Abstract
The Sindbis-group alphavirus S.A.AR86 encodes a threonine at nonstructural protein 1 (nsP1) 538 that is associated with neurovirulence in adult mice. Mutation of the nsP1 538 Thr to the consensus Ile found in nonneurovirulent Sindbis-group alphaviruses attenuates S.A.AR86 for adult mouse neurovirulence, while introduction of Thr at position 538 in a nonneurovirulent Sindbis virus background confers increased neurovirulence (M. T. Heise et al., J. Virol. 74:4207-4213, 2000). Since changes in the viral nonstructural region are likely to affect viral replication, studies were performed to evaluate the effect of Thr or Ile at nsP1 538 on viral growth, nonstructural protein processing, and RNA synthesis. Multistep growth curves in Neuro2A and BHK-21 cells revealed that the attenuated s51 (nsP1 538 Ile) virus had a slight, but reproducible growth advantage over the wild-type s55 (nsP1 538 Thr) virus. nsP1 538 lies within the cleavage recognition domain between nsP1 and nsP2, and the presence of the attenuating Ile at nsP1 538 accelerated the processing of S.A.AR86 nonstructural proteins both in vitro and in infected cells. Since nonstructural protein processing is known to regulate alphavirus RNA synthesis, experiments were performed to evaluate the effect of Ile or Thr at nsP1 538 on viral RNA synthesis. A combination of S.A.AR86-derived reporter assays and RNase protection assays determined that the presence of Ile at nsP1 538 led to earlier expression from the viral 26S promoter without affecting viral minus- or plus-strand synthesis. These results suggest that slower nonstructural protein processing and delayed 26S RNA synthesis in wild-type S.A.AR86 infections may contribute to the adult mouse neurovirulence phenotype of S.A.AR86.
Experimental Sindbis-group alphavirus infection of mice is a valuable system for studying virus and host determinants that contribute to disease. Previous studies have shown that both the genetics of the virus and the age of the mouse can affect the course of disease (reviewed in reference 11). Suckling mice infected with virulent Sindbis-group alphaviruses develop a lethal shock-like disease that is attenuated in older animals or if cell culture-adapted viruses are used (14, 15, 30). With a few notable exceptions, most Sindbis virus strains are unable to cause disease in mice greater than 10 to 14 days of age, even by the intracerebral (i.c.) route (reviewed in reference 11). However, S.A.AR86, a Sindbis-group alphavirus isolated in South Africa, does cause lethal disease in adult mice following i.c. inoculation (9, 28, 34). This ability to cause lethal disease in adult immunocompetent mice makes S.A.AR86 a valuable tool for studying viral and host determinants that contribute to viral encephalitis.
The alphavirus nonstructural proteins comprise the viral RNA-dependent RNA polymerase, and the individual nonstructural proteins have several known functions in viral RNA synthesis. nsP1 exhibits methyltransferase and guanyltransferase activity and is involved in viral RNA capping (23). nsP2 is a multifunctional protein that exhibits RNA helicase activity (5) and also plays a role in capping viral RNAs (33). Furthermore, nsP2 contains a protease domain in its C terminus that is responsible for processing of the mature nonstructural proteins from polyprotein precursors (1, 8). nsP3 is a phosphoprotein of unknown function, although it is essential for viral RNA replication (20), while nsP4 is the core viral RNA-dependent RNA polymerase (12). After viral entry and uncoating, the viral genome acts as an mRNA from which the nonstructural proteins are translated. The nonstructural proteins are produced as polyprotein precursors, which are then cleaved by the nsP2 protease to produce the mature nonstructural proteins nsP1, nsP2, nsP3, and nsP4. Most Sindbis-group alphaviruses have an opal termination codon at the 3′ end of nsP3, which truncates translation to produce the polyprotein precursor P123. Readthrough of the opal termination codon results in production of P1234, which includes all four nonstructural proteins and is present at approximately 20% of the level of P123 (reviewed in reference 29). S.A.AR86 is unique among the Sindbis-group alphaviruses in that it lacks an opal termination codon and should overproduce P1234 (28). The processing of the nonstructural polyprotein precursors into their mature forms plays an important role in regulating viral RNA synthesis (reviewed in reference 29). Early in infection, P123 plus nsP4, which is cleaved from P1234 immediately posttranslationally, is responsible for viral minus-strand synthesis (17-19, 27). Later in the course of infection, P123 is processed to produce mature nsP1, nsP2, and nsP3, which leads to a shutoff of minus-strand RNA synthesis and upregulation of plus-strand and 26S RNA synthesis (19, 27).
Although the viral nonstructural proteins play an essential role in viral RNA synthesis, their role in alphavirus pathogenesis has not been extensively investigated. Previous studies with the NSV strain of Sindbis virus, which causes 100% mortality following i.c. inoculation of 3- to 4-week-old mice (21), demonstrated a role for the viral glycoproteins in the pathogenesis of encephalitis (21). In particular, a histidine residue at position 55 of the E2 glycoprotein was shown to play a significant role in the neurovirulence of NSV (21). The actual mechanism of E2 His 55's action is not known, although it is likely that the mutation acts at the level of viral binding or entry into the host cell (31). In addition to His 55, other determinants within the viral glycoproteins clearly play a role in both neuroinvasion and neurovirulence (21). The 5′ untranslated region also contributes to neuroinvasion and the development of encephalitis (2, 16). Recent studies with S.A.AR86 (9) and Semliki Forest virus (32) demonstrated a role for the viral nonstructural proteins in adult mouse neurovirulence. In S.A.AR86, a single amino acid change at nsP1 position 538 dramatically reduces adult mouse neurovirulence (9). Replacement of Thr at nsP1 538 in S.A.AR86 with Ile attenuated the virus for neurovirulence in adult mice, while introduction of Thr at nsP1 538 in a nonneurovirulent Sindbis-group virus led to an increase in neurovirulence. The attenuating effect of Ile at nsP1 538 was not due to an overt defect in viral growth, since viruses containing Ile at nsP1 538 grew as well as wild-type S.A.AR86 both in vivo and in vitro. However, the mutant s51 (nsP1 538 Ile) virus did appear to be cleared more rapidly from the brains of infected mice than the wild-type s55 virus (9).
