Abstract
To address Sin Nombre (SN) virus persistence in deer mice, we sacrificed experimentally infected deer mice at eight time points from day 21 to day 217 postinoculation (p.i.) and examined their tissues for viral nucleocapsid (N) antigen expression and both negative-strand (genomic) and positive-strand (replicative/mRNA) viral S segment RNA titers. All the animals that we inoculated developed persistent infections, and SN virus could be isolated from tissues throughout the course of infection. The transition from an acute to a persistent pattern of infection appeared to occur between days 60 and 90 p.i. Beginning on day 60 p.i., the heart, brown adipose tissue (BAT), and lung retained antigen expression and genomic viral RNA the most frequently. We found a statistically significant association among the presence of replicative RNA in the heart, lung, and BAT, widespread antigen expression (in ≥5 tissues), and RNA viremia. Of these three tissues, the heart retained negative-strand RNA and viral N antigen the most consistently (in 25 of 26 animals). During persistence, there were two distinct patterns of infection: restricted versus disseminated tissue involvement. Mice with the restricted pattern exhibited N antigen expression in ≤3 tissues, an absence of viral RNA in the blood, neutralizing antibody titers of ≤1:1,280 (P = 0.01), and no replicative RNA in the heart, lung, or BAT. Those with the “disseminated” pattern showed N antigen expression in ≥5 tissues, neutralizing antibody titers of 1:160 to 1:20,480, replicative RNA in the heart, lung, and BAT at a high frequency, and RNA viremia. Virus could be isolated consistently only from mice that demonstrated the disseminated pattern. The heart, lung, and BAT are important sites for the replication and maintenance of SN virus during persistent infection.
Hantaviruses (Bunyaviridae, Hantavirus) are rodent-borne viruses with a worldwide distribution. Two distinct human illnesses are associated with hantavirus infection: hemorrhagic fever with renal syndrome (HFRS) and hantavirus cardiopulmonary syndrome (HCPS) (52). HFRS is almost entirely an Old World disease, with 150,000 to 200,000 cases occurring annually in Asia and Europe. The four viral species that cause HFRS are the Hantaan (HTN), Seoul (SEO), Puumala (PUU), and Dobrava-Belgrade viruses. HCPS occurs in North and South America. There are at least four etiologic agents of HCPS in North America: Sin Nombre (SN) virus, Bayou virus, New York virus, and Black Creek Canal (BCC) virus. Of these, SN virus is believed to be responsible for the large majority (>95%) of the more than 300 reported cases. The deer mouse is the predominant carrier of SN virus (17, 46).
Hantaviruses have a single stranded, negative-sense RNA genome consisting of three segments: the L, M, and S segments (8). The S segment encodes the viral nucleocapsid (N) protein, the M segment encodes the two viral envelope glycoproteins G1 and G2, and the L segment encodes the viral RNA-dependent RNA polymerase (RDRP). Each viral RNA segment is flanked by terminal consensus sequences that are believed to have a role in viral packaging and may be important for viral replication and transcription (32, 42). Viral entry into permissive cells is mediated through beta-3-integrin receptors, with replication and transcription occurring in the cytoplasm (8, 21). Using the negative-sense genomic RNAs (vRNAs) as templates, the RDRP carries out transcription and replication, generating positive-sense viral mRNAs and viral complementary RNAs (vcRNAs), respectively. The viral mRNAs are truncated at the 3′ end and have heterologous 5′ ends, presumably derived from host cellular RNAs, while the vcRNAs are full-length complements of the vRNAs. The viral mRNAs and vcRNAs are utilized for generating viral proteins and vRNAs, respectively.
HTN, PUU, and BCC virus infections are persistent in the rodents that serve as their natural reservoirs (5, 20, 29, 38), although in the case of SEO virus infection, only newborn rats, not adults, can become persistently infected experimentally (34). Provided that the natural host is used, persistent experimental infections are asymptomatic (no signs of clinical illness), although histopathological studies have been conducted only in the PUU virus model (19, 29, 55). These studies also demonstrated that infected mice are generally able to shed and/or transmit virus efficiently during the first 1 to 5 weeks of infection, and intermittently thereafter (5, 29, 33, 38, 55).
The tissue reservoirs for viral infection are not known for all of the hantavirus persistence models. Different studies have not uniformly examined a wide array of tissues. When tissues from infected rodents were examined at the latest points postinfection, the brain (BCC virus), lung (PUU virus, HTN virus, and SEO virus), and liver (PUU virus) retained one or more markers of infection (30, 34, 55). All investigations that have examined tissue distribution have agreed that of several tissues that were positive for viral markers during the acute phase, some became negative in the persistent phase. One study demonstrated that brown adipose tissue (BAT) retained PUU virus antigen in naturally infected, overwintering bank voles (20).
The mechanisms utilized by hantaviruses to establish persistent infections in host rodents are not understood. Previous animal model studies demonstrated that these viruses can persist in the host despite the presence of high titers of neutralizing antibodies (34, 38). The cell-mediated immune response during persistent infection with hantaviruses has been little studied. For this and other reasons, the interaction of the virus with the host immune system is rather poorly understood.
Based on field studies and experimental infections, SN virus is believed to cause a persistent infection in deer mice (9, 10, 44, 47). Using an experimental infection model, we examined deer mice persistently infected with Sin Nombre virus in order to investigate which tissues are involved in viral replication and maintenance, to characterize the temporal patterns of viral replication, and to investigate the relationship between neutralizing antibody titers and viral load (viral RNA and viral antigen).
MATERIALS AND METHODS
Animal handling and viral inoculations.
We adhered strictly to the biosafety recommendations of the U.S. Centers for Disease Control and Prevention in all aspects of this work (12, 15, 45). We wore respirators and personal protective gear during manipulations of infected animals in an outdoor quarantine facility as described previously (12). Animals were handled according to University of New Mexico animal research facility guidelines by using an approved protocol. Deer mice were obtained from an outbred colony from the Manzano Mountains of New Mexico and were used at the F1 to F3 generations (12, 13). For blood samples collected for nonterminal experiments, we anesthetized animals with methoxyflurane and collected blood from the retro-orbital sinus. For terminal experiments, we used tribromoethanol followed by exsanguination via cardiac puncture for euthanasia. For the time course study, 4- to 6-week-old deer mice received either 5 animal 50% infectious doses (ID50) of SN virus strain SN77734 or a sham tissue homogenate by intramuscular inoculation in order to generate infected mice and control mice, respectively (10). To detect infection, we assayed blood samples for antibodies to viral nucleocapsid (N) antigen by a strip immunoblot assay (SIA) on day 21 postinoculation (p.i.) or later (7, 10).
Nested RT-PCR.
We extracted RNA from solid tissues and blood by acid-phenol extraction in 4 M guanidinium chloride (10, 25). RNA was extracted from tissue culture supernatants by using the Qiamp Viral RNA Mini kit (Qiagen, Valencia, Calif.), which was designed for extracting RNA from cell-free materials. Total RNA derived from 2.5 mg of tissue, 7.5 μl of whole blood, or 25 μl of tissue culture supernatant was loaded in each PCR. We carried out reverse transcription-PCR (RT-PCR) with nested primers for the small (S) genomic segment of SN virus as described previously, except that we also included 0.4 U of RNasin per tube (10). The coordinates of the outer primers were positions 167 and 423 on the SN virus S segment, with inner primers at 190 and 401.
Quantitative TaqMan RT-PCR (QRT-PCR).
We used a Perkin-Elmer Applied Biosystems (Foster City, Calif.) 5700 sequence detection system and the two-step TaqMan Gold RT-PCR protocol as described by the manufacturer. Each sample was tested in triplicate. For RT, RNA derived from 2.5 mg of tissue was mixed with either the S-segment coordinate-167 sense primer 5′ AGCACATTACAGAGCAGACGGGC or the S-segment coordinate-423 antisense primer 5′ GGATAATCGGTAATGCAAAACT to determine total negative- and positive-strand S-segment RNA copies, respectively, in a volume of 100 μl at 25°C for 10 min, 48°C for 30 min, and 95°C for 5 min (10). Five microliters of cDNA was removed for subsequent PCRs. The S-segment primers used for the PCR were the coordinate-179 sense primer GCAGACGGGCAGCTGTG and the coordinate-245 antisense primer AGATCAGCCAGTTCCCGCT. The fluorescent probe was a positive-sense oligonucleotide at coordinate 198, TGCATTGGAGACCAAACTCGGAGAACTT. The probe was covalently labeled with the reporter dye FAM at the 5′ end and the quencher dye TAMRA at the 3′ end (Perkin-Elmer Applied Biosystems). All oligonucleotides were used at 200 nM.
