Abstract
We analyzed the transcription initiation activity of the left-end sequence (first 342 bp) of the adenovirus genome in the context of an adenovirus vector with E1 deleted in in vitro and in vivo gene transfer models. While nucleotide sequences 1 to 190 and 1 to 342 showed strong activity in three out of three lung cancer cell lines, nucleotide sequence 1 to 103 showed limited activity in H358, cells which show characteristics of type 2 alveolar cells. In vivo, the transcription initiation activities of nucleotide sequence 1 to 103 in the liver and the lung were minimal, while nucleotide sequences 1 to 190 and 1 to 342 showed strong activity comparable to that of the cytomegalovirus promoter. Further understanding of the transcription initiation activity of the left-end sequence of the adenovirus genome should lead to optimization of adenovirus vectors.
Adenovirus (Ad) vectors have been utilized for various gene therapy approaches, taking advantage of their high infectivity and easy viral preparation. However, selective expression in target tissue is still a problem (11). Especially in the context of promoter-based targeting, many tissue (or tumor)-specific promoters are known to lose their fidelity when they are configured in Ads with E1 deleted (ΔE1), which have been used frequently in Ad gene therapy (11, 20, 21, 23). For wild-type Ad, it has been reported that removal of the enhancer (Enh) which exists between the left inverted terminal repeat (ITR) and E1 region dramatically decreased the E1 RNA level (9, 10). This enhancer overlaps with the viral packaging signal and consists of one element that enhances the expression of all early viral genes and two repeated elements that enhance the expression of E1a regions (9). In the case of ΔE1 vectors, insertion of insulators between this enhancer and the transgene (20, 21) or altering the insertion position of the promoter (i.e., around E4) (17) can restore promoter fidelity. While these studies suggest that this enhancer and/or transcription initiation function existing at the left end (approximately 400 bp) of Ad sequence is a major reason for an exogenous promoter to lose its fidelity, the mechanisms of this fidelity loss in ΔE1 Ads have not been fully understood. In the context of a cryptic transcription initiation activity of the left-end sequence of the Ad genome, Hatfield and Hearing identified two transcription initiation sites (one in the ITR and the other between the ITR and Enh) in primer extension analysis of an in vitro transcription product (7), and Buvoli et al. identified another site within the Enh after infection of fetal cardiac myocytes with an Ad vector (3). These sites may contribute to the leakiness of an exogenous promoter placed in the ΔE1 region of an Ad vector. In this study, we focused on the transcription initiation activity of this region in the context of ΔE1 Ad vectors and analyzed it in a simplified experimental system in in vitro and in vivo gene transfer models.
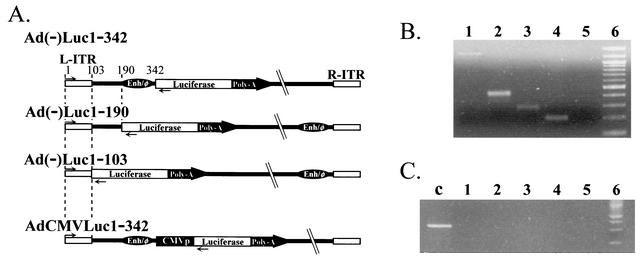
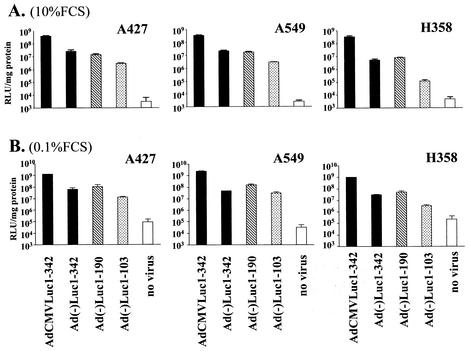
To analyze the transcription initiation activity of the Ad left-end sequence in ΔE1 Ads, we constructed four replication-defective recombinant Ads with E1 and E3 deleted, shown in Fig. 1A, by using the pAdEasy system (8). Each vector has the firefly luciferase cDNA and simian virus 40 polyadenylation signal derived from pGL3-Basic (Promega, Madison, Wis.) in a left-to-right direction. The three vectors of the Ad(−) series do not have any extrinsic promoter. The 5′ upstream region of the transgene cassettes of Ad(−)Luc1-342, Ad(−)Luc1-190, and Ad(−)Luc1-103 consists of the bp 1 to 342, 1 to 190, and 1 to 103 regions of Ad sequence, respectively. In Ad(−)Luc1-190 and Ad(−)Luc1-103, the Enh sequence was placed adjacent to the right ITR, as reported by Hearing and Shenk, to compensate for the loss of viral packaging function after the deletion of the Enh existing around the left end of the Ad genome (10). The positive-control vector AdCMV Luc1-342 has the same structure as does Ad(−)Luc1-342 except for a cytomegalovirus (CMV) immediate-early promoter driving luciferase expression. All the vectors were propagated in an E1-transcomplementing cell line, 293, and purified by double CsCl banding. The vectors had similar yields (1.1 × 1010 to 1.4 × 1010 viral particles/15-cm-diameter dish) and viral particle/PFU ratios (60 to 80 viral particles/PFU). Since recombination in the left-end sequence and the existence of replication-competent Ad could affect results, the structure of the vectors was confirmed by PCR with Taq DNA Polymerase (Qiagen, Valencia, Calif.), as described by the manufacturer, with the following primers: left end-sense (5′ CATCATCAATAATATACCTTATTTTGG 3′, corresponding to nucleotides [nt] 1 to 27 of ITR), left end-antisense (5′ GGTGGCTTTACCAACAGTACCGGAATG 3′, corresponding to the 5′ sequence of luciferase cDNA), E1-sense (5′ GAGACATATTATCTGCCACGGAGG 3′), and E1-antisense (5′ TTGGCATAGAAACCGGACCCAAGG 3′) (Fig. 1B and C). The results indicated the absence of replication-competent Ad and the correct structure of left-end sequence. With these vectors, we first analyzed the transcription initiation activity of Ad left-end sequence with cell lines. For this purpose we used three human lung cancer cell lines, A549, A427, and H358 (American Type Culture Collection CCL-185, HTB-53, and CRL-5807, respectively; Manassas, Va.). A549 and H358 were maintained with Roswell Park Memorial Institute (RPMI) medium 1640 containing 10% fetal calf serum (FCS), and A427 was maintained with minimum essential medium containing 10% FCS. One day after 50,000 cells per well were plated in a 24-well plate, the cells were infected at a multiplicity of infection (MOI) of 50 (PFU per cell) with three promoterless vectors and a positive-control vector in the infection medium (Dulbecco's modified Eagle's medium with 5% FCS). Two hours later, the medium was replaced with complete medium. After 48 h of cultivation, the cells were lysed with 100 μl of cell culture lysis buffer (Promega), and the resulting lysates were analyzed with the luciferase assay system (Promega) as described by the manufacturer. The protein concentration was determined with the Dc protein assay (Bio-Rad Laboratories, Hercules, Calif.). In A427 and A549, all three promoter-free constructs [Ad(−)Luc1-342, Ad(−)Luc1-190, and Ad(−)Luc1-103] showed high luciferase expression (Fig. 2A). In H358, the luciferase expression with Ad(−)Luc1-103 was clearly lower than that with Ad(−)Luc1-342 or Ad(−)Luc1-190. While the sequences 1 to 190 and 1 to 342 showed strong transcription initiation activity in three out of three lung cancer cell lines, the Ad ITR sequence (1 to 103) showed limited transcription initiation activity in H358, cells which show more characteristics of type 2 alveolar cells (based on ultrastructural findings of cytoplasmic structure, expression of the major lung surfactant-associated protein and cytochrome P-450, and incapability of lymph node metastasis or pleural dissemination in an intrabronchial orthotopic propagation model [2, 4, 6, 14]). The same experiment was also performed at a lower MOI (1 PFU/cell), and the tendency was the same as that at 50 PFU/cell. This indicated that the transcription initiation activity profile of these sequences was not affected by the dose of the virus (data not shown).
FIG. 1.
(A) Configuration of the vectors. Each vector has firefly luciferase cDNA and simian virus 40 polyadenylation signal in the left-to-right direction. Three vectors of the Ad(−) series, Ad(−)Luc1-342, Ad(−)Luc1-190, and Ad(−)Luc1-103, have base pair regions 1 to 342, 1 to 190, and 1 to 103 of Ad sequence upstream of the luciferase cassette, respectively. In Ad(−)Luc1-190 and Ad(−)Luc1-103, the enhancer-packaging signal sequence was placed adjacent to the right ITR to compensate for the loss of viral packaging function. Positive-control vector AdCMVLuc1-342 has the same structure as Ad(−)Luc1-342 except for a CMV promoter to drive luciferase expression. (B) Left-end sequences of the vectors. Virus sequences were analyzed by PCR with the primers left end-sense (→) and -antisense (←) shown in panel A. Amplified fragments were detected at the expected positions of each vector [bp 1289, 467, 313, and 226 for AdCMVLuc1-342, Ad(−)Luc1-342, Ad(−)Luc1-190, and Ad(−)Luc1-103, respectively]. Lane 1, AdCMVLuc1-342; lane 2, Ad(−)Luc1-342; lane 3, Ad(−)Luc1-190; lane 4, Ad(−)Luc1-103; lane 5, negative control (H2O); lane 6, 100-bp ladder. (C) Detection of replication-competent virus. To detect replication-competent Ads, viral DNA was analyzed by PCR with E1a-specific primers (E1a-sense and -antisense). Lane c, positive control (E1a-containing plasmid); lanes 1 to 6, same as defined for panel B.
