Abstract
Actinomycin D (ActD) is a transcription inhibitor and has been used in the treatment of certain forms of cancer. ActD has been reported to be a potential inhibitor of human immunodeficiency virus type 1 (HIV-1) replication due to its ability to inhibit reverse transcription. In contrast to what was expected, low concentrations of ActD (1 to 10 nM) upregulated HIV-1 replication 8- to 10-fold in MT-2 cells and had no effect on HIV-2 replication or on HIV-1 replication in MT-4, Jurkat, or peripheral blood mononuclear cells. The upregulation of HIV-1 replication was associated with an increase in HIV-1 transcription and a decrease in CD4 and CXCR4 expression. To further evaluate the effects of ActD on emergence of drug resistance in HIV-1 replication, a series of drug resistance assays were performed. Of interest, treatment of MT-2 cells with ActD also led to a high level of resistance to thymidine analogs (>1,000-fold increase in resistance to zidovudine and >250-fold to stavudine) but not to other nucleoside reverse transcriptases (RT), nonnucleoside RT, or protease inhibitors. This resistance appeared to be due to a suppression of host cell thymidine kinase-1 (TK-1) expression. These results indicate that ActD leads to a novel form of thymidine analog resistance by suppressing host cell TK-1 expression. These results suggest that administration of combination drugs to HIV-1-infected patients may induce resistance to antiretroviral compounds via a modification of cellular factors.
A characteristic feature of retroviruses is the presence of the enzyme reverse transcriptase (RT) (55). This enzyme catalyzes the production of a DNA copy of the RNA viral genome for subsequent integration into the host cell genome (55). The act of reverse transcription is a multistep process initially involving synthesis of the minus strand of DNA, using the viral genomic RNA as a template and a cellular tRNA as a primer. Minus-strand synthesis occurs in two stages (55). The initial synthesis of the minus strand in human immunodeficiency virus type-1 (HIV-1) reverse transcription begins near the 5′ end of genomic RNA. In this step, tRNA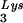 anneals to a primer-binding site and DNA synthesis proceeds for a short distance to the 5′ end of the viral genome. This is followed by minus-strand transfer, i.e., translocation of the initial minus-strand DNA product (minus-strand strong-stop DNA) to the 3′ terminus of genomic RNA. Minus-strand synthesis then continues until a full-length copy of the genome is made. During elongation of minus-strand DNA, the genomic RNA strand, once it has served its role as a template, is degraded by the intrinsic RNase H activity of the RT enzyme; this allows full-length minus-strand DNA to serve as the template for plus-strand DNA synthesis. Given the requirement for minus-strand transfer in the process of reverse transcription and the fact that this is a critical step in the retroviral life cycle that is not easily circumvented by viral mutation, agents that block this process, such as actinomycin D (ActD) (10, 15, 17, 26), are attractive candidates as potential therapeutic agents.
anneals to a primer-binding site and DNA synthesis proceeds for a short distance to the 5′ end of the viral genome. This is followed by minus-strand transfer, i.e., translocation of the initial minus-strand DNA product (minus-strand strong-stop DNA) to the 3′ terminus of genomic RNA. Minus-strand synthesis then continues until a full-length copy of the genome is made. During elongation of minus-strand DNA, the genomic RNA strand, once it has served its role as a template, is degraded by the intrinsic RNase H activity of the RT enzyme; this allows full-length minus-strand DNA to serve as the template for plus-strand DNA synthesis. Given the requirement for minus-strand transfer in the process of reverse transcription and the fact that this is a critical step in the retroviral life cycle that is not easily circumvented by viral mutation, agents that block this process, such as actinomycin D (ActD) (10, 15, 17, 26), are attractive candidates as potential therapeutic agents.
ActD is an inhibitor of transcription (43, 52) and is currently used in the treatment of certain forms of cancer (12, 33, 59). It binds to double-stranded DNA with a high affinity for GpC motifs (7, 16, 27), RNA-DNA hybrids (54), and single-stranded DNA (47, 57, 58). ActD has also been shown to be an inhibitor of the minus-strand transfer step (10, 15, 17, 26) in reverse transcription. By binding to minus-strand strong-stop DNA, the drug inhibits the activity of the viral nucleocapsid protein, which is critical for efficient strand transfer (10, 17). Thus, ActD blocks reverse transcription by a mechanism that is quite different from that of nucleoside RT inhibitors. It has been suggested that Act D may be able to serve as a novel therapeutic intervention in patients with HIV infection (10, 15, 17, 26) who have failed combination treatment (42).
To examine the potential of ActD to inhibit HIV-1 replication, we initiated a series of in vitro experiments to determine the impact of ActD on HIV-1 replication in different cell types. A high concentration of ActD was cytotoxic to cells and, as a consequence, viral replication was inhibited. To our surprise, however, at lower concentrations ActD led to an enhancement of HIV-1 replication in MT-2 cells, one of the cell lines used in our study. In these cells, ActD treatment resulted in a high level of resistance to thymidine analogs by interfering with the expression of thymidine kinase.
MATERIALS AND METHODS
Cells and reagents.
The MT-2 (18, 19), MT-4 (32, 41), and H9 (35, 44, 45) cell lines, the HIV-1 proviral DNA clone pNL4.3 (1), and the HIV-2-infected H9 cell line H9/HIV-2MVP15132 (3; L. Gürtler, J. Eberle, and F. Deinhardt, Abstr. 4th Int. Conf. AIDS, abstr. 1662, 1988) were obtained from the AIDS Research and Reference Reagent Program, National Institute of Allergy and Infectious Diseases (NIAID), National Institutes of Health (NIH); MT-2 and MT-4 were obtained from Douglas Richman; H9 was from Robert Gallo; pNL4.3 was from Malcolm Martin; and H9/HIV-2MVP15132 was from Lutz Gürtler and Friedrich Deinhard. Jurkat and RD cells (a human embryonal rhabdomyosarcoma cell line) were provided by the American Type Culture Collection (Rockville, Md.). MT-2, MT-4, and Jurkat cells were maintained in RPMI 1640 supplemented with 10% fetal bovine serum (HyClone Laboratories Inc., Logan, Utah), 10 mM l-glutamine, 100 U of penicillin/ml, and 100 μg of streptomycin/ml (RPMI-10) (24, 63). RD cells were maintained in Eagle's minimum essential medium supplemented with 10% fetal bovine serum, 100 U of penicillin/ml, and 100 μg of streptomycin/ml (EMEM-10) (24). Peripheral blood mononuclear cells (PBMCs) were isolated from heparinized whole blood from healthy donors using lymphocyte separation medium (ICN Biomedical Inc., Aurora, Ohio) (24, 63). PBMCs were stimulated using 5 μg of phytohemagglutinin (PHA) (Sigma, St. Louis, Mo.)/ml and 20 U of native interleukin-2 (Roche Molecular Biology, Indianapolis, Ind.)/ml in RPMI 1640 supplemented with 10% fetal calf serum and 50 U of gentamicin/ml (RPMI-10G) (2). The PHA-stimulated PBMCs were maintained in RPMI-10G in the presence of 20 U of interleukin-2/ml.
Generation of viral stocks.
To generate recombinant infectious HIV-1NL4.3 virus stocks, pNL4.3 was transfected into RD cells by using TransIT LT-1 (PanVera Corp., Madison, Wis.) following the manufacturer's protocol (24). Briefly, 75 × 103 RD cells in 2 ml of EMEM-10 in each well of a six-well plate was used for each transfection with the lipid and DNA complex (4 μl of TransIT LT-1 per 1 μg of the plasmid). After 24 h, 106 fresh MT-2 cells were added to the dishes, which were incubated at 37°C for an additional 24 h. The MT-2 cells thus infected were collected, washed, and cultured at 37°C for 3 days in 5 ml of RPMI-10. Cell-free culture supernatants were then obtained and stored at −80°C. The 50% tissue culture infectious dose (TCID50) of each stock was determined as previously described (24, 63). A stock of infectious HIV-2MVP15132 was produced by cocultivation of the chronically infected cell line H9/HIV-2MVP15132 with fresh uninfected H9 cells. HIV-2 virus production was determined by measurement of p27 antigen production, using a p27 antigen enzyme-linked immunosorbent assay kit (Zeptometrix Corp., Buffalo, N.Y.).
