Abstract
The first step in poliovirus (PV) RNA synthesis is the covalent linkage of UMP to the terminal protein VPg. This reaction can be studied in vitro with two different assays. The simpler assay is based on a poly(A) template and requires synthetic VPg, purified RNA polymerase 3Dpol, UTP, and a divalent cation. The other assay uses specific viral sequences [cre(2C)] as a template for VPg uridylylation and requires the addition of proteinase 3CDpro. Using one or both of these assays, we analyzed the VPg specificities and metal requirements of the uridylylation reactions. We determined the effects of single and double amino acid substitutions in VPg on the abilities of the peptides to serve as substrates for 3Dpol. Mutations in VPg, which interfered with uridylylation in vitro, were found to abolish viral growth. A chimeric PV containing the VPg of human rhinovirus 14 (HRV14) was viable, but substitutions of HRV2 and HRV89 VPgs for PV VPg were lethal. Of the three rhinoviral VPgs tested, only the HRV14 peptide was found to function as a substrate for PV1(M) 3Dpol in vitro. We also examined the metal specificity of the VPg uridylylation reaction on a poly(A) template. Our results show a strong preference of the RNA polymerase for Mn2+ as a cofactor compared to Mg2+ or other divalent cations.
Poliovirus (PV) is a small plus-stranded RNA virus belonging to the family Picornaviridae. The members of this virus family are characterized by the presence of a peptide (VPg) covalently linked to the 5′ end of the viral genome. Replication is a two-step process, beginning with the synthesis of a complementary minus strand (1). That strand, in turn, serves as a template for the production of progeny plus-strand RNAs. Although the basic steps of RNA replication are well established, very little information is available about the details of these processes. The enzyme primarily responsible for RNA synthesis is the viral RNA polymerase 3Dpol, which is both primer and template dependent and possesses two important synthetic activities in vitro (12, 37). The first activity, catalyzing the elongation of an oligonucleotide primer on an RNA template, was discovered >20 years ago and has been thoroughly characterized (4, 12). The second synthetic activity of 3Dpol was only recently identified as a reaction in which UMP is linked to the hydroxyl group of a tyrosine in VPg, yielding VPgpU and VPgpUpU (37, 39). These precursors, which can not only be found in PV-infected cells (8) but can also be made in crude replication complexes (49, 52), are believed to be the primers used by 3Dpol for both minus- and plus-strand RNA synthesis.
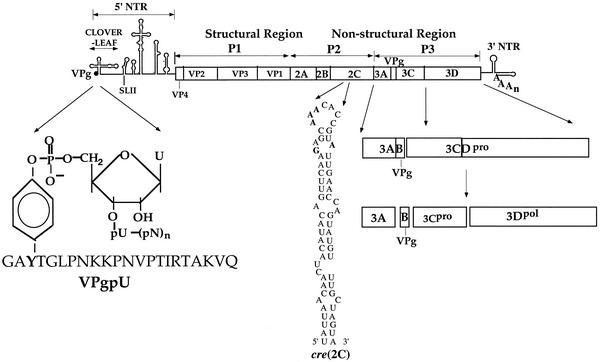
The RNA genome of PV (7,525 nucleotides [nt]) contains a long 5′ nontranslated region (NTR), a single open reading frame, a short 3′ NTR, and a poly(A) tail (Fig. 1) (20). VPg is attached to the 5′-terminal UMP of the RNA by a phosphodiester bond (2, 45). The linkage is between the 5′ phosphate of UMP and the hydroxyl group of a tyrosine in VPg (2, 26, 45). Translation of the RNA results in the synthesis of a polyprotein with one structural (P1) and two nonstructural (P2 and P3) domains. Of the nonstructural proteins of the virus, those that are most directly involved in RNA synthesis are 3Dpol; its precursor proteinase, 3CDpro; and VPg (37, 39, 44). In addition, RNA replication also requires important RNA elements located in the 5′ NTR and 3′ NTR (1, 3, 56). An essential cis replicating element (cre) is also present in the coding sequence of PV 2CATPase [cre(2C)] (Fig. 1) (15). This small RNA hairpin is similar in size and structure to internal cres of other picornaviruses [cre(VP1) of human rhinovirus 14 (HRV14) (29, 58), cre(VP2) of mengovirus and Theiler's virus (27), and cre(2A) of HRV2 (14)]. The PV and HRV hairpins all contain a conserved sequence (GXXXAAAXXXXXXA) either in the loop or in the loop and a nearby bulge (58; J. Yin, A. V. Paul, E. Wimmer, and E. Rieder, submitted for publication). We have identified the function of the first two As of the AAA sequence in PV1(M) cre(2C) (39, 44) and HRV2 cre(2A) (14) as providing templates for VPg uridylylation by 3Dpol in vitro. The other nucleotides in the motif are also important for optimal protein priming, RNA replication, and viral growth (58; Yin et al., submitted). The reactions catalyzed on transcripts of full-length viral RNAs or cres by both polymerases are strongly stimulated by the addition of their cognate 3CDpro proteins (14, 39, 44) as follows: PV VPg + PV 3Dpol + PV RNA + PV 3CDpro + Mg2+ → PV VPgpU + PV VPgpUpU and HRV2 VPg + HRV2 3Dpol + HRV2 RNA + HRV2 3CDpro + Mg2+ → HRV2 VPgpU + HRV2 VPgpUpU.
FIG. 1.
Structure of PV genomic RNA and processing of the P3 domain of the polyprotein. The single-stranded RNA genome of PV is shown with the terminal protein VPg (3B) at the 5′ end of the 5′ NTR and the 3′ NTR with the poly(A) tail (20). The 5′NTR consists of a cloverleaf and a large internal ribosomal entry site element. The attachment site of the 5′-terminal UMP of the RNA to the tyrosine of VPg (2, 45) is shown enlarged. Also shown enlarged is the predicted secondary structure of a cis replicating RNA element located in the coding sequence of 2CATPase [cre(2C)] (15, 39, 44). The nucleotides required for PV cre function are shown in boldface letters (44, 58). The polyprotein contains structural (P1) and nonstructural (P2 and P3) domains. The vertical lines within the polyprotein box represent the proteinase cleavage sites. Processing of the P3 domain of the polyprotein is shown enlarged.
In vitro, both the PV1(M) (37) and HRV2 (13) polymerases are also able to use poly(A) as a template instead of viral RNA: VPg + 3Dpol + poly(A) + Mn2+ (Mg2+) → VPgpU + VPgpUpU → VPg-poly(U). The final product of the reaction is VPg-linked poly(U), the 5′ end of minus-strand RNA (32). Although this reaction lacks the specificity of a viral RNA template, it does offer a simple assay to test VPg uridylylation when either viral RNA or 3CDpro is not available.
In vivo, the replication of PV RNA occurs on membranous vesicles (5), but the exact composition of the replication complex is not yet understood. The viral protein 3AB, the precursor of VPg, is predicted to be an important component of the replication complex because of its ability to stimulate elongation of RNA synthesis at low 3Dpol concentrations (36) and because of its ability to interact both with membranes (50) and with the RNA polymerase (18, 57). The hydrophobic region of this small basic RNA binding protein has been shown to associate with biological membranes in vitro (50). Evidence for a strong interaction between 3AB (18) or 3B (VPg) (57) and the RNA polymerase came from yeast two-hybrid studies, and the contact points between the two proteins were found to reside in the 3B domain of 3AB (57). Recently, Lyle et al. (28) identified four surface residues on the structure of 3Dpol which are required both for interaction with 3AB and for VPg uridylylation, suggesting that the binding sites for 3AB and the VPg primer overlap. These studies led to a model of RNA replication in which 3AB or one its precursors (40, 51) recruits the RNA polymerase to the membranes, where replication is then initiated. However, the observation that 3Dpol is able to uridylylate only 3B and not 3AB in vitro (A. V. Paul and E. Wimmer, unpublished data) suggests either that 3AB has to be processed prior to the initiation reaction or that a different precursor of VPg is the true substrate for the uridylylation reaction in vivo.
