Abstract
In eukaryotic cell nuclei, DNA associates with the core histones H2A, H2B, H3, and H4 to form nuclosomal core particles. DNA binding to histones is regulated by posttranslational modifications of N-terminal tails, e.g., acetylation and methylation of histones. These modifications play important roles in the epigenetic control of chromatin structure. Recently, evidence has been provided that biotinidase and holocarboxylase synthetase catalyze the covalent binding of biotin to histones. Primary aim of this study was to identify biotinylation sites in histone H2A and its variant H2AX. Secondary aims were to determine whether acetylation and methylation of histone H2A affect subsequent biotinylation, and to determine whether biotinidase and holocarboxylase synthetase localize to the nucleus in human cells. Biotinylation sites were identified using synthetic peptides as substrates for biotinidase. These studies provided evidence that K9 and K13 in the N-terminus of human histone H2A and H2AX are targets for biotinylation, and that K125, K127, and K129 in the C-terminus of histone H2A are targets for biotinylation. Biotinylation of lysine residues was decreased by acetylation of adjacent lysines, but was increased by dimethylation of adjacent arginines. The existence of biotinylated histone H2A in vivo was confirmed by using modification-specific antibodies. Antibodies to biotinidase and holocarboxylase synthetase localized primarily to the nuclear compartment, consistent with a role for these enzymes in regulating chromatin structure. Collectively, these studies have identified five novel biotinylation sites in human histones; histone H2A is unique among histones in that biotinylation sites include amino acid residues from the C-terminus.
Keywords: Biocytin, biotin, biotinidase, histone H2A, holocarboxylase synthetase, lysine
Abbreviations: DAPI, 4′, 6-diamidino-2-phenylindole; Fmoc, N-fluoren-9-ylmethoxycarbonyl; HCS, holocarboxylase synthetase; MALDI, matrix assisted laser desorption ionization
1. Introduction
Histones are quantitatively and qualitatively important DNA-binding proteins in eukaryotic cells. The following five major classes of histones have been identified: H1, H2A, H2B, H3, and H4 [1]. Histones carry a positive net charge due to the great abundance of arginine and lysine residues in these proteins [1]. The binding of negatively charged DNA to positively charged histones is mediated by electrostatic interactions. In chromatin, stretches of 146 base pairs of DNA are wrapped around octamers of core histones (one H3-H3-H4-H4 tetramer and two H2A-H2B dimers) to form nucleosomal core particles. Linker histone H1 associates with the DNA connecting two core particles to complete the nucleosomal assembly [1].
N-terminal tails of histones are exposed at the nucleosomal surface [1]. Lysine (K), arginine (R), serine (S), threonine (T), and glutamate residues in these tails are targets for posttranslational modifications such as acetylation, methylation, ubiquitination, phosphorylation, sumoylation, and poly(ADP-ribosylation) [2]. These covalent modifications play important roles in the epigenetic control of chromatin structure, genomic stability, and gene expression [3–5]. Some regions in C-terminal domains (e.g., hinge regions) are also exposed at the nucleosomal surface, and are potential targets for covalent modifications [1]. For example, K120 in histone H2B is a target for ubiquitination [6], and K108, K116, K120, and K125 in histone H2B are targets for acetylation [7]. Histone H2A is unique among core histones in having its C-terminal tail exposed at the nucleosomal surface [1,8]. Consistent with this observation, the following modifications have been identified in the C-terminus of histone H2A and its variant H2AX: ubiquitination of K119 [6,9] and phosphorylation of S139 [10,11], respectively.
Recently, a novel posttranslational modification of histones has been identified: biotinylation of lysine residues [12]. The covalent binding of biotin to histones is mediated by biotinidase and holocarboxylase synthetase (HCS) [13,14]. Biotinidase uses biocytin (biotinyl-ɛ-lysine) as a substrate for biotinylation of histones [13], whereas HCS uses biotin and ATP [14]. The following biotinylation sites have been identified in human histones: K4, K9, and K18 in histone H3 [15], and K8 and K12 in histone H4 [16]. Biotinylation of histones plays a role in the regulation of gene expression [17], cell proliferation [12,14], and the cellular response to DNA damage [17,18].
Previous studies suggested that histone H2A contains streptavidin-reactive material [12], consistent with biotinylation of this protein. Here we tested the hypothesis that specific lysine residues in histone H2A and its variant H2AX are targets for biotinylation by biotinidase. Histones H2A and H2AX were chosen as models for the following reasons. First, both histones H2A and H2AX contain biotinylation motifs in their N- and C-terminal domains (A. Kueh and J. Zempleni, unpublished observations). Second, the N- and C-terminal regions of histone H2A have important functions in telomeric silencing in yeast [19]. Third, phosphorylation of histone H2AX plays a role in the cellular response to DNA damage [11]. Fourth, various posttranslational modifications are known to occur in histone H2A, e.g., phosphorylation of S1 [20], acetylation of K5, K9 [21] and K13 [7], ubiquitination of K119 [6,9], phosphorylation T120 [22], and methylation of K125 or K127 [7]. Likely, these modifications affect subsequent biotinylation [16]. Collectively, identification of biotinylation sites in histones H2A and H2AX is likely to produce valuable insights into roles of these histones in chromatin structure and genomic stability.
This study addressed the following specific aims. First, we sought to identify the amino acid residues in histones H2A and H2AX that are targets for biotinylation by biotinidase. Second, we sought to determine whether acetylation, methylation, and phosphorylation of histone H2A affect its subsequent biotinylation by biotinidase. Third, we sought to determine whether biotinidase and HCS localize to the nucleus in human cells, consistent with a role of these enzymes in histone biotinylation in vivo.
