Abstract
We reported previously that the movement protein (MP) of tomato mosaic tobamovirus is phosphorylated, and we proposed that MP phosphorylation is important for viral pathogenesis. Experimental data indicated that phosphorylation enhances the stability of MP in vivo and enables the protein to assume the correct intracellular location to perform its function. A mutant virus designated 37A238A was constructed; this virus lacked two serine residues within the MP, which prevented its phosphorylation. In the present study, we inoculated plants with the 37A238A mutant, and as expected, it was unable to produce local lesions on the leaves. However, after an extended period, we found that lesions did occur, which were due to revertant viruses. Several revertants were isolated, and the genetic changes in their MPs were examined together with any changes in their in vivo characteristics. We found that reversion to virulence was associated first with increased MP stability in infected cells and second with a shift in MP intracellular localization over time. In one case, the revertant MP was not phosphorylated in vivo, but it was functional.
After an initial round of replication in primary infected cells, plant viruses spread to adjacent tissues via the plasmodesmata, thus establishing systemic infection (4, 6, 7, 16, 21, 26). In tobamoviruses (ToMV) the 30-kDa movement protein (MP) is essential for efficient intercellular transport (8, 22). Plant viruses lacking a functional MP cannot move from the primary infected cell (4, 7, 16, 21, 22). Despite its importance for viral pathogenesis, the molecular mechanism by which MP enables intercellular transport of ToMV in plants is unknown. Recently it was shown that the replicase-coding region is also involved in cell-to-cell movement by an as-yet-unknown mechanism (12).
In previous studies, we found that MP is produced transiently during the infection cycle (30) and is phosphorylated posttranslationally in infected cells (32). We have also reported that recombinant MP expressed in Escherichia coli can be phosphorylated within the C-terminal portion of the protein by casein kinase II (18). Citovsky et al. (5) and Waigmann et al. (29) also reported in vitro kinase activities that phosphorylate the C-terminal portion of MP and discussed its relevance to function.
We analyzed the phosphorylation sites in vivo by using MP mutants that were already available and additional artificially constructed alanine-scanned mutants (13). We isolated 32P-labeled MP tryptic peptides from protoplasts infected with the different mutants, compared the tryptic maps with that of wild-type MP, and thus narrowed down the candidate phosphorylation sites. The phosphorylation site identified first was serine 238. The serine residue was replaced with alanine to make a new mutant designated LQ238A. However, the MP produced by this mutant was still phosphorylated and functional.
We assumed that the serine residue or residues targeted for phosphorylation are likely to be more conserved than other residues. The sequences of several ToMV MPs were therefore aligned, and several possible phosphorylation sites were found, including serine 37. This residue was replaced with alanine to produce a ToMV designated 37A, which was unable to spread and cause necrotic lesions but could still produce progeny viral RNAs and other viral proteins within primary infected cells. We found that this virus was defective in terms of its ability to move between cells within the host plant.
As a result of the serine-to-alanine substitution at MP residue 37 we observed two phenotypic changes in the virus. First, after protoplast inoculation, the mutant virus did not show the usual shift in intracellular localization. The functional location of MP within the cell was visualized by using MP fused to the green fluorescent protein (GFP) of Aequorea victoria. When we analyzed the localization of MP-GFP by fluorescence microscopy, wild-type MP-GFP showed a temporal shift in localization both in inoculated protoplasts and in plants. The mutants with serine-to-alanine (or other) substitutions at residue 37, did not show the wild-type localization pattern. Such observations implied that MP was phosphorylated at Ser 37 or that Ser 37 was involved in secondary or tertiary structures important for targeting the appropriate intracellular structures, possibly including the plasmodesmata. Second, pulse-chase labeling experiments with a protoplast system showed that the mutant MP had less stability in vivo. The amount of MP synthesized during infection with mutant 37A was almost the same as that produced by wild-type virus, but the accumulation of mutant MP started to decline earlier than that of wild-type MP. These results indicated that phosphorylation at Ser 37 contributes to MP function, at least in part by maintaining its stability in vivo. When MP serine 37 was replaced with threonine, aspartic acid, or glutamic acid, such mutants were also unable to move between cells. Thus, it is likely that phosphorylation of serine 37 and/or its identity as a serine residue is essential for normal MP function.
Phosphorylation plays a key role in the function of MP in intercellular transport of ToMV and in their pathogenesis, perhaps by maintaining the correct structural conformation. We propose that a kinase activates the intercellular movement function of the MP. The aim of the present study was to gain further insights into the contributions made by Ser 37 phosphorylation to the function of wild-type MP. To this end, we isolated spontaneous revertants from the 37A238A mutants, characterized their genetic changes, and analyzed the contribution of each substitution to the virus phenotype.
MATERIALS AND METHODS
Plant materials, viruses, and in vitro transcription.
Tomato mosaic ToMV-L was used throughout this work (23); Nicotiana tabacum cv. Samsun was used as a systemic host, and N. tabacum cv. Xanthi-nc was used as a local lesion host. The plasmid pTLW3 contains ToMV cDNA downstream of the T7 promoter, from which infectious transcripts can be produced in vitro (9, 21). Mutants 37A238A and 37A (formerly LQ37A238A and LQ37A, respectively) were created by site-directed mutagenesis, as described in a previous report (13). The MP of the mutant virus 37A238A cannot be phosphorylated, and these virions are unable to infect tobacco or tomato plants (13).
The in vitro transcription reaction was performed with T7 RNA polymerase (Invitrogen) and m7GpppG (New England Biolabs) as the cap analog, as described previously (9, 21). Transcripts were subjected to virus reconstruction with purified coat protein, as described previously (24).
Isolation and sequence analysis of revertants.
Revertants were identified by their ability to form small, localized lesions on leaves inoculated with 37A238A, which had been reconstituted in vitro. Isolated local lesions were homogenized in 10 mM phosphate buffer (pH 7.0), and the sap was inoculated into the leaves of the local lesion host; this process was repeated three times. To propagate the virus, the homogenized local lesions were inoculated into the systemic host plants, and after 2 weeks, virus was purified as described previously (23). Each independent revertant was assigned a number from 1 to 9. However, during purification, we could not recover revertants 1 and 3.
Reverse transcription was performed with the purified RNA of each revertant as a template (10), and then DNA fragments spanning each MP gene were amplified by high-fidelity PCR with two oligonucleotide primers: 5′-MP-BamHI and 3′-MP-BamHI (Table 1) (1). The BamHI-restricted PCR fragment was ligated to pUC119 which had been linearized with BamHI. Recombinant plasmids underwent nucleotide sequence analysis with an ABI 377 or DSQ1000 (Shimadzu) sequencer.
TABLE 1.
