Abstract
Under the selective pressure of protease inhibitor therapy, patients infected with human immunodeficiency virus (HIV) often develop drug-resistant HIV strains. One of the first drug-resistant mutations to arise in the protease, particularly in patients receiving indinavir or ritonavir treatment, is V82A, which compromises the binding of these and other inhibitors but allows the virus to remain viable. To probe this drug resistance, we solved the crystal structures of three natural substrates and two commercial drugs in complex with an inactive drug-resistant mutant (D25N/V82A) HIV-1 protease. Through structural analysis and comparison of the protein-ligand interactions, we found that Val82 interacts more closely with the drugs than with the natural substrate peptides. The V82A mutation compromises these interactions with the drugs while not greatly affecting the substrate interactions, which is consistent with previously published kinetic data. Coupled with our earlier observations, these findings suggest that future inhibitor design may reduce the probability of the appearance of drug-resistant mutations by targeting residues that are essential for substrate recognition.
The development of the human immunodeficiency virus type 1 (HIV-1) protease inhibitors is regarded as a major success of structure-based drug design (16, 36, 62-64). Indeed, the protease inhibitors are considered the most potent drugs currently available for the treatment of AIDS (63). These drugs are often combined with other drugs to establish highly active antiretroviral therapy, which is credited with an approximately threefold drop in the death rate from AIDS since its introduction (38). Despite this remarkable success, the emergence of HIV-1 mutants that resist current drug regimes (1, 2, 14) remains a critical factor in clinical failure of antiviral therapy (9, 56). The relatively rapid appearance of resistant viral mutants among treated HIV-1 patients is attributable to the high rate of replication of the virus, coupled with a high intrinsic rate of mutation due to the infidelity of the HIV-1 reverse transcriptase (22, 46, 47).
The homodimeric HIV-1 protease is an effective therapeutic target because it allows viral maturation by sequentially cleaving at least 10 asymmetric and nonhomologous sequences in the Gag-Pol polyproteins (8, 19, 42). The six Food and Drug Administration-approved HIV-1 protease inhibitors, amprenavir (APV), indinavir (IDV), nelfinavir (NFV), saquinavir (SQV), ritonavir (RTV), and lopinavir (LPV), that are on the market are all competitive inhibitors (17), binding at the active site. Therefore, they compete directly with the enzyme's ability to recognize substrates (33, 34, 43, 44, 49, 60). These drugs are peptidomimetics that resulted from structure-based drug design efforts (7, 23-25, 55, 59). All of them have large, generally hydrophobic moieties that interact with the mainly hydrophobic S2-S2′ pockets in the active site (62). Despite chemical differences, these inhibitors occupy a similar space in the active site, and hence similar mutations in HIV-1 protease can cause multidrug resistance without substantially altering substrate binding (1, 45, 51).
Since the primary function of HIV-1 protease is to cleave its substrates rather than bind inhibitors, our laboratory analyzed the substrate recognition of this enzyme and attempted to arrive at a structural rationale for what constitutes a substrate. We determined the crystal structures of peptides bound to an inactive wild-type HIV-1 protease (44). Analysis of these complexes found that all the conserved protease-substrate hydrogen bonds involve only backbone atoms of the substrate and therefore are unlikely to be the primary determinant of substrate specificity; also, the conformation of the unprimed side of the peptide is conserved and asymmetric, resembling a toroid in the S1-S3 pockets, while the prime side of the peptide remains extended. Based on these two observations, we proposed that the major determinant of specificity for HIV-1 protease is that the conformation of the unprimed side of the substrate can form a toroid.
To further understand the substrate binding, drug resistance, and protein adaptability of HIV-1 protease (26, 43, 44, 53), we examined one of the first drug-resistant mutations to occur in HIV-1 protease among patients receiving antiviral therapy, V82A (54). This mutation occurs particularly in those patients receiving IDV or RTV (9, 10, 12, 39). Structurally, the Val82 residue is located at the P1-loops (Gly78-Asn83) near the active site (see Fig. 1a). The effect of this mutation on the binding to RTV, IDV, and NFV inhibitors is to significantly reduce their affinity (18, 27), whereas in contrast, the effect of this mutation on the ability of the enzyme to cleave substrates is smaller (18, 27). Understanding how the drug-resistant protease recognizes both substrates and inhibitors becomes crucial for future rational drug design. To address this issue, we have determined the crystal structures of the inactive drug-resistant variant D25N/V82A of HIV-1 protease in complex with either of three Gag peptide substrates—matrix-capsid (MA-CA), capsid-p2 (CA-p2), and p1-p6—and two clinically used drugs—SQV and RTV. We conducted structural analysis using these complexes in relevance to ligand-protease interaction, which includes shape complementarity, hydrogen bonds, van der Waals (VDW) packing, and estimation of VDW interaction energy. Since the wild-type structures for these ligands are already available in the Protein Data Bank (7, 23-25, 55), it allowed us to make a direct structural comparison. The findings of this study suggest that Val82 is not crucial for substrate recognition but is critical for inhibitor binding.
FIG. 1.
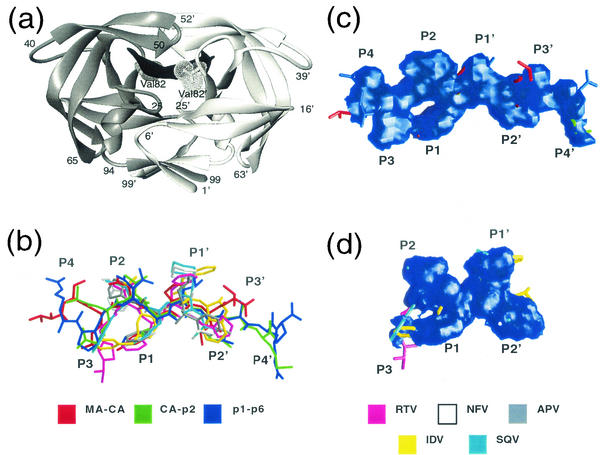
Overall structure of the complexes. (a) Ribbon diagram of HIV-1 protease dimer (PDB code 1F7A [43]) bound to the CA-p2 substrate highlights the position of Val82. (b) Superposition of the three substrateV82A complexes, MA-CA, CA-p2, and p1-p6, and five inhibitorWT compounds, APV (1HPV [25]), IDV (1HSG [7]), NFV (1OHR [23]), SQV (1HXB [55]), and RTV (1HXW [24]). (c and d) Consensus volume occupied by the three substrates (c) and the five inhibitors (d), generated using GRASP (40). The consensus volume is defined as the volume occupied by at least two of three substrates and three of five inhibitors for the substrateV82A and inhibitorWT complexes, respectively.
MATERIALS AND METHODS
Nomenclature.
Ligand acronyms with corresponding subscripts, such as V82A for the mutant or WT for wild-type protease, are used to distinguish HIV-1 protease variants throughout this article. For example, p1-p6V82A denotes p1-p6 cleavage site peptide complexed to the V82A mutant protease.
Mutagenesis, protein purification, and crystallization.
The protease gene containing the D25N modification was a gift from C. S. Craik, University of California San Francisco; the D25N mutation is required for complexing protease with its substrates without cleaving them. The alanine mutation at position 82 was introduced by using the QuickChange site-directed mutagenesis kit (Stratagene, La Jolla, Calif.). The protocol used for protein overexpression and purification has been described previously (21, 43, 48).