During the reaction, the quencher is dissociated from the reporter by endonucleolytic cleavage of the probe, and a sample is judged positive when the FAM fluorescence exceeds 0.05 reporter unit. In our experiments, all curves that exceeded that value progressed with the expected sigmoid amplification curve in later cycles. At the point that the absorption curve exceeds 0.05 U, the threshold cycle number (Ct), which is inversely related to the template copy number, is determined. PCR was conducted at 95°C for 10 min, followed by 40 repetitions of 95°C for 10 s, 50°C for 10 s, and 72°C for 30 s. A standard curve containing dilutions ranging from 50 to 5 × 105 copies of the template was used on each 96-well plate. All of our standard curves produced a −0.995 or better correlation coefficient between the quantity of template loaded and the log of the Ct value.
SIA for detection of anti-N antibodies in deer mouse serum.
In separate lanes, we loaded 2 ml of a solution of deer mouse serum (2 μl/ml) in phosphate-buffered saline (PBS) to produce a 3+ intensity control band, a 0.2-μl/ml dilution of serum for a 1+ intensity control, and 3 μg of recombinant, affinity-purified SN virus N antigen (7, 24). We suctioned the antigens onto a wetted nitrocellulose membrane under a vacuum. The membrane was cut lengthwise with a paper shredder into 1.6-mm-wide strips, and each strip was placed in a well in a Western blot tray containing 1 ml of milk-PBS buffer and 5 μl of rodent blood. The membranes were rocked gently overnight at room temperature. After the membranes were washed, we applied a 1:1,000 dilution of alkaline phosphatase-conjugated goat anti-Peromyscus leucopus immunoglobulin G antibodies (Kirkegaard and Perry, Gaithersburg, Md.) and rocked the membranes gently for 1 h. Bound alkaline phosphatase was then detected with nitroblue tetrazolium and 5-bromo-4-chloro-3-indolylphosphate substrate (31).
Focus reduction neutralization test.
Serially diluted (1:20 through 1:20,480) serum samples from infected mice were examined individually in 48-well tissue culture plates by a focus reduction neutralization test as described previously (7). Diluted sera were mixed with equal volumes of 45 focus-forming units of SN virus (strain SN77734) for 1 h at 37°C before incubation on Vero E6 cells. After adsorption for 4 h at 37°C, cells were overlaid with a medium containing 1.2% methylcellulose for 7 days. The methylcellulose layer was then removed by aspiration, and the cells were washed twice in PBS and fixed with methanol containing 0.5% H2O2. We then added rabbit anti-SN virus N protein serum (1:5,000), followed first by peroxidase-conjugated goat anti-rabbit immunoglobulin G and then by diaminobenzoine-metal substrate (Pierce). The neutralization titer of a serum was expressed as the maximum dilution that would reduce the number of foci by 80% (7).
IHC.
At necropsy, 20 tissues (heart, lung, kidney, liver, spleen, pancreas, thymus, brain, salivary gland, brown fat, white fat, testis or ovary, uterus, urinary bladder, skeletal muscle, gall bladder, adrenal gland, lymph node, and large intestine) were placed in Z-fix formalin (Anatech Ltd., Battle Creek, Mich.) for at least 24 h before they were embedded in paraffin. The paraffin-embedded tissues were cut into 4- to 6-μm-thick sections. We mounted the sections on glass slides coated with poly-l-Lys, deparaffinized them, and then stained them with anti-N antiserum (1:10,000) on an automated processor following antigen retrieval, as described previously (22). The serum was a hyperimmune polyclonal rabbit serum directed against the recombinant N antigen of SN virus strain 3H226 (6, 10, 25). The immune complexes were detected first with a biotinylated anti-rabbit secondary antibody, then with a horseradish peroxidase-avidin conjugate, and finally with an aminoethyl carbazole chromogen. The specific stain consisted of punctate, cytoplasmic granules. After applying hematoxylin as a counterstain, we mounted the slides with an aqueous mounting medium. Preimmune rabbit serum was extensively used initially to verify the specificity of the test during the development of the immunohistochemistry (IHC) procedure, which verified that the observed staining is specific for the viral N antigen.
Histopathological examination.
To determine whether infection caused any histopathologic changes, a veterinary pathologist blindly examined fixed tissue sections that had been stained with hematoxylin and eosin (22). Histopathological analysis was carried out on the same panel of 20 tissues examined in the IHC studies.
In vitro viral isolation.
Attempts to isolate SN virus in Vero E6 cells were conducted in 48-well plates. We obtained heart and lung samples from two infected mice at days 60, 120, 180, and 217 p.i. and created 1% tissue homogenates for each tissue individually as previously described (10). Our positive control for viral isolation was a 1% lung homogenate derived from an experimentally infected animal at day 13 p.i. (11). Each 1% tissue homogenate was serially diluted (10−2 through 10−7) by using supplemented minimal essential medium. A 200-μl volume of each dilution was incubated on Vero E6 cells (80 to 100% confluent) for 1 h at 37°C (11). After incubation, cells were washed with PBS and overlaid with 500 μl of medium. Supernatants were harvested at weekly intervals for the first 4 weeks after inoculation and were frozen for RNA analysis by nested RT-PCR. To detect viral amplification in cultures, nested RT-PCR was carried out on supernatants. Positive results were confirmed by the absence of amplification in controls without reverse transcriptase and by repeat RNA preparations.
Statistical analysis.
We utilized 2 × 2 contingency tables to determine if an association existed between the presence of positive-stranded viral RNA and either widespread antigen expression (in ≥5 tissues) or RNA viremia. Because values of <5 were present in each 2 × 2 table, we report P values from a two-tailed Fisher's exact text. To determine if a statistical difference existed between neutralizing antibody titers in animals with restricted versus widespread replication and antigen expression, we performed single-factor analysis-of-variance tests on the ranks of neutralization titers for the groups with widespread- versus restricted-infection patterns.
RESULTS
Sites of viral N antigen persistence and histopathology.
We inoculated groups of 26 and 15 juvenile deer mice with 5 animal ID50 of SN virus strain SN77734 or sham tissue homogenate, respectively, via the intramuscular route. At least five infected animals and three controls were sacrificed at each of the following days p.i.: days 60, 90, 120, 180, and 217. Blood obtained at the time of sacrifice was screened for anti-N antibodies by SIA. All mice that underwent viral inoculation seroconverted (data not shown). As with acute infection (10), histopathologic examinations of tissues revealed no evidence of histological abnormalities associated with experimental infection when infected animals were compared to controls (data not shown).
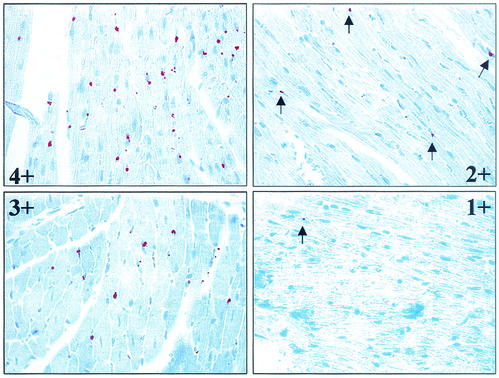
A panel of 20 tissues was examined for N-antigen expression by use of IHC (Table 1). Antigen expression was scored on a scale of 0+ to 4+ (Fig. 1). Antigen staining appeared to be restricted primarily to cells with the morphological characteristics of vascular endothelial cells in all tissues examined, with the exception of the liver, where Kupfer cell staining was frequently observed. There was a widespread distribution of N antigen on day 60 p.i., with many (8 to 12) of the tissues examined displaying antigen expression. This pattern of expression was similar to that seen in acutely infected mice examined between days 14 and 35 p.i. in an earlier study (10). Tissues that either did not display antigen expression or displayed antigen expression at a low frequency throughout the time course were the reproductive organs (uterus, ovary, and testis), brain, urinary bladder, large intestine, salivary gland, thymus, lymph node, and gall bladder. Beginning on day 90 p.i., the overall level of antigen expression and/or the proportion of mice displaying antigen expression declined compared to those for mice studied at day 60. Antigen expression was lost in the salivary gland, lymph node, gall bladder, and large intestine, and positive staining was reduced in frequency and intensity in the spleen, lung, kidney, pancreas, BAT, and white fat. The tissues that most consistently displayed antigen expression throughout the time course were the heart (25 of 26 samples analyzed), BAT (20 of 26), lung (19 of 26), white fat (18 of 26), and kidney (18 of 26), while the liver (13 of 26), pancreas (15 of 26), spleen (9 of 25), skeletal muscle (13 of 25), and adrenal gland (6 of 17) were more variable.
TABLE 1.
