Abstract
The replication proteins encoded in the P2 region of the poliovirus genome induce extensive rearrangement of cellular membranes into vesicles and are a required component of viral RNA replication complexes. To identify distinct viral protein(s) from the P2 region of the genome that were required to form functional RNA replication complexes, the P2 proteins were expressed in addition to P3 in HeLa S10 translation-RNA replication reactions. Membrane-associated preinitiation replication complexes were isolated from these reactions and used to measure negative-strand synthesis. The formation of replication complexes capable of initiating negative-strand synthesis was observed when either P23 or when P2 and P3 were expressed in the HeLa S10 translation-replication reactions. The amount of negative-strand RNA synthesized with P2 and P3 was approximately 50% of that observed with P23. Negative-strand synthesis was not observed when the processed forms of the P2 proteins (e.g., 2A, 2B, 2C, 2AB, and 2BC) were used in various combinations in place of P2. In contrast, the expression of 2A and 2BCP3 supported negative-strand synthesis at the same level observed with P23. Therefore, functional replication complexes were formed in reaction mixtures that contained either 2A and 2BCP3 or P2 and P3. Genetic complementation analysis of P23 RNA that contained a lethal mutation in 2C confirmed these results. The expression of 2BCP3 in trans restored the replication of P23-2C(P131N) RNA to wild-type levels. The expression of P2 and P3 also complemented the replication of this mutant RNA, although very inefficiently. Complementation was not observed in reactions that contained P2 alone, 2BC, or 2C. Based on these results, we propose that RNA replication complexes are initially formed with the primary cleavage products of P23 (i.e., P2 and P3 or 2A and 2BCP3), and that 2A and 2BCP3 are preferentially used in this process.
After poliovirion RNA is released into the cytoplasm of infected cells, it is translated by polyribosomes on the rough endoplasmic reticulum (rER) into a polyprotein that is subsequently processed by the viral proteases 2Apro and 3Cpro/3CDpro (31, 59). Primary cleavage of the viral polyprotein occurs at the P1-2A junction. This cleavage is mediated by 2Apro and results in the separation of the structural (P1) and nonstructural (P23) proteins (59). Functionally distinct precursors, intermediates, and individual proteins are formed after additional processing of P23 by 3Cpro/3CDpro. Two different proteolytic processing cascades have been proposed which are based on the initial site of cleavage in P23 (35). Proteolytic cleavage of P23 at the 2C-3A junction site generates P2 and P3 as the initial products. Alternatively, cleavage of P23 at the 2A-2B site results in the formation of 2A and 2BCP3. Further processing of P2, P3, and 2BCP3 into the individual replication proteins then follows. Viral RNA synthesis occurs in replication complexes that are assembled on membrane vesicles that proliferate in poliovirus-infected cells (15, 18, 19). The replication complexes are composed of viral and cellular proteins, replicative-intermediate RNAs, and newly synthesized progeny RNA (16, 17, 25, 60). Viral RNA replication initiates with the synthesis of negative-strand RNA on the input viral RNA. The newly synthesized negative-strand RNA then serves as a template for multiple rounds of positive-strand RNA synthesis.
Recent studies have advanced our understanding of membrane rearrangement into vesicles and have shown that proteins from the P2 region of the viral genome are required for the formation of replication complexes on the surface of membrane vesicles. Electron microscopy immunocytochemistry of isolated replication complexes and UV cross-linking studies have shown that 2C-containing proteins are bound to membrane-associated viral RNAs (17, 18). Viral proteins 2B, 2C, and 3A are membrane associated (15, 24, 42, 57), and 2BC, 2C, and 3A localize to ER membranes in the absence of other viral proteins (21, 23). Several viral proteins can induce membrane rearrangement and vesicle formation, although 2BC or 2BC in combination with 3A appears to induce rearrangements more closely resembling vesicles found in infected cells (2, 20, 52, 53). 2BC increases intracellular Ca2+ levels, presumably due to disruption of the ER (3), and alters membrane permeability (1). 2B, 2BC, and 3A sufficiently block protein transport from the ER to the Golgi to effectively disrupt the secretory system (7, 8, 21, 22). These proteins may also play a structural role and function to provide the spatial organization of the replication complex on the membranes required for RNA replication (18). Consistent with the results described above, RNA replication complexes isolated from HeLa S10 translation-replication reactions are enriched with the membrane-associated proteins 2BC, 2C, and 3AB (9).
In addition to membrane-association and RNA binding domains, 2C also contains motifs present in known nucleoside triphosphatase (NTPase) and helicase proteins (28-30). Recombinant 2C possesses both ATPase and GTPase activity (39, 45, 50). Guanidine HCl-dependent and -resistant mutants map near the NTP binding domain of 2C and appear to adversely affect ATPase activity (6, 45-48, 56). Guanidine specifically inhibits negative-strand initiation but does not inhibit negative-strand elongation or the initiation of positive-strand RNA synthesis (10). Several lethal mutations in the NTP binding motif have been described which inhibit viral RNA replication (38, 54) but do not affect the ability of 2C to bind to RNA or induce vesicle formation (20, 51). Guanidine also inhibits P2-containing proteins from associating with membranes (18), although 2C expressed in the absence of other viral proteins retains its ability to associate with membranes in the presence of guanidine (24). Overall, it is clear that 2C and P2-derived proteins are essential for the formation of functional membrane-associated RNA replication complexes, although the precise function of theses proteins in the RNA replication cycle as well as the exact processed form of these proteins remain unknown.
Recently, it has been shown that viral replication proteins can be provided in trans to support viral RNA synthesis in HeLa S10 translation-replication reactions (14, 58). Since optimal concentrations of the input RNAs are used in these reactions, translation begins at maximum rates at time zero. Therefore, individual viral proteins or viral precursor proteins can be expressed in high concentrations from different transcript RNAs and tested for their ability to assemble functional replication complexes with viral RNA templates in the same reaction. This contrasts with the situation in infected cells, where translation and RNA replication are codependent and multiple rounds of amplification of the input RNA are required to achieve maximum rates of translation. In this study, precursor proteins, processed intermediates, and individual proteins from the P2 region of the viral genome were expressed in cell-free reactions and tested for their ability to form functional replication complexes. The results showed that the expression of various combinations of 2AB, 2BC, 2A, 2B, and 2C did not result in the formation of active replication complexes. Only P2 plus P3 and 2A plus 2BCP3 (i.e., the primary cleavage products of P23) were able to form functional replication complexes in these assays. Since wild-type levels of negative-strand synthesis were only observed in reactions containing 2A plus 2BCP3, we propose that these proteins are preferentially used in the assembly of the membrane-associated replication complexes.
MATERIALS AND METHODS
Poliovirus cDNA clones.
