Abstract
Chimeric yellow fever virus (YF) RNAs were constructed in which the YF structural genes were replaced by the hepatitis C virus (HCV) structural genes or fusions between the YF and HCV structural genes. Interestingly, RNA replication required nucleotide complementarity between the 3′-located conserved sequence 1 and an RNA sequence located in the 5′ end of the YF capsid sequence. The (chimeric-)HCV structural proteins were efficiently expressed and processed, and the native E1/E2 heterodimer was formed. However, no indication for the production of HCV-like particles was obtained.
Yellow fever virus (YF) is the type member of the genus Flavivirus in the family Flaviviridae, a group of viruses that also contains the genera Pestivirus and Hepacivirus (for recent reviews, see references 5 and 23). The YF genome is a positive-stranded RNA molecule of approximately 11 kb that contains a 5′ end cap structure. The 3′ end is nonpolyadenylated but instead contains an extensive secondary RNA structure (17, 27, 28). The genome encodes for a single polyprotein of over 350 kDa that is co- and posttranslationally processed by host and viral proteases (23). The capsid (C) protein is released from the N-terminal part of this polyprotein by the viral protease NS2B-3. The envelope proteins prM and E are processed by signal peptidase cleavages, while the YF nonstructural proteins (NS1, NS2A, NS2B, NS3, NS4A, NS4B, NS5) are processed from the remainder of the polyprotein by host proteases and the viral NS2B-3 protease.
Hepatitis C virus (HCV) has been classified as a separate genus (Hepacivirus) in the family Flaviviridae. HCV encodes a polyprotein of approximately 3,000 amino acids. Co- and posttranslational cleavage of the polyprotein generates at least 10 polypeptides: the putative structural proteins C, E1, and E2 and several nonstructural proteins (NS2, NS3, NS4A, NS4B, NS5A, NS5B). In addition, a small hydrophobic polypeptide (p7) is encoded between the E2 and NS2 genes. The function of this polypeptide is unknown, but studies on the closely related bovine viral diarrhea virus p7 protein indicate that it has a function in virion formation (18). The C protein is the putative nucleocapsid protein, and E1 and E2 are the HCV envelope glycoproteins (15, 25) that form heterodimers stabilized by noncovalent interactions (11). These heterodimers are believed to constitute the native prebudding complex (11) and do not leave the endoplasmic reticulum (ER) due to retention signals present in the C termini of E1 and E2 (8, 9, 14).
Studies on HCV have been hampered by the lack of an efficient cell culture system. Recently, a system that allowed the replication of selectable full-length HCV genomes in a hepatoma cell line was established (19, 26). Despite the expression of HCV structural proteins, no evidence for the assembly of virions could be obtained.
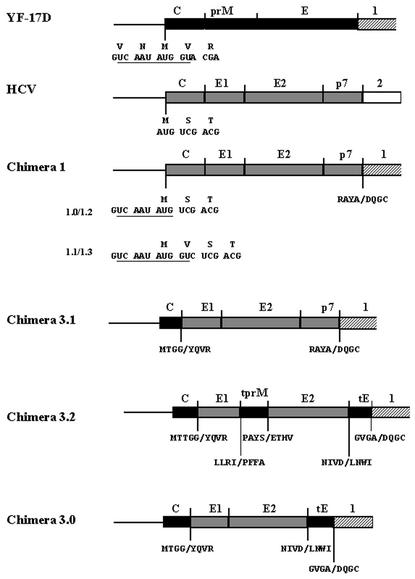
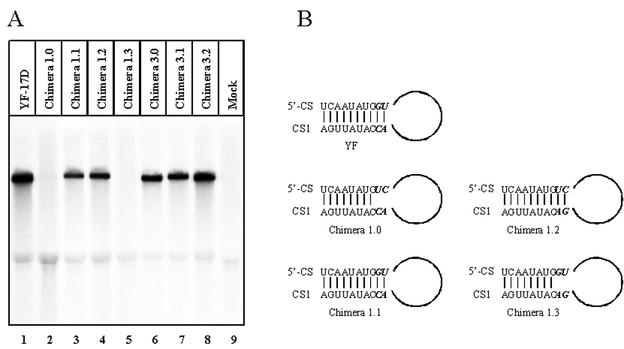
As a tool to study the expression of the HCV structural proteins and possibly HCV assembly, chimeric YF/HCV genomes were constructed in which the structural genes of YF-17D were replaced by the structural genes of a genotype 1a HCV strain. In the type 1 chimeras, the region encoding the YF structural proteins C, prM, and E (YF nucleotides [nt] 161 through 2452) was replaced by the region encoding the HCV structural proteins C, E1, and E2, as well as p7 (HCV nt 342 through 2768 [Fig. 1 ]). A major complication in the construction of these chimeras was the presence of a highly conserved sequence (5′ CS; nt 156 through 165) (17) overlapping with a downstream AUG codon in the YF-C gene (Fig. 1). This sequence is essential for flavivirus replication by interacting with a