The purified protein was concentrated to 2.5 mg ml−1 and equilibrated on ice for 30 min with a fivefold molar excess of the ligand. Crystals were obtained in hanging drops under more than one condition. The crystals used here were grown with a reservoir solution of 126 mM phosphate buffer (pH 6.2), 63 mM sodium citrate, and ammonium sulfate (28 to 31% and 22% for the substrates and inhibitors, respectively) (43, 57). The crystals were brick shaped, tiny, and colorless, with a maximum dimension between 0.1 and 0.2 mm; the protein concentration ranged between 0.4 and 1.9 mg ml−1.
Data collection.
Intensity data were collected by using a Rigaku R-axis image plate mounted on a Rigaku rotating-anode X-ray generator, with the latter operating at 50 kV and 100 mA. The Yale mirror system was used to focus the X-ray beam. Data for the CA-p2V82A, p1-p6V82A, and SQVV82A complexes were collected at 25°C, while data for the MA-CAV82A and RTVV82A complexes were collected at −80°C. The crystal system and cell dimensions were determined using the program DENZO (37, 40a). Indexing of reflections revealed that crystals for all of the complexes grew in a similar P212121 cell, except for the RTVV82A crystals, which grew in a P21 cell with β = 99.8°. However, its unit cell vectors, a, b, and c, were similar to those of the remaining complexes. The final data collection statistics for the five structures in this study are shown in Table 1.
TABLE 1.
Data collection and refinement statistics for the five proteaseV82A complexesa
| Variable | MA-CA | CA-p2 | p1-p6 | SQV | RTV |
|---|---|---|---|---|---|
| Ligand sequence | VSQNY-PIVQN | KARVL-AEAMS | RPGNF-LQSRP | See Fig. 3 | See Fig. 3 |
| Data collection | |||||
| Space group | P212121 | P212121 | P212121 | P212121 | P21 |
| a (Å) | 50.97 | 51.56 | 51.27 | 51.44 | 51.62 |
| b (Å) | 57.86 | 59.38 | 59.13 | 60.04 | 59.04 |
| c (Å) | 62.07 | 61.87 | 61.93 | 62.02 | 61.35 |
| β (°) | 99.8 | ||||
| Z | 4 | 4 | 4 | 4 | 4 |
| Temp (°C) | −80 | 25 | 25 | 25 | −80 |
| Resolution (Å) | 1.9 (1.97-1.9) | 2.15 (2.23-2.15) | 2.0 (2.07-2.0) | 2.5 (2.59-2.5) | 2.2 (2.28-2.5) |
| Total no. of reflections | 48,168 | 25,200 | 98,506 | 28,072 | 19,193 |
| No. of unique reflections | 14,278 (1086) | 10,094 (834) | 12,659 (1182) | 6,743 (496) | 13,097 (1183) |
| Rmerge (%) | 5.7 (19) | 7.4 (31) | 9.0 (33) | 8.0 (36) | 6.4 (27) |
| Completeness (%) | 94.9 (74) | 93.2 (76) | 95.4 (92) | 93.4 (71) | 71.2 (66) |
| I/σI | 5.8 | 7.5 | 7.7 | 7.0 | 7.6 |
| Refinement | |||||
| R value (%) | 20.1 (26) | 20.8 (32) | 17.9 (22) | 19.1 (30) | 22.3 (32) |
| Rfree (%) | 22.7 (32) | 24.3 (34) | 21.0 (29) | 25.7 (36) | 28.4 (37) |
| Sigma cutoff | 0 | 0 | 0 | 0 | 0 |
| RMSDb in: | |||||
| Bond lengths (Å) | 0.005 | 0.006 | 0.005 | 0.008 | 0.008 |
| Bond angles (°) | 1.3 | 1.4 | 1.8 | 1.3 | 1.4 |
Values in parentheses are the statistics corresponding to the highest-resolution shell.
RMSD, root mean square deviation.
Structure solution and crystallographic refinement.
Structure solution and crystallographic refinement for all five structures were carried out using the Crystallography & NMR System version 0.9 (5). A previously published inhibitor complex (PDB:1MTR) (35) was used as the starting model for solving substrate structures, while for inhibitor complexes, one of our substrateWT structures (PDB:1F7A) (43) was utilized. This interchange of substrate structures for refining inhibitor complexes and vice versa was followed to reduce model bias. For structures whose space group is P212121, one protease dimer was used for conducting isomorphous molecular replacement. However, for the RTVV82A complex, whose asymmetric unit contain two dimers, the correct structure solution was arrived at by using molecular replacement techniques. The cell dimensions that were originally computed for RTVV82A by DENZO exhibited a monoclinic P21 cell with β = 99.8°; the translation solution for one of the dimers was unambiguously obtained (θ1 = 3.0°, θ2 = 14.4°, θ3 = 35.7°; tx = 4.5 Å, ty = 0.2 Å, tz = 14.0 Å). By fixing the position of this dimer, the translation solution search was repeated and the solution for the second dimer in the asymmetric unit was also arrived at (θ1 = 180.2°, θ2 = 77.6°, θ3 = 178.3°; tx = 14.8 Å, ty = 16.7 Å, tz = −3.4 Å). The packing for individual dimers was approximately 35%, and the two dimers combined yielded a packing of ∼65%, indicating that the structure solution has yielded reliable solutions.
The strategy used here for refining the five crystal structures has been discussed elsewhere (43, 44). Each typical cycle of refinement included positional refinement, B-factor refinement, and visual inspection of the model in interactive graphics using the molecular modeling package CHAIN (50). Noncrystallographic symmetry restraints were applied only between dimers of the RTVV82A complex. No noncrystallographic symmetry restraints were imposed within the dimers of all five complexes because they are asymmetric. Before the start of crystallographic refinement, 10% of the reflections were set aside for cross-validation (Rfree) purposes; throughout the entire refinement, Rfree was monitored. The treatment of Rfree was carried out as elaborated in the literature (3, 28, 29). The locations of the ligands and protein were further reaffirmed by computing simulated annealed omit maps (4, 6). Final refinement statistics are also provided in Table 1.
Structural analysis. (i) Difference distance plots.
The difference distance plots are constructed to assess intramolecular relative shifts in response to ligand binding (26). Initially, Cαi-to-Cαj distances ([dij]A) were computed between all of the Cα atoms within a protease dimer of a complex, A, and repeated for a second complex, B, with which the first structure is compared. The difference in these distances ([Dij] = |[dij]A − [dij]B|) was plotted as a two-dimensional contour plot as a function of residue numbers (i and j) by using GNUPLOT (61).
(ii) Buried surface area.
The classic Lee and Richards program (31) was used to evaluate the surface area buried by the ligands on complexation. Accessible surface area (ASA) was computed for each of the ligand-bound protease structures. The ASA calculation was repeated for the unbound ligands in the absence of the protease coordinates as well. The difference in these two ASA values is defined as the surface area buried on complexation.
(iii) Shape complementarity.
