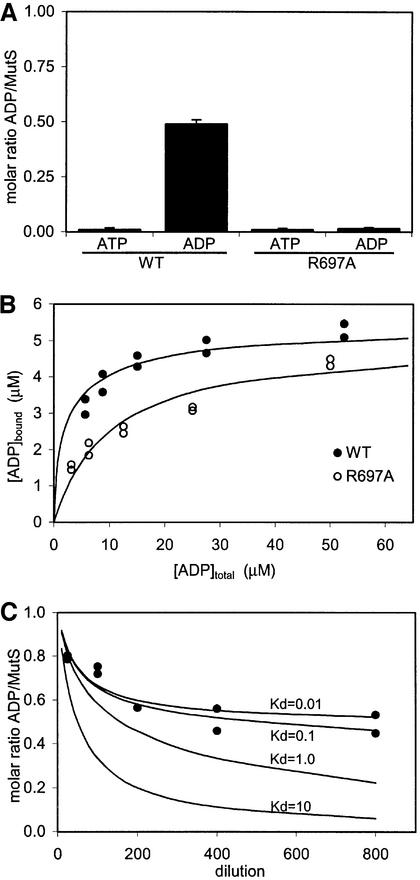
As a first step, we wanted to determine binding constants for the nucleotides. However, we had noticed previously that MutS retains a nucleotide after purification by two lines of evidence. First, in an ATPase assay in which ATP is regenerated from ADP, activity was found prior to addition of ATP to the reaction mixture. Secondly, when MutS was crystallized in the absence of nucleotides, electron density for ADP was clearly visible in the monomer binding to the mismatch (data not shown). This implies that MutS must bind the nucleotide with high affinity, enabling it to retain the nucleotide during purification. In order to determine the nature of the nucleotide, and to quantify the amount bound by purified MutS, the nucleotide was released from the protein through boiling and quantified in a bioluminescence assay. As shown in Figure 1A, no ATP could be observed in wild-type MutS. However, the amount of ADP in MutS was close to 50% of the protein (monomers MutS), suggesting that within the MutS dimer, only one of the two monomers retains ADP with high affinity. This finding is in agreement with earlier results obtained for human MutSα, where only ADP, but no ATP, could be detected (Blackwell et al., 1998). The R697A mutant was expressed and purified under conditions identical to the wild-type protein, showing no differences in multimerization as assayed by size exclusion chromatography (data not shown). However, R697A MutS no longer retains a nucleotide since neither ATP nor ADP could be detected in the bioluminescence assay (Figure 1).
Fig. 1. The MutS dimer has two non-equivalent ADP-binding sites. (A) Amount of ATP and ADP retained by purified MutS (wild-type and R697A) as measured in a luciferase assay. (B) Filter binding studies on wild-type and R697A MutS (5 µM) with increasing amounts of radiolabelled ADP. (C) Diluting out ADP from wild-type MutS. Black dots show the amount of radiolabelled ADP retained by MutS after dilution in a filter binding assay. Lines indicate theoretical dilution curves of a two-site binding model, with one site having a Kd of 10 µM and the second site as indicated in the graph.
To investigate whether both nucleotide-binding sites of the MutS dimer can bind ADP, we attempted to saturate both by adding increasing amounts of radiolabelled ADP in a filter binding assay. Figure 1B shows 100% saturation (monomers MutS), indicating that the second site can bind ADP, albeit with lower affinity than the first site. The binding curve in Figure 1B can be used for calculating approximate dissociation constants (Kd) but, since the high affinity site of wild-type MutS is already fully saturated when purified, no information about its affinity results from this experiment. An apparent dissociation constant (Figure 1C). could be calculated by diluting out the nucleotide. At intermediate dilutions (5–0.5 µM), the low affinity site rapidly loses the ADP. The high affinity site, however, retains the nucleotide even at very low concentrations (6.25 nM), leading to an estimated Kd of <0.1 µM. This value is in agreement with MutS retaining ADP during the purification, where it is never more dilute than 1 µM. When we use the value of 0.1 µM for the high affinity site to fit a two-site binding model to the binding curve of Figure 1B, we obtain a Kd for the second site of ∼10 µM (Table I).
Table I. ADP binding and ATP hydrolysis.
| Kd1 (µM) | Kd2 (µM) | Km (µM) | kcat (/min) | Hill coefficient | Inhibitiona | |
|---|---|---|---|---|---|---|
| Wild type | <0.1 | 10.3 ± 1.8 | 8.7 ± 1.0 | 13.2 ± 0.8 | 1.08 | 90% |
| R697A | 9.6 ± 2.0 | 9.6 ± 2.0 | 5.1 ± 0.7 | 1.1 ± 0.1 | 1.04 | 50% |
aInhibition of ATP hydrolysis at an equal molar ratio of ATP and AMPPNP.
In the R697A mutant, both monomers can bind ADP, but with a lower net affinity. Interestingly, fitting a two-site model to the binding curve of R697A MutS results in two sites with an equal affinity of ∼10 µM, again showing that in R697A MutS the high affinity site is lost.
The effect of asymmetry on ATP hydrolysis
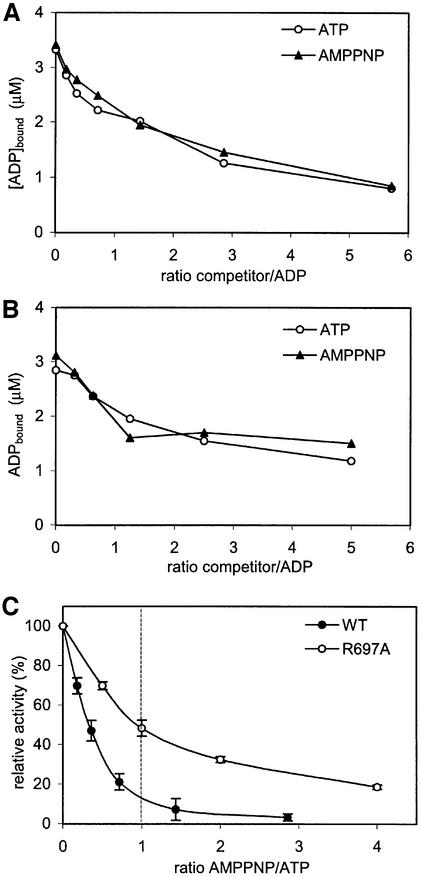
The data presented above strongly suggest that the two ATPase domains of the MutS dimer are asymmetric in nature. Next, we wanted to know whether there is an effect of the asymmetry on ATP hydrolysis. Analysis of the ATPase activity did not indicate cooperativity between the two domains, since the Hill coefficient is equal to 1. To see whether the two domains alternate during hydrolysis, we inhibited ATPase activity by the addition of the non-hydrolysable ATP analogue AMPPNP. In such an experiment, one of two outcomes can be expected. If only one monomer within the MutS dimer can hydrolyse ATP, or if both monomers hydrolyse independently from each other, 50% reduction in activity at 50% occupation of AMPPNP is expected. In contrast, if both monomers hydrolyse ATP, but in an alternating manner, occupation by AMPPNP in one monomer will also inhibit the other monomer, as it cannot progress without the first site completing hydrolysis. Thus, with an alternating ATPase, a much stronger reduction of activity at 50% occupation of AMPPNP is expected. For this experiment, the relative affinities for ATP and AMPPNP were determined by competing out radiolabelled ADP with unlabelled ATP or AMPPNP (Figure 2A and B). The two nucleotides competed with the bound ADP to the same extent, indicating that they are bound by MutS with equal affinities. In the ATPase inhibition experiments (Figure 2C), we found that at equal concentrations of ATP and AMPPNP, the activity of wild-type MutS was reduced by ∼90%. In contrast, the R697A mutant shows only 50% inhibition of ATPase activity at equimolar concentrations of ATP and AMPPNP.