Distribution of viral N antigen by tissue, neutralizing antibody titers, and viral RNA in blood
| Day p. i. and animal | IHC resulta
|
Blood resultb
|
||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Heart | Lung | Liver | Spleen | Kidney | Urinary bladder | Salivary gland | Intestine | Pancreas | Thymus | Brain | BAT | White fat | Uterus | Ovary | Testis | Lymph node | Skeletal muscle | Gall bladder | Adrenal gland | RT-PCR | Nab titer | |
| Day 60 | ||||||||||||||||||||||
| 2697 | 2+ | 2+ | 1+ | 1+ | 2+ | 0+ | 1+ | 0+ | 2+ | 0+ | 0+ | 2+ | 1+ | NA | NA | 0+ | ND | 1+ | 0+ | ND | + | 1,280 |
| 2698 | 3+ | 4+ | 2+ | 1+ | 3+ | 0+ | 2+ | 0+ | 1+ | 0+ | 0+ | 2+ | 3+ | NA | NA | 0+ | 1+ | 1+ | 0+ | 1+ | + | 1,280 |
| 2701 | 2+ | 1+ | 0+ | 1+ | 1+ | 0+ | 0+ | 0+ | 1+ | 0+ | 0+ | 2+ | 1+ | 0+ | 0+ | NA | ND | ND | 0+ | 1+ | + | 5,120 |
| 5184 | 3+ | 3+ | 3+ | 2+ | 2+ | 0+ | 0+ | 0+ | 2+ | 0+ | 0+ | 2+ | 2+ | NA | NA | 0+ | 0+ | 0+ | 0+ | ND | + | 160 |
| 5185 | 3+ | 3+ | 3+ | 3+ | 3+ | 0+ | 1+ | 1+ | ND | 0+ | 0+ | 3+ | 2+ | 0+ | 0+ | NA | 0+ | 0+ | 1+ | ND | + | 5,120 |
| Mean | 2.6+ | 2.6+ | 1.8+ | 1.6+ | 2.2+ | 0+ | 0.8+ | 0.2+ | 1.5+ | 0+ | 0+ | 2.2+ | 1.8+ | 0+ | 0+ | 0+ | 0.3+ | 0.5+ | 0.2+ | 1+ | ||
| Day 90 | ||||||||||||||||||||||
| 2689 | 1+ | 1+ | 1+ | 0+ | 1+ | 0+ | 0+ | 0+ | 1+ | 0+ | 0+ | 1+ | 1+ | NA | NA | 0+ | 0+ | 0+ | 0 | 0+ | + | 5,120 |
| 2690 | 1+ | 0 | 2+ | 0+ | 0+ | 0+ | 0+ | 0+ | 0+ | 0+ | 0+ | 0+ | 0+ | NA | NA | 0+ | ND | 0+ | ND | 0+ | − | 1,280 |
| 2691 | 2+ | 1+ | 0+ | 0+ | 1+ | 0+ | 0+ | 0+ | 1+ | 0+ | 0+ | 0+ | 0+ | NA | NA | 0+ | 0+ | 1+ | 0+ | 0+ | − | 5,120 |
| 2692 | 3+ | 3+ | 1+ | 0+ | 2+ | 0+ | 0+ | 0+ | 1+ | 0+ | 0+ | 2+ | 1+ | 0+ | ND | NA | ND | 1+ | 0+ | 3+ | + | 160 |
| 2699 | 2+ | 2+ | 1+ | 1+ | 1+ | 0+ | 0+ | 0+ | 2+ | 0+ | 0+ | 2+ | 1+ | NA | NA | 0+ | ND | 1+ | ND | ND | + | 1,280 |
| 2700 | 1+ | 0+ | 0+ | 0+ | 0+ | 0+ | 0+ | 0+ | 0+ | 0+ | 0+ | 1+ | 0+ | NA | NA | 0+ | ND | 0+ | 0+ | 0+ | − | 1,280 |
| Mean | 1.8+ | 1.2 | 0.8+ | 0.2+ | 0.8+ | 0+ | 0+ | 0+ | 0.8+ | 0+ | 0+ | 1+ | 0.5+ | 0+ | 0+ | 0+ | 0.5 | 0+ | 0.6+ | |||
| Day 120 | ||||||||||||||||||||||
| 2377 | 1+ | 0+ | 0+ | 0+ | 0+ | 0+ | 0+ | 0+ | 0+ | 0+ | 0+ | 0+ | 0+ | 0+ | 0+ | NA | 0+ | 0+ | 0+ | ND | − | 1,280 |
| 2679 | 1+ | 0+ | 0+ | 0+ | 0+ | 0+ | 0+ | ND | 0+ | 0+ | 0+ | 0+ | 0+ | NA | NA | 0+ | 0+ | 0+ | 0+ | ND | − | 1,280 |
| 2681 | 2+ | 1+ | 2+ | 0+ | 1+ | 0+ | 0+ | ND | 0+ | 0+ | 0+ | 1+ | 1+ | 0+ | 0+ | NA | 0+ | 0+ | 0+ | ND | + | 5,120 |
| 2674 | 3+ | 3+ | 3+ | 3+ | 1+ | 0+ | 0+ | 0+ | 2+ | 0+ | 0+ | 2+ | 1+ | 0+ | 0+ | NA | 0+ | 0+ | 0+ | 1+ | + | 20,480 |
| 2675 | 3+ | 3+ | 2+ | 3+ | 0+ | 0+ | 0+ | 0+ | 1+ | 0+ | 0+ | 1+ | 1+ | 0+ | 0+ | NA | 0+ | 0+ | 0+ | 0+ | + | 1,280 |
| Mean | 2+ | 1.4+ | 1.4+ | 1.2+ | 0.4+ | 0+ | 0+ | 0+ | 0.6+ | 0+ | 0+ | 0.8+ | 0.6+ | 0+ | 0+ | 0+ | 0+ | 0+ | 0+ | 0.5+ | ||
| Day 180 | ||||||||||||||||||||||
| 2671 | 2+ | 3+ | 1+ | 0+ | 1+ | 0+ | 0+ | 0+ | 2+ | 0+ | 0+ | 2+ | 2+ | NA | NA | 0+ | 0+ | 2+ | ND | ND | + | 1,280 |
| 2673 | 2+ | 3+ | 2+ | 0+ | 1+ | 0+ | 0+ | 0+ | 1+ | 0+ | 0+ | 1+ | 1+ | 0+ | 0+ | NA | 0+ | 1+ | 0+ | 0+ | + | 20,480 |
| 2676 | 2+ | 2+ | 0+ | 0+ | 0+ | 0+ | 0+ | 0+ | 1+ | 0+ | 0+ | 1+ | 1+ | 0+ | 0+ | NA | 0+ | 1+ | 0+ | 0+ | + | 5,120 |
| 2703 | 1+ | 0+ | 0+ | 0+ | 0+ | 0+ | 0+ | 0+ | 0+ | 0+ | 0+ | 1+ | 0+ | 0+ | 0+ | NA | 0+ | 1+ | ND | 0+ | − | 1,280 |
| 2704 | 2+ | 2+ | 0+ | 0+ | 1+ | 0+ | 0+ | 0+ | 1+ | 0+ | 0+ | 2+ | 1+ | 0+ | ND | NA | 0+ | 2+ | ND | ND | + | 5,120 |
| Mean | 1.8+ | 2+ | 0.6+ | 0+ | 0.6+ | 0+ | 0+ | 0+ | 1+ | 0+ | 0+ | 1.4+ | 1+ | 0+ | 0+ | 0+ | 0+ | 1.4+ | 0+ | 0+ | ||
| Day 217 | ||||||||||||||||||||||
| 2712 | 1+ | 0+ | 0+ | 0+ | 1+ | ND | 0+ | 0+ | 1+ | ND | 0+ | 0+ | 0+ | 0+ | 0+ | NA | 0+ | 0+ | 0+ | 0+ | + | 640 |
| 2714 | 0+ | 0+ | 0+ | 0+ | 0+ | 0+ | 0+ | 0+ | 0+ | 0+ | 0+ | 0+ | 0+ | 0+ | 0+ | NA | 0+ | 0+ | ND | 0+ | − | 40 |
| 2720 | 2+ | 1+ | 0+ | 0+ | 1+ | 0+ | 0+ | 0+ | 0+ | 0+ | 0+ | 1+ | 1+ | NA | NA | 0+ | 0+ | 2+ | ND | 1+ | + | 5,120 |
| 2725 | 2+ | 1+ | 2+ | 0+ | 1+ | 0+ | 0+ | 0+ | 0+ | 0+ | 0+ | 1+ | 1+ | NA | NA | 0+ | ND | 1+ | 0+ | 0+ | + | 1,280 |
| 2731 | 2+ | 1+ | 0+ | 2+ | 1+ | 0+ | 0+ | 0+ | 0+ | ND | 0+ | 2+ | 2+ | NA | NA | 0+ | 0+ | 2+ | ND | 1+ | + | 5,120 |
| Mean | 1.4+ | 0.6+ | 0.4+ | 0.4+ | 0.8+ | 0+ | 0+ | 0+ | 0.2+ | 0+ | 0+ | 0.8+ | 0.8+ | 0+ | 0+ | 0+ | 0+ | 1+ | 0+ | 0.4+ | ||
Antigen scoring: 0+, no positive cells; 1+, rare positive cells; 2+, few positive cells; 3+, moderate number of positive cells; 4+, many positive cells; ND, not done; NA, not applicable.
Nab, neutralizing antibody; +, viral RNA detected; −, no viral RNA detected.
FIG. 1.
