Abstract
The genomes of several poxviruses contain open reading frames with homology to the K3 and K5 genes of Kaposi's sarcoma-associated herpesvirus (KSHV) and the K3 gene of murine gammaherpesvirus 68, which target major histocompatibility complex class I (MHC-I) as well as costimulatory molecules for proteasomal or lysosomal degradation. The homologous gene product of myxomavirus (MV), M153R, was recently shown to reduce the cell surface expression of MHC-I. In addition, normal MHC-I surface expression was observed in cells infected with MV lacking M153R (J. L. Guerin, J. Gelfi, S. Boullier, M. Delverdier, F. A. Bellanger, S. Bertagnoli, I. Drexler, G. Sutter, and F. Messud-Petit, J. Virol. 76:2912-2923, 2002). Here, we show that M153R also downregulates the T-cell coreceptor CD4 and we study the molecular mechanism by which M153R achieves the downregulation of CD4 and MHC-I. Upon M153R expression, CD4 was rapidly internalized and degraded in lysosomes, whereas deletion of M153R from the genome of MV restored CD4 expression. The downregulation of both CD4 and MHC-I was dependent on the presence of lysine residues in their cytoplasmic tails. Increased ubiquitination of CD4 was observed upon coexpression with M153R in the presence of inhibitors of lysosomal acidification. Surface expression of CD4 was restored upon overexpression of Hrs, a ubiquitin interaction motif-containing protein that sorts ubiquitinated proteins into endosomes. Moreover, the purified PHD/LAP zinc finger of M153R catalyzed the formation of multiubiquitin adducts in vitro. Our data suggest that M153R acts as a membrane-bound ubiquitin ligase that conjugates ubiquitin to the cytoplasmic domain of substrate glycoproteins, with ubiquitin serving as a lysosomal targeting signal. Since a similar mechanism was recently proposed for KSHV K5, it seems that members of the unrelated families of gamma-2 herpesviruses and poxviruses share a common immune evasion mechanism that targets host cell immune receptors.
Viral immune escape mechanisms are central to the establishment and maintenance of viral infections in the presence of the highly developed innate and adaptive immune systems of vertebrate organisms (35). In particular, strategies leading to escape from the cellular immune response are widely found among different viral species (52). One way to escape detection by CD8 and CD4 lymphocytes is through the downregulation of major histocompatibility complex (MHC) class I (MHC-I) or class II (MHC-II) molecules. This process is accomplished by viral proteins dedicated to interfering with crucial steps of the antigen presentation pathway (16). Escape from T-cell recognition, however, can also be achieved through the downregulation of costimulatory molecules from the surface of infected cells. This process affects the activation of not only T cells but also NK cells (13, 24). Furthermore, viruses that directly infect T cells have been observed to downregulate the T-cell coreceptor CD4, thus hampering the ability of T cells to recognize their targets. CD4 downregulation has been described for human immunodeficiency virus (HIV) (11). The HIV protein Vpu targets newly synthesized CD4 for proteasomal destruction (47), whereas HIV Nef removes CD4 from the cell surface (41). Interestingly, both proteins also downregulate MHC-I molecules (29, 42).
CD4 downregulation has also been observed for myxomavirus (MV), a poxvirus that causes myxomatosis in European rabbits (2). T cells infected with MV show reduced surface expression of CD4 but not of other surface markers, such as CD18, CD43, and CD45. MV infection does not affect CD4 biosynthesis or CD4 association with p56lck (2). Instead, CD4 seems to be degraded in lysosomes, since CD4 cell surface expression is restored in the presence of lysosomal inhibitors. The MV gene product mediating CD4 downregulation is not known but seems to belong to the early-gene expression class, because CD4 downregulation is still observed even when late-gene expression is inhibited.
MHC-I molecules undergo a fate very similar to that of CD4 in MV-infected cells, since they are also rapidly internalized and degraded in lysosomes (54). The gene product responsible for MHC-I downregulation was recently found to be encoded by open reading frame M153R (18). The M153R protein belongs to a recently recognized family of viral proteins found in several poxviruses as well as in gamma-2 herpesviruses (15). This family is characterized by an amino-terminal PHD/LAP domain (a RING finger-like domain, also known as a RING-CH domain) (50) followed by two transmembrane domains. Various names have been proposed for this family, such as scrapins or MIR (18, 45). We refer to it as the K3 family of immune evasion proteins on the basis of the K3 genes found in both Kaposi's sarcoma-associated herpesvirus (KSHV) and murine gammaherpesvirus 68 (MHV68) (15). In human and mouse viruses, it was found that K3 downregulates MHC-I (12, 25, 49). In the presence of MHV68 K3, MHC-I is degraded by the proteasome (4). In KSHV K3-transfected cells, however, MHC-I exits the endoplasmic reticulum (ER) normally and reaches the cell surface, where it is rapidly internalized and degraded by lysosomal proteases (12, 25). KSHV contains a second copy of this gene family, K5, which downregulates MHC-I in a manner similar to that of K3 (12, 25). During this process of downregulation, MHC-I is dephosphorylated (38). In addition, K5 directs the internalization of the costimulatory molecules ICAM-1 and B7.2 (13, 24). The difference in substrate specificity between KSHV K3 and KSHV K5 has been localized to the transmembrane domains of the viral and host proteins (45). In addition, the cytoplasmic tails of the host target proteins require the presence of lysine residues (4, 14), which appear to be targeted for the addition of ubiquitin. For MHV68 K3, ubiquitination acts as a proteasomal degradation signal, while in the presence of KSHV K3 and KSHV K5, ubiquitin seems to act as an internalization signal. These findings suggest that the members of this protein family function as ubiquitin ligases (4, 14). This conclusion was also supported by in vitro ubiquitination assays demonstrating that the isolated PHD/LAP domain of K5 mediates the formation of multiubiquitin products in the presence of ubiquitin-activating (E1) and ubiquitin-conjugating (E2) enzymes (14).
The rapid internalization of MHC-I and costimulatory molecules followed by their lysosomal degradation in conjunction with the presence of a K3-family protein in the genome of MV led us to hypothesize that the MV M153R protein mediates the downregulation of CD4. In addition, we were interested in comparing the molecular mechanism of MV M153R-mediated substrate downregulation with that of KSHV K5. Our data reveal both similarities and differences between the downregulation of CD4 by MV M153R and the downregulation of MHC-I by KSHV K5. The results are consistent with a function of the K3 family as a widespread viral family of membrane-bound ubiquitin ligases that mediate the subcellular sorting and thus the degradation of a large variety of transmembrane glycoproteins involved in antiviral immune responses.
MATERIALS AND METHODS
Cell lines.
HeLa-Tet-Off cells, HeLa-CD4 cells, and GHOST cells were maintained in Dulbecco modified Eagle medium (Cellgro) with 10% fetal calf serum (HyClone).
Antibodies and reagents.
Anti-human CD4 (clone 13B8.2) used for flow cytometry experiments was purchased from Immunotech (Marseille, France). Anti-human CD4 (clone ED4-2) used for immunoprecipitation experiments was purchased from BioSource International (Camarillo, Calif.). Anti-human HLA-A-B-C W6/32 was purchased from Sigma (St. Louis, Mo.). Monoclonal antibody BB7.2 recognizing human HLA-A2.1 was obtained from BD PharMingen (San Diego, Calif.). Phycoerythrin (PE)-conjugated anti-mouse antibody was purchased from Sigma. Antiubiquitin monoclonal antibody P4D1 was obtained from Santa Cruz Biotechnology (Santa Cruz, Calif.).
Primary antibodies used for immunofluorescence were as follows: anti-EEA-1 (1:200; Transduction Labs), anti-Golgin-97 (1:100; Molecular Probes), anticalnexin (1:500; Stressgen), anti-gamma-AP-1 (1:200; Sigma), anti-CD4 (1:100; Immunotech), anti-transferrin receptor (TfR) (1:100; Zymed), and anti-Flag (1:400; Sigma).
Bafilomycin A and concanamycin A (both from Sigma), ammonium chloride (Fisher Scientific, Federal Way, Wash.), and MG132 (Boston Biochem, Cambridge, Mass.) were used at final concentrations of 2 μM, 50 nM, 50 mM, and 50 μM, respectively. Protein A or G beads were obtained from Santa Cruz Biotechnology. Leupeptin (Sigma) was used at 0.3 mM, and lactacystin (Boston Biochem) was used at 20 μM.
DNA constructs.
Plasmids A2.1/pUHD10.1 and A2.1d311/pUHD10.1 were described previously (38). For the construction of A2.1K311S, the codon for the residual lysine in the cytoplasmic tail of the A2.1d311 construct was converted to a serine codon by using site-directed mutagenesis. A 264-bp fragment was amplified from A2.1d311/pUHD10.1 template DNA by using the following primers: A, 5′-GAA GGC CTG CAG GGG ATG GAA CC; and B, 5′-CGC GCG GCA TGC TCA GGC GTA ATC TGG AAC ATC GTA TGG GTA ACT CCT CCT CCA. The resulting PCR product was digested with StuI and SphI and inserted into the corresponding sites in A2.1d311/pUHD10.1, thereby replacing the respective coding regions. The presence of the mutation in the resulting construct, A2.1K311S/pUHD10.1, was confirmed by sequencing. Plasmid A2.1K311S/pUHD10.1 was then used as a template to create the A2.1P317K construct, where a lysine was introduced into the sequence coding for the hemagglutinin (HA) epitope tag. Primer A was combined with primer C (5′-CGA ATT GCA TGC TCA GGC GTA ATC CTT AAC ATC GTA TGG) to amplify a 264-bp fragment by using A2.1K311S/pUHD10.1 as a template. The PCR fragment was digested with StuI and SphI and inserted into the corresponding sites in A2.1K311S/pUHD10.1. The conversion of the codon from proline to lysine was confirmed by sequencing.
K5-Flag/pUHD10-1 was constructed by inserting the coding region for K5-Flag from K5-Flag/pBI (38) into pUHD10-1 (17) by using the following primers: forward, 5′-CCT AGA ATT CAC CAT GGC GTC TAA GGA C-3′; and reverse, 5′-CCT AGG TAC CCT TGT CGT CGT CGT C-3′. The resulting PCR fragment was inserted between the EcoRI and KpnI sites in pUHD10-1. In addition, a BsiWI site was inserted prior to the first transmembrane domain by PCR-directed mutagenesis by using the following primers: forward, 5′-TGT GGC CGG AGA TGG AGC GTA CGG AAA TTT TTG AAC TGT-3′; and reverse, 5′-ACA GTT CAA AAA TTT CCG TAC GCT CCA CT CCG GCC ACA-3′. This mutagenesis resulted in a switch of one amino acid (Gln83→Thr). The restriction site introduced in this way was not used in this study.