Oligonucleotides used in this study
| Oligonucleotide | Nucleotides | Sequencea | Purpose | Corresponding ToMV nucleotidesb |
|---|---|---|---|---|
| 5′-MP-BamHI | 30 | gtttGGATCCTGATGGCTCTAGTTGTTAAAc | PCR amplification and sequencing of MP-coding region | 4904-4923 |
| 3′-MP-BamHI | 32 | gtaaGGATCCTTTAATACGAATCAGAATCCGCc | PCR amplification and sequencing of MP-coding region | c5701-5680 |
| 72I-AccIII | 41 | TTAGTCGGTaTTGTTGTGTCCGGaGAGTGGAATTTACCAGAd | Construction of 72Irev mutant | 5110-5150 |
| re72I-AccIII | 45 | TCCACTCtCCGGACACAACAAtACCGACTAAGCAAACATACCCACd | Construction of 72Irev mutant | c5140-5096 |
| L5850 | 21 | TGAACTGCTGTTGAACAGTAG | PCR amplification from MP downstream | c5850-5830 |
| L3600 | 23 | TATGTCTAAGTCTGTAGCTGCTC | PCR amplification from MP upstream | 3614-3636 |
| 126S-SfcI | 40 | AAAGTGGTCCCAAAcTAtaGTATTACAACAAAGGATGCAGd | Construction of 126Srev mutant | 5263-5302 |
| 219L-StyI | 33 | AATAATAAccTAGGTAAGGGGCGTTCAGGCGGAd | Construction of 219Lrev mutant | 5569-5601 |
| re219L-StyI | 42 | CCCTTACCTAggTTATTATTATTTTTCaGACCTCTTTTTGAGd | Construction of 219Lrev mutant | c5588-5547 |
| 244stop-BcIL | 28 | TGAAAAAtgaTcaGATAATTTGATTGAAd | Construction of 244FSrev mutant | 5631-5658 |
| re244stop-BcIL | 28 | ATTATCtgAtcaTTTTTCAACTTCATCAd | Construction of 244FSrev mutant | c5649-5622 |
| L2-Sac | 30 | tacatgagctcATCCGCGACCGACGTCTCGc | Construction of MP: Gfus mutants | c5685-5667 |
Underlined letters indicate restriction sites.
c, complementary sequence of the genome.
Lowercase letters indicate random nucleotides ensuring efficient restriction enzyme function.
Lowercase letters indicate nucleotides in the primers designed to change the amino acid sequence.
Protoplast isolation and inoculation of viral RNAs or transcripts.
An N. tabacum cv. BY-2 suspension culture was maintained as described previously (33). Protoplast isolation and the inoculation of viral RNA were performed as described previously (31). Western blotting was performed with an anti-ToMV MP antibody (20). Each protein sample, equivalent to about 105 protoplasts, was analyzed by sodium dodecyl sulfate (SDS)-10% polyacrylamide gel electrophoresis and blotted onto Immobilon nylon membranes (Millipore). Detection of coat protein accumulation was performed to determine the multiplication of each virus as described previously (31, 32).
Site-directed mutagenesis to create mutants.
Site-directed mutagenesis of the MP gene was performed by PCR with synthetic oligonucleotides. Template plasmid DNA used to direct the synthesis of each mutant was named with an initial letter p, while transcripts and viruses derived from the template DNA were named without this letter.
Plasmid p72Irev directs the synthesis of the transcript 72Irev in vitro. During infection, this produces a virus with a mutant MP containing a leucine-to-isoleucine substitution at position 72 of the 37A238A genome. Plasmid pTLQ37A238A was used as a template to create p72Irev. The first fragment, including the center of the MP gene and its 3′ region, was made by PCR with 72I-AccIII and L5850 as forward and reverse primers, respectively. To make the second fragment, which contained the center of the MP gene and its 5′ region, oligonucleotides L3700 and re72I-AccIII were used as forward and reverse primers. The larger KpnI (nucleotide 4390)-BstEII (nucleotide 5799) fragment of pTLW3 was ligated to the AccIII-BstEII-digested first fragment and the KpnI-AccIII-digested second fragment to create the plasmid p72Irev.
Plasmid p126Srev directs the synthesis of the transcript 126Srev in vitro, which produces mutant MP with a glycine-to-serine substitution at position 126 in the 37A238A genome. The first fragment was made by PCR with oligonucleotide 126S-SfcI and L5850 primers. The second PCR was performed with L3700 and the first fragment as a megaprimer. The KpnI-BstEII-restricted PCR fragment was replaced with the counterpart fragment of pTLW3 to obtain pTLQ126Srev.
p219Lrev directs the synthesis of the 219Lrev transcript in vivo, which during infection produces a mutant MP with a proline-to-leucine substitution at position 219 of the 37A238A genome. Plasmid p219Lrev was created by using oligonucleotide primers 219L-StyI and re219L-StyI. The first fragment, which includes a part of MP and its 3′ region, was made by PCR with 219L-StyI and L5850 as forward and reverse primers, respectively. The second fragment, spanning part of the MP gene and its 5′ region, was made with L3700 and re219L-StyI as forward and reverse primers, respectively. The KpnI-BstEII-digested pTLW3 was ligated to the StyI-BstEII-digested first fragment and the KpnI-StyI-digested second fragment to create p219Lrev.
p72I219Lrev directs the synthesis of the 72I219Lrev transcript in vivo, which produces a mutant MP with the both the substitutions of the 72Irev and 219Lrev mutants. Construction was performed basically in the same way as for p72Irev, except that the PCR was performed with p219Lrev as the template.
p244FSrev directs the synthesis of the 244FSrev transcript, which encodes a mutant MP containing a nonsense mutation at position 244 of the 37A238A genome. Primers 244stop-BclI and L5850 were used to create the first fragment, and L3700 and re244stop-BclI were used to create the second fragment. p244FSrev was produced by ligation of the large fragment between KpnI the BstEII sites in pTLW3 to the BclI-BstEII-restricted first fragment and the KpnI-BclI-restricted second fragment.
Construction of p72I244Fsrev, which directs the synthesis of the 72I244FSrev transcript, was performed as for p244FSrev, except that plasmid pTLQ72Irev DNA was used as the PCR template.
The LQwt:Gfus and LQ37A:Gfus viruses encode an MP with a 4-amino-acid truncation at the C terminus fused to GFP-S65A/Y145F (14), but they lack an intact coat protein gene (13). To construct mutant viruses 72Irev:Gfus and 126Srev:Gfus, which produce revertant MP-GFP fusion proteins, a PCR was performed with primers L3700 and L2-Sac (13) and plasmid p72Irev or p126Srev as the template. The KpnI-SacI fragment in LQwt:Gfus was replaced with a KpnI-SacI-restricted fragment including the revertant MP gene. The resultant viruses, 72Irev:Gfus and 126Srev:Gfus, encoded mutant MP-GFP fusion proteins with substitutions of leucine to isoleucine at position 72 or glycine to serine at position 126 of the MP, in addition to two serine-to-alanine substitutions.