A cDNA clone of the Mahoney strain of type I poliovirus, pT7-PV1(A)80 (14), was used as the parental clone for the following constructs: (i) pPV1-P1(FS) [pT7-PV1(A)80-P1(FS)] contained a 4-nucleotide (nt) insertion at nt 2474 in the capsid coding region (P1), resulting in a frameshift mutation. Translation of PV1-P1(FS) RNA resulted in the synthesis of a 64-kDa nonfunctional protein. There are no viral replication proteins synthesized from PV1-P1(FS). The addition of PV1-P1(FS) RNA to the indicated translation-replication reactions provided for equivalent amounts of RNA in all of the translation-replication reactions in an experiment. (ii) pDJB2 [pT7-PV1(A)80ΔC869-T6011] contained a 5,143-nt deletion of the polyprotein coding sequence (14) and was used as the parental construct to engineer viral protein expression clones. Expression clones were designed to express individual proteins from the P23 region of the viral genome (Fig. 1). pDJB2 was digested with MscI, which removed nt 630 to nt 1091, shrimp alkaline phosphatase treated, phenol extracted, and agarose gel purified. DNA oligonucleotides containing the coding sequence for specific viral proteins were PCR amplified from pT7-PV1(A)80 using the oligonucleotide primers listed in Table 1. The 5′ end of the oligonucleotides was designed to include Kozak's consensus sequence (34), an AUG initiation codon, and the start of the viral protein coding sequence. The 3′ end of the oligonucleotides contained the end of the viral protein coding sequences and two consecutive stop codons to prevent translation of the remaining C-terminal end of the 3D coding sequence that resulted from digestion of pDJB2 with MscI (Fig. 1D). (iii) pP23 [pT7-PV1(A)80P23] and pP3 [pT7-PV1(A)80P3] contained the normal stop codons found at the 3′ end of the viral protein coding sequence (Fig. 1B and C). The 3′ end of the oligonucleotide used to construct pP23 and pP3 was designed to end at the MscI site in the 3Dpol-coding region. After ligation into pDJB2, the coding sequence of 3Dpol was fully restored. (iv) p2BCP3 [pT7-PV1(A)802BCP3] was constructed by ligating the SpeI-to-FspI fragment of pP23 with the FspI-to-SpeI fragment of p2BC. (v) pP23-2C(P131N) [pT7-PV1(A)80P23-2C(P131N)] was constructed by mutating CC to AA at nt 1769 in pP23 by using the QuickChange site-directed mutagenesis kit (Stratagene) and the mutagenic oligonucleotides listed in Table 1. This created a pP23 expression clone with a lethal mutation in the NTPase domain of 2C that blocked negative-strand synthesis to below detectable levels in the in vitro replication assay. (vi) pPV1ΔGUA3 [pT7-PV1(A)80ΔG7418-A7422] was previously described and contained a deletion in the 3′ untranslated region that inhibits negative-strand RNA synthesis without impairing protein synthesis (14). PV1ΔGUA3 RNA was used as a helper RNA to provide all of the viral proteins in trans for complementation analysis. (vii) p2BCP3ΔGUA3 [pT7-PV1(A)802BCP3ΔG7418-A7422] was constructed by removing the AccI fragment from p2BCP3 and ligating in the AccI fragment from pRNA2(A)12ΔGUA3 (14). All plasmids constructed were verified by DNA sequencing.
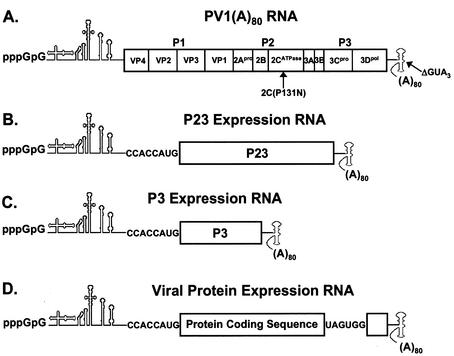
FIG. 1.
Poliovirus RNAs utilized in this study. (A) Diagram of poliovirus PV1(A)80 transcript RNA. Proteins encoded in the P1, P2, and P3 regions of the RNA are indicated. The locations of mutations used in this study are also shown. (B and C) Diagrams of the P23 and P3 expression RNAs. These RNAs contained the 5′ NTR, including the 5′ cloverleaf and IRES, an AUG initiation codon including Kozak's consensus sequence, the P23 (or P3) coding sequence, the 3′ NTR, and poly(A) tail. A 2BCP3 expression RNA was constructed using a similar strategy. (D) Diagram of a generic expression RNA containing the coding sequence for viral proteins from the P2 region of the viral genome. This RNA contains the 5′ cloverleaf and IRES, an AUG initiation codon, the coding sequence of one of the proteins listed in Table 1, two stop codons, the C-terminal end of the 3D coding sequence, the 3′ NTR, and poly(A) tail. The two stop codons were engineered at the end of the protein-coding sequence to terminate translation and block the translation of the C-terminal end of the 3D coding sequence.
TABLE 1.
Oligonucleotide primers used for cloning poliovirus expression constructsa
| Plasmid name | 5′ Oligonucleotide | 3′ Oligonucleotide |
|---|---|---|
| P23 | pGGGCCACCATGGGATTCGGACACCAAAAC | pCCAGCATAGTGGTCTACTGC |
| P2 | pGGGCCACCATGGGATTCGGACACCAAAAC | pCCCGGGTTACTATTGAAACAAAGCCTCCATAC |
| P3 | pCCCGGGCCACCATGGGACCACTCCAGTATAAAG | pCCAGCATAGTGGTCTACTGC |
| 2AB | pGGGCCACCATGGGATTCGGACACCAAAAC | pCCCGGGTTACTATTGCTTGATGACATAAGGTATC |
| 2BC | pCCCGGGCCACCATGGGCATCACCAATTACATAG | pCCCGGGTTACTATTGAAACAAAGCCTCCATAC |
| 2A | pGGGCCACCATGGGATTCGGACACCAAAAC | pCCCGGGTTACTATTGTTCCATGGCTTCTTC |
| 2B | pCCCGGGCCACCATGGGCATCACCAATTACATAG | pCCCGGGTTACTATTGCTTGATGACATAAGGTATC |
| 2C | pCCCGGGCCACCATGGGTGACAGTTGGTTGAAGAAG | pCCCGGGTTACTATTGAAACAAAGCCTCCATAC |
| P23-2C(P131N) | GCTAGTACATGGCAGCAACGGAACAGGTAAATC | GATTTACCTGTTCCGTTGCTGCCATGTACTAGC |
Nucleotides that were mutagenized are underlined.
RNA preparation.
Plasmid DNA was linearized using MluI and transcribed in vitro with bacteriophage T7 RNA polymerase as previously described (11). To synthesize RNA with a 7-methylguanosine cap analog, the final GTP concentration was lowered to 0.2 mM, and 0.8 mM m7G[5′]ppp[5′]G cap analog was added to the transcription reaction mixture. All transcript RNAs were purified by Sephadex G-50 gel filtration chromatography and precipitated in 0.2 M sodium acetate and three volumes of 100% ethanol. Final RNA concentration was determined by measuring UV absorption at 260 nm.
HeLa S10 translation-RNA replication reactions.
HeLa S10 extracts and HeLa cell translation initiation factors were prepared as previously described (11). HeLa S10 translation reaction mixtures contained 50% (by volume) HeLa S10 extract, 20% (by volume) HeLa cell translation initiation factors, 10% (by volume) 10× nucleotide reaction mix (10 mM ATP, 2.5 mM GTP, 2.5 mM UTP, 600 mM KCH3CO2, 155 mM HEPES-KOH [pH 7.4], 300 mM creatine phosphate, 4 mg of creatine phosphokinase/ml), 2 mM guanidine HCl, and transcript RNA. HeLa S10 translation reactions were incubated for 4 h at 34°C. Protein synthesis was monitored by adding 1.5 mCi of [35S]methionine/ml (1,500 Ci/mmol; Amersham) to a 10-μl portion of the HeLa S10 translation reaction mixture. After 3.5 h, 1 μl of the labeling reaction mixture was removed in duplicate and added to 100 μl of 0.1 N KOH-3% Casamino Acids. Labeled proteins were precipitated in 2 ml of 5% trichloroacetic acid, filtered, and quantitated by liquid scintillation counting. At the same time, 5 μl was removed and solubilized in 50 μl of 1× sodium dodecyl sulfate (SDS) sample buffer. A 25-μl portion was heated for 3 min at 100°C and analyzed by electrophoresis on an SDS-9-to-18% polyacrylamide gel. The gel was fixed for 30 min in 40% methanol, 10% CH3COOH, fluorographed in Amplify fluorographic reagent (Amersham) for 10 min, and dried.