The coding regions of M153R were amplified from their respective genomes and cloned into pcDNA3.1/Myc-His(+) version A (Invitrogen, Carlsbad, Calif.). Crude MV (strain Lausanne) DNA was incubated with primers having the sequences 5′-GCG AAG CTT ATG GCT ACT GTT GTA AAC-3′ (HindIII site is underlined) and 5′-GCG CTC GAG GCG GGT GAC TCC ACG ACG-3′ (XhoI site is underlined). An amplified 534-bp product was cleaned with a PCR purification kit (Gibco/Invitrogen), digested simultaneously with HindIII and XhoI, and cloned into those same sites in pcDNA3.1.
For M153R-Flag/pUHD10-1, the coding region for M153R was amplified from M153R/pcDNA3 by using the following primers: forward, 5′-ATC GGA ATT CAC CAT GGC TAC TGT TGT A-3′; and reverse (Flag), 5′-ATC GGG TAC CTC ACT TGT CAT CGT CAT CCT TGT AGT CAG CGG GTG ACT CCA CGA C-3′. The resulting PCR fragment was inserted between the EcoRI and KpnI sites of pUHD10-1 (17).
The pHIV-CD4 and pHIV-CD4KRcyto DNAs were described previously (47) and were obtained from K. Strebel (National Institutes of Health [NIH]). CD4-WT/pUHD10.1 and CD4KRcyto/pUHD10.1 were constructed by PCR with these plasmids as templates. The following primers were used to amplify the respective coding regions from pHIV-CD4 and pHIV-CD4KRcyto: forward, 5′-CGG AAT TCA CCA TGA ACC GGG GAG TCC CT-3′; and reverse, 5′-CGG GAT CCT TAT CAA ATG GGG CTA CAT GTC-3′. The resulting fragments were digested with EcoRI and BamHI and inserted into similar sites in plasmid pUHD10.1. The resulting constructs were confirmed by sequencing.
N-terminal glutathione S-transferase (GST) fusions of KSHV K5 PHD (amino acids 1 to 83) and M153R PHD (amino acids 1 to 94) were constructed by using pGEX-4T-1 (Amersham Pharmacia Biotech, Piscataway, N.J.). Restriction sites used for inserting the PHD-encoding regions were EcoRI and NotI. The following primers were purchased from Invitrogen: M153R-forward, 5′-GCG TCA CGG AAT TCA TGG CTA CTG TTG TAA ACA TGG AC-3′; M153R-reverse, 5′-TTA TCG ATA CGC GGC CGC TCA ACT ATC GTG GCA GTC GCG TTT ATA-3′; K5-forward, 5′-GCG TCA CGG AAT TCA TGG CGT CTA AGG ACG TAG AAG AG-3′; and K5-reverse, 5′-TTA TCG ATA CGC GGC CGC TCA TTC TTG GCG CTC CAT CTC CGG CCA-3′. These fusion proteins, as well as GST- ICP0-RING (amino acids 1 to 241 of ICP0) (obtained from R. Everett, Glasgow, Scotland) and GST-MmUbc6 and GST-MmUbc7 (obtained from A. Weissman), were expressed in either BL21 cells (Amersham) or Top10 cells (Invitrogen) and purified as previously described (5, 51). Purified E3 proteins were stored at −4°C, and E2 proteins were stored at −80°C in10% glycerol.
Expression plasmid Hrs/pcDNA3.1 was obtained from P. Woodman and was described previously (3).
Construction of recombinant poxviruses.
To generate recombinant vaccinia viruses (VVs) expressing M153R (VV-M153R), a HindIII-PmeI fragment was isolated from plasmid pcDNA3.1/M153R and cloned into vector pZVneo (20). The K5 gene was isolated from K5-Flag/pBI and cloned into pZVneo. Recombinant VVs were generated in CV-1 cells as described previously (37). VVs expressing CD4 (VV-CD4) and CD4 with a deletion of the cytoplasmic domain (VV-CD4Δcyto) (29) were generously provided by J. Yewdell (NIH).
To construct an MV with a disrupted copy of M153R, PCR primers that anchored within the coding region and amplified the flanking regions were designed by using viral genomic DNA as a template. The left flanking region was amplified by using JB27.01 (GCGGAGCTCATGGATATCTTTAATC) (SacI site in italic type) and JB33.01 (GCGACTAGTTCCAGGTTTACAACAGTG) (SpeI site in italic type). The right flanking region was amplified by using JB29.01 (GCGAAGCTTCTCGTGAACGTAATAACGATTC) (HindIII site in italic type) and JB30.01 (GCGCTCGAGATGGCGCGGTATATTATC) (XhoI site in italic type). The flanking regions were then cloned into pBluescript SKII. This step resulted in the loss of 450 of the 618 coding bases, or 73% of the M153R coding region. A green fluorescent protein (GFP) cassette under the control of a VV synthetic early-late promoter was cloned into the PstI and EcoRI sites between the two flanking regions. The resulting plasmid (pBS153KO) was transfected into baby green monkey kidney cells infected with MV (strain Lausanne) by following standard procedures. Recombinant virus was purified by three rounds of isolating fluorescent green foci.
Transfection.
Approximately 5 × 105 cells were transfected with a total of 1 μg of DNA in 6-cm dishes by using Effectene reagent (Qiagen, Valencia, Calif.). Plasmid pZsGreen1-C1 (Clontech) was used as a control for transfection. When necessary, samples were cotransfected with a vector plasmid (pUHD10-1) to keep the plasmid ratio constant. The best transfection results were obtained with 5 μl of Effectene reagent. The transfected cells were incubated at 37°C with 5% CO2 for 24 h before being analyzed.
Flow cytometry.
Cells were removed from tissue culture dishes with 0.05% trypsin-EDTA (Invitrogen), washed with ice-cold phosphate-buffered saline (PBS), and incubated with Cy-chrome-conjugated anti-HLA-A-B-C antibody W6/32 or with monoclonal antibody BB7.2 to HLA-A2.1 for 30 min on ice (at 1:100). Cells were washed with ice-cold PBS, either resuspended in ice-cold PBS or incubated with PE-conjugated anti-mouse secondary antibody (1:100 dilution), and washed again before analysis with a BD Biosciences (San Jose, Calif.) FACScalibur flow cytometer.
Immunofluorescence.
A total of 1.5 × 104 cells were plated on 15-mm coverslips (Fisher) and allowed to adhere overnight prior to infection or transfection. Following infection or transfection, cells were washed with PBS, fixed with 2% paraformaldehyde for 20 min at room temperature, and permeabilized with 0.2% Triton X-100 for 3 min at room temperature. Nonspecific binding sites were blocked with 3% bovine serum albumin and 0.5% fish gelatin in PBS (blocking solution) for 30 min at 37°C. Fixed cells were incubated overnight at 37°C with primary antibody diluted in blocking solution. Secondary and conjugated antibodies were diluted in blocking solution and incubated with cells for at least 30 min at 37°C. Cells were washed six times with PBS between all antibody treatments. Cells were fixed a second time with 2% paraformaldehyde after the final antibody treatment and washed twice with PBS. Coverslips were then mounted on slides and covered with Vectashield H-1200 plus 4′,6′-diamidino-2-phenylindole (Vector Laboratories, Burlingame, Calif.). Slides were visualized by using an Axioskop 2 light microscope (Zeiss, Thornwood, N.Y.). All pictures were taken in monochrome format, contrast enhanced, and artificially colored by using Openlab software (Improvision, Lexington, Mass.).
Uptake assay.
Cells were washed twice with cold PBS and transferred to 4°C. Anti-CD4 antibody was added to the cells and incubated for 30 min. Cells were fixed either immediately (0 min) or after the addition of fresh Dulbecco modified Eagle medium and incubation at 37°C for 2 h. Cells were stained with an Alexa Fluor:594 (1:300; Molecular Probes) secondary antibody and visualized as described for immunofluorescence.
Infection, metabolic labeling, and immunoprecipitation.
HeLa cells were grown to 90% confluence in 100-mm tissue culture dishes and infected with VV constructs at a multiplicity of infection of 8 PFU per cell. At 2.5 h postinfection, cells were incubated in serum-free and methionine-free medium for 30 min and metabolically labeled with [35S]cysteine-[35S]methionine (250 μCi; Amersham) for various times. After being labeled, cells were washed twice with PBS and lysed immediately in PBS containing 1% Triton X-100 and protease inhibitors (Roche, Indianapolis, Ind.); alternatively, the label was chased with excess (2 mM) unlabeled cysteine and methionine in the presence or absence of specific inhibitors for various times. Cell lysates were precleared with protein A or G-agarose beads for 1 to 16 h and incubated with 3 μg of antibody for 1 h and then with protein A or G beads for 1 h. Immunoprecipitated proteins were washed five times with 0.1% Triton X-100 in PBS. All samples were boiled in sample buffer and analyzed by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE). Some samples were treated with endoglycosidase H (endo H) (Roche) according to the manufacturer's instructions. Briefly, samples were initially denatured at 100°C in 40 μl of 0.3% SDS-150 mM 2-mercaptoethanol-50 mM sodium acetate (pH 5.5) and then incubated either without or with 4 mU of endo H overnight at 37°C. Gels were fixed, dried, and exposed to Kodak BioMax Transcreen-LE with an intensifying screen.
Ubiquitination assays.
In vitro reaction mixtures (20 μl total) for ubiquitination assays included 50 nM rabbit E1 (Boston Biochem), 28 μM ubiquitin, 10 mM ATP, 50 mM Tris-HCl (pH 7.5), 50 mM NaCl, and 2 mM dithiothreitol. E2 enzymes were used at concentrations ranging from 0.7 to 2.3 μM, and E3 enzyme concentrations ranged from 3 to 25 μM. After 90 min of incubation at 30°C, reactions were stopped by the addition of LDS reducing sample buffer (Invitrogen). Samples were electrophoresed through 4 to 12% Bis-Tris Novex gels (Invitrogen) and transferred to polyvinylidene difluoride membranes (Millipore) for Western blotting by using mouse antiubiquitin antibody P4D1 and a WesternBreeze chemiluminescence detection system (Invitrogen).
