Abstract
The translation initiation factor eIF4E is involved in the modulation of cellular growth. In the nucleus, where eIF4E is associated with PML nuclear bodies, eIF4E mediates nucleocytoplasmic transport of specific transcripts, and this contributes to its transformation activity. Surprisingly, we found that a trans cription factor, the proline-rich homeodomain protein PRH, is a negative regulator of eIF4E in myeloid cells, interacting with eIF4E through a conserved binding site typically found in translational regulators. Through this interaction, PRH inhibits eIF4E-dependent mRNA transport and subsequent transformation. These activities of PRH are independent of its transcriptional functions. Further, we found that 199 homeodomain proteins contain potential eIF4E-binding sites. Thus, there could be many tissue-specific regulators of eIF4E. These findings provide a model for regulation of a general factor, eIF4E, in tissue- specific contexts, and suggest that its regulation is important in differentiation and development.
Keywords: eIF4E/Hex/Hox11/PML/PRH
Introduction
The eukaryotic translation initiation factor 4E (eIF4E) is involved in modulation of cellular growth. eIF4E is essential to cellular survival. Expression of antisense oligonucleotides to eIF4E induces cell death in HeLa cells, and its disruption in Saccharomyces cerevisiae is lethal (Altmann et al., 1989; De Benedetti and Rhoads, 1990). Although essential, moderate overexpression of eIF4E leads to dysregulated cellular proliferation and malignant transformation (Lazaris-Karatzas et al., 1990, 1992; Lazaris-Karatzas and Sonenberg, 1992). In fact, eIF4E levels are prognostic indicators of clinical outcome in a variety of human cancers including breast cancer, head and neck squamous cell carcinoma and several non-Hodgkin B-cell lymphomas (Nathan et al., 1997a,b; De Benedetti and Harris, 1999; Wang et al., 1999).
The growth-promoting and transforming properties of eIF4E are thought to involve increased translation of mRNAs important to growth control (Sonenberg and Gingras, 1998). During cap-dependent translation, eIF4E binds the methyl-7-guanosine (m7G) cap present on the 5′ end of mRNAs and recruits the given transcript to the ribosome (Sonenberg and Gingras, 1998). eIF4E overexpression does not increase levels of synthesis uniformly for all proteins, with a subset of transcripts more sensitive to eIF4E levels (Sonenberg and Gingras, 1998). Many mRNAs involved in growth control have complex highly structured untranslated regions (UTRs), whereas housekeeping genes such as GADPH and actin have relatively short, unstructured UTRs. It is well established that the complexity of the UTR slows down translation rates. Thus, growth control mRNAs are not translated as readily as housekeeping mRNAs. Consistently, overexpression of eIF4E leads to increased translation of transcripts with highly structured UTRs. These messages are considered eIF4E sensitive and include transcripts such as ornithine decarboxylase, vascular endothelial growth factor (VEGF) and Pim-1 (Rousseau et al., 1996; Hoover et al., 1997).
A substantial fraction of eIF4E is found in the nucleus (up to 68%) where it localizes to discrete sites which we will refer to as eIF4E nuclear bodies (Lejbkowicz et al., 1992; Iborra et al., 2001). Sonenberg and co-workers showed that, in the nucleus, eIF4E functions in the selective transport of specific mRNAs from the nucleus to the cytoplasm (Rousseau et al., 1996). For instance, eIF4E increased production of cyclin D1 protein by promoting nucleo-cytoplasmic transport of cyclin D1 transcripts (Rousseau et al., 1996). In contrast, housekeeping genes such as GAPDH and actin are not affected. It has been suggested that specificity of eIF4E for certain transcripts is due to the complexity of UTRs in these transcripts (Rousseau et al., 1996). The molecular mechanism of how transcripts are transported, what the precise features that cause eIF4E sensitivity are, whether these are due to direct interactions with eIF4E and RNA or require accessory proteins, and how many transcripts are regulated in this way are currently not known. Interestingly, while the W73A eIF4E mutant can not form an active translation complex (Sonenberg and Gingras, 1998), it still functions in cyclin D1 mRNA transport and still transforms cells (Cohen et al., 2001). Thus, the ability of eIF4E to transform cells lies, at least in part, in its mRNA transport function.
eIF4E is associated with nuclear bodies in a variety of organisms including yeast, Drosophila, Xenopus, mice and humans (Lang et al., 1994; Cohen et al., 2001; Strudwick and Borden, 2002). In mammalian cells, eIF4E nuclear bodies coincide with those associated with the promyelocytic leukemia protein PML (Lai and Borden, 2000; Cohen et al., 2001; Topisirovic et al., 2002). Both eIF4E and PML nuclear bodies are disrupted by treatment with an analog of the m7G cap, but are not altered by treatment with RNase or DNase (Dostie et al., 2000; Cohen et al., 2001). The cap analog causes a structural rearrangement in eIF4E, and this is thought to underlie the sensitivity of eIF4E bodies to this treatment (Cohen et al., 2001). The RING domain of PML directly interacts with eIF4E, reducing its affinity for the m7G cap, and therefore mRNA, by >100-fold (Cohen et al., 2001; Kentsis et al., 2001). This loss of cap-binding activity is correlated with a disruption of the mRNA transport functions of eIF4E and a suppression of eIF4E-mediated transformation. Con sistently, high resolution electron microscopy studies indicate that there is RNA associated with the periphery but not inside the nuclear bodies (Boisvert et al., 2000). PML negatively regulates eIF4E functions in response to cellular stress (Topisirovic et al., 2002) and was the first negative regulator reported for the nuclear transport function of eIF4E (Cohen et al., 2001). Since PML is expressed in all mammalian cell types reported, it is positioned to be a general inhibitor of nuclear eIF4E function.
Although eIF4E is expressed in all eukaryotic cell types, recent studies suggest that, additionally, eIF4E functions in a tissue-specific manner. eIF4E expression during zebrafish development is dynamic and asymmetric, suggesting that its spatio-temporal concentration may be an important determinant in regulation of tissue- and mRNA-specific protein synthesis (Fahrenkrug et al., 1999). Studies in zebrafish suggest that eIF4E plays a role in regulating protein synthesis specifically during oogenesis, gastrulation and erythropoiesis (Fahrenkrug et al., 1999). In addition, the localization of eIF4E changes drastically during Xenopus development where, in stage 1–2 oocytes, eIF4E is localized diffusely throughout the cytoplasm; by stage 4, eIF4E has translocated to the nucleus and, in gastrula, eIF4E is found mainly in nuclear bodies (Strudwick and Borden, 2002). Thus, eIF4E appears to play a dynamic role in Xenopus development. Taken together, these data suggest that although eIF4E is a general factor, it may also, through interactions with tissue-specific regulators, act as a tissue-specific translation and/or mRNA transport enhancer.
Clearly, finding tissue-specific regulators of eIF4E activity is key to understanding how its functions could be modulated in this way. The association of eIF4E with PML led us to investigate whether a tissue-specific partner protein of PML, the proline-rich homeodomain PRH (Topcu et al., 1999), also associates with and modifies the activities of eIF4E. PRH, also known as the hemato poietically expressed homeodomain Hex, is expressed in limited tissues in adults including myeloid cells, lung, thyroid and liver (Hromas et al., 1993; Martinez Barbera et al., 2000). PRH functions in hematopoiesis in a variety of organisms including zebrafish, Xenopus, chicken, mice and humans (Crompton et al., 1992; Newman et al., 1997; Thomas et al., 1998; Yatskievych et al., 1999; Liao et al., 2000). We report that PRH inhibits the transformation and growth-promoting effects of eIF4E by inhibiting its mRNA transport function. These effects are a consequence of the direct interaction between these proteins. Further, we find that a substantial number of homeodomain proteins are positioned to be tissue-specific regulators of eIF4E. These findings allow us to develop a biochemical framework for tissue-specific modulation of nuclear eIF4E function.
Results
Endogenous PRH co-localizes with endogenous eIF4E nuclear bodies
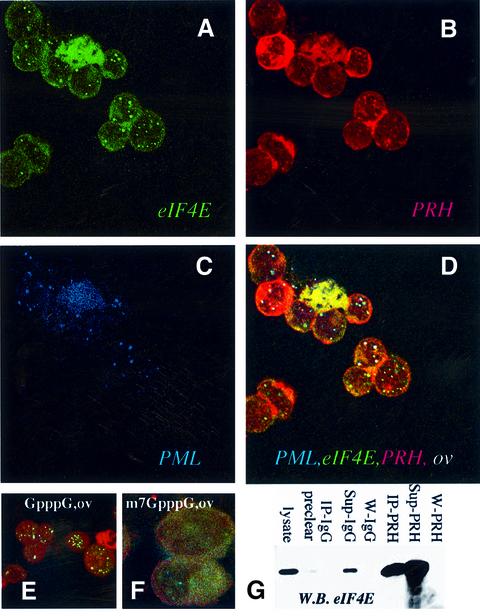
Previously we demonstrated that PML interacts separately with PRH and eIF4E (Lai and Borden, 2000; Cohen et al., 2001; Topisirovic et al., 2002). Unlike PML, which is expressed in all mammalian cell types reported (Melnick and Licht, 1999), PRH expression is limited (Hromas et al., 1993; Keng et al., 2000; Martinez Barbera et al., 2000). Using confocal microscopy, we examined whether PRH is positioned to be a tissue-specific modulator of eIF4E. PRH interacts with eIF4E in human promonocytic U937 cells, which express eIF4E, PRH and PML endogenously (Figure 1). PML is found nearly exclusively in nuclear bodies, whereas eIF4E and PRH have both punctate nuclear and cytoplasmic distributions, as reported (Lejbkowicz et al., 1992; Topcu et al., 1999; Lai and Borden, 2000; Cohen et al., 2001; Topisirovic et al., 2002). The nuclear and cytoplasmic localizations of PRH are consistent with putative nuclear localization signal (NLS) and nuclear export signal (NES) sequences found within the protein. In the overlay, nuclear bodies that contain PML, PRH and eIF4E are shown as white (Figure 1D). It is clear from these confocal micrographs that nearly all eIF4E nuclear bodies co-localize with PRH and PML. There are a few bodies, in red, that contain only PRH. These results indicate that PRH is positioned to modulate eIF4E activity in these cells.