FIG. 2.
(A) Transcription initiation activity of the left-end sequence in vitro (10% FCS). A549, A427, and H358 cells were maintained with proper medium containing 10% FCS. One day after plating, cells were infected at an MOI of 50 with vectors. Two hours later, the medium was replaced with the complete medium of each cell line. After 48 h of cultivation, the cells were analyzed for luciferase activity. The results are shown as relative light units per milligram of protein. (B) Transcription initiation activity of the left-end sequence under cell cycle arrest (0.1% FCS). The day after initial cell inoculation, the medium was replaced with medium containing 0.1% FCS. After 24 h, the cells were infected with each vector by using infection medium with 0.1% FCS for 2 h, followed by 48 h of incubation with medium containing 0.1% FCS. Under these conditions, all the cells failed to show detectable proliferation. Cell lysis and luciferase assays were performed in the same way as reported for panel A.
To rule out the influence of cell growth rate on transcription initiation activity, the same experiment was performed under cell cycle arrest conditions by serum starvation. Twenty-four hours after plating with complete medium, the medium was replaced with medium containing 0.1% FCS. After another 24 h, the cells were then infected with each vector by using infection medium with 0.1% FCS for 2 h, followed by 48 h of incubation with medium containing 0.1% FCS. Under these conditions, all cells failed to show detectable proliferation. The result of the luciferase assay showed nearly the same trend as in nonarrested cells (Fig. 2B).
Although the DNA replication of ΔE1 Ad vector without accompanying mature virus production may affect the result via the difference in DNA copy number, the difference in DNA copy number analyzed by real-time PCR was minimal and could not explain the wide difference of the luciferase assay results among the vectors (data not shown).
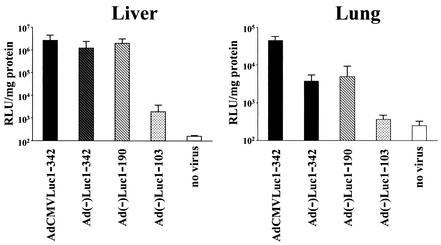
To clarify the transcription initiation activity in vivo, female BALB/c mice (Charles River Laboratories, Wilmington, Mass.) received 5 × 108 PFU of vectors intravenously via the tail vein. Two days later, the livers and lungs were harvested, rapidly frozen on dry ice, and stored at −80°C. On the day of analysis, tissues were ground into fine powder with a pestle and mortar in an ethanol-dry ice bath. The tissue powders were lysed with cell culture lysis buffer (approximately double the weight of the sample), and after three rounds of freezing and thawing followed by centrifugation, the recovered supernatants were analyzed for luciferase activity and protein concentration. Interestingly, in vivo results indicated the difference among the tested constructs more clearly (Fig. 3). Here, the fragments 1 to 342 and 1 to 190 of Ad showed the same transcription initiation activity as did the CMV promoter in liver, while the activities in the lung were 1 order of magnitude lower than that of the CMV promoter. On the other hand, fragment 1 to 103 showed much lower activity both in the liver and in the lung. In the liver, the activity of fragment 1 to 103 was more than 3 orders of magnitude lower than that of other vectors, and in the lung, the activity of fragment 1 to 103 was close to that of uninfected control. In these two organs, fragment 1 to 342 and fragment 1 to 190 showed strong transcription initiation activities but the activity of fragment 1 to 103 was minimal. Thus, the vector structure of Ad(−)Luc1-103 might be helpful to reduce the background expression from the transgene in the region where E1 is deleted.
FIG. 3.