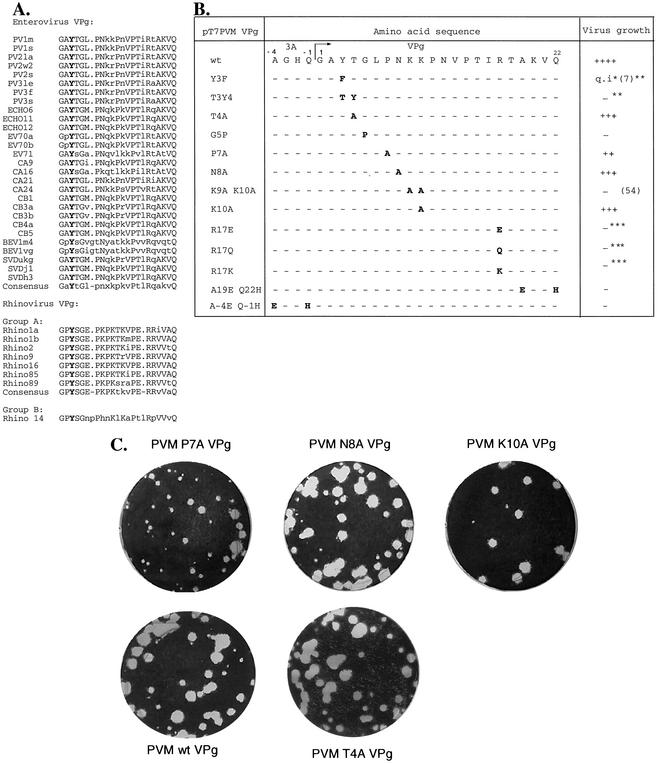
The enteroviral and rhinoviral VPgs are small basic peptides containing 21 to 23 amino acids; that of PV is 22 residues long (Fig. 2A) (56). They contain only a few amino acids, which are fully conserved in the VPgs of both genera. In addition to the N-terminal G and C-terminal Q (in the 3Cpro-specific Q-G cleavage sites), these residues include the tyrosine at position 3 (the attachment site to the RNA) and a glycine, a proline, and an arginine at positions 5, 14, and 17, respectively. The peptides also contain near their centers at least two positively charged amino acids, either two lysines or a lysine and an arginine. Interestingly, proline is the most abundant amino acid in the peptides, occurring at positions 7, 11, and 14 in the VPg of PV.
FIG. 2.
Effects of mutations in VPg on the viability of PV. (A) Enteroviral and rhinoviral VPg sequences. Conserved amino acids are indicated with uppercase letters. The fully conserved tyrosine, which provides the VPg-linking site, is shown in boldface letters. (B) Growth properties of PV VPg mutants. The table summarizes the growth properties of VPg mutants made in this study (Y3T T4Y, T4A, G5P, P7A, N8A, and K10A) and those previously described (Y3F, K9A K10A, R17E, R17Q, and R17K) (7, 21, 22, 43, 54). Also shown are two mutants in which either the N- or the C-terminal 3Cpro-specific cleavage site is eliminated (A−4E Q−1H in 3A and A19E Q22H in VPg). Mutations in the wt PV1(M) VPg sequence are indicated with boldface letters; the dashes represent conserved residues. *, quasi-infectious (q.i); **, Y3F and T3Y4 mutants were also made by Reuer et al. (43) and Kuhn et al. (21), respectively, but they contained an additional K10R mutation; ***, virus contained an additional K10R mutation in VPg, which had no effect on viability (22). The numbers in parentheses are references. ++++, wt-type growth; +++, slower than wt growth; ++, much slower than wt growth; −, no growth. (C) Plaque assay of viruses containing mutant VPgs. The plaque sizes of the three viable VPg mutants are compared to that of wt PV.
The central theme of this study was to further define the amino acid sequence requirements in VPg as it functions during PV genome replication. Previous mutational analyses of PV1(M) VPg have identified a number of amino acids in the peptide that can be replaced without loss of virus production in transfected cells (21, 22, 43). The generation of nonviable or quasi-infectious mutants demonstrated that certain amino acids are essential for viability (7, 21, 22, 43, 54). We have now extended these studies with the construction of six new VPg mutants and determined the effects of these amino acid substitutions on the growth properties of PV. By measuring the ability of wild-type (wt) and mutant VPg peptides to support VPg uridylylation in vitro, we show that viral growth is strictly dependent on the nucleotidylylation reaction. Of particular interest to us was testing whether VPgs of the genus Rhinovirus (those of HRV2, HRV89, and HRV14) that express subtle differences in the amino acid sequences could substitute for PV VPg. Only HRV14 VPg could substitute for PV VPg in vivo, yielding, however, a replication phenotype. The other two VPgs (those of HRV2 and HRV89) were incompatible with the PV replication machinery, for which we will offer a possible explanation.
In addition, yeast two-hybrid analyses have allowed us to map the determinants of the interaction between VPg and the RNA polymerase. Our results provide evidence that the formation of a complex between these two proteins is necessary for VPg uridylylation and consequently also for viral growth.
Two additional mutants were engineered in which the 3Cpro-dependent processing at the 3A-3B or the 3B-3C cleavage site was abolished. These mutants were tested in vitro for defects in the proteolytic processing cascade and in vivo for infectivity. The nonviable phenotype of these mutants suggests that the intact 3AB and 3BC precursors cannot replace VPg in the viral life cycle.
Although the enzymatic activity of PV 3Dpol in an oligonucleotide elongation reaction has been thoroughly investigated (4, 12), much less is known about the requirements of the enzyme for the uridylylation of VPg in vitro. Some of the biochemical parameters of the uridylylation reaction on a poly(A) template, such as pH and temperature optimum, have been determined (37). In the present study, we have examined the divalent-cation specificity of the RNA polymerase.
MATERIALS AND METHODS
Plasmids.
All PV cDNA sequences were derived from plasmid pT7PVM (53). The sequences listed for plasmids or oligonucleotides refer to the full-length plus-strand PV sequence, HRV14 sequence (48), HRV2 sequence (47), or HRV89 sequence (10). Nucleotide changes in oligonucleotides are underlined.
(i) pT7PVM HRV14 VPg.
A PCR fragment was made using the oligonucleotides a (5′ GGGGGTAACCCGCCTCACAATAAACTAAAAGCCCCAACTTTACGCCCAGTTGTGGTACAAGGACCAGGGTTCGATTACGCAGTGGC 3′; plus strand; nt 5179 to 5235 of the HRV14 VPg sequence and nt 5438 to 5463 of the pT7PVM 3Cpro sequence) and b [5′ CCTGCTTGATCTTCGAGCGC 3′; minus strand; nt 5618 to 5637 of the pT7PV1(M) sequence]. The fragment was cut with BglII and BstEII. Another PCR fragment was made using the oligonucleotides c (5′ GGGGGTTACCAGAATATGGTCCCTGGTGTCCAGCAAACAGTTTATACATGAC 3′; minus strand; nt 5167 to 5185 of the HRV14 VPg sequence and nt 5342 to 5371 of the pT7PVM 3A sequence) and d [5′ GGGTTGGATAGTTAACATCACCAGCC 3′; plus strand; nt 5230 to 5255 of the pT7PV1(M) sequence]. This fragment was digested with HpaI and BstEII, and the two PCR fragments were ligated into HpaI/BglII-restricted pT7PVM DNA.
(ii) pT7PVM HRV2 VPg.
A PCR fragment was made containing a BanII site using oligonucleotides e (5′ GGGGGGCCCTATTCAGGAGAACCAAAGCCCAAGACTAAAATCCCAGAAAGGCGTGTAGTAACACAAGGACCAGGGTTCGATTACGCAGTG 3′; plus strand; nt 5069 to 5131 of the HRV2 VPg sequence and nt 5438 to 5461 of the pT7PVM 3Cpro sequence) and b. The fragment was cut with BanII and BglII. Another PCR fragment was made containing a BanII site using the oligonucleotides f (5′ GGGGGGCCCCTGGTGTCCAGCAAACAGTTTATACATGAC 3′; minus strand; nt 5341 to 5371 of the pT7PVM sequence) and d. This fragment was digested with BanII and HpaI. The two PCR fragments were ligated into HpaI/BglII-cut pTVPVM.
(iii) pT7PVM HRV89 VPg.
The pT7PVM HRV89 VPg construct was made the same way as described for pT7PVM HRV2 VPg. The following two oligonucleotides were used for the synthesis of the first PCR fragment: g (5′ GGGGGGCCCTACTCAGGGGAACCTAAACCCAAAAGCAGAGCTCCAGAGAGAAGAGTAGTTACTCAGGGACCAGGGTTCGATTACGCAGTG 5′; plus strand; nt 5119 to 5181 of the HRV89 sequence and nt 5438 to 5461 of the pT7PVM 3Cpro sequence) and b. For the second PCR fragment, oligonucleotides f and d were used.
(iv) pT7PVM VPg and 3A mutants.
Seven VPg mutants (T4A, Y3T T4Y, G5P, P7A, N8A, K10A, and A19E Q22H) and one 3A mutant (A−4E Q−1H) were made by PCR mutagenesis. In the first step, two PCR fragments were made using the oligonucleotides listed in Table 1. The two fragments were then mixed and used as templates for the synthesis of a new fragment, which was cut with HpaI/BglII and cloned into similarly restricted pT7PVM.
TABLE 1.