Fig. 2. Inhibition of ATPase activity by AMPPNP. Competition of bound radiolabelled ADP with cold ATP or AMPPNP in (A) wild-type MutS and (B) R697A MutS. (C) Relative inhibition of ATPase activity by AMPPNP for wild-type and R697A MutS. The ATP concentration was kept constant, while the AMPPNP concentration was increased. Assay conditions were equal to those in (A) or (B).
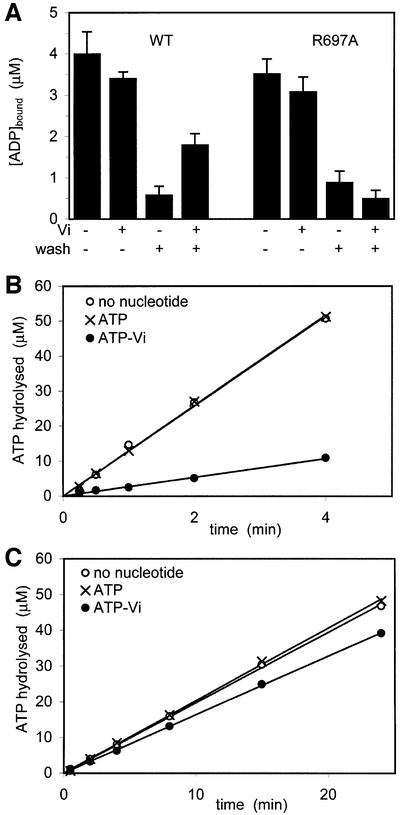
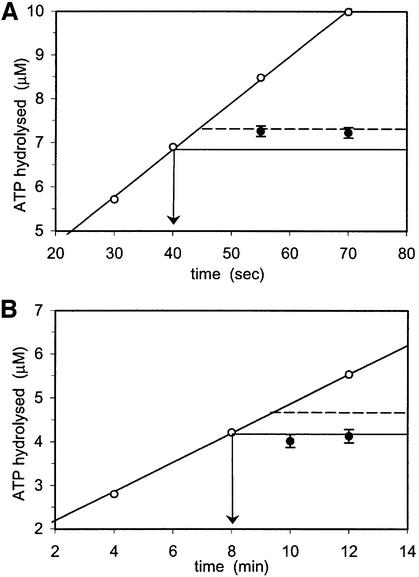
To confirm that inhibition of ATP hydrolysis in one site will prevent hydrolysis in the other nucleotide-binding site, we performed a vanadate (Vi) trapping experiment. Previously, vanadate was shown to inhibit ATP hydrolysis in the ABC ATPases, P-glycoprotein (Urbatsch et al., 1995b) and MalK (Sharma and Davidson, 2000), by forming a stable complex of ADP-Vi. In the trapping experiment, MutS was incubated with radiolabelled ATP in either the presence or absence of vanadate. Next, an excess of cold ATP was added to compete out the radiolabelled nucleotide. As shown in Figure 3A, in the presence of vanadate, both ATPase domains can bind a nucleotide, although with somewhat reduced affinity when compared with binding in the absence of Vi. After washing with cold ATP, most of the radiolabelled nucleotide is washed out after the pre-incubation with ATP only. However, after the pre-incubation with ATP and vanadate, only half of the bound nucleotide can be released with cold ATP. This is consistent with the observation that the two sites are asymmetric in nucleotide binding. Although both monomers can bind a nucleotide in the presence of Vi, only one site can be stably trapped by ADP-Vi. In a subsequent experiment, we analysed ATP hydrolysis after Vi trapping. For this, we used conditions identical to those used above, with the only difference that the pre-incubation step was performed with cold ATP, while the wash step was performed with radiolabelled ATP to monitor ATP hydrolysis afterwards (Figure 3B and C). Pre-incubation with ATP alone did not affect the rate of hydrolysis (∼12.5 µM ADP/min), but pre-incubation with vanadate resulted in a reduction up to 80% (∼2.7 µM ADP/min). This implies that Vi trapping in one monomer indeed inhibits hydrolysis in the other monomer. Interestingly, in R697A MutS, vanadate could not be stably trapped and, consequently, little inhibition of ATPase activity was observed.
Fig. 3. Vanadate trapping and ATPase inhibition. (A) MutS (5 µM) was incubated with radiolabelled ATP with or without vanadate (Vi). Samples were spotted directly onto nitrocellulose filters or washed first with a large excess of unlabelled ATP as indicated. (B and C) ATPase inhibition in wild-type and R697A MutS. In conditions identical to (A), MutS was incubated with unlabelled ATP with or without vanadate, after which labelled ATP was added to measure ATPase activity.
Finally, we used a pulse–chase experiment to analyse ATP hydrolysis during steady state. Here, radiolabelled ATP was incubated with MutS and subsequently chased with an excess of unlabelled ATP, after which the reaction was stopped with EDTA. As shown in Figure 4A, after addition of the ATP chase, a single turnover of 0.5 µM ATP was observed, equivalent to half the amount of MutS monomers. This indicates that during steady-state ATPase activity, hydrolysis takes place in only one monomer at a time. In contrast, for the R697A mutant, no additional hydrolysis is observed after the ATP chase (Figure 4B).
Fig. 4. Pulse–chase experiments on steady-state ATP hydrolysis for the wild type (A) and R697A MutS (B). During steady-state hydrolysis of radiolabelled ATP (35 µM), ATPase activity was quenched with EDTA at different time points (open circles), or first chased with a large excess (10 mM) of unlabelled ATP (arrow) before EDTA quenching (filled circles).