RNA synthesis.
Negative-strand RNA synthesis was measured using preinitiation RNA replication complexes (PIRCs) as previously described (9, 11). HeLa S10 translation-replication reaction mixtures containing the indicated transcript RNAs were incubated for 4 h at 34°C in the presence of 2 mM guanidine HCl. PIRCs were isolated by centrifugation and resuspended in 50 μl of RNA replication buffer containing 25 μl of S10 buffer [40 mM HEPES-KOH (pH 7.4), 120 mM KCH3CO2, 5.5 mM Mg(CH3CO2)2, 6 mM dithiothreitol, 10 mM KCl, 1 mM CaCl2, and 2 mM EGTA], 5 μl of 10× nucleotide reaction mix, 5 μM CTP, 30 μCi of [α-32P]CTP, and 50 μg of puromycin/ml, except where noted in the figure legends. The reactions were incubated for 1 h at 37°C and were terminated by the addition of 350 μl of 0.5% SDS buffer. The RNA was recovered by phenol extraction and ethanol precipitation and analyzed by CH3HgOH-agarose gel electrophoresis and autoradiography of the dried gel. RNA synthesis was quantitated by PhosphorImager (Molecular Dynamics) analysis using ImageQuant software. The relative molar amount of negative-strand RNA synthesized in each of the reactions was calculated.
RESULTS
Genetic complementation analysis of negative-strand RNA synthesis.
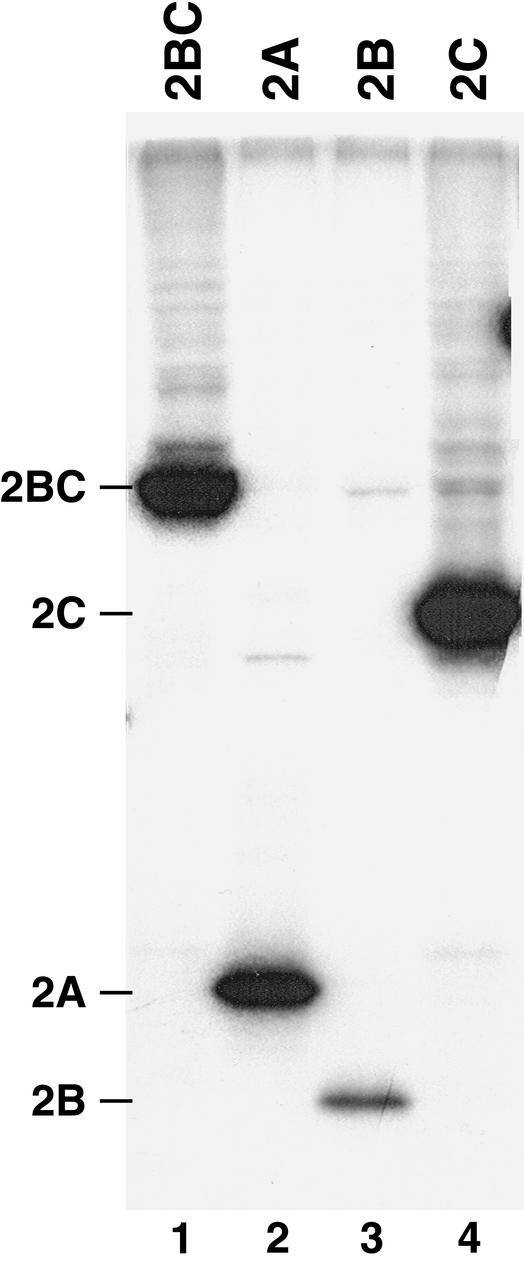
In vitro genetic complementation assays were performed to identify specific proteins encoded by the P2 region of the viral genome that were required to form functional RNA replication complexes. Modified poliovirus RNA transcripts that encoded specific viral proteins were constructed as described in Materials and Methods (Fig. 1). P23 RNA contained the 5′ cloverleaf and internal ribosomal entry site (IRES), an AUG start codon, the coding sequence for the P23 polyprotein, the 3′ nontranslated region (NTR), and the 3′ poly(A) tail (Fig. 1B). Additional viral RNA transcripts were engineered that encoded viral protein P3 (Fig. 1C) or viral proteins P2, 2AB, 2BC, 2A, 2B, or 2C (Fig. 1D and Table 1). The expected viral proteins were synthesized during in vitro translation reactions that contained 2A, 2B, 2C, 2BC, or P23 RNA (Fig. 2 and 3A) or P2, P3, or 2AB RNA (data not shown). One or more of the above viral RNAs were added to HeLa S10 translation-RNA replication reaction mixtures containing 2 mM guanidine HCl. PIRCs were isolated from these reactions and used to measure negative-strand RNA synthesis as previously described (9, 10). Only negative-strand synthesis was measured in these assays, since the transcript RNAs contained two nonviral Gs at their 5′ end which blocked positive-strand RNA synthesis to below detectable levels (12, 13, 32).
FIG. 2.
Expression of viral proteins in HeLa S10 translation-replication reactions. Each of the indicated RNAs at a final concentration of 50 nM were added to HeLa S10 translation-RNA replication reaction mixtures containing [35S]methionine. Protein synthesis was monitored as described in Materials and Methods. Labeled viral proteins were characterized by SDS-PAGE and were detected by fluorography of the dried gel.
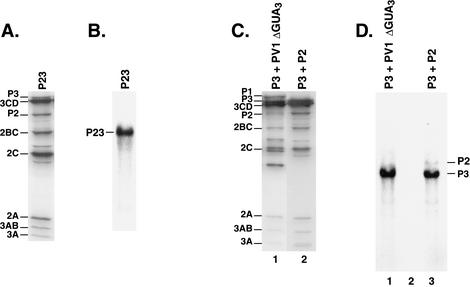
FIG. 3.
Negative-strand RNA synthesis on the P23 and P3 expression RNAs. (A) Protein synthesis was measured in a HeLa S10 translation-replication reaction mixtures containing 50 μg of P23 RNA/ml and [35S]methionine. The labeled viral proteins were analyzed by SDS-PAGE and fluorography as described in Materials and Methods. (B) Negative-strand RNA synthesis was measured using PIRCs isolated from the HeLa S10 translation-replication reaction described for panel A. 32P-labeled negative-strand RNA was analyzed by CH3HgOH-agarose gel electrophoresis and autoradiography. (C) Protein synthesis was measured in HeLa S10 translation-replication reaction mixtures containing 50 μg of P3 RNA/ml and an equimolar amount of either PV1ΔGUA3 RNA (lane 1) or P2 RNA (lane 2), as indicated. The labeled viral proteins were analyzed by SDS-PAGE as described for panel A. (D) Negative-strand RNA synthesis was measured using PIRCs isolated from HeLa S10 translation-replication reaction mixture as described for panel C. 32P-labeled negative-strand RNA was analyzed by gel electrophoresis, as described for panel B. The RNA replication reaction mixtures did not contain puromycin.
Identification of P2 proteins required for negative-strand synthesis.
P23 RNA expressed all of the viral replication proteins (Fig. 3A) and formed functional PIRCs that supported the synthesis of negative-strand RNA (Fig. 3B). The ability of P3 RNA to support negative-strand RNA synthesis in the presence of the viral replication proteins was also investigated. P3 RNA was added to HeLa S10 translation-replication reaction mixtures containing an equimolar amount of a nonreplicating full-length helper RNA, PV1ΔGUA3 (see Materials and Methods). PIRCs were isolated from these reactions and used to measure negative-strand RNA synthesis. The results showed that the P3 RNA was a functional template for negative-strand synthesis (Fig. 3D, lane 1) in the presence of the poliovirus replication proteins (Fig. 3C, lane 1). To determine if a precursor protein spanning the P2-P3 junction was required for negative-strand synthesis, we expressed the P2 and P3 proteins from separate RNAs and measured negative-strand synthesis. SDS-polyacrylamide gel electrophoresis (PAGE) analysis established that both the P2 and P3 proteins were synthesized and properly processed in this reaction (Fig. 3C, lane 2). Labeled negative-strand RNA was efficiently synthesized on P3 RNA in this reaction (Fig. 3D, lane 3). This demonstrated that an intact P23 precursor protein was not required to form functional replication complexes for negative-strand synthesis.