In vivo analysis of transcription initiation activity of the left-end sequence. Female BALB/c mice received 5 × 108 PFU of vectors intravenously via the tail vein. Two days later, the livers and lungs were harvested, rapidly frozen on dry ice, and stored at −80°C. On the day of analysis, tissues were ground into fine powder and lysed. After three rounds of freezing and thawing followed by centrifugation, the recovered supernatants were analyzed for luciferase activity and protein concentration.
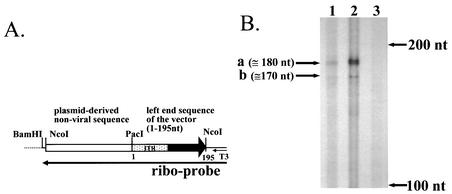
These data strongly suggest the existence of a transcription initiation site in the ITR region, but the site has not been identified in an analysis with a ΔE1 vector construct. Thus, we analyzed cellular RNA after infection with Ad(−)Luc1-103 by an RNase protection assay to determine the initiation site. A549 cells were infected at an MOI of 50 in the same way as the luciferase assay, and the total RNA and poly(A) RNA were extracted by using the RNeasy Kit (Qiagen) and Oligotex direct mRNA kit (Qiagen), respectively. The RNA probe was generated as follows: the NcoI fragment containing 195 nt of the left-end sequence of Ad(−)Luc1-103 and approximately 600 nt of nonviral plasmid-derived sequence was isolated from the shuttle plasmid that was used to generate this vector and cloned into the SmaI site of pBluescript II KS(+) (Stratagene, La Jolla, Calif.). The plasmid was cleaved with BamHI, and the RNA was transcribed with T3 polymerase with MAXIscript (Ambion, Austin, Tex.). The full-length probe was isolated by denaturing polyacrylamide electrophoresis. The RNase protection assay was performed with 5 μg of total RNA and 0.6 μg of poly(A) RNA as starting materials and employing RPA III (Ambion) as described by the manufacturer. The size of the protected fragment was determined with an RNA size marker transcribed from RNA Century marker template (Ambion). The results indicated that there are two protected fragments of approximately 180 and 170 nt, and this suggested that the transcription initiation site is 15 and 25 bp from the left end of the left-ITR sequence (Fig. 4). Although these initiation sites are different from the sites determined with in vitro transcription by Hatfield and Hearing (7), the difference might be due to differences in transcription conditions (in vitro transcription with HeLa whole-cell lysate versus infection of A549 cells with Ad vector).
FIG. 4.
Determination of transcription of the initiation site. RNA of A549 cells infected with Ad(−)Luc1-103 vector was analyzed by an RNase protection assay to determine the transcription initiation site. (A) The riboprobe for this assay was transcribed from the NcoI fragment isolated from the shuttle vector used in the construction of Ad(−)Luc1-103 by using T3 polymerase. This probe has 195 nt of the left-end sequence of Ad(−)Luc1-103 and approximately 600 nt of nonviral, plasmid-derived sequence. (B) In the RNase protection assay, two protected fragments were observed (a and b). These sites exist in the Ad ITR region. Lane 1, 5 μg of total RNA; lane 2, 0.6 μg of poly(A) RNA; lane 3, 5 μg of yeast RNA. Arrows on the right indicate fragment lengths determined with an RNA marker.
The phenomenon where specific promoters with a favorable profile can lose fidelity when inserted into the Ad ΔE1 region has been widely recognized (16). The insertion of an insulator between the Enh and the specific promoter has been reported elsewhere to reduce this promoter leakiness (20, 21). However, the effect of transcription initiation activity on the expression of a transgene in the ΔE1 region has not been clarified in vector constructs. In this study, we analyzed this function by using an ΔE1 vector construct in vitro and in vivo.
Our experiments using ΔE1 Ad constructs clearly indicated the strong transcription initiation activity of the left-ITR, comparable to that of the CMV promoter, which is known to be one of the strongest promoters in an Ad vector context. In plasmid-based analysis of transcription, the transcription initiation activity of left-end sequence has been reported to be much weaker than that of the original E1 initiation site (7). This discrepancy might be due to either the difference between the plasmid construct and the viral vector construct or the difference in experimental conditions (in vitro transcription and actual infection of viral vector). In the context of gene therapy vectors, our data, which correlate more strongly with an actual vector used in an in vivo administration setting, should be meaningful in determining better configurations of Ads.