In vivo 32P labeling and immunoprecipitation of MP.
BY-2 protoplasts were inoculated with wild-type or mutant transcripts (31). Protoplasts were subsequently cultured in the presence of [32P]orthophosphate (Amersham) at 7.4 MBq/ml. After collection, the protoplasts were lysed in radioimmunoprecipitation assay buffer containing pepstatin A (1 μg/ml), leupeptin (0.5 μg/ml), phenylmethylsulfonyl fluoride (1 mM), and β-glycerophosphate (100 mM) to reduce degradation and dephosphorylation. After the addition of anti-MP antiserum (20), the immunoprecipitates were collected with the aid of protein A-Sepharose (Pharmacia), washed with phosphate-buffered saline, and subjected to SDS-10% polyacrylamide gel electrophoresis (15). The gels were dried on filter paper, and radioactivity was detected with a Fuji Image Analyzer.
35S pulse-chase experiments with in vivo-synthesized MP.
BY-2 protoplasts were inoculated with wild-type and mutant transcripts and divided into aliquots sufficient to cover the required number of time points. During culture, actinomycin D (30 μg/ml) was added. Subsequently, [35S]methionine-cysteine protein-labeling mixture (NEN) at 1 MBq/ml was added to the culture medium and left for 10 min. Unlabeled methionine and cysteine were then added (each at a 1 mM final concentration) to stop the further incorporation of radioactivity. The protoplasts were harvested at 0, 4, 10, and 16 h after labeling, and the proteins were extracted and subjected to SDS-10% polyacrylamide gel electrophoresis. 35S-labeled MP bands were detected and traced quantitatively by using a Fuji Image Analyzer.
Observation by fluorescence microscopy.
Fluorescence microscopy was performed on protoplasts inoculated with transcripts of LQwt:Gfus, 72Irev:Gfus, 126Srev:Gfus, and LQ37A:Gfus. Inoculated protoplasts were cultured in the presence of 30 μg of actinomycin D per for the times indicated in the figures and fixed as described previously (13). Fluorescence micrographs were obtained by using Fujichrome 400 film with an Olympus BX60 microscope plus an epifluorescence attachment and a U-MWIB filter cube containing a BP 470-490 excitation filter, a DM505 dichroic mirror, and a BA-515IF emission barrier filter.
RESULTS
Wild-type ToMV produces MP which is phosphorylated at serine residues 37 and 238 (13). The ToMV mutant 37A238A was constructed experimentally to have two serine-to-alanine substitutions at these target phosphorylation sites. This mutant lacked these essential serine residues and therefore could not be phosphorylated when expressed during virus multiplication in plant cells (13). The MP of this mutant was also defective in promoting intercellular transport, and hence these virions were unable to cause local lesions on a hypersensitive host (Xanthi-nc tobacco) (13).
Isolation of revertants.
In one experiment, at about 4 days after the inoculation of mutant virus 37A238A into Xanthi-nc tobacco leaves, we observed the spontaneous appearance of small, local lesions (data not shown). Some of the local lesions were isolated from the leaves, the sap was extracted, and fresh Xanthi-nc leaves were inoculated; and this caused a similar number of local lesions as with the wild-type ToMV. This indicated that the sap contained revertant viruses which had regained the MP intercellular transport function by genetic changes in the 37A238A genome during primary replication.
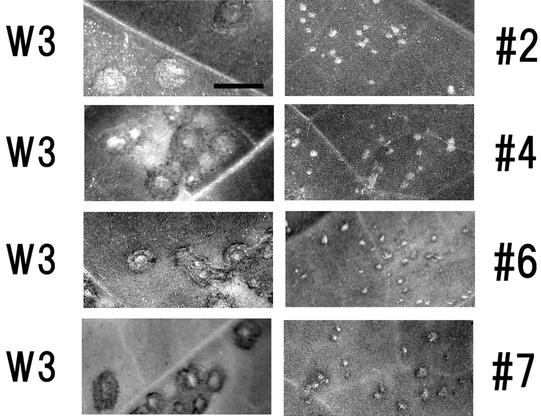
After three rounds of serial passage on Xanthi-nc leaves, we inoculated plants of the systemic host, Samsun tobacco. The revertants, designated 1 through 9, were each purified from the infected tobacco tissues by conventional procedures. However, after three rounds of passage, revertants 1 and 3 could not be recovered. The mosaic symptoms caused by the revertant virions were mild and appeared later than those caused by the wild-type virus. Local lesions caused by revertants 2, 4, 6 and 7 are shown in Fig. 1. To examine the relative functions of the MPs, we measured average lesion diameters to have an idea of relative movement of each virus. The average diameters of 30 to 40 lesions induced on Xanthi-nc plants by revertant viruses are shown in Table 2 relative to those induced by wild-type virus. This showed us that each revertant did acquire its movement function, but less efficiently than wild-type virus.
FIG. 1.
Local lesions caused by revertants isolated after three serial passages. The photographs show the local lesions caused by sap inoculation of each revertant (revertants 2, 4, 6, and 7), in parallel with those of wild-type ToMV (W3) inoculated into the same leaves, at 3 days postinoculation. Bar, 5 mm.
TABLE 2.
Sizes of local lesions of revertants and mutants
| Revertant or mutant | Lesion diameter (mm)a caused by:
|
Lesion diameter ratio | |
|---|---|---|---|
| Revertant or mutant | Wild type on the opposite side | ||
| 2 | 0.61 ± 0.14 | 2.91 ± 0.45 | 0.21 |
| 4 | 0.49 ± 0.11 | 2.55 ± 0.51 | 0.19 |
| 6 | 0.37 ± 0.11 | 2.13 ± 0.56 | 0.17 |
| 7 | 0.45 ± 0.10 | 2.31 ± 0.51 | 0.19 |
| 72Irev | 0.47 ± 0.06 | 2.04 ± 0.54 | 0.23 |
| 72I244FSrev | 0.40 ± 0.08 | 2.10 ± 0.57 | 0.19 |
| 126Srev | 0.54 ± 0.13 | 1.58 ± 0.24 | 0.34 |
Diameters of 30 to 40 lesions were measured after 6 days postinfection and are expressed as means and standard deviations.
Characterization of revertant behavior in protoplasts.