HIV replication assay.
The level of HIV-1 replication under various culture conditions was determined as follows. MT-2, MT-4, or Jurkat cells (4 × 106) were infected with 2,500 TCID50 of HIV-1NL4.3, HIVDH12, or HIV-2MVP15132 at 37°C for 2 h. PBMCs (5 × 106) were infected with 5,000 TCID50 of HIV-1NL4.3 for 2 h at 37°C. After infection, the infected cells were washed three times with warmed medium to remove unbound virus particles. The infected MT-2, MT-4, or Jurkat cells were cultured at 37°C for 7 days in 96-well flat-bottom plates at a density of 0.2 × 106 cells/ml in 0.2 ml of RPMI-10 in the presence or absence of various stimuli. The infected PBMCs were cultured at 106 cells/ml in 0.2 ml of RPMI-10G for 7 days at 37°C in 96-well flat-bottom plates. Each culture was performed in quadruplicate. HIV-1 or HIV-2 replication was determined by measuring the amount of p24 or p27 antigen in the culture supernatants at day 7 after infection by using a p24 antigen capture assay (Beckman-Coulter, Miami, Fla.) or p27 antigen capture assay (Zeptometrix Corp.).
HIV-1NL4.3 replication kinetic assays were performed as previously described (24). Briefly, HIV-1NL4.3-infected MT-2 cells were cultured in the absence or presence of 10 nM ActD (Sigma). Culture supernatants were collected every day and replaced with an equal volume of fresh complete medium. The concentration of ActD was maintained throughout the period of the culture. To monitor HIV-1 replication, the levels of p24 in tissue culture supernatants were measured by using a p24 antigen capture assay (Beckman-Coulter).
Flow cytometric analyses.
MT-2 cells were cultured for 3 days in the absence or presence of 10 nM Act D at 37°C. Cells (106 cells per sample) were washed with Dulbecco's phosphate-buffered saline (PBS) (Gibco, Grand Island, N.Y.) and then incubated with 10 μl of anti-CD4 immunoglobulin G1 (IgG1) conjugated with fluorescein isothiocyanate (FITC) (BD-PharMingen, San Diego, Calif.) and 10 μl of anti-CXCR4 IgG2a conjugated with phycoerythrin (PE) (BD-PharMingen) or 10 μl of anti-CCR5 IgG2a conjugated with PE (BD-PharMingen) at room temperature for 15 min in PBS supplemented with 2% bovine serum albumin (BSA) (Sigma). After incubation, the stained cells were washed with 2% BSA-PBS containing 0.5% NaN3 buffer and centrifuged (200 × g) for 5 min, and then cells were analyzed by using a Coulter XL flow cytometer (Beckman-Coulter). Events were collected by gating on forward and 90° light scatter. Positive fluorescence was determined using a matched control IgG1 conjugated with FITC or control IgG2a conjugated with PE (BD-PharMingen).
To analyze the cell cycle, MT-2 cells were cultured in the absence or presence of 10 nM ActD at 37°C for 1 or 3 days and then washed with PBS. Single-color flow cytometric analysis of DNA content was performed on both treated and control cell lines. The cells (106) were fixed by the addition of 1 ml of 70% ethanol. The cell pellets were washed with PBS and incubated in 1 ml of PBS containing 5 μl of DNase-free RNase (200 U/ml) (Roche Molecular Biology) at 37°C for 15 min, and then 100 μl of propidium iodide (0.5 mg/ml) was added to the cell suspension, followed by incubation on ice for at least 30 min. The stained cells were analyzed for red fluorescence (FL3) on a Coulter XL flow cytometer (Beckman-Coulter), with doublet discrimination achieved with an amorphous gate based on linear and peak FL3 signal. The distribution of cells in the G1, S, and G2/M phases of the cell cycle was calculated from the resulting DNA histogram by using Multicycle AV software, based on a zero-order polynomial S-phase model (Phoenix Flow Systems, San Diego, Calif.).
Virion-associated HIV RT assay.
Culture supernatants from HIV-1NL4.3-infected MT-2 cells were collected, and cell-free crude supernatants were clarified by low-speed centrifugation (500 × g for 5 min) and filtered through a 0.2-μm polyvinylidene difluoride membrane filter (Millipore, Bedford, MA) to remove cells. An ultracentrifugation (10,000 × g for 2.5 h at 4°C) was then performed, using 20% sucrose (wt/vol) in 150 mM NaCl-10 mM HEPES, pH 7.4, to pellet HIV-1 virions in the supernatant. The HIV-1 pellet was resuspended in the lysis buffer provided in the Colorimetric RT assay kit (Roche Molecular Biology). The reverse transcription assay was performed following the manufacturer's protocol, with the amount of cell lysate equivalent to 50 ng of p24.
Semiquantitative RT-PCR.
The HIV-1NL4.3-infected MT-2 cells (4 × 106) were cultured in T 25-cm2 flasks in the absence or presence of 10 nM ActD in 5 ml of RPMI-10 for 4 days. Cells were washed using cold PBS, and RNA was isolated from the cells using the RNeasy Isolation kit (Qiagen Inc., Valencia, Calif.). Total cellular RNA was treated with RNase-free DNase I (Invitrogen, Carlsbad, Calif.) following the vendor's protocol. The cDNA was synthesized using the Superscript First Strand Synthesis system for RT-PCR (Invitrogen) with oligo(dT) and a cDNA primer for HIV-1NL4.3 RNA (5′-TTGTTTTACATCATTAGTGTGGC-3′). The synthesized cDNA (1 μg) was serially diluted 1:4 (12 dilutions). Diluted cDNA was used as template for PCR to amplify thymidine kinase-1 (TK-1) or β-actin. PCR primer pairs were as follows: TK-1, 5′-GTGATTGGGGGAGCAGACAAG-3′ and 5′-ACTCAGCAGTGAAAGCCGCAG-3′; β-actin, 5′-GCTCGTCGTCGACAACGGCTC-3′ and 5′-CAACATGATCTGGGTCATCTTCTC-3′. The PCR mixtures (50 μl) containing 1× Expand HF buffer, oligo nucleotide pairs (400 nM), deoxynucleoside triphosphates (dNTPs) (200 nM), and 0.7 U of Expand High-Fidelity PCR System enzyme mix (Roche Molecular Biochemical) underwent 25 cycles at 95°C for 30 s, 55°C for 30 s, and 72°C for 1 min with the final extension at 72°C for 7 min. The reaction products were separated on a 1% agarose gel (Biowhittaker Molecular Applications, Walkersville, Md.) in the presence of 0.5 μg of ethidium bromide/ml and photographed.
Drug resistance assays.
Drug resistance assays were performed as previously described (24). Briefly, HIV-1-infected MT-2 cells were incubated in a 96-well flat-bottom plate in the presence or absence of various concentrations of drugs and cultured for 7 days at 37°C. The p24 levels in day 7-culture supernatants were measured by a p24 antigen capture assay kit (Beckman-Coulter). Each assay was performed in quadruplicate. 3′-azido-3′-deoxythymidine (AZT), 2′,3′-dideoxyinosine (ddI), 2′,3′-didehydro-3′-deoxythymidine (d4T), and 2′,3′-dideoxycytidine (ddC) were purchased from Sigma. Abacavir, lamivudine (3TC), and efavirenz (EFV) were obtained through the AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH. Indinavir was provided by Merck Research Laboratory (West Point, Pa.). 9-[2-(Phosphonylmethoxy)ethyl] adenine (PMEA) was provided by Gilead Science (Foster City, Calif.). Sensitivities were reported as the concentrations of the drugs that inhibited p24 production by 50% (IC50s) (24, 25).
DNA microarray analysis.