complementary sequence (CS1) at the 3′ end of the genome (20; J. P. Bredenbeek et al., submitted for publication). In view of this, four different type 1 chimeras were constructed (1.0, 1.1, 1.2, and 1.3) by fusion PCR. The authentic YF C gene AUG translation initiation codon was mutated in all four chimeric genomes, forcing translation to start at the downstream AUG codon overlapping the 5′ CS. The HCV-C gene was fused at this downstream AUG codon. In chimera 1.0, the YF 5′ CS lacked 2 nt (Fig. 2B), which could be detrimental to the replication of this chimeric genome. In chimera 1.1, the authentic 5′ CS was retained, resulting in an extra Val residue in HCV-C as the second amino acid (Fig. 1 and 2B). To circumvent adding amino acids to the HCV-C protein, chimera 1.2 was constructed by introducing compensating mutations in CS1 of chimera 1.0 (Fig. 2B). In this way, the putative 5′ CS-CS1 interaction as well as the authentic HCV-C protein sequence was retained. Finally, a construct (chimera 1.3) was created in which the (wild-type) 5′ CS was combined with a mutant CS1 (Fig. 2B). In this construct, the putative 5′ CS-CS1 interaction was partially disrupted, in addition to the introduction of an extra Val residue in the HCV-C protein. The replication of the in vitro-transcribed YF/HCV chimeric RNAs was analyzed by electroporating transcripts into BHK-21J cells (30). Viral RNA was labeled from 18 to 24 h postelectroporation with [3H]uridine in the presence of actinomycin D and analyzed on agarose gels (Bredenbeek et al., submitted). No viral RNA synthesis was detected in cells transfected with chimeras 1.0 and 1.3 (Fig. 2A, lanes 2 and 5). In contrast, chimeras 1.1 and 1.2 and the YF-17D control replicated efficiently (Fig. 2B, lanes 1, 3, and 4). In all lanes, a band corresponding to 28S rRNA was clearly detected, suggesting that the lack of chimera 1.0 and 1.3 RNA was not due to technical problems. These results clearly illustrate that the putative 5′ CS-CS1 interaction is crucial for efficient YF RNA replication. All further analyses were carried out with chimeras 1.1 and 1.2. Expression and subcellular localization of the HCV glycoproteins expressed from chimera 1.2 were analyzed by immunofluorescence assay (IFA) (Bredenbeek et al., submitted). When stained with monoclonal antibodies (MAbs) directed against HCV-E1 (MAb A4 [13]) and HCV-E2 (MAb H53 [9]), a specific perinuclear staining was observed, suggesting that the glycoproteins are localized in the ER (Fig. 3). These results were confirmed by counterstaining for the ER resident protein disulfide isomerase (data not shown). In various expression systems, a large fraction of E1 and E2 forms misfolded aggregates (7, 12, 13, 15). Since MAb H53 does not recognize misfolded E2 (9), the IFA staining with MAb H53 suggests that at least a fraction of the HCV glycoproteins is properly folded. Cells stained with HCV-C MAb MA1-080 (24) showed a clear membrane-associated signal for both chimeras. Western blot analysis of YF-NS1 and HCV-C, -E1, and -E2 indicated complete processing of the chimeric protein precursor (data not shown). Essentially similar results were obtained with chimera 1.1 (data not shown). To study the kinetics of synthesis and maturation of HCV-E1 and -E2, pulse-chase experiments were performed (9, 13) and the sensitivity of these proteins to endo=β-N-acetylglucosaminidase H (endo H) was analyzed. Proteins were labeled for 30 min and subsequently chased for up to 4 h. Immunoprecipitations (IP) were performed with the conformation-sensitive E2 MAb H53 (Fig. 4A). For both type 1 chimeras, coprecipitation of E1 was observed, indicating that native E1/E2 heterodimers were formed. No Endo H-resistant E1 and E2 proteins were