2. Methods and Materials
2.1. Identification of biotinylation sites
In previous studies, we developed a procedure to identify amino acid residues in histones that are targets for biotinylation [23]. Briefly, this procedure is based on the following analytical sequence: (i) short peptides (<20 amino acids in length) are synthesized chemically; amino acid sequences in these peptides are based on the sequence in a given region of a histone; (ii) peptides are incubated with biotinidase or HCS to conduct enzymatic biotinylation; and (iii) peptides are resolved by gel electrophoresis, and peptide-bound biotin is probed using streptavidin peroxidase. Amino acid substitutions (e.g., lysine-to-alanine substitutions) in synthetic peptides are used to corroborate identification of biotinylation sites. In addition, amino acid modifications (e.g., acetylation of lysines) in peptides can be used to investigate the cross-talk between biotinylation of histones and other known modifications of histones.
Here, peptides were synthesized based on the amino acid sequences in human H2A.1 (GenBank accession number M60752) and H2AX (GenBank accession number P16104). Peptides were synthesized using N-fluoren-9-ylmethoxycarbonyl (Fmoc)-activated L-isomers of amino acids [16]. One-letter annotation is used for denoting amino acids in this paper [24]. Chemically modified peptides were synthesized by using biotinylated, acetylated, and dimethylated ɛ -NH2-derivatives of Fmoc-lysine, dimethylated guanidino derivatives of Fmoc-arginine, and phosphorylated derivatives of Fmoc-serine. Peptides were quantified as described [16]. Identities of peptides were confirmed by matrix assisted laser desorption ionization (MALDI)-time of flight and by quadrupole-time of flight mass spectrometry in the Nebraska Center of Mass Spectrometry (University of Nebraska-Lincoln). Amino acid sequences of synthetic peptides are provided in the Results section.
Peptides from both the N- and C-terminal regions of histone H2A and H2AX were included in the analysis of biotinylation sites. Synthetic peptides were biotinylated enzymatically as described previously [16] with the following modifications. Five micrograms of a given peptide were dissolved in 100 μL of a mixture containing 15 μL of human plasma (as a source of biotinidase), 10 μL of biocytin solution (75 μmol/L final concentration, as a source of biotin), and 75 μL of Tris buffer (50 mmol/L final concentration, pH 8.0). Samples were incubated at 37°C for up to 45 minutes. Reactions were quenched by adding an equal volume of Tricine gel loading buffer (Invitrogen, Carlsbad, CA). Peptides were resolved by gel electrophoresis and peptide-bound biotin was probed by using streptavidin peroxidase [16].
2.2. Polyclonal antibodies
The following biotinylation sites were identified in histone H2A in the experiments described below: K9 and K13 in the N-terminal region, and K125, K127, and K129 in the C-terminal region. Here, we generated antibodies against K9-biotinylated histone H2A and K13-biotinylated histone H2A. In addition, we generated antibodies against the two human enzymes that mediate biotinylation of histones: biotinidase and HCS. Polyclonal antibodies were produced using a commercial facility (Cocalico Biologicals, Reamstown, PA). The following peptides were synthesized by AnaSpec, Inc. (San Jose, CA) and the University of Virginia Biomolecular Research Facility (Charlottesville, VA), respectively, for injection into rabbits: (i) N1–12bioK9 = SGRGKQGGK(biotin)ARAC (amino acids 1 – 12 in histone H2A plus a cysteine); (ii) N10–24bioK13 = ARAK(biotin)AKTRSSRAGLQC (amino acids 10–25 in histone H2A plus a cysteine); (iii) biotinidase (GenBank accession number NM_000060) = CLRKSRLSSGLVTAALYGRLYERD (amino acids 520 – 542 in biotinidase plus one cysteine); and (iv) HCS (GenBank accession number NM_000411) = EHVGRDDPKALGEEPKQRRGC (amino acids 58 – 77 in HCS plus one cysteine). Identities and purities of these peptides were confirmed by using high-performance liquid chromatography (HPLC) and MALDI (data not shown). Peptides were conjugated to keyhole limpet hemocyanin before injection into White New Zealand rabbits [16]. Rabbit serum was collected before (pre-immune serum) and after three injections with peptides mixed with Freund’s adjuvant over a period of 49 days. Immunoglobulin G was purified from serum by using the ImmunoPure (A) IgG Purification Kit (Pierce, Rockford, IL) according to the manufacturer’s protocol. Antibody specificities were investigated by using synthetic peptides and histone extracts from human cells as described [16].
2.3. Cell culture
Human-derived Jurkat lymphoma cells and JAr choriocarcinoma cells (ATCC, Manassas, VA) were cultured as described [25,26]. Acid extracts from Jurkat cell nuclei [12] were used for western blot analysis of biotinylated histone H2A [16], whereas JAr cells were used for analysis of biotinylated histone H2A by immunocytochemistry.
2.4. Immunocytochemistry
K9-biotinylated histone H2A, K13-biotinylated histone H2A, biotinidase, and HCS were visualized in JAr cells by using immunocytochemistry as described [27]. Primary antibodies were as described above. As a secondary antibody we used donkey anti-rabbit Cy2-labeled antibody (Jackson ImmunoResearch, West Grove, PA). Cytoplasmic and nuclear compartments were stained with rhodamine phalloidin and 4′, 6-diamidino-2-phenylindole (DAPI) (Sigma, St. Louis, MO) as described [27]. Images were obtained by using an Olympus FV500 confocal microscope (Microscopy Core Facility, University of Nebraska-Lincoln).
3. Results
3.1. Biotinylation sites in histones H2A and H2AX
Both the N- and C-termini of histone H2A contain targets for biotinylation by biotinidase. We synthesized the following five peptides based on the N- and C-termini of histone H2A: N1–9 = amino acid sequence SGRGKQGGK, N7–14 = GGKARAKA, N12–20 = AKAKTRSSR, C113–121 = AVLLPKKTE, and C122–129 = SHHKAKGK; subscript numbers denote the position of amino acid residues in histone H2A. These peptides were subjected to enzymatic biotinylation, and peptide-bound biotin was probed using gel electrophoresis and streptavidin peroxidase. Both N1–9 and N7–14 were good targets for biotinylation by biotinidase but N12–20 was not a good target (Fig. 1, lanes 1 – 3). Moreover, peptide C113–121 was not good target, but the C-terminal C122–129 was a good target for biotinylation (lanes 4 and 5). Previous studies are consistent with the hypothesis that lysine residues in these peptides are the most likely targets for biotinylation [16].