Semiquantitative scoring scale for SN virus N-antigen expression by IHC. Selected heart samples are shown at a magnification of ×630. Positive N-antigen expression is visible as a reddish, punctate stain. Scores: 1+, rare positive cells; 2+, few positive cells; 3+, a moderate number of positive cells; 4+, many positive cells. Arrows indicate sites of positive antigen staining.
The IHC studies also revealed that antigen expression patterns were not uniform among animals at each time point examined. Among subjects examined at day 90 through day 217 p.i., we observed 7 animals (animals 2690 and 2700 at day 90, animals 2677 and 2679 at day 120, animal 2703 at day 180, and animals 2712 and 2714 at day 217) that displayed restricted antigen expression (antigen expression in ≤3 tissues), while the remaining 19 animals had a relatively disseminated pattern of expression (in 5 to 9 tissues) (Table 1).
QRT-PCR.
QRT-PCR was carried out by first adding a single primer specific for either negative-strand (vRNA) or positive-strand (mRNA, vcRNA) viral S segment RNA in the RT reaction. Because positive-strand viral RNA is not packaged into mature virions, this method allowed us to distinguish those sites at which viral replication and/or transcription is occurring from those at which the virus is quiescent, through the quantification of positive-strand viral RNA (29).
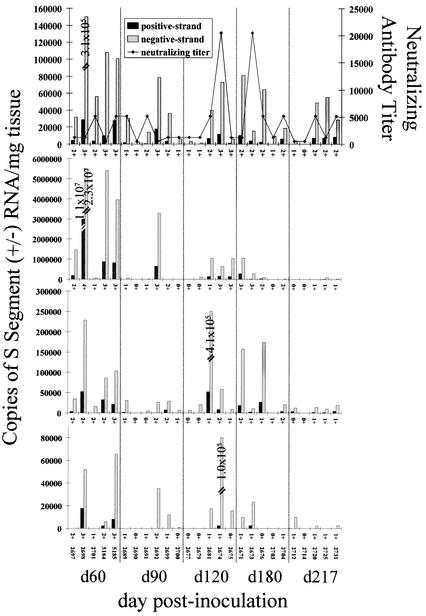
For the QRT-PCR studies, we focused on the heart, BAT, lung, and kidney because the IHC results demonstrated that these tissues stained positive for N antigen with the highest frequency, and therefore they seemed most likely to be serving as important sites for viral maintenance and replication during persistent infection. Consistent with the IHC results, we were able to detect negative-strand viral RNA in 25 of the 26 heart samples analyzed (Fig. 2). The single animal (animal 2714, at day 217 p.i.) that did not have viral RNA detectable by QRT-PCR also did not display antigen expression in any of the tissues examined (Fig. 2; Table 1). To confirm whether this animal had cleared its infection, we screened RNA extracted from the heart, lung, and BAT for total viral S segment RNA via nested RT-PCR, which is a more sensitive assay (sensitive to ≤10 copies of RNA in virions) than QRT-PCR (11). We found that viral RNA was present in all three tissues when this method was used (data not shown). Negative-strand RNA was detectable by QRT-PCR in BAT (23 of 26 samples analyzed) and the lung (20 of 25) at a higher frequency than N antigen. In contrast, we detected viral negative-strand RNA in the kidney (12 of 26 samples analyzed) at a lower frequency than was seen for N antigen expression. As with the antigen expression levels, we found that the quantities of both positive- and negative-strand RNA were quite variable between individual animals at each time point examined (Fig. 2).
FIG. 2.
Strand-specific TaqMan QRT-PCR titers, neutralizing antibody titers, and N-antigen expression in persistently infected mice. Negative- and positive-strand viral S segment RNA levels are expressed as copy numbers per milligram of tissue. Neutralizing antibodies are reported as the reciprocal of the end point dilution at which there was ≥80% reduction in the number of foci in Vero E6 cell monolayers. Semiquantitative scores (0 to 4+) for N antigen in IHC stains are listed on the x axis for each graph. Results obtained for each animal are reported in the same position in each graph. From top to bottom, graphs show results for the heart, lung, BAT, and kidney, respectively. Taqman QRT-PCR was not conducted on the lung sample from animal 2699.
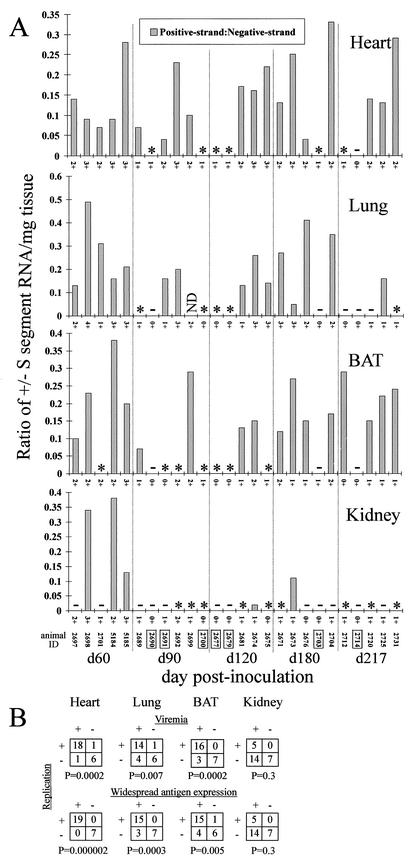
By screening for the presence of positive-strand viral RNA, we were able to determine which tissue samples were supporting active viral replication and/or transcription during persistence (Fig. 3A) (29). We found that positive-strand RNA was not detectable in 7 of the 26 heart samples analyzed (Fig. 2 and 3A). These heart samples were from the same seven animals that displayed a restricted antigen expression pattern (Table 1). The lung and BAT displayed a similar pattern of restricted viral replication in some animals; no replicative viral RNA was detectable in 10 animals for each of these tissues (Fig. 3A). This included lung or BAT samples from the seven or six of seven animals, respectively, that had restricted antigen expression. We were able to detect positive-strand RNA in only five of the kidney samples. In addition, there were six kidney samples that had stained positive for N antigen but did not have detectable negative- or positive-strand RNA by QRT-PCR (Fig. 2 and 3A). This pattern was not observed in any heart sample, while only one animal apiece displayed this phenotype in the lung and BAT. We found that the number of positive-strand copies never exceeded the level of negative-strand copies, nor were there any instances in which positive-strand RNA was detectable and negative-strand RNA was absent (Fig. 2 and 3). The ratio of positive-strand to negative-strand RNA copies was highly variable among individual tissue samples throughout the time course for each tissue examined (Fig. 3).
FIG. 3.
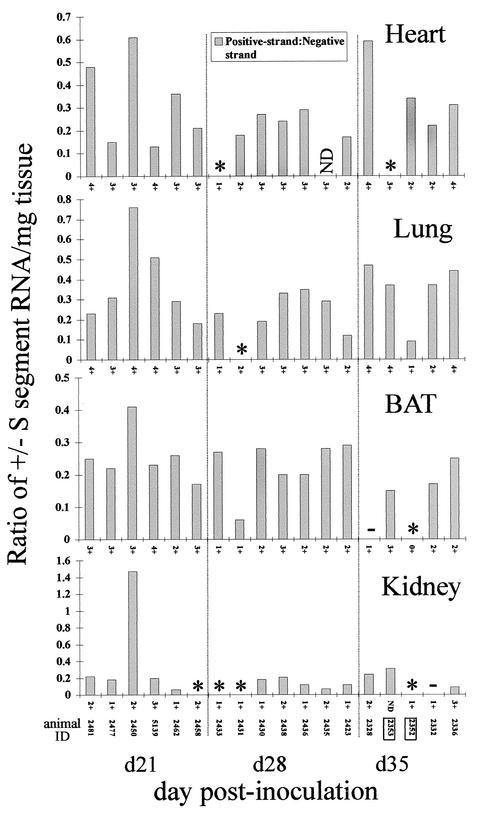
(A) Ratio of positive-strand (replicative) to negative-strand (genomic) SN virus S-segment RNA by tissue. An asterisk indicates that negative-strand RNA, but not positive-strand RNA, was detectable by QRT-PCR; a minus sign indicates that neither negative- nor positive-strand RNA was detectable by QRT-PCR. ND, not done. N-antigen scores (0 to 4+) are listed on the x axis for each graph. Identification numbers of animals whose blood samples tested negative for viral S segment RNA by nested RT-PCR are boxed. (B) Statistical analysis of the relationship between RNA viremia or widespread N-antigen expression and the presence of positive-strand RNA in persistently infected mice. A 2 × 2 contingency table was used to determine whether an association existed between the presence of positive-strand viral RNA in the heart, lung, BAT, or kidney and either the presence of RNA viremia or widespread antigen expression (defined as involvement of ≥5 tissues) in animals from day 60 to day 217 p.i.