We identified two transcription initiation sites around nt 15 and nt 25. Hatfield and Hearing identified one region between nt 148 and nt 158 (7), and Buvoli et al. identified one site in the enhancer-packaging signal region (around nt 230) (3). In comparison with these sites, the ones that we identified are very close to the end of the Ad genome, having a very short upstream region. While the nt 10 to 15 region (TAATAT) may serve as a TATA box for the nt 25 initiation site, the mechanism of the strong transcription initiation from this short region (especially from nt 15) is still unclear. One possible explanation might be a contribution of Ad terminal protein or preterminal protein that is bound to the end of the Ad DNA (19). Schaack et al. suggested that these proteins act in a manner similar to cellular DNA-binding proteins based on the amino acid homology to GCN4 and GAL4 (12, 13, 15, 18). The binding of terminal protein (or preterminal protein) onto common transcription factors (e.g., Oct-1) recruits the transcription complex regardless of the binding of the TATA binding protein onto the TATA box and may confer effective transcription initiation (22). This theory coincides with the fact that transcription from the nt 15 or 25 region was not detected in the extensive analyses done in a plasmid-based system (7).
In the context of transcription control, all Ad(−) series constructs showed relatively high transcription initiation activity in A549 and A427 cells. However, in H358 cells, the activity of the bp 1 to 103 region was much lower than that of the other two regions. Since Ad replication depends on cell cycling, there is a possibility that the transcription initiation activity is linked to the rate of cell cycling. However, the data obtained under cell cycle arrest by serum starvation showed the same profile, indicating that the difference was not due to the difference in cell cycling. In vivo, the differences between bp 1 to 103 and the other two vectors were more evident. Both in the liver and in the lung, bp 1 to 342 and 1 to 190 showed strong transcription activity comparable to that of the CMV promoter, but bp 1 to 103 showed minimal activity close to background. These data indicated that the bp 1 to 103 region has strong activity in cancer cells while it has minimal activity in normal organs in vivo. The ITR sequence of Ad includes many regions which bind to various transcription factors, i.e., SP1, NFI/CTF, NFIII/OCT1, and ATF (7), and major control elements of some promoters exist downstream of the transcription initiation site (5). Of note, this region has three SP1 sites, and SP1 is known to contribute to growth-related signal transduction, especially in the context of cancer (1). This may explain the minimal transcription initiation activity of the bp 1 to 103 region in fully developed organs. Also, our data suggest the possible application of the bp 1 to 103 region as a cancer-specific promoter.
This study revealed the strong transcription initiation activity of the left end of the Ad genome and the reduction of this activity in in vivo organs by minimizing the left-end sequence to the bp 1 to 103 region. Since the ectopic expression of the therapeutic gene in the liver caused by Ad hepatotropism is a limiting factor of Ad application, further understanding of the Ad vector system will lead to the development of optimized Ad vector systems for gene therapy applications.
Acknowledgments
This work was supported by grants from the National Institutes of Health (R01 HL67962, P50 CA89019, R01 CA86881, and U19 PR15015 to David T. Curiel) and the AVON Breast Cancer Research and Care Program (to Masato Yamamoto).
We thank Yasuo Adachi, Christina Balague, Peter Nagi, and Long Le for excellent advice.
REFERENCES
- 1.Black, A. R., J. D. Black, and J. Azizkhan-Clifford. 2001. Sp1 and kruppel-like factor family of transcription factors in cell growth regulation and cancer. J. Cell. Physiol. 188:143-160. [DOI] [PubMed] [Google Scholar]
- 2.Brower, M., D. N. Carney, H. K. Oie, A. F. Gazdar, and J. D. Minna. 1986. Growth of cell lines and clinical specimens of human non-small cell lung cancer in a serum-free defined medium. Cancer Res. 46:798-806. [PubMed] [Google Scholar]
- 3.Buvoli, M., S. J. Langer, S. Bialik, and L. A. Leinwand. 2002. Potential limitations of transcription terminators used as transgene insulators in adenoviral vectors. Gene Ther. 9:227-231. [DOI] [PubMed] [Google Scholar]