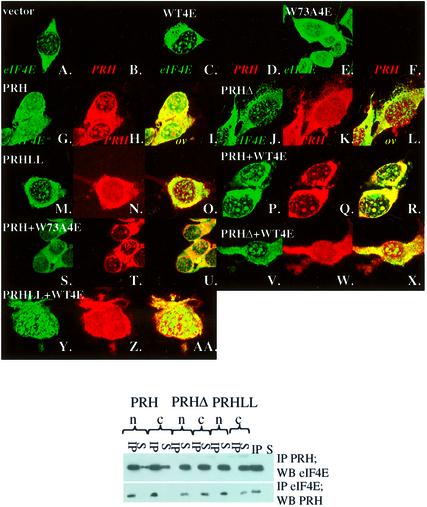
Fig. 1. eIF4E, PRH and PML interact in vivo. U937 cells were stained with eIF4E mAb conjugated directly to FITC, in green (A), an affinity-purified PRH polyclonal antibody, in red (B), and PML mAb 5E10, in blue (C). The eIF4E–PRH–PML overlay is shown in white (D). Each channel was recorded independently to avoid cross-talk. The objective is 100× with further 2-fold magnification. Experiments carried out with either mAb eIF4E-conjugated FITC or mAb eIF4E followed by FITC secondary antibody yielded identical results, as observed previously (Cohen et al., 2001). (E and F) K562 cells were triple stained as above and the overlay is shown for cells treated with GpppG or the cap analog, m7GpppG. These confocal micrographs (A–F) represent single optical sections through the plane of the cell. A further 2-fold magnification for (E) and 4-fold for (F). (G) K562 cell lysates were immunoprecipitated with PRH antibody (IP-PRH) or immunoglobulin (IgG), and the resulting western blot (W.B.) was probed as indicated. Sup indicates supernatant after immunoprecipitation; W, final wash of the beads; pre-clear as described previously in Carlile et al. (1998); and lysate, total cell lysate.
PRH bodies are dispersed by the m7G cap analog in vivo
eIF4E and PML nuclear bodies are disrupted by treatment with m7GpppG, whereas other nuclear structures such as splicing speckles or nucleoli are not (Dostie et al., 2000; Cohen et al., 2001). If PRH is part of the same nuclear structures, one would expect that it would also be sensitive to this treatment. To investigate this, K562 cells were permeabilized and incubated with m7GpppG cap analog, or with GpppG that does not bind eIF4E and does not disrupt its nuclear bodies (Figure 1E and F). Following treatment, cells were washed to remove proteins released from the nucleus and fixed. Incubation with GpppG does not affect the co-localization of PRH, eIF4E or PML. Strikingly, m7GpppG has dispersed the majority of all three proteins from bodies (Figure 1F). The same results were observed in U937 cells (data not shown). Thus, like eIF4E and PML, treatment with m7G mRNA cap analog disrupts PRH bodies. Further, treatment of cells with RNase or DNase does not significantly alter the subcellular distribution of PRH, PML or eIF4E (data not shown; Stuurman et al., 1992; Dostie et al., 2000; Cohen et al., 2001), consistent with previous findings that RNA is associated with the periphery of the PML nuclear body (Boisvert et al., 2000). Cap treatment causes a conformational rearrangement of eIF4E (McCubbin et al., 1988; von Der Haar et al., 2000; Cohen et al., 2001), and this is likely to be the cause of eIF4E, PML and PRH nuclear body disruption. These findings strongly suggest that the majority of PRH is part of the same nuclear structures as eIF4E and PML.
PRH associates with eIF4E in both the nucleus and the cytoplasm
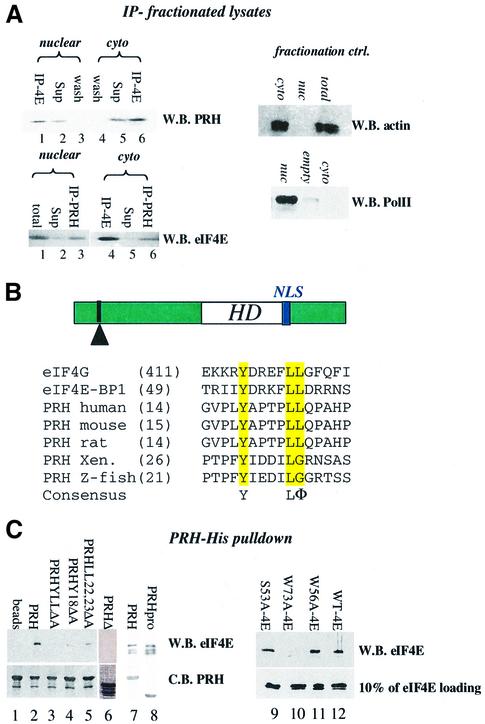
We examined whether PRH and eIF4E physically interact in vivo. Endogenous PRH and endogenous eIF4E co-immunopurified in lysates from K562 (Figure 1G) and U937 cells (Figure 2A). Both PRH and eIF4E have substantial nuclear and cytoplasmic distributions (Figure 1). To determine whether they interacted in both subcellular compartments, we fractionated cells and immunoprecipitated these fractions with PRH or eIF4E antibodies (Figure 2A). Clearly, eIF4E co-immunopre cipitated with PRH in both the nucleus (lane 1) and cytoplasm (lane 6). Conversely, PRH co-immunoprecipitated with eIF4E in both fractions (lanes 3 and 6, lower panel). The quality of the fractionation is demonstrated by monitoring the distribution of actin, which is mainly cytoplasmic, and of RNA polymerase II, which is nuclear. No PRH or eIF4E associated with IgG (data not shown; Figure 1G). Thus PRH is positioned to alter eIF4E activities in both subcellular compartments.
Fig. 2. PRH and eIF4E interact directly. (A) U937 cells were fraction ated, immunoprecipitated and analyzed by western analysis. IP refers to the immunoprecipitated fraction, Sup to supernatant after IP, and wash to last wash of protein A beads after IP. Fractionation controls are given in the adjacent panels, using actin as a cytoplasmic marker and RNA polymerase II as the nuclear marker. Total indicates total cell lysate; nuc, nuclear fraction; and cyto, cytoplasmic fraction. (B) Schematic of the PRH protein showing the position of the homeodomain (HD) relative to the eIF4E-binding site (indicated by the arrowhead) and the location of the NLS (in blue). The eIF4E-binding site in PRH from several organisms is shown. Accession numbers are as follows: NP-002720 (human), NP-032271 (mouse), NP-077361 (rat), AAB82335 (Xenopus) and NP-571009 (zebrafish). (C) His tag pull-down analysis of the PRH–eIF4E interaction. Wild-type (right panel) or mutant PRH proteins were immobilized on nickel–agarose beads and incubated with wild-type or mutant eIF4E as indicated. Loading of PRH mutants is shown in the bottom left panel. All constructs produced proteins at the expected molecular weight. Faster migrating bands indicate degradation products. eIF4E proteins used in lanes 7 and 8 are slightly degraded; the top band corresponds to full-length protein. Loading of eIF4E mutants is shown by western blot (W.B.) in the lower right panel. Equivalent amounts of eIF4E were used for experiments in the right panel where binding is ∼10% of eIF4E input. C.B. indicates Coomassie Blue-stained SDS–polyacrylamide gel.
PRH directly interacts with eIF4E using a conserved eIF4E-binding site
Inspection of the amino acid sequence of PRH revealed a consensus eIF4E-binding motif, similar to ones observed in eIF4G and 4EBP1 (Figure 2B). This sequence is defined by YXXXXLΦ, where X is any residue and Φ is a hydrophobic residue (Sonenberg and Gingras, 1998). We examined the possibility that PRH and eIF4E interacted directly through this site. Wild-type and mutant PRH-His and eIF4E proteins were produced in bacteria and purified to homogeneity. PRH-His protein, retained on nickel– agarose beads, binds free eIF4E. eIF4E is retained on the PRH–nickel–agarose beads (lane 2), but not on nickel– agarose beads alone (lane 1). Point mutations within the eIF4E consensus site in PRH abrogate its ability to bind eIF4E (lanes 3–5), as does deletion of a large portion of the N-terminal proline-rich region (residues 42–98, PRHΔ, lane 6). Although the eIF4E-binding site is still present in PRHΔ, this mutation unfolds the proline-rich region (data not shown). Thus, the eIF4E-binding site is necessary but not sufficient for association with eIF4E, requiring additional sequence for preservation of the three-dimensional structure of the N-terminal region. Consistently, other eIF4E-binding proteins such as eIF4G bind more effectively when the full-length eIF4G is used rather than a corresponding peptide containing only the consensus sequence (von Der Haar et al., 2000). Importantly, a construct containing only the proline-rich region of PRH (PRHpro, residues 1–136) binds eIF4E with the same affinity as wild-type PRH, indicating that the homeodomain and acidic regions of PRH do not contribute to binding in vitro (Figure 2C, lanes 7 and 8). Together, these data indicate that PRH uses a conserved eIF4E-binding site to interact directly with the eIF4E protein.
We extended these studies to establish which portion of eIF4E is required for this interaction. PRH binds the dorsal surface of eIF4E, since a W73A mutation abrogates binding (lane 10). This result is consistent with previous findings demonstrating that proteins which use a conserved eIF4E-binding site require W73 (Ptushkina et al., 1998; Sonenberg and Gingras, 1998). Mutation of either the cap-binding site, W56A, or of a putative phosphorylation site, S53A, does not alter binding to PRH (lanes 9 and 11). PRH associates with ∼10% of the eIF4E input (lower panel). Previous studies indicated that all of the eIF4E mutants used here were structured. W73A has wild-type affinity for m7GTP–Sepharose (Cohen et al., 2001) and, additionally, W73A, S53A and W56A all had secondary structure content indistinguishable from that of wild type, as monitored by circular dichroism studies (Kentsis et al., 2001; data not shown).
Thus, PRH interacts with the functionally important dorsal surface of eIF4E, where previous studies indicate that several positive and negative regulators of eIF4E bind. PRH uses a conserved eIF4E consensus sequence found in its N-terminal proline-rich region for this association. Further, this sequence is conserved amongst species from zebrafish to humans (Figure 2B), highlighting its potential functional importance.