Besides buried surface area, another crucial feature that has been suggested to determine the specificity and stability of protein-ligand association is shape complementarity (SC) (30). SC is related to the dot product of the normals of the apposing surfaces and the exponent of the distances between them (for the complete mathematical formulation, see reference 30). The protocol includes the following: construction of molecular surface for both ligands and protein, calculation of normal vectors (nligand and nprotein) over a user-designated grid space, and computation of the intermolecular distance matrix between each intermolecular grid point (xligand − xprotein). The SC by the ligand on the protein is now given by the relationship SCligand→protein = (nligand · nprotein)exp[−w(xligand − xprotein)]. The SCligandxprotein is then computed for each grid point, summed, and averaged. The calculation is repeated to obtain SCprotein→ligand. Finally, the average of the two SC values is the actual SC. In this analysis, we quote SC values as SCV82A/SCWT ratios.
(iv) VDW interaction energy.
Estimates of VDW interaction energy were computed to provide a theoretical quantitative assessment for the ligand-protease nonbonded interactions. VDW energy between ith and jth atoms is calculated using the Lennard-Jones potential function. The Crystallography & NMR System was used to perform this task; it utilizes data from reference 13 to obtain interatomic distances and incorporates the PROLSQ REPEL function (20) for treating nonbonded interactions.
RESULTS
Overall structural features.
Three decameric substrate peptides from the Gag cleavage sites of matrix-capsid (MA-CA: VSQNY-PIVQN), capsid-p2 (CA-p2: KARVL-AEAMS) and p1-p6 (RPGNF-LQSRP) and two protease inhibitors, SQV and RTV, were crystallized in complex with the an inactive (D25N) variant of HIV-1 protease with the drug resistant V82A mutation. The crystallographic statistics for these structures are listed in Table 1. The inhibitors and the substrate peptides were unambiguously located in the two difference Fourier maps, 2Fo - Fc and Fo - Fc, and each of the ligands was uniquely oriented within the protease dimer. This uniqueness in the orientation is crucial for accurately determining ligand-protease interactions.
As in wild-type HIV-1 protease-substrate complexes (43, 44) and protease-substrate analogue structures (33, 34, 60), the substrate peptides in the V82A complexes also assume a relatively extended conformation (Fig. 1a and b). Also preserved in the V82A complexes is the toroid shape on the unprimed side of the substrate (Fig. 1c). This shape is achieved by the packing of P3 and/or P4 against the P1 side chain. In spite of having a glycine at P3, as seen in the corresponding wild-type complex, the p1-p6V82A peptide also forms a toroid through changes in the peptide backbone conformation (Fig. 1b). In contrast, the shapes of the five commercially available drugs, as observed in the wild-type complexes (7, 23-25, 55) do not form this toroid (Fig. 1d). Thus, the inhibitors do not mimic the conserved structural motif exhibited by the substrate sequences.
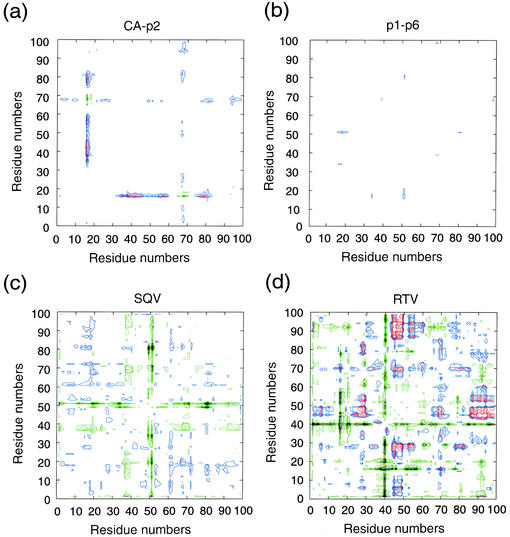
To probe the effect of the V82A mutation, we compared the structures of the ligandV82A complexes with their wild-type counterparts. To elucidate the distant structural alterations (26), difference distance plots were generated between the V82A and wild-type structures (24, 44, 55) of each ligand (Fig. 2). The plots involving substrate structures indicate only minor deviations (Fig. 2a and b), while the inhibitorV82A complexes, on the other hand, exhibit substantial shifts from the corresponding inhibitorWT structures, especially near the protease flaps (Lys45-Ile54) (Fig. 2c and d). Thus, the three substrateV82A complexes are very structurally similar to their substrateWT complexes, while inhibitorV82A complexes are not.
FIG. 2.
Double difference plots. Relative shifts in the V82A complexes in reference to the corresponding wild-type complexes. (a) CA-p2; (b) p1-p6; (c) SQV; (d) RTV. Each contour line represents a deviation by 0.25 Å. The different colors black, green, red, and blue distinguish the contour ranges −1.0 Å and below, −0.5 to −1.0 Å, 1.0 Å and above, and 0.5 to 1.0 Å, respectively.
Protease-ligand hydrogen bonds.
In protein-ligand interactions, hydrogen bonds play a crucial role in stabilizing the complex and, in many instances, also determining specificity. Analysis of the wild-type protease-ligand hydrogen bonds involving five inhibitors and six substrates suggests that the substrates form almost twice the number of hydrogen bonds with the protease (about 16) compared with the inhibitors (about 9) (Fig. 3a). Furthermore, the number of hydrogen bonds in each substrateWT complex is similar to that in the substrateV82A complexes whereas four hydrogen bonds were lost between the inhibitorWT and inhibitorV82A complexes. This difference (Fig. 3a) indicates that the mutation has specifically affected protease-inhibitor hydrogen bonding.
FIG. 3.
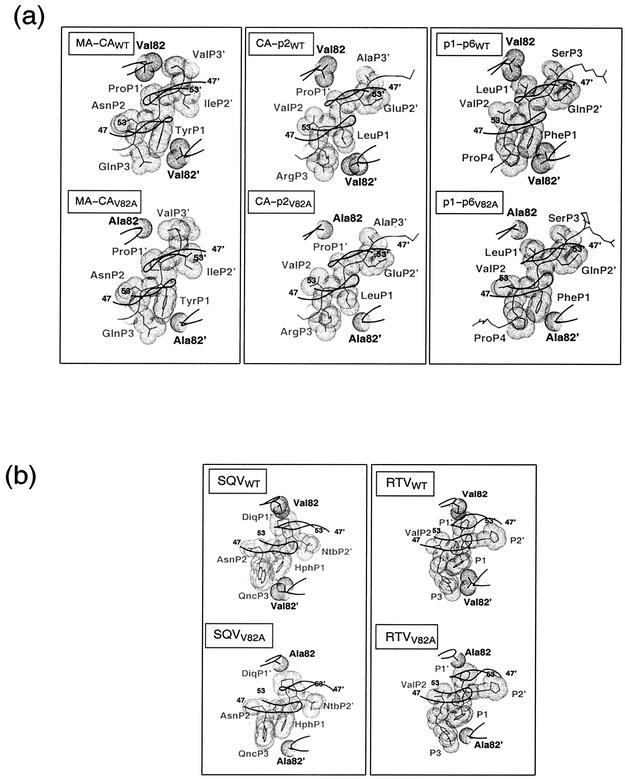
Protease-ligand hydrogen bonds. (a) Histogram representation of numbers of protease-ligand hydrogen bonds to elucidate differences between the substrate and inhibitor complexes. The numbers of hydrogen bonds in substrateWT and inhibitorWT complexes are in white; substrateV82A and inhibitorV82A complexes are colored black. (b) Ball-and-stick diagram of conserved hydrogen bonds (dotted lines) among the three substrateV82A complexes. Monomers are distinguished in black and gray, while the main-chain atoms of the CA-p2 peptide, shown as a representative substrate, are green. In this panel and the subsequent ones, nitrogen and oxygen atoms are highlighted by small spheres. (c) SQVV82A (black) and SQVWT (cyan): QncP3, 2-carbonylquinoline; HphP1, phenylalaninol group; DiqP1′, decahydro-1-methylsoquinoline-2-carbonyl; NtbP2′, tert-butylamino group. (d) RTVV82A (black) and RTVWT (cyan): P3, 2-isopropyl-4-thiazolyl; P2, valine; P1 and P1′, phenylalaninol groups; P2′, 5-thiazolyl.
