Abstract
Initiation codon context is an important determinant of translation initiation rates in both prokaryotes and eukaryotes. Such sequences include the Shine– Dalgarno ribosome-binding site, as well as other motifs surrounding the initiation codon. One proposed interaction is between the base immediately preceding the initiation codon (–1 position) and the nucleotide 3′ to the tRNAfMet anticodon, at position 37. Adenine is conserved at position 37, and a uridine at –1 has been shown in vitro to favor initiation. We have tested this model in vivo, by manipulating the chloroplast of the green alga Chlamydomonas reinhardtii, where the translational machinery is prokaryotic in nature. We show that translational defects imparted by mutations at the petA –1 position can be suppressed by compensatory mutations at position 37 of an ectopically expressed tRNAfMet. The mutant tRNAs are fully aminoacylated and do not interfere with the translation of other proteins. Although this extended base pairing is not an absolute requirement for initiation, it may convey added specificity to transcripts carrying non-standard initiation codons, and/or preserve translational fidelity under certain stress conditions.
Keywords: anticodon/Chlamydomonas/chloroplast/initiator tRNA
Introduction
The roles of sequences surrounding the initiation codon in prokaryotic translation initiation have been extensively studied (reviewed by McCarthy and Brimacombe, 1994). The completion of the Escherichia coli genome has facilitated global analysis of translation initiation context, either as a gene verification/discovery tool (Delamarche et al., 1999; Walker et al., 2002) or to study motifs that potentially modulate translation initiation rates (Stenstrom et al., 2001). The bulk of this analysis, however, has focused on the well-known Shine–Dalgarno (SD) ribosome-binding site or on nucleotides following the initiation codon, which may (Moll et al., 2001) constitute a regulatory ‘downstream box’. What has not been reassessed is the possibility of extended interactions between the mRNA and the initiator tRNA (tRNAfMet).
The question of extended tRNAfMet–mRNA base pairing was raised by the observation that residues complementary to nucleotides in the tRNAfMet anticodon loop are more prevalent immediately upstream of initiation codons than predicted by chance (Ganoza et al., 1985). In particular, a uridine at position –1 was proposed to allow a fourth base pair with the adenine at position 37 of the tRNAfMet (immediately downstream of the anticodon), a model consistent with several in vitro assays (see Discussion). Since many aspects of sequence context influence translation rates, and because in vitro experiments may not always fully reflect in vivo mechanisms, it is critical that the extended codon–anticodon model be tested in vivo. An accepted way to do so would be the demonstration that effects of –1 mutations on translation could be compensated by complementary mutations at position 37 in tRNAfMet. Such a strategy was useful in confirming the interaction of the SD sequence (Shine and Dalgarno, 1974) with a complementary sequence at the 3′ end of 16S rRNA (Hui and de Boer, 1987). Unfortunately, residue A37 was shown to be important for the aminoacylation of tRNAfMet in E.coli (Meinnel et al., 1993), rendering this approach problematic in bacteria.
Here we have chosen to test the extended base pairing model in the chloroplast, whose translational apparatus is derived from, and generally resembles that of, prokaryotes (Harris et al., 1994; Sugiura et al., 1998). In the chloroplast of the unicellular green alga Chlamydomonas reinhardtii, mutagenesis of the initiation codon of the petA mRNA, which encodes cytochrome f (cyt f), a component of the cytochrome b6/f complex in the photosynthetic electron transport chain, revealed that, as in prokaryotes, an AUG initiation codon is not required for the selection of the correct start site, but that AUU, ACG, ACC and ACU progressively reduce translation efficiency (Chen et al., 1995). Surprisingly, it was found in the same study that mutating the three nucleotides upstream of the initiation codon from AUU to UAA dramatically reduced cyt f synthesis, especially in the context of a mutant AUU initiation codon. This result raised the possibility that one or more of the –3 to –1 positions relative to the initiation codon had a functional importance. It was subsequently shown that mutation of the –1U residue severely reduced translation when combined with an AUU initiation codon, or at elevated temperature (35°C) in the context of an AUG initiation codon (Esposito et al., 2001). These observations, taken together with a 75% conservation of a uridine at position –1 in the chloroplast mRNAs of C.reinhardtii (the genome sequence is reported in Maul et al., 2002), led us to consider whether the E.coli extended interaction model could also be proposed for plastid translation. Here we show first that in the C.reinhardtii chloroplast, the adenine at position 37 of tRNAfMet is not important for aminoacylation. Then, using a series of mutations affecting the petA initiation codon and –1 uridine, we show that compensatory mutations introduced at position 37 of an ectopically expressed copy of the chloroplast initiator tRNA can partially restore the cyt f synthesis rate.