To screen for viremia (defined here as the presence of viral RNA, not infectious virus), we subjected samples of RNA that had been extracted from whole blood to nested RT-PCR for viral RNA. QRT-PCR was not used because an earlier study demonstrated that this technique was able to detect viral RNA from blood in only 1 of 19 samples that were positive by nested RT-PCR (10). Viral isolation assays were not carried out due to results from another study, in which we attempted to isolate infectious virus from blood obtained on day 13 (n = 3) and day 21 (n = 2) p.i. from experimentally infected deer mice (11). Although we were able to isolate virus from lung tissue from each animal, we did not detect infectious virus in any of the blood samples. We found that 19 of 26 mice had detectable viral RNA in blood during persistent infection (Table 1; Fig. 3A). To ascertain whether there was an association between replicative activity in the heart, lung, BAT, or kidney and either viremia or widespread antigen expression, or both, we carried out statistical analysis using 2 × 2 contingency tables. We found that there was a significant association between the presence of positive-strand viral RNA in the heart, lung, or BAT and either viremia or widespread antigen expression (Fig. 3B). We did not, however, find any such association for the kidney.
Acute infection.
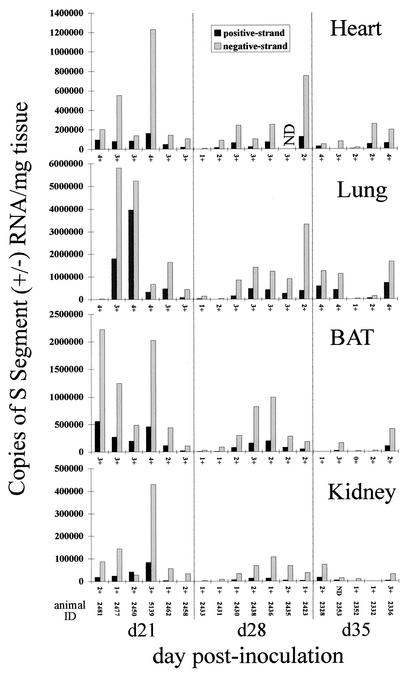
To address viral RNA titers and transcription/replication activity in acutely infected deer mice, we carried out strand-specific QRT-PCR on heart, lung, BAT, and kidney samples that were obtained from mice sacrificed on day 21, 28, or 35 p.i. in an earlier study (Fig. 4 and 5) (10). The highest negative- and positive-strand viral RNA titers (between days 21 and 217 p.i.) for the heart, BAT, and kidney were found on day 21 p.i., while peak titers for the lung were found on day 60 p.i. (Fig. 2, 4, and 6). Generally, the mean titers of viral RNA in samples of the heart, lung, BAT, and kidney obtained between days 90 and 217 p.i. were lower than those in samples obtained between days 21 and 60 p.i. (Fig. 6). Positive-strand RNA was detectable in all acute-phase samples examined, with the exception of two heart samples, one lung sample, two BAT samples, and five kidney samples (Fig. 4 and 5). For BAT and the kidney, the ratios of positive- to negative-strand RNA were fairly uniform between days 21 and 35 p.i. but became more variable beginning on day 60 p.i. (Fig. 3A and 5). The strand ratios in heart and lung samples were quite variable throughout the entire time course (day 21 to day 217 p.i.), with the exception of the day 28 p.i. lung samples.
FIG. 4.
Strand-specific TaqMan QRT-PCR titers and N-antigen expression in acutely infected mice. Negative- and positive-strand viral S segment RNA levels are expressed as copy numbers per milligram of tissue. N-antigen scores (0 to 4+) are listed on the x axis for each graph. The results obtained from each animal are reported in the same position along the x axis in each graph. ND, not done.
FIG. 5.
Ratio of positive-strand (replicative) to negative-strand (genomic) SN virus S-segment RNA by tissue in acutely infected mice. An asterisk indicates that negative-strand RNA, but not positive-strand RNA, was detectable by QRT-PCR; a minus sign indicates that neither negative- nor positive-strand RNA was detectable by QRT-PCR. ND, not done. N-antigen scores (0 to 4+) are listed on the x axis for each graph. Identification numbers of animals in whose blood no S-segment RNA was detectable by nested-RT-PCR are boxed.
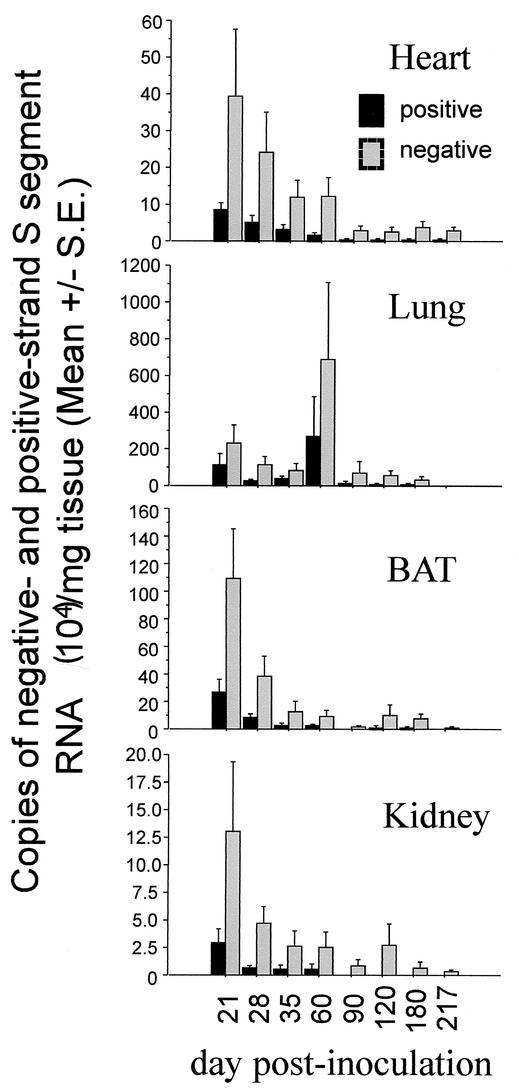
FIG. 6.
Strand-specific TaqMan QRT-PCR titers, reported as mean values for groups of animals rather than by individual animal as in Fig. 2 and 4. Negative- and positive-strand S-segment RNA copy numbers are expressed as means for all tissues of each kind examined at each time point.
Neutralizing antibodies.
The neutralizing antibody titers detectable in the persistently infected mice ranged from 1:40 to 1:20,480 (Table 1; Fig. 2). We were unable to find any correlation between the neutralizing antibody titer and the level of viral RNA and/or viral N antigen expression at the time of sacrifice, nor was there a relationship between antibody titers and the titer of infectious virus. We did, however, find a significant association (P = 0.01) between neutralizing antibody titers of ≤1:1,280 and restricted antigen expression.
Isolation of infectious virus.
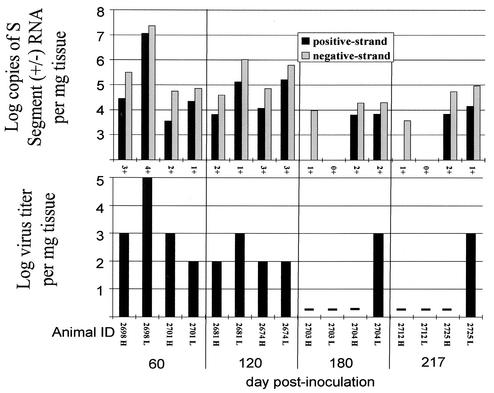
To determine whether infectious virus could be isolated from persistently infected mice, we prepared heart and lung tissue homogenates from two animals at each of the following time points after infection: days 60, 120, 180, and 217. Serial 10-fold dilutions, between 10−1 and 10−7, of these homogenates were inoculated onto Vero E6 cells for in vitro viral isolation and determination of end point titers. We were able to isolate SN virus from one or more tissue at each of the time points examined (Fig. 7). Infectious virus was recovered from six of eight lung samples and from four of eight heart samples. Titers of infectious virus ranged from 102 to 105 tissue culture infectious doses per mg of tissue. Each of the 10 samples from which virus was isolated also had both positive- and negative-strand viral RNA detectable by TaqMan RT-PCR. Of the six samples from which we could not isolate virus, four lacked positive-sense S-segment RNA. Viral RNA titers were 10- to 1,000-fold higher than infectious titers (Fig. 7).
FIG. 7.
Strand-specific TaqMan QRT-PCR titers, N-antigen expression, and infectious virus titers in persistently infected mice. (Top graphs) Log negative- and positive-strand viral S-segment RNA copy numbers, normalized per milligram of tissue. Semiquantitative scores (0 to 4+) for N-antigen expression in IHC stains are given along the x axis. (Bottom graphs) Log virus titers from the in vitro isolation experiment are reported per milligram of tissue. A minus sign indicates that no infectious virus was detected. Animal identification numbers and the tissue screened (H, heart; L, lung) are reported on the x axis. Results obtained from each animal are reported in the same position in each graph.