As previously observed in substrateWT complexes (44), the hydrogen bonds conserved among these three substrateV82A complexes involve only the backbone atoms of the peptides (Fig. 3b). Nine hydrogen bonds are conserved among the three substrateV82A complexes, eight of which are conserved among all six substrateWT complexes as well. Thus, the conserved hydrogen bonding pattern present among the substrateWT complexes is retained among the substrateV82A complexes as well, indicating that the protease-substrate hydrogen bonding is not perturbed by the V82A mutation.
In contrast, the inhibitorWT complexes, SQVWT and RTVWT, make 13 and 11 protease-inhibitor hydrogen bonds, respectively, 9 of which are conserved between them (Fig. 3c and d). In the SQVV82A and RTVV82A complexes, however, there are nine and seven protease-inhibitor hydrogen bonds, respectively, five of which are conserved between them. The catalytic aspartic acids, Asp25/25′, make four hydrogen bonds with one atom each of the inhibitor in the inhibitorWT complexes, and these four hydrogen bonds are also seen in the inhibitorV82A complexes with nearly identical interatomic distances. The conservation of these four hydrogen bonds supports the likelihood that the D25N mutation has not caused any change in the complex. A large-scale shift that is seen in the protease flaps (1.2 Å in the RTVV82A relative to the RTVWT complex) and that is not in the proximity of Asn25/25′ substantially alters hydrogen bonding patterns in the inhibitorV82A complexes. Thus, the differences in protease-inhibitor hydrogen bonding patterns between the inhibitorWT and the inhibitorV82A complexes unambiguously reveal that inhibitor binding has been substantially affected in the V82A drug-resistant protease.
VDW interactions.
The surface area buried by the ligands on binding the protease molecule, which is an indicative parameter of the extensiveness of the ligand-protein interface, was computed by the method of Lee and Richards (31) (Table 2). Typically, substrates buried 850 to 1000 Å2 of their accessible surface area while the inhibitors account for only 500 to 650 Å2. This amounts to a 30 to 40% reduction of buried surface area by the inhibitors relative to the substrates. Although the change in buried surface area due to mutation is not very significant (Table 2), the difference in area between the substrates and inhibitors indicates that the substrate-protease association is more extensive than the inhibitor-protease interface.
TABLE 2.
Comparison of the structural analysis performed on the five V82A complexes and their wild-type equivalentsa
| Structure | Surface area buried by ligand on complexation (Å2)
|
Ratio in shape complementarity V82A/WT | No. of ligand-protease VDWb contacts
|
Ligand-protease estimated VDW interaction energy (kcal/mole)
|
|||
|---|---|---|---|---|---|---|---|
| WT | V82A | WT | V82A | WT | V82A | ||
| MA-CAc | 928 | 867 | 0.83 | 174 (5) | 159 (3) | 4.6 | 3.5 |
| CA-p2c | 977 | 923 | 1.00 | 167 (1) | 164 (1) | 1.7 | 3.1 |
| p1-p6c | 1,029 | 1,038 | 1.07 | 197 (10) | 202 (3) | 0.3 | 0.1 |
| SQV | 612 | 630 | 0.82 | 165 (7) | 142 (1) | 18.9d | 6.7 |
| RTV | 526 | 500 | 0.72 | 165 (11) | 130 (2) | 7.9 | 15.2 |
The structural analysis includes surface area buried by the ligand on complexation, shape complementarity, number of ligand-protease VDW contacts, and ligand protease estimated VDW interaction energy. The numbers shown in parentheses highlight the number of atoms located within VDW radius distance of residue 82 in the corresponding structures.
The distance criterion used for the VDW contacts is 2.4 to 4.2 Å.
In the comparison of the peptide complexes, only equivalent residues were used, since the termini were disordered in some of the crystal structures.
The refinement of SQVWT was carried out using X-PLOR version 2.1 which had slightly different non-bonded energy parameters.
Complementation of shape by the VDW surface of protease residue 82 with the VDW surfaces of the ligands is likely to provide further evidence for the influence of mutation (Fig. 4a and b). The VDW surface of residue 82 (valine or alanine) does not make any contact with VDW surface on the primed side of the substrates, P1′-P3′ (Fig. 4a). On the unprimed side of the peptide, however, Ala82′ contacts all the substrates tangentially while Val82′ contacts only PheP1 in the p1-p6WT complex, closely resembling the pattern observed in the p1-p6V82A complex. While the SQVWT and RTVWT complexes exhibit a high degree of packing between the VDW surfaces of the inhibitor and Val82/82′ (Fig. 4b), the VDW surfaces of both Ala82 and Ala82′ fail to contact those of the inhibitor in the SQVV82A and RTVV82A complexes (Fig. 4b).
FIG. 4.
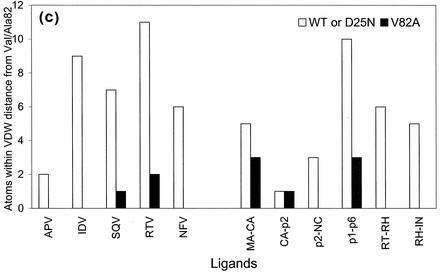
Protease-ligand VDW interactions. (a and b) VDW surfaces of residue 82, viewed down the dimer twofold axis, and the neighboring atoms of the corresponding substrates (a) and inhibitors (b). The ligands and the protease are shown in gray and black, respectively. A Cα trace of the overlying flaps is also shown. (c) Histogram representation of numbers of ligand atoms within VDW contact of residue 82 in the crystal structures determined here. The number of atoms in the substrateWT and inhibitorWT complexes are shown in white, and those in the substrateV82A and inhibitorV82A complexes are shown in black. Abbreviations are defined in the legend to Fig. 3.
In particular, VDW packing in the RTVWT complex is achieved by both side chain atoms of Val82′, CG1 and CG2, contacting the isopropyl group of the 2-isopropyl-4-thiazolyl at the P3 position of the drug. In addition, the isopropyl group extends the farthest at this site compared with any drug or substrate probed in this investigation (Fig. 1b). This isopropyl group appears to have been designed to target the pocket by the Val82 residue, and when an alanine mutation occurs, the P3 group is no longer complementary (Fig. 4b).
SQV, on the other hand, due to its similarities to the naturally occurring substrates, at least in the P2-P2′ region (Fig. 1b), is less vulnerable to V82A mutation. In fact, the computed shape complementarity (30) decreases by only 18% in SQV compared with 28% in RTV for the inhibitorV82A relative to corresponding wild-type complexes. However, with the exception of the MA-CA complexes, the substrate-protease shape complementarity between the wild type and V82A are very similar (Table 2).