An additional observation suggests that the two ATPase sites are coupled in hydrolysis. It has been shown that in MutS, the rate-limiting step in hydrolysis is the release of ADP (Gradia et al., 1999; Bjornson et al., 2000). With the Km of the ATPase activity being similar to the Kd of the low affinity site, the release of ADP apparently takes place in the low affinity site. Nevertheless, mutation of Arg697 did not affect the Km of the protein, but still resulted in a >10-fold decrease in activity. Since the R697A mutation primarily affects the high affinity site, this suggests that in addition to the low affinity site, the high affinity site also plays an important, but separate role in ATP hydrolysis.
Communication between the ATPase and DNA-binding domains
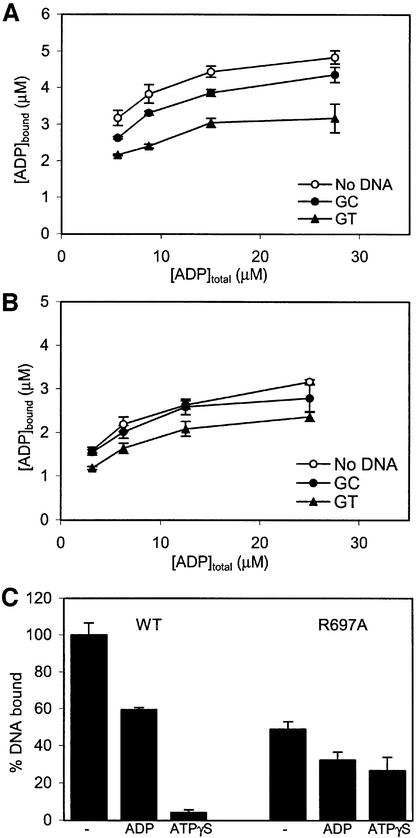
In the crystal structure, it was observed that the asymmetry in mismatch binding is ‘synchronized’ with nucleotide binding: the monomer binding to the mismatch also retains the nucleotide, while the other monomer does neither. Hence, we have studied the effect of asymmetry on the communication between the ATPase domain and the DNA-binding domain. For this purpose, ADP binding was measured in the presence and absence of homoduplex and heteroduplex DNA oligomers. As shown in Figure 5A, both types of DNA reduce the affinity for ADP, with the strongest effect observed for heteroduplex DNA. The R697A MutS behaves qualitatively similarly, but in this mutant the effect of homoduplex DNA is less (Figure 5B).
Fig. 5. Communication between ATPase and DNA-binding domains. (A) ADP binding in the absence or presence of homoduplex (GC) or heteroduplex (GT) DNA for wild-type MutS. (B) ADP binding for R697A MutS in the presence of DNA. (C) Relative amounts of DNA bound to MutS in the absence or presence of ADP or ATPγS. DNA bound by wild-type MutS in the absence of nucleotides was set to 100%.
In the opposite direction, the communication from ATPase domains to DNA-binding domains is strongly reduced in R697A MutS. R697A MutS still discriminates between homo- and heteroduplex DNA as proficiently as wild-type, albeit with somewhat reduced affinities (Table II). However, the ATP-induced release of DNA is nearly completely lost in the mutant MutS (Figure 5C), even though at the ATP concentration used both monomers will bind ATP (Figure 2B). This is consistent with the observed effects of the R697A mutation on nucleotide binding, where the high affinity site is lost. Apparently, in this mutant, a stable ATP-bound state cannot be achieved, resulting in the decreased DNA release.
Table II. DNA binding and in vivo repair.
| Kd-GT (µM) | Kd-GC (µM) | Kd-GC/Kd-GT | ATP releasea (%) | Rifampicin resistanceb | |
|---|---|---|---|---|---|
| Wild type | 0.44 ± 0.05 | 9.5 ± 0.9 | 21.6 | 4 | 0.2 ± 3.8 |
| R697A | 0.92 ± 0.10 | 17.8 ± 1.9 | 19.3 | 25 | 45 ± 12 |
| E694N | – | – | – | – | 45 ± 15 |
| mutS– | – | – | – | – | 29 ± 4 |
aRelative amount of DNA bound in the presence of ATPγS when compared with the amount bound in the absence of nucleotides (Figure 5C).
bMedian number of rifampicin-resistant colonies per 1 × 108 cells plated out.
In vivo repair: R697A MutS has a mutator phenotype
To analyse the importance of Arg697 in mismatch repair in general, we performed in vivo complementation assays to monitor the mutation frequency of R697A MutS (Table II). R697A MutS has a mutator phenotype similar to the control mutant E694N which is defective in ATP hydrolysis (data not shown), and which has a strong mutator phenotype (D.Georgijevic and N.de Wind, personal communications). As shown above, the loss of asymmetry of the ATPase domains affects both ATPase activity (Table I) and ATP-dependent DNA release (Figure 5C). Because these activities are crucial to the functioning of MutS, disruption of either one, or both, will impair mismatch repair in general, leading to the observed mutator phenotype in vivo.
Structural analysis of asymmetry in the MutS ATPase domains
Arg697 was mutated after detailed analysis of the interactions at the interface of the two ATPase domains (Figure 6). In the two monomers, this residue contributes most clearly to the asymmetry of the two domains (Figure 6B). It is located in a loop (residues 695–703) that immediately follows the DE motif (Walker B motif). These ‘DE-loops’ are in close proximity to each other and contact one another in an asymmetric fashion (Figure 6C, D and E). The DE-loop of monomer A is in a position that clashes with the P-loop of the opposing monomer B (residues 615–620), preventing it from binding a nucleotide. In contrast, the DE-loop of monomer B does not interfere with the nucleotide-binding site of its opposing monomer A, which is free to bind a nucleotide (Figure 6D). The two DE-loops are connected directly through Arg697 of monomer B (ArgB697) that hydrogen-bonds to the carbonyl oxygen of Gly698 in the DE-loop in monomer A. In contrast, the reverse contact (of Arg697 of monomer A to the DE-loop in monomer B) is absent, and this side chain makes no protein contacts.
The nucleotide-binding studies of MutS showed that in the absence of Arg697, the high affinity site is lost (Figures 1 and 3). This suggests that Arg697 promotes nucleotide binding in one monomer, rather than preventing binding in the other. By crossing over to the DE-loop of monomer A, ArgB697 pulls along its own DE-loop, thereby removing it from the P-loop of monomer A, which is now free to bind ADP. We have shown that although the two ATPase domains can bind ATP simultaneously, ATP hydrolysis can only take place in one site at a time (Figures 2, 3 and 4). In the low affinity site of monomer B, a residue of the Walker B motif, Glu694, has been displaced from the position that is necessary for coordinating the magnesium. This movement is brought about by ArgB697 that hydrogen bonds to GluB694 while it reaches over to the DE-loop of the opposing monomer (Figure 6E). Since Glu694 is essential for hydrolysis (Junop et al., 2001), displacement from its position is likely to affect ATP hydrolysis.