In this report, we have examined the proximal cause of the s51 phenotype. nsP1 538 is located in the cleavage recognition motif between nsP1 and nsP2 (reviewed in reference 29), suggesting that changes at this site might alter viral nonstructural protein processing and subsequently affect viral RNA synthesis. Although previous studies have demonstrated that the attenuating mutation at nsP1 538 does not adversely affect viral growth at a high multiplicity of infection (MOI) (9), we were interested in evaluating the effect of Ile or Thr at nsP1 538 on viral nonstructural protein processing and RNA synthesis. Using a combination of in vitro translation and immunoprecipitation of nonstructural proteins from S.A.AR86-infected cells, we demonstrated that the presence of the attenuating Ile at nsP1 538 accelerates processing of the nonstructural protein precursor into mature nonstructural proteins. This subsequently leads to the more rapid induction of viral 26S RNA synthesis by the attenuated mutant. Potential effects of this change in viral RNA synthesis on S.A.AR86 neurovirulence are discussed.
MATERIALS AND METHODS
Viruses and virus stocks.
Throughout, plasmids containing the viral cDNA are designated with the prefix “p.” This prefix is removed when referring to infectious virus derived from the cDNA clone: i.e., s55 represents virus derived from plasmid ps55. Clone ps55 represents a fully virulent infectious clone on the S.A.AR86 background, which has been described previously (28), and s55 exhibits all of the known biological characteristics of natural S.A.AR86 isolates (28, 34). Clone ps51 differs from clone ps55 at a single nucleotide (position 1672), where ps55 has cytidine and ps51 has thymidine. This nucleotide difference results in a single amino acid change at position 538 of nsP1, where ps51 encodes an isoleucine and ps55 encodes a threonine found in natural S.A.AR86 isolates (28). Clone pTR339 represents a consensus sequence based on the original Sindbis virus AR339 strain, in which cell culture-derived mutations have been corrected (14, 22). Clone p39ns1 was derived from pTR339 as described previously (9). p39ns1 differs from pTR339 at a single codon at nsP1 position 538, where p39ns1 encodes Thr, while pTR339 encodes an Ile.
Virus stocks were made as decribed previously (28). In brief, full-length transcripts were derived from linearized plasmids containing viral cDNA templates by using SP6-specific mMessage Machine in vitro transcription kits (Ambion). Transcripts were electroporated into BHK-21 cells grown in minimal essential medium (MEM; 10% fetal calf serum [Gibco], 10% tryptose phosphate broth [Sigma], and 0.29 mg of l-glutamine per ml [Biofluids]). Supernatants were harvested and frozen in 1-ml aliquots at 24 to 27 h after electroporation, and virus was titrated on BHK-21 cells as previously described (28).
S.A.AR86-derived replicons have been described previously (10). REP89-GFP refers to a replicon based on the wild-type S.A.AR86 genome with Thr at nsP1 position 538 and the gene for green fluorescent protein (GFP) expressed in place of the S.A.AR86 structural protein genes. REP91-GFP is identical to REP89-GFP, except for a single change at position 538 of nsP1, where REP91-GFP encodes an Ile. Replicons were packaged and titrated in BHK-21 cells as previously described (10).
Cell culture.
BHK-21 cells were maintained at 37°C in α-MEM (Gibco) supplemented with 10% donor calf serum (DCS), 10% tryptose phosphate broth, and 0.29 mg of l-glutamine per ml. Primary cultures of rat cortical neurons were harvested from fetal rats at 18 days postgestation. The cortex was isolated and incubated in 2.4 U of Dispase II per ml and 2 U of DNase per ml in calcium magnesium-free Hank's balanced salt solution for 15 min at 37°C. The neurons were then dissociated by several rounds of gentle trituration and transferred to complete medium (Dulbecco's MEM [DMEM] containing 10% fetal bovine serum and 20 μg of gentamicin per ml). The final suspension was plated at 105 cells per well in 24-well dishes (5.6 × 104 cells per cm2) coated with 0.1 mg of poly-d-lysine per ml. The neurons were maintained in complete medium and used in experiments at 7 days postharvest. Plaque assays of virus stocks and in vitro growth curve experiments were performed as previously described (28). For growth curves, BHK-21 cells were plated at 105 cells per well in 24-well plates (Sarstedt) for 14 to 16 h at 37°C. Medium was removed, and wells were infected with virus in triplicate at a multiplicity of infection (MOI) of 5.0 or 0.01. Alternatively, Neuro2A cells (ATCC CCL-131) were plated at 105 cells per well in 24-well plates for 48 h prior to virus infection at an MOI of 5.0 or 0.01. Cells were incubated at 37°C for 1 h. Wells were then washed three times with 0.5 to 1 ml of room temperature phosphate-buffered saline (PBS) supplemented with 1% DCS and Ca2+/Mg2+. One milliliter of growth medium was then added to each well, and cells were incubated at 37°C. Samples of supernatant were removed at various time points, with an equal volume of fresh medium added to maintain constant volume within each well. Samples were frozen at −80°C until analyzed by plaque assay as described previously (28).
RNA isolation and RNase protection assays.
BHK-21 cells were plated at 7.5 × 105 cells per well in 60-mm-diameter dishes for 14 h before infection with s55 or s51 at an MOI of 5.0. A 1-h attachment period preceded washing the cells three times with room temperature PBS (1% DCS and Ca2+/Mg2+). Cells were then incubated in growth media until time of RNA harvest. At 2, 3, 4, 5, or 6 h postinfection, the dishes were transferred to ice, and total cellular RNA was harvested as previously described by using the Ultraspec II RNA isolation system (Biotecx) (35). RNA from primary rat cortical neuron cultures was generated in the same manner; however, for BHK-21 cells, each RNA sample was derived from a single plate, while two wells were pooled for each sample of primary rat neuron RNA. RNase protection assays were performed as described previously with the RPAII system (Ambion) (35). A negative-sense probe specific for the S.A.AR86 plus and 26S RNA strands was generated from plasmid pDS-45 (9). Clone pDS-45 was linearized with EcoRV and transcribed in vitro with a Maxiscript SP6 transcription kit (Ambion) in the presence of [α-32P]UTP (Amersham) to yield a riboprobe approximately 500 nucleotides in length, of which 445 nucleotides were complementary to S.A.AR86 nucleotides 7371 to 7816. This probe generated protected fragments of 445 bp for the full-length plus-strand RNA and a 260-bp fragment for the 26S RNA. A positive-sense probe specific for the S.A.AR86 minus strand was generated by linearizing pDS-45 with XhoI and transcribing with a Maxiscript T7 transcription kit in the presence of an [α-32P]UTP. This probe protected a 445-bp fragment of the viral minus-strand RNA. A probe specific for mouse β-actin was generated with the mouse pTRI-β-actin template (Ambion) transcribed with the Maxiscript T7 transcription kit in the presence of [α-32P]UTP. The β-actin probe protected a 245-bp fragment of the mouse β-actin mRNA that served as a control for RNA loading. For the 2- and 3-h time points, 5 or 10 μg of total cellular RNA was used, while 0.5, 1, or 2 μg of total cellular RNA was used for the 4-, 5-, and 6-h time points to achieve probe excess. Twenty micrograms of total cellular RNA was used to detect viral minus strands in a two-step hybridization reaction as previously described (35). The RNA was hybridized with an excess of labeled probe (106 cpm) at 45°C overnight. Unhybridized probe was digested with RNase T1, and the intact RNA was precipitated. The protected radiolabeled fragments were analyzed on 6% acrylamide-8 M urea-Tris-borate-EDTA gels as previously described (35). The protected bands were quantified on a phosphoirmager, and viral RNA levels were normalized to β-actin.