Oligonucleotides used for mutagenesis of pT7PVM VPg
| Construct | Oligonucleotide sequenceb |
|---|---|
| pT7PVM VPg (T4A) | 5′ CACCAGGGAGCATACGCTGGTTTACCAAACAAAAAACCC 3′ (plus strand) |
| 5′ TGTTTGGTAAACCAGCGTATGCTCCCTGGTGTCCAGC 3′ (minus strand) | |
| pT7PVM VPg (Y3T T4Y) | 5′ CACCAGGGAGCAACCTATGGTTTACCAAACAAAAAACCC 3′ (plus strand) |
| 5′ GTTTGGTAAACCATAGGTTGCTCCCTGGTGTCCAGC 3′ (minus strand) | |
| pT7PVM VPg (G5P) | 5′ GCATACACTCCTTTACCAAACAAAAAACCC 3′ (plus strand) |
| 5′ GTTTGGTAAAGGAGTGTATGCTCCCTGGTG 3′ (minus strand) | |
| pT7PVM VPg (P7A) | 5′ TACACTGGTTTAGCGAACAAAAAACCCAACGTGCC 3′ (plus strand) |
| 5′ GGGTTTTTTGTTCGCTAAACCAGTGTATGCTCCC 3′ (minus strand) | |
| pT7PVM VPg (N8A) | 5′ ACTGGTTTACCAGCCAAAAAACCCAACGTGCCCAC 3′ (plus strand) |
| 5′ GTTGGGTTTTTTGGCTGGTAAACCAGTGTATGCTC 3′ (minus strand) | |
| pT7PVM VPg (K10A) | 5′ GGTTTACCAAACAAAGCACCCAACGTGCCCACC 3′ (plus strand) |
| 5′ GCACGTTGGGTGCTTTGTTTGGTAAACCAG 3′ (minus strand) | |
| pT7PVM VPg (A19E Q22H) | 5 ′ GACAGAGAAGGTACACGGACCAGGGTTCGATTAC 3′ (plus strand) |
| 5′ TGGTCCGTGTACCTTCTCTGTCCGAATGGTGG 3′ (minus strand) | |
| pT7PVM VPg (A − 4E Q − 1H)a | 5′ CTGTTTGAAGGACACCACGGAGCATACACTGG 3′ (plus strand) |
| 5′ CATATTTGACAAACTTCCTGTGGTGCCTCGT 3′ (minus strand) | |
| pT7PVM (nt 5230-5255) | 5′ GGGTTGGATAGTTAACATCACCAGCC 3′ (plus strand) |
| pT7PVM (nt 5618-5637) | 5′ CCTGCTTGATCTTCGAGCGC 3′ (minus strand) |
Mutations in 3A sequence.
Mutations from the wt sequence are underlined.
Enzymes.
PV 3Dpol was expressed in Escherichia coli from plasmid pT5T-3D (a gift of K. Kirkegaard) and purified as described before (33). PV 3CDpro (3Cpro H40A), containing a C-terminal His tag, was also expressed in E. coli and purified as described before (39).
VPg uridylylation assays.
Assay 1 measures the synthesis of VPgpU(pU) and of VPg-poly(U) on a poly(A) template (37). The reaction mixture (20 μl) contained 50 mM HEPES, pH 7.5, 8% glycerol, 0.5 mM manganese(II) acetate, 0.5 μg of poly(A), 1 μg of purified 3Dpol, 2 μg of synthetic PV VPg (9), 0.75 μCi of [α-32P]UTP (3,000 Ci/mmol; Perkin-Elmer), and 10 μM unlabeled UTP. The reaction mixtures were incubated at 34°C for 1 h, and the reaction was stopped by the addition of 5 μl of gel-loading buffer. The samples were analyzed by Tris-Tricine sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (Bio-Rad) with 13.5% polyacrylamide. The gels were dried without being fixed and were autoradiographed. The reaction products were quantitated with a PhosphorImager (Storm 860; Molecular Dynamics) by measuring the amount of [32P]UMP incorporated into the product. VPgpU(pU) represents the sum of VPgpU and VPgpUpU. Assay 2 is similar to assay 1 except that the poly(A) template is replaced by either 0.5 μg of cre(2C) RNA or 1 μg of PV1(M) transcript RNA, 0.5 mM manganese(II) acetate is replaced by 3.5 mM magnesium acetate, and 0.5 μM purified 3CDpro is added to the reaction mixture (39).
In vitro transcription and translation of mRNAs.
Plasmid DNAs were linearized with EcoRI and transcribed with phage T7 RNA polymerase (53). The RNAs were purified and translated in HeLa cell extracts at 30°C in the presence of [35S] Translabel (ICN Biochemicals) (30). Samples of the translation reaction mixture were analyzed on SDS-polyacrylamide gels (12.5%).
RNA transfection and plaque assays.
RNA transcripts were transfected into HeLa R19 monolayers by the DEAE-dextran method, as described previously (53). The viral lysates were collected, frozen, thawed three times, and centrifuged to remove cell debris. Virus titers were determined by plaque assays on HeLa R19 cell monolayers in six-well plates as described previously (30).
Yeast two-hybrid analysis.
The JG-3D plasmid carrying the wt PV 3Dpol sequence was constructed as described before (38). The EG3B plasmids containing the wt or mutant PV VPg sequences (R17E, Y3S, Y3F, G5P, and Y3T T4Y) were amplified by PCR from plasmid pT7PVM using primers (Table 2) designed to introduce an EcoRI site at the upstream (5′) end and two stop codons and an XhoI site at the downstream (3′) end of each coding sequence. The PCR fragments (EcoRI/XhoI cut) were ligated into the vector plasmid pEG202, which was also restricted with the same enzymes. Mutant EG3B plasmids (T4A, P7A, N8A, K9A K10A, R17K, and R17Q) were made by mutagenesis of the wt EG3B plasmid. PCR fragments, made using the corresponding primers listed in Table 2, were cut with EcoRI and XhoI and ligated into similarly restricted pEG202. The screening for interaction between 3Dpol and the wt or mutant VPgs was carried out as described before (38).
TABLE 2.
Oligonucleotides used for construction and mutagenesis of EG3B
| EG3B construct | Oligonucleotide sequencec |
|---|---|
| wt | 5′ GCGCGAATTCGGAGCATACACTGGTTTACCAAAC 3′ (plus strand)a |
| wt | 5′ GCGCCTCGAGTCATTATTGTACCTTTGCTG 3′ (minus strand)b |
| Y3S | 5′ GCGCGAATTCGGAGCATCCACTGGTTTACC 3′ (plus strand)a |
| Y3F | 5′ GCGCGAATTCGGAGCATTCACTGGTTTACC 3′ (plus strand)a |
| T4A | 5′ GGAGCATACGCTGGTTTACCAAAC 3′ (plus strand) |
| T4A | 5′ GTTTGGTAAACCAGCGTATGCTCC 3′ (minus strand) |
| Y3T T4Y | 5′ GCGCGAATTCGGAGCAACTTACGGTTTACC 3′ (plus strand)a |
| G5P | 5′ GCGCGAATTCGGAGCATACACTCCTTTACC 3′ (plus strand)a |
| P7A | 5′ GCATACACTGGTTTAGCAAACAAAAAACCC 3′ (plus strand) |
| P7A | 5′ GGGTTTTTTGTTTGCTAAACCAGTGTATGC 3′ (minus strand) |
| N8A | 5′ GCATACACTGGTTTACCAGCCAAAAAACCCAACGTGCCC 3′ (plus strand) |
| N8A | 5′ GGGCACGTTGGGTTTTTTGGCTGGTAAACCAGTGTATGC 3′ (minus strand) |
| K9A K10A | 5′ GGTTTACCAAACGCAGCACCCAACGTGCCCACC 3′ (plus strand) |
| K9A K10A | 5′ GGTGGGCACGTTGGGTGCTGCGTTTGGTAAACC 3′ (minus strand) |
| R17K | 5′ CGTGCCCACCATTAAGACAGCAAAGGTAC 3′ (plus strand) |
| R17K | 5′ GTACCTTTGCTGCTTAATGGTGGGCACG 3′ (minus strand) |
| R17Q | 5′ CGTGCCCACCATTCAGACAGCAAAGGTAC 3′ (plus strand) |
| R17Q | 5′ GTACCTTTGCTGTCTGAATGGTGGGCACG 3′ (minus strand) |
| R17E | 5′ GCGCCTCGAGTCATTATTGTACCTTTGCTGTCTCAAT 3′ (minus strand)b |
Oligonucleotide contains EcoRI site.
Oligonucleotide contains XhoI site.
Mutations from the wt sequence are underlined.
RESULTS
Effects of amino acid substitutions in VPg on the viability of PV.
Previous genetic and biochemical studies have revealed several mutations (Y3F, Y3T T4Y, K9A K10A, R17E, R17Q, and R17K) in PV VPg which either abolished viral growth or resulted in a quasi-infectious virus (Fig. 2B) (7, 21, 22, 43, 54). All but two of these, Y3F (7) and K9A K10A (54), contained an additional mutation (K10R), which, however, did not affect viability (21, 22, 43). In this study, we have selected five additional amino acids for mutagenesis. These amino acids are well conserved either among the VPgs of enteroviruses (T4 and N8) or within VPgs of both the genera Enterovirus and Rhinovirus (G5, P7, and K10) (Fig. 2A). In addition, we have reconstructed mutant Y3T T4Y without the additional K10R change. Transcript RNAs harboring these mutations were translated in vitro, and the translation products were compared to those obtained from wt pT7PVM. The products of polyprotein processing appeared to be normal for all the mutants, indicating an intact open reading frame (data not shown). The effects of the mutations (Y3T T4Y, T4A, G5P, P7A, N8A, and K10A) on viral growth were then tested (Fig. 2B). Transfection of HeLa cell monolayers with RNA transcripts derived from wt pT7PVM, pT7PVM T4A VPg, pT7PVM N8A VPg, and pT7PVM K10A VPg resulted in complete cytopathic effect (CPE) within 20 to 24 h. The plaque sizes of the resulting mutant viruses were about the same as that of the wt virus (Fig. 2C). The replacement of a proline with an alanine at position 7 (P7A) resulted in delayed CPE (36 h), and the virus had a reduced plaque size (Fig. 2C) compared to the wt virus. After five passages of the virus on HeLa cells, no revertants could be identified by genome sequencing. In contrast, there was no CPE following transfection with transcripts derived from either the G5P or Y3T T4Y constructs. Passaging the supernatants from transfected cells five times did not result in the appearance of any revertants.