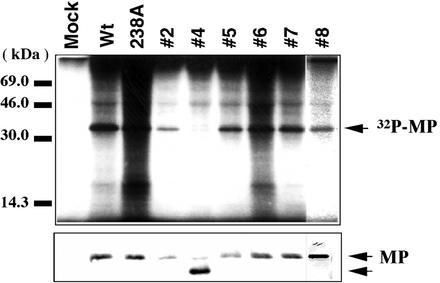
There are two possible explanations for the reversions. First, the mutant 37A238A genome may have reverted to a sequence close or identical to that of the wild type, thereby encoding functional MP. Second, the revertants may have acquired novel secondary mutations which complemented the lost original phosphorylation sites. To investigate these possibilities, BY-2 protoplasts were inoculated with each of the revertants. Northern blot analysis showed that their replication levels within the protoplasts were similar to that of wild-type ToMV and that each revertant also showed almost identical accumulation (data not shown). We confirmed the synthesis and assessed the accumulation of MP in the inoculated protoplasts as well. Using Western blotting, we demonstrated that most revertants produced MP of the same mobility and at almost the same level as wild-type virus and the 37A238A mutant. Only one revertant (revertant 4) produced an MP that migrated faster during electrophoresis than the others (Fig. 2, lower panel), indicating that this MP was truncated.
FIG. 2.
Characterization of MPs synthesized by revertants. (Upper panel) Autoradiograph of the in vivo phosphorylation of each virus. After metabolic labeling with 32P, MPs were immunoprecipitated from each lysate of infected protoplasts. As a positive control, wild-type (Wt) ToMV (W3) and LQ238A were used to show phosphorylated MP, as analyzed previously (13). (Lower panel) Immunodetection of MPs by anti-ToMV MP serum (20). Infection with revertant 4 produced a fast-migrating band, (indicating a truncated MP) as well as another minor band with the same mobility as the other MPs. The origin of the minor band was not examined in detail in this study, since artificially constructed MP mutants (72I244FSrev) supported the possibility that the truncated protein could support cell-to-cell movement function.
To examine the molecular mechanism causing the revertant phenotypes, revertant MPs were assayed for in vivo phosphorylation by using BY-2 protoplasts. Immunoprecipitation experiments with 32P-metabolically labeled proteins and anti-MP antiserum showed that the revertant MPs were phosphorylated in vivo, with the exception of revertant 4, which migrated faster and was not phosphorylated (Fig. 2, upper panel). There might be a 32P-lightly labeled band with the size of wild-type MP in samples of revertant 4 that was also seen in Western blotting. This might reflect the fact that the lesion isolation process could not sufficiently separate a mutant with truncated MP from wild-type virus. However, as shown below, we could construct and analyze an artificial MP mutant with a truncated MP sequence.
MP sequences of revertants.
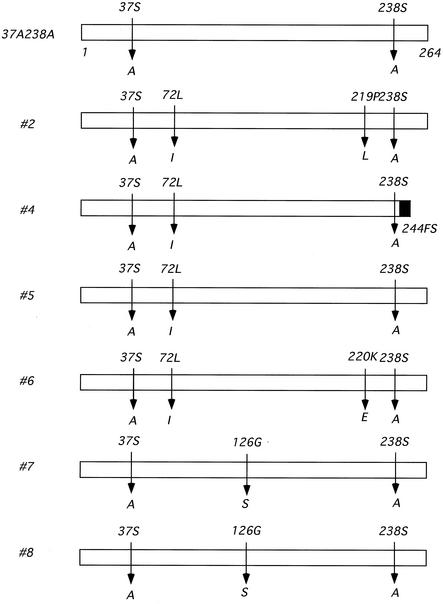
To understand the genetic background of the phenotypic changes in the revertant MPs, we amplified the sequence of each MP region by reverse transcription-PCR. The PCR fragments were cloned into the vector pBluescript (Stratagene), and the sequences of at least three independent clones were determined. The results are summarized in Table 3 and Fig. 3.
TABLE 3.
Summary of base changes found in revertant MPs
| Virus or revertant | Nucleotidea at the following position in the MP-coding region
|
||||||||
|---|---|---|---|---|---|---|---|---|---|
| 109 | 117 | 120 | 214 | 376 | 656 | 658 | 712 and 713 | 733 | |
| ToMV | U | U | U | Cuu | Ggu | cCg | Aaa | AG | G |
| 37A238A | G | C | C | Auu | G | C | A | GC | G |
| 2 | G | C | C | Auu | G | cUg | A | GC | G |
| 4 | G | C | C | Auu | G | C | A | GC | 10Ab |
| 5 | G | C | C | Auu | G | C | A | GC | G |
| 6 | G | C | C | Auu | G | C | Gaa | GC | G |
| 7 | G | C | C | C | Agu | C | A | GC | G |
| 8 | G | C | C | C | Agu | C | A | GC | G |
Underlining indicates the original substitutions introduced into 37A238A, boldface indicates base changes which cause amino acid substitutions (see Fig. 3), and lowercase indicate the triplets in which amino acid substitutions occur.
Ten adenylate bases were inserted and caused a coding frameshift and addition of KKKP.
FIG. 3.
Summary of the amino acid changes found in the revertants. The original amino acid is shown above each bar, and the substitutions found in each revertant are shown below. The black box shown for revertant 4 indicates attached residues irrelevant to the authentic MP sequence downstream from the frameshift mutation.
Rather unexpectedly, there were no examples of true revertants; all retained alanine at residues 37 and 238. Instead, they had acquired multiple base changes, including silent mutations. Among these changes, two substitutions were commonly observed. The first was a leucine-to-isoleucine substitution at residue 72, and the second was a glycine-to-serine substitution at residue 126.
To confirm whether or not such changes within the main MP-coding region are the major causes of reversion, we used site-directed mutagenesis to construct several viruses with mutant MPs. We assessed their virulence in plants to determine the phenotypic importance of each genetic change.
Molecular analysis of the substitutions involved in reversion.
We constructed several mutants by site-directed mutagenesis. At first we focused on the two substitutions which occurred in revertant 2: leucine to isoleucine at amino acid 72 and proline to leucine at 219. The former change also occurred in the genomes of revertants 4, 5, and 6. Thus, we suspected that this substitution alone could confer the revertant phenotype in the 37A238A background. To address this question, we constructed a mutant called 72Irev, which had acquired only the leucine-to-isoleucine substitution at amino acid residue 72. In parallel, we constructed mutants designated 219Lrev and 72I219Lrev, with only one proline-to-leucine substitution at amino acid residue 219 or with both substitutions, respectively (Table 4).
TABLE 4.