In vitro nonstructural protein processing.
Full-length s55, s51, TR339, and 39ns1 transcripts were generated with the Ambion mMessage mMachine SP6 transcription kit as described above. In vitro translation reactions with rabbit reticulocyte lysates (Promega) were performed by a modification of previously published procedures (1, 8). In brief, 0.64 μg of RNA was added to a 100-μl reticulocyte lysate reaction supplemented with [35S]methionine (Amersham-Pharmacia). Reaction mixtures were also supplemented with KCl to a final concentration of 0.05 M, and then incubated at 30°C for 40 min, at which point the chase was initiated by adding unlabeled methionine at a final concentration of 1 mM and cycloheximide at a final concentration of 0.6 mg/ml. Samples were then incubated at either 30 or 37°C for the duration of the chase. Five-microliter samples were removed at 0, 20, 40, 60, and 80 min into the chase and placed into 20 μl of gel loading buffer (8). Samples were heat denatured at 93°C for 3 min and analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) (10% polyacrylamide). Gels were analyzed by autoradiography or on a phosphorimager.
In vivo nonstructural protein processing.
BHK-21 cells were plated at 7.5 × 106 cells per dish in 60-mm dishes for 12 to 14 h prior to infection with s55 or s51 at an MOI of 5.0 in PBS supplemented with 1% DCS and Ca2+/Mg2+. One hour postinfection, the cells were washed three times with PBS and incubated for 1 h in methionine-free α-MEM supplemented with 1% DCS. [35S]methionine (Amersham-Pharmacia) was then added at 50 μCi/ml. The cells were incubated for 15 min, at which point the monolayers were washed, and the medium was replaced with α-MEM (10% DCS, 10% tryptose phosphate, 0.29 mg of l-glutamine per ml) supplemented with 4 mM nonradioactive methionine. Individual plates were harvested at 0, 10, 20, and 30 min into the chase by lysis in radioimmunoprecipitation assay (RIPA) buffer (0.5 ml/60-mm dish) (7). S.A.AR86 nonstructural proteins were immunoprecipitated with antibodies against nsP1 and nsP2 (gifts from Charles Rice) or normal rabbit sera as a nonspecific binding control according to previously described protocols (7). In brief, cell lysates were spun at 15,000 rpm in a microcentrifuge at 4°C for 5 min. One hundred to 200 μl of lysate was mixed with 40 μl of protein A-Sepharose (Sigma) and 1 μl of normal rabbit serum and agitated at 4°C overnight. The protein A was removed by centrifugation, and lysates were transferred to new tubes containing 2 μl of anti-nsP1, anti-nsP2, or normal rabbit antisera. The tubes were agitated for 2 h at 4°C and then transferred to new tubes containing 40 μl of protein A-Sepharose (Sigma). Following a 2-h incubation at 4°C, lysates were removed and the beads were washed three times in RIPA buffer and one time in 50 mM Tris (pH 7.6). Gel loading buffer was added to the beads before heating to 93°C for 3 min. The dissociated proteins were then analyzed by SDS-PAGE on 10% acrylamide gels, dried, and analyzed on a phosphorimager.
RESULTS
An attenuating mutation at nsP1 position 538 does not adversely affect S.A.AR86 growth in vitro.
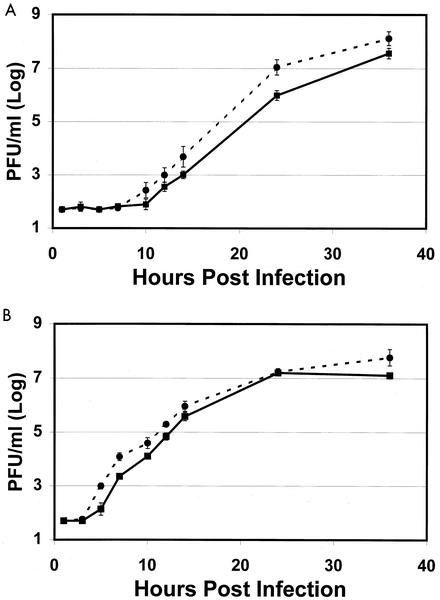
A single amino acid change at position 538 of the nsP1 protein significantly affects adult mouse neurovirulence of the Sindbis-group virus S.A.AR86. Since mutations in the nonstructural region might adversely affect viral RNA synthesis, studies were performed to evaluate the effect of the attenuating Ile or wild-type Thr at nsP1 538 on viral replication. The mutant virus grows equivalently to the wild-type virus in the brains of infected animals during the first few days of infection (9). Furthermore, both viruses exhibited similar growth kinetics when evaluated with single-step growth curves in vitro (9). While single-step grow curves provide useful information on viral growth kinetics, the high MOI used might have masked subtle defects in the growth of the attenuated s51 virus. Therefore, the growth kinetics of the mutant and wild-type viruses were evaluated by using multistep growth curves at a low MOI in BHK-21 and Neuro2A cells. Cells were infected with either the wild-type s55 (nsP1 538 Thr) or the mutant s51 (nsP1 538 Ile) virus at an MOI of 0.01, and virus production was evaluated by plaque assay. Consistent with earlier finding obtained with single-step growth curves (9), the mutant s51 virus grew at least as well as the wild-type s55 virus in multistep growth curves in both BHK-21 cells and Neuro2A cells (Fig. 1). Furthermore, in Neuro2A cells, s51 yields were approximately 1 log higher than s55 yields at late times in the growth curve (Fig. 1A). In BHK-21 cells, s51 yields were consistently higher than s55 yields at early times in the growth curve (Fig. 1B). The differences between BHK-21 and Neuro2A cells may reflect the accelerated rate of virus production in BHK-21 cells compared to that in Neuro2a cells. Most importantly, these results demonstrate that rather than causing a defect in virus growth, the attenuating Ile at nsP1 538 may actually confer a slight growth advantage to S.A.AR86 in both cultured fibroblasts and neuroblastoma-derived cells.