Results
Generation of trnfM mutants and chloroplast transformation
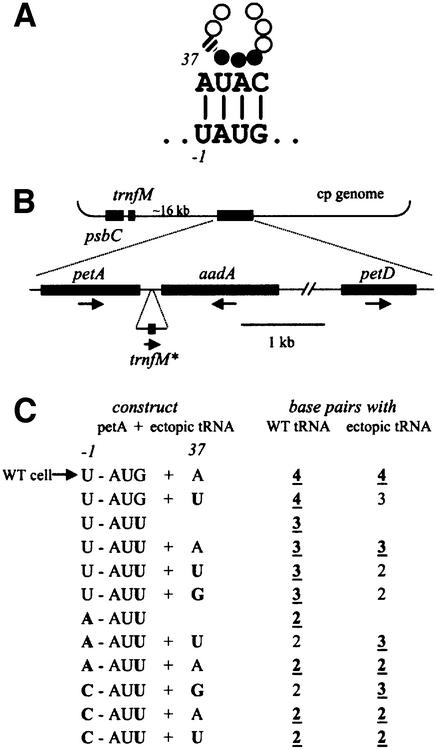
To test the possibility of functional base pairing between the –1 uridine of a messenger RNA and A37 of the initiator tRNA (Figure 1A), we stably transformed the chloroplast with a plasmid that simultaneously allows the introduction of mutations in the petA initiation region, and adds an ectopic copy of the trnfM gene (Figure 1B). These altered chloroplast genomes thus combined different mutations in the petA RNA and the initiator tRNA. Transformants were selected by resistance to spectinomycin (conferred by the aadA marker; Goldschmidt-Clermont, 1991) and screened for the presence of the desired mutations and absence of wild-type (WT) genomes by PCR, DNA filter hybridization analysis and genomic sequencing (data not shown).
Fig. 1. Expression of ectopic tRNAs in the chloroplast. (A) Extended interaction model between the initiator tRNA and mRNA –1U. The three closed circles denote the tRNA anticodon; the A37 residue is shown by a hatched circle. (B) Transformation strategy to mutate the petA gene and, for certain constructs, to introduce an additional tRNA gene into the chloroplast genome. The top part shows the location of the endogenous trnfM gene downstream of psbC, and its distance from the petA–petD region. The lower line shows the altered region in transformants (the 1.8 kb aadA/petD intergenic region is not to scale), with gene orientations indicated by arrows. The notation trnfM* indicates the ectopic copy. (C) Names of the strains created (left column), and the number of predicted base pairs that the particular version of the petA mRNA would form with the endogenous tRNAfMet (middle column) or with the ectopic tRNAfMet where present (right column). Strains are named by the petA mRNA sequences at –1 and the initiation codon (e.g. U-AUG), plus position 37 of the tRNAfMet (e.g. +A). Mutated residues relative to WT are in bold. In columns 2 and 3, the strongest base pairing possibility is bold and underlined.
The different combinations of mutations in the petA initiation region and the ectopic trnfM are represented in Figure 1C. Here, as throughout the manuscript, we show WT bases in normal type and mutated bases in bold. As shown in Figure 1C, for petA, the WT sequence of the initiation codon (–1U-AUG) was changed to AUU to reduce the translation rate, a context allowing easy analysis of the importance of the –1 residue (Chen et al., 1995; Esposito et al., 2001). Then, the WT –1 uridine was changed to A (A-AUU) or C (C-AUU). With these two (–1 and +3) mutations combined, only two base pairs of the four shown in Figure 1A can still occur with the initiator tRNA. When an ectopic copy of trnfM was introduced, we used either the WT sequence (A37, denoted +A) or the compensatory U37 (noted +U) or G37 (+G) mutations. According to the model, the compensatory effect of mutations in the ectopic trnfM should be revealed by an improved level of cyt f synthesis in the following two strains: A-AUU+U and C-AUU+G, respectively. In other words, as shown in Figure 1C, these two strains should express an initiator tRNA containing a mutation that restores a third base pair with the petA mRNA, whereas the endogenous wild-type tRNA would form only two base pairs. Furthermore, the U37 mutation should not suppress –1C, and G37 should not suppress –1A.
Expression of ectopic trnfM and aminoacylation levels
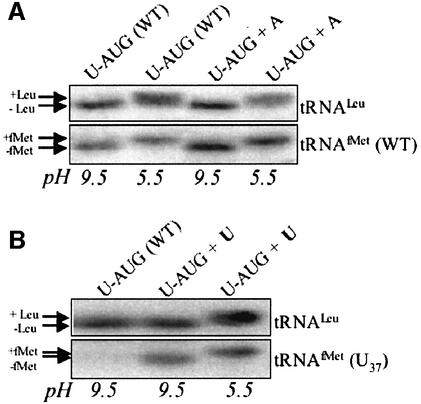
To substantiate the validity of our approach, we had to demonstrate that the ectopic trnfM gene was expressed (either from its own promoter or co-transcribed with petA, Figure 1B), and also that a mutation at position 37 of the initiator tRNA, unlike in E.coli (Meinnel et al., 1993), would not affect its aminoacylation. We used a RNA filter hybridization approach to address both questions, as shown in Figure 2. The first strains constructed for this analysis were U-AUG+A (expressing an ectopic WT trnfM gene) and U-AUG+U (expressing a U37 mutant tRNA). Comparison of lanes 1 and 3 in Figure 2A show the ∼70% increased accumulation of WT tRNAfMet in the U-AUG +A strain, demonstrating the expression of the ectopic WT trnfM gene, as compared with the control tRNALeu. To detect expression of the U37 mutant tRNA, we developed an oligonucleotide probe that allows one mismatch-specific recognition; this probe was a 19mer complementary to residues 29–46 of the mutant tRNA (allowing 10 G/C and 9 A/U base pairs with a complete match). As shown in Figure 2B, this gives no signal with the endogenously expressed WT tRNAfMet present in every strain (lane 1), but identifies the U37 mutant tRNA (lane 2), which accumulated to an estimated 60–70% of the endogenous WT tRNAfMet level, based on comparisons with tRNALeu.