The effect of mutation was further investigated by computing ligand-protease VDW interaction energy (Table 2). A comparison of the estimated VDW interaction energy between the wild-type and V82A substrate structures shows a difference of less than 1.5 kcal/mol. The SQVV82A complex, surprisingly, decreased VDW interaction energy by approximately 12 kcal/mol. However, this is probably because SQVWT complex (55) was refined with an earlier force field in X-PLOR 2.1 and even small differences in VDW radii can lead to substantial differences in the estimates of energies if different force fields are used. In the RTVV82A structure, where the refinement force field is the same for as that used to refine the RTVWT complex (24), the increase in computed VDW interaction energy, by nearly 10 kcal/mol, indicates that RTVV82A is less favorable than the RTVWT complex.
The number of substrate atoms within a VDW radius of residue 82 was compared in nine substrate complexes (three substrateV82A and six substrateWT complexes [43, 44]) and seven inhibitor structures (two inhibitorV82A and five inhibitorWT structures [7, 23-25, 55]) (Fig. 4c). In all drug complexes except APV, six or more atoms of the drug molecule surround the side chain atoms of Val82 (Fig. 4c), explaining the importance of Val82 for inhibitor binding. In contrast, five or fewer atoms of the substrate peptides surround Val82 in most of the substrateWT complexes. The only substrate complex for which this is an exception is the p1-p6WT complex, where 10 substrate atoms lie within the VDW contact distance of the Val82 side chain. The p1-p6V82A is also the only complex where contact of seven atoms within VDW radii of Ala82 is lost, thus reducing the VDW interaction considerably (Fig. 4c) compared with the p1-p6WT complex. However, this reduction seems to be compensated in the p1-p6V82A complex by the conservation of the total number of substrate-protease VDW contacts (Table 2). This compensation is achieved by subtle side chain alterations undergone by Asn at the P2 position of the p1-p6 substrate in combination with slight changes in Val32 and Ile47 of the protease. Thus, as seen in the CA-p2WT structure (43), the protease as well as its substrates are highly adaptable to each other.
In comparison with the inhibitorWT complexes, the inhibitorV82A complexes have far fewer atoms making VDW contacts with Ala82 (Fig. 4c). Only one atom comes within VDW contact of Ala82 in the SQVV82A complex, while seven atoms surround Val82 in the SQVWT complex. The decrease is even larger in the RTVV82A complex, where only 2 atoms come within the VDW distance of Ala82, while 11 atoms surround Val82 in the RTVWT complex. This clustering of atoms around Val82 in the inhibitor complexes suggests that the inhibitors were designed to contact the valine residue.
As an extension to VDW contacts by Val82 (or Ala82) with ligands, assessment of VDW contacts between the ligands and the entire protein is also crucial. The change in the number of substrate-protease VDW contacts was either absent (as in Ca-p2 and p1-p6) or nominal (as in MA-CA) (Table 2). However, significant loss in the number of inhibitor-protease VDW contacts was observed between the SQV complexes (165 in SQVWT and 142 in SQVV82A) (Table 2) and the RTV complexes (165 in RTVWT and 130 in RTVV82A). Thus, the overall effect of V82A mutation in the substrate complexes is minimal compared to its affect on the inhibitor complexes.
DISCUSSION
Our results reveal high structural similarity among the three V82A substrate complexes, CA-p2V82A, p1-p6V82A, and MA-CAV82A, and their previously reported wild-type counterparts (43, 44), particularly around the active-site region, suggesting that the V82A mutation has not influenced substrate binding. Conformational changes were observed, however, at the active site between the inhibitorV82A structures and their previously published wild-type counterparts (24, 55), suggesting that the protease mutation has indeed influenced inhibitor binding. In particular, the movement by the protease flaps in the RTVV82A complex by more than 1 Å in comparison to the RTVWT complex (24) is another indication of modified inhibitor binding. Difference distance plots show almost no shifts in the backbone as a result of V82A on substrate recognition (Fig. 2a and b) but show significant shifts in the inhibitor complexes (Fig. 2c and d). Thus, the V82A mutation appears to minimally perturb substrate binding while strongly affecting inhibitor binding.
Protease-ligand hydrogen bonds are crucial for retaining the stability of the complexes. The lack of difference in protease-substrate hydrogen bonds between the substrateV82A and substrateWT complexes (Fig. 3a) suggests that the specificity and stability of protease-substrate complexes is unaltered. The inhibitorV82A complexes, on the other hand, exhibit large-scale shifts with respect to their wild-type complexes, resulting in a loss of several hydrogen bonds. The substrates, which are longer than inhibitors, form more extensive hydrogen bond network with the protease, involving a variety of its residues and encompassing the entire active site (Fig. 3b). This network is probably a key factor in allowing the protease to maintain its function despite having many mutations. Because inhibitors in the inhibitorWT complexes have so few hydrogen bond acceptors and donors, the loss of hydrogen bonds due to the V82A mutation further decreases the specificity and affinity of the complex.
The asymmetric consensus volume occupied by the substrates compared to the relatively symmetric inhibitors illustrates the crucial differences between them (Fig. 1c and d). Coupled with our earlier findings with the wild-type protease-substrate complexes (44), this observation supports the hypothesis that the asymmetric toroidal shape on the unprimed side of the substrate is important for substrate specificity. The inhibitors, however, do not possess this shape (Fig. 1c). The toroidal shape of the substrates is the only conserved structural motif exhibited by the otherwise nonhomologous cleavage site sequences and could be utilized in designing the next generation of inhibitors. In fact, the positions corresponding to the P3-P1′ sites of the substrates may be used to model inhibitors instead of using the positions of P2-P2′. A recent study has revealed that inhibitors with a macrocyclic group connecting the P3 and P1 residues are likely to be more efficient (35).
Since hydrophobic residues in HIV-1 protease have a higher propensity for conferring drug-resistant mutations (54), a study of changes in VDW interactions around the mutation site is essential for understanding drug resistance. Lack of tight packing between the VDW surfaces of Val82/82′ and the substrate, seen in the substrateWT complexes, suggests that the valine residue may not be extensively utilized during substrate binding (Fig. 4a). The similarity between the substrateWT and substrateV82A complexes, particularly in relation to the VDW packing of Ala82/82′ with the substrates, is further supported by the conservation of the estimated VDW interaction energy (Table 2). On the other hand, the apposition of the VDW surfaces of the Val82/82′ residue and inhibitors (Fig. 4b) in the inhibitorWT complexes suggests that the alanine mutation at residue 82 will probably affect inhibitor binding. A previous study using the V82A mutant in complex with a C2-symmetry-based diol analogue, A-77003, pointed out that the protease is highly adaptable to accommodating the valine-to-alanine alteration (1). The present study, however, reveals substantial loss of VDW packing on both sides of the active-site cleft when residue 82 is an alanine in the inhibitorV82A complexes.