FIG. 1.
The attenuated mutant s51 (nsP1 538 Ile) grows to higher levels than the wild-type s55 (nsP1 538 Thr) virus in multistep in vitro growth curves. Neuro2A (A) or BHK-21 (B) cells were infected with s55 (solid squares) or s51 (solid circles) at an MOI of 0.01. Samples of supernatant were removed at the indicated time points and evaluated for virus yield by plaque assay. The data shown are from one of three representative experiments. Each point represents the average titer of three independent wells with the standard deviation.
The attenuating Ile at nsP1 538 accelerates accumulation of mature nsP1 in infected cells in vitro.
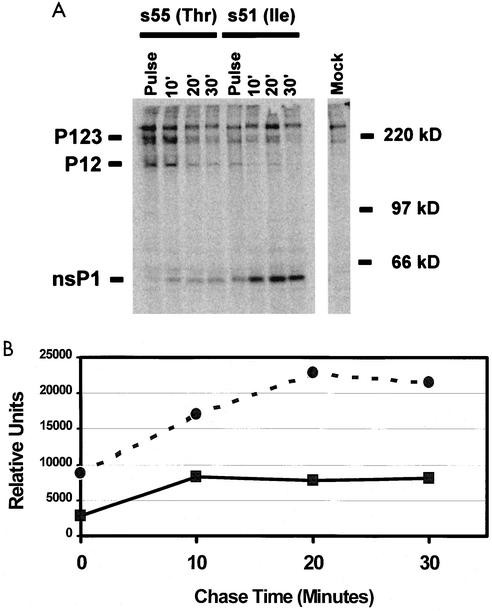
Position 538 lies within the cleavage recognition domain between nsP1 and nsP2, suggesting that the presence of Ile or Thr at this position might modulate processing of the nonstructural protein polyprotein precursor, which could have downstream effects on viral RNA synthesis and growth. In order to evaluate whether the presence of Ile or Thr at nsP1 538 altered maturation of the viral nonstructural proteins, BHK-21 cells were infected with either the wild-type s55 (nsP1 538 Thr) or the attenuated s51 (nsP1 538 Ile) virus, pulsed with [35S]methionine, and then chased with excess cold methionine. Cell lysates were harvested during the pulse and chase, nsP1 was immunoprecipitated with nsP1-specific antisera, and the precipitated proteins were analyzed by SDS-PAGE as previously described (7). The 200-kDa P123 nonstructural protein precursor and the 135-kDa P12 cleavage intermediate and mature nsP1 were evident in both s55- and s51-infected cell lysates (Fig. 2). A nonspecific band apparent in uninfected lysates comigrated at slightly less than 200 kDa, making analysis of the 200 kDa P123 precursor difficult. However, clear differences existed between s55 and s51 with regard to the presence of the P12 cleavage intermediate and the accumulation of mature nsP1 (Fig. 2). The P12 cleavage intermediate was present at higher levels in s55-infected cell lysates over the course of the chase, while mature nsP1 accumulated to higher levels at earlier times in the s51-infected cell lysates (Fig. 2B). Similar results were observed when nsP2 was immunoprecipitated from s55- and s51-infected cells (data not shown). These results suggest that the attenuating Ile at nsP1 538 altered processing of the viral nonstructural proteins, resulting in more rapid accumulation of mature nonstructural proteins within the infected cells.
FIG. 2.
Accelerated nonstructural protein processing by the attenuated s51 virus in infected cells. BHK-21 cells were infected with the wild-type s55 (solid squares) virus (nsP1 538 Thr) or the attenuated s51 (solid circles) virus (nsP1 538 Ile) at an MOI of 5.0 for 2 h. The infected cells were then pulsed with [35S]methionine for 15 min followed by a chase with an excess of cold methionine for 10, 20, or 30 min, at which points, cell lysates were generated. (A) NsP1 was immunoprecipitated from the cell lysates and analyzed by SDS-PAGE on 10% polyacrylamide gels. Normal rabbit serum served as a control for nonspecific antibody interactions. (B) Plot of the relative levels of nsP1 from s55- or s51-infected cells from panel A. Shown are the results of one of four representative experiments.
The attenuating Ile at nsP1 538 accelerates nonstructural protein processing in vitro.