A microarray gene expression analysis was conducted using the Affymetrix GeneChip system (Affymetrix, Inc., Santa Clara, Calif.). HuFL and U95A GeneChips containing probes to ∼7,000 and ∼12,500 human genes, respectively, were used. Isolation and preparation of the RNAs for GeneChip hybridization and quantitation were performed using the manufacturer's recommended protocols (Affymetrix). In short, MT-2 cells were cultured for 3 or 4 days in the absence or presence of 10 nM Act D at 37°C. Total RNA was isolated using the RNeasy kit (Qiagen) and mRNA was converted to double-stranded cDNA (ds-cDNA) by using the SuperScript Choice system for cDNA synthesis (Invitrogen) with an oligo(dT) primer containing a T7 RNA polymerase promoter to prime the first-strand synthesis. Biotin-labeled cRNA was generated by in vitro transcription (Enzo Biochem, New York, N.Y.) by the addition of T7 RNA polymerase and biotinylated nucleotides to the ds-cDNA. The labeled cRNA was hybridized to the probes on the GeneChips, which were then washed and stained with strepavidin-conjugated PE by using the Affymetrix GeneChip Fluid Station 400 (Affymetrix). Gene expression levels were determined following laser scanning of the GeneChip at 570 nm. The average intensities for each chip were scaled to 150, and differentially expressed genes were identified by comparing the chips using the Microarray Suite 4.0 software (Affymetrix). Genes differentially expressed at levels greater than threefold were considered candidates for further studies.
PCR amplification and direct DNA sequencing.
Genomic DNA was extracted from HIV-1NL4.3-infected MT-2 cells (106) by using the QIAamp DNA Blood Mini kit (Qiagen). The nucleotide sequence of a 1,685-bp fragment of the HIV-1 genome containing gag (p7/p1/p6), protease, and part of RT was amplified by PCR with the Expand High-Fidelity PCR system (Roche Molecular Biochemical) with the following primer pair: forward primer (nucleotides [nt] 1881 to 1904 of pNL4.3), 5′-GAAGCAATGAGCCAAGTAACAAAT-3′; reverse primer (nt 3543 to 3566), 5′-GATATGTCCATTGGCCTTGCCCCT-3′. The reaction mixtures (50 μl) containing 1× Expand HF buffer, oligonucleotide pairs (400 nM), dNTPs (200 nM) (Roche Molecular Biochemical), and 1.75 U of Expand High-Fidelity PCR system enzyme mix were subjected to 25 cycles of 95°C for 30 s, 55°C for 30 s, and 72°C for 2 min, with the final extension at 72°C for 7 min. The PCR products were purified with the QIAquick spin PCR purification kit (Qiagen). Sequencing reactions were performed with the ABI PRISM BigDye Terminator cycle sequencing kit (PE Biosystems, Foster City, Calif.) with the sequencing primers 5′-AGAAGCAGGAGCCGATAGACAAGG-3′, 5′-AAGCCAGGAATGGATGG-3′, and 5′-ATAATACACTCCATGTACTGGTTC-3′. The reaction products were purified using a DyeEx Spin kit (Qiagen), resolved by electrophoresis on 6.0% polyacrylamide gels, and analyzed with an Applied Biosystems 377 automated sequencing system (PE Biosystems,); the protease and the RT gene were translated and aligned with the Sequence Navigator (PE Biosystems) (63). Changes in the protease and RT regions were compared with the HIV-1 clade B consensus sequence as a reference (38).
Western blot analysis.
HIV-1NL4.3-infected MT-2 cells were cultured for 4 days in the absence or presence of 10 nM Act D. The cells (5 × 106) were washed with PBS and resuspended in radioimmunoprecipitation assay (RIPA) lysis buffer (50 mM Tris-HCl [pH 7.5]-150 mM NaCl-1% NP-40 [Roche Molecular Biochemical]-0.5% sodium deoxycholate [Sigma]-0.1% sodium dodecyl sulfate [Sigma]-1× protease inhibitor cocktail [Sigma]) (48). Total cellular protein content was determined using the BCA protein assay kit (Pierce, Rockford, Ill.). The lysate (50 μg of protein) was loaded onto a 12% bis-Tris gel (Invitrogen) and transferred to a nitrocellulose membrane (Invitrogen). Blots were then probed with 10 μg of mouse anti-human TK monoclonal antibody (QED Bioscience, San Diego, Calif.)/ml. The primary antibody was detected with a horseradish peroxidase-conjugated anti-mouse Ig (Amersham Life Science, Piscataway, N.J.). A positive signal was detected with the ECL Plus Western blotting detection system (Amersham). The membrane was then stripped with the Restore Western blot stripping buffer (Pierce). Blots were reprobed with 0.4 μg of mouse anti-human actin (Santa Cruz Biotechnology, Santa Cruz, Calif.)/ml. The signal was detected as described above.
Statistical analysis.
Differences between HIV variants in comparative replicative abilities were calculated by using the unpaired t test using the StatView program (Abacus Concepts, Berkeley, Calif.).
RESULTS
Effect of ActD on HIV-1 replication.
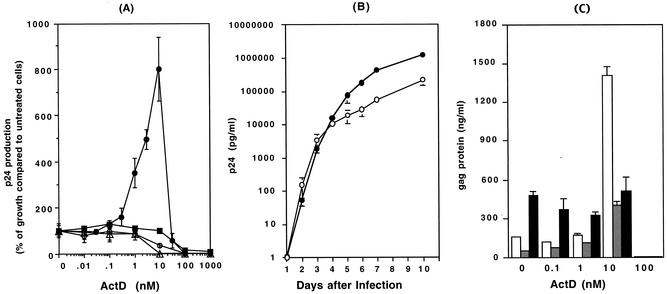
Previous studies have demonstrated that ActD inhibits the minus-strand transfer step of reverse transcription, and it has been speculated that ActD may be a potent anti-HIV-1 reagent (10, 15, 17, 26). To evaluate the potential inhibitory effects of ActD on HIV-1 replication in tissue culture, MT-2, MT-4, or Jurkat cells or PHA-stimulated PBMCs infected with recombinant HIV-1NL4.3 were cultured in the presence of various concentrations of ActD (Fig. 1A). Concentrations of ActD greater than 100 nM inhibited HIV replication in all tested cells. Trypan blue staining revealed that these inhibitory effects were likely to be a consequence of the cellular toxicity of ActD (<1% viable cells at more than 100 nM ActD). Interestingly and unexpectedly, however, low concentrations (1 to 10 nM) of ActD upregulated HIV-1 replication by 8- to 10-fold in MT-2 cells. This ability of ActD to enhance HIV-1NL4.3 replication was not seen using Jurkat, MT-4, or PHA-stimulated PBMCs. To determine if the increase in p24 production in MT-2 cells was associated with an increase in viral titers or was merely an increase in the release of p24 antigens from infected cells, TCID50 titers and p24 levels of tissue culture supernatants from control and ActD (10 nM)-treated cultures were compared. The TCID50 titers of day 7 culture supernatants in the absence and presence of 10 nM ActD were 460 ± 76 and 329 ± 63 TCID50 per ng of p24, respectively (P > 0.05). Thus, the increase in p24 levels in ActD-treated cultures was reflective of enhanced HIV-1 replication rather than merely being due to increased release of p24 protein from MT-2 cells.
FIG. 1.