Nucleotide sequence accession number.
The corrected sequence for MV strain Lausanne M153R (see Results) was deposited in GenBank under accession number NC_001132.
RESULTS
M153R downregulates CD4 surface expression.
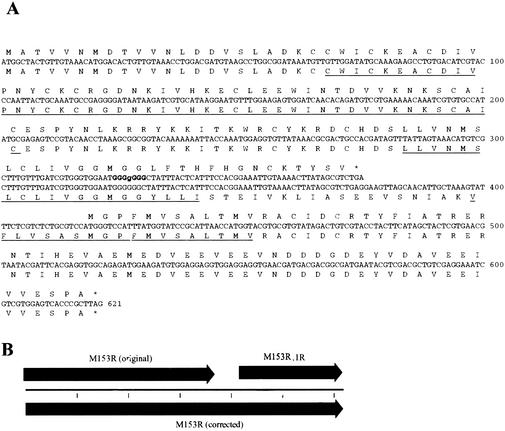
Several poxvirus family members contain open reading frames that are similar to the gamma-2 herpesvirus K3 genes (15, 18). The gamma-2 herpesvirus K3 family is characterized by a conserved PHD/LAP domain followed by two transmembrane domains. This topology has also been predicted for all K3-related poxvirus proteins, except for that encoded by open reading frame M153R of MV strain Lausanne, which lacks a second transmembrane domain (7). To determine whether this truncation was due to a mutation in this strain or a sequencing error, we resequenced this part of the MV strain Lausanne genome. A sequencing error was found that resulted in a predicted premature stop codon (Fig. 1B). The predicted protein encoded by the corrected MV M153R sequence contains two putative transmembrane domains (Fig. 1A).
FIG. 1.
Sequence of MV Lausanne M153R. A correction to the original MV strain Lausanne sequence (GenBank accession number AF170726) at position 148855 resulted in the collapse of two predicted open reading frames (ORFs) into a single CDS. (A) The original putative amino acid sequences for M153R and M153R.1R are presented above the nucleotide sequences. An extra guanine in a stretch of six G's (bold type) resulted in the incorrect identification of the termination of the original M153R ORF and the prediction of a second ORF denoted M153R.1R. The corrected predicted amino acid sequence is presented below the nucleotide sequence. The predicted PHD domain and transmembrane domains are underlined. (B) Schematic of the change in the ORF map.
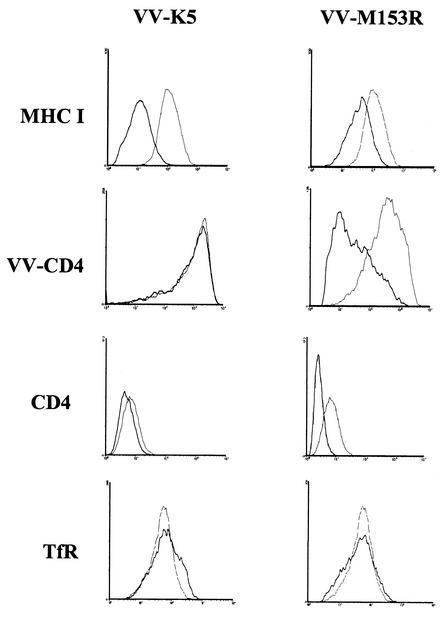
MV Lausanne was previously shown to downregulate the T-cell coreceptor CD4 in infected rabbit T cells (2). Moreover, the M153R gene of strain T1 was shown to downregulate MHC-I as well as the Fas ligand CD95 (18). To examine whether M153R is responsible for CD4 downregulation, we generated recombinant VV-M153R. For comparison, KSHV K5 was also expressed from VV (VV-K5). We monitored the surface expression of human CD4 in HeLa cells either coinfected with VV-CD4 or constitutively expressing CD4 by flow cytometry with anti-CD4 antibody. In addition, MHC-I surface expression was measured by flow cytometry with HLA-A-B-C-specific monoclonal antibody W6/32. A reduction in MHC-I surface expression was observed upon infection with VV-M153R compared to the results for cells infected with wild-type VV, whereas no downregulation of TfR was observed (Fig. 2). These results are consistent with previous observations in human cells transfected with M153R of strain T1 (18). The M153R-mediated MHC-I downregulation was somewhat less pronounced than that in cells infected with VV-K5. This difference might have been caused by the divergence of human MHC sequences from rabbit MHC sequences. In addition, CD4 surface expression was clearly reduced in VV-M153R-infected cells, whereas VV-K5 had only a minor effect on CD4 levels. A reduction in CD4 surface expression was also observed in HeLa-CD4 cells infected with VV-M153R. We conclude that M153R, but not KSHV K5, interferes with CD4 surface expression.
FIG. 2.
M153R downregulates MHC-I and CD4. HeLa or HeLa-CD4 cells infected with wild-type VV (light line), VV-K5 (left panel, dark line), or VV-M153R (right panel, dark line) were analyzed by flow cytometry. Surface expression of endogenous MHC-I was visualized with Cy-chrome-labeled monoclonal antibody W6/32. CD4 expression was achieved by coinfection with VV-CD4 or constitutive expression in HeLa-CD4 cells. Surface expression of CD4 was detected with anti-CD4 monoclonal antibody. TfR expression was detected with anti-TfR monoclonal antibody, and nontransfected cells served as a control (light line) in this experiment. Note that both CD4 and MHC-I are downregulated by M153R, whereas K5 downregulates MHC-I only. TfR is downregulated by neither M153R nor K5.
Deletion of M153R from MV restores CD4 expression.
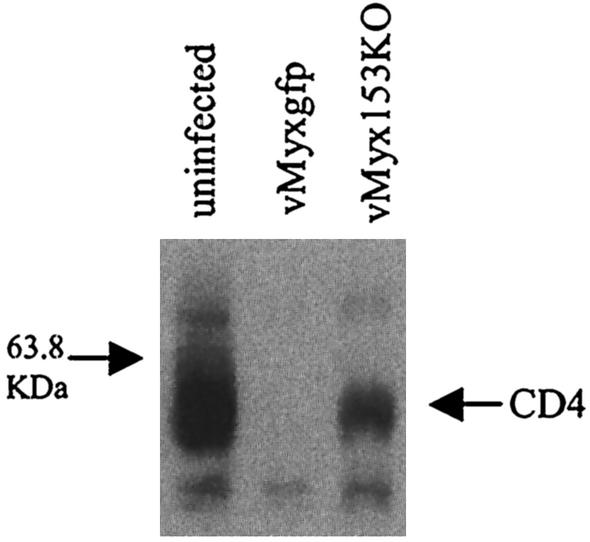
The results presented above suggested that M153R was responsible for the previously described downregulation of CD4 by MV (2). To examine whether deletion of M153R restores the expression of CD4 in MV-infected cells, we generated a deletion mutant (see Materials and Methods). Deletion of M153R was confirmed by PCR of genomic DNA (data not shown). CD4 downregulation was studied in human GHOST cells. This cell line is derived from HOS human osteosarcoma cells, and the growth of MV in GHOST cells was established previously (J. Barrett and G. McFadden, unpublished observations). GHOST cells are transfected with CD4 and HIV coreceptors (9) and therefore express high levels of CD4, as confirmed by immunoblotting of total lysates (Fig. 3). However, CD4 was completely absent 24 h after infection with wild-type MV (Fig. 3, second lane). In contrast, deletion of M513R restored CD4 expression. These data clearly show that MV requires M153R to completely inhibit CD4 expression and confirm that M153R is responsible for the specific downregulation of CD4 by MV.
FIG. 3.
Deletion of M153R from MV restores CD4 expression. A total of 106 GHOST cells were either mock infected or infected with GFP-expressing MV (vMyxgfp) or M153 knockout virus (vMyx153KO). Infections were allowed to proceed overnight, and samples were collected 24 h postinfection. Cells were lysed in 1% Triton X-100 lysis buffer, and 50-μg samples of whole-cell lysates were run on a 10% polyacrylamide gel, blotted, and probed with a rabbit polyclonal anti-human CD4 antibody.
CD4 is degraded in lysosomes in the presence of M153R.
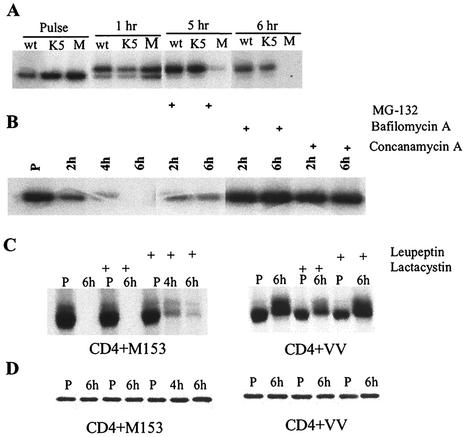
The molecular mechanism used by M153R to inhibit the surface expression of its target proteins is not known. The M153R-related proteins of KSHV, K3 and K5, target their substrates (MHC-I or the costimulatory molecules B7.2 and ICAM-1) to lysosomes for destruction (12, 13, 24, 25). In contrast, in MHV68 K3-transfected cells, MHC-I is degraded by proteasomes (4, 49). To determine the fate of CD4 in M153R-expressing cells, we studied the biosynthesis of CD4 by metabolic labeling and immunoprecipitation. HeLa cells were coinfected with VV-CD4 and VV-M153R or infected with VV-CD4 alone. As an additional control, we coinfected cells with VV-K5 and VV-CD4. At 2.5 h postinfection, cells were labeled for 30 min and then the label was chased for 1 to 6 h. After detergent lysis and immunoprecipitation with anti-CD4 antibody, samples were digested with endo H to determine the maturation of the N-linked glycosylation of CD4 during the chase. As shown in Fig. 4A, CD4 was endo H sensitive during the pulse and became endo H resistant during the chase in all samples, indicating that CD4 exited the ER. However, CD4 molecules disappeared from lysates of VV-M153R-infected samples toward the end of the chase, indicating that CD4 is degraded in VV-M153R-infected cells after exiting the ER.
FIG. 4.