observed, indicating that at least the majority of the glycoproteins were retained in the ER or cis-Golgi compartment. Chase periods of up to 8 h gave similar results (data not shown). Essentially similar data were obtained with the E1-specific MAb A4 (data not shown). The lack of Endo H-resistant E1 and E2 suggested that HCV-like particles were not produced by type 1 chimeras. The assembly of HCV-like particles is likely dependent on RNA encapsidation sequences for the formation of intracellular HCV capsids. The type 1 chimeric genomes could lack such sequences. Therefore, an alternative approach would be to construct chimeric genomes that express the YF-C protein to ensure the formation of YF capsid structures. These capsid structures are supposed to interact during virus assembly with the cytoplasmic domains of the YF, or possibly HCV, prebudding complex. Studies on YF/DEN and YF/JE chimeras indicate that intratypic interactions between the capsid and prebudding complex of flaviviruses are, to some extent, conserved (6, 16, 29). To pursue this approach, a set of YF/HCV chimeric genomes (type 3) was constructed in which the C gene of YF was present, in addition to various fusions between (parts of) the glycoprotein genes of YF and HCV. Studies using flavivirus replicons (21; E. A. Kooi and P. J. Bredenbeek, unpublished data) demonstrated that all sequences required for assembly are located outside the structural genes. Type 3 chimeric RNAs should thus contain all sequences required for the assembly of YF capsids. In chimera 3.1, the HCV E1, E2, and p7 coding sequences were replaced by the YF-prM and YF-E genes (Fig. 1), allowing for the assembly of YF capsids and an HCV prebudding complex. In chimera 3.2 (Fig. 1), the sequences for YF-prM (YF nt 482 through 866) and -E (YF nt 974 through 2318) ectodomains were replaced by the putative ectodomains of HCV-E1 (HCV nt 915 through 1365) and HCV-E2 (HCV nt 1491 through 2436), respectively (10). These glycoprotein domain fusions were aimed at generating glycoprotein complexes that would resemble HCV E1/E2 complexes at the luminal side of the ER while resembling the YF prebudding complex at the cytosolic side. Finally, as a control, chimera 3.0 (Fig. 1) was constructed. In chimera 3.0, the sequences encoding YF-prM and the ectodomain of E (YF nt 482 through 2318) were replaced by the HCV-E1 coding sequence and the putative ectodomain coding sequences of HCV-E2 (HCV nt 915 through 2436), respectively. This chimeric genome is likely unable to produce E1/E2 heterodimer due to the absence of interaction between the HCV-E1 and YF-E transmembrane domains (TMDs). All of the type 3 chimeric RNAs replicated at similar efficiencies compared to the YF-17D control (Fig. 2A, lanes 6, 7, and 8). IFA revealed that HCV-E1 and -E2 were efficiently expressed (Fig. 3). To study the maturation of HCV glycoproteins, we performed pulse-chase experiments. Upon IP with MAb H53 (Fig. 4B), E2 was detected but no coprecipitation of E1 was observed for chimeras 3.0 and 3.2. In contrast, for chimera 3.1, clear coprecipitation of E1 was detected. These results suggest that chimeras 3.0 and 3.2 are unable to form the noncovalently linked E1/E2 heterodimers. Upon IP with MAb A4 (Fig. 4C), coprecipitation of E2 was observed for all type 3 chimeras. This observation is probably due to precipitation of the cysteine-linked E1/E2 aggregates with MAb A4. The glycoproteins and glycoprotein complexes produced by all type 3 chimeric replicons did not acquire Endo H resistance during a 4-h chase period, demonstrating that at least the majority of the glycoproteins were retained in the ER or cis-Golgi compartment. Again, these data suggest that no HCV-like particles were produced.