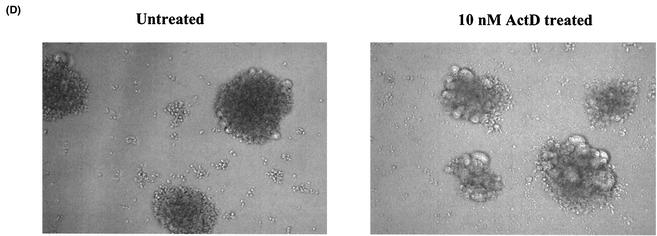
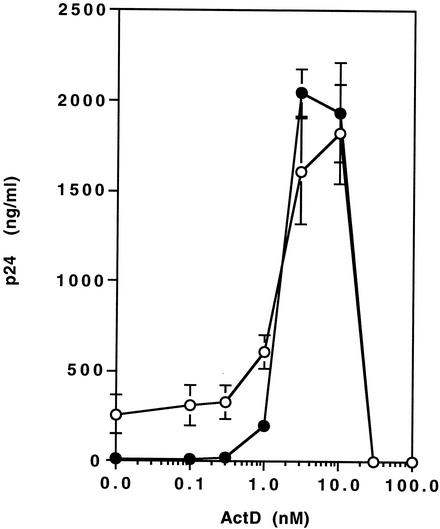
ActD upregulates HIV-1 replication but not HIV-2 replication. (A) MT-2 (closed circles), MT-4 (open circles), Jurkat (open triangles), or PHA-stimulated PBMCs (closed squares) were infected with HIV-1NL4.3 for 2 h at 37°C. The infected cells were cultured for 7 days in the presence of various concentrations of ActD (0 to 1,000 nM). HIV-1 replication was measured by a p24 antigen capture assay. Results show the percentage of virus growth in ActD-treated cells compared to that in untreated cells. In this experiment, the p24 concentrations in the culture supernatant of MT-2, MT-4, Jurkat, and PBMCs in the absence of ActD were 159 ± 33, 56 ± 11, 9.1 ± 2.1, and 10 ± 1.4 ng/ml, respectively. (B) HIV-1NL4.3-infected MT-2 cells were cultured in 10 ml of RPMI-10 in T-25 cm2 culture flasks in the presence (closed circles) or absence (open circles) of 10 nM ActD, and culture supernatants were collected every day. The assay was performed in triplicate, and the p24 levels in culture supernatants were measured by the p24 antigen capture assay. (C) MT-2 cells were infected with HIV-1NL4.3 (white bar), HIV-1DH12 (gray bar), or HIV-2MVP15132 (black bar) for 2 h at 37°C. After washing, the MT-2 cells were cultured for an additional 7 days at 37°C in the presence of 0 to 100 nM ActD. The p24 (for HIV-1) or p27 (for HIV-2) antigen levels in the culture supernatants were measured by p24 or p27 antigen capture assays. (D) MT-2 cells were infected with HIV-1NL4.3 and cultured for 4 days in the absence or presence of 10 nM ActD.
To assess if the difference in virus replication was secondary to a shift in replication kinetics, levels of virus were measured on a daily basis for 10 days (Fig. 1B). For the first 3 days after infection, there was no difference between the ActD-treated culture and the control; however, after day 4, higher levels of virus were seen in the ActD-treated culture (p24 concentrations on day 7 in the absence and presence of ActD were 569 ± 10 and 4,522 ± 72 ng/ml, respectively; P < 0.01). Therefore, ActD appeared to be acting by prolonging the time of productive infection rather than by enhancing infectivity. To further understand this enhancement of HIV-1 replication by ActD, the impact of ActD on the replication of two other viruses, HIV-1DH12 (51) and HIV-2MVP15132, was also investigated. Replication of HIV1DH12 increased from 53 ± 2.3 to 403 ± 50 ng/ml in the presence of 10 nM ActD on day 7 (P < 0.01). Replication of HIV-2MVP15132 was not enhanced by ActD (p27 concentrations on day 7 in the absence and presence of 10 nM ActD were 460 ± 35 and 513 ± 119 ng/ml, respectively [P > 0.05]) (Fig. 1C).
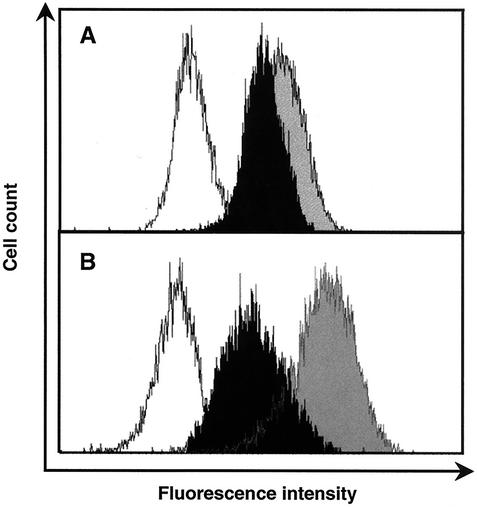
To determine whether these changes were a consequence of alterations in receptor or coreceptor expression, levels of CD4 and CXCR4 expression on MT-2 cells were also analyzed. Fresh uninfected MT-2 cells were cultured for 3 days in the presence or absence of 10 nM ActD and were then analyzed by flow cytometry for CD4 and CXCR4 expression. Of note, the expression of CD4 and of CXCR4 on ActD-treated MT-2 cells was downregulated by 30 and 80%, respectively (Fig. 2). Therefore, the increase in HIV-1 replication induced by ActD after 4 days of infection was not due to an increase in receptor or coreceptor expression leading to enhanced infectibility in later rounds of infection. To define the cell specificity of this phenomenon, MT-4 or Jurkat cells and PBMCs were also treated with 10 nM ActD for 3 days and the expression levels of CD4 and CXCR4 were monitored. The CD4 expression on MT-4 cells and Jurkat cells was upregulated by 1.5- and 2-fold, respectively; however, CXCR4 expression was not influenced by the treatment. ActD had no effect on the expression of CD4 or CXCR4 on PBMCs (data not shown). To evaluate whether or not the prolongation of time of productive infection was due to decreased syncytium formation and enhanced survival of infected cells, cultures were monitored for syncytium production. As seen in Fig. 1D, ActD treatment led to an increase in syncytium formation.
FIG. 2.
ActD downregulates expression of CD4 and CXCR4 on MT-2 cells. MT-2 cells were cultured for 3 days in the presence of 0 (gray) or 10 nM (black) ActD. The treated cells were stained with fluorescence-conjugated anti-CD4 or -CXCR4. The staining patterns for the isotype control antibodies are shown in white. Fluorescence intensity was measured by a Coulter XL flow cytometer. (A) The mean fluorescence intensities (MFI) for CD4 staining on control and ActD-treated cells were 16.6 and 11.3, respectively. (B) The MFI for CXCR4 on control and ActD-treated cells were 41.6 and 8.29. Results are representative of three independent experiments.
MT-2 cells are transformed with human T-cell leukemia virus type 1 (HTLV-1) and produce infectious HTLV-1 virions (11, 31). HTLV-1 has been reported to enhance HIV replication in vitro (4, 9, 34, 36, 62). To determine whether or not the enhancement of HIV production by ActD was due to enhanced HTLV-1 production, p19 levels in culture supernatants were measured with a p19 antigen capture kit (Zeptometrix Corp.). No difference in HTLV-1 production was seen in the absence or presence of 10 nM Act D (the amounts of p19 on day 4 in the absence and presence of 10 nM ActD were 290 ± 33 and 227 ± 23 ng/ml, respectively [P > 0.05]). Therefore, the activation of HIV-1 was not associated with HTLV-1 activation.
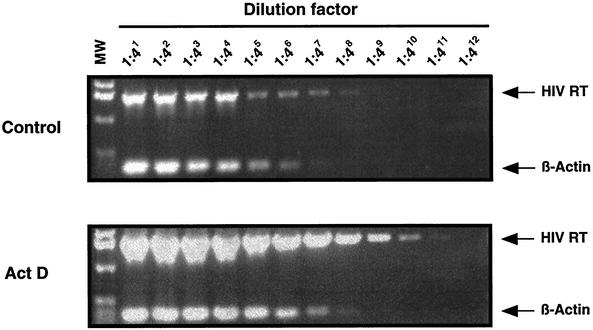
To determine the direct effects of ActD on HIV-1 RT activity in MT-2 cells, RT enzyme assays were performed using lysates of HIV-1-infected MT-2 cells. Various concentrations of ActD (0 to 100 μM) had no direct effects on RT activity (data not shown). To examine the effect of ActD on HIV-1 gene transcription, a semiquantitative RT-PCR was utilized to compare the levels of HIV-1 RNA and β-actin in control and ActD-treated cells (Fig. 3). To confirm the absence of contaminating DNA, control cDNA reactions were prepared in which RT was omitted from each reaction mixture. Amplified products were not seen in these controls (data not shown). In the presence of 10 nM ActD, HIV-1 RNA transcription increased 16-fold. Therefore, despite its well-known ability to inhibit transcription in most systems (43, 52), ActD enhanced HIV-1 RNA transcription in MT-2 cells.
FIG. 3.