DISCUSSION
To establish a persistent infection in a host, a virus must be able to regulate its cytopathic effect on host cells, to maintain functionally intact genomes in cells, and to avoid elimination by the host's immune system (2). We have shown that SN virus establishes long-term maintenance of its genome in the host for at least 8 months, at least in part in cells that morphologically resemble vascular endothelial cells. We did not find evidence that SN virus infection results in histopathologic changes in host tissues. These findings support the widely accepted premise that the coevolution of SN virus and the deer mouse has resulted in a relationship that supports a lifelong viral infection devoid of any severely deleterious consequences to the host (26).
One review suggests that in other hantavirus animal models, the transition from acute to persistent infection occurs after the first 3 to 4 weeks of infection, in part because it is at this time that infectious titers, viral antigen expression, and/or RNA levels peak and begin to decline (43). Furthermore, after this time, there are periodic episodes of viral recrudescence, suggesting that cyclical bursts of replication may take place during persistent infection. Our findings may also be demonstrating that pattern. When we examine the mean viral RNA titers from the heart, lung, BAT, and kidney from day 21 to day 217 p.i., we observe that the highest titers of viral RNA are found 21 days p.i. and decline thereafter (Fig. 6). The exception to this pattern is seen in lung tissue, where mean viral RNA titers peaked at day 21 and subsequently declined until day 60, at which time titers peaked again. This mean value reflected an apparent recrudescence of viral RNA in just two animals in which viral RNA loads were extraordinarily high.
The expression and distribution of viral antigen does not follow the same kinetics seen for viral RNA. We did not see a restriction in the number of tissues expressing antigen or the overall levels of antigen expression until day 90 p.i. Taken together, these data suggest that the transition between acute and persistent infection occurs some time between day 60 and day 90 p.i.
The levels and distribution patterns of viral RNA or N antigen were very variable among our animals, which may in part be explained by their outbred status (13). Between days 90 and 217 p.i., there appeared to be two phenotypes, those of restricted versus disseminated viral replication and antigen expression. The restricted phenotype, seen in 7 of the 26 animals, was characterized by N-antigen expression confined to ≤3 tissues, absence of viral RNA in the blood (n = 6), neutralizing antibody titers of ≤1:1,280 (P = 0.01), and an absence of the replicative form of RNA in the heart, lung, and kidney. Taken together, these data suggest that SN virus may be able to enter into a period of functional “latency” during persistence, during which replication is diminished and the negative-sense genomic RNA, N antigen, and infectivity are maintained at low levels predominantly in the heart, lungs, and/or BAT.
The disseminated pattern of infection was observed in 19 animals. This pattern was characterized by a widespread distribution of N-antigen expression involving ≥5 tissues, neutralizing antibody titers ranging from 1:160 to 1:20,480, positive-strand RNA in the heart (n = 19), lung (n = 15), and BAT (n = 15), and viral RNA in the blood (n = 18). This pattern was present in three or more mice at each time point. In comparisons of mice with the disseminated phenotype that were taken on day 60 p.i. or earlier to those taken on day 90 p.i. or later, the latter group had lower mean viral RNA titers (both positive- and negative-strand RNA) in the heart, lung, and BAT (Fig. 6).
Several lines of evidence suggest that the heart, lung, and BAT are important sites for viral maintenance and replication during persistent infection. These tissues, in addition to the kidney, retain N-antigen staining most frequently at day 60 p.i. and later. More animals still had viral genomic RNA in the same three tissues. In the kidney, however, negative-strand RNA was detected less frequently (12 of 26 samples analyzed) than antigen staining (18 of 26). Only 5 of the 26 kidney samples had positive-strand RNA, while 6 samples had detectable levels of N antigen but no detectable viral RNA forms. It may be that immune complexes or other extra-virion sources of N antigen collect passively in the kidney in some cases. The statistical analysis demonstrated a significant association between the presence of positive-strand RNA in the heart, lung, and BAT and either RNA viremia or a widespread distribution of N antigen (≥5 tissues), while no such associations were identified for the kidney. The fact that the heart most consistently retains negative-strand RNA (25 of 26 samples analyzed) and N antigen (25 of 26) in persistent infection may indicate that infection of this tissue is critical not only in preventing clearance of the virus but also for maintaining viral replication, which is necessary for seeding secondary sites of infection throughout persistence. The lung and BAT may also represent important viral reservoirs during persistence; in fact, the lung was a more reliable source of infectious virus (Fig. 7). Because each mouse in this study was examined at only a single time point, we were not able to determine whether SN virus undergoes cyclical bursts of replication in many deer mice or if, instead, persistent infection results in distinct but relatively static phenotypes, perhaps reflecting the different genetic backgrounds of individual deer mice.
We have shown that SN virus persistently maintains its genome and N antigen in host endothelial cells and that infectious virus persists through day 217 p.i. Previously, Hutchinson et al. demonstrated that genomic viral S segment titers mirrored viral isolation titers when blood samples from cotton rats infected with BCC virus were examined (29). We found that viral RNA titers exceeded infectious titers 10- to 1,000-fold, regardless of whether we measured genomic or antigenomic RNA species. This observed difference may indicate an accumulation of defective RNA- or neutralizing antibody-bound (inactivated) forms and/or an accumulation of other noninfectious forms such as nucleocapsid inclusions bearing viral RNA. In fact, some fraction of the positive N-antigen staining we observed could be due to nucleocapsid inclusions, such as are sometimes seen with other members of the genus Hantavirus (36).
Other negative-stranded RNA viruses maintain vRNA consistently throughout persistent infection, while the abundances of mRNA and vcRNA are variable. Another member of the family Bunyaviridae, La Crosse virus, expresses vRNA in mosquito midguts more frequently than either mRNA or vcRNA during persistent infection (16). This pattern of selective persistence of vRNA is also seen with enterovirus and influenza C virus (3, 39). Collectively, these studies suggest that restricting viral replication and/or transcription may be a common strategy of RNA viruses for the establishment and maintenance of persistent infection in the host. As a consequence, one might anticipate that the virus should reduce its recognition by the immune system.
The La Crosse virus study also demonstrated that replicative activity (generating vcRNA) and transcription (generating mRNA) are dependent on the host cell biosynthetic status, as persistently infected mosquitoes receiving blood meals actively produced replicative RNA at a significantly higher frequency than controls that did not receive a blood meal (16). In addition to being an important tissue during SN virus persistence, BAT has also been observed as a prominent site of viral antigen in overwintering bank voles naturally infected with PUU virus (20). In order to maintain viable core body temperatures in winter months, rodents rely, in part, on BAT for nonshivering thermogenesis (37, 41). Nonshivering thermogenesis is accomplished through the uncoupling of cellular respiration from ATP synthesis in the mitochondria via the uncoupling protein (UCP) (40, 41). Because cellular heat shock proteins (HSPs) are required to shuttle UCP from the cytosol into the mitochondria, their expression is increased during nonshivering thermogenesis. Viruses from a wide variety of diverse families have evolved many unique ways of engaging, utilizing, or regulating the expression of the various foldases and chaperones that comprise the heat shock gene family (1, 23, 27, 35, 48-51). We have obtained preliminary evidence that cold stress may be capable of inducing SN virus replication in vivo, a finding similar to that observed for other persistent RNA virus infections (4, 28; Botten et al., unpublished). On the basis of these observations, we speculate that one way in which SN virus replication may be regulated by environmental factors is that the virus that overwinters in BAT may be subject to regulation by the availability of chaperones such as hsp70, which could be used to facilitate reactivation in the spring.
The mechanisms by which viral replication is regulated during hantavirus infection are not clearly understood. It is unlikely that viral infectivity and transmissibility are regulated solely through immune mechanisms. We favor instead a model wherein both immune responses and an evolved intracellular interaction between virus and host determine whether SN virus is down-regulated or undergoes recrudescence. A study of molecular correlates of hantavirus persistence in vitro showed a correlation between the productivity of infectious units and the absence of a particular group of segment-terminal deletions in SEO virus (42). While this study showed a pattern of down-regulation followed by periodic reactivation that superficially resembled that seen in natural rodent infection, the relevance of such in vitro models to infections of rodents will remain open to question in the absence of confirmation in an in vivo model. It is of some interest, then, that a similar phenomenon has been described for in vivo and in vitro models of infection with lymphocytic choriomeningitis virus (43).
In addition to the NC gene, several viruses in the family Bunyaviridae have a second open reading frame on the viral S segment that encodes the nonstructural protein (NSs) (8). In Bunyamwera virus infection, an NSs protein inhibits the viral RDRP, and deletion mutants may have a reduced ability to induce transcription of gamma interferon, resulting in reduced pathogenicity (53, 54). NSs-deficient Rift Valley fever virus showed a reduced ability to induce expression of alpha/beta interferon (14). In the genomes of SN virus and its close relatives, including all vole-borne hantaviruses sequenced to date, a reading frame for a putative NSs protein is universally open in dozens of sequenced isolates (B. Hjelle, unpublished data). Identification of this putative protein and analysis of its in vivo activities should clarify whether it serves a role similar to that of the Bunyamwera virus or Rift Valley Fever virus NSs protein.