CD4 exits the ER and is degraded in lysosomes in the presence of M153R. (A) Intracellular transport of CD4. HeLa cells were infected with wild-type VV (wt), VV-K5 (K5), or VV-M153R (M). Cells were labeled for 30 min (pulse) and chased for the indicated times. CD4 molecules were immunoprecipitated and treated with endo H. (B) HeLa cells were coinfected with VV-CD4 and VV-M153. Stabilization of CD4 was tested in the presence or absence of the indicated inhibitors. (C) HeLa cells were infected with VV-CD4 and VV-M153R or with VV-CD4 and wild-type VV, labeled for 30 min (P), and chased for up to 6 h in the presence or absence of the indicated inhibitors. (D) One-third of the same cell lysate as that used in panel C was immunoprecipitated with antivimentin as a loading control.
To determine whether proteasomal or lysosomal proteolysis was responsible for CD4 degradation, VV-M153R- and VV-CD4-coinfected cells were treated with inhibitors of either proteasomal or lysosomal degradation. Infected HeLa cells were pulse-labeled with [35S]methionine for 30 min at 3 h postinfection. During the following chase period of 4 to 6 h, we applied either proteasomal inhibitor MG132 or inhibitors of lysosomal acidification: bafilomycin A, concanamycin A, and ammonium chloride. Both concanamycin A and bafilomycin A, specific inhibitors of vesicular H+-ATPases, have been reported to interfere with early to late endosomal transport (6, 53). Immunoprecipitation and SDS-PAGE showed that CD4 was degraded in the absence of inhibitors, as in the previous experiment (Fig. 4B). The proteasomal inhibitor MG132 did prevent some degradation, but complete inhibition of CD4 degradation was observed only in cells treated with the inhibitors of vesicular H+-ATPases. The peptide-aldehyde inhibitor MG132 is also a potent inhibitor of calpains and lysosomal cysteine proteases, e.g., cathepsins (31).
In order to determine whether the observed stabilization of CD4 by MG132 was the result of proteasome inhibition or inhibition of lysosomal proteases, we treated the cells with the specific proteosome inhibitor lactacystin. In addition, we used leupeptin, an inhibitor of lysosomal serine and cysteine proteases. As shown in Fig. 4C, no stabilization of CD4 was observed with lactacystin, even upon overexposure of the autoradiograph. In contrast, leupeptin had a partial stabilizing effect. Together with the acquisition of endo H resistance prior to degradation, these data indicate that CD4 degradation occurs mainly in lysosomes. These data are consistent with the lysosomal degradation of CD4 observed in MV-infected cells (2).
M153R localizes to endosomes and the trans-Golgi network.
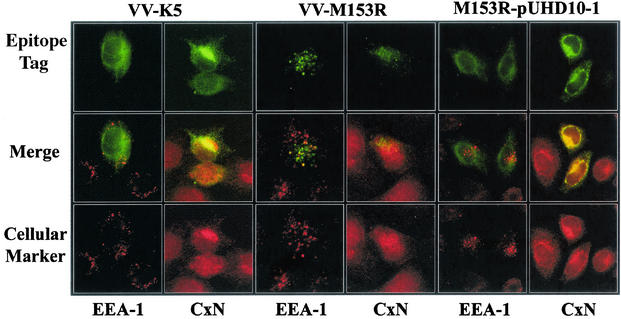
Both murine and human gamma-2 herpesvirus homologs of M153R localize to the ER (4, 12, 38). The primary structure of M153R reveals two putative transmembrane domains, suggesting that M153R is a type III transmembrane protein. To determine the location of M153R, we analyzed HeLa cells infected with VV-M153R by immunofluorescence (Fig. 5). In contrast to K5, M153R did not colocalize with calnexin but showed a punctuate staining pattern that partially overlapped the staining pattern for early endosomes (EEA-1). This observation is in contrast to the previous report that M153R of strain T1 localizes to the ER (18). Since M153R expressed from plasmids was used in the previous experiment, we examined the staining pattern for M153R upon transfection with an expression plasmid. As shown in Fig. 5, plasmid-expressed M153R colocalized with calnexin but not with EEA-1. Thus, it seems that M153R is an ER-resident protein. During viral infection, however, it is targeted to endosomes. This effect could be a by-product of membrane rearrangements occurring during viral envelopment (48). Importantly, K5 does not seem to share this fate, suggesting differences in their subcellular targeting signals.
FIG. 5.
Subcellular localization of M153R. HeLa cells were infected with VV-K5 (left panels) or VV-M153R (middle panels) at a multiplicity of infection of 0.1. At 18 h postinfection, cells were stained with antibodies against the cellular marker EEA-1 (early endosomes) or calnexin (CxN) (ER). The right panels show HeLa cells transfected with M153R expressed from expression vector pUHD10-1 (17) and stained as described above. (Bottom row) The binding of primary antibodies was visualized with an Alexa Fluor:568 secondary antibody (red). (Top row) VV-M153R carries a Myc tag and was stained with an FITC-conjugated monoclonal anti-Myc antibody (green). VV-K5 and M153R-pUHD10-1 are Flag tagged. Colocalization is visualized as yellow in the merge panels.
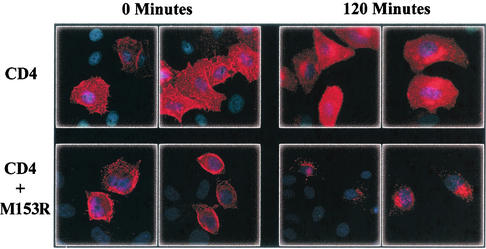
Next, we wanted to know whether the intracellular location of M153R was essential for its function in targeting CD4 to the lysosomal compartment. Therefore, we studied the fate of CD4 in an uptake assay with M153R-cotransfected cells. CD4-cotransfected HeLa cells were incubated with anti-CD4 antibody for 30 min on ice and then incubated for 2 h at 37°C. In cells transfected with CD4 only, CD4 staining remained uniform during this treatment, consistent with cell membrane localization or the rapid recycling described for CD4 expression in HeLa cells (39) (Fig. 6). In contrast, in HeLa cells cotransfected with M153R and CD4, many cells showed punctuate staining after incubation at 37°C, indicating that CD4 was removed from the cell surface by endocytosis (Fig. 6). Thus, M153R induces the rapid removal of CD4 from the cell surface in a manner very similar to the uptake of MHC-I molecules observed in KSHV K3- and KSHV K5-transfected cells (12, 24). Our data also suggest that this function can be performed by M153R at different locations along the exocytic and endocytic routes.
FIG. 6.
M153R enhances endocytosis of CD4. HeLa cells were transfected with either pHIV-CD4 or pHIV-CD4 together with M153R-Flag/pUHD10-1. At 24 h posttransfection, cells were transferred to 4°C and stained with anti-CD4 antibody. Cells were then either fixed or transferred to 37°C for 120 min to allow for internalization of antibody bound to CD4. Localization of CD4 was visualized after fixation with an Alexa Fluor:594 secondary antibody (red).
Lysines in the cytoplasmic tails of target molecules are required for M153R-mediated downregulation.
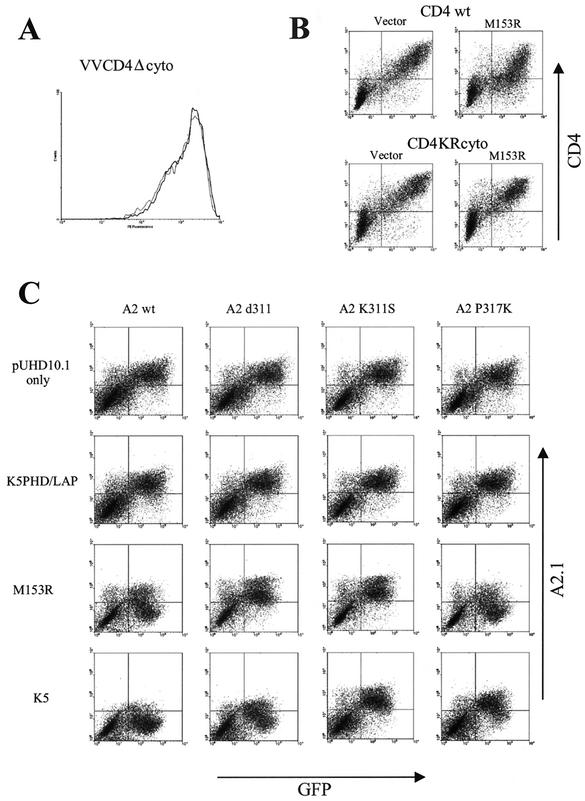
The resemblance of the fate of CD4 in the presence of M153R to that of MHC-I and costimulatory molecules B7.2 and ICAM-1 in KSHV K5-transfected cells suggested that M153R might act by a mechanism similar to that of K5. For KSHV K5, it was shown that the downregulation of MHC-I as well as B7.2 required the presence of lysine residues in their cytoplasmic tails (14). To examine whether lysines in the cytoplasmic domains of CD4 and MHC-I are required for downregulation by M153R, we monitored the cell surface expression seen with VV-CD4Δcyto infection in the presence of VV-M153R in HeLa cells. No difference in the level of surface expression was observed relative to that seen with VV-CD4Δcyto infection in the absence of VV-M153R (Fig. 7A; the control experiment with VV-CD4 [wild type] is shown in Fig. 2). Thus, the tail of CD4 is required for downregulation by M153R.
FIG. 7.
M153R requires lysine residues to downregulate CD4 and HLA-A2.1. (A) HeLa cells were infected with VV-CD4Δcyto in the presence (dark line) or absence (light line) of M153R. (B) (Top) HeLa cells were transfected with GFP together with CD4 only (left) or with CD4 and M153R (right). (Bottom) HeLa cells were transfected with GFP together with CD4KRcyto only (left) or with CD4KRcyto and M153R (right). Surface expression of CD4 was visualized by using anti-CD4 antibody together with PE-conjugated anti-mouse antibody. CD4 staining is plotted against GFP fluorescence. (C) The indicated HLA-A2.1 constructs were cotransfected into HeLa cells with GFP and either a vector control; K5PHD/LAP, a nonfunctional mutant of K5 (38); M153R-Flag/pUHD10.1; or K5/pUHD10.1. HLA-A2.1 surface expression was monitored by using HLA-A2.1-specific antibody BB7.2. HLA-A2.1 staining is plotted against GFP fluorescence. wt, wild type.