Two double mutants were designed to test the requirement in viral growth for proteolytic cleavage between VPg and either 3A or 3Cpro at the N and the C terminus of VPg, respectively. Site-specific mutations were introduced into the nucleotides encoding the Q in the Q-G processing sites. The alanines at position −4 in 3A and VPg (Fig. 2B), which are known to enhance cleavage at Q-G cleavage sites (22), were mutated to E to ascertain a complete block in processing by 3Cpro-3CDpro. Translation reactions of full-length PV RNA transcripts carrying the A−4E Q−1H mutations in 3A (Fig. 2B) contained uncleaved 3AB but lacked 3A (data not shown). Similarly, translation reactions of mutant A19E Q22H in VPg contained unprocessed P3 and 3BCD but no detectable 3AB (data not shown). Both cleavage site mutations led to a nonviable growth phenotype, and no revertants could be isolated during passaging of transfection supernatants.
Comparison of wt and mutant VPg peptides as substrates for uridylylation by 3Dpol.
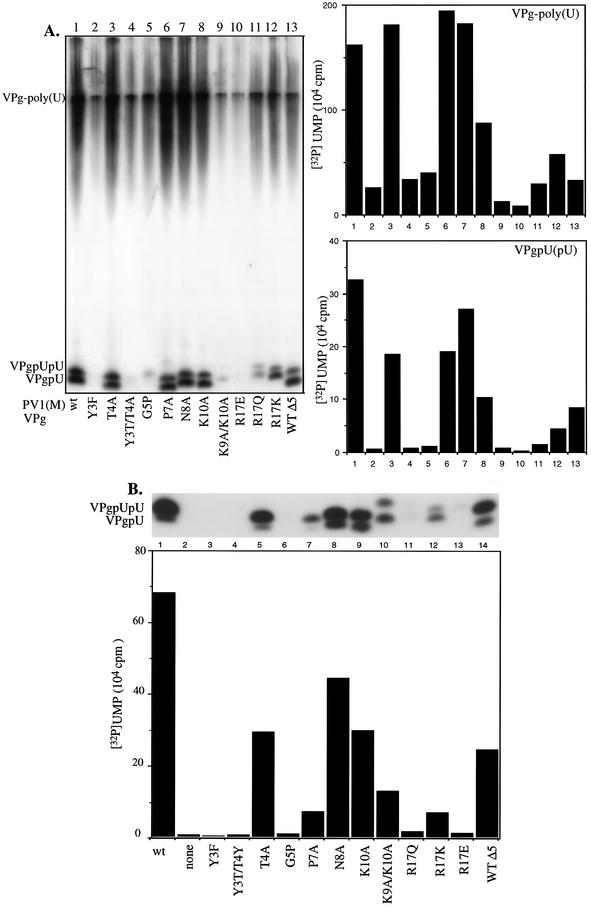
It has already been shown that the Y3F and R17E mutations in VPg interfere with uridylylation in vitro (37). In order to assess the effects of the other amino acid substitutions in PV1(M) VPg, we have now tested 10 additional synthetic mutant peptides in the in vitro reaction. Figure 3A shows a comparison of the wt and mutant VPg peptides as substrates for 3Dpol on a poly(A) template. In addition to the Y3F mutant, the Y3T T4Y mutant VPg peptide was also totally inactive in the reaction (Fig. 3A, compare lanes 2 and 4 with lane 1), confirming the importance of a tyrosine at the third position. Of the other mutant peptides, G5P, K9A K10A, R17E, and R17Q had <10% of wt activity (Fig. 3A, compare lanes 5, 9, 10, and 11 with lane 1). The mutant peptides K10A and R17K (Fig. 3A, compare lanes 8 and 12 with lane 1) exhibited significantly reduced activities compared to wt VPg. The substrate activities of the remaining mutant VPgs, T4A, P7A, and N8A (Fig. 3A, compare lanes 3, 6, and 7 with lane 1), were only slightly lower than that of the wt. Deletion of five amino acids from the C terminus of VPg (WT Δ5) was also found to be deleterious to VPg uridylylation (Fig. 3A, compare lane 13 with lane 1). Overall, the results of VPg-poly(U) synthesis paralleled those observed in the uridylylation reaction itself (Fig. 3A). This was expected, since VPgpU and VPgpUpU are precursors of the final product, VPg-linked poly(U) (37). The small amount of polymeric product made in the absence of VPg (data not shown) or with the inactive VPgs (Fig. 3A, lanes 2, 4, 9, and 10) is most likely due to the terminal uridylyltransferase activity of 3Dpol (41).
FIG. 3.
Effects of mutations in PV1(M) VPg on uridylylation in vitro. (A) Assay 1 (see Materials and Methods) was used with poly(A) as a template, except that the synthetic wt VPg peptide was replaced with a mutant peptide, as indicated. (B) Assay 2 (see Materials and Methods) was used with cre(2C) RNA as a template in the presence of 3CDpro, except that wt VPg was replaced by a mutant peptide, as indicated.
To determine if there is a change in the VPg specificity of 3Dpol when different templates are used for uridylylation, we repeated the experiments with transcripts of cre(2C) RNA in the presence of 3CDpro (Fig. 3B). Overall, the results obtained with the two templates were very similar except when VPg (P7A) was the substrate. This mutant peptide exhibited significantly lower activity in the assay with a cre(2C) template than with poly(A) (Fig. 3A, compare lane 6 with lane 1, and B, compare lane 7 with lane 1). The reason for this is not known. We also have no explanation for the aberrant migration of the upper band when the K9A K10A peptide is used as a substrate on the cre(2C) template. The unexpectedly slow movement of this band in the gel suggests that the product is a VPg-linked trinucleotide (VPgpUpUpU) rather than VPgpUpU.
Analysis of PV 3Dpol-VPg interaction by the yeast two-hybrid system.
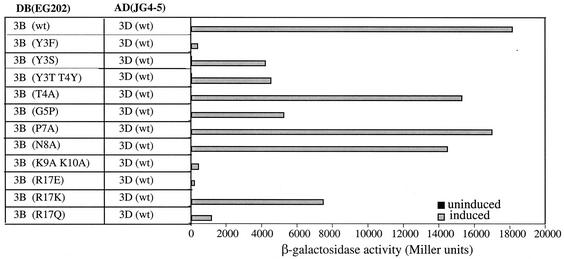
Strong interactions between PV 3AB and 3Dpol (18, 57) and between wt VPg and 3Dpol (57) in the yeast two-hybrid system have been demonstrated previously. To test the possibility that the deleterious effects of some of the VPg mutations on uridylylation were related to impaired interaction between 3Dpol and VPg molecules, we used the yeast two-hybrid system. The coding regions of 3Dpol and 3B were cloned into plasmids JG4-5 and EG202, respectively, to produce fusion proteins. Western blot analyses indicated that the proteins were expressed in comparable quantities (data not shown). As depicted in Fig. 4, the mutants fell into three groups. The first group of mutations (T4A, P7A, and N8A) had very little deleterious effect on the protein-protein interaction. The second group (Y3S, Y3T T4Y, G5P, and R17K) exhibited a two- to threefold reduction in the interaction between polymerase and VPg molecules. The final group included those amino acid changes which reduced the interaction by >90% (Y3F, K9A K10A, R17E, and R17Q). Overall, these results indicate a strong correlation between the abilities of the mutant VPg peptides to interact with 3Dpol and to function as substrates for VPg uridylylation (compare Fig. 4 with Fig. 3). One of the mutant VPgs (Y3S) shown in Fig. 4 was not tested in the in vitro uridylylation assay because the synthetic peptide was not available for analysis.
FIG. 4.
Yeast two-hybrid analysis of the interaction between wt PV 3Dpol and wt or mutant VPgs. Yeast strain EGY48 was transformed with the indicated DNA binding domain (DB)- and transcriptional activation domain (AD)-viral protein fusion plasmids (see Materials and Methods). Protein-protein interaction was quantitated by analysis of β-galactosidase activity.
Replacement of PV VPg with VPgs of HRVs.