A low level of negative-strand synthesis was also observed on P2 RNA in this reaction (Fig. 3D, lane 3). The reason for the reduced level of negative-strand synthesis observed on P2 RNA templates was not directly determined, but most likely resulted from the change in the sequence in the 3′ region of this RNA (Fig. 1D). Other viral expression RNAs that were constructed using a similar strategy were also very poor templates for negative-strand synthesis. The efficient synthesis of negative-strand RNA observed on P3 RNA relative to P2 RNA did not appear to result from the translation of the P3 proteins in cis from the P3 RNA templates. In a separate experiment, we showed that a frameshift mutation in P3 RNA, which blocked its translation after the sixth amino acid, had no significant effect on its ability to serve as a template for negative-strand synthesis in reactions where all of the viral proteins were provided in trans (N. Sharma and J. B. Flanegan, unpublished results). Therefore, P3 RNA served as a good template to assay for negative-strand synthesis in assays containing various combinations of the P2 and P3 proteins.
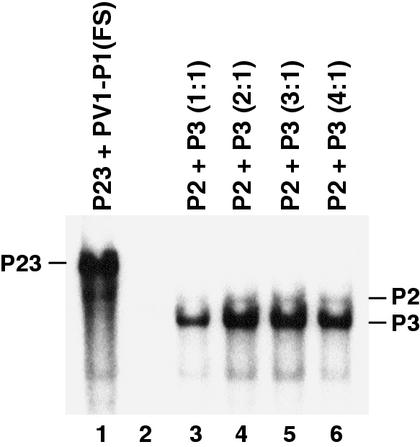
The level of negative-strand RNA synthesis on P23 RNA was compared to that observed on P3 RNA in reactions that contained an equivalent or excess molar amount of P2 RNA (Fig. 4). With a 1:1 ratio of the P2 and P3 RNAs, the molar amount of negative-strand RNA synthesized on P3 RNA was about 50% of the amount synthesized on P23 RNA (Fig. 4, compare lanes 1 and 3). Increasing the P2/P3 RNA ratio to 2:1 resulted in an increase in negative-strand synthesis on P3 RNA to 100% of the amount observed with P23 RNA (Fig. 4, compare lanes 1 and 4). An additional increase in the ratio to 3:1 did not improve negative-strand synthesis, and increasing the ratio to 4:1 was slightly inhibitory (Fig. 4, lanes 5 and 6). These results clearly indicated that a viral protein spanning the P2-P3 junction of the polyprotein was not required for the formation of functional RNA replication complexes.
FIG. 4.
Effect of the P2/P3 RNA ratio on negative-strand RNA synthesis. Negative-strand RNA synthesis was measured using PIRCs isolated from HeLa S10 translation-replication reaction mixtures containing either P23 RNA plus PV1-P1(FS) RNA (lane 1) or P2 RNA plus P3 RNA (lanes 3 to 6) at the indicated molar ratios. The total RNA concentration was 30 nM in lane 1 and 40 nM in lanes 3 to 6. PV1-P1(FS) RNA was added to the reaction shown in lane 1 to provide for equivalent reaction conditions as described in Materials and Methods. 32P-labeled negative-strand RNA synthesized was analyzed by CH3HgOH-agarose gel electrophoresis and autoradiography of the dried gel.
P2 is required for negative-strand synthesis on P3 RNA.
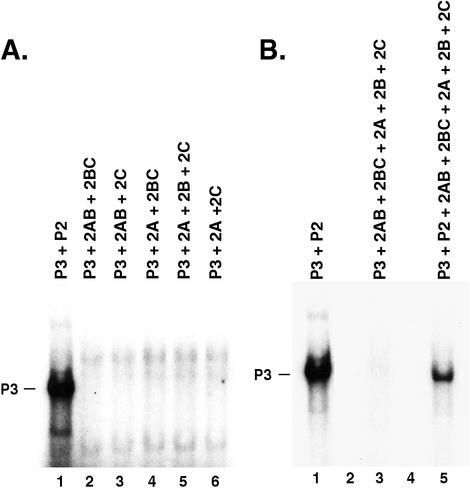
To determine if the processed forms of viral protein(s) from the P2 region of the viral genome could form functional RNA replication complexes, 2AB, 2BC, 2A, 2B, and 2C were expressed in all possible combinations with P3 in HeLa S10 translation-replication reactions. Viral RNA synthesis was then measured in PIRCs isolated from these reactions (Fig. 5A). Proteins 2AB and 2BC expressed together with P3 were not able to synthesize negative-strand RNA (Fig. 5A, lane 2). None of the other protein combinations (i.e., 2AB plus 2C, 2A plus 2BC, or 2A plus 2B plus 2C) when expressed with P3 were able to synthesize negative-strand RNA (Fig. 5A, lanes 3 to 6). Therefore, only when intact P2 was expressed with P3 was it possible to form functional RNA replication complexes that synthesized negative-strand RNA.
FIG. 5.
Negative-strand synthesis on P3 RNA in the presence of P2 or processed forms of the P2 proteins. (A) Negative-strand RNA synthesis was measured using PIRCs isolated from 150 μl of HeLa S10 translation-replication reaction mixtures containing each of the RNAs indicated at the top of each lane at a concentration of 25 nM (lane 1), 17 nM (lanes 2 to 4 and lane 6), and 13 nM (lane 5). The RNA replication reaction mixtures did not contain puromycin. 32P-labeled RNA synthesized was analyzed by CH3HgOH-agarose gel electrophoresis and autoradiography of the dried gel. (B) Negative-strand RNA synthesis was measured using PIRCs isolated from 150 μl of HeLa S10 translation-replication reaction mixtures that contained a 13 nM concentration of each of the P3, P2, 2AB, and 2BC RNAs as indicated. In lanes 3 and 5, 8 nM 2A RNA, 9 nM 2B RNA, and 7 nM 2C RNA were present in the reaction mixtures. Protein synthesis was monitored as described in Materials and Methods. 32P-labeled RNA synthesized was analyzed by CH3HgOH-agarose gel electrophoresis and autoradiography of the dried gel.
Further analysis of the viral proteins synthesized in HeLa S10 translation-replication reactions by SDS-PAGE suggested that proteolytic processing of 2AB and 2BC into 2A, 2B, and 2C was less efficient than what was observed in reactions with P2 RNA. To determine if this had any effect on RNA replication, we added the 2A, 2B, and 2C RNAs to the HeLa S10 translation-replication reactions containing the 2AB and 2BC RNAs (Fig. 5B, lane 3). This change, however, did not result in any detectable negative-strand synthesis in the PIRCs isolated from these reactions (Fig. 5B, lane 3). However, the further addition of P2 RNA to this reaction restored negative-strand synthesis on P3 RNA (Fig. 5B, lane 5). This demonstrated that even in the presence of seven different viral RNAs and their translation products, functional RNA replication complexes were formed as long as P2 and P3 were expressed together in the HeLa S10 translation-replication reaction. The reason for the reduced level of negative-strand synthesis on P3 RNA that was observed in Fig. 5B, lane 5, was not determined. This may have been the result of the combined effect of a reduction in the translation of the P2 and P3 RNAs and an additional reduction in the replication of P3 RNA due to direct competition with the five other viral RNAs present in this reaction. Whether or not the presence of the additional P2 cleavage products had any direct inhibitory effect on RNA replication will require additional investigation.