To evaluate whether lysines in the tail of CD4 were necessary, we took advantage of a previously described CD4 construct in which all cytoplasmic lysines were replaced with arginines (CD4KRcyto) (47) (kindly provided by K. Strebel). Both wild-type CD4 and CD4KRcyto were subcloned into expression vector pUHD10.1 and either cotransfected with M153R expressed from the same vector or cotransfected with the vector only. For identification of transfectants, cells were also cotransfected with a plasmid expressing GFP. After staining with anti-CD4 antibody, the levels of surface expression in the presence or absence of M153R were compared. As shown in Fig. 7B, the levels of surface expression were reduced for wild-type CD4 but not for the lysine mutant, suggesting that tail lysines are important for CD4 downregulation.
In a second series of experiments (Fig. 7C), the role of cytoplasmic lysines was examined by using a panel of derivatives of the human MHC-I molecule HLA-A2.1. A truncated version of HLA-A2.1, A2.1d311, which was still downregulated by KSHV K5 but not by KSHV K3, despite lacking most of the HLA-A2.1 cytoplasmic tail, was previously described (38). This construct was cotransfected into HeLa cells with either KSHV K5 or M153R. An HLA-A2.1 construct that contained most of the cytoplasmic tail followed by an HA epitope tag (A2.1d331) and a mutant KSHV K5 construct that was nonfunctional were included as controls (38). Surface expression of HLA-A2.1 was monitored with the HLA-A2.1-specific monoclonal antibody BB7.2. The intensities of HLA-A2.1 fluorescence versus the fluorescence of cotransfected GFP are depicted in Fig. 7C. The truncated version was not downregulated by M153R, whereas surface expression for A2.1d331, containing an additional 20 amino acids in its tail, was reduced. The A2.1d311 construct contains the cytoplasmic tail sequence RRK followed by the HA epitope tag sequence YDYDVPDYA. The remaining lysine in this construct was replaced by a serine to generate A2.1K311S. A lysine (underlined) was then introduced into the HA epitope tag of A2.1K311S to generate the cytoplasmic tail sequence RRSDYDVKYA in construct A2.1P317K. Removal of the lysine from A2.1d311 rendered the resulting molecule resistant to KSHV K5-mediated downregulation, and the introduction of a lysine into the HA epitope tag restored HLA-A2.1 downregulation by KSHV K5. Interestingly, A2.1P317K was also downregulated by M153R. This result suggests that the presence of at least one lysine in the tails of M153R substrates is an essential prerequisite for downregulation. However, since A2.1d311, which also contains a single lysine, was not downregulated by M153R, we further conclude that the presence of a lysine is not sufficient. It is possible that the position of the lysine in the cytoplasmic tail plays a role. For instance, the distance of the lysine residues from the transmembrane domain could be a deciding factor, since the lysine in the A2.1d311 construct is separated from the transmembrane domain by only two residues. At present, we do not know the exact sequence or spatial requirements. However, these experiments provide clear evidence for a crucial role for lysines in the tails of M153R substrates.
Increased ubiquitination of CD4 by M153R.
The finding that the introduction of a lysine into the HA epitope tag, a sequence unrelated to the cytoplasmic tail of HLA-A2.1, rendered the resulting molecule susceptible to M153R downregulation suggested that lysines in the tails of substrate molecules are required for M153R downregulation, not as part of a recognition motif but for a different reason. One possibility is that the lysines represent targets for posttranslational modifications, such as ubiquitination of the ɛ-amino group of lysines. A hypothetical role for M153R as a ubiquitin ligase was further suggested by the similarity of the PHD/LAP domain to the RING finger domain, a motif frequently found in ubiquitin ligases (26). The corresponding domain of KSHV K5 has been shown to catalyze ubiquitination in vitro, and MHC molecules have been observed to be ubiquitinated in the presence of KSHV K5 (14). For these reasons, we examined whether CD4 is ubiquitinated in the presence of M153R and whether the purified PHD domain of M153R acts as a ubiquitin ligase in vitro.
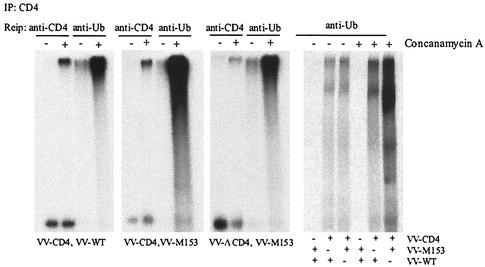
Ubiquitination of CD4 was studied by coinfection of cells with VV-CD4 and VV-M153R. Controls were VV-CD4 coinfected with wild-type VV or tailless VV-CD4Δcyto coinfected with VV-M153R. We also treated cells with concanamycin A to prevent the degradation of CD4. At 3 h postinfection, cells were metabolically labeled for 4.5 h, lysed, and immunoprecipitated with anti-CD4 antibody. The resulting immunoprecipitate was reimmunoprecipitated with either anti-CD4 antibody or antiubiquitin antibody P4D1. The results are shown in Fig. 8. As observed above, the amount of CD4 was reduced in VV-M153R-infected cells in comparison to that of tailless CD4 or CD4 expressed in the absence of M153R (Fig. 8; compare the fifth lane to the first and ninth lanes). Reimmunoprecipitation with antiubiquitin antibody revealed the presence of a high-molecular-weight species that was observed regardless of the presence of M153R or the cytoplasmic tail (Fig. 8; compare the 7th lane with the 3rd and 11th lanes). In addition, a high-molecular-weight protein species that barely entered the SDS gel was coimmunoprecipitated with anti-CD4 antibody in concanamycin A-treated cells in each infection. Reimmunoprecipitation with antiubiquitin antibody yielded a similar high-molecular-weight protein, whereas the lower-molecular-weight CD4 was not reimmunoprecipitated with P4D1. This high-molecular-weight complex was not precipitated from HeLa cells that were not coinfected with CD4 (Fig. 8, right panel), suggesting that a polyubiquitinated CD4 conjugate was stabilized in the presence of concanamycin A. Moreover, a significantly larger amount of ubiquitinated, full-length CD4 was recovered from M153R-coinfected cells in comparison to either tailless CD4 or CD4 expressed in the absence of M153R. Increased CD4 ubiquitination was observed in several independent experiments. One of the repeat experiments is shown in the right panel of Fig. 8. This increased ubiquitination of CD4 in the presence of M153R is consistent with a potential function of M153R as a ubiquitin ligase.
FIG. 8.
Increased CD4 ubiquitination in VV-M153R-infected cells. HeLa cells were infected with VV-CD4 and wild-type (WT) VV, with VV-CD4 and VV-M153R, with VV-CD4Δcyto and VV-M153R, or with VV-M153R and WT VV. CD4 molecules were immunoprecipitated (IP) from lysates of cells labeled for 4 h in the presence or absence of concanamycin A. CD4 was dissociated from protein A-Sepharose beads in 1% SDS followed by reimmunoprecipitation (Reip) with either anti-CD4 or antiubiquitin (anti-Ub) antibody. The right panel is from an independent experiment.
The PHD/LAP domain of M153R acts as a ubiquitin ligase in vitro.
Ubiquitination involves a cascade of proteins that activate and transfer ubiquitin (for a review, see reference 21). Ubiquitin-activating or E1 enzymes transfer ubiquitin to ubiquitin-conjugating or E2 enzymes, which in turn transfer ubiquitin to substrate molecules. The latter step is facilitated by ubiquitin ligases or E3 enzymes in two ways. First, ubiquitin ligases facilitate ubiquitination by bringing the E2 enzyme and the substrate into close proximity. Second, ubiquitin ligases catalyze, by an unknown mechanism, the ubiquitination reaction, as shown by the formation of polyubiquitinated proteins by coincubation with E1, E2, E3, ubiquitin, and ATP (27). For RING E3 enzymes, it is sufficient to include the RING-domain-containing fragment of the protein in this reaction. For instance, an N-terminal GST fusion of the RING domain of the herpes simplex virus immediate-early transactivator, ICP0, was shown in vitro to act as an E3 ligase in the presence of the E2 enzyme UbcH5a (5, 19). Furthermore, an N-terminal GST fusion of the KSHV K5 PHD domain catalyzed the formation of high-molecular-weight ubiquitin adducts in the presence of UbcH5a (14). To ascertain the ability of the PHD domain of M153R to act as a ubiquitin ligase in vitro, we fused GST to the N-terminal 94 amino acids of M153R. As positive controls, we used GST-ICP0-RING (amino acids 1 to 241) and a fusion generated between the KSHV K5 PHD domain (amino acids 1 to 83) and GST. Immunoblotting with antiubiquitin antibody of ICP0 incubated with UbcH5a (as well as ubiquitin, E1, and ATP) resulted in ubiquitinated protein bands as well as a high-molecular-weight smear similar to the one reported previously (5, 19) (Fig. 9, right panel). These high-molecular-weight ubiquitinated proteins were not observed in the presence of UbcH5a alone, whereas low-molecular-weight monoubiquitin and presumably diubiquitin were observed in all reactions. We observed both antiubiquitin antibody-reactive protein and high-molecular-weight smears in the presence of GST-M153R PHD, similar to those seen with GST-ICP0-RING. These data clearly support an E3 ligase activity for M153R.
FIG. 9.
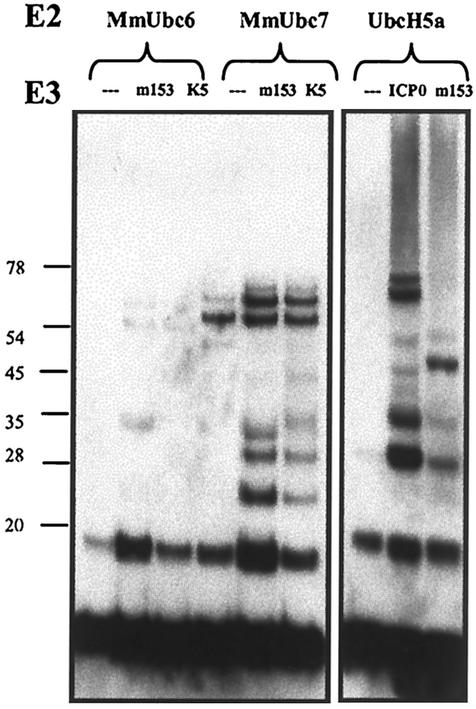
Ubiquitin ligase activity of the M153R PHD/LAP domain in vitro. (Right panel) Comparison of the ubiquitination activities of the ICP0-RING domain and the M153R PHD domain. The ubiquitination reaction mixture containing E1, UbcH5a, ubiquitin, and ATP, together with purified GST fusions with the ICP0-RING domain or M153R PHD domain, was separated by SDS-PAGE and immunoblotted with antiubiquitin antibody. High-molecular-weight ubiquitinated products were observed in the presence of ICP0-RING as well as M153R PHD but not in their absence. (Left panel) M153R- and KSHV K5-specific ubiquitination with MmUbc6 and MmUbc7. E2 enzymes MmUbc6 (first through third lanes) and MmUbc7 (fourth through sixth lanes) were incubated with E1, ATP, and ubiquitin (first and fourth lanes), in the second and fifth lanes M153R PHD was added, and in the third and sixth lanes K5 PHD was added. The resulting products were analyzed by immunoblotting with antiubiquitin antibody. Molecular weights of marker proteins are shown to the left (in thousands). When compared with samples containing no E3 (first and fourth lanes), GST fusions with the PHD domains of M153R and K5 were found to have E3 ligase activity with MmUbc7 and minimal activity with MmUbc6.