PRH negatively regulates cyclin D1 production post-transcriptionally
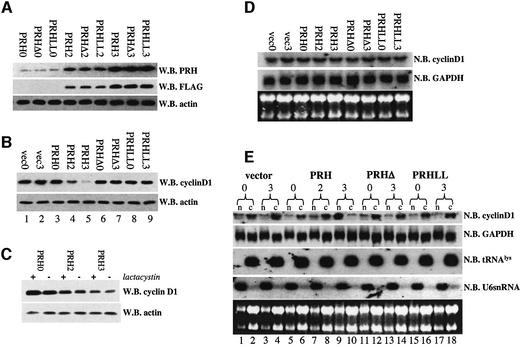
Overexpression of eIF4E results in increased levels of cyclin D1 protein due to increased nucleo-cytoplasmic transport of cyclin D1 transcripts (Rousseau et al., 1996; Lai and Borden, 2000; Cohen et al., 2001). The direct interaction of PRH with eIF4E led us to investigate whether PRH overexpression alters cyclin D1 protein production. In this way, PRH could be a tissue-specific regulator of the growth-promoting properties of eIF4E. We constructed a series of tet-inducible U937 cell lines with either wild-type PRH, or with mutants that disrupt the direct interactions with eIF4E: PRHΔ or PRHLL2324AA (Figure 2C). All of these constructs had a FLAG(M2) tag to distinguish transfected from endogenous PRH. The time course and levels of induction are nearly identical for the wild-type and mutant proteins, allowing direct comparisons to be made amongst cell lines (Figure 3A). Overexpressed PRH immunoprecipitates and co-localizes with endogenous PRH (http://atlas.physbio.mssm.edu/∼kbgroup/supplementary/PRH). Further, overexpression of PRH does not alter eIF4E or PML protein levels (Figure 5).
Fig. 3. PRH suppresses cyclin D1 protein production and cyclin D1 mRNA transport. (A) Induction of PRH, PRHΔ and PRHLL cells is the same in the U937 system as observed by both PRH polyclonal antibody and FLAG tag antibody. PRHLL refers to the PRHLL2324AA mutant and PRHΔ to the deletion mutant. (B) Cyclin D1 levels are suppressed by overexpression of PRH wild-type but not mutant constructs. (C) Addition of 10 µM lactacystin does not reverse the effects of PRH on cyclin D1 protein levels. (D) Northern analysis of RNA isolated from total cell lysates of PRH-overexpressing cells indicates that there are no alterations in the total levels of cyclin D1 mRNA. (E) Fractionation studies in conjunction with northern analysis reveal that PRH overexpression leads to retention of cyclin D1 mRNA in the nucleus. Cells transfected with vector or overexpressing PRH mutants do not have these effects. n indicates the nuclear, and c the cytoplasmic fraction. tRNALys is a marker for the cytoplasmic fraction, and U6snRNA for the nuclear fraction. The numbers 0, 2 and 3 indicate days post-tetracycline withdrawal. W.B., western blot; N.B., northern blot.
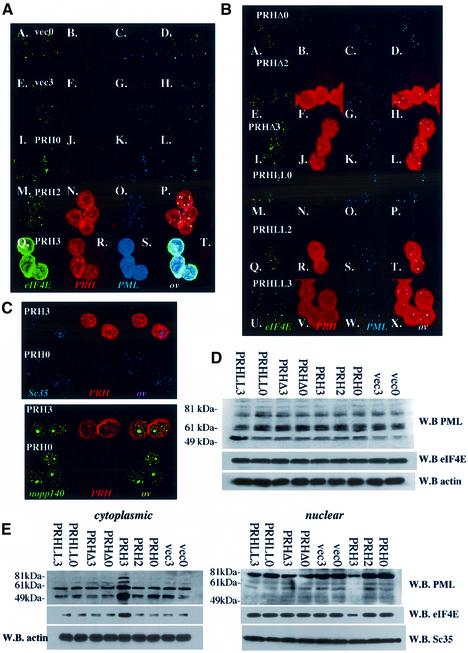
Fig. 5. PRH overexpression leads to disruption of eIF4E and PML nuclear bodies. (A) Confocal micrographs show that PRH overexpression leads to disruption of nuclear bodies as indicated. Cells are triple stained for eIF4E (green), PRH (red) and PML (blue). FLAG antibody is used to detect PRH, so only exogenous PRH is observed. The numbers 0 and 3 refer to days post-tetracycline withdrawal. (B) Mutant PRH that does not bind to eIF4E does not disrupt eIF4E nuclear bodies. PRHLL refers to the PRHLL2324AA mutation, and PRHΔ the deletion mutant. (C) The subcellular distribution of Sc35 splicing speckles (Sc35), Cajal bodies and nucleoli (nopp140) does not change upon PRH overexpression. Confocal micrographs represent single sections through the plane of the cells. (D) Protein levels in total cell lysates from cells overexpressing PRH, PRHLL, PRHΔ or vector-transfected controls. (E) Fractionation studies reveal that PRH overexpression leads to re-localization of eIF4E and PML to the cytoplasm. W.B. indicates western blot. The PML antibody mAb 5E10 indicates that there is a difference in isoform distribution between the two subcellular compartments, consistent with previous studies (Stuurman et al., 1992; Flenghi et al., 1995). Further, it is possible that some bands represent degradation products rather than different isoforms of PML. Actin is used for the loading control in the cytoplasmic fraction, and Sc35 (splicing speckles) in the nuclear fraction. Note that staining for PRH and eIF4E looks more intense after the bodies are disrupted (A), but by western analysis these levels do not change (D). This is due to differences in antibody availability increasing in the fixed cells after dispersal of the nuclear bodies, whereas this is not an issue on the denaturing gels used for western analysis.
We examined the effects of PRH overexpression on cyclin D1 protein levels (Figure 3B). PRH overexpression substantially reduces cyclin D1 protein levels (lanes 4 and 5) relative to vector controls, but does not alter production of the housekeeping protein actin, consistent with eIF4E’s specificity (Rousseau et al., 1996). PRHΔ and PRHLL2324AA, which do not interact with eIF4E, do not alter cyclin D1 protein levels (lanes 6–9) relative to vector controls. Several studies indicate that cyclin D1 levels can be regulated by proteosomal degradation (Langenfeld et al., 1997). However, addition of 10 µM lactacystin did not alter PRH’s repression of cyclin D1 protein levels (Figure 3C). Northern analysis of cyclin D1 transcript levels indicated that PRH, PRHΔ or PRHLL2324AA overexpression did not substantially alter mRNA levels of either cyclin D1 or GAPDH (Figure 3D). Thus PRH modulates cyclin D1 levels post-transcriptionally but not through proteosomal degradation, and does not modulate steady-state transcript stability. Since GAPDH and actin levels are unaffected, these effects of PRH demonstrate the same specificity as observed for eIF4E. These mutational analyses demonstrate that the ability of PRH to modulate cyclin D1 levels is strongly linked to its interaction with eIF4E.
PRH inhibits eIF4E-dependent mRNA transport of cyclin D1 and growth
We examined the possibility that PRH inhibits eIF4E-dependent transport of cyclin D1 transcripts. Using the tet-inducible system, cells were fractionated into nuclear and cytoplasmic compartments, and RNAs were isolated from each fraction and analyzed by northern methods. The quality of the fractionation was assessed by monitoring tRNALys, a mainly cytoplasmic RNA, and U6snRNA, a mainly nuclear RNA (Figure 3E).
Prior to withdrawal of tetracycline, the amounts of cyclin D1 found in the nucleus and cytoplasm are approximately equal (lanes 5 and 6) in cells transfected with PRH, similar to observations in vector controls (lanes 1 and 2). However, when PRH expression is high (PRH3, Figure 3A), it is clear that the majority of cyclin D1 mRNA is in the nucleus whereas there is no change in the vector controls (lanes 3 and 4 versus 9 and 10). Concomitantly, there is a reduction of cyclin D1 mRNA in the cytoplasm. In contrast to wild-type PRH, expression of PRHΔ or PRHLL2324AA does not alter cyclin D1 mRNA distribution (lanes 11 and 12 versus 13 and 14, and lanes 15 and 16 versus 17 and 18). Notably, the compartmentalization of GAPDH transcripts, isolated from the same experiments as shown for cyclin D1, is not altered by expression of any of the constructs. Thus, PRH modulates cyclin D1 protein levels through retention (either directly or indirectly) of cyclin D1 transcripts in the nucleus. Further, the ability to retain these transcripts is linked to PRH’s ability to interact directly with eIF4E.
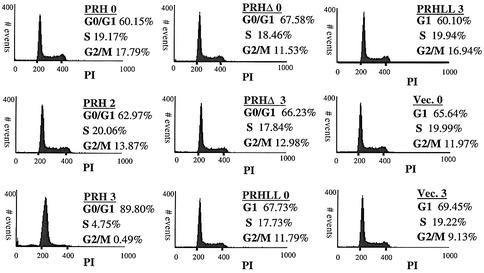
Fluorescence-activated cell sorting (FACS) analysis revealed that PRH overexpression and subsequent suppression of cyclin D1 protein production are correlated with G1 arrest (Figure 4). DNA content was monitored by propidium iodide. In contrast to wild type, PRHΔ and PRHLL2324AA do not alter the cell cycle relative to vector controls, consistent with the requirement for a direct interaction between PRH and eIF4E.
Fig. 4. PRH overexpression induces G1 arrest in U937 cells. Tet-inducible U937 cells expressing wild-type PRH, PRHΔ and PRHLL2324AA (PRHLL) mutants were stained with propidium iodide (PI), and the DNA content was measured by FACS. Cell phase distributions were determined using the CellQuest software. The numbers 0, 2 and 3 indicate days after tetracycline withdrawal.