Crystal structure of R697A MutS
To see how the R697A mutation causes an uncoupling of the ATPase domains, we crystallized the R697A MutS mutant in complex with mismatched DNA, in both the absence and presence of 100 µM ADP. Crystals grown in the absence of ADP appeared to be twinned and could not be used for structure determination. The crystals obtained in the presence of ADP, however, were not twinned and resulted in a structure to 2.6 Å (Table III). Outside the ATPase domains, no significant changes could be observed in the structure when compared with the wild-type protein. However, within the two ATPase domains, several changes are apparent (Figure 6F). The most obvious difference is that under conditions identical to the wild-type MutS crystallization (100 µM ADP), both monomers now bind ADP. In addition, the glutamate of the DE motif of monomer B (GluB694) is no longer displaced, and is now free to coordinate the Mg2+. Finally, with Arg697 no longer present, there are no contacts between the DE-loops of monomer A and B.
Table III. Data collection and refinement statistics.
| Data set | |
| Space group | P212121 |
| Cell dimensions (Å) | 89.9, 92.4, 261.6 |
| Resolution (Å) | 30–2.6 |
| Rmerge (%) | 12.4 (65.7) |
| I/σI | 4.6 (1.1) |
| Redundancy | 3.5 |
| Completeness (%) | 98.7 (99.1) |
| Refinement | |
| Reflections | 65273 |
| Rfree reflections | 1240 |
| Rfinal (%) | 21.5 |
| Rfree (%) | 24.9 |
| No. of atoms | |
| Total | 13420 |
| Protein | 12286 |
| DNA | 714 |
| Ligands | 64 |
| Water | 356 |
| R.m.s.d. bonds (Å) | 0.010 |
| R.m.s.d. angles (°) | 1.133 |
Numbers in parentheses refer to the highest resolution shell (2.74–2.60 Å).
Surprisingly, this did not result in a complete restoration of symmetry. The conformation of the DE-loop in monomer B is still in the same position as in the wild-type protein. The DE-loop of monomer A is no longer fixed by ArgB697, but its conformation is not identical to the other monomer, but in between the DE-loop of wild-type monomer A and monomer B. These differences may be due to the binding of the mismatched DNA that affects ADP binding in both wild-type and R697A MutS as observed in the filter binding studies (Figure 5). The twinning of the nucleotide-free crystals, however, suggests that in the absence of ADP, the two domains could be symmetric in nature.
Discussion
Asymmetry of the ATPase domains in MutS homologues
In this work, we have shown that the two chemically identical ATPase domains of the E.coli MutS dimer are asymmetric in nucleotide binding and are coupled in ATP hydrolysis. The data presented on the ATPase activities of the two domains strongly favour an alternating ATP hydrolysis mechanism. Inhibition of one site by AMPPNP or ADP-Vi simultaneously inhibits the other site, which cannot be explained simply by negative cooperativity as no such behaviour was observed for ATP hydrolysis (Table I). In addition, it was shown that during steady-state ATPase activity, hydrolysis takes place in only one monomer at a time. At the heart of this asymmetry lies a conserved arginine, Arg697, located at the interface of the two ATPase domains. Analysis of the crystal structure showed that Arg697 of each monomer contacts the opposing monomer in an asymmetric manner, promoting nucleotide binding in one monomer, while preventing hydrolysis in the other. Mutation of this single residue leads to loss of the high affinity site. As a result, the transition state analogue ADP-Vi can no longer be trapped, radiolabelled ATP can be washed out before it is hydrolysed, and ATP binding no longer releases DNA as efficiently as in the wild-type protein. In addition, the mutation uncouples the two ATPase domains in hydrolysis, as binding of AMPPNP or ADP-Vi in one site no longer inhibits hydrolysis in the other subunit.
In the crystal structures of Taq MutS (Obmolova et al., 2000; Junop et al., 2001), the equivalent of Arg697 (Arg666) does not have any contacts with the opposing monomer and, accordingly, no asymmetry of the ATPase domains was observed. The reason for the difference from the E.coli MutS structure is not clear, but may be caused by different crystal contacts. Nevertheless, sequence alignment (Figure 6G) shows that Arg697 is strictly conserved in all MutS homologues, with the exception of MSH5 where the arginine has been replaced by a lysine. In contrast, the MutS2 proteins (Eisen, 1998) (also called MutS paralogues, MSP; Culligan et al., 2000) that are believed not to be involved in DNA repair, do not show conservation of the arginine. The conservation of the arginine and adjacent residues (GRGT/G) suggests that asymmetry of the ATPase domains is a conserved feature of all MutS homologues. Indeed, a mutation was found in the DE-loop of MSH6 (GRGT→GRGI) in a Dutch HNPCC patient (Berends et al., 2002). Also, a mutation at the same position in yeast MSH6 (GRGG→GRGD) was shown to cause a dominant mutator phenotype, with abnormal stimulation of ATPase activity by DNA, and a strongly decreased ATP-induced release of DNA (Hess et al., 2002). Moreover, the ATPase domains of the yeast and human MSH2–MSH6 heterodimers were also found to be asymmetric in function (Iaccarino et al., 1998; Studamire et al., 1998). Hence, it seems likely that the asymmetry of the ATPase domains and the alternating ATP hydrolysis are conserved features of all MutS homologues, and essential for mismatch repair.
The DE-loop in other ATPases
Asymmetry of ATPase domains is not a unique feature of MutS among the ABC ATPases; several others have been shown to be asymmetric and/or alternate their ATPase activity. These include the CFTR (Carson et al., 1995), the multidrug resistance protein Pgp (Urbatsch et al., 1995a), the histidine transporter HisP (Kreimer et al., 2000) and the maltose transporter MalK (Sharma and Davidson, 2000). However, Arg697 is only conserved in the MutS homologues, and is absent in the other ABC ATPases (Figure 6G). They do, however, all have a loop in a position similar to the DE-loop of MutS. In this view, it is interesting to note that in the structurally related ATPase domains of RecA-like ATPases (Murzin et al., 1995), an insertion is found at the position of the DE-loop that is involved in regulation of ATP binding and hydrolysis (Figures 6G and 7). In RecA (Story et al., 1992) and the hexameric helicases gp4 (Sawaya et al., 1999) and RepA (Niedenzu et al., 2001), this insertion referred to as loop 1 makes direct contacts with the DNA, a strong effector of ATPase activity. In a similar fashion, in the F1 synthetase, the insertion makes contact with the central stalk, or γ-subunit, that couples the protonmotive force to ADP binding and ATP synthesis in the ATPase domains (Abrahams et al., 1994). Thus, although Arg697 of MutS does not have any structural homologues in closely related ATPases, the DE-loop on which it is located is important in other ATPases. Being directly connected to the Walker B motif that is crucial for the coordination of a magnesium ion, it is an ideal tool for the control of hydrolysis. Subtle changes in its position will remove the residues of the Walker B motif from their position, and hence prevent hydrolysis. The way in which the tool is controlled (by DNA in the helicases, by the γ-subunit in F1 synthetase, or the opposing monomer in MutS) has determined the shape and position of the loop during evolution. Because asymmetry is also present in several ABC transporters, it will be interesting to see whether these are also controlled through the DE-loop or whether nature has developed other means to control asymmetry.