Role of 2BCP3 in viral RNA replication.
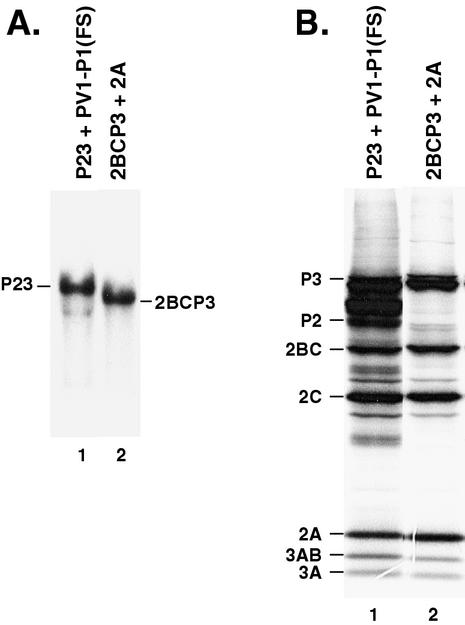
The results described above suggest that intact P2 was required for the initiation of negative-strand RNA synthesis. In contrast, the results of a previous study showed that a dicistronic poliovirus RNA construct, which contained an encephalomyocarditis virus (EMCV) IRES inserted at the 2A-2B junction site, was viable and formed virus in transfected HeLa cells (40, 43). The primary translation products of this RNA would be P1-2A and 2BCP3. This suggests that a viral protein spanning the 2A-2B junction and intact P2 were not used to form replication complexes with this RNA. To investigate this point further, we examined negative-strand synthesis in PIRCs formed in reactions in which 2A and 2BCP3 were expressed separately. Negative-strand RNA synthesis was measured in PIRCs isolated from HeLa S10 translation-replication reactions that contained either P23 RNA or 2A RNA plus 2BCP3 RNA. SDS-PAGE showed that equivalent amounts of the viral replication proteins, including protein 2A, were synthesized in each reaction (Fig. 6B). Under these conditions, we observed that similar molar amounts of negative-strand RNA were synthesized on both P23 RNA and 2BCP3 RNA (Fig. 6A). This result suggests that a viral protein spanning the 2A-2B junction is not required for negative-strand synthesis as long as 2BC is expressed as part of a 2BCP3 precursor protein.
FIG. 6.
Negative-strand RNA synthesis on 2BCP3 RNA in the presence of 2A. (A) Negative-strand RNA synthesis was measured using PIRCs isolated from 100 μl of HeLa S10 translation-replication reaction mixtures containing either 20 nM P23 RNA and 10 nM PV1-P1(FS) RNA (lane 1) or 20 nM 2BCP3 RNA and 20 nM 2A RNA (lane 2). 32P-labeled negative-strand RNA was analyzed by CH3HgOH-agarose gel electrophoresis and autoradiography. (B) The labeled viral proteins from the HeLa S10 translation-replication reaction mixtures shown in panel A were analyzed by SDS-PAGE and fluorography as described in Materials and Methods.
Complementation analysis of P23-2C(P131N) RNA.
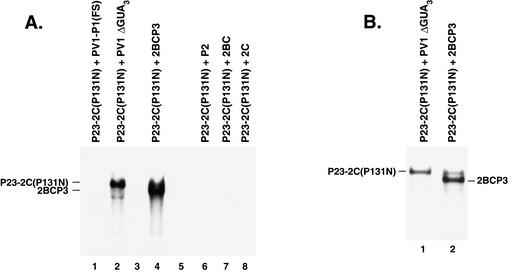
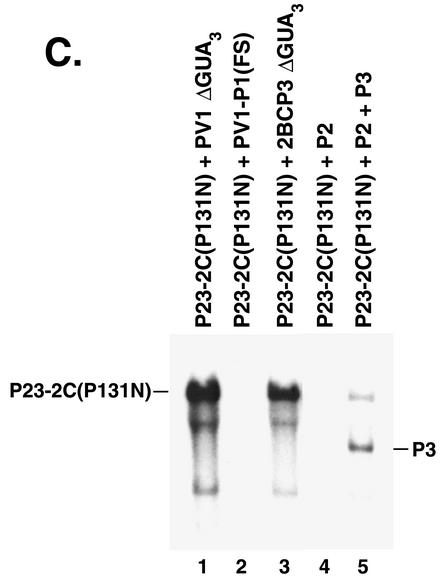
As an alternate approach to identify proteins in the P2 coding region that were required to form functional RNA replication complexes, complementation analysis was performed using a 2C mutant RNA. A previously characterized lethal mutation, 2C(P131N), which is located in the NTP binding domain of 2C was selected. This mutation was chosen since it inhibits the NTPase activity of 2C and RNA replication (54). Negative-strand synthesis was measured in PIRCs isolated from HeLa S10 translation-replication reactions containing P23-2C(P131N) RNA and either PV1ΔGUA3 RNA or PV1-P1(FS) RNA. Labeled negative-strand RNA was only synthesized when all of the wild-type replication proteins were provided in trans by the PV1 ΔGUA3 helper RNA (Fig. 7A, lanes 1 and 2). Labeled negative-strand RNA was not synthesized in the presence of PV1-P1(FS) RNA, indicating that the 2C(P131N) mutation inhibited negative-strand synthesis, as expected. To identify the 2C-containing protein that was responsible for the observed complementation, RNAs which encoded 2BCP3, P2, 2BC, or 2C were added to HeLa S10 translation-replication reactions containing P23-2C(P131N) RNA. Negative-strand RNA synthesis was then measured in PIRCs isolated from these reactions (Fig. 7A, lanes 4 to 8). Detectable levels of negative-strand synthesis were only observed with the 2BCP3 RNA (Fig. 7A, lane 4). In this case, labeled negative-strand RNA was synthesized on both the P23-2C(P131N) RNA and the 2BCP3 helper RNA. To further quantitate the levels of negative-strand synthesis on P23-2C(P131N) RNA in the presence of PV1ΔGUA3 or 2BCP3 helper RNAs, the labeled product RNAs were analyzed on a 0.8% agarose gel. Similar amounts of negative-strand RNA were synthesized on the P23-2C(P131N) RNA in both reactions (Fig. 7B). Negative-strand synthesis on P23-2C(P131N) RNA in the presence of 2BCP3 RNA was complicated by the synthesis of labeled negative-strand RNA on the 2BCP3 RNA (Fig. 7A, lane 4, and B, lane 2). To overcome this problem, a replication-incompetent 2BCP3 RNA containing a 3′ ΔGUA3 mutation was constructed and was used as a helper RNA to complement the 2C(P131N) mutation. Negative-strand synthesis on P23-2C(P131N) RNA in the presence of 2BCP3ΔGUA3 helper RNA was comparable to the levels observed in reactions containing PV1ΔGUA3 helper RNA (Fig. 7C, compare lanes 1 and 3).
FIG. 7.
Genetic complementation analysis of P23-2C(P131N) RNA. (A) Negative-strand RNA synthesis was measured using PIRCs isolated from 150 μl of HeLa S10 translation-replication reaction mixtures containing 13 nM P23-2C(P131N) RNA and 13 nM helper RNA as indicated above each lane. Protein synthesis was monitored as described in Materials and Methods. 32P-labeled negative-strand RNA synthesized in these reactions was analyzed by 1% CH3HgOH-agarose gel electrophoresis and autoradiography. (B) Negative-strand RNA synthesis was measured as described for panel A in reactions containing P23-2C(P131N) RNA and a helper RNA as indicated above each lane. The labeled product RNAs were analyzed by electrophoresis on a 0.8% CH3HgOH-agarose gel to enhance their separation. (C) The synthesis of negative-strand RNA was measured as described for panel A in reactions containing P23-2C(P131N) RNA and a helper RNA as indicated above each lane. The labeled product RNAs were characterized by 1% CH3HgOH-agarose gel electrophoresis and autoradiography.