PRH overexpression disrupts eIF4E and PML nuclear bodies
The marked effects on cyclin D1 transport led us to determine whether PRH modulated eIF4E function by altering its subcellular distribution. We monitored the subcellular distributions of endogenous PML, endogenous eIF4E and exogenous PRH. Prior to withdrawal of tetracycline, no expression of PRH-FLAG is observed, and the PML and eIF4E proteins co-localize in nuclear bodies (Figure 5A, I–L) as expected and as observed in vector controls (Figure 5A, A–H). However, when substantial exogenous PRH is produced (PRH3), the localization of PML and eIF4E is altered substantially (Figure 5A, Q–T). Overexpressed PRH is found in some nuclear bodies, but the majority is found diffusely throughout the cytoplasm. The majority of eIF4E and PML are redistributed to the cytoplasm, with nearly a complete loss of nuclear bodies. PRH co-localizes with the remaining eIF4E and PML bodies. PRH mutants that do not bind eIF4E in vitro have a more diffuse pattern than wild-type PRH and do not alter the distribution of either PML or eIF4E (Figure 5B and D). Importantly, overexpression of wild-type or mutant PRH does not alter PML or eIF4E protein levels (Figure 5D). Fractionation studies confirm that PML and eIF4E are redistributed to the cytoplasm by PRH, whereas PRH mutants have no effect (Figure 5E). Several known isoforms of PML are present, consistent with previous studies (Flenghi et al., 1995). In addition, there may also be some degradation products of some PML isoforms present. These data suggest that PRH inhibits eIF4E’s mRNA transport function by disrupting eIF4E nuclear bodies. Importantly, localization of splicing speckles or nucleoli and Cajal bodies was not altered by PRH (Figure 5C). Thus, PRH must interact directly with eIF4E in order to disrupt eIF4E and PML nuclear bodies, and this disruption is specific, leaving other nuclear organelles intact. The exact mechanism of how overexpression results in disruption of PML and eIF4E nuclear bodies is currently not known.
For comparison, we monitored the effects of overexpression of PML, since it also inhibits eIF4E-dependent mRNA transport. Using a tet-inducible system, we observed that overexpression of PML leads to larger PML nuclear bodies (http://atlas.physbio.mssm.edu/∼kbgroup/supplementary/PRH) as observed in other cell types (e.g. Topisirovic et al., 2002). Endogenous PRH and eIF4E still localize with these bodies where PML does not alter the subcellular distribution of these proteins or levels of expression. Thus, the effects of PRH are specific, where overexpression of PML or eIF4E does not lead to re-localization of body constituents to the cytoplasm (Topisirovic et al., 2002). Moreover, PRH disrupts the eIF4E nuclear body and subsequently eIF4E’s ability to transport mRNAs.
PRH and eIF4E associate independently of PML
In the above studies, we monitored the ability of PRH to modulate eIF4E-dependent cyclin D1 mRNA transport and growth in U937 cells that endogenously express PML, PRH and eIF4E. Since both PRH and eIF4E associate directly with the PML protein (Topcu et al, 1999; Cohen et al., 2001), we set out to establish whether these proteins associate in cells in the absence of PML and, in this way, if PRH modulates eIF4E function independently. Initially, we monitored the subcellular localization of PRH and eIF4E proteins in PML–/– cells (Figure 6). Note that PRH is not expressed endogenously in these cells, but eIF4E is. Overexpression of eIF4E resulted in a pattern similar to that observed for endogenous eIF4E in these cells, where eIF4E is found both in nuclear bodies and diffusely throughout the cytoplasm (Figure 6A versus C). In PRH-expressing cells, PRH is found throughout the cytoplasm and in nuclear bodies (Figure 6G–I), similar to the pattern observed for endogenous PRH in U937 cells. PRH expression leads to the apparent disruption of endogenous eIF4E nuclear bodies, with some bodies remaining, and an additional diffuse pattern similar to observations in U937 cells (Figure 5A), suggesting that the relative levels of PRH and eIF4E are important for localization. In cells overexpressing both PRH and eIF4E, these two proteins co-localize in nuclear bodies, as well as being found diffusely throughout the cytoplasm and nucleoplasm (Figure 6P–R). Clearly, no proteins specific to the myeloid lineage are required to mediate this interaction in vivo as it occurs readily in the PML–/– mouse embryo fibroblast (MEF)-derived cell line.
Fig. 6. The PRH–eIF4E interaction is independent of PML. Upper panel: PML–/– fibroblasts were stably transfected with eIF4E or PRH constructs as indicated. Confocal micrographs represent single sections through the plane of the cells. Lower panel: PML–/– cells transfected as indicated, immunoprecipitated (IP) with the Xpress antibody (IP PRH) and western blotted for eIF4E (W.B. eIF4E), or vice versa. n, nuclear fraction; c, cytoplasmic fraction; S = supernatant after IP. IP and S in the last two lanes on the right are positive controls for IP, where in the upper panel it is IP eIF4E, and in the lower panel IP PRH, and were probed as indicated.
Importantly, mutations in PRH that disrupt association with eIF4E result in the inability of PRH to associate with eIF4E nuclear bodies, leaving both endogenous and exogenous eIF4E nuclear bodies intact (Figure 6J–L and M–O). Finally, expression of PRH and W73A eIF4E results in the expected pattern, where PRH is found in some nuclear bodies, presumably through association with endogenous eIF4E (Figure 6S–U). W73A eIF4E forms very large bodies, as observed previously (Topisirovic et al., 2002). Immunofluorescence results were confirmed by immunoprecipitation studies (Figure 6, lower panel). Here, mutants that could not bind eIF4E (PRHΔ and PRHLL2324AA) did not co-immunoprecipitate, whereas wild-type proteins did.
We examined whether PML interferes with the PRH eIF4E association in cells in which PRH is not normally expressed but PML is. Experiments in NIH-3T3 cells yielded results identical to those observed in PML–/– cells (http://atlas.physbio.mssm.edu/∼kbgroup/supplementary/PRH). Similarly to U937 cells, overexpression of PRH resulted in re-localization of PML and eIF4E to the cytoplasm. Consistently, mutagenesis studies indicated that this reorganization required a direct interaction between eIF4E and PRH. Thus the presence or absence of PML, even in cellular contexts where PRH is not normally expressed, does not alter PRH’s association with eIF4E. Thus, the association of PRH and eIF4E in cells, as in vitro, is independent of PML.
PRH inhibits eIF4E-dependent transformation
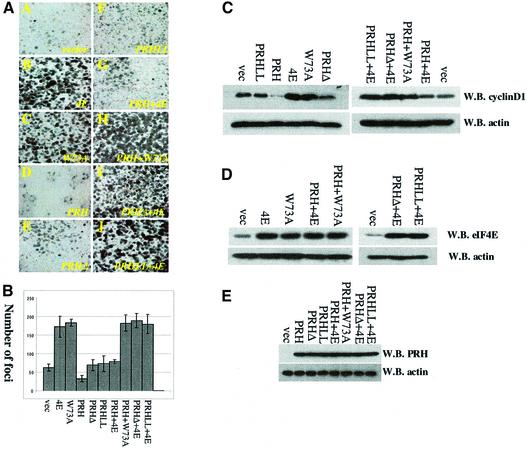
eIF4E overexpression transforms immortalized cell lines (Lazaris-Karatzas et al., 1990; Sonenberg and Gingras, 1998). Thus, we examined the ability of PRH to modulate eIF4E-mediated transformation in anchorage-dependent foci formation assays in NIH-3T3 and PML–/– fibroblasts using stable transfection. It is clear from Figure 7A that eIF4E-overexpressing cells produce so many foci that it is difficult to quantitate as foci coalesce, as observed previously (Lazaris-Karatzas et al., 1990; Sonenberg and Gingras, 1998). For these experiments, only distinct foci of a minimal size and above were counted, so that the number of foci in eIF4E-overexpressing cells may be underestimated. Importantly, overexpression of PRH wild-type or mutant proteins does not affect levels of eIF4E expression, and vice versa (Figure 7D and E), in either cell line.
Fig. 7. PRH suppresses oncogenic transformation of eIF4E and expression of cyclin D1. (A) Anchorage-dependent foci formation assays in PML–/– cells. Foci stained with Giemsa formed as a result of different transfections. Cells were transfected as indicated: 4E (wild-type eIF4E), PRH (wild-type PRH), PRHLL (PRHLL2324AA), PRHΔ (PRH ApaI deletion), W73A (eIF4E W73A mutation) and vector. Identically sized areas were taken from representative regions of each Petri dish. The results were quantitated in (B). Foci were counted in five dishes per treatment, and values are ± SD. (C) Cyclin D1 levels. Western blots (W.B.) of the indicated experiments after transfection. (D and E) Western analysis indicated that overexpression of eIF4E and PRH did not alter each other’s levels.
Results in these two cell lines were analogous; therefore, only those from PML–/– cells will be discussed in detail (Figure 7A). For NIH-3T3 results, see http://atlas.physbio.mssm.edu/∼kbgroup/supplementary/PRH. The number of foci per experiment is shown in Figure 7B. In vector-transformed cells, some foci are present, consistent with the inherent transformed phenotype reported for PML–/– cells. However, substantially more foci are visible in eIF4E-overexpressing cells than in the vector controls (Figure 7A, panels A and B). PRH expression reduced the number of foci relative to both the vector, presumably acting on the endogenous eIF4E, and the eIF4E-overexpressing cells. Experiments using either the PRHΔ or PRHLL2324AA mutants revealed more foci than those using wild-type PRH, similar in number to those observed in vector controls (Figure 7A, panels E and J versus A). Co-expression of eIF4E and wild-type PRH resulted in a significant reduction in the number of foci relative to cells overexpressing eIF4E alone (Figure 7A, panel B versus G). As reported previously, the W73A eIF4E mutant, which is deficient in translation activity (Sonenberg and Gingras, 1998), transforms cells as readily as wild-type eIF4E (Figure 7A, panel C), consistent with previous results (Cohen et al., 2001). Co-expression of PRH and the W73A eIF4E mutant resulted in approximately the same number of foci as with W73A eIF4E alone (Figure 7A, panel C versus H). Further, co-expression of either PRHLL2324AA or PRHΔ with wild-type eIF4E resulted in the same number of foci as experiments using wild-type eIF4E alone (Figure 7A, panels I and J versus B). Thus, PRH’s ability to inhibit eIF4E-mediated transformation is linked to its ability to interact directly with eIF4E and disrupt eIF4E nuclear bodies.