Characterization of original revertants and mutants constructed in this study
| Virus | Amino acid at the following position in MP:
|
Phosphor- ylationa | Cell-to-cell movementb | |||||
|---|---|---|---|---|---|---|---|---|
| 37 | 72 | 126 | 219 | 238 | 244 | |||
| Wild type | S | L | G | P | A | K | + | + |
| Revertant 2 | A | I | L | A | + | + | ||
| 721rev | A | I | A | + | + | |||
| 219Lrev | A | L | A | − | − | |||
| 721219Lrev | A | I | L | A | + | + | ||
| Revertant 4 | A | I | A | FSc | − | + | ||
| 72I 244FSrev | A | I | A | Stopd | − | + | ||
| 244FSrev | A | A | Stop | − | − | |||
| Revertant 8 | A | S | A | + | + | |||
| 126Srev | A | S | A | + | + | |||
Results of assay of in vivo phosphorylation of the MP.
Ability to cause local lesions on Xanthi-nc tobacco.
FS, base insertions caused a frameshift mutation at residue 244.
Base changes caused a nonsense mutation at residue 244.
When Xanthi-nc plants were inoculated with the transcripts from these mutants, both 72Irev and 72I219Lrev caused local lesions, indicating that they were both viable. Therefore, it was confirmed that the L72I substitution could convert a nonfunctional 37A238A MP into a functional one. In contrast, 219Lrev was unable to cause lesions, suggesting that the 219L substitution did not complement the lost phosphorylation sites of the defective MP. We confirmed that L72I did not obtain an additional mutation(s) in progeny viruses, which results in restored function (data not shown).
Revertant 4 produced a smaller MP due to a C-terminal truncation. When we introduced a nonsense mutation into the 37A238A MP to create 244FSrev, the resultant transcript did not cause local lesions on Xanthi-nc tobacco leaves. This suggested that the truncation did not contribute to the phenotypic reversion. It was likely that the 72I substitution helped revertant 4 to recover its viability (72I244FSrev in Table 4).
The mutations found commonly in revertants 7 and 8 were glycine-to-serine substitutions at residue 126. We introduced the glycine-to-serine substitution at residue 126 of the 37A238A virus genome to create 126Srev. When Xanthi-nc plants were inoculated with the transcript, local lesions appeared, indicating that the G126S amino acid substitution could convert a nonfunctional 37A238A MP into a functional one. We confirmed that 126S did not obtain an additional mutation(s) in progeny viruses, which results in restored function (data not shown).
The fact that both 72Irev and 126Srev could cause local lesions but the 37A238A mutant could not indicates that certain single-amino-acid substitutions could restore the intercellular transport function to 37A238A MP. On the basis of these independently occurring changes among the revertants, we suspected that the other substitutions observed in the original revertants contributed little, if at all, to the revertant phenotype (Fig. 1). It was at least confirmed that dysfunctional intercellular transport caused by the 37A238A substitution was compensated for, or masked considerably by, the 72I or 126S substitution. To examine the relative functions of the MPs, we measured average lesion diameters caused by virus (Table 2). The average diameters of 30 to 40 lesions induced on Xanthi-nc plants by mutants are shown in Table 2 relative to those induced by wild-type virus. This showed us that each mutant reacquired its movement function, but less efficiently than wild-type virus.
Phosphorylation analysis of mutant MPs.
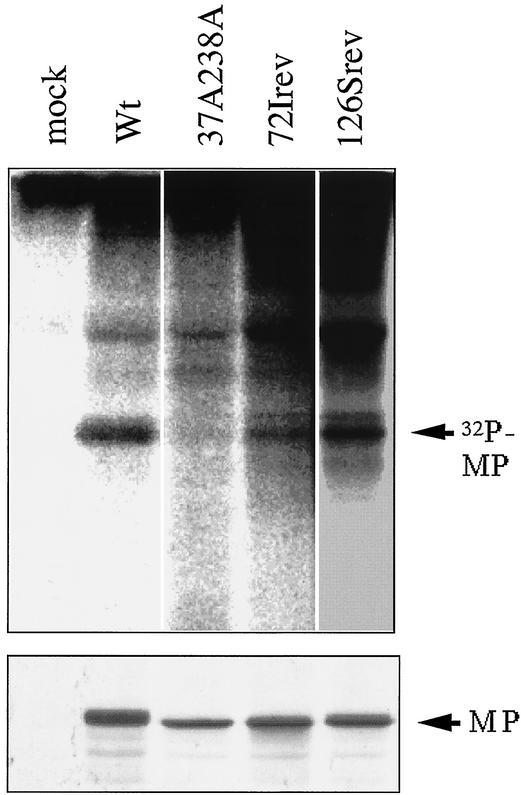
The MP phosphorylation of these additional mutants was assessed in vivo following the inoculation of each transcript into BY-2 protoplasts. The experiment (Fig. 4) demonstrated that the MPs of 72Irev and 126Srev were phosphorylated in vivo, while that of 37A238A was expressed but not phosphorylated, as described previously (13).
FIG. 4.
Characterization of MPs synthesized by 72Irev and 126Srev. (Upper panel) Autoradiograph showing the in vivo phosphorylation of each virus. After metabolic labeling with 32P, MPs were immunoprecipitated from each lysate of infected protoplasts. Wild-type (Wt) ToMV (W3) and 37A238A were used as positive and negative controls to show phosphorylated MP, as analyzed previously (13). (Lower panel) Immunodetection of MPs by anti-ToMV MP serum (20).
It appeared that the 72Irev MP was phosphorylated, but at a lower level than 126Srev and the wild-type virus. The wild-type MP underwent phosphorylation at two residues, as shown in previous work. Assuming that the wild-type phosphorylation occurred stoichiometrically and that its intensity reflected both residues, the band obtained for 72Irev suggested that its MP was phosphorylated, but less than the wild-type MP. The reason for this is not known. In the case of LQ126Srev, the band suggested one or two phosphorylation sites. However, we did not investigate this point further.
We also checked phosphorylation of other mutants constructed in this study (Table 4). The MP of 72I219Lrev was phosphorylated, while that of 219Lrev was not. It was suggested that the 72I substitution has a positive effect on susceptibility to phosphorylation even in the presence of the 219L substitution.
The MPs of 244FSrev and 72I244FSrev were not phosphorylated, but the 72I244FSrev MP was functional (Table 3). This was consistent with the result for revertant 4 (Fig. 2) and with our previous data, where LQD233 MP was not phosphorylated but was functional (32).
Stability of mutant MPs in vivo.
In our previous study, we found that MP instability or a reduced in vivo half-life was caused by serine-to-alanine substitutions at residues 37 and 238, lowering the protein's susceptibility to phosphorylation (13). The data suggested that MP with a short half-life could not function adequately. We analyzed the half-lives of the 72Irev and 126Srev MPs constructed in this study to understand why the revertants regained their function.