Fig. 7. The DE-loop in different ATPases showing the different interactions with effectors. The sequences aligned in Figure 6G are coloured in black. (A) Escherichia coli MutS dimer binding to mismatch DNA (coloured in dark grey), with the DE-loop contacting the other ATPase domain. (B) Top view from the bovine F1-ATPase, with the DE-loop in contact with the central stalk (γ-subunit) (also coloured in black). (C) Bacteriophage T7 gp4 helicase, and (D) E.coli RepA helicase in which the DE-loop is thought to contact a centrally located DNA.
Function of asymmetry in DNA mismatch repair
The alternating activity of the ATPase domains has diverse functions in the different ATPase families. In the F1 synthetase, it is used for the sequential binding of ADP-Pi, formation of ATP and ATP release (Abrahams et al., 1994). In the ABC membrane transporters, it is coupled to substrate transport (Kreimer et al., 2000; van Veen et al., 2000), while, in the helicases, alternating hydrolysis is suggested to propel the helicase along the DNA during DNA unwinding (Patel and Picha, 2000; Singleton et al., 2000). However, it is not directly clear what the function of the alternating ATPase in DNA mismatch repair could be. The strong effect of the loss of asymmetry on DNA release suggests that it is needed for proper cooperation of mismatch and ATP binding in order to attract MutL. In this way, the asymmetry of the ATPase domains is used to emphasize the asymmetry in the mismatch binding and could help to orient MutL binding, much like the model described in Junop et al. (2001). In higher species, this asymmetric MutL binding could have been the origin of the heterodimeric MutL dimers found in eukaryotes. It is interesting to note that the asymmetry of ATPase domains is already present prior to DNA binding. Hence, it is possible that the nucleotide state of the ATPase domains controls the DNA binding, rather than vice versa. In other words, before a mismatch is bound, it is already decided by the nucleotide state of each monomer, which monomer will bind the mismatch. This is similar to the eukaryotic heterodimers, where only one of the two monomers is designated to bind a mismatch.
In addition, it is likely that asymmetry of the ATPase domains is also used at later stages in repair. If only needed to emphasize asymmetry for MutL binding, a single functional ATPase domain would suffice. However, in eukaryotes, both ATPase domains are necessary, since the double mutation of both ATPase domains leads to a more severe phenotype than either single mutation alone (Iaccarino et al., 1998; Studamire et al., 1998). Also, mutation of the ATPase domain of MSH2 or MSH6 appeared to affect different stages of mismatch repair (Bowers et al., 2000). The repair process is a complex cascade of events, which involve recognition of the mismatch by MutS and assembly of the MutS–MutL complex, activation of MutH, loading and guiding of exonucleases and helicases to remove the correct stretch of DNA, and resynthesis of the daughter strand. We propose that rather than only being needed for release of mismatch DNA at the beginning of the repair process, as is put forward in the existing models, the ATPase activities of MutS also function in later stages of repair. To ensure proper timing of the different events, the alternating ATPase activities of the MutS dimer would be well suited to form the centre of coordination. Binding of ATP in the correct ATPase domain would invoke one stage of repair, while hydrolysis of ATP would allow for the other monomer to bind ATP and initiate the next stage in repair.
Materials and methods
Reagents and sample preparation
All chemicals were purchased from Merck, unless stated otherwise. ATP, ADP, AMPPNP and ATPγS were obtained from Fluka, and α- and γ-32P-labelled ATP from Pharmacia-Amersham. R697A MutS was constructed through site-directed mutagenesis of wild-type MutS plasmid pMQ372 (kindly provided by M.Marinus) or pM800 for ΔC800 MutS (Lamers et al., 2000). E694N MutS plasmid was kindly provided by N.de Wind. All MutS proteins were purified as described in Lamers et al. (2000). The heteroduplex oligomer contains a G:T mismatch at position 9 (Lamers et al., 2000). The homoduplex oligomer contains a G:C base pair at this position. All reactions were in MutS buffer containing 250 mM NaCl, 20 mM HEPES pH 7.5 and 10 mM MgCl2.
Luciferase assay
Concentrations of ATP and ADP in purified MutS were determined as described in Blackwell et al. (1998) using the ATP bioluminescent assay kit (Sigma). Dilute concentrations of MutS (0.5, 1 and 2 µM, 100 µl) were boiled for 10 min at 100°C to release the bound nucleotide. For calibration, similar amounts of pure ATP and ADP were treated in the same way. ADP was measured after conversion to ATP by pyruvate kinase (Roche). All luciferase activity was measured in a TD-20/20 luminometer (Turner Designs).
ADP filter binding assay
Filter binding studies were performed using a Schleicher & Schuell Minifold II slot-blot apparatus and a 0.45 µm nitrocellulose filter (BA-85). MutS (5 µM) was incubated with different amounts (1–60 µM) of radiolabelled ATP for >30 min at room temperature to achieve complete hydrolysis of ATP to ADP. The extent of hydrolysis was monitored by separating ATP and ADP using thin-layer chromatography (TLC). For the diluting-out experiment, 5 µM MutS was incubated with 50 µM [α-32P]ATP to saturate both sites. Subsequently, 10 µl aliquots were diluted in MutS buffer to a final volume of 10–800 µl ([MutS] = 5–0.062 µM) before spotting on the filter. For the DNA effect on ADP binding, 8 µM of hetero- or homoduplex DNA was included in the reaction mixture containing 5 µM MutS and 3.125–25 µM ATP. For the competition assays, 6.25 µM radiolabelled ATP was incubated with 5 µM MutS. Subsequently, cold ADP, ATP or AMPPNP were mixed briefly with the MutS mixture and spotted immediately on the filter. In the vanadate trapping experiment, 5 µM MutS, 35 µM [α-32P]ATP and 100 µM vanadate were incubated for 1 h. Samples were spotted directly onto the filter, or washed first with 1 vol. of 1 mM unlabelled ATP. Sodium ortho-vanadate (Sigma) stock solution was prepared as described in Goodno (1982), and boiled before use. Dried nitrocellulose filters were exposed to an imaging plate, read-out with a BAS 1000 phosphoimager (Fuji) and quantified with Tina software (Raytest).