Since there is a strong correlation between eIF4E’s ability to increase cyclin D1 protein levels and its ability to transform cells (Cohen et al., 2001), we examined whether PRH suppression of foci formation is correlated with suppression of cyclin D1 protein levels. PRH reduces cyclin D1 protein levels relative to vector controls in both PML–/– and NIH-3T3 cells (Figure 7C; http://atlas.physbio.mssm.edu/∼kbgroup/supplementary/PRH), presumably by acting on endogenous eIF4E. However, PRH mutants that do not interact directly with eIF4E, i.e. PRHΔ and PRHLL2324AA, have cyclin D1 levels similar to those observed for vector controls. As expected, cyclin D1 levels are elevated relative to vector controls in cells overexpressing eIF4E or W73A eIF4E. Co-expression of PRH and eIF4E results in lower levels of cyclin D1 relative to cells expressing eIF4E. Mutagenesis studies indicate that as for foci formation, there is a strong correlation between the ability of PRH to inhibit cyclin D1 protein production and to interact directly with eIF4E. Thus, there is a correlation between the ability of PRH to bind eIF4E directly, to associate in cells, to suppress eIF4E-dependent cyclin D1 transport and subsequently to suppress eIF4E-mediated oncogenic transformation. Furthermore, these activities of PRH are independent of PML.
The nuclear fraction of PRH is critical for its roles in modulating eIF4E mRNA transport and transformation
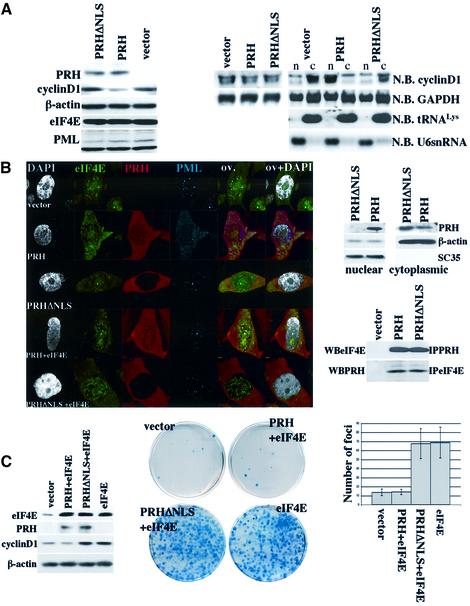
To assess the importance of the nuclear localization of PRH to its ability to modulate eIF4E activity, we mutated its NLS. We identified a putative NLS sequence in PRH (Figure 2B), and subsequently mutated arginines 188 and 189 to alanine. Mutations in this region (PRHΔNLS) resulted in a complete loss of the nuclear localization of PRH, as observed by confocal microscopy and by fractionation analysis (Figure 8B). The loss of nuclear localization was confirmed by fractionation methods in conjunction with western analysis (Figure 8B, right panel). Further, the distribution of PML and eIF4E nuclear bodies is unchanged by this mutant. PRH and PRHΔNLS both interact with the cytoplasmic fraction of eIF4E (Figure 8B, bottom right panel).
Fig. 8. Nuclear localization of PRH is necessary for its function in the suppression of eIF4E-mediated transport and transformation. All experiments were carried out in NIH-3T3 cells. (A) Left panel: western analysis of total cell lysates overexpressing PRH, PRHΔNLS and vector controls. Right panel: northern analysis of total mRNA and (far right) RNA from nuclear (n) and cytoplasmic (c) fractions. (B) Left panel: confocal micrographs of cells transfected as described. Cells were triple stained for eIF4E (green), PRH (red) and PML (Cy5 shown in blue). In addition, DAPI staining is shown in gray. Overlays are as described in Figure 1. The objective is 100× with a further 2-fold magnification. Right panel: western analysis of the nuclear and cytoplasmic fraction where β-actin and Sc35 are markers for the cytoplasmic and nuclear fraction, respectively. Below, co-immunoprecipitation analysis shows that exogenous PRH and PRHΔNLS interact with endogenous eIF4E. Total cell lysates were immunoprecipitated (IP) with eIF4E or PRH antibody. (C) Left panel: western analysis of cells transfected as described. Right panel: anchorage-dependent foci formation assays transfected as described. On the far right, foci were counted in five dishes per treatment and values are ± SD. WB indicates western blot, and N.B. northern blot.
We monitored the ability of PRHΔNLS to modulate cyclin D1 levels. In contrast to experiments with the wild-type protein, PRHΔNLS overexpression did not alter cyclin D1 protein levels where cyclin D1 levels were similar to those observed in vector control experiments (Figure 8A, left panel). Further, PRHΔNLS did not repress cyclin D1 protein production in the presence of eIF4E, in contrast to wild-type PRH (Figure 8C, left panel). Importantly, overexpression of PRHΔNLS did not alter cyclin D1 transcript levels or steady-state stability of these transcripts (Figure 8A, right panel). Fractionation studies in conjunction with northern analysis indicate that PRHΔNLS does not alter the nucleo-cytoplasmic distribution of cyclin D1 transcripts relative to vector controls (Figure 8A, right panel). In contrast, PRH overexpression caused nuclear retention of transcripts (Figures 3 and 8A).
We extended these studies to determine whether PRHΔNLS alters eIF4E-mediated transformation as described above for wild-type PRH (Figure 8C). Unlike wild-type PRH, overexpression of PRHΔNLS resulted in the same number of foci as observed for vector controls (data not shown). PRHΔNLS did not suppress eIF4E-mediated transformation where there were the same number of foci as observed in cells only expressing eIF4E, whereas wild-type PRH substantially reduces the number of foci in cells overexpressing eIF4E (Figure 8C). In summary, the nuclear localization of PRH is required for its ability to disrupt PML and eIF4E nuclear bodies and to suppress eIF4E-mediated cyclin D1 mRNA transport, cyclin D1 protein production and transformation.
Other homeodomain proteins are potential modulators of eIF4E
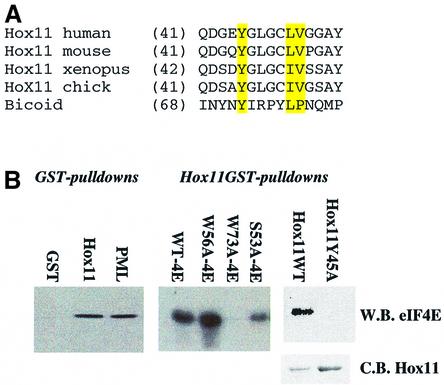
Our results demonstrate that PRH is a tissue-specific modulator of eIF4E. This leaves open the question of whether, in those tissues and organisms where PRH is not expressed, there are other regulators of eIF4E function. In particular, could PRH regulation of eIF4E represent a general phenomenon where other homeodomain proteins, with limited expression, modulate eIF4E activity? We carried out database searches of the 803 homeodomain proteins found in the Swissprot database for the presence of the YXXXXLΦ motif using PROSE (http://bioweb.pasteur.fr/seqanal/interfaces/prose.html). A total of 199 homeodomain proteins contain at least one of these potential binding sites, including Hox11, a close relative of PRH and the Drosophila Bicoid protein (http://icb.mssm.edu/borden/hd_tbl.html). In 100 of these, the binding site is found N-terminal to the homeodomain, as in PRH (Figure 2B). The binding site in Hox11 was conserved between human and mouse, and has a conservative substitution of L to I in Xenopus and chicken. To assess if these sites were involved in interactions with eIF4E, we produced Hox11–GST (Figure 9). Hox11 binds eIF4E with similar affinities to those observed for PRH and for the RING of PML. Like other proteins that utilize this binding motif, the W73A mutation of eIF4E eliminates Hox11 binding, but mutation of distal sites on eIF4E does not. Mutation of the consensus binding site in Hox11 (Y45A) results in a loss of association with eIF4E. Further, the Bicoid protein uses its consensus sequence to bind eIF4E directly (Niessing et al., 2002). Thus, other homeodomain proteins are potential modulators of eIF4E, and this type of regulation is likely to be a general phenomenon and not one limited to cell types that express PRH.
Fig. 9. Other homeodomains utilize conserved eIF4E-binding sites to bind eIF4E. (A) The eIF4E-binding site in Hox11 is conserved from chicken to human. Conserved residues are highlighted in yellow. Accession numbers are: XP-046733 (human), P43345 (mouse), AA14453 (Xenopus) and 093366 (chicken). (B) Purified Hox11 binds to purified wild-type eIF4E with the same affinity as PML and PRH (Figure 2), binding ∼10% of eIF4E input. Hox11 and PML RING are immobilized GST fusion proteins, and were incubated with purified wild-type or mutant eIF4E, as indicated. Hox11Y45A mutation does not interact with eIF4E. Loading for Hox11 wild-type and mutants is shown by the Coomassie Blue gel (C.B. Hox11). W.B. indicates western blot.
Discussion
Although eIF4E is required for survival in all eukaryotic cells, its subcellular localization and levels of expression are altered during zebrafish and Xenopus development (Sonenberg and Gingras, 1998; Fahrenkrug et al., 1999; Strudwick and Borden, 2002). These observations led us to investigate whether these activities of eIF4E were modulated in a tissue-specific manner, allowing eIF4E to be regulated during development and differentiation in a variety of contexts. Here we report the first tissue-specific modulator of eIF4E activity, PRH. Unlike eIF4E, which is expressed ubiquitously in eukaryotes, PRH expression is limited to only a few cell types. We demonstrate that PRH inhibits eIF4E’s function in mRNA transport through a direct interaction using a conserved eIF4E-binding site. Other parts of the N-terminal proline-rich region are also required for eIF4E binding, by preserving the structural integrity of this part of the protein. The PRH–eIF4E interaction leads to disruption of eIF4E nuclear bodies, disruption of eIF4E-mediated transport of cyclin D1 mRNA, subsequent retention of cyclin D1 mRNA in the nucleus and reduction in cyclin D1 protein levels. eIF4E undoubtedly regulates the transport of other, as yet unidentified, transcripts. Whether it is cyclin D1 mediating the observed effects on growth or other transcripts is a focus of future studies. However, monitoring cyclin D1 allows us to monitor the transport process.
These molecular events are correlated with the ability of PRH to repress the growth-promoting and transforming properties of eIF4E in a variety of cell lines. In addition, PRH and eIF4E interact in the cytoplasm through its dorsal surface (W73), suggesting that PRH also modulates the translation activity of eIF4E, since the dorsal surface is required for eIF4E’s assembly into the translation pre-initiation complex. Our preliminary data suggest that PRH inhibits eIF4E-dependent translation in vitro (unpublished observations). Thus, PRH may alter other physiological activities of eIF4E through modulation of eIF4E-dependent translation. Further, PRH does not require PML, which binds both PRH and eIF4E, for any of these activities. Thus, PRH is positioned to be a powerful regulator of eIF4E in specific tissues and in a variety of organisms. This interaction could be key to the balance between proliferation and differentiation.