ActD enhances HIV-1 RNA transcription. MT-2 cells were infected with HIV-1NL4.3 for 2 h at 37°C. The infected cells were cultured for 4 days in the presence or absence of 10 nM ActD. Cells were washed and RNA was isolated. The RNA was treated with DNase and subsequently used to synthesize cDNA. The cDNA (1 μg) was serially diluted 1:4 and then served as template for subsequent PCR to amplify HIV-1 RT or β-actin. To confirm the absence of contaminating DNA, control cDNA reactions were prepared in which RT was omitted from each reaction. Amplified products were not seen in these controls (data not shown). β-Actin served as an internal control to monitor the efficiency of amplification of each of the RNAs. The PCR product sizes of HIV RT and β-actin are 1,540 and 353 bp, respectively. MW refers to a molecular marker consisting of a 1 kbp ladder (Invitrogen).
Effect of ActD on drug susceptibility.
Given the enhancement of HIV-1 replication in MT-2 cells by ActD, we were interested in determining whether or not this increase in replication had an effect on the rate of emergence of drug-resistant variants. To address this question, a series of drug resistance assays was performed to determine the IC50s for a series of drugs, utilizing control or ActD-treated cells (Table 1). In the presence of 10 nM ActD, HIV grown in MT-2 cells showed a high level of resistance to thymidine analogs (>1,000-fold increase in AZT IC50, >250-fold for d4T). In contrast, only a modest increase (two- to fivefold) in the IC50s of indinavir, ddI, ddC, 3TC, abacavir (ABC), PMEA, and EFV was observed in the presence of ActD.
TABLE 1.
Effect of ActD on drug sensitivity in MT-2 cells
| ActD (nM) | IC50 (nM)a
|
||||||||
|---|---|---|---|---|---|---|---|---|---|
| IDV | AZT | d4T | ddI | ddC | 3TC | ABC | PMEA | EFV | |
| 0 | 15 ± 5.4 (1.0) | 9.3 ± 3.7 (1.0) | 40 ± 16 (1.0) | 251 ± 60 (1.0) | 101 ± 18 (1.0) | 131 ± 96 (1.0) | 95 ± 20 (1.0) | 233 ± 42 (1.0) | 0.7 ± 0.14 (1.0) |
| 10 | 51 ± 20 (3.3) | >10,000 (>1,075) | >10,000 (>250) | 391 ± 27 (1.6) | 471 ± 73 (4.7) | 262 ± 85 (2.0) | 362 ± 82 (3.8) | 586 ± 70 (2.5) | 2.4 ± 0.6 (3.4) |
Data are means ± standard errors. Each construct was independently studied at least three times. Values in parentheses represent the fold differences in IC50s compared to that of 0 nM ActD.
To better evaluate the impact of ActD on resistance to AZT, MT-2 cells were infected with HIV-1NL4.3 and cultured for 7 days in the presence of 1 μM AZT with various concentrations of ActD (Fig. 4). In the absence of ActD, HIV-1 replication was inhibited 96% by the presence of AZT. The inhibitory effect of AZT on HIV replication was completely suppressed by the addition of ActD at concentrations of 3 to 10 nM.
FIG. 4.
ActD treatment reverses the inhibition of HIV replication by RT inhibitors. MT-2 cells were infected with HIV-1 NL4.3 for 2 h at 37°C. The infected cells were cultured for 7 days in the presence of various concentrations of ActD (0 to 100 nM) without AZT (open circles) or with 1 μM AZT (closed circles). HIV-1 replication was measured by a p24 antigen capture assay.
To understand the mechanism of this high level of resistance to thymidine analogs, we first performed proviral DNA sequencing of the HIV RT gene to look for known mutations associated with drug resistance (49). No resistance-associated mutations were found in the protease and RT regions (data not shown). These data suggested that ActD led to thymidine analog resistance as a consequence of changes at the cellular level.
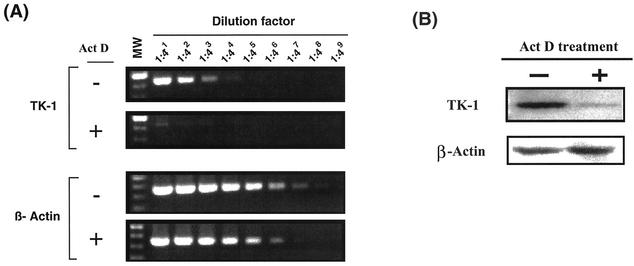
To examine this possibility, a DNA microarray analysis was performed with MT-2 cells in the presence and absence of ActD. The results of this analysis revealed that ActD treatment led to a 27.3-fold downregulation in transcription of the TK-1 gene, a gene that catalyzes the initial phosphorylation of thymidine analogs (14). Similar changes were not observed in other nucleoside kinases, including adenosine (no change), cytidine (1.3-fold increase), and guanosine (1.4-fold increase) kinases. To confirm the downregulation of transcription of TK-1, a semiquantitative RT-PCR analysis was performed. The cells treated with 10 nM ActD had lower expression of the TK-1 gene in a setting of no changes in transcription of β-actin (Fig. 5A).
FIG. 5.
ActD treatment downregulates transcription of TK-1. MT-2 cells were infected with HIV-1NL4.3 for 2 h at 37°C. The infected cells were cultured for 4 days in the absence or presence of 10 nM ActD. (A) Cells were washed and RNA was isolated. The RNA was treated with DNase and subsequently used to synthesize cDNA; the cDNA (1 μg) was serially diluted 1:4. The diluted cDNA served as template for subsequent PCRs to amplify TK-1 and β-actin. To confirm the absence of contaminating DNA, control cDNA reactions were prepared in which RT was omitted from each reaction. Amplified products were not seen in these controls (data not shown). β-Actin served as an internal control to monitor the efficiency of amplification. The PCR product sizes of TK-1 and β-actin were 540 and 353 bp, respectively. MW refers to a molecular marker consisting of a 100-bp ladder (Invitrogen). (B) The infected cells were washed and cellular protein was extracted using RIPA buffer (see Materials and Methods). For each blot, 50 μg of total cellular protein was loaded on a 12% bis-Tris sodium dodecyl sulfate-acrylamide gel, and Western blotting was performed using an anti-TK-1 antibody. Subsequently, the antibody was stripped and then the blot was reprobed using anti-actin antibody. Images were quantitated using the Epi chemi II Darkroom and LabWork software (Ultra-violet Products, Ltd., Upland, Calif.).
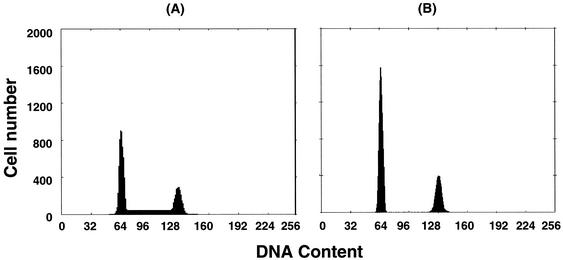
To define the impact of this decrease in RNA transcription at the protein level, Western blotting was performed using anti-TK-1 and anti-β-actin antibodies. MT-2 cells treated with 10 nM ActD showed an 85% decrease in the expression of TK-1 and no change in β-actin protein expression (Fig. 5B). TK-1 is a cell cycle-regulated enzyme and its activity fluctuates with DNA synthesis, being high in dividing and malignant cells and low in quiescent cells (50). To assess the effect of ActD on cell cycle progress in MT-2 cells, a cell cycle analysis utilizing propidium iodide was performed (Fig. 6). The percentage of cells in S phase in control and ActD-treated cells was 30.6 and 1.6%, respectively (P < 0.01). To assess the impact of HIV-1 infection on cell cycle progression in ActD-treated MT-2 cells, HIV-infected MT-2 cells were also analyzed. HIV-1 infection had no effect on the decrease in the percentage of cells in S phase observed after ActD treatment of uninfected cells (data not shown).
FIG. 6.