Fig. 1.
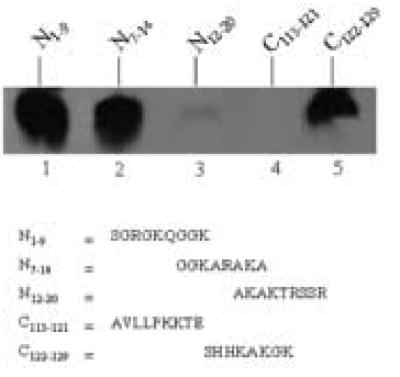
Both N- and C-terminal regions of histone H2A are targets for biotinylation by biotinidase. Synthetic peptides based on histone H2A were incubated with biotinidase and biocytin for enzymatic biotinylation. Peptides were resolved by gel electrophoresis and peptide-bound biotin was probed using streptavidin peroxidase.
We verified that peptide biotinylation approached maximal levels under the conditions described in Methods and Materials. First, the time course of biotinylation of peptide N1–9 was monitored at timed intervals for up to 45 minutes; concentrations of peptide, biocytin, and biotinidase were kept constant as described above. Biotinylation of peptide N1–9 was detectable 15 minutes after starting the incubation with biotinidase and reached maximal levels after 45 minutes (data not shown), consistent with previous studies [15]. Second, we tested effects of substrate (biocytin) availability. Peptide N1–9 was incubated with biotinidase at various concentrations of biocytin (7.5, 37.5, 75, 112.5, and 150 μmol/L) for 45 minutes. Biotinylation of N1–9 reached a plateau at 75 μmol/L of biocytin (data not shown), consistent with previous studies [13]. Finally, we varied the concentration of peptide N1–9 in the biotinylation reaction. The biotinylation signal paralleled the amount of N1–9 added to incubation mixtures (data not shown).
Next, we identified biotinylation targets in the N-terminus of histone H2A. A first series of experiments suggested that K9 is a biotinylation target, based on the following lines of evidence. Peptide N1–9 (containing both K5 and K9) was heavily biotinylated in response to incubation with biotinidase (Fig. 2, lane 1). If K9 was substituted with alanine (peptide K9A1–9) no biotinylation was detectable (lane 2). In contrast, substitution of K5 with alanine residues (peptide K5A1–9) did not decrease biotinylation (lane 3). If both lysine residues in peptide N1–9 were substituted with alanines (peptide K5,9A1–9) no biotinylation was detectable (lane 4). Biotinylation of K9 by biotinidase was further corroborated using the following control. Peptide N7–14 contains both K9 and K13 from histone H2A, and was heavily biotinylated in response to incubation with biocytin and biotinidase (data not shown). If K13 in N7–14 was substituted with alanine, the biotinylation signal decreased only moderately; in contrast, if K9 was substituted with an alanine the biotinylation signal decreased substantially (data not shown).
Fig. 2.
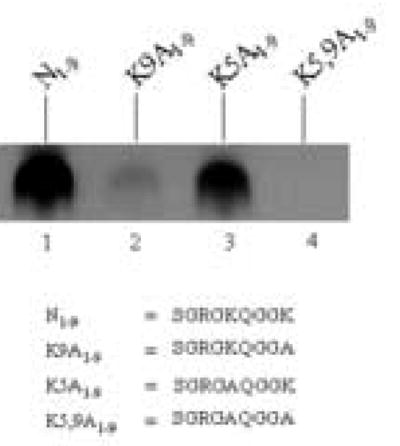
K9 in histone H2A is a good target for biotinylation by biotinidase. Synthetic peptides based on histone H2A were incubated with biotinidase and biocytin for enzymatic biotinylation. Peptides were resolved by gel electrophoresis and peptide-bound biotin was probed using streptavidin peroxidase.
A second series of experiments suggested that K13 in the N-terminus of histone H2A becomes a target for biotinylation if the neighboring K15 is modified. This notion is based on the following lines of evidence. Peptide N12–20 contains both K13 and K15 and was a poor target for biotinylation by biotinidase (Fig. 3, lane 1). However, if K15 was substituted with an alanine (peptide K15A12–20), K13 became a good target for biotinylation (lane 2). Substitution of K13 with an alanine (peptide K13A12–20) did not render K15 a good target for biotinylation (lane 3). If both lysine residues in peptide N12–20 were substituted with alanine residues (peptide K13,15A12–20) no biotinylation was detectable (lane 4). Note that the lysine-to-alanine substitutions used here are an artificial system that does not necessarily represent histones from human cells. However, findings described below suggest that naturally occurring variations in amino acid sequences (see histone variant H2AX) and posttranslational modifications of amino acids (see cross-talk among histone modifications) render K13 a good target for biotinylation.
Fig. 3.
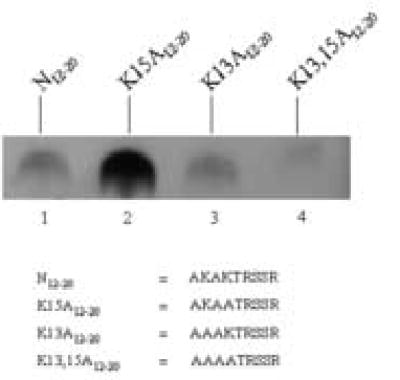
Substitution of K15 in histone H2A with alanine renders K13 a good target for biotinylation by biotinidase. Synthetic peptides based on histone H2A were incubated with biotinidase and biocytin for enzymatic biotinylation. Peptides were resolved by gel electrophoresis and peptide-bound biotin was probed using streptavidin peroxidase.