Radioactive ATPase assay
For the effect of vanadate on ATPase activity, 5 µM MutS was incubated with 35 µM ATP and 100 µM vanadate for 1 h. Then, 1 vol. of 1 mM radiolabelled ATP was added to the mixture, the reaction was quenched by the addition of 1 vol. of 0.5 M EDTA and placed on ice. In the pulse–chase experiment (Hingorani et al., 1997), 1 µM MutS was incubated with 35 µM radiolabelled ATP and quenched at different time points, or first chased with 1 vol. of 10 mM unlabelled ATP before quenching. A 0.5 µl aliquot of the reaction mixtures was spotted onto a PEI-cellulose TLC plate (Merck) and developed in 0.5 M ortho-phosphoric acid/KOH pH 3.8. Dried TLC plates were exposed to an imaging plate and quantified as described above.
Spectrophotometric ATPase assay
ATPase activity of MutS was measured by coupling ATP hydrolysis to oxidation of NADH (Panuska and Goldthwait, 1980). Wild-type MutS (5 µM) or R697A MutS (10 µM) were mixed with 1–50 µM ATP and hydrolysis measured during 5 min. For inhibition of ATPase activity by AMPPNP, reactions were performed with 6.25 µM ATP and 1.6–25 µM AMPPNP (wild-type MutS), or 25 µM ATP and 3.1–50 µM AMPPNP (R697A MutS).
DNA filter binding assay
DNA affinity studies were performed in a similar fashion to the ADP filter binding studies. MutS was incubated for ∼30 min with different amounts of 32P-radiolabelled hetero- or homoduplex DNA and spotted onto nitrocellulose filters. For heteroduplex DNA, 1 µM MutS and 0.5–4 µM DNA were used. For homoduplex DNA, 5 µM MutS and 5–40 µM DNA were mixed. For ATP-induced release of DNA, 1 µM MutS was incubated with 1 µM heteroduplex DNA and 150 µM ADP or ATPγS for 15 min before spotting onto the filter.
Modelling of substrate binding and ATPase activity
All binding data were fitted to either a single site binding model
or a two-site binding model
where Sbound is the amount of bound ligand (i.e. ADP or DNA), Stotal is the total amount of ligand, Bmax is the total amount of the binding sites and Kd is the dissociation constant.
For the kinetic data, the following equation was used:
where Vmax is the maximal activity, S is the concentration of the substrate (ATP), n is the Hill coefficient, and Km is the substrate concentration where V = 1/2 × Vmax. For all modelling of wild-type MutS, it was taken into account that 50% of the MutS population retained ADP. Model fitting was performed by least-square minimization using the Solver add-in of Excel (Microsoft).
Rifampicin resistance assay
Spontaneous mutation rates were monitored by rifampicin resistance assay as described by Wu and Marinus (1994). In brief, a MutS-deficient strain RK1517 (kindly provided by P.Modrich) was transformed with wild-type MutS, R697A MutS, E694N MutS or empty vector. Overnight colonies were picked, and grown in LB to near saturation (OD600 ∼1). All cells were found to grow at similar rates, implying that the mutations in MutS are not toxic to the cells. Subsequently, 1 × 108 cells were plated out on LB plates containing 50 µg/ml rifampicin and incubated at 37°C overnight. All strains were plated out in eight different dishes and each experiment was performed twice.
Structure determination of R697A MutS
Crystals of R697A ΔC800 were grown in either the absence or presence of 100 µM ADP under conditions described earlier (Lamers et al., 2000). X-ray diffraction data were collected at 100 K on beamline ID14-EH2 at the ESRF synchrotron in Grenoble. All data were processed using MOSFLM (Leslie, 1992) and SCALA (Evans, 1997). The model of wild-type MutS (PDB accession code 1E3M) was used for rigid-body refinement in Refmac5 (Murshudov et al., 1997); manual model building was performed in O (Jones et al., 1991), and final refinement was done with Refmac5 using the TLS option (21 domains), and ARP/wARP (Lamzin and Wilson, 1997) to build the solvent atoms. Details of crystallographic analysis are given in Table III. The final structure of R697A ΔC800 was submitted to the protein data bank under accession number 1N69.
Structure-based sequence alignment of ATPases
A structural homology search for the ATPase domain of MutS (residues 568–741) was performed using the DALI server (Holm and Sander, 1993). The first 15 hits were all found to be part of one of three ATPase families of the SCOP database (Murzin et al., 1995) ‘RecA protein-like (ATPase domains)’, ‘ABC transporter ATPase domains-like’ and ‘extended AAA-ATPase domain’. Several members of each family were used for structural comparison, using lsqkab (Kabsch, 1976) for superposition of the Walker A motif. Initial sequence alignments were created with ClustalX (Thompson, 1997), and subsequently modified manually according to the crystal structures.
Acknowledgments
Acknowledgements
We thank N.de Wind for the plasmid of E694N MutS, M.Marinus for plasmid pMQ372, P.Modrich for strain RK1517. O.Weichenrieder, R.Persson, J.Lebbink and N.de Wind for critically reading the manuscript, A.Perrakis, O.Weichenrieder and group members for discussion, and staff at the ESRF for support in data collection. This work was supported by the Dutch Cancer Society (Grants NKI 96-1279 and NKI 2001-2479).