ActD treatment decreases the percentage of MT-2 cells in S phase. MT-2 cells were cultured for 3 days in the absence (A) or presence (B) of 10 nM ActD. The treated cells were stained with propidium iodide. The stained cells were analyzed for red fluorescence. The left peaks constitute cells in G0/G1, the right peaks constitute cells in G2/M; cells in S phase are between the G0/G1 and G2/M peaks. The distribution of cells in the G0/G1, S, and G2/M cell cycle phases was calculated from the resulting DNA histogram by using Multicycle AV software (Phoenix Flow Systems). Results are representative of three independent experiments.
DISCUSSION
ActD has been reported to be a potential inhibitor of HIV-1 replication due to its inhibitory effect on minus-strand transfer during the process of reverse transcription (10, 15, 17, 26). Despite the strong prior data on inhibition of HIV-1 reverse transcription in vitro, the present study has demonstrated that low concentrations of ActD (1 to 10 nM) actually enhance replication of HIV-1 in MT-2 cells. Of note, this effect was limited to HIV-1. Similar effects were not seen with HIV-2MVP15132 or with HIV-1 replication in Jurkat, MT-4, or PBMCs.
In an effort to determine the mechanism(s) underlying this phenomenon, we examined the effect of ActD on HTLV-1 production, HIV-1 replication kinetics, receptor and coreceptor expression, transcription of cellular genes, and effects on cell cycle. As mentioned above, MT-2 and MT-4 cells are HTLV-1-transformed cell lines. HTLV-1-transformed cells produce infectious HTLV-1 virions (11, 31) and a number of cytokines (21). HTLV-1 has been reported to enhance HIV-1 replication in vitro (4, 9, 34, 36, 62). MT-4 cells produce low or undetectable levels of HTLV-1, while MT-2 cells are high-level producers of the virus (31). Since ActD enhanced HIV-1 replication in MT-2 cells but not in MT-4 cells, it was speculated that the mechanism of HIV-1 activation by ActD might be mediated via the activation of HTLV-1 production from ActD-treated MT-2 cells. However, this possibility seems unlikely, since we were unable to find evidence that ActD enhances HTLV-1 production
Our evaluation of HIV-1 replication kinetics revealed that replication was enhanced only after day 4 of infection (Fig. 1B). In addition, ActD treatment downregulated the expression of CD4 and CXCR4 on MT-2 cells by 30 and 80%, respectively (Fig. 2). Therefore, ActD does not appear to be acting by extending the time of productive infection or enhancing the infectibility of MT-2 cells. The data are more consistent with the influence of ActD on the magnitude of productive infection. Since ActD downregulated the expression of receptors for HIV-1 infection, it was speculated that ActD might result in less syncytium formation and prolonged survival of infected cells. However, as shown in Fig. 1D, ActD enhanced syncytium formation in MT-2 cells. As demonstrated in Fig. 3, ActD leads to an increase in HIV-1 transcription. Thus, the present data suggest that this effect of ActD on HIV-1 replication is most likely a result of alterations in virus gene transcription. Whether this is due to increased transcription or increased transcript stability is the focus of ongoing experiments.
ActD enhanced HIV-1NL4.3 and HIV-1DH12 replication by 8- to 10-fold in MT-2 cells but did not stimulate HIV-2 MVP15132 replication in these cells (Fig. 1C). HIV-1 and HIV-2 both require the trans-activator Tat protein for viral RNA transcription and viral replication in host cells (46). Since ActD enhanced HIV-1 replication without modifying HIV-2, it is unlikely that the effect on HIV-1 is mediated via Tat.
Semiquantitative RT-PCR and cell cycle analyses revealed that ActD treatment enhanced transcription of HIV-1 DNA by 16-fold (Fig. 3). Thus, this ActD effect in MT-2 cells appears to be related to an increase in RNA production, despite the well-described activity of ActD to inhibit transcription. Using a transient promoter function assay, it has been demonstrated that ActD activates the HIV-1 promoter in HeLa cells via an enhancement of phosphorylation of RNA polymerase II (Pol II) (6). Further studies of the mechanism of this effect have revealed that ActD leads to a dissociation of the complex between 7SK RNA (a small nuclear RNA) and P-TEFb (Tat-specific transcription elongation factor complex) (39, 61). Once dissociated, the inhibitory effects on this complex on the phosphorylation of Pol II are removed. Thus, it is possible that ActD leads to an increase in HIV-1 replication by a similar mechanism. However, given the fact that ActD enhanced HIV-1 replication only in MT-2 cells (Fig. 1A), an additional mechanism may be involved. The precise mechanism is still unclear and is now the focus of additional work.
An additional unexpected finding was the development of multidrug resistance in the presence of ActD (Table 1). We speculate that the broad low-level resistance was not truly resistance but was a reflection of the high level of viral replication, requiring a high concentration of drug. In contrast, 10 nM ActD treatment led to high-level resistance to AZT and d4T without induction of nucleotide substitutions associated with drug resistance. Western blotting and RT-PCR pointed to a downregulation of TK-1 as the likely mechanism for this phenomenon (Fig. 5). TK-1 is a key enzyme in the salvage synthesis of thymidine monophosphate (TMP) from thymidine. Intracellular TMP is quickly phosphorylated to dTDP (TDP) and dTTP (TTP). Since TTP is an allosteric effector of ribonucleotide reductase, imbalances in the TTP pool disturb the supply of both purines and pyrimidines for DNA synthesis and repair. Therefore, imbalanced dNTP pools increase the mutation rate (37, 40). Even though ActD suppressed the expression of TK1 in MT-2 cells, we have not seen any drug resistance mutations during short-term culture. However, it is possible that ActD might facilitate the rate of emergence of HIV-1 mutations associated with drug resistance during long-term passage in MT-2 cells.
TK-1 is a cell cycle-regulated enzyme. Its activity fluctuates with DNA synthesis, being highest in dividing and malignant cells and lowest in quiescent cells (50). The expression of TK-1 is meticulously controlled at the transcriptional and posttranscriptional level (28, 53). Its activity increases markedly after the G1/S transition and then declines rapidly in G2 (50). Replication of retroviruses depends on cycling cells. Passage through the G1/S phase of the cell cycle is essential for efficient reverse transcription (8, 13, 20, 22, 23, 56). ActD is known as a cell cycle synchronizer that arrests the G1 phase in the cell cycle (5, 29). However, in MT-2 cells, cell cycle analysis revealed that ActD decreased the number of cells in S phase (Fig. 6). Therefore, the described decrease in the TK-1 level in MT-2 cells following ActD treatment may be a consequence of having fewer cells in S phase.
Here we demonstrated that ActD treatment downregulated TK-1 expression in MT-2 cells. The initial phosphorylation of AZT and d4T by TK-1 is a critical step in their metabolism, leading to chain termination of DNA synthesis during the RT phase of HIV replication (14). Resistance to nucleoside analogs, e.g., AZT, has been well studied at the level of the virus. Amino acid substitutions (49), insertions (49), or deletions (24, 25, 30, 60) can lead to high-level resistance to AZT. In addition, as reported here, it appears that AZT resistance can also be seen following chemotherapeutic manipulation of the host cell by downregulation of TK-1 expression.
Acknowledgments
We thank Tatsuhiko Igarashi and Malcolm Martin for providing HIVDH12 and Quan-En Yang and Shizuko Sei for valuable assistance in taking photomicrographs. We also thank Robin Derwar for critical reading of the manuscript and Anthony S. Fauci for guidance and support.
This work was supported by NIAID under contract N01-CO-12400 with SAIC-Frederick, Inc.
REFERENCES
- 1.Adachi, A., H. E. Gendelman, S. Koenig, T. Folks, R. Willey, A. Rabson, and M. A. Martin. 1986. Production of acquired immunodeficiency syndrome-associated retrovirus in human and nonhuman cells transfected with an infectious molecular clone. J. Virol. 59:284-291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.AIDS Clinical Trials Group. 1997. Virology manual for HIV laboratories. Division of AIDS, National Institute of Allergy and Infectious Diseases, Bethesda, Md. [Online.] http://www.niaid.nih.gov./daids/vir_manual.