FIG. 1.
Schematic outline of the structural gene region of YF, HCV, and the chimeric genomes used in this study. YF structural genes are indicated by black boxes, and HCV structural genes are indicated by shaded boxes. Hatched boxes represent the YF-NS1 gene. The amino acid sequences of the constructed gene fusions are shown below the fusion points. The nucleotide sequence at the C fusion of the type 1 chimeras is also shown. The 5′ CS is underlined. tprM, TMD and C terminus of YF-prM; tE, TMD and C terminus of YF-E.
FIG. 2.
(A) Analysis of viral RNA replication. Cells were electroporated with the indicated chimeric RNAs and labeled with [3H]uridine in the presence of actinomycin D. RNA was isolated, denaturated with glyoxal, and separated on an agarose gel. (B) Putative cyclization motifs in YF and type 1 YF/HCV chimeras. Mutated sequences are shown in bold italics. In chimeras 1.0 and 1.3, the complementarity between 5′ CS and CS1 is partially destroyed whereas the complementarity is restored in chimeras 1.1 and 1.2. See text for details.
FIG. 3.
Indirect immunofluorescence analysis of HCV protein expression by YF/HCV chimeras. Typical results are shown for YF-17D, chimera 1.2, and chimera 3.2. Specific immunofluorescence was detected with MAbs recognizing HCV-E1 (MAb A4), HCV-E2 (MAb H53), and HCV-C (MAb MA1-080).
FIG. 4.
Pulse-chase analysis of HCV glycoprotein maturation and glycosylation. (A) Cells electroporated with type 1 chimeric RNA were pulse-labeled and subsequently chased for 4 h. Lysates were immunoprecipitated with anti-E2 (MAb H53) and treated with Endo H (+) or left untreated (−). (B and C) Cells electroporated with type 3 chimeric RNA were pulse-labeled and subsequently chased for 4 h. Lysates were immunoprecipitated with anti-E2 (MAb H53) (panel B) or anti-E1 (MAb A4) (panel C) and treated with Endo H (+) or left untreated (−).
For tick-borne encephalitis virus, it has been demonstrated that de novo-synthesized prM and E form heterodimers (1, 31). The interaction between the TMDs of the prM and E proteins is required for this heterodimer formation (1, 2). The design of chimera 3.2 was based on these apparently similar characteristics of the HCV and flavivirus envelope protein TMDs. However, the results obtained with chimera 3.2 demonstrated that the YF-prM and YF-E TMDs could not substitute for the function of the HCV-E1 and HCV-E2 TMDs in the formation of E1/E2 heterodimers. Exchanging the E2 TMD for the TMD of CD4 demonstrated that this E2-CD4 protein had lost the ability to interact with E1 (9). However, it could be argued that the rapid transport of the E2-CD4 protein to the cell surface prevented interactions required for the heterodimerization of E1 and E2-CD4. Our results with chimeras 3.0 and 3.2 show that, even when the TMDs of E1 and E2 are replaced by similar TMDs from proteins normally retained in the ER, no heterodimerization between E1 and E2 takes place. These results provide further evidence for a direct role of the HCV-E1 and -E2 TMDs in the formation of the native glycoprotein complex. Since a cell culture system in which HCV particles are produced is not available, it is unknown whether E1 and E2 acquire Endo H resistance during secretion of the virus. When the E2 ectodomain was fused to the TMD of CD4, the chimeric protein did acquire Endo H resistance (9). CD4 is a protein that is normally transported to the cell surface via the secretory pathway, which is also proposed to be the pathway used for the release of flavivirus particles. We therefore