Abstract
In recent years, several laboratories have reported on the cloning of herpes simplex virus type 1 (HSV-1) genomes as bacterial artificial chromosomes (BACs) in Escherichia coli and on procedures to manipulate these genomes by using the bacterial recombination machinery. However, the HSV-BACs reported so far are either replication incompetent or infectious, with a deletion of one or more viral genes due to the BAC vector insertion. For use as a multipurpose clone in research on HSV-1, we attempted to generate infectious HSV-BACs containing the full genome of HSV-1 without any loss of viral genes. Our results were as follows. (i) E. coli (YEbac102) harboring the full-length HSV-1 genome (pYEbac102) in which a BAC flanked by loxP sites was inserted into the intergenic region between UL3 and UL4 was constructed. (ii) pYEbac102 was an infectious molecular clone, given that its transfection into rabbit skin cells resulted in production of infectious virus (YK304). (iii) The BAC vector sequence was almost perfectly excisable from the genome of the reconstituted virus YK304 by coinfection of Vero cells with YK304 and a recombinant adenovirus, AxCANCre, expressing Cre recombinase. (iv) As far as was examined, the reconstituted viruses from pYEbac102 could not be phenotypically differentiated from wild-type viruses in vitro and in vivo. Thus, the viruses grew as well in Vero cells as did the wild-type virus and exhibited wild-type virulence in mice on intracerebral inoculation. (v) The infectious molecular clone pYEbac102 is in fact useful for mutagenesis of the HSV-1 genome by bacterial genetics, and a recombinant virus carrying amino acid substitutions in both copies of the α0 gene was generated. pYEbac102 will have multiple applications to the rapid generation of genetically engineered HSV-1 recombinants in basic research into HSV-1 and in the development of HSV vectors in human therapy.
Herpes simplex viruses (HSVs) are ubiquitous and important human pathogens associated with a variety of diseases, including encephalitis, keratitis, mucocutaneous diseases, and genital diseases (45). Since Post and Roizman first reported a generalized technique for the construction of genetically engineered recombinant HSVs with site-specific mutations (33), HSV mutants have significantly contributed to the understanding of viral gene functions and the evaluation of their roles in HSV pathogenesis (35). There is also a growing body of evidence that vectors based on HSV are potent gene delivery vehicles for gene therapy and potential antitumor agents for treatment of malignancy (28, 43, 46). Recently, two groups explored two HSV recombinant viruses as therapeutic agents for malignant gliomas in phase I trials and successfully completed safety studies in which the viruses were inoculated intracranially into human subjects (27, 34). In the research field of human therapy, manipulation of the viral genome is also a key to developing and improving HSV vectors.
Recently, several laboratories reported the cloning of HSV type 1 (HSV-1) genomes into an F plasmid as bacterial artificial chromosomes (BACs) (17, 36, 41). This technique allows the stable maintenance of HSV genomes as BACs in Escherichia coli and mutagenesis of the viral genome in E. coli by using the bacterial recombination machinery (6). The reconstitution of infectious virus is achieved by transfection of the BAC plasmid into mammalian cells, thereby allowing the generation of desired mutant viruses more easily and quickly (6). However, the HSV-BACs reported so far are either replication incompetent or infectious, with a deletion of one or more viral genes because of the BAC vector insertion. Thus, Stavropoulos and Strathdee (41), and Saeki et al. (36) cloned HSV-1 genomes as BACs in E. coli from which the viral packaging elements were deleted. While these HSV-BACs allow for the efficient packaging of helper-dependent HSV vectors (commonly known as HSV amplicons [40]) into virions, they cannot be used for the generation of infectious mutant viruses. Subsequently, Horsburgh et al. (17) constructed a replication-competent HSV-BAC in which the viral thymidine kinase (TK) gene was destroyed due to the BAC insertion. Because the HSV-1 TK gene is dispensable for viral replication in cell cultures (32), transfection of the HSV-BAC into mammalian cells resulted in production of infectious viruses. However, any recombinant viruses reconstituted from the HSV-BACs are negative for the viral TK activity, which is required for virulence in vivo (13), and therefore the recombinant viruses are not testable in most animal studies. Furthermore, the TK activity is also required for hypersensitivity to commonly used antiherpetic drugs, including acyclovir and ganciclovir (9), which is a critical safeguard for replication-competent HSV vectors used in onclolytic viral therapy and human gene therapy. To overcome these problems and for multipurpose utility in the field of basic research on HSVs and development of HSV vectors in human therapy, improvement of HSV-BACs is required. In considering the evidence that all genetically engineered mutant HSVs and HSV vectors generated to date were originally derived from wild-type viruses, the requirements of ideal HSV-BACs applicable to any type of HSV research are (i) that the cloned HSV-BAC genome resembles as much as possible that of the wild-type virus, meaning that the BAC should be infectious and contain a full-length HSV genome, and (ii) that reconstituted virus from the HSV-BAC is phenotypically indistinguishable from wild-type virus in vitro and in vivo. In this study we attempted to generate infectious HSV-BAC clones that satisfy these requirements, and we report the construction of a BAC containing a full-length infectious clone of HSV-1 strain F [HSV-1(F)] in which the BAC vector was inserted into the intergenic region between UL3 and UL4. We also provide evidence that (i) the BAC vector sequence is efficiently removed by a Cre/Lox site-specific recombination system using a recombinant adenovirus expressing Cre recombinase, (ii) the viruses reconstituted from the HSV-BAC reported here grew as well as wild-type virus in Vero cells and maintained the wild-type virulence in a mouse model when inoculated intracerebrally, and (iii) the BAC-cloned HSV-1 infectious genome can be manipulated by mutagenesis in E. coli α0 recombinant mutant virus with amino acid substitutions in both copies of a diploid viral gene, the α0 gene, was generated.