The N-terminal tail of human histone H2AX differs from the tail in histone H2A in two positions [19]: glutamine in position 6 is substituted with threonine, and threonine in position 16 is substituted with serine in histone H2AX. First, we synthesized the following two peptides based on the N-terminus of histone H2AX: Q6T1–9 = amino acid sequence SGRGKTGGK, and T16S12–20 = AKAKSRSSR. Both peptides were good targets for biotinylation by biotinidase (Fig. 4, lanes 1 and 3). Peptide N7–14 represents a moderate target for biotinylation (see above) and was used as a control (lanes 2 and 5). In fact, the N-terminus of histone H2AX was a better target for biotinylation than the N-terminus of histone H2A (compare lanes 1 – 3 with lanes 4 – 6). Specifically, a peptide containing both K9 and K13 (Q6T1–9) was a good target for biotinylation (lane 1), whereas a peptide containing K13 and K15 (T16S12–20) was biotinylated only moderately in response to incubation with biotinidase (lane 3). Peptide K5,9A1–9 does not contain any lysine residues and was used as a negative control (lane 7); no biotinylation was detectable after incubation with biotinidase.
Fig. 4.
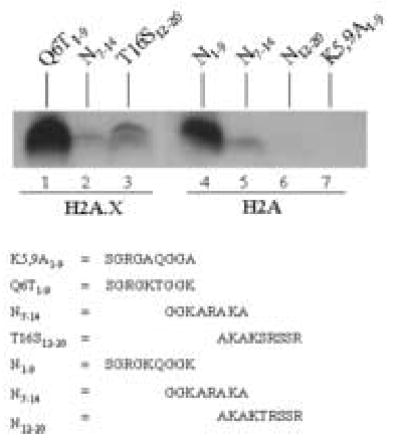
The N-terminus of H2AX is a good substrate for biotinylation by biotinidase. Synthetic peptides based on histone H2AX were incubated with biotinidase and biocytin for enzymatic biotinylation. Peptides were resolved by gel electrophoresis and peptide-bound biotin was probed using streptavidin peroxidase.
In a next series of experiments we confirmed that K9 and K13 in variant H2AX are specifically targeted by biotinylation in analogy to the findings described for histone H2A. Overall, we observed the same trends for peptides based on histone H2AX compared with histone H2A. Peptide Q6T1–9 contains both K9 and K13 from histone H2AX and was biotinylated in response to incubation with biotinidase (Fig. 5, lane 1). Substitution of K9 with alanine (peptide Q6T,K9A1–9) substantially decreased biotinylation (lane 2), whereas substitution of K5 with alanine (peptide Q6T,K5A1–9) decreased biotinylation only moderately (lane 3). Peptide T16S12–20 contains both K13 and K15 and was biotinylated in response to incubation with biotinidase (lane 4). If K15 was substituted with alanine (peptide K15A,T16S12–20) biotinylation decreased moderately (lane 5). No biotinylation was detectable if K13 was substituted with alanine (peptide K13A,T16S12–20; lane 6). If both lysine residues in peptide T16S12–20 were substituted with alanine (peptide K13,15A,T16S12–20) no biotinylation was detectable (lane 7).
Fig. 5.
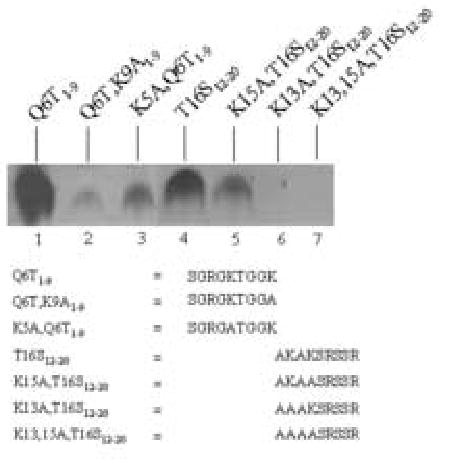
K9 and K13 in histone H2AX are targets for biotinylation by biotinidase. Synthetic peptides based on histone H2A and H2AX were incubated with biotinidase and biocytin for enzymatic biotinylation. Peptides were resolved by gel electrophoresis and peptide-bound biotin was probed using streptavidin peroxidase.
Lysines in the C-terminus of histone H2A were targeted for biotinylation by biotinidase. The C-terminus of histone H2A contains three lysine residues in positions 125, 127, and 129. A synthetic peptide including all three of these lysines (C122–129) was a good substrate for biotinylation by biotinidase (Fig. 6, lane 1). Biotinylation decreased only moderately, if K125 and K127 were substituted with alanine residues (peptide K125,127A122–129; lane 2), suggesting that K129 is a good target for biotinylation. Consistent with this hypothesis, substitution of K125 and K129 (peptide K125,129A122–129), and K127 and K129 (peptide K127,129A122–129) with alanine residues caused a considerable decrease of biotinylation (lanes 3 and 4, respectively). If all lysine residues in peptide C122–129 were substituted with alanine residues (peptide K125,127,129A122–129) no biotinylation was detectable (lane 5).
Fig. 6.

K125, K127, and K127 in the C-terminus of histone H2A are targets for biotinylation by biotinidase. Synthetic peptides based on histone H2A were incubated with biotinidase and biocytin for enzymatic biotinylation. Peptides were resolved by gel electrophoresis and peptide- bound biotin was probed using streptavidin–peroxidase.
The C-terminus of histone H2AX was not a good target for biotinylation. Note, that N-terminal sequences are highly conserved between histones H2A and H2AX, but that the C-terminal sequences of these two histones are unique [19]. Here, we synthesized the following three peptides based on the C-terminus of histone H2AX: C113–121 = AVLLPKKTS, C122–131 = ATVGPKAPSG, and C132–142 = GKKATQASQEY. Theses peptides were not biotinylated in response to incubation with biotinidase (data not shown).