In addition, except for APV, all the drugWT complexes contain six or more inhibitor atoms within VDW contact of the Val82/82′ residues (Fig. 4c). A drastic drop in this number is observed near Ala82/82′ in the case of the inhibitorV82A complexes, while, with the exception of the p1-p6V82A complex, almost no change is exhibited in the substrate structures. The significant reduction in the p1-p6V82A complex (Fig. 4c) seems to be compensated by new interactions triggered by subtle side chain alterations by Asn (P2 of the p1-p6 substrate) and Val32 and Ile47 of the V82A protease. There is some indication, although not an absolute correlation, that when the computed buried surface area is large, as is the case for p1-p6, for the six substrateWT complexes (44), the measured kcat/Km values is small (11, 15, 52, 58). However, the relatively small kcat/Km of p1-p6 (11, 15, 52, 58) of the isolated peptide is not critical for viral maturation since the cleavage between nucleocapsid and p1 (NC-p1) is observed to be the rate-determining step in the processing of the polyproteins (42, 65). Among the inhibitors, RTV in particular contains an isopropyl group at the tip of the P3 site, which forms packing interactions with both CG1 and CG2 of Val82 and, unlike all the other drugs and substrates, is left substantially exposed when alanine is substituted for valine (Fig. 4b). Also, the reduction in shape complementarity (30) and the steep increase in the estimated VDW interaction energy (Table 2) lend further support to the notion that the V82A mutation more probably affects inhibitor binding. The fact that the corresponding values are almost unaltered in the substrate structures, especially for the p1-p6 complexes (Table 2), provides further evidence that Val82 need not be extensively utilized for substrate binding.
Kinetic assays for substrate cleavage and inhibitor binding for both wild-type and V82A variants have been performed extensively in other laboratories (11, 15, 18, 27, 32, 41, 52, 58). A comparison of kinetic parameters for these protease variants indicates only a moderate decrease in enzyme activity. The kcat,V82A/kcat,WT ratios range from 1 to 0.5, while the Km,V82A/Km,WT ratios range from 1 to 2 (18, 27). In contrast, the binding of the inhibitors RTV, IDV, and NFV is more severely affected by the mutation. The Ki,V82A/Ki,WT ratios range from 4.3 to 21.1, indicating that inhibition of V82A variant by these drugs, relative to their inhibition of wild-type protease, is substantially reduced (18, 27). These observations thus suggest that substrate cleavage by the V82A variant is relatively less compromised than the relative binding of the inhibitors IDV, NFV, and RTV. These findings are consistent with the conclusions of our structural analysis.
The structural analysis performed in this study of five ligand complexes of HIV-1 protease V82A in comparison with their wild-type counterparts elucidates the extent of adaptability that is possible when a mutation confers drug resistance. As drug therapy provides a selective pressure, HIV-1 protease will mutate in such a manner as to maintain activity while resisting the drugs. The implications of this study for future inhibitor design are that it is crucial to use only residues that are vital to substrate recognition as contact residues for new inhibitors. Perhaps this type of directed inhibitor-design will make it more difficult for further drug resistance to evolve.
Acknowledgments
Saquinavir and ritonavir were obtained through the NIH AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH. We thank Charles Craik from UCSF for the original D25N plasmid of HIV-1 protease. We also thank Balaji Bhyravbhatla for technical support during data collection, Vincent Chou and Nicole M. King for graphics assistance, and Claire Baldwin for editorial advice.
This research was supported by the American Cancer Society (RPG-99-213-01-MBC) and the National Institutes of Health (GM64849).
REFERENCES
- 1.Baldwin, E. T., T. N. Bhat, B. Liu, N. Pattabiraman, and J. W. Erickson. 1995. Structural basis of drug resistance for the V82A mutant of HIV-1 proteinase. Nat. Struct. Biol. 2:244-249. [DOI] [PubMed] [Google Scholar]
- 2.Boden, D., and M. Markowitz. 1998. Resistance to human immunodeficiency virus type 1 protease inhibitors. Antimicrob. Agents Chemother. 42:2775-2783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brunger, A. T. 1992. The free R value: a novel statistical quantity for assessing the accuracy of crystal structures. Nature 355:472-474. [DOI] [PubMed] [Google Scholar]
- 4.Brunger, A. T. 1991. Simulated annealing in crystallography. Annu. Rev. Phys. Chem. 42:197-223. [Google Scholar]
- 5.Brunger, A. T., P. D. Adams, G. M. Clore, W. L. DeLano, P. Gros, R. W. Grosse-Kunstleve, J. S. Jiang, Kuszewski, J., M. Nilges, N. S. Pannu, R. J. Read, L. M. Rice, T. Simonson, and G. L. Warren. 1998. Crystallography & NMR System: a new software suite for macromolecular structure determination. Acta Crystallogr. D54:905-921. [DOI] [PubMed]
- 6.Brunger, A. T., A. Krukowski, and J. Erickson. 1990. Slow-cooling protocols for crystallographic refinement by simulated annealing. Acta Crystallogr. A46:585-593. [DOI] [PubMed]
- 7.Chen, Z., Y. Li, E. Chen, D. L. Hall, P. L. Darke, C. Culberson, J. A. Shafer, and L. C. Kuo. 1994. Crystal structure at 1.9-Å resolution of human immunodeficiency virus (HIV) II protease complexed with L-735,524, an orally bioavailable inhibitor of the HIV proteases. J. Biol. Chem. 269:26344-26348. [PubMed] [Google Scholar]
- 8.Chou, K.-C., A. G. Tomasselli, I. M. Reardon, and R. L. Heinrickson. 1996. Predicting human immunodeficiency virus protease cleavage sites in proteins by a discriminant function method. Proteins 24:51-72. [DOI] [PubMed] [Google Scholar]
- 9.Condra, J. H., W. A. Schleif, O. M. Blahy, L. J. Gabryelski, D. J. Graham, J. C. Quintero, A. Rhodes, H. L. Robbins, E. Roth, M. Shivaprakash, D. Titus, T. Yang, H. Teppler, K. E. Squires, P. J. Deutsch, and E. Emini. 1995. In vivo emergence of HIV-1 variants resistant to multiple protease inhibitors. Nature 374:569-571. [DOI] [PubMed] [Google Scholar]
- 10.Deeks, S. G., R. M. Grant, G. W. Beatty, C. Horton, J. Detmer, and S. Eastman. 1998. Activity of a ritonavir plus saquinavir-containing regimen in patients with virologic evidence of indinavir or ritonavir failure. AIDS Res. Hum. Retroviruses 12:F97-F102. [DOI] [PubMed] [Google Scholar]
- 11.Dunn, B. M., A. Gustchina, A. Wlodawer, and J. Kay. 1994. Subsite preferences of retroviral proteinases. Methods Enzymol. 241:254-278. [DOI] [PubMed] [Google Scholar]
- 12.Eastman, P. S., J. Mittler, R. Kelso, C. Gee, E. Boyer, J. Kolberg, M. Urdea, J. M. Leonard, D. W. Norbeck, H. Mo, and M. Markowitz. 1998. Genotypic changes in human immunodeficiency virus type 1 associated with loss of suppression of plasma viral RNA levels in subjects treated with ritonavir (Norvir) monotherapy. J. Virol. 72:5154-5164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Engh, R. A., and R. Huber. 1991. Accurate bond and angle parameters for X-ray protein-structure refinement. Acta Crystallogr. A47:392-400.
- 14.Erickson, J. W., and K. B. Stanley. 1996. Structural mechanisms of HIV drug resistance. Annu. Rev. Pharmacol. Toxicol. 36:545-571. [DOI] [PubMed] [Google Scholar]
- 15.Ermolieff, J., X. Lin, and J. Tang. 1997. Kinetic properties of saquinavir-resistant mutants of human immunodeficiency virus type I protease and their implications in drug resistance in vivo. Biochemistry 36:12364-12370. [DOI] [PubMed] [Google Scholar]
- 16.Flexner, C. 1998. HIV-protease inhibitors. N. Engl. J. Med. 338:1281-1292. [DOI] [PubMed] [Google Scholar]
- 17.Forstenlehner, M. 2000. AIDS: new FDA approved agents AIDS. Pharm. Unserer Zeit 29:58.. [PubMed] [Google Scholar]
- 18.Gulnik, S. V., L. I. Suvorov, B. Liu, B. Yu, B. Anderson, H. Mitsuya, and J. W. Erickson. 1995. Kinetic characterization and cross-resistance patterns of HIV-1 protease mutants selected under drug pressure. Biochemistry 34:9282-9287. [DOI] [PubMed] [Google Scholar]
- 19.Henderíon, L. E., T. D. Copeland, R. C. Sowder, A. M. Schultz, and S. Oraszlan. 1988. Human retroviruses, cancer and AIDS: approaches to prevention and therapy. Alan R. Liss, New York, N.Y.