While M153R is active with UbcH5a in vitro, it is unclear whether or not this association is biologically significant. UbcH5a was originally described as the E2 enzyme interacting with the HECT-domain E3 enzyme E6AP (46). Since M153R and all other K3-family PHD/LAP proteins localize to intracellular membranes, we surmised that these proteins might function in conjunction with membrane-bound E2 enzymes. Two mammalian E2 enzymes that localize to intracellular membranes are the murine homologs of the yeast E2 enzymes Ubc6 and Ubc7, designated MmUbc6 and MmUbc7, respectively (51). We examined whether fusions of GST with these E2 enzymes would support the formation of multiubiquitin adducts by the M153R PHD domain or by the KSHV K5 PHD domain in vitro. As shown in Fig. 9, left panel, we observed only minimal activity of either M153R or KSHV K5 with MmUbc6. This activity was not due to the E1-E2 reaction, since the ubiquitination of MmUbc6 was detected readily in nonreducing gels (data not shown). In contrast, additional ubiquitin-reactive protein bands were observed in the presence of MmUbc7 for both M153R and KSHV K5. In contrast to what was seen with UbcH5a, however, we did not observe a high-molecular-weight smear of bands. These novel bands were smaller than the bands for the fusions of GST with the E2 or E3 enzymes. Therefore, it seems that both M153R and KSHV K5 catalyzed the formation of multiubiquitin chains in the presence of MmUbc7. We conclude that M153R acts as a ubiquitin ligase and that M153R is also able to function with membrane-associated E2 enzymes such as Ubc7.
CD4 is internalized via the multivesicular body pathway.
Ubiquitination-mediated endocytosis of cell surface receptors involves the sorting of receptors that recognize ubiquitin and sort protein cargo along the endsome/multivesicular body/lysosome pathway (28). In yeast cells, class E vacuolar protein sorting (Vps) mutants fail to sort hydrolases to the vacuole, the yeast equivalent of the lysosome. Instead, proteins accumulate in a perivacuolar class E compartment (44). Several human homologs of yeast class E proteins have been shown to be involved in the subcellular sorting of ubiquitinated proteins (28). The hepatocyte growth factor receptor substrate (Hrs) is thought to be the human homolog of Vps27 (30). Hrs is a cytosolic protein that is recruited to early endosomal membranes. Targeted disruption of Hrs in mice leads to abnormally enlarged early endosomes resembling the class E compartment (30). Human Hrs and Drosophila Hrs contain a ubiquitin interaction motif that is required for binding to ubiquitin in vitro (3, 33). In cells, Hrs binds to TfR fused to ubiquitin but not to wild-type TfR (43). The overexpression of Hrs has a dominant-negative effect and was shown to inhibit the internalization of ubiquitinated TfR (43) and epidermal growth factor receptor (EGFR) (3, 10, 33). Therefore, it is thought that Hrs plays a central role in the sorting of ubiquitinated cell surface receptors.
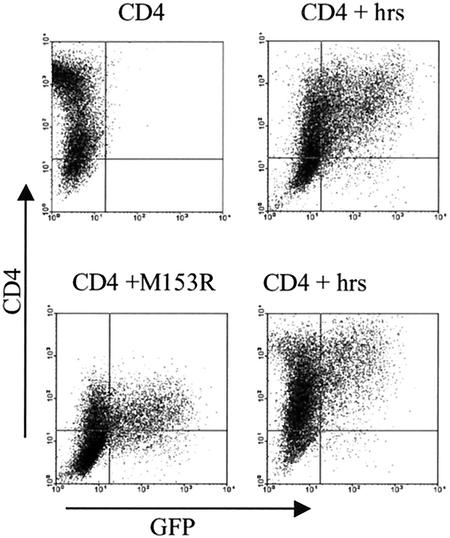
To examine whether Hrs is involved in M153R-mediated internalization, we cotransfected CD4 and M153R with or without Hrs into HeLa cells. As a control, cells were transfected with CD4 with or without Hrs. The results, shown in Fig. 10 (note that the CD4 control was not cotransfected with GFP), indicate that Hrs was able to restore normal surface levels of CD4 in VV-M153R-transfected cells. In contrast, Hrs had no effect on wild-type CD4. The latter is an important control, since CD4 is known to be rapidly internalized in HeLa cells (39). Since surface levels of wild-type CD4 did not change, we conclude that the internalization of transfected CD4 occurs independently of Hrs. However, M153R-mediated endocytosis of CD4 seems to depend on Hrs, consistent with a molecular mechanism that involves the ubiquitination of CD4 (Fig. 11).
FIG. 10.
Overexpression of Hrs (Vps27) restores CD4 surface expression in VV-M153R-transfected cells. HeLa cells cotransfected with CD4 (top left panel), CD4, Hrs, and GFP (top right panel), CD4, M153R, and GFP (bottom left panel), and CD4, Hrs, M153R, and GFP (bottom right panel) were analyzed by flow cytometry. Surface expression of CD4 was detected with anti-CD4 monoclonal antibody. CD4 staining is plotted against GFP fluorescence. Note that the CD4 control did not contain GFP. While CD4 is downregulated in VV-M153R-transfected cells, Hrs appears to reverse the effect of M153R in cells cotransfected with Hrs and M153R.
FIG. 11.
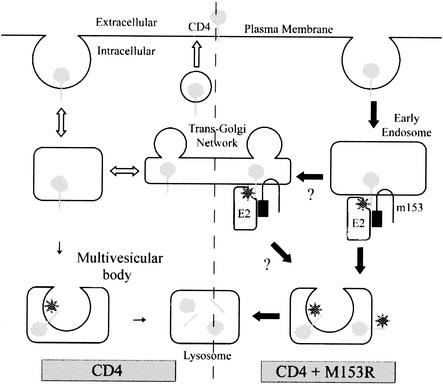
Model for CD4 downregulation by M153R. Under normal conditions (open arrows), CD4 is endocytosed from the cell surface and recycled back to the cell surface. This process may involve the trans-Golgi network, and a small percentage is trafficked to the lysosomes via multivesicular bodies. In the presence of M153R (solid arrows), however, CD4 is not recycled efficiently. Instead, it interacts with M153R in the endosomes and/or trans-Golgi network. M153R recruits an unknown ubiquitin-activating (E2) enzyme which results in the ubiquitination of CD4 (stars). Ubiquitinated CD4 is then efficiently trafficked to the lysosomes via multivesicular bodies.
DISCUSSION
Our data suggest the following order of events for the downregulation of MHC-I as well as CD4 by the MV M153R protein (Fig. 11). CD4 is synthesized and exocytosed normally and can be detected on the cell surface, at least for a brief period of time. Once at the cell surface, CD4 is rapidly internalized and sorted to intracellular vesicles. At present, we do not know whether M153R induces this internalization or simply inhibits CD4 recycling to the cell surface. It is known that CD4 expressed in HeLa cells is endocytosed quite efficiently and recycled to the cell surface (39). It is thus possible that CD4 meets M153R during spontaneous recycling. Also, MHC-I molecules are known to be internalized, so the same model could apply to MHC-I. Since the rates of MHC-I internalization differ from cell type to cell type, it would be interesting to investigate how efficiently M153R downregulates MHC-I in different cell types (8). CD4 endocytosis is slowed down in the presence of the kinase Lck, which is present in T cells but not in HeLa cells (40). We did not examine Lck in our cell system. However, the association of Lck with CD4 is not altered in MV-infected cells, yet CD4 is still internalized and degraded in lysosomes (2). These findings suggest that M153R also mediates the lysosomal targeting of CD4 in the presence of Lck.
So far, a direct interaction between CD4 and M153R has not been detected (M. Mansouri, unpublished data). It is thus possible either that M153R does not interact directly with its substrate or that this interaction is very weak or transient, similar to the transient interaction recently reported for KSHV K3 and MHC-I (22). Indirect evidence for interactions of K3-family proteins with their targets also comes from domain-swapping experiments showing that the transmembrane domains of K5 can transfer to K3 the ability to downregulate B7.2 (45). Moreover, transfer of the transmembrane domain and carboxy terminus from MHC-I to CD8 renders CD8 susceptible to downregulation by K3 and K5 (25). The most likely explanation for these observations is that the transmembrane regions of K3-family proteins interact with the transmembrane regions of their substrates. These interactions may also define substrate specificity. However, the transmembrane regions of the M153R substrates MHC-I, CD95, and CD4 do not share any significant homology. Similarly, the transmembrane regions of MHC-I, ICAM-1, and B7.2, the substrates of KSHV K5, are very different. Therefore, it remains to be shown how this family distinguishes its substrates. The contribution of the cytoplasmic domain seems to be limited to providing lysine residues, since lysines restored HLA-A2.1 downregulation irrespective of the sequence context (Fig. 7) (14). However, the spacing of lysines from the membrane may play a role.
We further propose that M153R, upon interacting with CD4 or MHC-I, instigates the ubiquitination of the cytoplasmic tails of the substrates. The addition of ubiquitin can then serve as the lysosomal targeting signal. An example of this kind of receptor downregulation is Cbl-mediated ubiquitination of the cytoplasmic tail of receptor tyrosine kinases, such as EGFR, which targets the EGFR to lysosomes (27). In this example, ubiquitin serves as a lysosomal targeting signal (23). EGFR targeting is also dependent on Hrs (3, 10) The RING-domain protein Cbl can thus serve as a model for the molecular mechanism by which M153R as well as other K3-family members downregulate their substrates. The RING domain of Cbl acts as ubiquitin ligase in vitro, similar to the isolated PHD/LAP domain of M153R or KSHV K5 (Fig. 9) (14).