- 3.Beyl, W., K. Nehring, L. Gürtler, J. Eberle, and F. Deinhardt. 1987. AIDS Verursacht durch HIV-2. Münch. Med. Wochenschr. 129:895-896. [Google Scholar]
- 4.Boehnlein, E., M. Siekevitz, D. W. Ballard, J. W. Lowenthal, L. Rimsky, H. Bogerd, J. Hoffman, Y. Wano, B. R. Franza, and W. C. Greene. 1988. Stimulation of the HIV-1 enhancer by the HTLV-I tax gene product involves the action of inducible cellular proteins. J. Virol. 63:1578-1586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bonin, L. R., and J. K. McDougall. 1997. Human cytomegalovirus IE2 86-kilodalton protein binds p53 but does not abrogate G1 checkpoint function. J. Virol 71:5861-5870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cassé, C., F. Giannoni, V. T. Nguyen, M.-F. Dubois, and O. Bensaude. 1999. The transcriptional inhibitors, actinomycin D and α-amanitin activate the HIV-1 promoter and favor phosphorylation of the RNA polymerase II C-terminal domain. J. Biol. Chem. 274:16097-16106. [DOI] [PubMed] [Google Scholar]
- 7.Chen, F. M. 1988. Binding specificities of actinomycin D to self-complementary tetranucleotide sequences -XGCY-. Biochemistry 27:6393-6397. [DOI] [PubMed] [Google Scholar]
- 8.Chen, I. S. Y., and H. M. Temin. 1982. Establishment of infection by spleen necrosis virus: inhibition in stationary cells and the role of secondary infection. J. Virol. 41:183-191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cheng, H., J. Tarnok, and W. Parks. 1998. Human immunodeficiency virus type 1 genome activation induced by human T-cell leukemia virus type 1 Tax protein is through cooperation of NF-κB and Tat. J. Virol. 72:6911-6916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Davis, W. R., S. Gabbara, D. Hupe, and J. A. Peliska. 1998. Actinomycin D inhibition of DNA strand transfer reactions catalyzed by HIV-1 reverse transcriptase and nucleocapsid protein. Biochemistry 37:14213-14221. [DOI] [PubMed] [Google Scholar]
- 11.Dhib-Jalbut, S., P. M. Hoffman, T. Yamabe, D. Sun, J. Xia, H. Eisenberg, G. Berger, and F. W. Ruscetti. 1994. Extracellular human T-cell lymphotropic virus type I Tax protein induces cytokine production in adult human microglial cells. Ann. Neurol. 36:787-790. [DOI] [PubMed] [Google Scholar]
- 12.Farber, S. 1966. Chemotherapy in the treatment of leukemia and Wilms' tumor. JAMA 198:826-836. [PubMed] [Google Scholar]
- 13.Fritsch, E. F., and H. M. Temin. 1977. Inhibition of viral DNA synthesis in stationary chicken embryo fibroblasts infected with avian retroviruses. J. Virol. 24:461-469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Furman, P. A., J. A. Fyfe, M. H. Clair, Sr., K. Weinhold, J. L. Rideout, G. A. Freeman, S. N. Lehrman, D. P. Bolognesi, S. Broder, H. Mitsuya, and D. W. Barry. 1986. Phosphorylation of 3′-azido-3′-deoxythymidine and selective interaction of the 5′-triphosphate with human immunodeficiency virus reverse transcriptase. Proc. Natl. Acad. Sci. USA 83:8333-8337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gabbara, S., W. R. Davis, L. Hupe, D. Hupe, and J. A. Peliska. 1999. Inhibitors of DNA strand transfer reactions catalyzed by HIV-1 reverse transcriptase. Biochemistry 38:13070-13076. [DOI] [PubMed] [Google Scholar]
- 16.Goodisman, J., R. Rehfuss, B. Ward, and J. C. Dabrowiak. 1992. Site-specific binding constants for actinomycin D on DNA determined from footprinting studies. Biochemistry 31:1046-1058. [DOI] [PubMed] [Google Scholar]
- 17.Guo, J., T. Wu, J. Bess, L. E. Henderson, and J. G. Levin. 1998. Actinomycin D inhibits human immunodeficiency virus type 1 minus-strand transfer in in vitro and endogenous reverse transcriptase assays. J. Virol. 72:6716-6724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Haertle, T., C. J. Carrera, D. B. Wasson, L. C. Sowers, D. D. Richman, and D. A. Carson. 1988. Metabolism and anti-human immunodeficiency virus-1 activity of 2-halo-2′,3′-dideoxyadenosine derivatives. J. Biol. Chem. 263:5870-5875. [PubMed] [Google Scholar]
- 19.Harada, S., Y. Koyanagi, and N. Yamamoto. 1985. Infection of HTLV-III/LAV in HTLV-carrying cells MT-2 and MT-4 and application in a plaque assay. Science 229:563-566. [DOI] [PubMed] [Google Scholar]
- 20.Harel, J., E. Rassart, and P. Jolicoeur. 1981. Cell cycle dependence of synthesis of unintegrated viral DNA in mouse cells newly infected with murine leukemia virus. Virology 110:202-207. [DOI] [PubMed] [Google Scholar]
- 21.Hollsberg, P., and D. A. Hafler. 1993. Pathogenesis of diseases induced by human lymphotropic virus type I infection. N. Engl. J. Med. 328:1173-1182. [DOI] [PubMed] [Google Scholar]
- 22.Hsu, T. W., and J. M. Taylor. 1982. Effect of aphidicolin on avian sarcoma virus replication. J. Virol. 44:493-498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Humphries, E. H., and H. M. Temin. 1974. Requirement for cell division for initiation of transcription of Rous sarcoma virus RNA. J. Virol. 14:531-546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Imamichi, T., S. C. Berg, H. Imamichi, J. C. Lopez, J. A. Metcalf, J. Fallon, and H. C. Lane. 2000. Relative replication fitness of a high-level 3′-azido-3′-deoxythimidine-resistant variant of human immunodeficiency virus type-1 possessing an amino acid deletion at codon 67 and a novel substitution (Thr to Gly) at codon 69. J. Virol. 74:10958-10964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Imamichi, T., T. Sinha, H. Imamichi, Y. M. Zhang, J. A. Metcalf, J. Falloon, and H. C. Lane. 2000. High-level resistance to 3′-azido-3′-deoxythimidine due to a deletion in the reverse transcriptase gene of human immunodeficiency virus type 1. J. Virol. 74:1023-1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jeeninga, R. E., H. T. Huthoff, A. P. Gultyaev, and B. Berkhout. 1998. The mechanism of actinomycin D-mediated inhibition of HIV-1 reverse transcription. Nucleic Acids Res. 26:5472-5479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kamitori, S., and F. Takusagawa. 1992. Crystal structure of the 2:1 complex between d(GAAGCTTC) and the anticancer drug actinomycin D. J. Mol. Biol. 225:445-456. [DOI] [PubMed] [Google Scholar]
- 28.Kauffman, M. G., and T. J. Kelly. 1991. Cell cycle regulation of thymidine kinase: residues near the carboxyl terminus are essential for the specific degradation of the enzyme at mitosis. Mol. Cell. Biol. 11:2538-2546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Khan, Q. A., and A. Dipple. 2000. Diverse chemical carcinogens fail to induce G1 arrest in MCF-7 cells. Carcinogenesis 21:1611-1618. [PubMed] [Google Scholar]
- 30.Kim, E.-Y., M. A. Winters, R. M. Kagan, and T. C. Merigan. 2001. Functional correlates of insertion mutations in the protease gene of human immunodeficiency virus type 1 isolates from patients. J. Virol. 75:11227-11233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Koyanagi, Y., Y. Hinuma, J. Schneider, T. Chosa, G. Hunsmann, N. Kobayashi, M. Hatanaka, and N. Yamamoto. 1984. Expression of HTLV-specific polypeptides in various human T-cell lines. Med. Microbiol. Immunol. 173:127-140. [DOI] [PubMed] [Google Scholar]
- 32.Larder, B. A., G. Darby, and D. D. Richman. 1989. HIV with reduced sensitivity to zidovudine (AZT) isolated during prolonged therapy. Science 243:1731-1734. [DOI] [PubMed] [Google Scholar]
- 33.Lewis, J. L., Jr. 1972. Chemotherapy of gestational choriocarcinoma. Cancer 30:1517-1521. [DOI] [PubMed] [Google Scholar]
- 34.Lindholm, P. F., S. J. Marriott, S. D. Gitlin, C. A. Bohan, and J. N. Brady. 1990. Induction of nuclear NF-κB DNA binding activity after exposure of lymphoid cells to soluble Tax1 protein. New Biol. 2:1034-1043. [PubMed] [Google Scholar]
- 35.Mann, D. L., S. J. O'Brien, D. A. Gilbert, Y. Reid, M. Popovic, E. Read-Connole, R. C. Gallo, and A. Gazdar. 1989. Origin of the HIV-susceptible human CD4+ cell line H9. AIDS Res. Hum. Retrovir. 5:253-255. [DOI] [PubMed] [Google Scholar]
- 36.Marriott, S. J., P. F. Lindholm, R. L. Reid, and J. N. Brady. 1991. Soluble HTLV-I Tax1 protein stimulates proliferation of human peripheral blood lymphocytes. New Biol. 3:678-686. [PubMed] [Google Scholar]
- 37.Meuth, M. 1989. The molecular basis of mutations induced by deoxyribonucleoside triphosphate pool imbalances in mammalian cells. Exp. Cell. Res. 181:305-316. [DOI] [PubMed] [Google Scholar]
- 38.Myers, G., B. Korber, S. Wain-Hobson, K.-T. Jeang, L. E. Henderson, and G. N. Pavlakis. 1994. Human retroviruses and AIDS. Los Alamos National Laboratory, Los Alamos, N.Mex.