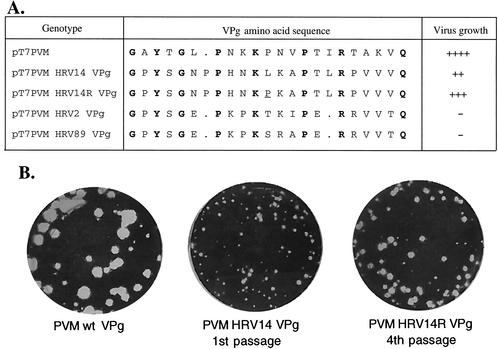
We have also determined the effects of multiple amino acid substitutions in VPg on viral growth and uridylylation by replacing the VPg of PV with those of three different HRVs (HRV14, HRV2, and HRV89). These viruses are members of the genus Rhinovirus in the family Picornaviridae. The VPg of HRV14 contains 23 amino acids, 1 more than the VPg of PV, and of these only 10 are common to both peptides (Fig. 5A) (20, 48). Transfection of HeLa cells with transcripts of a chimeric PV (pT7PVM HRV14 VPg) which contained the HRV14 VPg instead of the PV1(M) VPg resulted in delayed CPE (48 h) compared to the wt (20 h). A viable virus was produced which had a small-plaque phenotype (Fig. 5B). After several passages of this virus on HeLa cells, midsize plaque variants could be observed (Fig. 5B). Five of these variants from two different transfections were isolated and expanded on HeLa cells. The RNA was isolated, and following reverse transcription-PCR, the VPg sequences of the viruses were determined. All of the viruses contained a single nucleotide change, C→T, resulting in a leucine (CTA)-to-proline (CCA) substitution. This mutation regenerated the proline at position 11 of the wt PV1(M) VPg sequence (Fig. 5A).
FIG. 5.
Growth properties of wt and chimeric PVs containing rhinoviral VPgs. (A) The sequences of HRV14, HRV2, and HRV89 VPgs are compared to that of PV. Amino acids conserved among all four peptides are shown with boldface letters. Transfections with transcripts derived from pT7PVM HRV14 VPg, pT7PVM HRV2 VPg, and pT7PVM HRV89 VPg were carried out as described in Materials and Methods. Virus derived from two transfections with pT7PVM HRV14 VPg was passaged four times; several large-plaque variants were isolated, and their VPg sequences were determined. All revertants contained an amino acid change (L12P) in HRV14 VPg (underlined). (B) Plaque size of wt PV1(M) compared to those of PVM HRV14 VPg and its revertant. ++++, wt-type growth; +++, slower than wt growth; ++, much slower than wt growth; −, no growth. Dots indicate missing amino acids relative to the HRV14 VPg sequence.
There are only eight amino acids common to the VPgs of PV1(M) and of HRV2 and HRV89 (Fig. 5A) (10, 20, 47). The two rhinoviral VPgs are 21 amino acids long, and they differ from each other only in 3 amino acids at positions 11 to 13. They contain two negatively charged glutamic acids, which are absent in the other two viral VPgs. Transfection of HeLa cells with transcripts of the chimeric pT7PVM HRV2 VPg and pT7PVM HRV89 VPg did not yield viable virus, and no viable revertants could be isolated after five passages of the transfection supernatants on HeLa cell monolayers (data not shown).
Comparison of PV1(M) and HRV VPgs as substrates for PV1(M) 3Dpol in VPg uridylylation.
PV 3Dpol is able to utilize as a substrate, in addition to its own VPg, the VPg of HRV14 (39) (Fig. 6). The yield of VPgpU(pU) with HRV14 VPg is about threefold lower than with PV1(M) VPg when either poly(A) or cre(2C) RNA is used as a template. In contrast to HRV14 VPg, the VPgs of HRV2 and HRV89 have barely detectable substrate activities on both templates.
FIG. 6.
Comparison of PV and rhinovirus VPgs as substrates for uridylylation. (A) Assay 1 (see Materials and Methods) was used with poly(A) as a template, except that PV VPg was replaced with those of HRV14, HRV2, and HRV89, as indicated. (B) Assay 2 (see Materials and Methods) with cre(2C) RNA as a template was used in the presence of 3CDpro to test uridylylation with the viral VPgs, as indicated.
It was previously shown that both the free and uridylylated forms of viral VPgs [those of PV1(M), HRV2, HRV14, and HRV89] exhibit aberrant migration patterns in SDS-polyacrylamide gels (8, 13, 14, 26, 37, 39). Although the molecular masses of these peptides are only 2 kDa, their migration resembles that of a 7- to 12-kDA peptide. The reason for the aberrant migration pattern of the VPg peptides is not yet known.
Divalent-cation specificity of VPg uridylylation on a poly(A) template.
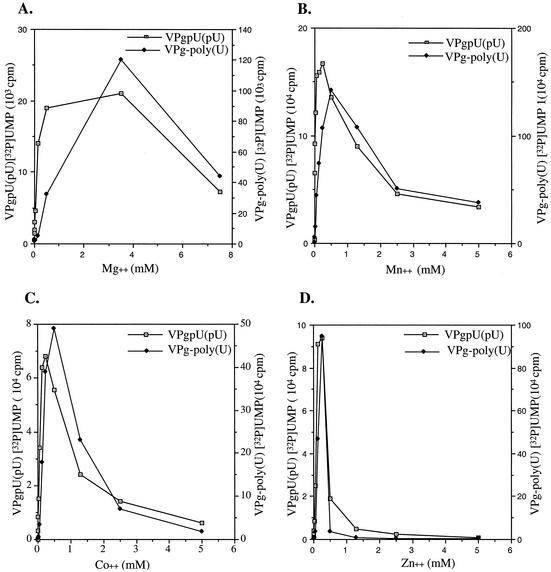
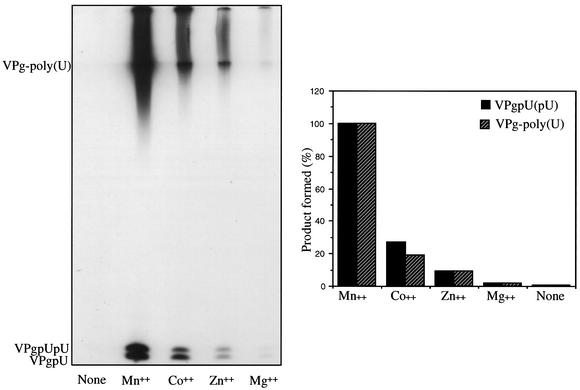
In our continuing studies of the biochemical characterization of the VPg uridylylation reaction, we have now carried out a detailed examination of the divalent-cation requirements of 3Dpol. Previous reports have dealt with the metal specificity of 3Dpol only in an oligonucleotide elongation reaction (4). As shown in Fig. 7, a number of transition metals function as activators of 3Dpol on a poly(A) template, whereas no products are formed in the absence of a divalent cation (Fig. 8). In the presence of Mg2+, optimal VPgpU(pU) synthesis occurs at 0.5 to 3.5 mM (Fig. 7A). The other cations exhibit optimal activity in a relatively narrow concentration range. The optimal concentrations of Mn2+ (Fig. 7B), Co2+ (Fig. 7C), and Zn2+ (Fig. 7D) are 0.1 to 0.25 mM. Overall, somewhat higher cation concentrations were required for the elongation of VPgpU(pU) into VPg-linked poly(U) than for the synthesis of the precursors. Some divalent cations, such as Ca2+, Fe2+, and Ni2+, are essentially inactive in the uridylylation reaction, and Ni2+ strongly inhibits stimulation by Mn2+ (data not shown). Measured at the optimal concentrations, the best activator of 3Dpol in the synthesis of both precursors and polymer on a poly(A) template is Mn2+, followed by Co2+, Zn2+, and Mg2+ (Fig. 8).
FIG. 7.
Effect of divalent cation concentration on the uridylylation of VPg. Assay 1 (see Materials and Methods) with a poly(A) template was used to determine the optimal divalent concentration required for VPg uridylylation. (A) Mg2+; (B) Mn2+; (C) Zn2+; (D) Co2+.
FIG. 8.
Metal specificity of VPg uridylylation. Assay 1 (see Materials and Methods) was used for VPg uridylylation, except that Mn2+ (0.5 mM) was replaced with Co2+ (0.5 mM), Zn2+ (0.25 mM), or Mg2+ (3.5 mM), as indicated. The amount of product obtained with Mn2+ was taken as 100%.
DISCUSSION
Previous work with mutants of PV identified several amino acid changes in the peptide that have very little or no effect on viral growth (L6M, K9Q, K10R, N12K, V13M, I16L, T18Q, and K20L), while other substitutions were lethal (Y3F, Y3S, T4Y, Y3T T4Y, R17E, R17Q, R17K, and A19K K20A) (21, 22, 43). It should be pointed out that all of these early mutants, which were made using a mutagenesis cartridge, contained an additional K10R mutation. Since the K10R change did not appear to have an effect on viral growth, it was assumed that the phenotype observed was due only to the mutations listed above. Subsequently, a new Y3F mutant without additional changes was made by Cao and Wimmer (7) and was found to be quasi-infectious. More recently, Xiang et al. (54) observed that the double mutation K9A K10A in VPg resulted in a nonviable growth phenotype. We have continued these studies with the construction of six new PV VPg mutants, which contained changes in amino acids highly conserved in the VPgs of either enteroviruses alone or both enteroviruses and rhinoviruses (Fig. 2A). Three of the mutants (T4A, N8A, and K10A) exhibited growth properties similar to the those of wt virus, one was growth impaired (P7A), and two were nonviable (G5P and Y3T T4Y). Glycine 5, which was replaced with a proline in one of the nonviable mutants, is one of the few amino acids fully conserved in the VPgs of both the genera Enterovirus and Rhinovirus. It is interesting that this glycine is part of a potential myristoylation signal in VPg (GXXXS/T/G) (35), although no myristoylated VPg has ever been observed in PV-infected HeLa cells. Further investigations are necessary to examine this phenomenon.