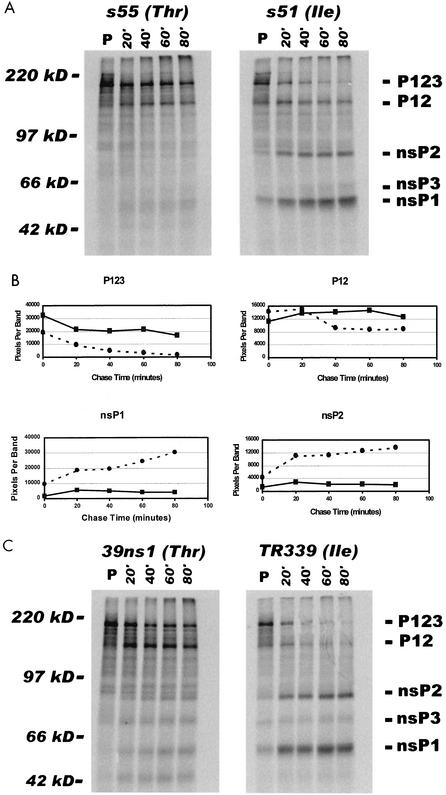
In order to further evaluate the effect of the attenuating Ile or wild-type Thr on nonstructural protein processing, in vitro nonstructural protein translation and processing assays were performed. The alphavirus nsP2 protease is active in vitro, and nonstructural protein processing will occur in rabbit reticulocyte lysates (1, 8), making it possible to evaluate the processing kinetics of the S.A.AR86 nonstructural proteins containing Thr or Ile at nsP1 position 538. Full-length viral RNA transcripts from S.A.AR86 infectious clones with the wild-type Thr (ps55) or mutant Ile (ps51) at nsP1 position 538 were generated with SP6 polymerase. These transcripts were then used as templates for the production of 35S-labeled nonstructural proteins by using rabbit reticulocyte lysates. In vitro translation reaction mixtures were incubated for 40 min, at which point, excess unlabeled methionine and cycloheximide were added to stop the translation reactions and permit analysis of the processing kinetics of the translated polyproteins by SDS-PAGE (Fig. 3). Consistent with previous reports (1, 8), a 250-kDa band corresponding to the P1234 precursor was evident in both s55 and s51 translation reactions following the 40-min pulse, and this band had largely disappeared after the first 15 min of the chase. The 200-kDa p123 precursor was also evident for both viruses, as well as the 135-kDa P12 cleavage intermediate. Over the course of the chase, both the P123 and P12 precursors were processed to produce mature nonstructural proteins; however, this processing occurred more rapidly for the mutant s51 virus nonstructural proteins. Mature nsP1 and nsP2 were readily apparent for the s51-derived nonstructural proteins by 20 min into the chase. In fact, mature nsP1 and nsP2 were detectable even at time zero in the chase, suggesting that nonstructural protein processing occurred for the s51-derived proteins during the 40-min pulse. In contrast, mature nonstructural proteins were barely detectable from the wild-type s55 in vitro translation reactions, even at 80 min into the chase (Fig. 3A and B). Consistent with the increased accumulation of mature nsP1 and nsP2, the s51 translation reactions also showed a corresponding decrease in the levels of the P123 precursor and P12 cleavage intermediate, while P123 and P12 appeared more stable for s55. In a representative experiment, the levels of P123 had decreased by 83% for s51, but only 34% for s55 by 60 min into the chase. To further test the effect of Ile or Thr at nsP1 538 on nonstructural protein processing, the in vitro nonstructural processing kinetics of the Sindbis virus TR339 and its mutant, 39ns1, were analyzed. TR339 and 39ns1 differ only at nsP1 538, where TR339 encodes Ile and 39ns1 encodes Thr (9). The presence of Thr at position 538 in 39ns1 confers increased neurovirulence in 3-week-old mice (9). Analysis of the processing of the in vitro-translated nonstructural proteins from these two viruses demonstrated that the presence of Thr at nsP1 538 of 39ns1 delayed processing of the nonstructural protein precursors (Fig. 3C). The kinetics of 39ns1 (nsP1 538 Thr) nonstructural protein processing were similar to those for s55, while processing of the TR339 (nsP1 538 Ile)-derived nonstructural proteins was very similar to that of the s51 virus (Fig. 3A and B). These results, combined with the results from virally infected cells, suggest that the presence of the attenuating Ile at nsP1 538 accelerated cleavage of the viral nonstructural proteins, leading to more rapid accumulation of mature nonstructural proteins.
FIG. 3.
The attenuating Ile at nsP1 538 accelerates nonstructural protein processing in vitro. (A) Full-length RNA transcripts for s55 (solid squares) or s51 (solid circles) were in vitro translated in the presence of [35S]methionine by using rabbit reticulocyte lysates. Translation reactions were incubated for 40 min at 30°C. At this point, an excess of unlabeled methionine and cycloheximide was added to stop translation, and reactions were shifted to 37°C. Samples were removed for analysis by SDS-PAGE at 0, 20, 40, 60, and 80 min after stopping translation and increasing the temperature. (B) Levels of the P123 and P12 cleavage intermediates as well as mature nsP1 and nsP2 for the s55 or s51 translation reactions were analyzed over time on a phosphorimager. The data represent the total number of pixels for the relevant band at each time point. The data shown are from the gels pictured in panel A. (C) Full-length RNA transcripts for the attenuated TR339 (nsP1 538 Ile) and neurovirulent 39ns1 (nsP1 538 Thr) were translated as described for Fig. 2A. Shown are the results of one of three representative experiments.
The attenuating Ile at nsP1 538 upregulates expression from the S.A.AR86 26S promoter.
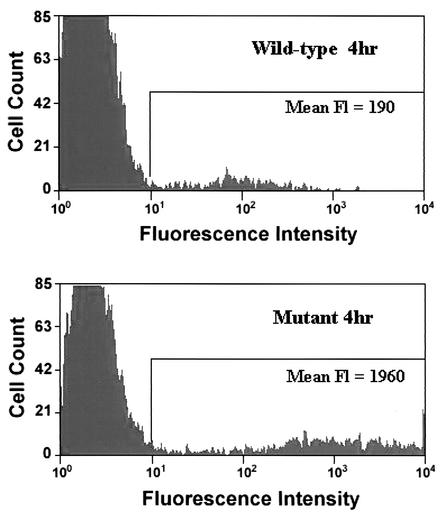
Nonstructural protein processing regulates alphavirus RNA synthesis (reviewed in reference 29); therefore, we hypothesized that the accelerated nonstructural protein processing by the attenuated s51 virus would affect viral RNA synthesis in terms of kinetics and/or steady-state levels. To test this, BHK-21 cells were infected with S.A.AR86-derived replicon particles expressing GFP and containing either Thr or Ile at nsP1 538. GFP expression, as measured by flow cytometry, was then used as a surrogate readout for gene expression from the S.A.AR86 26S promoter. Cells infected with the mutant replicon with Ile at nsP1 538 (REP91-GFP) exhibited 4- to 10-fold-higher levels of fluorescent intensity than cells infected with the wild-type replicon with Thr at nsP1 538 (REP89-GFP) at 4 h postinfection (Fig. 4). In a representative experiment, the mean fluorescent intensity of REP91-GFP-infected cells was 1,960, compared to an intensity of 190 for REP89-GFP at 4 h postinfection. This effect was also observed in Neuro2A cells, as well as primary rat cortical neurons (data not shown), suggesting that the attenuated s51 virus exhibits more rapid and higher-level expression from the viral 26S promoter than the wild-type s55 virus in a cell type-independent manner.
FIG. 4.
The attenuating Ile at nsP1 538 increases expression from the S.A.AR86 26S promoter. BHK-21 cells were infected with S.A.AR86-derived replicon particles with the wild-type Thr (REP89-GFP) or the mutant Ile (REP91-GFP) at nsP1 538 at an MOI of 0.1. These replicons express GFP from the viral 26S promoter, which was used as a surrogate marker of 26S expression. At 4 h postinfection, cells were harvested and analyzed for GFP expression (fluorescence [Fl]) by flow cytometry. Shown are representative histograms from one of three experiments with BHK-21 cells.