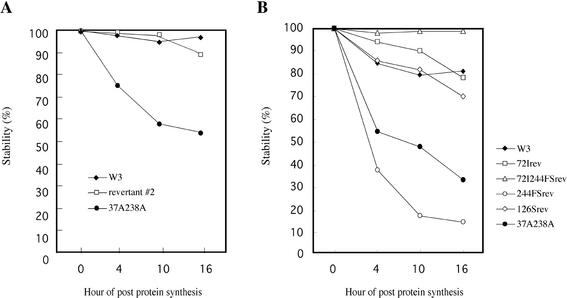
Protoplasts were inoculated with each virus, and at 8 h postinfection they were labeled for 10 min with [35S]methionine-[35S]cysteine. Unlabeled methionine and cysteine were then added to the culture medium, and the protoplasts were harvested immediately and at intervals of several hours. The proteins were separated by SDS-polyacrylamide gel electrophoresis, and the gels were scanned with a BAS1000 Image Analyzer (Fuji Film). After the detection of band images, their densities were scanned. The amounts of 35S-labeled MP were normalized to the density of the MP bands obtained at the start of the chase. The experiment showed that the movement-competent revertant 2 produced a relatively stable MP, while the parental 37A238A mutant produced a much less stable MP, as reported earlier (Fig. 5A) (13). It is likely that changes in the 37A238A MP protein structure and stability can occur as a result of a single amino acid substitution.
FIG. 5.
Half-lives of MPs synthesized in vivo by revertant 2 (A) and some mutant viruses constructed in this study (B), analyzed by pulse-chase experiments.
In separate experiments, we also examined the stabilities of 72Irev, 72I244FSrev, and 244FSrev. 244FSrev MP was expressed initially at a level similar to that of the wild type, but it had a very short half-life, with a relatively rapid decline in accumulation over time (Fig. 5B). In contrast, 72I244FSrev MP showed in vivo stability equal to or greater than those of the wild-type and 72I MPs (Fig. 5B). Thus, especially in revertant 4, the 72I substitution caused the reversion phenotype and increased MP stability in vivo, even though the protein was not phosphorylated.
We also characterized the stability of 126Srev MP in parallel to those of the wild-type, 37A238A, and 72Irev MPs (Fig. 5B). The data demonstrated that 126Srev MP had a longer half-life than 37A238A MP and that its stability was equivalent to those of the wild-type and 72Irev MPs.
According to our observations, the recovery of MP function was always associated with a gain of stability. The possibility exists that if the virus could produce an MP with an extended half-life compared to the original 37A238A MP, it could move between the cells.
Intracellular localization of mutant MPs.
It has been reported that the MP of tobamoviruses is associated with plasmodesmata, the cytoskeleton, microtubules, and the endoplasmic reticulum (2, 11, 13, 17, 19, 25). This intracellular localization is believed to be closely associated with MP function. In our previous study identifying the MP phosphorylation sites, one of our aims was to understanding the intracellular behavior of MP in relation to its phosphorylation status (13). To achieve this, we fused the GFP gene of A. victoria to the C-terminal ends of MP mutants. Nonfunctional MPs had different localization patterns than wild-type MP (13). The loss of the phosphorylation sites apparently prevented the typical wild-type MP localization pattern that was seen in protoplasts.
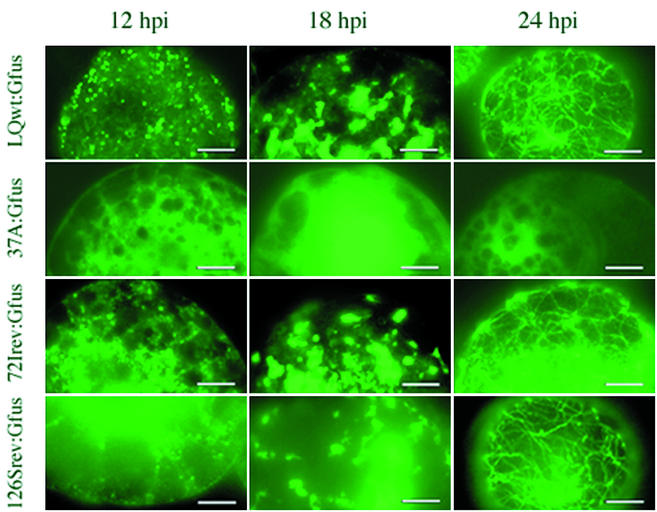
To examine the present mutants in the same way, we constructed several new viruses which produce mutant MP fused C-terminally with GFP. One of these viruses, 72Irev MP-GFP, encoded a chimeric 72Irev MP fused C-terminally with the sequence of GFP. Several localization patterns of MP-GFP were produced after the inoculation of mutant and wild-type MP viruses into BY-2 protoplasts, and these are shown in Fig. 6 at various time points after infection. Wild-type, 72Irev, and 126Srev MP-GFPs showed punctate fluorescent structures (dots) at 12 h postinoculation, irregular fluorescent structures and fluorescent patches between 12 and 18 h postinoculation, and filamentous structures after 24 h p.i. postinoculation. Thus, 72Irev and 126Srev showed a localization pattern similar to that of wild-type virus (Fig. 6), but 37A MP-GFP showed scattered fluorescence and lacked such a distinct localization pattern. Therefore, the phenotypic reversion of 72Irev and 126Srev mutant MPs was accompanied by an intracellular localization pattern closely resembling that of the wild-type MP. This difference was caused by the single amino acid substitution 72I or 126S.
FIG. 6.
Observation of MP-GFP localization in BY-2 protoplasts by fluorescence microscopy. Wild-type, 37A, 72Irev, and 126Srev MPs fused with GFP were expressed during LQwt:Gfus, 37A:Gfus (13), 72Irev:Gfus, and 126Srev:Gfus infections. Protoplasts were collected at 12, 18, and 24 h postinfection, fixed, and observed. Bars, 10 μm.
DISCUSSION
When we first constructed the 37A238A mutant and inoculated plants with the virus, no local lesions were observed on the leaves. However, local lesions did appear when we left the inoculated plants for an extended period of time. The sap extracted from these late-occurring lesions caused new local lesions on freshly inoculated leaves, at a level comparable to that for the wild-type virus. At first, we expected that such viruses would be true revertants, with an MP that had reacquired the original phosphorylation sites, leading to the recovery of function. It was anticipated that nucleotide sequence analysis of such revertants would give us clues as to the significance of MP phosphorylation in its intercellular transport function in the wild-type virus.