References
- Abrahams J.P., Leslie,A.G., Lutter,R. and Walker,J.E. (1994) Structure at 2.8 Å resolution of F1-ATPase from bovine heart mitochondria. Nature, 370, 621–628. [DOI] [PubMed] [Google Scholar]
- Allen D.J., Makhov,A., Grilley,M., Taylor,J., Thresher,R., Modrich,P. and Griffith,J.D. (1997) MutS mediates heteroduplex loop formation by a translocation mechanism. EMBO J., 16, 4467–4476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aquilina G. and Bignami,M. (2001) Mismatch repair in correction of replication errors and processing of DNA damage. J. Cell Physiol., 187, 145–154. [DOI] [PubMed] [Google Scholar]
- Bellacosa A. (2001) Functional interactions and signaling properties of mammalian DNA mismatch repair proteins. Cell Death Differ., 8, 1076–1092. [DOI] [PubMed] [Google Scholar]
- Berends M.J. et al. (2002) Molecular and clinical characteristics of MSH6 variants: an analysis of 25 index carriers of a germline variant. Am. J. Hum. Genet., 70, 26–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjornson K.P., Allen,D.J. and Modrich,P. (2000) Modulation of MutS ATP hydrolysis by DNA cofactors. Biochemistry, 39, 3176–3183. [DOI] [PubMed] [Google Scholar]
- Blackwell L.J., Martik,D., Bjornson,K.P., Bjornson,E.S. and Modrich,P. (1998) Nucleotide-promoted release of hMutSα from heteroduplex DNA is consistent with an ATP-dependent translocation mechanism. J. Biol. Chem., 273, 32055–32062. [DOI] [PubMed] [Google Scholar]
- Blackwell L.J., Wang,S. and Modrich,P. (2001) DNA chain length dependence of formation and dynamics of hMutSα·hMutLα heteroduplex complexes. J. Biol. Chem., 276, 33233–33240. [DOI] [PubMed] [Google Scholar]
- Bowers J., Tran,P.T., Liskay,R.M. and Alani,E. (2000) Analysis of yeast MSH2–MSH6 suggests that the initiation of mismatch repair can be separated into discrete steps. J. Mol. Biol., 302, 327–338. [DOI] [PubMed] [Google Scholar]
- Bowers J., Tran,P.T., Joshi,A., Liskay,R.M. and Alani,E. (2001) MSH–MLH complexes formed at a DNA mismatch are disrupted by the PCNA sliding clamp. J. Mol. Biol., 306, 957–968. [DOI] [PubMed] [Google Scholar]
- Carson M.R., Travis,S.M. and Welsh,M.J. (1995) The two nucleotide-binding domains of cystic fibrosis transmembrane conductance regulator (CFTR) have distinct functions in controlling channel activity. J. Biol. Chem., 270, 1711–1717. [DOI] [PubMed] [Google Scholar]
- Chang G. and Roth,C.B. (2001) Structure of MsbA from E.coli: a homolog of the multidrug resistance ATP binding cassette (ABC) transporters. Science, 293, 1793–1800. [DOI] [PubMed] [Google Scholar]
- Culligan K.M., Meyer-Gauen,G., Lyons-Weiler,J. and Hays,J.B. (2000) Evolutionary origin, diversification and specialization of eukaryotic MutS homolog mismatch repair proteins. Nucleic Acids Res., 28, 463–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diederichs K., Diez,J., Greller,G., Muller,C., Breed,J., Schnell,C., Vonrhein,C., Boos,W. and Welte,W. (2000) Crystal structure of MalK, the ATPase subunit of the trehalose/maltose ABC transporter of the archaeon Thermococcus litoralis. EMBO J., 19, 5951–5561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisen J.A. (1998) A phylogenomic study of the MutS family of proteins. Nucleic Acids Res., 26, 4291–4300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans P.R. (1997) Scala. Joint CCP4 and ESF-EAMCB Newslett., 33, 22–24. [Google Scholar]
- Galio L., Bouquet,C. and Brooks,P. (1999) ATP hydrolysis-dependent formation of a dynamic ternary nucleoprotein complex with MutS and MutL. Nucleic Acids Res., 27, 2325–2331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodno C.C. (1982) Myosin active-site trapping with vanadate ion. Methods Enzymol., 85, 116–123. [DOI] [PubMed] [Google Scholar]
- Gorbalenya A.E. and Koonin,E.V. (1990) Superfamily of UvrA-related NTP-binding proteins. Implications for rational classification of recombination/repair systems. J. Mol. Biol., 213, 583–591. [DOI] [PubMed] [Google Scholar]
- Gradia S., Acharya,S. and Fishel,R. (1997) The human mismatch recognition complex hMSH2–hMSH6 functions as a novel molecular switch. Cell, 91, 995–1005. [DOI] [PubMed] [Google Scholar]
- Gradia S., Subramanian,D., Wilson,T., Acharya,S., Makhov,A., Griffith,J. and Fishel,R. (1999) hMSH2–hMSH6 forms a hydrolysis-independent sliding clamp on mismatched DNA. Mol. Cell, 3, 255–261. [DOI] [PubMed] [Google Scholar]
- Hall M.C. and Matson,S.W. (1999) The Escherichia coli MutL protein physically interacts with MutH and stimulates the MutH-associated endonuclease activity. J. Biol. Chem., 274, 1306–1312. [DOI] [PubMed] [Google Scholar]
- Hess M.T., Gupta,R.D. and Kolodner,R.D. (2002) Dominant Saccharomyces cerevisiae msh6 mutations cause increased mispair binding and decreased dissociation from mispairs by Msh2–Msh6 in the presence of ATP. J. Biol. Chem., 277, 25545–25553. [DOI] [PubMed] [Google Scholar]
- Hingorani M.M., Washington,M.T., Moore,K.C. and Patel,S.S. (1997) The dTTPase mechanism of T7 DNA helicase resembles the binding change mechanism of the F1-ATPase. Proc. Natl Acad. Sci. USA, 94, 5012–5017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holm L. and Sander,C. (1993) Protein structure comparison by alignment of distance matrices. J. Mol. Biol., 233, 123–138. [DOI] [PubMed] [Google Scholar]
- Hopfner K.P., Karcher,A., Shin,D.S., Craig,L., Arthur,L.M., Carney,J.P. and Tainer,J.A. (2000) Structural biology of Rad50 ATPase: ATP-driven conformational control in DNA double-strand break repair and the ABC-ATPase superfamily. Cell, 101, 789–800. [DOI] [PubMed] [Google Scholar]
- Hung L.W., Wang,I.X., Nikaido,K., Liu,P.Q., Ames,G.F. and Kim,S.H. (1998) Crystal structure of the ATP-binding subunit of an ABC transporter. Nature, 396, 703–707. [DOI] [PubMed] [Google Scholar]
- Iaccarino I., Marra,G., Palombo,F. and Jiricny,J. (1998) hMSH2 and hMSH6 play distinct roles in mismatch binding and contribute differently to the ATPase activity of hMutSα. EMBO J., 17, 2677–2686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iaccarino I., Marra,G., Dufner,P. and Jiricny,J. (2000) Mutation in the magnesium binding site of hMSH6 disables the hMutSα sliding clamp from translocating along DNA. J. Biol. Chem., 275, 2080–2086. [DOI] [PubMed] [Google Scholar]
- Jones P.M. and George,A.M. (1999) Subunit interactions in ABC transporters: towards a functional architecture. FEMS Microbiol. Lett., 179, 187–202. [DOI] [PubMed] [Google Scholar]