Since PRH is required for blood differentiation in zebrafish, Xenopus and mice, and for liver and thyroid organogenesis in mice (Yatskievych et al., 1999; Keng et al., 2000; Liao et al., 2000; Martinez Barbera et al., 2000), its negative regulation of eIF4E may be important to these processes. Interestingly, eIF4E is highly overexpressed in patients with a variety of non-Hodgkin B-cell lymphomas (Wang et al., 1999) where PRH levels increase upon differentiation from early pre-B cells to late pre-B cells (Manfioletti et al., 1995). There are other examples of PRH modulation during differentiation. For example, PRH levels increase after differentiation of HL60 cells (myeloblasts–promyelocytes) to granulocytes but decrease upon differentiation of HL60s to the monocyte–macrophage lineage (Manfioletti et al., 1995). Recent data indicate that Bicoid inhibits translation of caudal mRNA through association with eIF4E (Niessing et al., 2002). Thus, regulation in all tissue types is not as simple as the levels of PRH relative to eIF4E and probably involves additional factors such as PML or Hox11 and their relative activities in these cell types. Further, it is not clear that all homeodomain proteins that bind eIF4E will be inhibitory. Clearly eIF4E can be regulated by a variety of unrelated mechanisms in addition to those described here. For instance, the 4E-binding proteins inhibit its translational activities in response to extracellular signals (Sonenberg and Gingras, 1998). These modes of regulation are also undoubtedly important to roles that eIF4E plays in differentiation and development.
Although PML and PRH both act as negative regulators of the mRNA transport function of eIF4E, their mechanisms of action are distinct. PML utilizes its RING domain to bind the same region of eIF4E as PRH (Cohen et al., 2001). Direct binding by the RING reduces eIF4E’s affinity for the m7G cap by >100-fold (Cohen et al., 2001; Kentsis et al., 2001). This biochemical activity of PML is correlated with its ability to inhibit eIF4E-mediated transformation of fibroblasts. Unlike PRH, overexpression of PML and eIF4E results in larger nuclear bodies that still localize with the respective partners, consistent with previous transfection experiments in other cell lines (Borden et al., 1995; Topisirovic et al., 2002). There is no apparent disruption of the bodies or re-localization of PML, PRH or eIF4E in these cells. This is in stark contrast to PRH overexpression in a variety of cell lines (shown here), which results in disruption of most bodies and substantial re-localization of eIF4E and PML to the cytoplasm (Figures 5 and 6). Unlike PML, PRH only modestly inhibits eIF4E cap-binding activity by ∼10-fold (our unpublished observations). Although PRH and PML utilize different mechanisms to inhibit eIF4E-mediated transport, their potencies as inhibitors of cyclin D1 protein production appear quite similar (data not shown). Since PML, PRH and eIF4E can bind one another independently of the third component, it seems likely that PML and PRH could have synergistic effects on eIF4E function.
Identification of PRH as the second negative regulator of nuclear eIF4E function may help to address the question of why PML–/– mice do not get more cancers than littermate controls (Wang et al., 1998). Our previous studies with PML and eIF4E led us to hypothesize that PML is a mammalian modulator of an evolutionarily older organelle comprised, at least in part, of eIF4E nuclear bodies (Cohen et al., 2001). In this model, PML and eIF4E balance each other’s contrasting effects on growth. The data presented here suggest that in the absence of PML, there are other negative regulators of these eIF4E functions, and the presence of these additional inhibitors, which may or may not act synergistically, could regulate eIF4E. We found that a substantial number of homeodomain proteins contain eIF4E-binding sites and demonstrated that Hox11 utilizes this site to interact with eIF4E on its dorsal surface (W73). Thus, there are potentially many tissue-specific proteins that regulate eIF4E in this manner.
To our knowledge, this is the first report of a homeodomain protein regulating mRNA transport. Other homeodomain proteins are negative regulators of growth, such as Cdx1 which negatively regulates cyclin D1 levels transcriptionally (Lynch et al., 2000). Alternatively, Msx1 post-transcriptionally stimulates cyclin D1 protein production and cell growth (Hu et al., 2001). Other groups have focused on the potential transcriptional activities of PRH typical of homeodomain proteins. Mutation of the homeodomain does not alter the ability of PRH to repress reporter constructs, but deletion of the proline-rich region does (Tanaka et al., 1999; Guiral et al., 2001). Inter estingly, the parts of the proline-rich region required for repression of reporter genes correspond to those required for association with eIF4E, which are deleted in the PRHΔ mutant. It is formally possible that the reporter assays monitored an alteration in transport and not directly transcription. Alternatively, PRH may have additional functions in transcription. For instance, the few PRH bodies in the nucleus that do not localize with eIF4E nuclear bodies in U937 cells may be sites for PRH transcriptional activity.
The Bicoid homeodomain protein also controls gene expression post-transcriptionally as well as transcriptionally (Niessing et al., 1999). Bicoid mediates control of gene expression in the Drosophila embryo to allow proper anterior–posterior patterning by activating transcription of zygotic segmentation genes. Interestingly, PRH plays a role in anterior–posterior patterning, but in the mouse (Martinez Barbera et al., 2000). Interestingly, Bicoid uses a consensus eIF4E-binding site to interact with eIF4E and repress translation of caudal mRNA (Niessing et al., 2002). The transcriptional activation and translational repression activities of Bicoid involve different parts of the protein, although both activities require the homeodomain for DNA–RNA binding.
In summary, we demonstrate that the mRNA transport function of eIF4E is modulated in a tissue-specific manner by PRH. This is the first evidence that the integrity of this nuclear structure is required for eIF4E-dependent mRNA transport. PRH thereby inhibits the growth-promoting and transforming properties of eIF4E. PRH appears to be the first homeodomain protein reported that modulates mRNA transport, and these activities are independent of its role as a transcription factor. Apart from PRH, there are a wide variety of other possible homeodomain protein modulators of eIF4E, including Hox11. These findings position homeodomain proteins in a new role as modulators of eIF4E. Further, we provide a biochemical framework for the tissue-specific regulation of a general factor, eIF4E, and its involvement in processes such as differentiation and development.
Materials and methods
Reagents
Full-length cDNA constructs for bacterial (PRH-His tag) and mammalian overexpression (pcDNA3.1 PRH) were described previously in Topcu et al. (1999). For the tet-inducible system, PRH-pTRE was a kind gift of Robert Hromas. PRHΔ was made by digestion with ApaI, which deletes residues 42–98. We identified a putative NLS in PRH. Mutation of R188 and R189 to alanine resulted in no nuclear PRH (Figure 8) and is referred to as PRHΔNLS in the text. PRHpro contains only the proline-rich region of PRH residues 1–136. Mammalian and bacterial expression constructs for eIF4E and PML were described in Cohen et al. (2001). Full-length Hox11 was subcloned into pGEX6T. Point mutations were engineered using the Stratagene Quikchange kit. DNA sequencing was used to verify the integrity of all constructs.
Polyclonal antibodies to full-length PRH were produced and gave similar results to those obtained with a fragment of the proline-rich region (Topcu et al., 1999). The PRH antibody was affinity purified against purified PRH-His protein. Antibodies used against PML include monoclonal antibody (mAb) 5E10 which recognizes human PML (Stuurman et al., 1992), polyclonal antibodies which recognize both human and mouse PML, kind gifts of Gerd Maul and Paul Freemont, described previously in Borden et al. (1995) and Topisirovic et al. (2002), and the polyclonal antibody against nopp140 (Meier, 1996). Additional antibodies used include mouse anti-eIF4E mAb (BD Transduction Laboratories), mouse anti-cyclinD1 mAb (BD PharMingen), mouse anti-FLAG-M2 mAb (Sigma), mouse anti-Xpress mAb (Invitrogen), mouse anti-SC35 mAb (BD PharMingen) and rabbit polyclonal anti-PolII antibody [sc-899 (Santa Cruz Biotechnology)].
Cell culture
U937 and K562 cells were maintained as described previously in Cohen et al. (2001) and NIH-3T3 and PML–/– cells as in Topisirovic et al. (2002). Tet-inducible U937T cells were grown in 0.3 µg/ml tetracycline–HCl and, when indicated, were washed four times with PBS and resuspended in tetracycline–HCl-free medium. Where indicated, cells were treated with 10 µM lactacystin for 24 h.
Cell transfection
Tet-inducible U937T cells (2 × 107 cells) (Boer et al., 1998) were co-transfected with 10 µg of pIND (neomycin selectable marker gene) and 10 µg of pTRE-PRH or mutants by electroporation. Transfected cells were seeded at 1 × 105 on methylcellulose plates supplemented with full growth medium and selected with 1 mg/ml G418. The plates were incubated until macroscopic colonies were visible. Single colonies were dispensed into 24-well plates containing full growth medium, and individual clones were tested for induction of pTRE constructs. Positive clones were tested, with ‘leaking’ clones excluded.
NIH-3T3 and PML–/– cells were plated at 1.5 × 105 cells/35 mm tissue culture dish 24 h prior to transfection. Transfections were performed using GeneJammer Transfection Reagent (Stratagene) according to the manufacturer’s instructions. NIH-3T3 and PML–/– cells were transfected with 3 µg of pMV, pMV-eIF4E wild-type or mutants, or pcDNA 3.1, pcDNA 3.1-PRH wild-type or mutants. At 48 h post-transfection, cells were selected in 1 mg/ml G418-containing medium.
Foci growth assay
Anchorage-dependent foci formation assays were as described previously (Cohen et al., 2001). Briefly, stably transfected NIH-3T3 and PML–/– cells were seeded at 105 cells per 100 mm dish and maintained for 7 days with addition of fresh medium containing 1 mg/ml G418 every 3 days. After the incubation, cells were dried, fixed in methanol at room temperature, stained with Giemsa and counted. The numbers represent the average of five plates, and experiments were repeated independently at least three times.