- 4.Falzon, M., J. B. McMahon, and H. M. Schuller. 1986. Xenobiotic-metabolizing enzyme activity in human non-small-cell derived lung cancer cell lines. Biochem. Pharmacol. 35:563-568. [DOI] [PubMed] [Google Scholar]
- 5.Farnham, P. J., and A. L. Means. 1990. Sequences downstream of the transcription initiation site modulate the activity of the murine dihydrofolate reductase promoter. Mol. Cell. Biol. 10:1390-1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gazdar, A. F., R. I. Linnoila, Y. Kurita, H. K. Oie, J. L. Mulshine, J. C. Clark, and J. A. Whitsett. 1990. Peripheral airway cell differentiation in human lung cancer cell lines. Cancer Res. 50:5481-5487. [PubMed] [Google Scholar]
- 7.Hatfield, L., and P. Hearing. 1991. Redundant elements in the adenovirus type 5 inverted terminal repeat promote bidirectional transcription in vitro and are important for virus growth in vivo. Virology 184:265-276. [DOI] [PubMed] [Google Scholar]
- 8.He, T. C., S. Zhou, L. T. da Costa, J. Yu, K. W. Kinzler, and B. Vogelstein. 1998. A simplified system for generating recombinant adenoviruses. Proc. Natl. Acad. Sci. USA 95:2509-2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hearing, P., and T. Shenk. 1986. The adenovirus type 5 E1A enhancer contains two functionally distinct domains: one is specific for E1A and the other modulates all early units in cis. Cell 45:229-236. [DOI] [PubMed] [Google Scholar]
- 10.Hearing, P., and T. Shenk. 1983. The adenovirus type 5 E1A transcriptional control region contains a duplicated enhancer element. Cell 33:695-703. [DOI] [PubMed] [Google Scholar]
- 11.Hitt, M. M., C. L. Addison, and F. L. Graham. 1997. Human adenovirus vectors for gene transfer into mammalian cells. Adv. Pharmacol. 40:137-206. [DOI] [PubMed] [Google Scholar]
- 12.Hope, I. A., and K. Struhl. 1986. Functional dissection of a eukaryotic transcriptional activator protein, GCN4 of yeast. Cell 46:885-894. [DOI] [PubMed] [Google Scholar]
- 13.Ma, J., and M. Ptashne. 1987. Deletion analysis of GAL4 defines two transcriptional activating segments. Cell 48:847-853. [DOI] [PubMed] [Google Scholar]
- 14.Mase, K., T. Iijima, N. Nakamura, T. Takeuchi, M. Onizuka, T. Mitsui, and M. Noguchi. 2002. Intrabronchial orthotopic propagation of human lung adenocarcinoma—characterizations of tumorigenicity, invasion and metastasis. Lung Cancer 36:271-276. [DOI] [PubMed] [Google Scholar]
- 15.Mitchell, P. J., and R. Tjian. 1989. Transcriptional regulation in mammalian cells by sequence-specific DNA binding proteins. Science 245:371-378. [DOI] [PubMed] [Google Scholar]
- 16.Ring, C. J., J. D. Harris, H. C. Hurst, and N. R. Lemoine. 1996. Suicide gene expression induced in tumour cells transduced with recombinant adenoviral, retroviral and plasmid vectors containing the ERBB2 promoter. Gene Ther. 3:1094-1103. [PubMed] [Google Scholar]
- 17.Rubinchik, S., D. Wang, H. Yu, F. Fan, M. Luo, J. S. Norris, and J. Y. Dong. 2001. A complex adenovirus vector that delivers FASL-GFP with combined prostate-specific and tetracycline-regulated expression. Mol. Ther. 4:416-426. [DOI] [PubMed] [Google Scholar]
- 18.Schaack, J., W. Y. Ho, P. Freimuth, and T. Shenk. 1990. Adenovirus terminal protein mediates both nuclear matrix association and efficient transcription of adenovirus DNA. Genes Dev. 4:1197-1208. [DOI] [PubMed] [Google Scholar]
- 19.Shenk, T. 1996. Adenoviridae: the viruses and their replication, p. 2111-2148. In B. N. Fields, D. M. Knipe, and P. M. Howley (ed.), Fields virology, 3rd ed., vol. 2. Lippincott-Raven Publishers, Philadelphia, Pa.
- 20.Steinwaerder, D. S., and A. Lieber. 2000. Insulation from viral transcriptional regulatory elements improves inducible transgene expression from adenovirus vectors in vitro and in vivo. Gene Ther. 7:556-567. [DOI] [PubMed] [Google Scholar]
- 21.Vassaux, G., H. C. Hurst, and N. R. Lemoine. 1999. Insulation of a conditionally expressed transgene in an adenoviral vector. Gene Ther. 6:1192-1197. [DOI] [PubMed] [Google Scholar]
- 22.White, R. J., and S. P. Jackson. 1992. Mechanism of TATA-binding protein recruitment to a TATA-less class III promoter. Cell 71:1041-1053. [DOI] [PubMed] [Google Scholar]
- 23.Zhang, J., and S. J. Russell. 1996. Vectors for cancer gene therapy. Cancer Metastasis Rev. 15:385-401. [DOI] [PubMed] [Google Scholar]