3.2. Cross-talk among histone modifications
Previous studies suggested that acetylation and methylation of histone H4 affect subsequent biotinylation [16]. Of note, K5, K9, K13, and other lysine residues in human histone H2A are targets for acetylation [7]. Here, we provide evidence that acetylation and methylation of histone H2A are likely to affect subsequent biotinylation. Peptide N1–9 (containing both K5 and K9) was heavily biotinylated in response to incubation with biotinidase, and was used as a positive control (Fig. 7, lane 1). Acetylation of K5 caused a moderate decrease in the decreased the biotinylation of K9 (lane 2). A peptide containing acetylated K9 and free K5 was not a target for biotinylation (lane 3). This is consistent with our observation that K9 but not K5 is a target for biotinylation (see above). Moreover, dimethylation of R3 did not cause a change in the biotinylation signal (data not shown), because the adjacent K5 is not a biotinylation target. Peptide N7–14 (containing both K9 and K13) was a good target for biotinylation (lane 4). Again, acetylation of K9 decreased the biotinylation signal substantially (data not shown). Moreover, both dimethylation and acetylation of K13 decreased the biotinylation of K9 (lanes 5 and 6). On the other hand, dimethylation R11 considerably increased the enzymatic biotinylation of K9 or K13 (or both) by biotinidase (data not shown). Peptide N12–20 (containing both K13 and K15) was a poor target for biotinylation, but dimethylation of R17 substantially increased the biotinylation of K13 or K15 (or both) (data not shown). Increased biotinylation of lysine residues in response to biotinylation of adjacent arginine residues is consistent with previous studies [16].
Fig. 7.
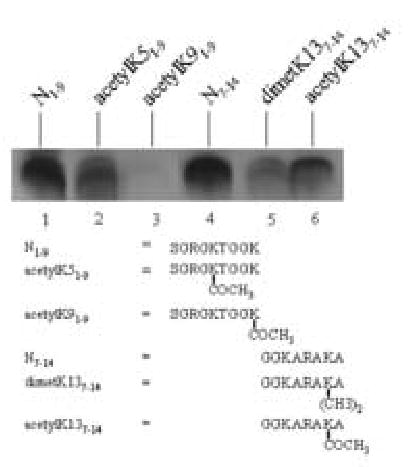
Methylation and acetylation of amino acids in the N-terminus of histone H2A affect the subsequent biotinylation of adjacent lysine residues by biotinidase. Synthetic peptides based on histone H2A were incubated with biotinidase and biocytin for enzymatic biotinylation. Peptides were resolved by gel electrophoresis and peptide-bound biotin was probed using streptavidin peroxidase.
3.3 Biotinylation of histone H2A in human cells
Human cells contain biotinylated histone H2A, as judged by using novel biotinylation site-specific antibodies. In a first series of experiments we raised antibodies to K9-biotinylated and K13-biotinylated histone H2A, and validated the specificity of these antibodies by using synthetic peptides. We ran the same two peptides that were used for injections into rabbits (N1–12bioK9 and N10–25bioK13) on a polyacrylamide gel, and probed the biotin tag by using streptavidin peroxidase. The two peptides produced a similar signal (Fig. 8A, compare lane 1 and 2), suggesting that biotinylation of peptides and loading of peptides on gels was similar. Second, the two peptides and a non-biotinylated control (N1–20) were probed with antibodies to K9- and K13-biotinylated histone H2A. Anti-K9bio antibody did not bind to peptide N1–20 (lanes 3) but cross-reacted with both biotinylated peptides: N10–25bioK9and N10–25bioK13 (lanes 4 and 5); pre-immune serum did not produce a detectable signal (data not shown). In contrast, anti-K13bio antibody did not bind to N1–20 and peptide N10–25bioK9 (lanes 6 and 7), but was specific for peptide N10–25bioK13 containing biotinylated K13 (lane 8); pre-immune serum did not produce a detectable signal (data not shown). Moreover, antibodies to biotinylated histone H2A did not cross-reacted with biotinylated peptides based on histone H4: N6–15bioK8 = GGK(biotin)GLGKGGA and N6–15bioK12 = GGKGLGK(biotin)GGA (data not shown). Collectively, these data suggest that both anti-K9bio and anti-K13bio are specific for biotinylated histone H2A peptides and are unlikely to cross-react with other biotinylated histones (see below). These data also suggest that antibody anti-K13bio is biotinylation site-specific, whereas antibody anti-K9bio cross-reacts with both biotinylated K9 and K13.
Fig. 8.
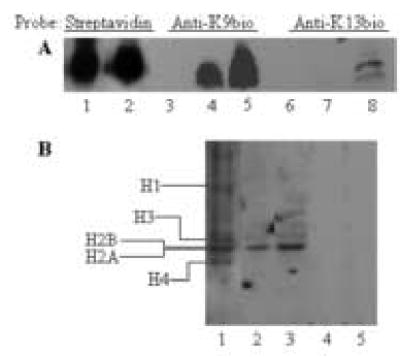
Western blot analysis of biotinylated histones. Panel A: Synthetic peptides based on histone H2A were probed with streptavidin peroxidase or antibodies to histone H2A, biotinylated in position K9 or K13. Lanes 1, 4, and 7 = peptide N1–12bioK9 [SGRGKQGGK(biotin)ARAC]; lanes 2, 5, and 8 = peptide N10–24bioK13 [ARAK(biotin)AKTRSSRAGLQC]; and lanes 3 and 6 = peptide N1–20 (SGRGKQGGKARAKAKTRSSR). Panel B: Jurkat cells contain biotinylated histone H2A. Nuclear histones were purified by acid extraction. Histones were resolved by gel electrophoresis and probed as follows: lane 1 = streptavidin peroxidase; lane 2 = antibody anti- K9bio; lane 3 = antibody anti-K13bio, lane 4 = pre-immune serum for antibody anti-K9bio; and lane 5 = pre-immune serum for antibody anti-K13bio.