Little is known about the T-cell immune response to hantavirus infection. This is partially because most natural host rodents are exotic and outbred, so inbred lines of rodents and antigen-presenting cells and the necessary immunological reagents needed to characterize the T-cell response are not always available. Both anti-N and neutralizing anti-glycoprotein antibodies are present at high titers in persistently infected host rodents (34, 38). In naturally infected wild-caught deer mice, SN virus is maintained as a quasispecies, which may provide a mechanism for avoiding host immunity and promoting viral persistence (18). It is not yet known whether such genomic variants are expressed or if variation occurs in epitopes of immunologic importance in rodents.
In order to place our present findings in context, future studies are needed to (i) gain an understanding of how SN virus is restricted to tissue-specific endothelial cells during persistence, (ii) define the cell-mediated immune response, (iii) determine how the SN virus is able to avoid host immunity, and (iv) determine the mechanisms by which SN virus replication is regulated during infection.
Acknowledgments
We thank J. Summers, M. Buchmeier, M. Saavedra, J. Berger, K. Gottlieb, C. Schmaljohn, and R. Ricci for helpful comments and/or technical assistance. We also thank the U.S. Fish and Wildlife Service (Sevilleta National Wildlife Refuge) for its cooperation in this study.
This work was supported by U.S. Public Health Service grants RO1 A1 41692 and R21 AI50763 and by the Center for Emerging Infectious Disease grant, funded by the Centers for Disease Control and Prevention. J.B. was an Infectious Diseases and Inflammation Training Grant Fellow (Public Health Service grant T32 A107538) during the course of this work.
REFERENCES
- 1.Agostini, I., S. Popov, J. Li, L. Dubrovsky, T. Hao, and M. Bukrinsky. 2000. Heat-shock protein 70 can replace viral protein R of HIV-1 during nuclear import of the viral preintegration complex. Exp. Cell Res. 259:398-403. [DOI] [PubMed] [Google Scholar]
- 2.Ahmed, R., L. A. Morrison, and D. M. Knipe. 1997. Viral persistence, p. 181-205. In N. Nathanson, R. Ahmed, F. Gonzalez-Scarano, D. E. Griffin, K. V. Holmes, F. A. Murphy, and H. L. Robinson (ed.), Viral pathogenesis. Lippincott-Raven Publishers, Philadelphia, Pa.
- 3.Andreoletti, L., T. Bourlet, D. Moukassa, L. Rey, D. Hot, Y. Li, V. Lambert, B. Gosselin, J. F. Mosnier, C. Stankowiak, and P. Wattre. 2000. Enteroviruses can persist with or without active viral replication in cardiac tissue of patients with end-stage ischemic or dilated cardiomyopathy. J. Infect. Dis. 182:1222-1227. [DOI] [PubMed] [Google Scholar]
- 4.Ben Nathan, D., S. Lustig, and H. D. Danenberg. 1991. Stress-induced neuroinvasiveness of a neurovirulent noninvasive Sindbis virus in cold or isolation subjected mice. Life Sci. 48:1493-1500. [DOI] [PubMed] [Google Scholar]
- 5.Bernshtein, A. D., N. S. Apekina, T. V. Mikhailova, Y. A. Myasnikov, L. A. Khlyap, Y. S. Korotkov, and I. N. Gavrilovskaya. 1999. Dynamics of Puumala hantavirus infection in naturally infected bank voles (Clethrionomys glareolus). Arch. Virol. 144:2415-2428. [DOI] [PubMed] [Google Scholar]
- 6.Bharadwaj, M., J. Botten, N. Torrez-Martinez, and B. Hjelle. 1997. Rio Mamore virus: genetic characterization of a newly recognized hantavirus of the pygmy rice rat, Oligoryzomys microtis, from Bolivia. Am. J. Trop. Med. Hyg. 57:368-374. [DOI] [PubMed] [Google Scholar]
- 7.Bharadwaj, M., R. Nofchissey, D. Goade, F. Koster, and B. Hjelle. 2000. Humoral immune responses in the hantavirus cardiopulmonary syndrome. J. Infect. Dis. 182:43-48. [DOI] [PubMed] [Google Scholar]
- 8.Bishop, D. H. L. 1996. Biology and molecular biology of Bunyaviruses, p. 19-62. In R. M. Elliot (ed.), The Bunyaviridae. Plenum Press, New York, N.Y.
- 9.Borucki, M. K., J. D. Boone, J. E. Rowe, M. C. Bohlman, E. A. Kuhn, R. DeBaca, and S. C. St Jeor. 2000. Role of maternal antibody in natural infection of Peromyscus maniculatus with Sin Nombre virus. J. Virol. 74:2426-2429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Botten, J., K. Mirowsky, D. Kusewitt, M. Bharadwaj, J. Yee, R. Ricci, R. M. Feddersen, and B. Hjelle. 2000. Experimental infection model for Sin Nombre hantavirus in the deer mouse (Peromyscus maniculatus). Proc. Natl. Acad. Sci. USA 97:10578-10583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Botten, J., K. Mirowsky, C. Ye, K. A. Gottlieb, M. Saavedra, L. Ponce, and B. Hjelle. 2002. Shedding and intracage transmission of Sin Nombre hantavirus in the deer mouse (Peromyscus maniculatus) model. J. Virol. 76:7587-7594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Botten, J., R. Nofchissey, H. Ahern, P. Rodriguez-Moran, I. A. Wortman, D. Goade, T. Yates, and B. Hjelle. 2000. Outdoor facility for quarantine of wild rodents infected with hantavirus. J. Mammal. 81:250-259. [Google Scholar]
- 13.Botten, J., R. Ricci, and B. Hjelle. 2001. Establishment of a deer mouse (Peromyscus maniculatus rufinus) breeding colony from wild-caught founders: comparison of reproductive performance of wild-caught and laboratory-reared pairs. Comp. Med. 51:314-318. [PubMed] [Google Scholar]
- 14.Bouloy, M., C. Janzen, P. Vialat, H. Khun, J. Pavlovic, M. Huerre, and O. Haller. 2001. Genetic evidence for an interferon-antagonistic function of Rift Valley Fever virus nonstructural protein NSs. J. Virol. 75:1371-1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Centers for Disease Control and Prevention. 1994. Laboratory management of agents associated with hantavirus pulmonary syndrome: interim biosafety guidelines. Morb. Mortal. Wkly. Rep. 43:1-7. [PubMed] [Google Scholar]
- 16.Chandler, L. J., L. P. Wasieloski, C. D. Blair, and B. J. Beaty. 1996. Analysis of La Crosse virus S-segment RNA and its positive-sense transcripts in persistently infected mosquito tissues. J. Virol. 70:8972-8976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Childs, J. E., T. G. Ksiazek, C. F. Spiropoulou, J. W. Krebs, S. Morzunov, G. O. Maupin, K. L. Gage, P. E. Rollin, J. Sarisky, and R. E. Enscore. 1994. Serologic and genetic identification of Peromyscus maniculatus as the primary rodent reservoir for a new hantavirus in the southwestern United States. J. Infect. Dis. 169:1271-1280. [DOI] [PubMed] [Google Scholar]
- 18.Feuer, R., J. D. Boone, D. Netski, S. P. Morzunov, and S. C. St Jeor. 1999. Temporal and spatial analysis of Sin Nombre virus quasispecies in naturally infected rodents. J. Virol. 73:9544-9554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gavrilovskaya, I. N., N. S. Apekina, A. Bernshtein, V. T. Demina, N. Okulova, Y. A. Myasnikov, and M. P. Chumakov. 1990. Pathogenesis of hemorrhagic fever with renal syndrome virus infection and mode of horizontal transmission of hantavirus in bank voles. Arch. Virol. 110(Suppl. 1):57-62.