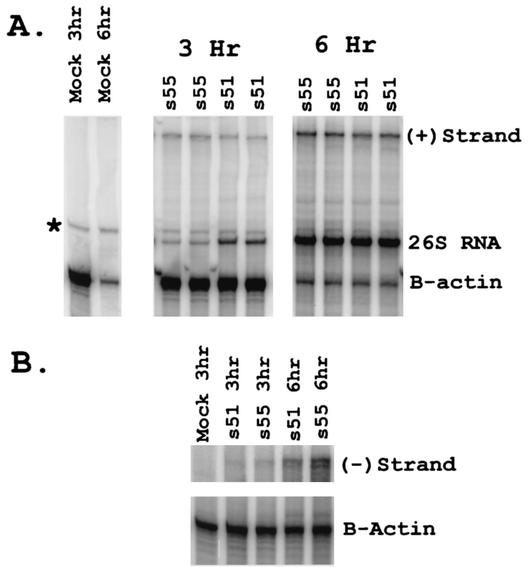
In order to more directly assess the effects of Ile or Thr at nsP1 538 on viral RNA synthesis, BHK-21 cells were infected with either the mutant s51 virus (nsP1 538 Ile) or wild-type s55 (nsP1 538 Thr), and total cellular RNA was harvested at various times postinfection. RNase protection assays with probes specific for the genomic plus-strand and 26S RNA or the genomic minus-strand RNA were performed. A probe specific for mouse β-actin was used as a control to normalize for RNA loading. After normalizing for β-actin levels, we saw minimal differences in minus- and plus-strand RNA levels between the wild-type and mutant viruses at all time points analyzed. However, at early times postinfection (3 h), s51 consistently exhibited a three- to fourfold increase in 26S RNA compared to s55 (Fig. 5 and Table 1), which is consistent with the GFP data from the replicon infected cells (Fig. 4). However, this difference was transient, since s55 and s51 26S RNA levels were equivalent by 5 to 6 h postinfection (Table 1). Similar results were obtained following s55 or s51 infection of primary rat neuron cultures (Table 1), demonstrating that this effect is not restricted to fibroblasts. These results demonstrate that rather than impairing viral replication, the attenuating Ile at nsP1 538 altered the kinetics of viral nonstructural protein processing and accelerated the induction of viral 26S RNA synthesis.
FIG. 5.
The attenuating Ile leads to faster induction of 26S RNA synthesis, but does not affect minus- or plus-strand synthesis. BHK-21 cells were infected with the wild-type s55 or mutant s51 viruses at an MOI of 5.0. Total cellular RNA was harvested at 3 or 6 h postinfection and analyzed by RNase protection assays using probes protecting fragments from the viral plus and 26S RNAs, the viral minus-strand RNA, or mouse β-actin. Representative RNase protection assay showing the 3- and 6-h time points for s55- or s51-infected cells. Each lane represents RNA from an independent sample. Five micrograms of total cellular RNA was used for each sample at the 3-h time point, and 1 μg per sample was used for the 6-h time point. Minus-strand and β-actin bands are shown at different intensities due to the excess of β-actin signal compared to minus-strand levels.
TABLE 1.
Ratios of s51 versus s55 RNA in infected cells
| Cell type | Time post- infection (h) | s51/s55 RNA ratioa
|
||
|---|---|---|---|---|
| Minus strand | Plus strand | 26S RNA | ||
| BHK-21 | 3 | 0.8, 0.8 | 1.1, 0.6, 1.0 | 4.2, 3.1, 4.2 |
| 4 | 1.3 | 1.2 | 1.4 | |
| 5 | 0.8 | 1.0 | 1.0 | |
| 6 | 0.9 | 1.2, 0.9 | 1.0, 1.0 | |
| Primary neurons | 3 | 0.8 | 2.7 | |
| 6 | 0.6 | 1.0 | ||
Each data point represents the ratio of s51 to s55 RNA for a single experiment.
DISCUSSION
We have previously demonstrated that a single amino acid change from Thr to Ile at nsP1 position 538 attenuates the Sindbis-group alphavirus S.A.AR86 for adult mouse neurovirulence (9). The alphavirus nonstructural proteins comprise the viral RNA-dependent RNA polymerase, suggesting that the attenuating mutation would affect viral replication in some manner. Therefore, a series of experiments were performed to assess the effect of Ile or Thr at nsP1 538 on viral growth, nonstructural protein synthesis and processing, and viral RNA synthesis. Molecular analysis of viruses with Ile or Thr at nsP1 538 demonstrated that the attenuating Ile accelerated nonstructural protein processing and caused more rapid induction of 26S RNA synthesis in infected cells. While it remains to be proven whether accelerated nonstructural protein processing and 26S RNA synthesis are directly responsible for the attenuated phenotype of s51, these experiments demonstrate that a simple defect in viral replication does not provide an explanation. Since Ile is found at position 538 in several nonneurovirulent Sindbis-group alphaviruses, these studies raise the possibility that the wild-type S.A.AR86 virus has evolved to down-regulate or delay induction of 26S RNA synthesis and that this contributes to adult mouse neurovirulence.
The simplest explanation for the attenuated phenotype of the s51 virus was that the mutant virus replicated less efficiently than the wild-type virus. However the s51 mutant did not exhibit growth defects in vitro and may have a slight growth advantage compared to the wild-type s55 virus (Fig. 1) (9). s55 and s51 also grow to similar levels within the brains of infected animals (9), further supporting the idea that a simple growth defect does not contribute to the attenuation of s51.
nsP1 538, where wild-type S.A.AR86 encodes a unique Thr residue, is located at position P3 within the 8-amino-acid cleavage recognition motif between nsP1 and nsP2 (reviewed in reference 29). The high level of conservation of Ile at nsP1 538 among Sindbis-group alphaviruses had led to the suggestion that the P3 position of the cleavage domain is important in regulating nonstructural protein cleavage (29). This suggestion is experimentally supported by our finding that the presence of the wild-type S.A.AR86-encoded Thr at this position slows the rate of nonstructural protein processing (Fig. 2 and 3). Altered processing at the nsP1/nsP2 junction by the wild-type or mutant S.A.AR86 viruses might also affect the generation of mature nsP2 and nsP3, since the nsP2 protease is more active at the nsP2/nsP3 cleavage site after nsP1 is cleaved from nsP2 (26). This is supported by the observation that Thr at nsP1 538 increased the stability of the P123 precursor in the in vitro processing assays (Fig. 3). It is also possible that the presence of the wild-type Thr at nsP1 538 affects the stability of mature nsP1 in infected cells, since mature nsP1 levels in s55-infected cells did not reach those observed in s51-infected cells, even at late times postchase, when the majority of the P12 cleavage intermediate had disappeared (Fig. 2). However, P12 was clearly more stable in s55-infected cells and in the s55 in vitro translation reactions (Fig. 3), suggesting that the major effect of Ile or Thr at nsP1 538 is on processing of the nonstructural proteins at the nsP1/nsP2 junction.