Fig. 2. Expression and aminoacylation of tRNAs. RNAs from the strains indicated above the gels were extracted at pH 5.5 or 9.5, as noted below the gels, and fractionated by electrophoresis through a 15% polyacrylamide gel buffered to pH 5.5. Gels were transferred to membranes and probed with oligonucleotides complementary to (A) tRNALeu and WT tRNAfMet, as indicated at the right, or (B) tRNALeu and the ectopic U37 tRNAfMet. The positions of charged (+Leu and +fMet) and uncharged (–Leu and –fMet) tRNAs are indicated at the left.
To analyze the aminoacylation level of the tRNAfMet in these strains, the RNAs were extracted either at pH 5.5 or 9.5, where the aminoacylation state is maintained or lost, respectively. Should an amino acid be present on tRNAs extracted at pH 5.5, a visible shift would occur during electrophoretic fractionation. As shown in Figure 2A, the quantitative shift in migration between lanes 1 and 2 shows that in a WT strain, tRNAfMet is 100% aminoacylated. Lanes 3 and 4 thus show that the expression of an ectopic trnfM gene does not affect the aminoacylation level, which remains at or near 100%. Similarly, lanes 2 and 3 of Figure 2B show that in the U-AUG+U strain, the mutant tRNA is fully aminoacylated. The chloroplast tRNALeu was used in all cases as a control for successful retention or removal of the charged amino acid. Finally, to determine whether the insertion of a trnfM gene downstream of petA affected petA mRNA processing or stability, RNA filter hybridization analysis was performed. These experiments revealed no differences in petA mRNA processing and no reduction in its accumulation (data not shown).
In summary, ectopic trnfM genes are expressed, do not affect aminoacylation of the endogenous WT tRNAfMet and, importantly, mutating residue 37 does not interfere with aminoacylation. In other words, the strains represented in Figure 1C are appropriate for examining a possible extended codon–anticodon interaction.
Chloroplast protein synthesis in strains expressing ectopic trnfM genes
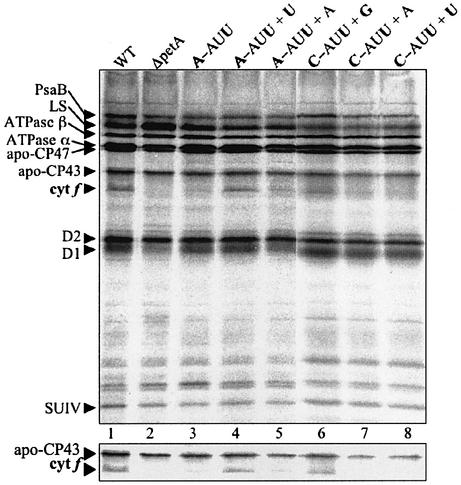
To visualize either specific or global effects of the ectopic tRNAs in different petA mRNA contexts, we analyzed chloroplast protein synthesis rates by labeling cells for 5 min with [14C]acetate in the presence of cycloheximide, a cytosolic translation inhibitor. As shown in Figure 3 (lane 1), a well-defined set of proteins is labeled in WT cells, including cyt f, as evidenced by the specific absence of this species in strain ΔpetA, in which the petA gene is deleted (lane 2). Apart from these controls, the results showed that the ectopic expression of either the WT-A37, U37 or G37 tRNAfMet (lanes 4–8) had no general effect on chloroplast translation rates, probably because of the presence of an equal or greater amount of the WT tRNAfMet transcribed from the endogenous gene. This important observation means that variations in cyt f expression should result only from a gene-specific mechanism.
Fig. 3. Chloroplast protein pulse labeling in the presence of a cytoplasmic translation inhibitor. Strains are indicated across the top with nomenclature as described in the legend to Figure 1; ΔpetA is a strain lacking the petA gene. Clearly visible chloroplast proteins are indicated at the left: PsaB, core subunit B of photosystem I (PSI); LS, large subunit of Rubisco; ATPase β, β subunit of the ATP synthase; ATPase α, α subunit of the ATP synthase; apo-CP47: 47 kDa subunit of PSII; apo-CP43, 43 kDa PSII subunit; cytf, cytochrome f; D2, PSII reaction center; D1, PSII reaction center; SUIV, subunit IV of the cytochrome b6/f complex. The lower panel shows a section of the same gel, with the exposure adjusted to emphasize cytf synthesis.
Figure 3 also implies that for mRNAs which have their WT sequence in the initiation codon region (i.e. other than petA), mutating residue 37 of the initiator tRNA has no major consequence on translation. This result is predictable for mRNAs which, like petA, have the conserved U-AUG sequence, but also no change was observed for mRNAs, which have a WT A-AUG sequence, such as rbcL (encoding the Rubisco large subunit), psbC (encoding the photosystem I CP43 protein) and psbD (encoding the photosystem II D2 protein). Thus, at least under optimal laboratory growth conditions, a mutant U37 tRNA does not measurably stimulate translation of mRNAs with a WT A-AUG sequence by allowing the putative fourth base pair.