To determine whether the effect of these mutations on viral growth can be correlated with VPg uridylylation in vitro, we used synthetic VPg peptides as substrates for 3Dpol with either poly(A) or cre(2C) RNA as a template. Our results clearly show that viral growth is strictly dependent on VPg uridylylation. As expected, no VPgpU(pU) was formed in the absence of VPg or with the Y3F and Y3T T4Y peptides, in which the RNA-linking site was eliminated. Furthermore, the inability of T3 and Y4 in the Y3T T4Y peptide to function as an acceptor of UMP demonstrated the strict specificity of the polymerase for both the identity and location of the amino acid used for the linkage. The lethal growth phenotypes of the PV VPg mutants G5P, K9A K10A, R17E, R17K, and R17Q paralleled a lack of substrate activity of the corresponding mutant peptides in the in vitro assay. The reduced growth properties of virus derived from pT7PVM P7A VPg reflected the observed inhibitory effect of the P7A change in vitro. Three of the VPg variants (T4A, N8A, and K10A), which had growth properties comparable to the wt virus, exhibited nearly wt substrate activities.
In addition to the VPg mutants described above, we have also tested in the in vitro assay a peptide (WT Δ5) that lacked the five C-terminal amino acids (TAKVQ) of PV1(M) VPg. This shortened version exhibited only about one-half of the wt VPg activity, indicating either that the overall length of the peptide cannot be changed or that one or more of the deleted amino acids are important for function. The latter possibility is supported by the observation that a K20E substitution in VPg results in a virus with defective growth properties (24).
It was previously reported that the VPg of PV can be replaced with the VPg of an enterovirus, echovirus 9, and the resulting virus exhibits growth properties similar to those of wt PV1(M) (22). The VPg of echovirus 9 differs in only four amino acids (M6, K12, L16, and Q18) from PV VPg (L6, N12, I16, and T18). In this study, we replaced the VPg of PV with those of three different HRVs (HRV14, HRV2, and HRV89) belonging to the genus Rhinovirus. The VPg of HRV14 has only 45% homology with the VPg of PV, but it does contain the well-conserved amino acids of enterovirus VPgs (Y3, G5, P8, K11, K13, P15, and R18) located at either the same or nearby positions in the peptide (Fig. 2A). Surprisingly, such limited homology is sufficient for the functioning of HRV14 VPg both in vitro in VPg uridylylation with PV 3Dpol and in vivo in a PV chimera. The small-plaque virus produced reverted to a variant with an increased plaque size. This virus contained a single-nucleotide change, C→T, substituting a leucine (CTA) for a proline (CCA) at position 12 of the HRV14 VPg sequence. Although this observation might suggest that P11 has an important role in PV VPg function, that does not appear to be the case. A P11A VPg mutant of PV has properties similar to those of wt virus (Paul and Wimmer, unpublished). Therefore, either a P11L change is more detrimental than the P11A mutation or the presence of P11 is important for the growth of PV only in the background of HRV14 VPg.
The VPgs of HRV2 and HRV89 differ from each other only at positions 11 to 13, and they both share eight amino acids with PV VPg (Fig. 2A). Although they do contain the essential amino acids of PV (Y3, G5, K10, K12, and R17), the two rhinoviral VPgs are nonfunctional both in vitro as substrates for PV1(M) 3Dpol and in vivo in a PV chimera. This is most likely due to the presence of two negatively charged glutamic acids at positions 6 and 15, which are absent in both the PV and HRV14 VPgs.
HRVs have been divided recently into serotypes A and B, a taxonomy based on sequences rather than receptor use (46). All serotype A rhinoviruses (e.g., HRV2 and HRV89) appear to carry two glutamic acid residues in their VPgs (Fig. 2), which makes their VPgs incompatible with uridylylation by PV proteins. At present, the complete sequence of only one serotype B rhinovirus (HRV14) is known. It remains to be seen whether the VPgs of this subgroup are all substrates for PV 3Dpol-catalyzed uridylylation.
It is interesting that the RNA polymerase of HRV2 exhibits less strict substrate specificity than PV1 3Dpol. Unlike PV 3Dpol, the rhinoviral enzyme is functional in the uridylylation of all four viral VPgs [those of PV1(M), HRV2, HRV89, and HRV14] (13, 14). When wt or mutant PV1(M) VPg is used as a substrate, the two RNA polymerases have very similar substrate specificities and they require the same amino acids (Y3, G5, K9 K10, and R17) to function (13, 14).
Our results indicate that of the 15 amino acids tested thus far in PV VPg, only a few (Y3, G5, K9 K10, and R17) are essential for viral growth, and these are also required for VPg uridylylation in vitro. Although the exact roles of these amino acids for VPg function in uridylylation are not yet known, one might predict that they are involved in an interaction with either the polymerase or the UTP, or with both. It has been previously shown that both VPg (57) and its precursor, 3AB (18), interact with the polymerase and that the 3B domain of the protein is the primary determinant of the interaction (57). The yeast two-hybrid analyses reported in this study indicate that at least one of the functions of these essential amino acids in VPg is to promote an interaction with the polymerase. All of the VPg mutants impaired in interaction with 3Dpol are also defective in VPg uridylylation. That the formation of a complex between these proteins is a prerequisite for optimal uridylylation has also been shown by Lyle et al. (28), who identified four amino acids on the surface of the polymerase that are required both for 3AB binding and for VPg uridylylation.
An interaction between 3Dpol or 3CDpro and 3AB is known to have at least five other functions in vitro besides VPg uridylylation: (i) 3AB stimulates the oligonucleotide elongation activity of the polymerase (23, 36), (ii) 3AB promotes the formation of a specific complex with 3CDpro on the 5′-terminal cloverleaf (17, 55), (iii) purified 3AB enhances the self-processing of proteinase 3CDpro (31), (iv) 3CDpro cleaves membrane-bound 3AB to 3A and VPg (23), and (v) 3AB binds 3Dpol to the membranes (28). It is interesting that in the first two of these functions the K9 K10 and R17 residues of the VPg domain of 3AB are required not only for interaction with the polymerase but also for the protein's RNA binding activity (54).
The question of which of these amino acids in VPg are involved in binding UTP is not yet answered. We have previously suggested the possibility that the positively charged guanidinium group of R17 might serve this function (37), since this amino acid is known to be an important binding site for anionic ligands in different enzymes. Indeed, a mutation, R17K, proved lethal for PV replication (22). It has been shown that the protein-priming reaction catalyzed by the DNA polymerase of phage PRD1 requires an arginine of the terminal protein for binding the phosphate groups of the donor triphosphate (19).
Some amino acids of VPg that are essential for viral growth might have functions that are not directly involved in RNA replication. It is known that the N-terminal G and the C-terminal Q of VPg, which constitute part of the 3Cpro-specific Q-G cleavage site, and the A at the P4 position (A19) are expected to be required for normal processing of the 3AB and 3BC precursors.
During the past few years, a great deal of information has accumulated about the protein and RNA factors involved in the initiation of PV RNA replication. The role of cre(2C) as a template for VPg uridylylation has been firmly established (39, 44; Yin et al., submitted), and the specific binding of 3CDpro to the upper stem of this RNA element in vitro has now also been demonstrated (Yin et al., submitted). Our studies reported in this paper and those of Lyle et al. (28) clearly show that the formation of a complex between 3Dpol and VPg is a prerequisite for VPg uridylylation. In addition, recent results of Pathak et al. (34) indicate that 3Cpro can replace 3CDpro in the reaction, suggesting that an interaction between 3Cpro and 3Dpol may also be involved in the process. Taken together, these findings are consistent with a basic model for the initiation reaction in which 3CDpro (3Cpro) interacts with a complex of 3Dpol-VPg and enhances its binding to the cre(2C) RNA structure. The RNA polymerase then uridylylates VPg, using the first A of the conserved AAA sequence in the loop of the hairpin as a template (39, 40, 44; Yin et al., submitted). Whether the VPgpU precursors produced in this reaction are translocated to the 3′ end of the poly(A) tail (39, 40) to be used as the primers for minus-strand RNA synthesis or serve as primers during plus-strand synthesis remains to be determined.