believe that, if the YF/HCV genomes produced HCV-like particles, at least a fraction of the glycoprotein complexes should have acquired Endo H resistance. Since it is unknown if E1 or E2 present in virus particles undergoes conformational changes during transport through the Golgi apparatus, it might have been possible that an Endo H-resistant E1/E2 complex was no longer recognized by the conformation-sensitive E2 MAb H53. However, pulse-chase experiments using conformation-independent E2 MAbs (data not shown) yielded similar results. Additional indications for the lack of HCV-like particle production were obtained by transmission and immuno-electron microscopy studies on transfected cells and reverse transcription-PCR analysis of the medium harvested from these cells (data not shown). Interestingly, intracellular HCV-like particles were observed by expression of the HCV structural proteins in the baculovirus expression system (3) or by Semliki Forest virus replicons (4). However, these particles were not secreted into the medium. Compared to the YF replicons used in this study, the baculovirus and Semliki Forest virus expression systems produce a much larger amount of proteins. It still has to be established whether the particles produced in these (over-)expressing systems truly resemble native HCV particles.
We can envisage several explanations for the lack of HCV-like particle production using YF replicons. (i) An important HCV-specific RNA signal required for the encapsidation of the viral RNA might be lacking in our type 1 chimeras. Assembly of HCV capsids at the site of the prebudding complex might be required for the assembly of HCV virions. If YF capsids could substitute for HCV capsids, it should be expected that the 3.1 chimera produces HCV-like particles. However, it might be possible that the evolutionary relationship between HCV and YF is not close enough for the conservation of intratypic capsid-prebudding complex interactions. Since the chimera types 3.0 and 3.2 do not produce the E1/E2 heterodimer, it is unlikely that in these cases a functional HCV prebudding complex can be formed. (ii) A cellular factor required for the morphogenesis of HCV-like particles might be lacking in the BHK-21J cells. Also in HepG2 and Huh-7 cells, no evidence has been obtained for HCV-like particles (R. Molenkamp and P. J. Bredenbeek, unpublished data), suggesting that, if such a factor exists, it is lacking in all tested cell types. Alternatively, the studied cell lines might express factors that have an inhibitory effect on HCV-like particle formation. (iii) An HCV-encoded factor not present in our YF/HCV chimeric RNAs might be required for the formation of infectious virus. Recent data suggest that YF NS2A plays a crucial role in the formation of infectious particles (22). However, the recently established full-length selectable HCV RNAs (19, 26) containing the entire HCV open reading frame also do not produce virus particles in Huh-7 cells. The absence of HCV-like particle production by our YF/HCV chimeras might actually be due to a combination of the factors mentioned above.
Acknowledgments
We thank Nicolette Huijkman for technical assistance during the initial stages of this work. We are grateful to Harry Greenberg, Jean Dubuisson, and Jacob Schlessinger for MAbs A4, H53, and 1A5.
R.M. was supported by EU grant QLK2-CT-1999-00356. E.A.K. was supported by grant 901-02-173 from the Council for Medical and Health Research of The Netherlands Organization for Scientific Research (NWO-MW).