Finally, Figure 3 shows the effects of mutant ectopic tRNAs on expression of mutant petA mRNAs. Lanes 3–5 show that in A-AUU, cyt f synthesis is undetectable, but is substantial in the presence of mutant U37 tRNA, but not with an extra copy of the WT A37 tRNA. Lanes 6–8 show similarly that C-AUU can be suppressed by mutant G37 tRNA, but not by the wild-type or mutant U37 tRNAs. These data strongly suggest that allele-specific interactions can occur between the mRNA –1 base pair and position 37 of tRNAfMet.
Cytochrome f accumulation in trnfM ectopic mutants
The pulse labeling experiments of Figure 3 have limited quantitative value because cyt f is poorly labeled relative to other proteins. Therefore, immunoblot analysis was used, since previous work has shown that the level of cyt f accumulation in Chlamydomonas accurately reflects its synthesis rate (Kuras and Wollman, 1994; Chen et al., 1995). Figure 4A shows the effects of different ectopic tRNAsfMet on cyt f translation in the A-AUU mutant. In the presence of only the WT endogenous tRNA, the context of an AUU initiation codon shows that the –1U to A mutation reduces translation from 25% (in U-AUU, lane 3) to <5% (in A-AUU, lane 4). However, the presence of an ectopic tRNAfMet harboring the U37 mutation (in A-AUU+U, lane 5) constitutes a compensatory mutation (see Figure 1C), and results in the increase of accumulation to ∼50%, thus suppressing the effect of the –1 mutation. As a control, we show that an ectopic WT tRNAfMet does not restore the translation level (lane 6), demonstrating that the effect observed in lane 5 is not due to the overaccumulation of an initiator tRNA, but rather to the specific presence of a uridine at position 37 of this tRNA. This result clearly shows that the –1 residues of the mRNA and position 37 of the tRNA can base pair according to the model proposed in Figure 1A.
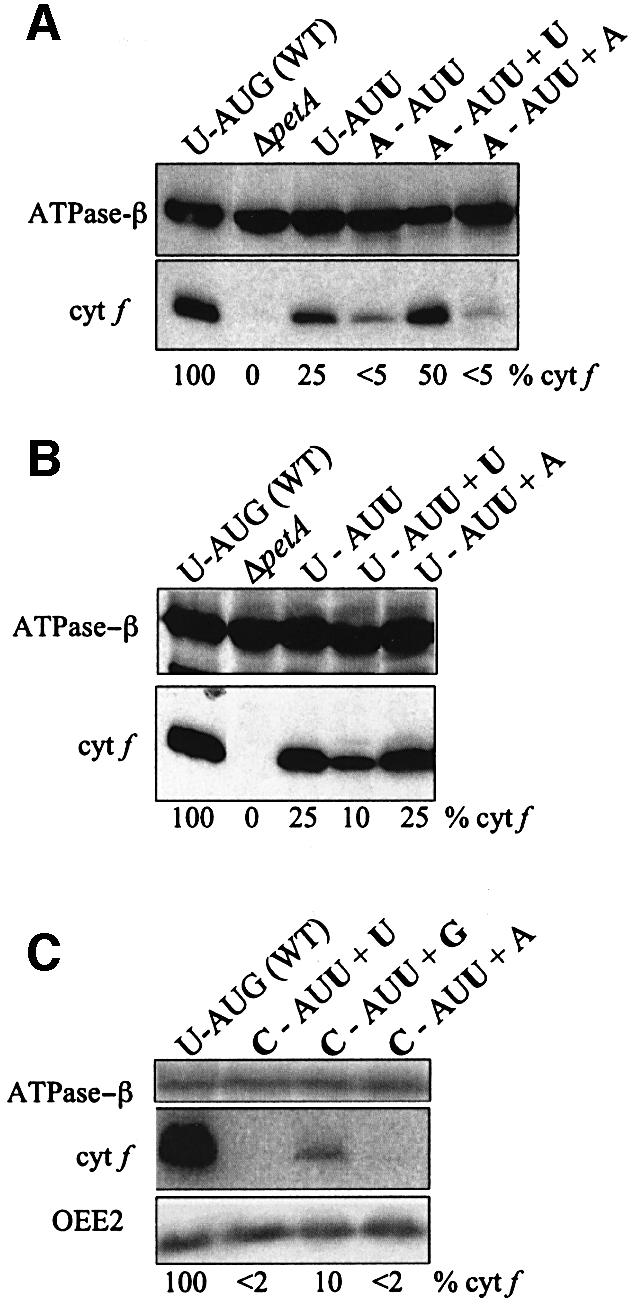
Fig. 4. Analysis of cyt f accumulation. Immunoblot analysis was used to measure the accumulation of cyt f (34 kDa) in the indicated strains, with the ATPase β subunit (55 kDa) as a loading control. The values indicated for cyt f accumulation, shown beneath each gel and relative to WT, were estimated from multiple blots. (A) Control strains compared with those with mRNA –1 mutations. (B) Control strains compared with those with a WT –1 sequence in petA, but expressing ectopic tRNAs. (C) A WT control compared with strains carrying –1C mutations in petA mRNA. Here the indicated cyt f accumulation levels are mean values obtained for four independent clones for each strain, quantified against both the ATPase β subunit and the OEE2 protein. OEE2 is the nucleus-encoded PSII oxygen-evolving enhancer protein 2 (Mayfield et al., 1987).