- 20.Hendrickson, W. A., and J. H. Konnert. 1980. In R. Diamond, S. Ramaseshan, and K. Venkatesan (ed.), Computing in crystallography, p. 13.01-13.23. Indian Academy of Sciences, Bangalore, India.
- 21.Hui, J. O., A. G. Tomasselli, I. M. Reardon, J. M. Lull, D. P. Brunner, C.-S. C. Tomich, and R. L. Heinrikson. 1993. Large scale purification and refolding of HIV-1 protease from Escherichia coli inclusion bodies. J. Protein Chem. 12:323-327. [DOI] [PubMed] [Google Scholar]
- 22.Ji, J. P., and L. A. Loeb. 1992. Fidelity of HIV-1 reverse transcriptase copying RNA in vitro. Biochemistry 31:954-958. [DOI] [PubMed] [Google Scholar]
- 23.Kaldor, S. W., V. J. Kalish, J. F. Davies, Jr., B. V. Shetty, J. E. Fritz, K. Appelt, J. A. Burgess, K. M. Campanale, N. Y. Chirgadze, D. K. Clawson, B. A. Dressman, S. D. Hatch, D. A. Khalil, M. B. Kosa, P. P. Lubbehusen, M. A. Muesing, A. K. Patick, S. H. Reich, K. S. Su, and J. H. Tatlock. 1997. Viracept (nelfinavir mesylate, AG1343): a potent, orally bioavailable inhibitor of HIV-1 protease. J Med. Chem. 40:3979-3985. [DOI] [PubMed] [Google Scholar]
- 24.Kemf, D. J., K. C. Marsh, J. F. Denissen, E. McDonald, S. Vasavanonda, C. A. Flentge, B. E. Green, N. L. Fino, C. H. Park, X. P. Kong, N. E. Wideburg, A. Saldivar, L. Ruiz, W. M. Kati, L. Sham, T. Robins, D. Stewart, A. Hsu, J. J. Plattner, J. M. Leonard, and W. Norbeck. 1995. ABT-538 is a potent inhibitor of human immunodeficiency virus protease and has high oral bioavailability in humans. Proc. Natl. Acad. Sci. USA 92:2484-2488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim, E. E., C. T. Baker, M. D. Dwyer, M. A. Murcko, B. G. Roa, R. D. Tung, and M. A. Navia. 1995. Crystal structure of HIV-1 protease in complex with VX-478, a potent and orally bioavailable inhibitor of the enzyme. J Am. Chem. Soc. 117:1181. [Google Scholar]
- 26.King, N. M., L. Melnick, M. Prabu-Jeyabalan, E. A. Nalivaika, S.-S. Yang, Y. Gao, X. Nie, C. Zepp, D. L. Heefner, and C. A. Schiffer. 2002. Lack of synergy for inhibitors targeting a multi-drug-resistant HIV-1 protease. Protein Sci. 11:418-429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Klabe, R. M., L. T. Bacheler, P. J. Ala, S. Erickson-Viitanen, and J. L. Meek. 1998. Resistance to HIV protease inhibitors: a comparison of enzyme inhibition and antiviral potency. Biochemistry 37:8735-8742. [DOI] [PubMed] [Google Scholar]
- 28.Kleywegt, G. J. 2000. Validation of protein crystal structures. Acta Crystallogr. D56:249-265. [DOI] [PubMed]
- 29.Kleywegt, G. J., and T. A. Jones. 1995. Where freedom is given liberties are taken. Structure 3:535-540. [DOI] [PubMed] [Google Scholar]
- 30.Lawrence, M. G., and P. M. Colman. 1993. Shape complementarity at protein/protein interfaces. J Mol. Biol. 234:946-950. [DOI] [PubMed] [Google Scholar]
- 31.Lee, B., and F. M. Richards. 1971. The interpretation of protein structures: estimation of static accessibility. J Mol. Biol. 55:379-400. [DOI] [PubMed] [Google Scholar]
- 32.Lin, Y., X. Lin, L. Hong, S. Foundling, R. L. Heinrikson, S. Thaisrivongs, W. Leelamanit, D. Raterman, M. Shah, B. M. Dunn, and J. Tang. 1995. Effect of point mutations on the kinetics and the inhibition of human immunodeficiency virus type I protease: relationship to drug resistance. Biochemistry 34:1143-1152. [DOI] [PubMed] [Google Scholar]
- 33.Mahalingam, B., J. Louis, C. Reed, J. Adomat, J. Krouse, Y. Wang, R. Harrison, and I. T. Weber. 1999. Structural and kinetic analysis of drug resistant mutants of HIV-1 protease. Eur. J Biochem. 263:238-245. [DOI] [PubMed] [Google Scholar]
- 34.Mahalingam, B., J. M. Louis, J. Hung, R. W. Harrison, and I. T. Weber. 2001. Structural implications of drug-resistant mutants of HIV-1 protease: high-resolution crystal structures of the mutant protease/substrate analogue complexes. Proteins 43:455-464. [DOI] [PubMed] [Google Scholar]
- 35.Martin, J. L., J. Begun, A. Schindeler, W. A. Wickramasinghe, D. Alewood, P. F. Alewood, D. A. Bergman, R. I. Brinkworth, G. Abbenante, D. R. March, R. C. Reid, and D. P. Fairlie. 1999. Molecular recognition of macrocyclic peptidomimetic inhibitors by HIV-1 protease. Biochemistry 38:7978-7988. [DOI] [PubMed] [Google Scholar]
- 36.McDonald, C. K., and D. R. Kuritkzkes. 1997. Human immunodeficiency virus type 1 protease inhibitors. Arch. Intern. Med. 157:951-959. [PubMed] [Google Scholar]
- 37.Minor, W. 1993. XDISPLAYF program. Purdue University, West Lafayette, Ind.