It was surprising that P2 RNA was not able to complement the replication of the P23-2C(P131N) mutant RNA in the above experiments (Fig. 7A, lane 6, and C, lane 4). However, in experiments where both P2 and P3 RNAs were added to reactions containing P23-2C(P131N) RNA, a very low but detectable level of labeled negative-strand synthesis was observed on the P3 RNA and the P23-2C(P131N) RNA (Fig. 7C, lane 5). Therefore, 2BCP3 appears to be the preferred precursor of protein 2C that is used to assemble viral RNA replication complexes.
DISCUSSION
The viral replication proteins and their immediate precursors perform essential functions at specific stages of poliovirus RNA replication. Proteins encoded in the P2 region of the viral genome provide enzymatic activities that are required for viral replication, are integral to the structural integrity of the replication complex, and are believed to mediate the attachment of viral RNA in replication complexes to cellular membranes (18). In this study, cell-free reactions were used to identify the processed forms of the P2 proteins that were able to form functional RNA replication complexes. Since the poliovirus replication proteins can function in trans in HeLa S10 translation-replication reactions to support negative-strand RNA synthesis (14, 58), we expressed the individual P2 region proteins from separate transcript RNAs and assayed for negative-strand synthesis in PIRCs isolated from these reactions. The results showed that only two precursor forms of the P2 region proteins, intact P2 and 2BCP3, were able to form functional replication complexes that supported negative-strand RNA synthesis.
P2 and 2BCP3 are required to form functional RNA replication complexes.
By expressing P2 and P3 separately, we found that the P2 proteins were properly processed and formed functional replication complexes with P3 proteins and synthesized negative-strand RNA, although at about 50% of the level observed with P23 RNA. Negative-strand RNA synthesis was not observed when the processed forms of P2 were expressed with P3. Expression of a larger P2 precursor, 2BCP3, in the presence of 2A expressed from a separate RNA, resulted in even higher levels of negative-strand RNA synthesis that were comparable to that observed with P23 RNA. This suggested that 2BCP3 was the preferred precursor required to assemble replication complexes needed to initiate negative-strand RNA synthesis. As an alternative experimental approach, a genetic complementation analysis of a lethal mutation in the NTP binding domain of 2C that specifically inhibits RNA replication (54) was also performed. Negative-strand RNA synthesis was not observed with P23-2C(P131N) RNA, demonstrating that this mutation completely inhibited negative-strand RNA synthesis. Proteins P2, 2BC, and 2C were unable to complement this mutation, suggesting that these proteins were unable to replace the mutant 2C-related proteins in PIRCs. This was consistent with the results of a previous study in which complementation of the 2C(P131N) mutation was not observed in transfected cells expressing 2C or 2BC (55). Although a detectable level of complementation was observed when P2 was expressed in combination with P3, the complementation observed was very inefficient.
Previously, a complementation analysis of a viral RNA containing a mutation in the hydrophobic domain of 3A was performed to identify the 3A-containing protein that was required to restore RNA replication (58). The results from that study showed that viral protein P3, with an active 3C protease (3Cpro), was required for efficient complementation of this 3A mutant. Wild-type 3AB and P3 with a mutation in the 3Cpro domain were not able to complement the 3A mutant. This suggests that intact P3 is required in the assembly of functional replication complexes. Results from the present study also suggest that functional replication complexes are only formed with precursor forms of the viral replication proteins.
Primary products of viral polyprotein processing of P23 are required for negative-strand RNA synthesis.
These results are consistent with the polyprotein processing cascades previously proposed by Lawson and Semler (35). They suggested that after primary cleavage at the P1-P2 site by 2Apro, alternate processing cascades then proceed as determined by the initial site of cleavage of P23 by 3Cpro. Initial cleavage of P23 at the 2A-2B junction followed by rapid cleavage at the 2C-3A junction results in the formation of the membrane-associated replication proteins. This generates 2BCP3 as the first precursor protein formed after initial cleavage of P23. 2BCP3 is then rapidly processed to form 2BC and P3 in both in vitro translation reactions and virus-infected cells (35). Alternatively, cleavage of P23 at the 2C-3A junction generates soluble P2 and P3 proteins as the first precursors formed after initial cleavage of P23. Slow processing of P2 and P3 occurs later to generate 2A, 2BC, 3AB, and 3CD. The predominate cleavage cascade was the membrane-bound pathway starting with cleavage at 2A-2B, which is consistent with a membrane requirement for the initiation of RNA replication.
Our results demonstrated that the replication proteins formed after the initial cleavage of P23 by 3Cpro (i.e., 2A plus 2BCP3 and P2 plus P3) were required for negative-strand RNA synthesis. The proteins formed after additional processing were unable to form functional replication complexes. Consistent with the membrane-bound processing cascade, 2A plus 2BCP3 synthesized a greater amount of negative-strand RNA than did P2 plus P3 and complemented P23-2C(P131N) RNA more efficiently than P2 plus P3. Therefore, 2BCP3, which is a primary product in the membrane-associated processing pathway, is also the protein that is required for the assembly of functional membrane-associated RNA replication complexes.
Our results were also consistent with previous studies with dicistronic viral RNAs that contained the EMCV IRES inserted at various processing sites in the viral polyprotein. Translation of these RNAs resulted in the independent synthesis of viral proteins that are encoded by different regions of the viral genome (40, 43). Insertion of the EMCV IRES between 2A and 2B, such that 2A is expressed separately from 2BCP3, resulted in a viral RNA that produced viable virus after transfection into HeLa cells, consistent with the results described here (40). In contrast, insertion of the EMCV IRES between 2C and 3A, such that P1-P2 is expressed separately from P3, resulted in a viral RNA that was unable to replicate and produce virus after transfection into HeLa cells (43). The primary defect appeared to be the initial cleavage event by 2Apro, separating P1 from P2. In our experiments, however, P2 and P3 were expressed from separate viral RNAs without the complication of processing at the P1-P2 junction by 2Apro. This difference most likely explains why significant levels of negative-strand synthesis were observed in the cell-free reactions that contained P2 plus P3, whereas viable virus was not observed in cells transfected with the dicistronic construct that expressed the P2 and P3 proteins separately.
Formation of PIRCs on cytoplasmic membrane vesicles.
The observation that expression of 2BC with 3A results in vesicles more closely resembling those found in poliovirus-infected cells (52) appears to be consistent with a 2BCP3 requirement for formation of functional PIRCs that efficiently synthesize negative-strand RNA. In recent studies, preformed membrane vesicles were tested for their ability to support poliovirus replication (26). Vesicle formation in HeLa cells was induced by expressing viral membrane-associated proteins 2BC, 2C, or 3AB, or by expressing the full complement of viral replication proteins from a replication-incompetent RNA. These induced membrane vesicles were unable to support the replication of supertransfected poliovirus RNA. Supertransfected poliovirus RNA formed distinct membrane vesicles that contained active replication complexes, which were separate from the previously induced vesicles, suggesting that the formation of a functional replication complex occurs in cis by a coupled process of viral translation, membrane proliferation, and rearrangement into vesicles, and RNA replication (26). Furthermore, certain mutations in 2B, 2C, and 3A are not complemented by wild-type virus in vivo. This suggests that specific functions of these proteins are required in cis or that these mutations disrupted higher order structures in the viral RNA that are required for RNA replication (reviewed in reference 61). The inability to complement mutant virus in vivo may also result in part because 2BCP3, which is the precursor protein that is preferentially used to form functional replication complexes, is a short-lived intermediate (35). Once 2BCP3 undergoes proteolytic processing, it would no longer be able to form functional replication complexes in trans with a mutant RNA. This may be another reason why translation and RNA replication are tightly linked in virus-infected cells (41). This linkage between translation and replication in combination with the cis-active replication elements found in the viral genome (4, 5, 27, 33, 36, 37, 44, 49) insures that viral RNA replication in infected cells is both efficient and highly specific for functional viral RNAs.