In a similar manner, we studied the effect of an ectopic WT tRNAfMet with a U-AUU context for the petA mRNA, the sequence upstream of the initiation codon being unchanged this time. The expression of an ectopic WT tRNAfMet has no effect on cyt f synthesis (Figure 4B, lane 5), whereas the mutated U37 tRNA decreases protein accumulation from 25 to 10% (lane 4). This likely results from the mutated tRNA competing with the endogenous tRNA at an early step in translation, and ultimately decreasing the rate, since it can form only two base pairs with the U-AUU mRNA, instead of three for the endogenous tRNA.
To further confirm the model and our initial results, we analyzed the effect of a –1C mutation in the mRNA and the possible compensatory effect that could be conferred by a G37 mutation in the initiator tRNA. The –1C mutation has a more dramatic effect on translation than the –1A mutation and, given the rarity of C residues at position –1 of chloroplast mRNAs (4 out of 64 in C.reinhardtii), this indicates that it is unfavorable for translation initiation. This effect could be compensated neither by an ectopic WT tRNAfMet (Figure 4C, lane 4) nor by the U37 mutation (lane 2). However, the G37 mutation in the tRNA partially restored translation initiation (from <2 to ∼10%), thus confirming the extended base pairing model. This compensatory effect is equal in magnitude to an A–U restored base pair (Figure 4A), i.e. about 5-fold, although the absolute levels are lower with a –1C.
Discussion
We have used a compensatory mutation strategy to test the hypothesis of base pairing between the –1 nucleotide of a chloroplast mRNA and the anticodon loop of the initiator tRNA. This work was made possible in the chloroplast of C.reinhardtii by showing that an ectopic copy of the trnfM gene can be expressed without globally affecting protein synthesis rates, and that mutating position 37 does not affect its aminoacylation. The ectopic trnfM gene was used, in part, because we were unable to obtain strains in which the endogenous trnfM gene had been replaced by a mutated copy (data not shown). The ectopically expressed versions of tRNAfMet carrying anticodon mutations were able to increase translation of petA –1 mutant mRNAs with an AUU initiation codon. In contrast, cyt f translation in the U-AUU strain was reduced in the presence of a mutant tRNA (Figure 4B). This result can be interpreted by considering that the 30S subunit can bind IF-2 and the initiator tRNA before binding the mRNA (Gualerzi and Pon, 1990; Wu et al., 1996). 30S subunits containing the mutant initiator tRNA would bind this mRNA relatively poorly in this scenario.
Our in vivo analysis supports earlier in vitro work, suggesting that the initiation codon–anticodon interaction may be greater than three base pairs, although the –1 position is only one determinant of translational efficiency (Hui et al., 1984). For example, short oligoribonucleotides with a U upstream of the AUG initiation codon allowed 5-fold more efficient formation of an initiation complex with 70S ribosomes than those with a purine upstream of AUG (Ganoza et al., 1978). Moreover, 30S and 70S ribosomes bound fMet–tRNA more efficiently in response to UAUG than AAUG (Ganoza et al., 1982) or AUG (Eckhardt and Luhrmann, 1981) in vitro. Dipeptide synthesis was also 2-fold more efficient with UAUGUUU than AUGUUU or AAUGUUU (Ganoza et al., 1982). Sequence analysis also offers some support for the extended base pairing hypothesis. Prokaryotic initiation codons are preferentially preceded by a U compared with internal AUG codons (Ganoza et al., 1985) and, in C.reinhardtii, 75% of chloroplast genes known to be expressed possess a U at the –1 position. Moreover, a U at the –1 position is optimal for expression of foreign genes in E.coli (Gross et al., 1990; Morelle et al., 1991). Although we have focused here on base pairing, the –1 position may have other types of interactions with the translational machinery. Evidence for this includes the protection from chemical modification of the T4 gene 32 mRNA –1 position when studied in vitro in ternary complexes (Hüttenhofer and Noller, 1994).
While this work describes an extended codon– anticodon interaction in initiation, four base-pair codon– anticodon interactions have been proposed for tRNA frameshift suppressors with an expanded anticodon loop (reviewed in Culbertson et al., 1990). Although recent results suggest that an extended interaction may not always occur, rather that the expanded anticodon loop inhibits its use in translation forcing a near-cognate tRNA to be used (Qian et al., 1998), artificially created expanded anticodon loops in some cases clearly support frameshifting (e.g. Magliery et al., 2001). This case is not strictly analogous to our findings, however, since we have altered a base without changing normal tRNA size or structure.