The polyprotein precursor P123 plus mature nsP4 is essential for minus-strand synthesis during the first few hours of the viral infection (17-19, 27). Although the P123-nsP4 combination is capable of a low level of full-length plus-strand synthesis (19), highly efficient plus-strand genomic and 26S RNA synthesis is dependent on the presence of mature nonstructural proteins (27). Processing of the P123 precursor into the mature nsP1, nsP2, and nsP3 proteins is coincident with shutoff of minus-strand synthesis and an increase in plus-strand genomic and 26S RNA synthesis (27). Furthermore, several temperature-sensitive mutants with defects in the nsP2 proteinase exist that exhibit specific defects in 26S RNA synthesis (6, 13, 24, 25). Therefore, delayed cleavage of the nonstructural protein precursor due to the presence of Thr at nsP1 position 538 would be expected to delay induction of 26S RNA synthesis, which is consistent with our observations that the wild-type s55 virus exhibits a delay in 26S synthesis compared to the mutant s51 virus (Fig. 4 and 5).
Although their role in the attenuated phenotype of the s51 virus (nsP1 538 Ile) is yet to be determined, altered nonstructural protein processing and/or accelerated induction of 26S RNA synthesis might be attenuating through several mechanisms, including (i) increased induction of antiviral factors, such as type I interferon; (ii) enhanced cytopathic effect on infected cells due to earlier expression of viral structural genes; or (iii) differential effects on host cell macromolecular synthesis by nonstructural protein precursors compared to those of mature nonstructural proteins. The presence of Thr at nsP1 538 in the neurovirulent s55 virus may delay 26S RNA synthesis until after host cell macromolecular synthesis has been shut off, thereby limiting induction of type I interferon or other immune mediators by the infected cell. It is also possible that Ile at nsP1 538 is attenuating due to altered cytopathic effect or shutoff of host cell macromolecular synthesis. The structural proteins of Sindbis-group alphaviruses are at least partly responsible for viral cytopathic effect (4), while the nonstructural proteins are thought to mediate shutoff of host cell macromolecular synthesis (4). Rapid induction of 26S RNA by the attenuated mutant would lead to earlier production of the viral structural proteins, which may kill the host cell more rapidly and result in an overall decrease in virus production. However, it is also possible that the mutant virus may in fact be less cytopathic, since mutations at nsP2 position 726 of Sindbis virus that result in a noncytopathic phenotype also exhibit hyperprocessing of the viral nonstructural proteins (3). In this scenario, the mutant virus may not kill infected cells as effectively as the wild-type virus, which might lead to decreased pathology within the brains of infected animals. Alternatively, altered nonstructural protein processing may affect shutoff of host macromolecular synthesis, resulting in less-efficient virus production due to competition for resources within the infected cell. However, the present results argue against a defect in virus production by the attenuated mutant, since this has not been observed either in vivo or in vitro (Fig. 1) (9).
In conclusion, we have presented data demonstrating that an attenuating mutation in nsP1 of the adult mouse neurovirulent Sindbis-group virus S.A.AR86 accelerates cleavage of the viral nonstructural proteins into their mature forms, which leads to more rapid induction of viral 26S RNA synthesis. Additional studies are under way to determine whether this alteration in protein processing and viral RNA synthesis plays a direct role in the attenuated phenotype of the s51 (nsP1 538 Ile) virus.
Acknowledgments
This research was supported by NIH research grant R01 AR47190. These studies were initiated while M.T.H. was supported by an NIH postdoctoral fellowship (F32 AI10146).
We thank Nancy Davis, Kate Ryman, and William Klimstra for helpful discussions regarding this research. We also thank Charles Rice for providing antibodies against nsP1 and nsP2. Dwayne Muhammed provided excellent technical support with cell culture.
REFERENCES
- 1.Ding, M. X., and M. J. Schlesinger. 1989. Evidence that Sindbis virus NSP2 is an autoprotease which processes the virus nonstructural polyprotein. Virology 171:280-284. [DOI] [PubMed] [Google Scholar]
- 2.Dubuisson, J., S. Lustig, N. Ruggli, Y. Akov, and C. M. Rice. 1997. Genetic determinants of Sindbis virus neuroinvasiveness. J. Virol. 71:2636-2646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Frolov, I., E. Agapov, T. A. Hoffman, Jr., B. M. Pragai, M. Lippa, S. Schlesinger, and C. M. Rice. 1999. Selection of RNA replicons capable of persistent noncytopathic replication in mammalian cells. J. Virol. 73:3854-3865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Frolov, I., and S. Schlesinger. 1994. Comparison of the effects of Sindbis virus and Sindbis virus replicons on host cell protein synthesis and cytopathogenicity in BHK cells. J. Virol. 68:1721-1727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gomez-de Cedron, M., N. Ehsani, M. L. Mikkola, J. A. Garcia, and L. Kaariainen. 1999. RNA helicase activity of Semliki Forest virus replicase protein NSP2. FEBS Lett. 448:19-22. [DOI] [PubMed] [Google Scholar]
- 6.Hardy, W. R., Y. S. Hahn, R. J. De Groot, E. G. Strauss, and J. H. Strauss. 1990. Synthesis and processing of the nonstructural polyproteins of several temperature-sensitive mutants of Sindbis virus. Virology 177:199-208. [DOI] [PubMed] [Google Scholar]
- 7.Hardy, W. R., and J. H. Strauss. 1988. Processing the nonstructural polyproteins of Sindbis virus: study of the kinetics in vivo by using monospecific antibodies. J. Virol. 62:998-1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hardy, W. R., and J. H. Strauss. 1989. Processing the nonstructural polyproteins of Sindbis virus: nonstructural proteinase is in the C-terminal half of nsP2 and functions both in cis and in trans. J. Virol. 63:4653-4664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Heise, M. T., D. A. Simpson, and R. E. Johnston. 2000. A single amino acid change in nsP1 attenuates neurovirulence of the Sindbis-group alphavirus S.A.AR86. J. Virol. 74:4207-4213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Heise, M. T., D. A. Simpson, and R. E. Johnston. 2000. Sindbis-group alphavirus replication in periosteum and endosteum of long bones in adult mice. J. Virol. 74:9294-9299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Johnston, R. E., and C. J. Peters. 1996. Alphaviruses, p. 843-898. In B. N. Fields, D. M. Knipe, and P. M. Howley (ed.), Fields virology, 3rd ed. Lippincott-Raven, Philadelphia, Pa.