Cellular fractionation and northern analysis
Fractionation and RNA isolation were described previously in Lai and Borden (2000) and Topisirovic et al. (2002). Briefly, U937T cells were rinsed twice in ice-cold 1× PBS pH 7.4 and resuspended in lysis buffer B [10 mM Tris pH 8.4, 140 mM NaCl, 1.5 mM MgCl2, 0.5% NP-40, 1 mM dithiothreitol and RNasin (100 U/ml) (Promega)] with slow pipeting. Nuclear suspensions were centrifuged at 1000 g for 3 min at 4°C and the supernatant saved as the cytoplasmic fraction. Nuclear pellets were resuspended in lysis buffer B. A one-tenth volume of the detergent [3.3% (w/v) sodium deoxycholate and 6.6% (v/v) Tween-40] was added under slow vortexing, and the nuclear suspension was incubated on ice for 5 min. Nuclei were pelleted by centrifugation at 1000 g for 3 min at 4°C, and supernatant (post-nuclear fraction) was saved and added to the cytoplasmic fraction. Nuclei were rinsed once in lysis buffer B. This protocol yielded intact nuclei as determined by light microscopy, with no significant cytoplasmic contamination as seen from the tRNALys content.
Total or fractionated RNAs were isolated by the TRizol (Gibco) procedure. The RNA from the nuclear fraction was treated additionally with RNase-free DNase I (Promega). A 5 µg aliquot of RNA was loaded on a 1% formaldehyde/agarose gel and transferred onto positively charged nylon membrane (Roche). Membranes were pre-hybridized in ULTRAhyb buffer (Ambion) and probed with cyclin D1 cDNA probe (20 pM), GAPDH cDNA probe (5 pM) (Ambion), biotinylated tRNALys antisense oligoprobe (30 pM) or U6 antisense oligoprobe (30 pM). cDNA probes were biotinylated using the BrightStar Psoralen–Biotin kit (Ambion) and signals were detected using CDP Star chemiluminescence (Ambion).
Western blot and co-immunoprecipitation studies
For western analysis, cells were lysed in RIPA buffer supplemented with complete protease inhibitors (Roche) on ice. A 20 µg aliquot of whole-cell extracts was analyzed. All primary antibodies except 5E10 and affinity-purified PRH antibody (1:100) were used at 1:4000. Signals were revealed by chemiluminescence (SuperSignal). Fractionation for co-immunoprecipitations was as described previously (Carlile et al., 1998; Lai and Borden, 2000; Topisirovic et al., 2002).
Immunofluorescence and laser scanning confocal microscopy
Experiments were as described previously (Cohen et al., 2001; Topisirovic et al., 2002). Briefly, cells were rinsed twice in 1× PBS pH 7.4, fixed for 20 min in 100% methanol at –20°C or in 3.7% paraformaldehyde at room temperature, blocked and permeabilized with blocking buffer (10% fetal bovine serum, 0.1% Tween-20 in 1× PBS pH 7.4). Both fixation protocols gave identical results. Secondary antibodies were: fluorescein isothiocyanate (FITC)-conjugated donkey anti-rabbit antibody, Texas red-conjugated donkey anti-rabbit antibody, Texas red-conjugated donkey anti-mouse antibody, Cy5-conjugated donkey anti-mouse antibody (Jackson ImmunoResearch Laboratories) or Alexa 350-conjugated donkey anti-mouse antibody (Molecular Probes). For triple staining, cells were additionally fixed with 3.7% paraformaldehyde for 10 min at room temperature, washed and incubated with a 1:20 dilution of FITC-conjugated mouse anti-eIF4E mAb (BD Transduction Laboratories) at 4°C overnight. Fluorescence was observed using 100× magnification with a zoom of 2, unless indicated otherwise, on a Leica TCS-SP (UV) confocal microscope exciting at 488, 568 or 351/364 nm. All channels were detected separately, with no cross-talk between them. Micrographs represent single sections with a thickness of 300 nm. Experiments were repeated three times with >500 cells in each sample. Cap treatments were performed as in Dostie et al. (2000).
Flow cytometric analysis
For flow cytometry, ∼108 U937 cells were washed twice in PBS and fixed by the addition of ice-cold ethanol to 70% at 4°C for 20 min. After fixation, cells were pelleted, resuspended in the staining solution (10 µg/ml propidium iodide, 30 U/ml RNase A in PBS) and incubated for 30 min at 37°C. Data were collected using a FACScalibur apparatus (Becton Dickinson) and results were analyzed with Becton Dickinson Cell Quest software. For each sample, 10 000 events were collected, and clumped cells were gated out.
GST pull-down assays
GST fusion proteins (PML, Hox11, eIF4E and mutants) or PRH-His tag proteins were expressed in Escherichia coli BL21 (DE-3) cells, induced with IPTG and purified as described in Cohen et al. (2001) for GST fusions and according to the manufacturer’s instructions for PRH-His. IF4E–GST was cleaved with thrombin (Amersham) to release it from GST. The purity and concentrations of all isolated proteins were verified by SDS–PAGE and UV spectroscopy using calculated extinction coefficients, respectively. GST fusion proteins bound to glutathione– Sepharose or PRH-His fusions bound to nickel–agarose were incubated with purified eIF4E in 0.5 ml of binding buffer (PBS supplemented with 0.5 M KCl and 1% NP-40) for 1 h at room temperature while tumbling. Beads were washed three times with binding buffer, sedimented by centrifugation and analyzed by western blotting.
Acknowledgments
Acknowledgements
We are grateful for the kind gifts of antibodies, constructs and cell lines from Paul Freemont, Gerd Maul, L.de Jong, Thomas Meier, Nahum Sonenberg, Terrence Rabbitts, Melanie McConnell, P.P.Pandolfi and Rob Hromas. We thank Ling Shao for help with FACS data collection and analysis. We are grateful for technical assistance from Allan Capili, Elizabeth Covingtion and Katherine Foster. Confocal laser scanning microscopy was performed at the MSSM-LCSM core facility, supported with funding from the NIH (1 S10 RR0 9145-01) and NSF (DBI-9724504). K.L.B.B. is a scholar of the Leukemia and Lymphoma Society. Financial support was provided by the NIH (CA 80728 and CA 88991) and the Charlotte Geyer Foundation.
References
- Altmann M., Muller,P.P., Pelletier,J., Sonenberg,N. and Trachsel,H. (1989) A mammalian translation initiation factor can substitute for its yeast homologue in vivo. J. Biol. Chem., 264, 12145–12147. [PubMed] [Google Scholar]
- Boer J., Bonten-Surtel,J. and Grosveld,G. (1998) Overexpression of the nucleoporin CAN/NUP214 induces growth arrest, nucleocytoplasmic transport defects and apoptosis. Mol. Cell. Biol., 18, 1236–1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boisvert F.M., Hendzel,M.J. and Bazett-Jones,D.P. (2000) Promyelocytic leukemia (PML) nuclear bodies are protein structures that do not accumulate RNA. J. Cell Biol., 148, 283–292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borden K.L.B., Boddy,M.N., Lally,J., O’Reilly,N.J., Martin,S., Howe,K., Solomon,E. and Freemont,P.S. (1995) The solution structure of the RING finger domain from the acute promyelocytic leukaemia proto-oncoprotein PML. EMBO J., 14, 1532–1541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlile G.W., Tatton,W.G. and Borden,K.L. (1998) Demonstration of a RNA-dependent nuclear interaction between the promyelocytic leukaemia protein and glyceraldehyde-3-phosphate dehydrogenase. Biochem. J., 335, 691–696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen N., Sharma,M., Kentsis,A., Perez,J.M., Strudwick,S. and Borden,K.L. (2001) PML RING suppresses oncogenic trans formation by reducing the affinity of eIF4E for mRNA. EMBO J., 20, 4547–4559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crompton M.R., Bartlett,T.J., MacGregor,A.D., Manfioletti,G., Buratti,E., Giancotti,V. and Goodwin,G.H. (1992) Identification of a novel vertebrate homeobox gene expressed in haematopoietic cells. Nucleic Acids Res., 20, 5661–5667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Benedetti A. and Harris,A.L. (1999) eIF4E expression in tumors: its possible role in progression and malignacies. Int. J. Biochem. Cell Biol., 31, 59–72. [DOI] [PubMed] [Google Scholar]
- De Benedetti A. and Rhoads,R.E. (1990) Overexpression of eukaryotic protein synthesis initiation factor 4E in HeLa cells results in aberrant growth and morphology. Proc. Natl Acad. Sci. USA, 87, 8212–8216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dostie J., Lejbkowicz,F. and Sonenberg,N. (2000) Nuclear eukaryotic initiation factor 4E (eIF4E) colocalizes with splicing factors in speckles. J. Cell Biol., 148, 239–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fahrenkrug S.C., Dahlquist,M.O., Clark,K.J. and Hackett,P.B. (1999) Dynamic and tissue-specific expression of eIF4E during zebrafish embryogenesis. Differentiation, 65, 191–201. [DOI] [PubMed] [Google Scholar]
- Flenghi L. et al. (1995) Characterization of a new monoclonal antibody (PG-M3) directed against the aminoterminal portion of the PML gene product: immunocytochemical evidence for high expression of PML proteins on activated macrophages, endothelial cells and epithelia. Blood, 85, 1871–1880. [PubMed] [Google Scholar]
- Guiral M., Bess,K., Goodwin,G. and Jayaraman,P.S. (2001) PRH represses transcription in hematopoietic cells by at least two independent mechanisms. J. Biol. Chem., 276, 2961–2970. [DOI] [PubMed] [Google Scholar]