Next, histone extracts from Jurkat cell nuclei were probed with antibodies to biotinylated histone H2A. The histone extracts contained biotinylated histones H1, H2A, H2B, H3 and H4, as judged by staining with streptavidin peroxidase (Fig. 8B, lane 1). The polyclonal antibodies raised in this study were specific for histone H2A and did not cross-react with other classes of histones (lanes 2 and 3). If biotinylated histones were probed with pre-immune serum, no detectable signal was produced (lanes 4 and 5).
Biotinylated histone H2A localized to the nucleus in JAr choriocarcinoma cells, as judged by confocal microscopy and antibodies against biotinylated histone H2A. First, the subcellular localization of K9-biotinylated histone H2A was visualized using antibody anti-K9bio (Fig. 9A). Nuclear and cytoplasmic compartment were stained with DAPI and rhodamine phalloidin, respectively (Fig. 9A). Merged images are consistent with nuclear localization of K9- biotinylated histone H2A. Pre-immune serum did not generate a detectable signal. Analogous experiments were conducted for K13-biotinylated histone H2A. Antibody anti-K13bio localized primarily to the cell nucleus (Fig. 9B).
Fig. 9.
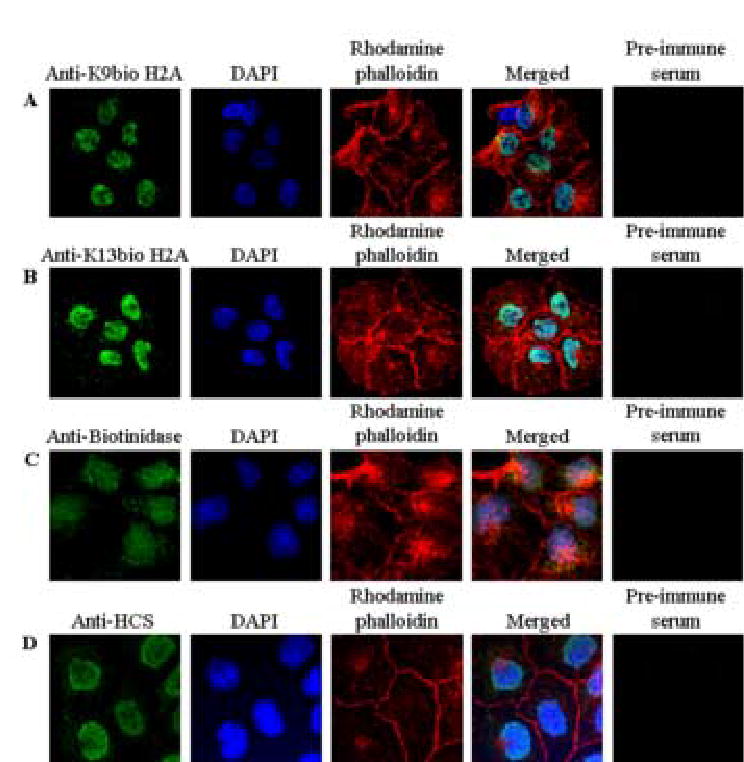
Biotinylated histone H2A, biotinidase, and HCS localize primarily to the nucleus in JAr choriocarcinoma cells. Cells were stained with antibodies to K9-biotinylated histone H2A (panel A), K13-biotinylated histone H2A (panel B), biotinidase (panel C), and HCS (panel D). The nuclear and cytoplasmic compartments were stained using DAPI and rhodamine phalliodin, respectively. Images entitled “Merged” were created by overlaying images obtained by staining with antibody, DAPI, and rhodamine phalliodin. Pre-immune sera were used as negative controls.
Both biotinidase and HCS showed considerable nuclear localization in JAr cells. First, we validated the specificity of antibodies against biotinidase and HCS using synthetic peptides as described for histone antibodies (data not shown). The subcellular localization of biotinidase in JAr cells was visualized using confocal microscopy and anti-biotinidase (Fig. 9C). Nuclear and cytoplasmic compartment were stained with DAPI and rhodamine phalloidin, respectively (Fig. 9C). Merged images were consistent with nuclear localization of biotinidase. Pre-immune serum did not generate a detectable signal (Fig. 9C). Analogous experiments were conducted for HCS. Anti-HCS also localized to the cell nucleus; pre-immune serum (negative control) did not generate a detectable signal (Fig. 9D).
4. Discussion
This study provides evidence that (a) K9 and K13 in the N-terminus of histones H2A and H2AX are targets for biotinylation by biotinidase; (b) that K125, K127, and K129 in the C-terminus of histone H2A are targets for biotinylation by biotinidase; (c) that K9- and K13- biotinylated histone H2A reside in human cell nuclei; (d) that acetylation and dimethylation of lysine residues in histones decrease subsequent biotinylation of adjacent lysine residues; (e) that dimethylation of arginine residues increases subsequent biotinylation of adjacent lysine residues; and (f) that both HCS and biotinidase reside primarily in the nuclear compartment.