- 20.Gavrilovskaya, I. N., N. S. Apekina, Y. Myasnikov, A. D. Bernshtein, E. V. Ryltseva, E. A. Gorbachkova, and M. P. Chumakov. 1983. Features of circulation of hemorrhagic fever with renal syndrome (HFRS) virus among small mammals in the European U.S.S.R. Arch. Virol. 75:313-316. [DOI] [PubMed] [Google Scholar]
- 21.Gavrilovskaya, I. N., M. Shepley, R. Shaw, M. H. Ginsberg, and E. R. Mackow. 1998. β3 Integrins mediate the cellular entry of hantaviruses that cause respiratory failure. Proc. Natl. Acad. Sci. USA 95:7074-7079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Green, W., R. Feddersen, O. Yousef, M. Behr, K. Smith, J. Nestler, S. Jenison, T. Yamada, and B. Hjelle. 1998. Tissue distribution of hantavirus antigen in naturally infected humans and deer mice. J. Infect. Dis. 177:1696-1700. [DOI] [PubMed] [Google Scholar]
- 23.Gurer, C., A. Cimarelli, and J. Luban. 2002. Specific incorporation of heat shock protein 70 family members into primate lentiviral virions. J. Virol. 76:4666-4670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hjelle, B., S. Jenison, N. Torrez-Martinez, B. Herring, S. Quan, A. Polito, S. Pichuantes, T. Yamada, C. Morris, F. Elgh, H. W. Lee, H. Artsob, and R. Dinello. 1997. Rapid and specific detection of Sin Nombre virus antibodies in patients with hantavirus pulmonary syndrome by a strip immunoblot assay suitable for field diagnosis. J. Clin. Microbiol. 35:600-608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hjelle, B., S. Jenison, N. Torrez-Martinez, T. Yamada, K. Nolte, R. Zumwalt, K. MacInnes, and G. Myers. 1994. A novel hantavirus associated with an outbreak of fatal respiratory disease in the southwestern United States: evolutionary relationships to known hantaviruses. J. Virol. 68:592-596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hjelle, B., and T. Yates. 2001. Modeling hantavirus maintenance and transmission in rodent communities, p. 77-90. In C. S. Schmaljohn and S. T. Nichol (ed.), Hantaviruses. Springer-Verlag, Berlin, Germany. [DOI] [PubMed]
- 27.Hu, J., and D. Anselmo. 2000. In vitro reconstitution of a functional duck hepatitis B virus reverse transcriptase: posttranslational activation by hsp90. J. Virol. 74:11447-11455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hughes, J. V., S. C. Doll, and T. C. Johnson. 1985. Hypothermia-inducing peptide promotes recovery of vesicular stomatitis virus from persistent animal infections. J. Virol. 53:781-785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hutchinson, K. L., P. E. Rollin, and C. J. Peters. 1998. Pathogenesis of a North American hantavirus, Black Creek Canal virus, in experimentally infected Sigmodon hispidus. Am. J. Trop. Med. Hyg. 59:58-65. [DOI] [PubMed] [Google Scholar]
- 30.Hutchinson, K. L., P. E. Rollin, W. J. Shieh, S. Zaki, P. W. Greer, and C. J. Peters. 2000. Transmission of Black Creek Canal virus between cotton rats. J. Med. Virol. 60:70-76. [DOI] [PubMed] [Google Scholar]
- 31.Jenison, S., T. Yamada, C. Morris, B. Anderson, N. Torrez-Martinez, N. Keller, and B. Hjelle. 1994. Characterization of human antibody responses to Four Corners hantavirus infections among patients with hantavirus pulmonary syndrome. J. Virol. 68:3000-3006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jonsson, C. B., and C. S. Schmaljohn. 2001. Replication of hantaviruses. Curr. Top. Microbiol. Immunol. 256:15-32. [DOI] [PubMed] [Google Scholar]
- 33.Kariwa, H., M. Fujiki, K. Yoshimatsu, J. Arikawa, I. Takashima, and N. Hashimoto. 1998. Urine-associated horizontal transmission of Seoul virus among rats. Arch. Virol. 143:365-374. [DOI] [PubMed] [Google Scholar]
- 34.Kariwa, H., M. Kimura, S. Yoshizumi, J. Arikawa, K. Yoshimatsu, I. Takashima, and N. Hashimoto. 1996. Modes of Seoul virus infections: persistency in newborn rats and transiency in adult rats. Arch. Virol. 141:2327-2338. [DOI] [PubMed] [Google Scholar]
- 35.Khandjian, E. W., and H. Turler. 1983. Simian virus 40 and polyoma virus induce synthesis of heat shock proteins in permissive cells. Mol. Cell. Biol. 3:1-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kobayashi, K., C. Itakura, J. Arikawa, and N. Hashimoto. 1991. Renal lesions in rats infected with Rattus serotype hantavirus (SR-11 strain). Zentbl. Veterinarmed. B 38:657-664. [DOI] [PubMed] [Google Scholar]
- 37.LeBlanc, J. 1992. Mechanisms of adaptation to cold. Int. J. Sports Med. 13(Suppl. 1):S169-S172. [DOI] [PubMed] [Google Scholar]
- 38.Lee, H. W., P. W. Lee, L. J. Baek, C. K. Song, and I. W. Seong. 1981. Intraspecific transmission of Hantaan virus, etiologic agent of Korean hemorrhagic fever, in the rodent Apodemus agrarius. Am. J. Trop. Med. Hyg. 30:1106-1112. [DOI] [PubMed] [Google Scholar]
- 39.Marschall, M., A. Schuler, and E. Meier. 1996. Influenza C virus RNA is uniquely stabilized in a steady state during primary and secondary persistent infections. J. Gen. Virol. 77:681-686. [DOI] [PubMed] [Google Scholar]
- 40.Matz, J. M., M. J. Blake, H. M. Tatelman, K. P. LaVoi, and N. J. Holbrook. 1995. Characterization and regulation of cold-induced heat shock protein expression in mouse brown adipose tissue. Am. J. Physiol. 269:R38-R47. [DOI] [PubMed] [Google Scholar]
- 41.Matz, J. M., K. P. LaVoi, R. J. Moen, and M. J. Blake. 1996. Cold-induced heat shock protein expression in rat aorta and brown adipose tissue. Physiol. Behav. 60:1369-1374. [DOI] [PubMed] [Google Scholar]
- 42.Meyer, B. J., and C. Schmaljohn. 2000. Accumulation of terminally deleted RNAs may play a role in Seoul virus persistence. J. Virol. 74:1321-1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Meyer, B. J., and P. J. Southern. 1997. A novel type of defective viral genome suggests a unique strategy to establish and maintain persistent lymphocytic choriomeningitis virus infections. J. Virol. 71:6757-6764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mills, J. N., T. G. Ksiazek, B. A. Ellis, P. E. Rollin, S. T. Nichol, T. L. Yates, W. L. Gannon, C. E. Levy, D. M. Engelthaler, T. Davis, D. T. Tanda, J. W. Frampton, C. R. Nichols, C. J. Peters, and J. E. Childs. 1997. Patterns of association with host and habitat: antibody reactive with Sin Nombre virus in small mammals in the major biotic communities of the southwestern United States. Am. J. Trop. Med. Hyg. 56:273-284. [DOI] [PubMed] [Google Scholar]
- 45.Mills, J. N., T. L. Yates, J. E. Childs, R. R. Parmenter, T. G. Ksiazek, P. E. Rollin, and C. J. Peters. 1995. Guidelines for working with rodents potentially infected with hantavirus. J. Mammal. 76:716-722. [Google Scholar]
- 46.Nerurkar, V. R., J. W. Song, K. J. Song, J. W. Nagle, B. Hjelle, S. Jenison, and R. Yanagihara. 1994. Genetic evidence for a hantavirus enzootic in deer mice (Peromyscus maniculatus) captured a decade before the recognition of hantavirus pulmonary syndrome. Virology 204:563-568. [DOI] [PubMed] [Google Scholar]
- 47.Netski, D., B. H. Thran, and S. C. St Jeor. 1999. Sin Nombre virus pathogenesis in Peromyscus maniculatus. J. Virol. 73:585-591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nevins, J. R. 1982. Induction of the synthesis of a 70,000 Dalton mammalian heat shock protein by the adenovirus E1A gene product. Cell 29:913-919. [DOI] [PubMed] [Google Scholar]
- 49.Niewiarowska, J., J. C. D'Halluin, and M. T. Belin. 1992. Adenovirus capsid proteins interact with HSP70 proteins after penetration in human or rodent cells. Exp. Cell Res. 201:408-416. [DOI] [PubMed] [Google Scholar]
- 50.Parks, C. L., R. A. Lerch, P. Walpita, M. S. Sidhu, and S. A. Udem. 1999. Enhanced measles virus cDNA rescue and gene expression after heat shock. J. Virol. 73:3560-3566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sainis, I., C. Angelidis, G. Pagoulatos, and I. Lazaridis. 1994. The hsc70 gene which is slightly induced by heat is the main virus inducible member of the hsp70 gene family. FEBS Lett. 355:282-286. [DOI] [PubMed] [Google Scholar]
- 52.Schmaljohn, C., and B. Hjelle. 1997. Hantaviruses: a global disease problem. Emerg. Infect. Dis. 3:95-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Weber, F., A. Bridgen, J. K. Fazakerley, H. Streitenfeld, N. Kessler, R. E. Randall, and R. M. Elliott. 2002. Bunyamwera bunyavirus nonstructural protein NSs counteracts the induction of alpha/beta interferon. J. Virol. 76:7949-7955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Weber, F., E. F. Dunn, A. Bridgen, and R. M. Elliott. 2001. The Bunyamwera virus nonstructural protein NSs inhibits viral RNA synthesis in a minireplicon system. Virology 281:67-74. [DOI] [PubMed] [Google Scholar]
- 55.Yanagihara, R., H. L. Amyx, and D. C. Gajdusek. 1985. Experimental infection with Puumala virus, the etiologic agent of nephropathia epidemica, in bank voles (Clethrionomys glareolus). J. Virol. 55:34-38. [DOI] [PMC free article] [PubMed] [Google Scholar]