REFERENCES
- 1.Allison, S. L., K. Stadler, C. W. Mandl, C. Kunz, and F. X. Heinz. 1995. Synthesis and secretion of recombinant tick-borne encephalitis virus protein E in soluble and particulate form. J. Virol. 69:5816-5820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Allison, S. L., K. Stiasny, K. Stadler, C. W. Mandl, and F. X. Heinz. 1999. Mapping of functional elements in the stem-anchor region of tick-borne encephalitis virus envelope protein E. J. Virol. 73:5605-5612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baumert, T. F., S. Ito, D. T. Wong, and T. J. Liang. 1998. Hepatitis C virus structural proteins assemble into viruslike particles in insect cells. J. Virol. 72:3827-3836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blanchard, E., D. Brand, S. Trassard, A. Goudeau, and P. Roingeard. 2002. Hepatitis C virus-like particle morphogenesis. J. Virol. 76:4073-4079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Burke, D. S., and T. P. Monath. 2001. Flaviviruses, p. 1043-1125. In D. M. Knipe, and P. M. Howley (ed.), Fields virology. Lippincott-Raven Publishers, Philadelphia, Pa.
- 6.Chambers, T. J., A. Nestorowicz, P. W. Mason, and C. M. Rice. 1999. Yellow fever/Japanese encephalitis chimeric viruses: construction and biological properties. J. Virol. 73:3095-3101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Choukhi, A., A. Pillez, H. Drobecq, C. Sergheraert, C. Wychowski, and J. Dubuisson. 1999. Characterization of aggregates of hepatitis C virus glycoproteins. J. Gen. Virol. 80:3099-3107. [DOI] [PubMed] [Google Scholar]
- 8.Cocquerel, L., S. Duvet, J. C. Meunier, A. Pillez, R. Cacan, C. Wychowski, and J. Dubuisson. 1999. The transmembrane domain of hepatitis C virus glycoprotein E1 is a signal for static retention in the endoplasmic reticulum. J. Virol. 73:2641-2649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cocquerel, L., J. C. Meunier, A. Pillez, C. Wychowski, and J. Dubuisson. 1998. A retention signal necessary and sufficient for endoplasmic reticulum localization maps to the transmembrane domain of hepatitis C virus glycoprotein E2. J. Virol. 72:2183-2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cocquerel, L., C. Wychowski, F. Minner, F. Penin, and J. Dubuisson. 2000. Charged residues in the transmembrane domains of hepatitis C virus glycoproteins play a major role in the processing, subcellular localization, and assembly of these envelope proteins. J. Virol. 74:3623-3633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Deleersnyder, V., A. Pillez, C. Wychowski, K. Blight, J. Xu, Y. S. Hahn, C. M. Rice, and J. Dubuisson. 1997. Formation of native hepatitis C virus glycoprotein complexes. J. Virol. 71:697-704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dubuisson, J. 2000. Folding, assembly and subcellular localization of hepatitis C virus glycoproteins. Curr. Top. Microbiol. Immunol. 242:135-148. [DOI] [PubMed] [Google Scholar]
- 13.Dubuisson, J., H. H. Hsu, R. C. Cheung, H. B. Greenberg, D. G. Russell, and C. M. Rice. 1994. Formation and intracellular localization of hepatitis C virus envelope glycoprotein complexes expressed by recombinant vaccinia and Sindbis viruses. J. Virol. 68:6147-6160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Duvet, S., L. Cocquerel, A. Pillez, R. Cacan, A. Verbert, D. Moradpour, C. Wychowski, and J. Dubuisson. 1998. Hepatitis C virus glycoprotein complex localization in the endoplasmic reticulum involves a determinant for retention and not retrieval. J. Biol. Chem. 273:32088-32095. [DOI] [PubMed] [Google Scholar]