The findings presented here are likely to generate novel insights into biological functions of histone biotinylation. Ongoing studies in our laboratory have provided evidence that biotinylation of distinct lysine residues in various classes of histones might play unique roles in the cellular response to double strand breaks of DNA, chromatin condensation, telomere structure, and nucleoli structure (unpublished observations). Specifically, we propose that biotinylation of histone H2A might participate in the following processes. First, biotinylation of histone H2A targets K9, which also is a target for acetylation; these two modifications are mutually exclusive. It has been proposed that acetylation of K9 in histone H2A might be associated with transcriptionally active chromatin [28]. We speculate that biotinylation of K9 decreases the transcriptional activity of chromatin, mediated by decreased acetylation of K9. Second, phosphorylation of histone H2AX is known to participate in DNA repair events, mediated by accumulation at sites of DNA damage [11]. Biotinylation of histones is known to change in response to DNA damage [17,18], but it remains to be determined whether biotinylation of K9 and K13 in histone H2A plays a role in repair events. Third, biotinylation of lysines in the C-terminus of histone H2A might affect histone-histone interactions in nucleosomes, based on the following lines of reasoning. Histone H2A is unique among core histones in having its C-terminal tail exposed at the nucleosomal surface [1,8]. However, the larger part of the C-terminal domain of histone H2A and other histones is buried inside the nucleosomes [1]. The C-terminal histone fold domain is predominantly α-helical with a long central helix bordered on each side by a loop segment (β-bridge, hinge region) and a shorter helix [1]. The long helix acts as a dimerization interface between histones [1]. We speculate that the biotinylation of C-terminal lysine residues in histone H2A affects the dimerization of histones. Note that K125 or K127 are also targets for methylation [7]. Effects of lysine methylation in the C-terminus of histone H2A are uncertain, but interactions between biotinylation and methylation are likely to occur in vivo.
We shall point out the following uncertainty of the studies presented here. Whereas the existence of K9- and K13-biotinylated histone H2A has been confirmed by using antibodies, no such antibodies have yet been raised against histone H2A biotinylated in the C-terminus. Hence, while this study suggests that K125, K127, and K129 are targets for biotinidase, the existence of these modifications in vivo awaits formal confirmation.
Previous studies suggested that dimethylation of arginine residues in histone H4 increases biotinylation of adjacent lysine residues [16]. We observed a similar pattern for histone H2A: dimethylation of R11 increased the biotinylation of K9 or K13 (or both) by biotinidase, and dimethylation of R17 increased the biotinylation of K13 or K15 (or both). Note that arginine residues in histones can be converted to citrulline and ornithine by deimination [29,30]. Citrulline and ornithine residues are good targets for biotinylation by biotinidase (Y. C. Chew and J. Zempleni; unpublished observation). Collectively, posttranslational modifications of arginine residues are likely to play important roles in histone biotinylation.
Finally, this study provides evidence that significant fractions of cellular biotinidase and HCS localize to the nuclear compartment, consistent with a role of these enzymes in chromatin structure. Previous studies are consistent with a nuclear localization of HCS [14]. These previous studies suggested that the majority of HCS localized to the nuclear periphery rather than the nucleoplasm. The functional significance of this observation is currently being investigated. The amino acid residues in histones that are targets for biotinylation by HCS await identification, whereas targets for biotinidase have been characterized [15,16]. Unlike for HCS, the cellular distribution of biotinidase is controversial. This study and a previous study [31] are consistent with nuclear localization of biotinidase; in contrast, Wolf and co-workers suggested that biotinidase localizes to the cytoplasm but not to the nucleus [32]. The reasons for these apparently conflicting observations are unknown.
Acknowledgments
This work was supported by NIH grants DK 60447 and DK 063945, and by NSF EPSCoR grant EPS-0346476. This paper is a contribution of University of Nebraska-Agricultural Research Division, Lincoln, NE 68583 (Journal Series No. 14986).
References
- 1.Wolffe, A. Chromatin. San Diego, CA: Academic Press; 1998.
- 2.Fischle W, Wang Y, Allis CD. Histone and chromatin cross-talk. Curr Opin Cell Biol. 2003;15:172–183. doi: 10.1016/s0955-0674(03)00013-9. [DOI] [PubMed] [Google Scholar]
- 3.D’Amours D, Desnoyers S, D’Silva I, Poirier GG. Poly(ADP-ribosyl)ation reactions in the regulation of nuclear functions. Biochem J. 1999;342:249–268. [PMC free article] [PubMed] [Google Scholar]
- 4.Grewal SI, Moazed D. Heterochromatin and epigenetic control of gene expression. Science. 2003;301:798–802. doi: 10.1126/science.1086887. [DOI] [PubMed] [Google Scholar]
- 5.Cheng, X, Collins, RE, and Zhang, X. Structural and sequence motifs of protein (histone) methylation enzymes. Annu Rev Biophys Biomol Struct 2004 (in press). [DOI] [PMC free article] [PubMed]
- 6.Fischle W, Wang Y, CD A. Binary switches and modification cassettes in histone biology and beyond. Nature. 2003;425:475–479. doi: 10.1038/nature02017. [DOI] [PubMed] [Google Scholar]