Acknowledgments
We thank Joan Morasco and Brian O'Donnell for excellent technical assistance and Sushma Abraham Ogram and Nidhi Sharma for critical reading of the manuscript.
This work was supported by Public Health Service grant AI 32123 from the National Institute of Allergy and Infectious Diseases.
REFERENCES
- 1.Aldabe, R., A. Barco, and L. Carrasco. 1996. Membrane permeabilization by poliovirus proteins 2B and 2BC. J. Biol. Chem. 271:23134-23137. [DOI] [PubMed] [Google Scholar]
- 2.Aldabe, R., and L. Carrasco. 1995. Induction of membrane proliferation by poliovirus proteins 2C and 2BC. Biochem. Biophys. Res. Commun. 206:64-76. [DOI] [PubMed] [Google Scholar]
- 3.Aldabe, R., A. Irurzun, and L. Carrasco. 1997. Poliovirus protein 2BC increases cytosolic free calcium concentrations. J. Virol. 71:6214-6217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Andino, R., G. E. Rieckhof, P. L. Achacoso, and D. Baltimore. 1993. Poliovirus RNA synthesis utilizes an RNP complex formed around the 5′-end of viral RNA. EMBO J. 12:3587-3598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Andino, R., G. E. Rieckhof, and D. Baltimore. 1990. A functional ribonucleoprotein complex forms around the 5′ end of poliovirus RNA. Cell 63:369-380. [DOI] [PubMed] [Google Scholar]
- 6.Baltera, R. F., Jr., and D. R. Tershak. 1989. Guanidine-resistant mutants of poliovirus have distinct mutations in peptide 2C. J. Virol. 63:4441-4444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Barco, A., and L. Carrasco. 1995. A human virus protein, poliovirus protein 2BC, induces membrane proliferation and blocks the exocytic pathway in the yeast Saccharomyces cerevisiae. EMBO J. 14:3349-3364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Barco, A., and L. Carrasco. 1998. Identification of regions of poliovirus 2BC protein that are involved in cytotoxicity. J. Virol. 72:3560-3570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Barton, D. J., E. P. Black, and J. B. Flanegan. 1995. Complete replication of poliovirus in vitro: preinitiation RNA replication complexes require soluble cellular factors for the synthesis of VPg-linked RNA. J. Virol. 69:5516-5527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Barton, D. J., and J. B. Flanegan. 1997. Synchronous replication of poliovirus RNA: initiation of negative-strand RNA synthesis requires the guanidine-inhibited activity of protein 2C. J. Virol. 71:8482-8489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Barton, D. J., B. J. Morasco, and J. B. Flanegan. 1996. Assays for poliovirus polymerase, 3D(Pol), and authentic RNA replication in HeLa S10 extracts. Methods Enzymol. 275:35-57. [DOI] [PubMed] [Google Scholar]
- 12.Barton, D. J., B. J. Morasco, and J. B. Flanegan. 1999. Translating ribosomes inhibit poliovirus negative-strand RNA synthesis. J. Virol. 73:10104-10112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Barton, D. J., B. J. Morasco, L. E. Smerage, and J. B. Flanegan. 2002. Poliovirus RNA replication and genetic complementation in cell-free reactions, p. 461-469. In B. L. Semler and E. Wimmer (ed.), Molecular biology of picornaviruses. American Society for Microbiology, Washington, D.C.
- 14.Barton, D. J., B. J. O'Donnell, and J. B. Flanegan. 2001. 5′ cloverleaf in poliovirus RNA is a cis-acting replication element required for negative-strand synthesis. EMBO J. 20:1439-1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bienz, K., D. Egger, and L. Pasamontes. 1987. Association of polioviral proteins of the P2 genomic region with the viral replication complex and virus-induced membrane synthesis as visualized by electron microscopic immunocytochemistry and autoradiography. Virology 160:220-226. [DOI] [PubMed] [Google Scholar]
- 16.Bienz, K., D. Egger, and T. Pfister. 1994. Characteristics of the poliovirus replication complex. Arch. Virol. Suppl. 9:147-157. [DOI] [PubMed] [Google Scholar]
- 17.Bienz, K., D. Egger, T. Pfister, and M. Troxler. 1992. Structural and functional characterization of the poliovirus replication complex. J. Virol. 66:2740-2747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bienz, K., D. Egger, M. Troxler, and L. Pasamontes. 1990. Structural organization of poliovirus RNA replication is mediated by viral proteins of the P2 genomic region. J. Virol. 64:1156-1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Caliguiri, L. A., and I. Tamm. 1969. Membranous structures associated with translation and transcription of poliovirus RNA. Science 166:885-886. [DOI] [PubMed] [Google Scholar]
- 20.Cho, M. W., N. Teterina, D. Egger, K. Bienz, and E. Ehrenfeld. 1994. Membrane rearrangement and vesicle induction by recombinant poliovirus 2C and 2BC in human cells. Virology 202:129-145. [DOI] [PubMed] [Google Scholar]
- 21.Doedens, J. R., T. H. Giddings, Jr., and K. Kirkegaard. 1997. Inhibition of endoplasmic reticulum-to-Golgi traffic by poliovirus protein 3A: genetic and ultrastructural analysis. J. Virol. 71:9054-9064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Doedens, J. R., and K. Kirkegaard. 1995. Inhibition of cellular protein secretion by poliovirus proteins 2B and 3A. EMBO J. 14:894-907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Echeverri, A., R. Banerjee, and A. Dasgupta. 1998. Amino-terminal region of poliovirus 2C protein is sufficient for membrane binding. Virus Res. 54:217-223. [DOI] [PubMed] [Google Scholar]
- 24.Echeverri, A. C., and A. Dasgupta. 1995. Amino terminal regions of poliovirus 2C protein mediate membrane binding. Virology 208:540-553. [DOI] [PubMed] [Google Scholar]
- 25.Egger, D., L. Pasamontes, R. Bolten, V. Boyko, and K. Bienz. 1996. Reversible dissociation of the poliovirus replication complex: functions and interactions of its components in viral RNA synthesis. J. Virol. 70:8675-8683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Egger, D., N. Teterina, E. Ehrenfeld, and K. Bienz. 2000. Formation of the poliovirus replication complex requires coupled viral translation, vesicle production, and viral RNA synthesis. J. Virol. 74:6570-6580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Goodfellow, I., Y. Chaudhry, A. Richardson, J. Meredith, J. W. Almond, W. Barclay, and D. J. Evans. 2000. Identification of a cis-acting replication element within the poliovirus coding region. J. Virol. 74:4590-4600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gorbalenya, A. E., V. M. Blinov, A. P. Donchenko, and E. V. Koonin. 1989. An NTP-binding motif is the most conserved sequence in a highly diverged monophyletic group of proteins involved in positive strand RNA viral replication. J. Mol. Evol. 28:256-268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gorbalenya, A. E., and E. V. Koonin. 1989. Viral proteins containing the purine NTP-binding sequence pattern. Nucleic Acids Res. 17:8413-8440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gorbalenya, A. E., E. V. Koonin, A. P. Donchenko, and V. M. Blinov. 1988. A conserved NTP-motif in putative helicases. Nature 333:22.. [DOI] [PubMed] [Google Scholar]