- 15.Grakoui, A., C. Wychowski, C. Lin, S. M. Feinstone, and C. M. Rice. 1993. Expression and identification of hepatitis C virus polyprotein cleavage products. J. Virol. 67:1385-1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guirakhoo, F., J. Arroyo, K. V. Pugachev, C. Miller, Z. X. Zhang, R. Weltzin, K. Georgakopoulos, J. Catalan, S. Ocran, K. Soike, M. Ratterree, and T. P. Monath. 2001. Construction, safety, and immunogenicity in nonhuman primates of a chimeric yellow fever-dengue virus tetravalent vaccine. J. Virol. 75:7290-7304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hahn, C. S., Y. S. Hahn, C. M. Rice, E. Lee, L. Dalgarno, E. G. Strauss, and J. H. Strauss. 1987. Conserved elements in the 3′ untranslated region of flavivirus RNAs and potential cyclization sequences. J. Mol. Biol. 198:33-41. [DOI] [PubMed] [Google Scholar]
- 18.Harada, T., N. Tautz, and H. J. Thiel. 2000. E2-p7 region of the bovine viral diarrhea virus polyprotein: processing and functional studies. J. Virol. 74:9498-9506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ikeda, M., M. Yi, K. Li, and S. M. Lemon. 2002. Selectable subgenomic and genome-length dicistronic RNAs derived from an infectious molecular clone of the HCV-N strain of hepatitis C virus replicate efficiently in cultured Huh7 cells. J. Virol. 76:2997-3006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Khromykh, A. A., H. Meka, K. J. Guyatt, and E. G. Westaway. 2001. Essential role of cyclization sequences in flavivirus RNA replication. J. Virol. 75:6719-6728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Khromykh, A. A., A. N. Varnavski, and E. G. Westaway. 1998. Encapsidation of the flavivirus kunjin replicon RNA by using a complementation system providing Kunjin virus structural proteins in trans. J. Virol. 72:5967-5977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kummerer, B. M., and C. M. Rice. 2002. Mutations in the yellow fever virus nonstructural protein NS2A selectively block production of infectious particles. J. Virol. 76:4773-4784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lindenbach, B. D., and C. M. Rice. 2001. Flaviviridae: the viruses and their replication, p. 991-1041. In D. M. Knipe, and P. M. Howley (ed.), Fields virology. Lippincott-Raven Publishers, Philadelphia, Pa.
- 24.Moradpour, D., T. Wakita, K. Tokushige, R. I. Carlson, K. Krawczynski, and J. R. Wands. 1996. Characterization of three novel monoclonal antibodies against hepatitis C virus core protein. J. Med. Virol. 48:234-241. [DOI] [PubMed] [Google Scholar]
- 25.Op De Beeck, A., L. Cocquerel, and J. Dubuisson. 2001. Biogenesis of hepatitis C virus envelope glycoproteins. J. Gen. Virol. 82:2589-2595. [DOI] [PubMed] [Google Scholar]
- 26.Pietschmann, T., V. Lohmann, A. Kaul, N. Krieger, G. Rinck, G. Rutter, D. Strand, and R. Bartenschlager. 2002. Persistent and transient replication of full-length hepatitis C virus genomes in cell culture. J. Virol. 76:4008-4021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Proutski, V., E. A. Gould, and E. C. Holmes. 1997. Secondary structure of the 3′ untranslated region of flaviviruses: similarities and differences. Nucleic Acids Res. 25:1194-1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rice, C. M., E. M. Lenches, S. R. Eddy, S. J. Shin, R. L. Sheets, and J. H. Strauss. 1985. Nucleotide sequence of yellow fever virus: implications for flavivirus gene expression and evolution. Science 229:726-733. [DOI] [PubMed] [Google Scholar]
- 29.van der Most, R. G., K. Murali-Krishna, R. Ahmed, and J. H. Strauss. 2000. Chimeric yellow fever/dengue virus as a candidate dengue vaccine: quantitation of the dengue virus-specific CD8 T-cell response. J. Virol. 74:8094-8101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.van Dinten, L. C., J. A. den Boon, A. L. Wassenaar, W. J. Spaan, and E. J. Snijder. 1997. An infectious arterivirus cDNA clone: identification of a replicase point mutation that abolishes discontinuous mRNA transcription. Proc. Natl. Acad. Sci. USA 94:991-996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wengler, G., and G. Wengler. 1989. Cell-associated West Nile flavivirus is covered with E+pre-M protein heterodimers which are destroyed and reorganized by proteolytic cleavage during virus release. J. Virol. 63:2521-2526. [DOI] [PMC free article] [PubMed] [Google Scholar]