- 7.Zhang L, Eugeni EE, Parthun MR, Freitas MA. Identification of novel histone posttranslational modifications by peptide mass fingerprinting. Chromosoma. 2003;112:77–86. doi: 10.1007/s00412-003-0244-6. [DOI] [PubMed] [Google Scholar]
- 8.Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 9.Ausio J, Abbott DW, Wang X, Moore SC. Histone variants and histone modifications: a structural perspective. Biochem Cell Biol. 2001;79:693–708. [PubMed] [Google Scholar]
- 10.Downs JA, Allard S, Jobin-Robitaille O, Javaheri A, Auger A, Bouchard N, Kron SJ, Jackson SP, Cote J. Binding of chromatin-modifying activities to phosphorylated histone H2A at DNA damage sites. Mol Cell. 2004;16:979–990. doi: 10.1016/j.molcel.2004.12.003. [DOI] [PubMed] [Google Scholar]
- 11.Paull TT, Rogakou EP, Yamazaki V, Kirchgessner CU, Gellert M, Bonner WM. A critical role for histone H2AX in recruitment of repair factors to nuclear foci after DNA damage. Curr Biol. 2000;10:886–895. doi: 10.1016/s0960-9822(00)00610-2. [DOI] [PubMed] [Google Scholar]
- 12.Stanley JS, Griffin JB, Zempleni J. Biotinylation of histones in human cells: effects of cell proliferation. Eur J Biochem. 2001;268:5424–5429. doi: 10.1046/j.0014-2956.2001.02481.x. [DOI] [PubMed] [Google Scholar]
- 13.Hymes J, Fleischhauer K, Wolf B. Biotinylation of histones by human serum biotinidase: assessment of biotinyl-transferase activity in sera from normal individuals and children with biotinidase deficiency. Biochem Mol Med. 1995;56:76–83. doi: 10.1006/bmme.1995.1059. [DOI] [PubMed] [Google Scholar]
- 14.Narang MA, Dumas R, Ayer LM, Gravel RA. Reduced histone biotinylation in multiple carboxylase deficiency patients: a nuclear role for holocarboxylase synthetase. Hum Mol Genet. 2004;13:15–23. doi: 10.1093/hmg/ddh006. [DOI] [PubMed] [Google Scholar]
- 15.Sarath G, Kobza K, Rueckert B, Camporeale G, Zempleni J, Haas E. Biotinylation of human histone H3 and interactions with biotinidase. FASEB J. 2004;18:A103. (abstract) [Google Scholar]
- 16.Camporeale G, Shubert EE, Sarath G, Cerny R, Zempleni J. K8 and K12 are biotinylated in human histone H4. Eur J Biochem. 2004;271:2257–2263. doi: 10.1111/j.1432-1033.2004.04167.x. [DOI] [PubMed] [Google Scholar]
- 17.Peters DM, Griffin JB, Stanley JS, Beck MM, Zempleni J. Exposure to UV light causes increased biotinylation of histones in Jurkat cells. Am J Physiol Cell Physiol. 2002;283:C878–C884. doi: 10.1152/ajpcell.00107.2002. [DOI] [PubMed] [Google Scholar]
- 18.Kothapalli N, Zempleni J. Double strand breaks of DNA decrease biotinylation of lysine-12 in histone H4 in JAr cells. FASEB J. 2004;18:A103–104. (abstract) [Google Scholar]
- 19.Wyatt HR, Liaw H, Green GR, Lustig AJ. Multiple roles for Saccharomyces cerevisiae histone H2A in telomere position effect, Spt phenotypes and double-strand-break repair. Genetics. 2003;164:47–64. doi: 10.1093/genetics/164.1.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pantazis P, Bonner WM. Quantitative Determination of Histone Modification H2A Acetylaton and Phosphorylation. J Biol Chem. 1981;256:4669–4675. [PubMed] [Google Scholar]
- 21.Goll MG, Bestor TH. Histone modification and replacement in chromatin activation. Genes Dev. 2002;16:1739–1742. doi: 10.1101/gad.1013902. [DOI] [PubMed] [Google Scholar]
- 22.Aihara H, Nakagawa T, Yasui K, Ohta T, Hirose S, Dhomae N, Takio K, Kaneko M, Takeshima Y, Muramatsu M, Ito T. Nucleosomal histone kinase-1 phosphorylates H2A Thr 119 during mitosis in the early Drosophila embryo. Genes Dev. 2004;18:877–888. doi: 10.1101/gad.1184604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Camporeale, G, Chew, YC, Kueh, A, Sarath, G, and Zempleni, J. Use of synthetic peptides for identifying biotinylation sites in human histones. In: McMahon, RJ, McMahon, RJs. Avidin-Biotin Technology in the Life Sciences. Totowa, N.J.: Humana Press; 2005 (in press).
- 24.Garrett, RH, and Grisham, CM. Biochemistry. Fort Worth, TX: Saunders College Publishing; 1995.
- 25.Manthey KC, Griffin JB, Zempleni J. Biotin supply affects expression of biotin transporters, biotinylation of carboxylases, and metabolism of interleukin-2 in Jurkat cells. J Nutr. 2002;132:887–892. doi: 10.1093/jn/132.5.887. [DOI] [PubMed] [Google Scholar]
- 26.Crisp SERH, Camporeale G, White BR, Toombs CF, Griffin JB, Said HM, Zempleni J. Biotin supply affects rates of cell proliferation, biotinylation of carboxylases and histones, and expression of the gene encoding the sodium-dependent multivitamin transporter in JAr choriocarcinoma cells. Eur J Nutr. 2004;43:23–31. doi: 10.1007/s00394-004-0435-9. [DOI] [PubMed] [Google Scholar]
- 27.Cheung WL, Ajiro K, Samejima K, Kloc M, Cheung P, Mizzen CA, Beeser A, Etkin LD, Chernoff J, Earnshaw WC, Allis CD. Apoptotic phosphorylation of histone H2B is mediated by mammalian sterile twenty kinase. Cell. 2003;113:507–517. doi: 10.1016/s0092-8674(03)00355-6. [DOI] [PubMed] [Google Scholar]
- 28.Turner BM. Cellular memory and the histone code. Cell. 2002;111:285–291. doi: 10.1016/s0092-8674(02)01080-2. [DOI] [PubMed] [Google Scholar]
- 29.Bannister AJ, Schneider R, Kouzarides T. Histone methylation: dynamic or static? Cell. 2002;109:801–806. doi: 10.1016/s0092-8674(02)00798-5. [DOI] [PubMed] [Google Scholar]
- 30.Cuthbert GL, Daujat S, Snowden AW, Erdjument-Bromage H, Hagiwara T, Yamada M, Schneider R, Gregory PD, Tempst P, Bannister AJ, Kouzarides T. Histone deimination antagonizes arginine methylation. Cell. 2004;118:545–553. doi: 10.1016/j.cell.2004.08.020. [DOI] [PubMed] [Google Scholar]
- 31.Pispa J. Animal biotinidase. Ann Med Exp Biol Fenniae. 1965;43:4–39. [PubMed] [Google Scholar]
- 32.Stanley CM, Hymes J, Wolf B. Identification of alternatively spliced human biotinidase mRNAs and putative localization of endogenous biotinidase. Mol Genet Metab. 2004;81:300–312. doi: 10.1016/j.ymgme.2003.12.006. [DOI] [PubMed] [Google Scholar]