Unexpectedly, nucleotide sequence analysis of the revertants showed that they had acquired secondary mutations. These were unpredictable amino acid changes, such as 72I and 126S, which were commonly found among the MP sequences of independently isolated revertants. In the present study, we confirmed that these substitutions are the main reason for the regain of function in revertants from the 37A238A MP mutant. The MP of 72Irev is phosphorylated less than that of the wild type, but that of 126Irev is similar to that of the wild type (Fig. 4). Both mutants still retained 37A and 238A. They must therefore be phosphorylated on as-yet-uncharacterized residues. It was also shown that their cell-to-cell movement function is inferior to that of the wild type by the comparison of local lesion sizes caused by each virus (Table 1).
New phosphorylation sites could not be predicted by examining the primary sequences of the revertants. The leucine-to-isoleucine substitution of 72I itself cannot serve as a new phosphorylation target site, but the glycine-to-serine substitution of 126S could be a potential target for serine-threonine kinase. However, we did not address this issue further, partly because we found one movement-competent mutant that produced nonphosphorylated MP (72I244FSrev MP, reported in this study). Another mutant MP lacking the C-terminal 31 amino acids (constructed in vitro and designated LQD233) was not phosphorylated in vivo but could still retain some, if not full, competence for cell-to-cell movement (32).
The MP amino acid sequences of 72Irev and 126Srev have cryptic phosphorylation sites. Such sites could be used in adaptive evolution, especially when the wild-type sequence has acquired harmful substitutions during propagation. However, we cannot explain why we do not find 72I or 126S in the wild-type virus population. Citovsky et al. and Waigmann et al. also reported analyses of regulatory effects of MP phosphorylation (5, 29). They analyzed the phosphorylation sites of Ser258, Thr261, and Ser 265 of MP of tobacco mosaic virus, a similar but different ToMV. They focused on the C-terminal part of MP and suggested that the effects of phosphorylation on plasmodesmatal permeability were host dependent. We thus suspect that susceptibility to phosphorylation in different parts of MP is involved in such host adaptation through mutations. At least, the revertants and mutants constructed in this study could perform cell-to-cell movement less efficiently than wild-type virus, taking assays of relative sizes of local lesions as a base of comparison (Table 2).
Since structural information is not available for the MP at present, we cannot discuss in detail the link between each genetic change and the functional reversion. However, we speculate that such changes enhance the stability of the 37A238A MP in vivo and confer the proper intracellular localization and/or function. Phosphorylation in the wild-type MP appears to increase its stability in vivo.
Saito et al. reported that domains I (residues 56 to 96) and II (residues 125 to 164) are conserved among tobamovirus MPs (27). The former region includes amino acid 72, and the latter includes amino acid 126. Residue 72 is highly conserved, but amino acid 126 is not. Domain I is roughly and tentatively assigned to be a cytoplasmic, transmembrane domain (3). The significance of the 72I substitution in the context of the MP amino acid sequence is an interesting point.
It was possible that the 72I substitution favored the recovery of MP function only when it was introduced into the 37A238A MP sequence. To address this issue, we introduced the 72I mutation to the MP gene of wild-type virus. Preliminary experiments showed that the new 72I virus could cause symptoms in plants. This excluded the possibility that the 72I amino acid change was detrimental to and impaired the function of wild-type MP. It was possible that the 72I substitution contributed to the movement function of MP not specifically but cooperatively with the 37A and 238A mutations. The competitive fitness of viruses with such mutations within the virus population is an interesting issue to be addressed in the future.
Recently Toth et al. (28) reported a tobacco mosaic virus vector that was improved in terms of cell-to-cell movement and host range through the DNA-shuffling method. The resulting vectors have acquired amino acid substitutions in the MP-coding regions. One of these substitutions, L72V, was shown to be deeply involved. Replacement of leucine 72 is commonly observed in both their and our experiments. The importance of this coincidence awaits further analysis.
We proposed earlier that the presence of serine at position 37 or the phosphorylation of 37S is essential for intracellular stability and the correct localization of the MP to achieve its normal function (13). However, both 72Irev and 126Srev MPs maintained the serine-to-alanine substitutions at positions 37 and 238, and they were able to transport the virus RNA. Thus, the presence of serine at position 37 is not always essential for the function of the protein. Instead, it is more likely that the MP has to preserve an amino acid sequence which can achieve the stability and/or the conformation changes needed for intercellular movement. In certain cases where the MP acquired mutations and could not be phosphorylated, it still retained a degree of stability in vivo, showed the correct intracellular localization, and could perform the intercellular movement function. However, mutations such as this were very rare. Therefore, we can conclude that in the wild-type virus, phosphorylation of the MP is an important, but not necessary, prerequisite for function.
Acknowledgments
We thank Munenori Sato, Shohei Kaneko, Yuichi Kuwabara, Masaru Kakinuma, and Yoshihisa Kamekawa for propagating revertants at the Department of Biosciences, Teikyo University.
S.K. was supported by a Research Fellowship from The Japanese Society for the Promotion of Science for Young Scientists. This work was supported in part by “Molecular Mechanisms of Plant-Pathogenic Microbe Interaction,” a grant-in-aid for scientific research on priority area (A) from the Ministry of Education, Culture, Sports, Science, and Technology of Japan (no. 08680733) to Y.W.