Taken together, in vitro, sequence analysis, and now in vivo data strongly support a 5′ extended codon– anticodon interaction in translation initiation. Further, it can be argued that this situation, at least for some mRNAs, is likely to be widespread among bacteria and chloroplasts. In chloroplasts, the A37 residue is highly conserved in the initiator tRNA anticodon loop (Sprinzl et al., 1998), as is the –1U residue upstream of mRNA initiation codons. On the other hand, position 37 of the initiator tRNA is variable in mitochondrial DNAs (Sprinzl et al., 1998). While ostensibly arguing against further generalization of the mechanism described here, a closer look reveals apparent co-evolution of tRNAfMet position 37 with mRNA –1 positions. For example, mitochondrial tRNAfMet of the lower fungus Hyaloraphidium curvatum carries a C at position 37 and, at the same time, 12 of 14 AUG initiation codons carry a –1G (Forget et al., 2002). On the other hand, its close relative Spizellomyces punctatus (DDBJ/EMBL/GenBank accession No. AF404303) features a position 37 A in the tRNAfMet and 0/13 –1 positions are G, but 5/13 are U, including all three cox genes. Thus, this type of interaction may be evolutionarily favored both in prokaryotes and organelles, but like other initiation elements, including the SD and AUG codon itself, is undoubtedly not universal.
The advantage of an extended codon–anticodon interaction during prokaryotic-type translation initiation remains to be elucidated, and must take into account the variability of –1 residues and other regulatory motifs. One hypothesis is that the possibility of having four base pairs can improve synthesis of certain proteins under environmental stress conditions, or where mRNA or tRNA levels are limiting. Alternatively, this additional base pair may be part of the fundamental mechanism favoring the formation of the initiation complex, especially for non-AUG codons, thus conferring a more subtle evolutionary advantage.
Materials and methods
Construction of trnfM transforming plasmids
A 1280 bp EcoRI–SalI fragment containing trnfM was subcloned from the plasmid p578 (Chlamydomonas Genetics Center, Duke University) into pBluescript-SK– (Stratagene) to create the plasmid pfmetsk-. An SspI site was introduced 18 bp downstream of the trnfM gene using site-directed mutagenesis (Kunkel, 1985), creating the fM-CAUAsspI mutation (CAUA is the sequence of the anticodon and first nucleotide downstream). The resulting plasmid containing the SspI site was subjected to a second round of site-directed mutagenesis yielding fM-CAUUsspI and fM-CAUGsspI. To create plasmids containing an ectopic copy of trnfM, a 450 bp HincII fragment from pfmetsk- containing either trnfM fM-CAUAsspI, fM-CAUUsspI or fM-CAUGsspI was inserted into the StuI site of the plasmid pQMAD, which contains the aadA gene (Goldschmidt-Clermont, 1991) inserted in the EcoRV site between petA and petD. pQMAD had been altered to contain one of several mutations at the initiation region of petA (Esposito et al., 2001).
Chlamydomonas strains, culture conditions and chloroplast transformation
P17 (Stern et al., 1991), a wild-type Chlamydomonas strain derived from CC373 (mt+, ac-u-c-2–21; Shepherd et al., 1979) by bombardment with the atpB gene, was used as a recipient for chloroplast transformation. The ΔpetA strain was described previously (Kuras and Wollman, 1994). Chlamydomonas strains were grown in TAP medium (Harris, 1989) at 25°C under constant light (70 µE/m2/s). Modified genes were introduced into the chloroplast genome by tungsten-particle bombardment (Kindle et al., 1991). Transformants expressing the aadA cassette were selected on TAP medium containing spectinomycin (100 µg/ml).
In vivo aminoacylation levels
In vivo aminoacylation levels were analyzed by RNA extraction and filter hybridization analysis according to Varshney et al. (1991). Total RNAs were extracted from liquid cultures of C.reinhardtii either with solutions buffered at pH 8.5, to obtain extracts containing deacylated tRNAs, or at pH 5.5 to obtain extracts where aminoacylation of tRNAs was preserved. Extraction at pH 8.5 was not sufficient to obtain complete deacylation of tRNAfMet and extracts were further treated for 1 h at room temperature in 20 mM Tris–HCl pH 9.5. Extracts were then fractionated by electrophoresis in 15% polyacrylamide gels with the running buffer and gel buffered at pH 5.5 to maintain the aminoacylated state of tRNA extracts obtained at pH 5.5. After electroblotting onto Hybond N+ membranes (Amersham), we used [32P]5′-labeled oligonucleotides as probes. For tRNALeu, the probe was complementary to positions 18–32, with numbering according to Sprinzl et al. (1998). For tRNAfMet, we used oligonucleotides that matched either the WT sequence or the U37 mutation (one mismatch with the WT sequence), complementary to positions 29–46.
Protein pulse labeling
Experiments were carried out as described previously (Drapier et al., 1992) with 5 µCi/ml of [14C]acetate (50 mCi/mmol, Amersham) in the presence of an inhibitor of cytoplasmic translation (6.6 µg/ml cycloheximide, Sigma). Proteins of solubilized cells were then separated in 12–18% polyacrylamide/SDS–urea gels as described previously (Piccioni et al., 1981).
Protein preparation and immunoblotting
Total protein isolation and immunoblotting were as described previously (Drager et al., 1998). Blots were reacted with primary antibodies directed against the Chlamydomonas chloroplast-encoded cyt f (Chen et al., 1995), ATPase β subunit (a gift from F.-A.Wollman), the nuclear-encoded OEE2 protein and a secondary anti-rabbit IgG HRP-conjugated antibody (Promega, 1:2 500 dilution), and visualized by enhanced chemiluminescence (Durant, 1990) or with 125I-labeled protein A (Figure 4C only).