Another important unanswered question about the initiation of RNA replication is the identity of the protein being uridylylated in vivo. Although VPg functions well as a substrate in vitro, it is required at a molar concentration about 50-fold higher than that of 3Dpol (37, 39). Therefore, it is possible that a precursor of VPg with higher affinity for 3Dpol, 3CDpro, or the cre(2C) RNA than the peptide itself is the true substrate for uridylylation in vivo. Thus far our attempts to uridylylate purified 3AB or 3ABC in vitro have failed (Paul and Wimmer, unpublished). The other possible VPg precursor, 3BC(D), remain to be tested in vitro. In any case, our results with the 3A-3B and 3B-3C cleavage site mutants indicate that linkage of 3AB or 3BC to PV RNA is not compatible with viral growth. It is interesting, however, that a 3B-3C cleavage site mutant of encephalomyocarditis virus (EMCV), a cardiovirus, was found to exhibit a low level of RNA synthesis after transfection, and either 3BC or 3ABC could be detected associated with the progeny virus RNA. The difference between the two sets of results might be related to a different processing cascade of P3 in EMCV- and PV-infected cells. In the case of EMCV, the processing of P3 results primarily in the formation of a 3ABC precursor (16), while in PV-infected cells, the predominant processing products of P3 are 3AB and 3CD (25).
It has been known for many years that DNA and RNA polymerases require divalent cations for optimal activity. The metal believed to be the true cofactor for polymerases in vivo is Mg2+, which is also the cation most frequently used in assays measuring the elongation of oligonucleotides. Recently, Arnold et al. (4) determined the metal specificity of PV 3Dpol in the elongation of oligonucleotide primers on short homopolymeric templates. These studies indicated the following preferences of the enzyme: Mn2+ > Co2+ > Ni2+ > Fe2+ > Mg2+ > Ca2+ > Cu2+ > Zn2+. The enzyme's activity with this template-primer is 10 times higher with Mn2+ than with Mg2+, and this was attributed to a reduced Km of 3Dpol for the template-primer. The VPg-uridylylating activity of 3Dpol is quite different from its oligonucleotide elongation activity in that in the former case the enzyme uses the hydroxyl group of a tyrosine instead of the hydroxyl group of a sugar as the nucleotide acceptor. Therefore, it is not surprising that the metal specificity of the enzyme in the protein-priming reaction on a poly(A) template is different: Mn2+ > Co2+ > Zn2+ > Mg2+. Other transition metals, such as Ni2+, Fe2+, and Cu2+, had no detectable stimulatory activities in the in vitro assay. The stimulatory activity of Mn2+ compared to Mg2+ is even more striking in the VPg uridylylation reaction and VPg-poly(U) synthesis than in the oligonucleotide elongation assay. The very large difference in the activity of 3Dpol with the two cations is similar to what has been observed in other protein-priming systems, such as HRV2 3Dpol (13), or the polymerases of DNA viruses, such as adenovirus (42), bacteriophage Ф29 (11), or PRD1 (6). In contrast to this reaction, in which poly(A) is the template, Mg2+ and Mn2+ are about equally active as cofactors when VPg is uridylylated on the cre(2C) RNA in the presence of 3CDpro (39).
ADDENDUM IN PROOF
Our recent studies indicate that the change in PV VPg of proline 14, which is fully conserved in both enteroviruses and rhinoviruses, to an alanine, resulted in quasi-infectious RNA. Progeny virus had regained the wt sequence in five independent isolates.
Acknowledgments
We are grateful to K. Kirkegaard for the generous gift of plasmid pT5T3D. We thank F. Maggiore and the late J. Owen for expert technical assistance.
This work was supported by grant AI15122 from the National Institute of Allergy and Infectious Diseases.
REFERENCES
- 1.Agol, V. I., A. V. Paul, and E. Wimmer. 1999. Paradoxes of the replication of picornaviral genomes. Virus Res. 62:129-147. [DOI] [PubMed] [Google Scholar]
- 2.Ambros, V., and D. Baltimore. 1978. Protein is linked to the 5′ end of poliovirus RNA by a phosphodiester linkage to tyrosine. J. Biol. Chem. 253:5263-5266. [PubMed] [Google Scholar]
- 3.Andino, R., G. E. Rieckhof, P. L. Achacoso, and D. Baltimore. 1993. Poliovirus RNA synthesis utilizes an RNP complex formed around the 5′-end of viral RNA. EMBO J. 12:3587-3598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Arnold, J. J., S. K. B. Ghosh, and C. E. Cameron. 1999. Poliovirus RNA-dependent RNA polymerase (3Dpol). Divalent cation modulation of primer, template, and nucleotide selection. J. Biol. Chem. 274:37060-37069. [DOI] [PubMed] [Google Scholar]
- 5.Bienz, K., D. Egger, T. Pfister, and M. Troxler. 1992. Structural and functional characterization of the poliovirus replication complex. J. Virol. 66:2740-2747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Caldentey, J., L. Blanco, H. Savilahti, D. H. Bamford, and M. Salas. 1992. In vitro replication of bacteriophage PRD1 DNA. Metal activation of protein-primed initiation and DNA elongation. Nucleic Acids Res. 20:3971-3976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cao, X., and E. Wimmer. 1995. Intergenomic complementation of a 3AB mutant in dicistronic polioviruses. Virology 209:315-326. [DOI] [PubMed] [Google Scholar]
- 8.Crawford, N. M., and D. Baltimore. 1983. Genome-linked protein VPg of poliovirus is present as free VPg and VPgpUpU in poliovirus-infected cells. Proc. Natl. Acad. Sci. USA 80:7452-7455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dreef-Tromp, C. M., H. van der Elst, J. E. van den Boogaart, G. A. van der Marel, and J. H. van Boom. 1992. Solid-phase synthesis of an RNA nucleopeptide fragment from the nucleoprotein of poliovirus. Nucleic Acids Res. 20:2435-2439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Duechler, M., T. Skern, W. Sommergruber, C. Neubauer, P. Gruendler, I. Fogy, D. Blaas, and E. Kuechler. 1987. Evolutionary relationships within the human rhinovirus genus: comparison of serotypes 89, 2 and 14. Proc. Natl. Acad. Sci. USA 84:2605-2609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Esteban, J. A., A. Bernad, M. Salas, and L. Blanco. 1992. Metal activation of synthetic and degradative activities of Ф29 DNA polymerase, a model enzyme for protein-primed DNA replication. Biochemistry 31:350-359. [DOI] [PubMed] [Google Scholar]
- 12.Flanegan, J. B., and D. Baltimore. 1977. Poliovirus-specific primer-dependent RNA polymerase able to copy poly(A). Proc. Natl. Acad. Sci. USA 74:3677-3680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gerber, K., E. Wimmer, and A. V. Paul. 2001. Biochemical and genetic studies of the initiation of human rhinovirus 2 RNA replication: Purification and enzymatic analysis of the RNA-dependent RNA polymerase 3Dpol. J. Virol. 75:10969-10978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gerber, K., E. Wimmer, and A. V. Paul. 2001. Biochemical and genetic studies of the initiation of human rhinovirus 2 RNA replication: identification of a cis-replicating element in the coding sequence of 2Apro. J. Virol. 75:10979-10990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Goodfellow, I., Y. Chaudhry, A. Richardson, J. Meredith, J. W. Almond, W. Barclay, and D. J. Evans. 2000. Identification of a cis-acting replication element within the poliovirus coding region. J. Virol. 74:4590-4600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hall, D. J., and A. C. Palmenberg. 1996. Cleavage site mutations in the encephalomyocarditis virus P3 region lethally abrogate the normal processing cascade. J. Virol. 70:5954-5961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Harris, K. S., W. Xiang, L. Alexander, A. V. Paul, W. S. Lane, and E. Wimmer. 1994. Interaction of the polioviral polypeptide 3CDpro with the 5′ and 3′ termini of the poliovirus genome: identification of viral and cellular cofactors necessary for efficient binding. J. Biol. Chem. 269:27004-27014. [PubMed] [Google Scholar]
- 18.Hope, D. A., S. E. Diamond, and K. Kirkegaard. 1997. Genetic dissection of interaction between poliovirus 3D polymerase and viral protein 3AB. J. Virol. 71:9490-9498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hsieh, J.-C., S.-K. Yoo, and J. Ito. 1990. An essential arginine residue for initiation of protein-primed DNA replication. Proc. Natl. Acad. Sci. USA 87:8665-8669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kitamura, N., B. L. Semler, P. G. Rothberg, G. R. Larsen, C. J. Adler, A. J. Dorner, E. A. Emini, R. Hanecak, J. J., Lee, S. van der Werf, C. W. Anderson, and E. Wimmer. 1981. Primary structure, gene organization and polypeptide expression of poliovirus RNA. Nature 291:547-553. [DOI] [PubMed] [Google Scholar]