REFERENCES
- 1.Barnes, W. M. 1994. PCR amplification of up to 35-kb DNA with high fidelity and high yield from lambda bacteriophage templates. Proc. Natl. Acad. Sci. USA 91:2216-2220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boyco, V., J. Ferralli, J. Ashby, P. Schellenbaum, and M. Heinlein. 2000. Function of microtubules in intercellular transport of plant virus RNA. Nat. Cell Biol. 2:826-832. [DOI] [PubMed] [Google Scholar]
- 3.Brill, L. M., R. S. Nunn, T. W. Kahn, M. Yeager, and R. N. Beachy. 2000. Recombinant tobacco mosaic virus movement protein is an RNA-binding, alpha-helical membrane protein. Proc. Natl. Acad. Sci. USA 97:7112-7117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Carrington, J. C., K. D. Kasschau, S. K. Mahajan, and M. C. Schaad. 1996. Cell-to-cell and long-distance transport of viruses in plants. Plant Cell 8:1669-1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Citovsky, V., B. G. McLean, J. R. Zupan, and P. Zambryski. 1993. Phosphorylation of tobacco mosaic virus cell-to-cell movement protein by a developmentally regulated plant cell wall-associated protein kinase. Genes Dev. 7:904-910. [DOI] [PubMed] [Google Scholar]
- 6.Dawson, W. O. 1992. Tobamovirus-plant interactions. Virology 186:359-367. [DOI] [PubMed] [Google Scholar]
- 7.Deom, C. M., M. Lapidot, and R. N. Beachy. 1992. Plant virus movement proteins. Cell 69:221-224. [DOI] [PubMed] [Google Scholar]
- 8.Deom, C. M., M. J. Oliver, and R. N. Beachy. 1987. The 30-kilodalton gene product of tobacco mosaic virus potentiates virus movement. Science 237:389-394. [DOI] [PubMed] [Google Scholar]
- 9.Hamamoto, H., Y. Sugiyama, N. Nakagawa, E. Hashida, Y. Matsunaga, S. Takemoto, Y. Watanabe, and Y. Okada. 1993. A new tobacco mosaic virus vector and its use for the systemic production of angiotensin-I-converting enzyme inhibitor in transgenic tobacco and tomato. Bio/Technology 11:930-932. [DOI] [PubMed] [Google Scholar]
- 10.Hamamoto, H., Y. Watanabe, H. Kamada, and Y. Okada. 1997. Amino acid changes in the putative replicase of tomato mosaic tobamovirus that overcome resistance in Tm-1 tomato. J. Gen. Virol. 78:461-464. [DOI] [PubMed] [Google Scholar]
- 11.Heinlein, M., B. L. Epel, H. S. Padgett, and R. N. Beachy. 1995. Interaction of tobamovirus movement proteins with the plant cytoskeleton. Science 270:1983-1985. [DOI] [PubMed] [Google Scholar]
- 12.Hirashima, K., and Y. Watanabe. 2001. Tobamovirus replicase coding region is involved in cell-to-cell movement. J. Virol. 75:8831-8836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kawakami, S., H. S. Padgett, D. Hosokawa, Y. Okada, R. N. Beachy, and Y. Watanabe. 1999. Phosphorylation and/or presence of serine 37 in the movement protein of tomato mosaic tobamovirus is essential for intracellular localization and stability in vivo. J. Virol. 73:6831-6840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kawakami, S., and Y. Watanabe. 1997. Use of green fluorescent protein as a molecular tag of protein movement in vivo. Plant Bio/Technol. 14:127-130. [Google Scholar]
- 15.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 16.Lucas, W. J., and R. L. Gilbertson. 1994. Plasmodesmata in relation to viral movement within leaf tissues. Annu. Rev. Phytopathol. 32:387-411. [Google Scholar]
- 17.Mas, P., and R. N. Beachy. 2000. Role of microtubules in the intracellular distribution of tobacco mosaic virus movement protein. Proc. Natl. Acad. Sci. USA 97:12345-12349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Matsushita, Y., K. Hanazawa, K. Yoshioka, T. Oguchi, S. Kawakami, Y. Watanabe, M. Nishiguchi, and H. Nyunoya. 2000. In vitro phosphorylation of the movement protein of tomato mosaic tobamovirus by a cellular kinase. J. Gen. Virol. 81:2095-2102. [DOI] [PubMed] [Google Scholar]
- 19.McLean, B. G., J. Zupan, and P. C. Zambryski. 1995. Tobacco mosaic virus movement protein associates with the cytoskeleton in tobacco cells. Plant Cell 7:2101-2114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Meshi, T., D. Hosokawa, M. Kawagishi, Y. Watanabe, and Y. Okada. 1992. Reinvestigation of intracellular localization of the 30K protein in tobacco protoplasts infected with tobacco mosaic virus RNA. Virology 187:809-813. [DOI] [PubMed] [Google Scholar]
- 21.Meshi, T., Y. Watanabe, and Y. Okada. 1992. Molecular pathology of tobacco mosaic virus revealed by biologically active cDNAs, p. 149-186. In T. M. A. Wilson and J. W. Davies (ed.), Genetic engineering with plant viruses. CRC Press, Boca Raton, Fla.
- 22.Meshi, T., Y. Watanabe, T. Saito, A. Sugimoto, T. Maeda, and Y. Okada. 1987. Function of the 30kd protein of tobacco mosaic virus: involvement in cell-to-cell movement and dispensability for replication. EMBO J. 6:2557-2563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ohno, T., M. Aoyagi, Y. Yamanashi, H. Saito, S. Ikawa, T. Meshi, and Y. Okada. 1984. Nucleotide sequence of the tobacco mosaic virus (tomato strain) genome and comparison with the common strain genome. J. Biochem. 96:1915-1923. [DOI] [PubMed] [Google Scholar]
- 24.Otsuki, Y., I. Takebe, T. Ohno, M. Fukuda, and Y. Okada. 1977. Reconstitution of tobacco mosaic virus rods occurs bidirectionally from an internal initiation region: demonstration by electron microscopic serology. Proc. Natl. Acad. Sci. USA 75:1727-1730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Padgett, H. S., B. L. Epel, W. T. Kahn, M. H. Heinlein, Y. Watanabe, and R. N. Beachy. 1996. Distribution of tobamovirus movement protein in infected leaves and implications for cell-to-cell spread of infection. Plant J. 10:1079-1088. [DOI] [PubMed] [Google Scholar]
- 26.Robards, A. W., and W. J. Lucas. 1990. Plasmodesmata. Annu. Rev. Plant Physiol. Plant Mol. Biol. 41:369-419. [Google Scholar]
- 27.Saito, T., Y. Imai, T. Meshi, and Y. Okada. 1988. Interviral homologies of the 30K proteins of tobamoviruses. Virology 167:653-656. [PubMed] [Google Scholar]
- 28.Toth, R. L., G. P. Pogue, and S. Chapman. 2002. Improvement of the movement and host range properties of a plant virus vector through DNA shuffling. Plant J. 30:593-600. [DOI] [PubMed] [Google Scholar]
- 29.Waigmann, E., M.-H. Chen, R. Bachmaier, S. Ghoshroy, and V. Citovsky. 2000. Regulation of plasmodesmatal transport by phosphorylation of tobacco mosaic virus cell-to-cell movement protein. EMBO J. 19:4875-4884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Watanabe, Y., Y. Emori, I. Ooshika, T. Meshi, T. Ohno, and Y. Okada. 1984. Synthesis of TMV-specific RNAs and proteins at the early stage of infection in tobacco protoplasts: transient expression of the 30K protein and its mRNA. Virology 133:18-24. [DOI] [PubMed] [Google Scholar]
- 31.Watanabe, Y., T. Meshi, and Y. Okada. 1987. Infection of tobacco protoplasts with in vitro transcribed tobacco mosaic virus RNA using an improved electroporation method. FEBS Lett. 219:65-69. [Google Scholar]
- 32.Watanabe, Y., T. Ogawa, and Y. Okada. 1992. In vivo phosphorylation of the 30-kDa protein of tobacco mosaic virus. FEBS Lett. 313:181-184. [DOI] [PubMed] [Google Scholar]
- 33.Watanabe, Y., T. Ohno, and Y. Okada. 1982. Virus multiplication in tobacco protoplasts inoculated with tobacco mosaic virus RNA encapsulated in large unilamellar vesicle liposomes. Virology 120:478-480. [DOI] [PubMed] [Google Scholar]