Acknowledgments
Acknowledgements
We thank Karen Kindle, Francis-André Wollman and Yves Choquet for useful discussions. This work was supported by NSF grants MCB-9723274 and INT-9603351 to D.B.S. D.E. was supported by a Howard Hughes Medical Institute Predoctoral Fellowship. S.E. is a recipient of a doctoral fellowship from the Ministère de l’Education Nationale (France).
References
- Chen X., Kindle,K.L. and Stern,D.B. (1995) The initiation codon determines the efficiency but not the site of translation initiation in Chlamydomonas chloroplasts. Plant Cell, 7, 1295–1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Culbertson M., Leeds,P., Sandbaken,M. and Wilson,P. (1990) Frameshift suppression. In Hill,W., Dahlberg,A., Garrett,P., Moore,P., Schlessinger,D. and Warner,J. (eds), The Ribosome: Structure, Function and Evolution. American Society for Microbiology, Washington, DC, pp. 559–570.
- Delamarche C., Guerdoux-Jamet,P., Gras,R. and Nicolas,J. (1999) A symbolic–numeric approach to find patterns in genomes. Application to the translation initiation sites of E.coli. Biochimie, 81, 1065–1072. [DOI] [PubMed] [Google Scholar]
- Drager R.G., Girard-Bascou,J., Choquet,Y., Kindle,K.L. and Stern,D.B. (1998) In vivo evidence for 5′→3′ exoribonuclease degradation of an unstable chloroplast mRNA. Plant J., 13, 85–96. [DOI] [PubMed] [Google Scholar]
- Drapier D., Girard-Bascou,J. and Wollman,F.-A. (1992) Evidence for nuclear control of the expression of the atpA and atpB chloroplast genes in Chlamydomonas. Plant Cell, 4, 283–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durant I. (1990) Light-based detection of biomolecules. Nature, 346, 297–298. [DOI] [PubMed] [Google Scholar]
- Eckhardt H. and Luhrmann,R. (1981) Recognition by initiator transfer ribonucleic acid of a uridine 5′ adjacent to the AUG codon: different conformational states of formylatable methionine-accepting transfer ribonucleic acid at the ribosomal peptidyl site. Biochemistry, 20, 2075–2080. [DOI] [PubMed] [Google Scholar]
- Esposito D., Hicks,A.J. and Stern,D.B. (2001) A role for initiation codon context in chloroplast translation. Plant Cell, 13, 2373–2384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forget L., Ustinova,J., Wang,Z., Huss,V.A. and Franz Lang,B. (2002) Hyaloraphidium curvatum: a linear mitochondrial genome, tRNA editing and an evolutionary link to lower fungi. Mol. Biol. Evol., 19, 310–319. [DOI] [PubMed] [Google Scholar]
- Ganoza M.C., Fraser,A.R. and Neilson,T. (1978) Nucleotides contiguous to AUG affect translational initiation. Biochemistry, 17, 2769–2775. [DOI] [PubMed] [Google Scholar]
- Ganoza M.C., Sullivan,P., Cunningham,C., Hader,P., Kofoid,E.C. and Neilson,T. (1982) Effect of bases contiguous to AUG on translation initiation. J. Biol. Chem., 257, 8228–8232. [PubMed] [Google Scholar]
- Ganoza M.C., Marliere,P., Kofoid,E.C. and Louis,B.G. (1985) Initiator tRNA may recognize more than the initiation codon in mRNA: a model for translational initiation. Proc. Natl Acad. Sci. USA, 82, 4587–4591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldschmidt-Clermont M. (1991) Transgenic expression of aminoglycoside adenine transferase in the chloroplast: a selectable marker for site-directed transformation of Chlamydomonas. Nucleic Acids Res., 19, 4083–4090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross G., Mielke,C., Hollatz,I., Blocker,H. and Frank,R. (1990) RNA primary sequence or secondary structure in the translational initiation region controls expression of two variant interferon-β genes in Escherichia coli. J. Biol. Chem., 265, 17627–17636. [PubMed] [Google Scholar]
- Gualerzi C.O. and Pon,C.L. (1990) Initiation of mRNA translation in prokaryotes. Biochemistry, 29, 5881–5889. [DOI] [PubMed] [Google Scholar]
- Harris E.H. (1989) The Chlamydomonas Sourcebook: A Comprehensive Guide to Biology and Laboratory Use. Academic Press, San Diego, CA. [DOI] [PubMed]
- Harris E.H., Boynton,J.E. and Gillham,N.W. (1994) Chloroplast ribosomes and protein synthesis. Microbiol. Rev., 58, 700–754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hui A. and de Boer,H.A. (1987) Specialized ribosome system: preferential translation of a single mRNA species by a subpopulation of mutated ribosomes in Escherichia coli. Proc. Natl Acad. Sci. USA, 84, 4762–4766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hui A., Hayflick,J., Dinkelspiel,K. and de Boer,H.A. (1984) Mutagenesis of the three bases preceding the start codon of the β-galactosidase mRNA and its effect on translation in Escherichia coli. EMBO J., 3, 623–629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hüttenhofer A. and Noller,H.F. (1994) Footprinting mRNA–ribosome complexes with chemical probes. EMBO J., 13, 3892–3901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kindle K.L., Richards,K.L. and Stern,D.B. (1991) Engineering the chloroplast genome: techniques and capabilities for chloroplast transformation in Chlamydomonas reinhardtii. Proc. Natl Acad. Sci. USA, 88, 1721–1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunkel T.A. (1985) Rapid and efficient site-specific mutagenesis without phenotypic selection. Proc. Natl Acad. Sci. USA., 82, 488–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuras R. and Wollman,F.