A function of all K3-family proteins as ubiquitin ligases also fits with the finding that homologous K3 proteins of MHV68 and human KSHV target their substrates to proteasomes and lysosomes, respectively. It seems plausible that for MHV68 K3, ubiquitination leads to proteasomal destruction, whereas for KSHV K5 and KSHV K3, ubiquitination leads to lysosomal targeting. However, what determines the targeting of ubiquitinated substrates for either lysosomal or proteasomal destruction still needs to be elucidated. For M153R, we observed that CD4 was targeted exclusively for destruction along the endosomal/lysosomal route, similar to the situation for KSHV proteins. In contrast to recent observations with KSHV K5 (14), we did not observe a difference in the levels of ubiquitination of CD4 and MHC-I (data not shown) in the presence or absence of M153R unless cells were treated with concanamycin A. Interestingly, significant ubiquitination of CD4 was also observed in the absence of M153R, and this ubiquitination appeared to be independent of the presence of the cytoplasmic tail.
Recent work suggests that ubiquitination plays a central role in sorting transmembrane proteins to multivesicular bodies (28), a prerequisite of lysosomal destruction. Since concanamycin A inhibits early to late endosomal transport (6, 53), the most likely explanation for this observation is that the inhibition of lysosomal acidification stabilizes ubiquitinated intermediates at the level of multivesicular bodies. Since tailless CD4 was also ubiquitinated, it seems that M153R-independent ubiquitination also occurs at lysines in the extracellular domain of CD4. Importantly, CD4 did not seem to be internalized via the Hrs-dependent pathway in the absence of M153R, suggesting that the CD4 ubiquitination observed in the absence of M153R does not play a role in subcellular targeting but is a by-product of CD4 turnover.
Our model for CD4 downregulation by M153R (Fig. 11) is different in some aspects from and is similar in other aspects to the mechanism for two other viral proteins that are known to downregulate CD4: the HIV proteins Vpu and Nef. HIV Vpu localizes to the ER membrane, facing the cytoplasm (29, 47). A cytosolic orientation is likely for both termini of M153R as well, since both MHV68 K3 and KSHV K5 have this orientation (4, 45). Vpu interacts with CD4 as well as the cytosolic multimolecular SCF ubiquitin ligase complex (34). Moreover, the cytoplasmic lysines of CD4 are also required for Vpu-mediated degradation (47). Thus, Vpu also acts as a ubiquitin ligase, albeit in a more indirect manner, by recruiting a RING-domain-containing cellular E3 complex. Moreover, Vpu sends its substrates, CD4 and MHC-I, to the proteasome for destruction (47). In contrast, HIV Nef induces the rapid internalization of both CD4 and MHC-I from the cell surface (reviewed in reference 11). MHC-I is targeted to the trans-Golgi network whereas CD4 is targeted to lysosomes in Nef-transfected cells (41, 42). However, neither mechanism seems to involve ubiquitin, since the lysines in the cytoplasmic tails of CD4 and MHC-I do not seem to represent the crucial determinants of Nef-mediated downregulation (1, 32). Thus, it seems that CD4 as well as MHC-I downregulation by M153R is distinct from that by either Nef or Vpu.
It was recently shown for KSHV K3-transfected cells that MHC-I is targeted to lysosomes via the trans-Golgi network (36). We did not study whether the trans-Golgi network is involved in CD4 uptake. Trans-Golgi network involvement seems possible, however, since M153R was found to colocalize with trans-Golgi network markers in VV-infected cells (data not shown).
The conservation of MHC-I immune evasion sequences across viral species is unusual. Most other MHC-I evasion proteins, such as herpes simplex virus ICP47 or human cytomegalovirus US6-family members, are highly species specific. In contrast, the K3 family, as defined by a PHD/LAP domain followed by two transmembrane domains, is found in several different gamma-2 herpesviruses as well as in a number of poxviruses (15, 18). Therefore, the question arises as to how these proteins were acquired by such diverse virus families. One possible explanation is that the K3 family is of viral origin and has been transmitted laterally to other viruses during coinfection. An alternative hypothesis is that these proteins are homologs of host proteins that have been independently acquired by each virus family. The integration of host genes into viral genomes is a common practice for both the herpesvirus and the poxvirus families. We favor the latter hypothesis, since similar proteins can be found in many eukaryotic genomes, including the human genome. Moreover, preliminary results suggest that the human homologs act as ubiquitin ligases and are bound to intracellular membranes (E. Bartee, unpublished observations). Most intriguingly, Swanson and colleagues recently reported that a yeast protein, Doa10, is required for the degradation of hydrophobic proteins by the Ubc6/Ubc7-dependent ER degradation pathways (50). The Doa10 protein contains an N-terminal PHD/LAP domain followed by 10 transmembrane domains. This PHD/LAP domain was also shown to have E3 ligase activity in vitro. It is therefore tempting to speculate that a Doa10-like molecule gave rise to a series of membrane-bound ubiquitin ligases involved in the degradation of transmembrane proteins. These putative Doa10 derivatives may have been copied independently into the genomes of gamma-2 herpesviruses and poxviruses, where they evolved to serve as immune evasion molecules.
Acknowledgments
Mandana Mansouri and Eric Bartee contributed equally to this work.
We thank Roger Everett for generous gifts of reagents and help with setting up ubiquitination assays. We are also indebted to Jon Yewdell for providing several VV constructs used in this study. We also thank Klaus Strebel and Richard Axel for CD4 constructs, Alan Weissman for MmUbc6 and MmUbc7, and Philip Woodman for Hrs. We thank Scott Hansen for help with VV growth and cell culturing and Hunter Metcalf for assisting with some of the cloning projects. GHOST (3) parental cells from Vineet KewalRamani and Dan Littman were obtained through the AIDS Research and Reference Reagent Program of the Division of AIDS, NIAID, NIH.
This work was funded by RO1 AI 48585 and AI 49793 for G.T. and L.T. and RO1 CA94011-01A1 for K.F.
REFERENCES
- 1.Aiken, C., J. Konner, N. R. Landau, M. E. Lenburg, and D. Trono. 1994. Nef induces CD4 endocytosis: requirement for a critical dileucine motif in the membrane-proximal CD4 cytoplasmic domain. Cell 76:853-864. [DOI] [PubMed] [Google Scholar]
- 2.Barry, M., S. F. Lee, L. Boshkov, and G. McFadden. 1995. Myxoma virus induces extensive CD4 downregulation and dissociation of p56lck in infected rabbit CD4+ T lymphocytes. J. Virol. 69:5243-5251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bishop, N., A. Horman, and P. Woodman. 2002. Mammalian class E vps proteins recognize ubiquitin and act in the removal of endosomal protein-ubiquitin conjugates. J. Cell Biol. 157:91-101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boname, J. M., and P. G. Stevenson. 2001. MHC class I ubiquitination by a viral PHD/LAP finger protein. Immunity 15:627-636. [DOI] [PubMed] [Google Scholar]
- 5.Boutell, C., S. Sadis, and R. D. Everett. 2002. Herpes simplex virus type 1 immediate-early protein ICP0 and its isolated RING finger domain act as ubiquitin E3 ligases in vitro. J. Virol. 76:841-850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bowman, E. J., A. Siebers, and K. Altendorf. 1988. Bafilomycins: a class of inhibitors of membrane ATPases from microorganisms, animal cells, and plant cells. Proc. Natl. Acad. Sci. USA 85:7972-7976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cameron, C., S. Hota-Mitchell, L. Chen, J. Barrett, J. X. Cao, C. Macaulay, D. Willer, D. Evans, and G. McFadden. 1999. The complete DNA sequence of myxoma virus. Virology 264:298-318. [DOI] [PubMed] [Google Scholar]
- 8.Capps, G. G., M. Van Kampen, C. L. Ward, and M. C. Zuniga. 1989. Endocytosis of the class I major histocompatibility antigen via a phorbol myristate acetate-inducible pathway is a cell-specific phenomenon and requires the cytoplasmic domain. J. Cell Biol. 108:1317-1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cecilia, D., V. N. KewalRamani, J. O'Leary, B. Volsky, P. Nyambi, S. Burda, S. Xu, D. R. Littman, and S. Zolla-Pazner. 1998. Neutralization profiles of primary human immunodeficiency virus type 1 isolates in the context of coreceptor usage. J. Virol. 72:6988-6996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chin, L. S., M. C. Raynor, X. Wei, H. Q. Chen, and L. Li. 2001. Hrs interacts with sorting nexin 1 and regulates degradation of epidermal growth factor receptor. J. Biol. Chem. 276:7069-7078. [DOI] [PubMed] [Google Scholar]
- 11.Collins, K. L., and D. Baltimore. 1999. HIV's evasion of the cellular immune response. Immunol. Rev. 168:65-74. [DOI] [PubMed] [Google Scholar]
- 12.Coscoy, L., and D. Ganem. 2000. Kaposi's sarcoma-associated herpesvirus encodes two proteins that block cell surface display of MHC class I chains by enhancing their endocytosis. Proc. Natl. Acad. Sci. USA 97:8051-8056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Coscoy, L., and D. Ganem. 2001. A viral protein that selectively downregulates ICAM-1 and B7-2 and modulates T cell costimulation. J. Clin. Investig. 107:1599-1606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Coscoy, L., D. J. Sanchez, and D. Ganem. 2001. A novel class of herpesvirus-encoded membrane-bound E3 ubiquitin ligases regulates endocytosis of proteins involved in immune recognition. J. Cell Biol. 155:1265-1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Früh, K., E. Bartee, K. Gouveia, and M. Mansouri. 2002.. Immune evasion by a novel family of viral PHD/LAP-finger proteins of gamma-2 herpesviruses and poxviruses. Virus Res. 88:55-68. [DOI] [PubMed]