- 39.Nguyen, V. T., T. Kiss, A. A. Michels, and O. Bensaude. 2001. 7SK small nuclear RNA binds to and inhibits the activity of CDK9/cyclin T complexes. Nature 414:322-325. [DOI] [PubMed] [Google Scholar]
- 40.Oliver, F. J., M. K. Collins, and A. Lopez-Rivas. 1997. Overexpression of a heterologous thymidine kinase delays apoptosis induced by factor deprivation and inhibitors of deoxynucleotide metabolism. J. Biol. Chem. 272:10624-10630. [DOI] [PubMed] [Google Scholar]
- 41.Pauwels, R., E. De Clercq, J. Desmyter, J. Balzarini, P. Goubau, P. Herdewijn, H. Vanderhaeghe, and M. Vandeputte. 1987. Sensitive and rapid assay on MT-4 cells for detection of antiviral compounds against the AIDS virus. J. Virol. Methods 16:171-185. [DOI] [PubMed] [Google Scholar]
- 42.Perrin, L., and A. Telenti. 1998. HIV treatment failure: testing for HIV resistance in clinical practice. Science 280:1871-1873. [DOI] [PubMed] [Google Scholar]
- 43.Perry, R. P., and D. E. Kelley. 1970. Inhibition of RNA synthesis by actinomycin D: characteristic dose-response of different RNA species. J. Cell. Physiol. 76:127-139. [DOI] [PubMed] [Google Scholar]
- 44.Popovic, M., E. Read-Connole, and R. C. Gallo. 1984. T4 positive human neoplastic cell lines susceptible to and permissive for HTLV-III. Lancet ii:1472-1473. [DOI] [PubMed]
- 45.Popovic, M., M. G. Sarngadharan, E. Read, and R. C. Gallo. 1984. Detection, isolation, and continuous production of cytopathic retroviruses (HTLV-III) from patients with AIDS and pre-AIDS. Science 224:497-500. [DOI] [PubMed] [Google Scholar]
- 46.Rabson, A. B., and B. J. Graves. 1997. Synthesis and processing of viral RNA, p. 205-261. In J. M. Coffin, S. H. Hughes, and H. E. Varmus (ed.), Retroviruses. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y. [PubMed]
- 47.Rill, R. L., and K. H. Hecker. 1996. Sequence-specific actinomycin D binding to single-stranded DNA inhibits HIV reverse transcriptase and other polymerases. Biochemistry 35:3525-3533. [DOI] [PubMed] [Google Scholar]
- 48.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed., p. 18.38. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 49.Schinazi, R. F., B. A. Larder, and J. W. Mellors. 2000. Mutations in retroviral genes associated with drug resistance. Int. Antivir. News 8:65-91. [Google Scholar]
- 50.Sherley, J. L., and J. K. Thomas. 1988. Regulation of human thymidine kinase during the cell cycle. J. Biol. Chem. 263:8350-8358. [PubMed] [Google Scholar]
- 51.Shibata, R., M. D. Hoggan, C. Broscius, G. Englund, T. S. Theodore, A. Buckler-White, L. O. Arthur, Z. Israel, A. Schultz, H. C. Lane, and M. A. Martin. 1995. Isolation and characterization of a syncytium-inducing, macrophage/T-cell line-tropic human immunodeficiency virus type 1 isolate that readily infects chimpanzee cells in vitro and in vivo. J. Virol. 69:4453-4462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sobell, H. M. 1985. Actinomycin and DNA transcription. Proc. Natl. Acad. Sci. USA 82:5328-5331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Stewart, C. J., M. Ito, and S. E. Conrad. 1987. Evidence for transcriptional and post-transcriptional control of the cellular thymidine kinase gene. Mol. Cell. Biol. 7:1156-1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Takusagawa, F., K. T. Takusagawa, R. G. Carlson, and R. F. Weaver. 1997. Selectivity of F8-actinomycin D for RNA:DNA hybrids and its anti-leukemia activity. Bioorg. Med. Chem. 5:1197-1207. [DOI] [PubMed] [Google Scholar]
- 55.Telesnitsky, A., and S. P. Goff. 1997. Reverse transcriptase and the generation of retroviral DNA, p. 121-160. In J. M. Coffin, S. H. Hughes, and H. E. Varmus (ed.), Retroviruses. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y. [PubMed]
- 56.Varmus, H. E., T. Padgett, S. Heasley, G. Simon, and J. M. Bishop. 1977. Cellular functions are required for synthesis and integration of avian sarcoma virus-specific DNA. Cell 11:307-319. [DOI] [PubMed] [Google Scholar]
- 57.Wadkins, R. M., and T. M. Jovin. 1991. Actinomycin D and 7-aminoactinomycin D binding to single-stranded DNA. Biochemistry 30:9469-9478. [DOI] [PubMed] [Google Scholar]
- 58.Wadkins, R. M., E. A. Jares-Erijman, R. Klement, A. Rudiger, and T. M. Jovin. 1996. Actinomycin D binding to single-stranded DNA: sequence specificity and hemi-intercalation model from fluorescence and 1H NMR spectroscopy. J. Mol. Biol. 262:53-68. [DOI] [PubMed] [Google Scholar]
- 59.Waring, M. J. 1981. DNA modification and cancer. Annu. Rev. Biochem. 50:159-192. [DOI] [PubMed] [Google Scholar]
- 60.Winters, M. A., J. M. Schapiro, J. Lawrence, and T. C. Merigan. 1998. Human immunodeficiency virus type 1 protease genotypes and in vitro protease inhibitor susceptibilities of isolates from individuals who were switched to other protease inhibitors after long-term saquinavir treatment. J. Virol. 72:5303-5306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yang, Z., Q. Zhu, K. Luo, and Q. Zhou. 2001. The 7SK small nuclear RNA inhibits the CDK9/cyclin T1 kinase to control transcription. Nature 414:317-322. [DOI] [PubMed] [Google Scholar]
- 62.Zack, J. A., A. J. Cann, J. P. Lugo, and I. S. Y. Chen. 1988. HIV-1 production from infected peripheral blood T cells after HTLV-I induced mitogenic stimulation. Science 240:1026-1029. [DOI] [PubMed] [Google Scholar]
- 63.Zhang, Y.-M., H. Imamichi, T. Imamichi, H. C. Lane, J. Falloon, M. B. Vasudevachari, and N. P. Salzman. 1997. Drug resistance during indinavir therapy is caused by mutations in the protease gene and in its Gag substrate cleavage sites. J. Virol. 71:6662-6670. [DOI] [PMC free article] [PubMed] [Google Scholar]