- Jones T.A., Zou,J.-Y., Cowan,S.W. and Kjeldgaard,M. (1991) Improved methods for the building of protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A, 47, 110–119. [DOI] [PubMed] [Google Scholar]
- Junop M.S., Obmolova,G., Rausch,K., Hsieh,P. and Yang,W. (2001) Composite active site of an ABC ATPase: MutS uses ATP to verify mismatch recognition and authorize DNA repair. Mol. Cell, 7, 1–12. [DOI] [PubMed] [Google Scholar]
- Kabsch W. (1976) A solution for the best rotation to relate two sets of vectors. Acta Crystallogr. A, 32, 922–923. [Google Scholar]
- Kreimer D.I., Chai,K.P. and Ferro-Luzzi Ames,G. (2000) Nonequivalence of the nucleotide-binding subunits of an ABC transporter, the histidine permease and conformational changes in the membrane complex. Biochemistry, 39, 14183–14195. [DOI] [PubMed] [Google Scholar]
- Lamers M.H., Perrakis,A., Enzlin,J.H., Winterwerp,H.H., de Wind,N. and Sixma,T.K. (2000) The crystal structure of DNA mismatch repair protein MutS binding to a G x T mismatch. Nature, 407, 711–717. [DOI] [PubMed] [Google Scholar]
- Lamzin V.S. and Wilson,K.S. (1997) Automated refinement for protein crystallography. Methods Enzymol., 277, 269–305. [DOI] [PubMed] [Google Scholar]
- Leslie A.G.W. (1992) Recent changes to the MOSFLM package for processing film and image plate data. Joint CCP4 and ESF-EAMCB Newslett., 26. [Google Scholar]
- Locher K.P., Lee,A.T. and Rees,D.C. (2002) The E.coli BtuCD structure: a framework for ABC transporter architecture and mechanism. Science, 296, 1091–1098. [DOI] [PubMed] [Google Scholar]
- Lynch H.T. and de la Chapelle,A. (1999) Genetic susceptibility to non-polyposis colorectal cancer. J. Med. Genet., 36, 801–818. [PMC free article] [PubMed] [Google Scholar]
- Mannering D.E., Sharma,S. and Davidson,A.L. (2001) Demonstration of conformational changes associated with activation of the maltose transport complex. J. Biol. Chem., 276, 12362–12368. [DOI] [PubMed] [Google Scholar]
- Marti T.M., Kunz,C. and Fleck,O. (2002) DNA mismatch repair and mutation avoidance pathways. J. Cell Physiol., 191, 28–41. [DOI] [PubMed] [Google Scholar]
- Murshudov G.N., Vagin,A.A. and Dodson, E.J. (1997) Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D, 53, 240–255. [DOI] [PubMed] [Google Scholar]
- Murzin A.G., Brenner,S.E., Hubbard,T. and Chothia,C. (1995) SCOP: a structural classification of proteins database for the investigation of sequences and structures. J. Mol. Biol., 247, 536–540. [DOI] [PubMed] [Google Scholar]
- Niedenzu T., Roleke,D., Bains,G., Scherzinger,E. and Saenger,W. (2001) Crystal structure of the hexameric replicative helicase RepA of plasmid RSF1010. J. Mol. Biol., 306, 479–487. [DOI] [PubMed] [Google Scholar]
- Obmolova G., Ban,C., Hsieh,P. and Yang,W. (2000) Crystal structures of mismatch repair protein MutS and its complex with a substrate DNA. Nature, 407, 703–710. [DOI] [PubMed] [Google Scholar]
- Panuska J.R. and Goldthwait,D.A. (1980) A DNA-dependent ATPase from T4-infected Escherichia coli. Purification and properties of a 63,000-dalton enzyme and its conversion to a 22,000-dalton form. J. Biol. Chem., 255, 5208–5214. [PubMed] [Google Scholar]
- Parker B.O. and Marinus,M.G. (1992) Repair of DNA heteroduplexes containing small heterologous sequences in Escherichia coli. Proc. Natl Acad. Sci. USA, 89, 1730–1734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel S.S. and Picha,K.M. (2000) Structure and function of hexameric helicases. Annu. Rev. Biochem., 69, 651–697. [DOI] [PubMed] [Google Scholar]
- Qu Q. and Sharom,F.J. (2001) FRET analysis indicates that the two ATPase active sites of the P-glycoprotein multidrug transporter are closely associated. Biochemistry, 40, 1413–1422. [DOI] [PubMed] [Google Scholar]
- Sawaya M.R., Guo,S., Tabor,S., Richardson,C.C. and Ellenberger,T. (1999) Crystal structure of the helicase domain from the replicative helicase-primase of bacteriophage T7. Cell, 99, 167–177. [DOI] [PubMed] [Google Scholar]
- Schofield M.J., Nayak,S., Scott,T.H., Du,C. and Hsieh,P. (2001) Interaction of Escherichia coli MutS and MutL at a DNA mismatch. J. Biol. Chem., 276, 28291–28299. [DOI] [PubMed] [Google Scholar]
- Sharma S. and Davidson,A.L. (2000) Vanadate-induced trapping of nucleotides by purified maltose transport complex requires ATP hydrolysis. J. Bacteriol., 182, 6570–6576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singleton M.R., Sawaya,M.R., Ellenberger,T. and Wigley,D.B. (2000) Crystal structure of T7 gene 4 ring helicase indicates a mechanism for sequential hydrolysis of nucleotides. Cell, 101, 589–600. [DOI] [PubMed] [Google Scholar]
- Story R.M., Weber,I.T. and Steitz,T.A. (1992) The structure of the E.coli recA protein monomer and polymer. Nature, 355, 318–325. [DOI] [PubMed] [Google Scholar]
- Studamire B., Quach,T. and Alani,E. (1998) Saccharomyces cerevisiae Msh2p and Msh6p ATPase activities are both required during mismatch repair. Mol. Cell. Biol., 18, 7590–7601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su S.S. and Modrich,P. (1986) Escherichia coli mutS-encoded protein binds to mismatched DNA base pairs. Proc. Natl Acad. Sci. USA, 83, 5057–5061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson J.D., Gibson,T.J., Plewniak,F., Jeanmougin,F. and Higgins,D.G. (1997) The ClustalX windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res., 25, 4876–4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urbatsch I.L., Sankaran,B., Bhagat,S. and Senior,A.E. (1995a) Both P-glycoprotein nucleotide-binding sites are catalytically active. J. Biol. Chem., 270, 26956–26961. [DOI] [PubMed] [Google Scholar]
- Urbatsch I.L., Sankaran,B., Weber,J. and Senior,A.E. (1995b) P-glycoprotein is stably inhibited by vanadate-induced trapping of nucleotide at a single catalytic site. J. Biol. Chem., 270, 19383–19390. [DOI] [PubMed] [Google Scholar]
- vanVeen H.W., Margolles,A., Muller,M., Higgins,C.F. and Konings,W.N. (2000) The homodimeric ATP-binding cassette transporter LmrA mediates multidrug transport by an alternating two-site (two-cylinder engine) mechanism. EMBO J., 19, 2503–2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu T.H. and Marinus,M.G. (1994) Dominant negative mutator mutations in the mutS gene of Escherichia coli. J. Bacteriol., 176, 5393–5400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi M., Dao,V. and Modrich,P. (1998) MutS and MutL activate DNA helicase II in a mismatch-dependent manner. J. Biol. Chem., 273, 9197–9201. [DOI] [PubMed] [Google Scholar]