- 31.Harris, K. S., C. U. T. Hellen, and E. Wimmer. 1990. Proteolytic processing in the replication of picornaviruses. Semin. Virol. 1:323-333. [Google Scholar]
- 32.Herold, J., and R. Andino. 2000. Poliovirus requires a precise 5′ end for efficient positive-strand RNA synthesis. J. Virol. 74:6394-6400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jacobson, S. J., D. A. Konings, and P. Sarnow. 1993. Biochemical and genetic evidence for a pseudoknot structure at the 3′ terminus of the poliovirus RNA genome and its role in viral RNA amplification. J. Virol. 67:2961-2971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kozak, M. 1986. Point mutations define a sequence flanking the AUG initiator codon that modulates translation by eukaryotic ribosomes. Cell 44:283-292. [DOI] [PubMed] [Google Scholar]
- 35.Lawson, M. A., and B. L. Semler. 1992. Alternate poliovirus nonstructural protein processing cascades generated by primary sites of 3C proteinase cleavage. Virology 191:309-320. [DOI] [PubMed] [Google Scholar]
- 36.Lyons, T., K. E. Murray, A. W. Roberts, and D. J. Barton. 2001. Poliovirus 5′-terminal cloverleaf RNA is required in cis for VPg uridylylation and the initiation of negative-strand RNA synthesis. J. Virol. 75:10696-10708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mirmomeni, M. H., P. J. Hughes, and G. Stanway. 1997. An RNA tertiary structure in the 3′ untranslated region of enteroviruses is necessary for efficient replication. J. Virol. 71:2363-2370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mirzayan, C., and E. Wimmer. 1992. Genetic analysis of an NTP-binding motif in poliovirus polypeptide 2C. Virology 189:547-555. [DOI] [PubMed] [Google Scholar]
- 39.Mirzayan, C., and E. Wimmer. 1994. Biochemical studies on poliovirus polypeptide 2C: evidence for ATPase activity. Virology 199:176-187. [DOI] [PubMed] [Google Scholar]
- 40.Molla, A., A. V. Paul, M. Schmid, S. K. Jang, and E. Wimmer. 1993. Studies on dicistronic polioviruses implicate viral proteinase 2Apro in RNA replication. Virology 196:739-747. [DOI] [PubMed] [Google Scholar]
- 41.Novak, J. E., and K. Kirkegaard. 1994. Coupling between genome translation and replication in an RNA virus. Genes Dev. 8:1726-1737. [DOI] [PubMed] [Google Scholar]
- 42.Paul, A. V., A. Molla, and E. Wimmer. 1994. Studies of a putative amphipathic helix in the N-terminus of poliovirus protein 2C. Virology 199:188-199. [DOI] [PubMed] [Google Scholar]
- 43.Paul, A. V., J. Mugavero, A. Molla, and E. Wimmer. 1998. Internal ribosomal entry site scanning of the poliovirus polyprotein: implications for proteolytic processing. Virology 250:241-253. [DOI] [PubMed] [Google Scholar]
- 44.Paul, A. V., E. Rieder, D. W. Kim, J. H. van Boom, and E. Wimmer. 2000. Identification of an RNA hairpin in poliovirus RNA that serves as the primary template in the in vitro uridylylation of VPg. J. Virol. 74:10359-10370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pfister, T., and E. Wimmer. 1999. Characterization of the nucleoside triphosphatase activity of poliovirus protein 2C reveals a mechanism by which guanidine inhibits poliovirus replication. J. Biol. Chem. 274:6992-7001. [DOI] [PubMed] [Google Scholar]
- 46.Pincus, S. E., D. C. Diamond, E. A. Emini, and E. Wimmer. 1986. Guanidine-selected mutants of poliovirus: mapping of point mutations to polypeptide 2C. J. Virol. 57:638-646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pincus, S. E., H. Rohl, and E. Wimmer. 1987. Guanidine-dependent mutants of poliovirus: identification of three classes with different growth requirements. Virology 157:83-88. [DOI] [PubMed] [Google Scholar]
- 48.Pincus, S. E., and E. Wimmer. 1986. Production of guanidine-resistant and -dependent poliovirus mutants from cloned cDNA: mutations in polypeptide 2C are directly responsible for altered guanidine sensitivity. J. Virol. 60:793-796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rieder, E., A. V. Paul, D. W. Kim, J. H. van Boom, and E. Wimmer. 2000. Genetic and biochemical studies of poliovirus cis-acting replication element cre in relation to VPg uridylylation. J. Virol. 74:10371-10380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rodriguez, P. L., and L. Carrasco. 1993. Poliovirus protein 2C has ATPase and GTPase activities. J. Biol. Chem. 268:8105-8110. [PubMed] [Google Scholar]
- 51.Rodriguez, P. L., and L. Carrasco. 1995. Poliovirus protein 2C contains two regions involved in RNA binding activity. J. Biol. Chem. 270:10105-10112. [DOI] [PubMed] [Google Scholar]
- 52.Suhy, D. A., T. H. Giddings, Jr., and K. Kirkegaard. 2000. Remodeling the endoplasmic reticulum by poliovirus infection and by individual viral proteins: an autophagy-like origin for virus-induced vesicles. J. Virol. 74:8953-8965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Teterina, N. L., A. E. Gorbalenya, D. Egger, K. Bienz, and E. Ehrenfeld. 1997. Poliovirus 2C protein determinants of membrane binding and rearrangements in mammalian cells. J. Virol. 71:8962-8972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Teterina, N. L., K. M. Kean, A. E. Gorbalenya, V. I. Agol, and M. Girard. 1992. Analysis of the functional significance of amino acid residues in the putative NTP-binding pattern of the poliovirus 2C protein. J. Gen. Virol. 73:1977-1986. [DOI] [PubMed] [Google Scholar]
- 55.Teterina, N. L., W. D. Zhou, M. W. Cho, and E. Ehrenfeld. 1995. Inefficient complementation activity of poliovirus 2C and 3D proteins for rescue of lethal mutations. J. Virol. 69:4245-4254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tolskaya, E. A., L. I. Romanova, M. S. Kolesnikova, A. P. Gmyl, A. E. Gorbalenya, and V. I. Agol. 1994. Genetic studies on the poliovirus 2C protein, an NTPase. A plausible mechanism of guanidine effect on the 2C function and evidence for the importance of 2C oligomerization. J. Mol. Biol. 236:1310-1323. [DOI] [PubMed] [Google Scholar]
- 57.Towner, J. S., T. V. Ho, and B. L. Semler. 1996. Determinants of membrane association for poliovirus protein 3AB. J. Biol. Chem. 271:26810-26818. [DOI] [PubMed] [Google Scholar]
- 58.Towner, J. S., M. M. Mazanet, and B. L. Semler. 1998. Rescue of defective poliovirus RNA replication by 3AB-containing precursor polyproteins. J. Virol. 72:7191-7200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Toyoda, H., M. J. Nicklin, M. G. Murray, C. W. Anderson, J. J. Dunn, F. W. Studier, and E. Wimmer. 1986. A second virus-encoded proteinase involved in proteolytic processing of poliovirus polyprotein. Cell 45:761-770. [DOI] [PubMed] [Google Scholar]
- 60.Troxler, M., D. Egger, T. Pfister, and K. Bienz. 1992. Intracellular localization of poliovirus RNA by in situ hybridization at the ultrastructural level using single-stranded riboprobes. Virology 191:687-697. [DOI] [PubMed] [Google Scholar]
- 61.Wimmer, E., C. U. Hellen, and X. Cao. 1993. Genetics of poliovirus. Annu. Rev. Genet. 27:353-436. [DOI] [PubMed] [Google Scholar]