- Hoover D.S., Wingett,D.G., Zhang,J., Reeves,R. and Magnuson,N.S. (1997) Pim-1 protein expression is regulated by its 5′-untranslated region and translation initiation factor elF-4E. Cell Growth Differ., 8, 1371–1380. [PubMed] [Google Scholar]
- Hromas R., Radich,J. and Collins,S. (1993) PCR cloning of an orphan homeobox gene (PRH) preferentially expressed in myeloid and liver cells. Biochem. Biophys. Res. Commun., 195, 976–983. [DOI] [PubMed] [Google Scholar]
- Hu G., Lee,H., Price,S.M., Shen,M.M. and Abate-Shen,C. (2001) Msx homeobox genes inhibit differentiation through upregulation of cyclin D1. Development, 128, 2373–2384. [DOI] [PubMed] [Google Scholar]
- Iborra F.J., Jackson,D.A. and Cook,P.R. (2001) Coupled transcription and translation within nuclei of mammalian cells. Science, 293, 1139–1142. [DOI] [PubMed] [Google Scholar]
- Keng V.W. et al. (2000) Homeobox gene Hex is essential for onset of mouse embryonic liver development and differentiation of the monocyte lineage. Biochem. Biophys. Res. Commun., 276, 1155–1161. [DOI] [PubMed] [Google Scholar]
- Kentsis A., Dwyer,E.C., Perez,J.M., Sharma,M., Chen,A., Pan,Z.Q. and Borden,K.L. (2001) The RING domains of the promyelocytic leukemia protein PML and the arenaviral protein Z repress translation by directly inhibiting translation initiation factor eIF4E. J. Mol. Biol., 312, 609–623. [DOI] [PubMed] [Google Scholar]
- Lai H.K. and Borden,K.L. (2000) The promyelocytic leukemia (PML) protein suppresses cyclin D1 protein production by altering the nuclear cytoplasmic distribution of cyclin D1 mRNA. Oncogene, 19, 1623–1634. [DOI] [PubMed] [Google Scholar]
- Lang V., Zanchin,N.I., Lunsdorf,H., Tuite,M. and McCarthy,J.E. (1994) Initiation factor eIF-4E of Saccharomyces cerevisiae. Distribution within the cell, binding to mRNA and consequences of its overproduction. J. Biol. Chem., 269, 6117–6123. [PubMed] [Google Scholar]
- Langenfeld J., Kiyokawa,H., Sekula,D., Boyle,J. and Dmitrovsky,E. (1997) Posttranslational regulation of cyclin D1 by retinoic acid: a chemoprevention mechanism. Proc. Natl Acad. Sci. USA, 94, 12070–12074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazaris-Karatzas A. and Sonenberg,N. (1992) The mRNA 5′ cap-binding protein, eIF-4E, cooperates with v-myc or E1A in the transformation of primary rodent fibroblasts. Mol. Cell. Biol., 12, 1234–1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazaris-Karatzas A., Montine,K.S. and Sonenberg,N. (1990) Malignant transformation by a eukaryotic initiation factor subunit that binds to mRNA 5′ cap. Nature, 345, 544–547. [DOI] [PubMed] [Google Scholar]
- Lazaris-Karatzas A., Smith,M.R., Frederickson,R.M., Jaramillo,M.L., Liu,Y.L., Kung,H.F. and Sonenberg,N. (1992) Ras mediates translation initiation factor 4E-induced malignant transformation. Genes Dev., 6, 1631–1642. [DOI] [PubMed] [Google Scholar]
- Lejbkowicz F., Goyer,C., Darveau,A., Neron,S., Lemieux,R. and Sonenberg,N. (1992) A fraction of the mRNA 5′ cap-binding protein, eukaryotic initiation factor 4E, localizes to the nucleus. Proc. Natl Acad. Sci. USA, 89, 9612–9616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao W., Ho,C.Y., Yan,Y.L., Postlethwait,J. and Stainier,D.Y. (2000) Hhex and scl function in parallel to regulate early endothelial and blood differentiation in zebrafish. Development, 127, 4303–4313. [DOI] [PubMed] [Google Scholar]
- Lynch J., Suh,E.R., Silberg,D.G., Rulyak,S., Blanchard,N. and Traber,P.G. (2000) The caudal-related homeodomain protein Cdx1 inhibits proliferation of intestinal epithelial cells by down-regulation of D-type cyclins. J. Biol. Chem., 275, 4499–4506. [DOI] [PubMed] [Google Scholar]
- Manfioletti G., Gattei,V., Buratti,E., Rustighi,A., De Iuliis,A., Aldinucci,D., Goodwin,G.H. and Pinto,A. (1995) Differential expression of a novel proline-rich homeobox gene (Prh) in human hematolymphopoietic cells. Blood, 85, 1237–1245. [PubMed] [Google Scholar]
- Martinez Barbera J.P., Clements,M., Thomas,P., Rodriguez,T., Meloy,D., Kioussis,D. and Beddington,R.S. (2000) The homeobox gene Hex is required in definitive endodermal tissues for normal forebrain, liver and thyroid formation. Development, 127, 2433–2445. [DOI] [PubMed] [Google Scholar]
- McCubbin W.D., Edery,I., Altmann,M., Sonenberg,N. and Kay,C.M. (1988) Circular dichroism and fluorescence studies on protein synthesis initiation factor eIF-4E and two mutant forms from the yeast Saccharomyces cerevisiae. J. Biol. Chem., 263, 17663–17671. [PubMed] [Google Scholar]
- Meier U.T. (1996) Comparison of the rat nucleolar protein nopp140 with its yeast homology SRP40. Differential phosphorylation in vertebrates and yeast. J. Biol. Chem., 271, 19376–19384. [PubMed] [Google Scholar]
- Melnick A. and Licht,J.D. (1999) Deconstructing a disease: RARα, its fusion partners and their roles in the pathogenesis of acute promyelocytic leukemia. Blood, 93, 3167–3215. [PubMed] [Google Scholar]
- Nathan C.A., Carter,P., Liu,L., Li,B.D., Abreo,F., Tudor,A., Zimmer,S.G. and De Benedetti,A. (1997a) Elevated expression of eIF4E and FGF-2 isoforms during vascularization of breast carcinomas. Oncogene, 15, 1087–1094. [DOI] [PubMed] [Google Scholar]
- Nathan C.A., Liu,L., Li,B.D., Abreo,F.W., Nandy,I. and De Benedetti,A. (1997b) Detection of the proto-oncogene eIF4E in surgical margins may predict recurrence in head and neck cancer. Oncogene, 15, 579–584. [DOI] [PubMed] [Google Scholar]
- Newman C.S., Chia,F. and Krieg,P.A. (1997) The XHex homeobox gene is expressed during development of the vascular endothelium: overexpression leads to an increase in vascular endothelial cell number. Mech. Dev., 66, 83–93. [DOI] [PubMed] [Google Scholar]
- Niessing D., Dostatni,N., Jackle,H. and Rivera-Pomar,R. (1999) Sequence interval within the PEST motif of Bicoid is important for translational repression of caudal mRNA in the anterior region of the Drosophila embryo. EMBO J., 18, 1966–1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niessing D., Blanke,S. and Jackle,H. (2002) Bicoid associates with the 5′ cap bound complex of caudal mRNA and represses translation. Genes Dev., 16, 2576–2582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ptushkina M., von der Haar,T., Vasilescu,S., Frank,R., Birkenhager,R. and McCarthy,J.E. (1998) Cooperative modulation by eIF4G of eIF4E-binding to the mRNA 5′ cap in yeast involves a site partially shared by p20. EMBO J., 17, 4798–4808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rousseau D., Kaspar,R., Rosenwald,I., Gehrke,L. and Sonenberg,N. (1996) Translation initiation of ornithine decarboxylase and nucleocytoplasmic transport of cyclin D1 mRNA are increased in cells overexpressing eukaryotic initiation factor 4E. Proc. Natl Acad. Sci. USA, 93, 1065–1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonenberg N. and Gingras,A.C. (1998) The mRNA 5′ cap-binding protein eIF4E and control of cell growth. Curr. Opin. Cell Biol., 10, 268–275. [DOI] [PubMed] [Google Scholar]
- Strudwick S. and Borden,K.L. (2002) The emerging roles of translation factor eIF4E in the nucleus. Differentiation, 70, 10–22. [DOI] [PubMed] [Google Scholar]
- Stuurman N., de Graaf,A., Floore,A., Josso,A., Humbel,B., de Jong,L. and van Driel,R. (1992) A monoclonal antibody recognizing nuclear matrix-associated nuclear bodies. J. Cell Sci., 101, 773–784. [DOI] [PubMed] [Google Scholar]
- Tanaka T., Inazu,T., Yamada,K., Myint,Z., Keng,V.W., Inoue,Y., Taniguchi,N. and Noguchi,T. (1999) cDNA cloning and expression of rat homeobox gene, Hex and functional characterization of the protein. Biochem. J., 339, 111–117. [PMC free article] [PubMed] [Google Scholar]
- Thomas P.Q., Brown,A. and Beddington,R.S. (1998) Hex: a homeobox gene revealing peri-implantation asymmetry in the mouse embryo and an early transient marker of endothelial cell precursors. Development, 125, 85–94. [DOI] [PubMed] [Google Scholar]
- Topcu Z., Mack,D.L., Hromas,R.A. and Borden,K.L. (1999) The promyelocytic leukemia protein PML interacts with the proline-rich homeodomain protein PRH: a RING may link hematopoiesis and growth control. Oncogene, 18, 7091–100. [DOI] [PubMed] [Google Scholar]
- Topisirovic I., Capili,A.D. and Borden,K.L. (2002) γ Interferon and cadmium treatments modulate eukaryotic initiation factor 4E-dependent mRNA transport of cyclin D1 in a PML-dependent manner. Mol. Cell. Biol., 22, 6183–6198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Der Haar T., Ball,P.D. and McCarthy,J.E. (2000) Stabilization of eukaryotic initiation factor 4E binding to the mRNA 5′-cap by domains of eIF4G. J. Biol. Chem., 275, 30551–30555. [DOI] [PubMed] [Google Scholar]
- Wang S., Rosenwald,I.B., Hutzler,M.J., Pihan,G.A., Savas,L., Chen,J.J. and Woda,B.A. (1999) Expression of the eukaryotic translation initiation factors 4E and 2α in non-Hodgkin’s lymphomas. Am. J. Pathol., 155, 247–255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z.G., Delva,L., Gaboli,M., Rivi,R., Giorgio,M., Cordon-Cardo,C., Grosveld,F. and Pandolfi,P.P. (1998) Role of PML in cell growth and the retinoic acid pathway. Science, 279, 1547–1551. [DOI] [PubMed] [Google Scholar]
- Yatskievych T.A., Pascoe,S. and Antin,P.B. (1999) Expression of the homebox gene Hex during early stages of chick embryo development. Mech. Dev., 80, 107–109. [DOI] [PubMed] [Google Scholar]