-A. (1994) The assembly of cytochrome b6/f complexes: an approach using genetic transformation of the green alga Chlamydomonas reinhardtii. EMBO J., 13, 1019–1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magliery T.J., Anderson,J.C. and Schultz,P.G. (2001) Expanding the genetic code: selection of efficient suppressors of four-base codons and identification of ‘shifty’ four-base codons with a library approach in Escherichia coli. J. Mol. Biol., 307, 755–769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maul J.E., Lilly,J.W., Cui,L., de Pamphilis,C.W., Harris,E.H. and Stern,D.B. (2002) The Chlamydomonas reinhardtii plastid chromosome: islands of genes in a sea of repeats. Plant Cell, 14, 1–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayfield S., Rahire,M., Frank,G., Zuber,H. and Rochaix,J.-D. (1987) Expression of the nuclear gene encoding oxygen-evolving enhancer protein 2 is required for high levels of photosynthetic oxygen evolution in Chlamydomonas reinhardtii. Proc. Natl Acad. Sci. USA, 84, 749–753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy J.E.G. and Brimacombe,R. (1994) Prokaryotic translation: the interactive pathway leading to initiation. Trends Genet., 10, 402–407. [DOI] [PubMed] [Google Scholar]
- Meinnel T., Mechulam,Y., Lazennec,C., Blanquet,S. and Fayat,G. (1993) Critical role of the acceptor stem of tRNAs(Met) in their aminoacylation by Escherichia coli methionyl-tRNA synthetase. J. Mol. Biol., 229, 26–36. [DOI] [PubMed] [Google Scholar]
- Moll I., Huber,M., Grill,S., Sairafi,P., Mueller,F., Brimacombe,R., Londei,P. and Blasi,U. (2001) Evidence against an interaction between the mRNA downstream box and 16S rRNA in translation initiation. J. Bacteriol., 183, 3499–3505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morelle G., Frank,R. and Meyerhans,A. (1991) Restructuring the translation initiation region of the human parathyroid hormone gene for improved expression in Escherichia coli. Biochim. Biophys. Acta, 1089, 320–324. [DOI] [PubMed] [Google Scholar]
- Piccioni R.G., Bennoun,P. and Chua,N.H. (1981) A nuclear mutant of Chlamydomonas reinhardtii defective in photosynthetic photophosphorylation. Characterization of the algal coupling factor ATPase. Eur. J. Biochem., 117, 93–102. [DOI] [PubMed] [Google Scholar]
- Qian Q., Li,J.N., Zhao,H., Hagervall,T.G., Farabaugh,P.J. and Bjork,G.R. (1998) A new model for phenotypic suppression of frameshift mutations by mutant tRNAs. Mol. Cell, 1, 471–482. [DOI] [PubMed] [Google Scholar]
- Shepherd H.S., Boynton,J.E. and Gillham,N.W. (1979) Mutations in nine chloroplast loci of Chlamydomonas affecting photosynthetic functions. Proc. Natl Acad. Sci. USA, 76, 1353–1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shine J. and Dalgarno,L. (1974) The 3′-terminal sequence of Escherichia coli 16S ribosomal RNA: complementarity to nonsense triplets and ribosome binding sites. Proc. Natl Acad. Sci. USA, 71, 1342–1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sprinzl M., Horn,C., Brown,M., Ioudovitch,A. and Steinberg,S. (1998) Compilation of tRNA sequences and sequences of tRNA genes. Nucleic Acids Res., 26, 148–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenstrom C.M., Jin,H., Major,L.L., Tate,W.P. and Isaksson,L.A. (2001) Codon bias at the 3′-side of the initiation codon is correlated with translation initiation efficiency in Escherichia coli. Gene, 263, 273–284. [DOI] [PubMed] [Google Scholar]
- Stern D.B., Radwanski,E.R. and Kindle,K.L. (1991) A 3′ stem/loop structure of the Chlamydomonas chloroplast atpB gene regulates mRNA accumulation in vivo. Plant Cell, 3, 285–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugiura M., Hirose,T. and Sugita,M. (1998) Evolution and mechanism of translation in chloroplasts. Annu. Rev. Genet., 32, 437–459. [DOI] [PubMed] [Google Scholar]
- Varshney U., Lee,C. and RajBandhary,U. (1991) Direct analysis of aminoacylation levels of tRNAs in vivo. Application to studying recognition of Escherichia coli initiator tRNA mutants by glutaminyl-tRNA synthetase. J. Biol. Chem., 266, 24712–24718. [PubMed] [Google Scholar]
- Walker M., Pavlovic,V. and Kasif,S. (2002) A comparative genomic method for computational identification of prokaryotic translation initiation sites. Nucleic Acids Res., 30, 3181–3191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X.Q., Iyengar,P. and RajBhandary,U.L. (1996) Ribosome-initiator tRNA complex as an intermediate in translation initiation in Escherichia coli revealed by use of mutant initiator tRNAs and specialized ribosomes. EMBO J., 15, 4734–4739. [PMC free article] [PubMed] [Google Scholar]