- 12.Kamer, G., and P. Argos. 1984. Primary structural comparison of RNA-dependent polymerases from plant, animal and bacterial viruses. Nucleic Acids Res. 12:7269-7282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Keränen, S., and L. Kääriäinen. 1979. Functional defects of RNA-negative temperature-sensitive mutants of Sindbis and Semliki Forest viruses. J. Virol. 32:19-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Klimstra, W. B., K. D. Ryman, and R. E. Johnston. 1998. Adaptation of Sindbis virus to BHK cells selects for use of heparan sulfate as an attachment receptor. J. Virol. 72:7357-7366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Klimstra, W. B., K. D. Ryman, K. A. Bernard, K. B. Nguyen, C. A. Biron, and R. E. Johnston. 1999. Infection of neonatal mice with Sindbis virus results in a systemic inflammatory response syndrome. J. Virol. 73:10387-10398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kobiler, D., C. M. Rice, C. Brodie, A. Shahar, J. Dubuisson, M. Halevy, and S. Lustig. 1999. A single nucleotide change in the 5′ noncoding region of Sindbis virus confers neurovirulence in rats. J. Virol. 73:10440-10446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lemm, J. A., and C. M. Rice. 1993. Assembly of functional Sindbis virus RNA replication complexes: requirement for coexpression of P123 and P34. J. Virol. 67:1905-1915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lemm, J. A., and C. M. Rice. 1993. Roles of nonstructural polyproteins and cleavage products in regulating Sindbis virus RNA replication and transcription. J. Virol. 67:1916-1926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lemm, J. A., T. Rümenapf, E. G. Strauss, J. H. Strauss, and C. M. Rice. 1994. Polypeptide requirements for assembly of functional Sindbis virus replication complexes: a model for the temporal regulation of minus- and plus-strand RNA synthesis. EMBO J. 13:2925-2934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li, G. P., M. W. La Starza, W. R. Hardy, J. H. Strauss, and C. M. Rice. 1990. Phosphorylation of Sindbis virus nsP3 in vivo and in vitro. Virology 179:416-427. [DOI] [PubMed] [Google Scholar]
- 21.Lustig, S., A. C. Jackson, C. S. Hahn, D. E. Griffin, E. G. Strauss, and J. H. Strauss. 1988. Molecular basis of Sindbis virus neurovirulence in mice. J. Virol. 62:2329-2336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McKnight, K. L., D. A. Simpson, S.-C. Lin, T. A. Knott, J. M. Polo, D. F. Pence, D. B. Johannsen, H. W. Heidner, N. L. Davis, and R. E. Johnston. 1996. Deduced consensus sequence of Sindbis virus strain AR339: mutations contained in laboratory strains which affect cell culture and in vivo phenotypes. J. Virol. 70:1981-1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mi, S., R. Durbin, H. V. Huang, C. M. Rice, and V. Stollar. 1989. Association of the Sindbis virus RNA methyltransferase activity with the nonstructural protein nsP1. Virology 170:385-391. [DOI] [PubMed] [Google Scholar]
- 24.Sawicki, D. L., and S. G. Sawicki. 1985. Functional analysis of the A complementation group mutants of Sindbis HR virus. Virology 144:20-34. [DOI] [PubMed] [Google Scholar]
- 25.Scheele, C. M., and E. R. Pfefferkorn. 1969. Inhibition of interjacent ribonucleic acid (26S) synthesis in cells infected by Sindbis virus. J. Virol. 4:117-122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shirako, Y., and J. H. Strauss. 1990. Cleavage between nsP1 and nsP2 initiates the processing pathway of Sindbis virus nonstructural polyprotein P123. Virology 177:54-64. [DOI] [PubMed] [Google Scholar]
- 27.Shirako, Y., and J. H. Strauss. 1994. Regulation of Sindbis virus RNA replication: uncleaved P123 and nsP4 function in minus-strand RNA synthesis, whereas cleaved products from P123 are required for efficient plus-strand RNA synthesis. J. Virol. 68:1874-1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Simpson, D. A., N. L. Davis, L. Seh-Ching, D. Russell, and R. E. Johnston. 1996. Complete nucleotide sequence and full-length cDNA clone of S.A.AR86, a South African alphavirus related to Sindbis. Virology 222:464-469. [DOI] [PubMed] [Google Scholar]
- 29.Strauss, J. H., and E. G. Strauss. 1994. The alphaviruses: gene expression, replication, and evolution. Microbiol. Rev. 58:491-562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Trgovcich, J., J. F. Aronson, and R. E. Johnston. 1996. Fatal Sindbis virus infection of neonatal mice in the absence of encephalitis. Virology 224:73-83. [DOI] [PubMed] [Google Scholar]
- 31.Tucker, P. C., S. H. Lee, N. Bui, D. Martinie, and D. E. Griffin. 1997. Amino acid changes in the Sindbis virus E2 glycoprotein that increase neurovirulence improve entry into neuroblastoma cells. J. Virol. 71:6106-6112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tuittila, M. T., M. G. Santagati, M. Röyttä, J. A. Määttä, and A. E. Hinkkanen. 2000. Replicase complex genes of Semliki Forest virus confer lethal neurovirulence. J. Virol. 74:4579-4589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vasiljeva, L., A. Merits, P. Auvinen, and L. Kaariainen. 2000. Identification of a novel function of the alphavirus capping apparatus. RNA 5′-triphosphatase activity of nsP2. J. Biol. Chem. 275:17281-17287. [DOI] [PubMed] [Google Scholar]
- 34.Weinbren, M. P., R. H. Kokernot, and K. C. Smithburn. 1956. Strains of Sindbis-like virus isolated from Culicine mosquitoes in the Union of South Africa. S. Afr. Med. J. 30:631-636. [PubMed] [Google Scholar]
- 35.White, L. J., J.-G. Wang, N. L. Davis, and R. E. Johnston. 2001. Role of alpha/beta interferon in Venezuelan equine encephalitis virus pathogenesis: effect of an attenuating mutation in the 5′ untranslated region. J. Virol. 75:3706-3718. [DOI] [PMC free article] [PubMed] [Google Scholar]