- 38.Molla, A., G. R. Granneman, E. Sun, and D. J. Kempf. 1998. Recent developments in HIV protease inhibitor therapy. Antiviral Res. 39:1-23. [DOI] [PubMed] [Google Scholar]
- 39.Molla, A., M. Korneyeva, Q. Gao, S. Vasavanonda, P. J. Schipper, H. M. Mo, M. Markowitz, T. Chernyavskiy, P. Niu, N. Lyons, A. Hsu, G. R. Granneman, D. D. Ho, C. A. Boucher, J. M. Leonard, D. W. Norbeck, and D. J. Kempf. 1996. Ordered accumulation of mutations in HIV protease confers resistance to ritonavir. Nat. Med. 2:760-766. [DOI] [PubMed] [Google Scholar]
- 40.Nicholls, A., K. Sharp, and B. Honig. 1991. Protein folding and association: insights from the interfacial and thermodynamic properties of hydrocarbons. Proteins 11:281-296. [DOI] [PubMed] [Google Scholar]
- 40a.Otwinowski, Z., and W. Minor. 1997. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276:307-326. [DOI] [PubMed] [Google Scholar]
- 41.Pazhanisamy, S., C. M. Stuver, A. B. Cullinan, N. Margolin, B. G. Rao, and D. J. Livingston. 1996. Kinetic characterization of human immunodeficiency virus type-1 protease-resistant variants. J. Biol. Chem. 271:17979-17985. [DOI] [PubMed] [Google Scholar]
- 42.Pettit, S. C., N. Sheng, R. Tritch, S. Erickson-Viitanen, and R. Swanstrom. 1998. The regulation of sequential processing of HIV-1 Gag by the viral protease. Adv. Exp. Med. Biol. 436:15-25. [DOI] [PubMed] [Google Scholar]
- 43.Prabu-Jeyabalan, M., E. Nalivaika, and C. A. Schiffer. 2000. How does a symmetric dimer recognize an asymmetric substrate? A substrate complex of HIV-1 protease. J. Mol. Biol. 301:1207-1220. [DOI] [PubMed] [Google Scholar]
- 44.Prabu-Jeyabalan, P., E. Nalivaika, and C. A. Schiffer. 2002. Substrate shape determines specificity of recognition for HIV-1 protease: analysis of crystal structures of six substrate complexes. Structure 10:369-381. [DOI] [PubMed] [Google Scholar]
- 45.Ridky, T., and J. Leis. 1995. Development of drug resistance to HIV-1 protease inhibitors. J. Biol. Chem. 270:29621-29623. [DOI] [PubMed] [Google Scholar]
- 46.Roberts, J. D., K. Bebenek, and T. A. Kunkel. 1988. The accuracy of reverse transcriptase from HIV-1. Science 242:1171-1173. [DOI] [PubMed] [Google Scholar]
- 47.Roberts, J. D., B. D. Preston, and L. A. Johnston. 1989. Fidelity of two retroviral reverse transcriptases during DNA-dependent DNA synthesis in vitro. Mol. Cell. Biol. 9:469-476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rosé, J. R., L. M. Babe., and C. S. Craik. 1995. Defining the level of human immunodeficiency virus type (HIV-1) protease activity required for HIV-1 particle maturation and infectivity. J Virol. 69:2751-2758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rosé, R. B., C. S. Craik, N. L. Douglas, and R. M. Stroud. 1996. Three-dimensional structures of HIV-1 and SIV protease product complexes. Biochemistry 35:12933-12944. [DOI] [PubMed] [Google Scholar]
- 50.Sack, J. S. 1988. CHAIN—a crystallographic modeling program. J Mol. Graphics 6:224-225. [DOI] [PubMed] [Google Scholar]
- 51.Schinazi, R. F., B. A. Larder, and J. W. Mellors. 1997. Mutations in retroviral genes associated with drug resistance. Int. Antiviral News 5:129-142. [Google Scholar]
- 52.Schock, H. B., V. M. Garsky, and L. C. Kuo. 1996. Mutational anatomy of an HIV-1 protease variant conferring cross-resistance to protease inhibitors in clinical trials. J. Biol. Chem. 271:31957-31963. [DOI] [PubMed] [Google Scholar]
- 53.Scott, W. R., and C. A. Schiffer. 2000. Curling of flap tips in HIV-1 protease as a mechanism for substrate entry and tolerance of drug resistance. Struct. Fold. Des. 8:1259-1265. [DOI] [PubMed] [Google Scholar]
- 54.Shafer, R. W., D. Stevenson, and B. Chan. 1999. Human immunodeficiency virus reverse transcriptase and protease sequence database. Nucleic Acids Res. 27:348-352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Spinelli, S., Q. Liu, Z., P. M. Alzari, P. H. Hirel, and R. J. Poljak. 1991. The three-dimensional structure of the aspartyl protease from the HIV-1 isolate BRU. Biochemie 73:1391-1396. [DOI] [PubMed] [Google Scholar]
- 56.Swanstrom, R., and J. Eron. 2000. Human immunodeficiency virus type-1 protease inhibitors: therapeutic successes and failures, supression and resistance. Pharmacol. Ther. 86:145-170. [DOI] [PubMed] [Google Scholar]
- 57.Thanki, N., J. K. M. Rao, S. I. Foundling, J. Howe, J. B. Moon, J. O. Hui, A. G. Tomasselli, R. L. Heinrikson, S. Thaisrivongs, and A. Wlodawer. 1992. Crystal structure of a complex of HIV-1 protease with dihydroxyethylene-containing inhibitor: comparisons with molecular modeling. Protein Sci. 1:1061-1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tozser, J., I. Blaha, T. D. Copeland, E. M. Wondrak, and S. Oroszlan. 1991. Comparison of the HIV-1 and HIV-2 proteinases using oligopeptide substrates representing cleavage sites in Gag and Gag-Pol polyproteins. FEBS Lett. 281:77-80. [DOI] [PubMed] [Google Scholar]
- 59.Vacca, J. P., B. D. Dorsey, W. A. Schleif, R. B. Levin, S. L. McDaniel, P. L. Darke, J. Zugay, J. C. Quintero, O. M. Blahy, E. Roth, V. V. Sardana, A. J. Schlabach, P. I. Graham, J. H. Condra, L. Gotlib, M. K. Hollloway, J. Lin, I.-W. Chen, K. Vastag, D. Ostovic, P. S. Anderson, E. A. Emini, and J. R. Huff. 1994. L-735,524: an orally bioavailable human immunodeficiency virus type 1 protease inhibitor. Proc. Natl. Acad. Sci. USA 91:4096-4100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Weber, I. T., J. Wu, J. Adomat, R. W. Harrison, A. R. Kimmel, E. M. Wondrak, and J. M. Louis. 1997. Crystallographic analysis of human immunodeficiency virus 1 protease with an analog of the conserved CA-p2 substrate-interactions with frequently occurring glutamic acid residue at P2′ position of substrates. Eur. J Biochem. 249:523-530. [DOI] [PubMed] [Google Scholar]
- 61.Williams, T., and C. Kelley. 1998. GNUPLOT © 1986-1993, 1998 (contact for further information http://www.gnuplot.info).
- 62.Wlodawer, A., and J. W. Erickson. 1993. Structure-based inhibitors of HIV-1 protease. Annu. Rev. Biochem. 62:543-585. [DOI] [PubMed] [Google Scholar]
- 63.Wlodawer, A., and A. Gustchina. 2000. Structural and biochemical studies of retroviral proteases. Biochim. Biophys. Acta 1477:16-34. [DOI] [PubMed] [Google Scholar]
- 64.Wlodawer, A., M. Miller, B. K. Sathyanarayana, E. Baldwin, I. T. Weber, L. M. Selk, L. Clawson, J. Schneider, and S. B. Kent. 1989. Conserved folding in retroviral proteases: crystal structure of a synthetic HIV-1 protease. Science 245:616-621. [DOI] [PubMed] [Google Scholar]
- 65.Zhang, Y., H. Qian, Z. Love, and E. Barklis. 1998. Analysis of the assembly function of the human immunodeficiency virus type 1 gag protein nucleocapsid domain. J. Virol. 72:1782-1789. [DOI] [PMC free article] [PubMed] [Google Scholar]