- 16.Früh, K., A. Gruhler, R. M. Krishna, and G. J. Schoenhals. 1999. A comparison of viral immune escape strategies targeting the MHC class I assembly pathway. Immunol. Rev. 168:157-166. [DOI] [PubMed] [Google Scholar]
- 17.Gossen, M., and H. Bujard. 1992. Tight control of gene expression in mammalian cells by tetracycline-responsive promoters. Proc. Natl. Acad. Sci. USA 89:5547-5551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guerin, J. L., J. Gelfi, S. Boullier, M. Delverdier, F. A. Bellanger, S. Bertagnoli, I. Drexler, G. Sutter, and F. Messud-Petit. 2002. Myxoma virus leukemia-associated protein is responsible for major histocompatibility complex class I and Fas-CD95 down-regulation and defines scrapins, a new group of surface cellular receptor abductor proteins. J. Virol. 76:2912-2923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hagglund, R., C. Van Sant, P. Lopez, and B. Roizman. 2002. Herpes simplex virus 1-infected cell protein 0 contains two E3 ubiquitin ligase sites specific for different E2 ubiquitin-conjugating enzymes. Proc. Natl. Acad. Sci. USA 99:631-636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hayflick, J. S., W. J. Wolfgang, M. A. Forte, and G. Thomas. 1992. A unique Kex2-like endoprotease from Drosophila melanogaster is expressed in the central nervous system during early embryogenesis. J. Neurosci. 12:705-717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hershko, A., and A. Ciechanover. 1998. The ubiquitin system. Annu. Rev. Biochem. 67:425-479. [DOI] [PubMed] [Google Scholar]
- 22.Hewitt, E. W., L. Duncan, D. Mufti, J. Baker, P. G. Stevenson, and P. J. Lehner. 2002. Ubiquitylation of MHC class I by the K3 viral protein signals internalization and TSG101-dependent degradation. EMBO J. 21:2418-2429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hicke, L. 1997. Ubiquitin-dependent internalization and down-regulation of plasma membrane proteins. FASEB J. 11:1215-1226. [DOI] [PubMed] [Google Scholar]
- 24.Ishido, S., J. K. Choi, B. S. Lee, C. Wang, M. DeMaria, R. P. Johnson, G. B. Cohen, and J. U. Jung. 2000. Inhibition of natural killer cell-mediated cytotoxicity by Kaposi's sarcoma-associated herpesvirus K5 protein. Immunity 13:365-374. [DOI] [PubMed] [Google Scholar]
- 25.Ishido, S., C. Wang, B. S. Lee, G. B. Cohen, and J. U. Jung. 2000. Downregulation of major histocompatibility complex class I molecules by Kaposi's sarcoma-associated herpesvirus K3 and K5 proteins. J. Virol. 74:5300-5309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Joazeiro, C. A., and A. M. Weissman. 2000. RING finger proteins: mediators of ubiquitin ligase activity. Cell 102:549-552. [DOI] [PubMed] [Google Scholar]
- 27.Joazeiro, C. A., S. S. Wing, H. Huang, J. D. Leverson, T. Hunter, and Y. C. Liu. 1999. The tyrosine kinase negative regulator c-Cbl as a RING-type, E2-dependent ubiquitin-protein ligase. Science 286:309-312. [DOI] [PubMed] [Google Scholar]
- 28.Katzmann, D. J., M. Babst, and S. D. Emr. 2001. Ubiquitin-dependent sorting into the multivesicular body pathway requires the function of a conserved endosomal protein sorting complex, ESCRT-I. Cell 106:145-155. [DOI] [PubMed] [Google Scholar]
- 29.Kerkau, T., I. Bacik, J. R. Bennink, J. W. Yewdell, T. Hunig, A. Schimpl, and U. Schubert. 1997. The human immunodeficiency virus type 1 (HIV-1) Vpu protein interferes with an early step in the biosynthesis of major histocompatibility complex (MHC) class I molecules. J. Exp. Med. 185:1295-1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Komada, M., and P. Soriano. 1999. Hrs, a FYVE finger protein localized to early endosomes, is implicated in vesicular traffic and required for ventral folding morphogenesis. Genes Dev. 13:1475-1485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee, D. H., and A. L. Goldberg. 1998. Proteasome inhibitors: valuable new tools for cell biologists. Trends Cell Biol. 8:397-403. [DOI] [PubMed] [Google Scholar]
- 32.Le Gall, S., L. Erdtmann, S. Benichou, C. Berlioz-Torrent, L. Liu, R. Benarous, J. M. Heard, and O. Schwartz. 1998. Nef interacts with the mu subunit of clathrin adaptor complexes and reveals a cryptic sorting signal in MHC I molecules. Immunity 8:483-495. [DOI] [PubMed] [Google Scholar]
- 33.Lloyd, T. E., R. Atkinson, M. N. Wu, Y. Zhou, G. Pennetta, and H. J. Bellen. 2002. Hrs regulates endosome membrane invagination and tyrosine kinase receptor signaling in Drosophila. Cell 108:261-269. [DOI] [PubMed] [Google Scholar]
- 34.Margottin, F., S. P. Bour, H. Durand, L. Selig, S. Benichou, V. Richard, D. Thomas, K. Strebel, and R. Benarous. 1998. A novel human WD protein, h-beta TrCp, that interacts with HIV-1 Vpu connects CD4 to the ER degradation pathway through an F-box motif. Mol. Cell 1:565-574. [DOI] [PubMed] [Google Scholar]
- 35.McFadden, G., and P. M. Murphy. 2000. Host-related immunomodulators encoded by poxviruses and herpesviruses. Curr. Opin. Microbiol. 3:371-378. [DOI] [PubMed] [Google Scholar]
- 36.Means, R. E., S. Ishido, X. Alvarez, and J. U. Jung. 2002. Multiple endocytic trafficking pathways of MHC class I molecules induced by a herpesvirus protein. EMBO J. 21:1638-1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Molloy, S. S., L. Thomas, J. K. VanSlyke, P. E. Stenberg, and G. Thomas. 1994. Intracellular trafficking and activation of the furin proprotein convertase: localization to the TGN and recycling from the cell surface. EMBO J. 13:18-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Paulson, E., C. Tran, C. Collins, and K. Früh. 2001. KSHV-K5 inhibits phosphorylation of the major histocompatibility complax class I tail. Virology 288:369-378. [DOI] [PubMed] [Google Scholar]
- 39.Pelchen-Matthews, A., J. E. Armes, and M. Marsh. 1989. Internalization and recycling of CD4 transfected into HeLa and NIH3T3 cells. EMBO J. 8:3641-3649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pelchen-Matthews, A., I. Boulet, D. R. Littman, R. Fagard, and M. Marsh. 1992. The protein tyrosine kinase p56lck inhibits CD4 endocytosis by preventing entry of CD4 into coated pits. J. Cell Biol. 117:279-290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Piguet, V., Y. L. Chen, A. Mangasarian, M. Foti, J. L. Carpentier, and D. Trono. 1998. Mechanism of Nef-induced CD4 endocytosis: Nef connects CD4 with the mu chain of adaptor complexes. EMBO J. 17:2472-2481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Piguet, V., L. Wan, C. Borel, A. Mangasarian, N. Demaurex, G. Thomas, and D. Trono. 2000. HIV-1 Nef protein binds to the cellular protein PACS-1 to downregulate class I major histocompatibility complexes. Nat. Cell Biol. 2:163-167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Raiborg, C., K. G. Bache, D. J. Gillooly, I. H. Madshus, E. Stang, and H. Stenmark. 2002. Hrs sorts ubiquitinated proteins into clathrin-coated microdomains of early endosomes. Nat. Cell Biol. 4:394-398. [DOI] [PubMed] [Google Scholar]
- 44.Raymond, C. K., I. Howald-Stevenson, C. A. Vater, and T. H. Stevens. 1992. Morphological classification of the yeast vacuolar protein sorting mutants: evidence for a prevacuolar compartment in class E vps mutants. Mol. Biol. Cell 3:1389-1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sanchez, D. J., L. Coscoy, and D. Ganem. 2001. Functional organization of MIR2, a novel viral regulator of selective endocytosis. J. Biol. Chem. 18:18-22. [DOI] [PubMed] [Google Scholar]
- 46.Scheffner, M., J. M. Huibregtse, and P. M. Howley. 1994. Identification of a human ubiquitin-conjugating enzyme that mediates the E6-AP-dependent ubiquitination of p53. Proc. Natl. Acad. Sci. USA 91:8797-8801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schubert, U., L. C. Anton, I. Bacik, J. H. Cox, S. Bour, J. R. Bennink, M. Orlowski, K. Strebel, and J. W. Yewdell. 1998. CD4 glycoprotein degradation induced by human immunodeficiency virus type 1 Vpu protein requires the function of proteosomes and the ubiquitin-conjugating pathway. J. Virol. 72:2280-2288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sodeik, B., and J. Krijnse-Locker. 2002. Assembly of vaccinia virus revisited: de novo membrane synthesis or acquisition from the host? Trends Microbiol. 10:15-24. [DOI] [PubMed] [Google Scholar]
- 49.Stevenson, P. G., S. Efstathiou, P. C. Doherty, and P. J. Lehner. 2000. Inhibition of MHC class I-restricted antigen presentation by gamma 2-herpesviruses. Proc. Natl. Acad. Sci. USA 97:8455-8460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Swanson, R., M. Locher, and M. Hochstrasser. 2001. A conserved ubiquitin ligase of the nuclear envelope/endoplasmic reticulum that functions in both ER-associated and Matalpha2 repressor degradation. Genes Dev. 15:2660-2674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tiwari, S., and A. M. Weissman. 2001. Endoplasmic reticulum (ER)-associated degradation of T cell receptor subunits. Involvement of ER-associated ubiquitin-conjugating enzymes (E2s). J. Biol. Chem. 276:16193-16200. [DOI] [PubMed] [Google Scholar]
- 52.Tortorella, D., B. E. Gewurz, M. H. Furman, D. J. Schust, and H. L. Ploegh. 2000. Viral subversion of the immune system. Annu. Rev. Immunol. 18:861-926. [DOI] [PubMed] [Google Scholar]
- 53.Woo, J. T., C. Shinohara, K. Sakai, K. Hasumi, and A. Endo. 1992. Isolation, characterization and biological activities of concanamycins as inhibitors of lysosomal acidification. J. Antibiot. (Tokyo) 45:1108-1116. [DOI] [PubMed] [Google Scholar]
- 54.Zuniga, M. C., H. Wang, M. Barry, and G. McFadden. 1999. Endosomal/lysosomal retention and degradation of major histocompatibility complex class I molecules is induced by myxoma virus. Virology 261:180-192. [DOI] [PubMed] [Google Scholar]