- 21.Kuhn, R. J., H. Tada, M. F. Ypma-Wong, J. J. Dunn, B. L. Semler, and E. Wimmer. 1988. Construction of a “mutagenesis cartridge” for poliovirus genome-linked viral protein: isolation and characterization of viable and nonviable mutants. Proc. Natl. Acad. Sci. USA 85:519-523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kuhn, R. J., H. Tada, M. F. Ypma-Wong, B. L. Semler, and E. Wimmer. 1988. Mutational analysis of the genome-linked protein VPg of poliovirus. J. Virol. 62:4207-4215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lama, J., A. V. Paul, K. S. Harris, and E. Wimmer. 1994. Properties of purified recombinant poliovirus protein 3AB as substrate for viral proteinases and as a cofactor for viral polymerase 3Dpol. J. Biol. Chem. 269:66-70. [PubMed] [Google Scholar]
- 24.Lama, J., M. A. Sanz, and P. L. Rodriguez. 1995. A role for 3AB protein in poliovirus genome replication. J. Biol. Chem. 270:14430-14438. [DOI] [PubMed] [Google Scholar]
- 25.Lawson, M. A., and B. L. Semler. 1992. Alternate poliovirus nonstructural protein processing cascades generated by primary sites of 3C proteinase cleavage. Virology 191:309-320. [DOI] [PubMed] [Google Scholar]
- 26.Lee, Y. F., A. Nomoto, B. M. Detjen, and E. Wimmer. 1977. A protein covalently linked to poliovirus genome RNA. Proc. Natl. Acad. Sci. USA 74:59-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lobert, P. E., N. Escriou, J. Ruelle, and T. Michiels. 1999. A coding RNA sequence acts as a replication signal in cardioviruses. Proc. Natl. Acad. Sci. USA 96:11560-11565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lyle, J. M., A. Clewell, K. Richmond, O. C. Richards, D. A. Hope, S. C. Schultz, and K. Kirkegaard. 2002. Similar structural basis for membrane localization and protein priming by an RNA-dependent RNA polymerase. J. Biol. Chem. 277:16324-16331. [DOI] [PubMed] [Google Scholar]
- 29.McKnight, K. L., and S. M. Lemon. 1998. The rhinovirus type 14 genome contains an internally located RNA structure that is required for viral replication. RNA 4:1569-1584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Molla, A., A. V. Paul, and E. Wimmer. 1991. Cell-free, de novo synthesis of poliovirus. Science 254:1647-1651. [DOI] [PubMed] [Google Scholar]
- 31.Molla, A., K. S. Harris, A. V. Paul, S. H. Shin, J. Mugavero, and E. Wimmer. 1994. Stimulation of 3Cpro-related proteolysis by the genome-linked protein VPg and its precursor 3AB. J. Biol. Chem. 269:27015-27020. [PubMed] [Google Scholar]
- 32.Nomoto, A., B. Detjen, R. Pozzatti, and E. Wimmer. 1977. The location of the polio genome protein in viral RNAs and its implication for RNA synthesis. Nature 268:208-213. [DOI] [PubMed] [Google Scholar]
- 33.Pata, J. D., S. C. Schultz, and K. Kirkegaard. 1995. Functional oligomerization of poliovirus RNA-dependent RNA polymerase. RNA 1:466-477. [PMC free article] [PubMed] [Google Scholar]
- 34.Pathak, H. B., S. K. B. Ghosh, A. W. Roberts, S. D. Sharma, J. D. Yoder, J. J. Arnold, D. W. Gohara, D. J. Barton, A. V. Paul, and C. Cameron. 2002. Structure-function relationships of the RNA-dependent RNA polymerase from poliovirus (3Dpol). J. Biol. Chem. 277:31551-31562. [DOI] [PubMed] [Google Scholar]
- 35.Paul, A. V., A. Schultz, S. E. Pincus, S. Oroszlan, and E. Wimmer. 1987. Capsid protein of VP4 of poliovirus is N-myristoylated. Proc. Natl. Acad. Sci. USA 84:7827-7831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Paul, A. V., X. Cao, K. S. Harris, J. Lama, and E. Wimmer. 1994. Studies with poliovirus polymerase 3Dpol: stimulation of poly(U) synthesis in vitro by purified poliovirus protein 3AB. J. Biol. Chem. 269:29173-29181. [PubMed] [Google Scholar]
- 37.Paul, A. V., J. H. van Boom., D. Filippov, and E. Wimmer. 1998. Protein-primed RNA synthesis by purified poliovirus RNA polymerase. Nature 393:280-284. [DOI] [PubMed] [Google Scholar]
- 38.Paul, A. V., J. Mugavero, J. Yin, S. Hobson, S. Schultz, J. H. van Boom, and E. Wimmer. 2000. Studies on the attenuation phenotype of polio vaccines: poliovirus RNA polymerase derived from Sabin type 1 sequence is temperature sensitive in the uridylylation of VPg. Virology 272:72-84. [DOI] [PubMed] [Google Scholar]
- 39.Paul, A. V., E. Rieder, D. W. Kim, J. H. van Boom, and E. Wimmer. 2000. Identification of an RNA hairpin in poliovirus RNA that serves as the primary template in the in vitro uridylylation of VPg. J. Virol. 74:10359-10370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Paul, A. V. 2002. Possible unifying mechanism of picornavirus genome replication, p. 227-246. In B. L. Semler and E. Wimmer (ed.), Molecular biology of picornaviruses. ASM Press, Washington, D.C.
- 41.Plotch, S. J., O. Palant, and Y. Gluzman. 1989. Purification and properties of poliovirus RNA polymerase expressed in Escherichia coli. J. Virol. 63:216-225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pronk, R., W. van Dreil, and P. C. van der Vliet. 1994. Replication of adenovirus DNA in vitro is ATP-independent. FEBS Lett. 337:33-38. [DOI] [PubMed] [Google Scholar]
- 43.Reuer, Q., R. J. Kuhn, and E. Wimmer. 1990. Characterization of poliovirus clones containing lethal and nonlethal mutations in the genome-linked protein VPg. J. Virol. 64:2967-2975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rieder, E., A. V. Paul, D. W. Kim, J. H. van Boom, and E. Wimmer. 2000. Genetic and biochemical studies of poliovirus cis-acting replication element cre in relation to VPg uridylylation. J. Virol. 74:10371-10380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rothberg, P. G., T. J. R. Harris, A. Nomoto, and E. Wimmer. 1978. O4-(5′-uridylyl)tyrosine is the bond between the genome-linked protein and the RNA of poliovirus. Proc. Natl. Acad. Sci. USA 75:4868-4872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Savolainen, C., S. Blomqvist, M. N. Mulders, and T. Hovi. 2002. Genetic clustering of all 102 human rhinovirus prototype strains: serotype 87 is close to human enterovirus 70. J. Gen. Virol. 83:333-340. [DOI] [PubMed] [Google Scholar]
- 47.Skern, T., W. Sommergruber, D. Blaas, P. Gruendler, F. Fraundorfer, C. H. Pieler, I. Fogy, and E. Kuechler. 1985. Human rhinovirus 2: complete nucleotide sequence and proteolytic processing signals in the capsid protein region. Nucleic Acids Res. 13:2111-2126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Stanway, G., P. J. Hughes, R. C. Mountford, P. D. Minor, and J. W. Almond. 1984. The complete nucleotide sequence of a common cold virus: human rhinovirus 14. Nucleic Acids Res. 12:7859-7875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Takegami, T., R. J. Kuhn, C. W. Anderson, and E. Wimmer. 1983. Membrane-dependent uridylylation of the genome-linked protein VPg of poliovirus. Proc. Natl. Acad. Sci. USA 80:7447-7451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Towner, J. S., T. V. Ho, and B. L. Semler. 1996. Determinants of membrane association for poliovirus protein 3AB. J. Biol. Chem. 271:26810-26818. [DOI] [PubMed] [Google Scholar]
- 51.Towner, J. S., M. M. Mazanet, and B. L. Semler. 1998. Rescue of defective poliovirus RNA replication by 3AB-containing precursor polyproteins. J. Virol. 72:7191-7200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Toyoda, H., C. F. Yang., N. Takeda, A. Nomoto, and E. Wimmer. 1987. Analysis of RNA synthesis of type 1 poliovirus by using an in vitro molecular genetic approach. J. Virol. 61:2816-2822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Van der Werf, S., J. Bradley, E. Wimmer, F. W. Studier, and J. J. Dunn. 1986. Synthesis of infectious poliovirus RNA by purified T7 RNA polymerase. Proc. Natl. Acad. Sci. USA 83:2330-2334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Xiang, W., A. Cuconati, A. V. Paul, X. Cao, and E. Wimmer. 1995. Molecular dissection of the multifunctional poliovirus RNA-binding protein 3AB. RNA 1:892-904. [PMC free article] [PubMed] [Google Scholar]
- 55.Xiang, W., K. S. Harris, L. Alexander, and E. Wimmer. 1995. Interaction between the 5′-terminal cloverleaf and 3AB/3CDpro of poliovirus is essential for RNA replication. J. Virol. 69:3658-3667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Xiang, W., A. V. Paul, and E. Wimmer. 1997. RNA signals in entero- and rhinovirus genome replication. Semin. Virol. 8:256-273. [Google Scholar]
- 57.Xiang, W., A. Cuconati, D. Hope, K. Kirkegaard, and E. Wimmer. 1998. Complete protein linkage map of poliovirus P3 proteins: interaction of polymerase 3Dpol with VPg and with genetic variants of 3AB. J. Virol. 72:6732-6741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yang, Y., R. Rijnbrand, K. L. McKnight, E. Wimmer, A. Paul, A. Martin, and S. M. Lemon. 2002. Sequence requirements for viral RNA replication and VPg uridylylation directed by the internal cis-acting replication element (cre) of human rhinovirus type 14. J. Virol. 76:7485-7494. [DOI] [PMC free article] [PubMed] [Google Scholar]