MATERIALS AND METHODS
Cells and viruses.
Vero, rabbit skin, and 143TK− cells were kindly provided by Bernard Roizman and were maintained as described previously (22, 23). 293 cells (ATCC CRL1573) were maintained in Dulbecco's modified Eagle medium (DMEM) containing 10% fetal calf serum (FCS). A recombinant adenovirus, AxCANCre (19), was constructed, propagated, and titrated in 293 cells according to the instructions of the manufacturer (Takara). Wild-type HSV-1(F), a limited-passage isolate, is the prototype strain used in these laboratories. HSV-1(F), the TK-negative recombinant HSV-1(F)Δ305 and the recombinant R7205 were kindly provided by B. Roizman. The properties of these viruses were described previously (3, 12, 32).
Plasmids.
pRB4867 (15) contains the BamHI Q fragment of HSV-1(F). pRB442 (3) contains a 6.3-kbp KpnI-HindIII fragment of HSV-1(F) which encompasses the left end of the UL region, including the region between 0.0415 and 0.084 map units. To construct pRB5198, an approximately 270-bp XhoI-KpnI fragment of pRB5160 (23) containing the bidirectional polyadenylation [poly(A)] signals of HSV-1 UL21 and UL22 was cloned into XhoI and KpnI sites of pBluescript II KS(+) (Stratagene). To generate BAC-pRB5198, a 6.5-kbp NotI fragment of pBeloBAC11 (Research Genetics) (NotI sites were blunt ended by treatment with T4 DNA polymerase) was cloned into the HincII site of pRB5198. To construct loxP-pRB442, oligonucleotide 5′-GCGGATCCATAACTTCGTATAATGTATGCTATACGAAGTTATAGATCTGC-3′, annealed withits complement, was digested with BamHI and BglII and inserted into the BamHI site of pRB442. loxP-HO-pRB442 was generated by cloning the HindIII O fragment of HSV-1(F) into loxP-pRB442. To construct pB2, a BamHI-SphI fragment of BAC-pRB5198 (both sites were blunt ended by treatment with T4 DNA polymerase) containing the BAC vector and the bidirectional poly(A) signals was cloned into the blunt-ended BamHI site of loxP-HO-pRB442. pKO5M was generated by cloning the zeocin resistance gene as a blunt-ended BclI-FokI fragment of pcDNA4/HisMax C (Invitrogen) (both sites were blunt ended by treatment with T4 DNA polymerase) into the NaeI site of pKO3 (25) and deleting the chloramphenicol resistance gene by digesting with BstBI and religating. pKO3 was kindly provided by George Church. pRB4986 (23) carries an HSV-1(F) DNA fragment encoding α0 codons 111 to 241 cloned in pGBT9 (Clontech). Codons 165 to 167 of pRB4986 were replaced with the alanine codons to yield pGBT9α0:165-167A by using the QuickChange site-directed mutagenesis kit (Stratagene) according to the manufacturer's directions. The MfeI-DraIII fragment of pGBT9α0:165-167A containing the sequence carrying the mutations was ligated into an MfeI-DraIII-digested pRB3710 (30), which contains the SacI-PstI fragment carrying the entire ICP0-coding sequence. The resultant plasmid was designated pRB3710/α0:165-167A. pKO5M/α0:165-167A was generated by cloning the SacI-PstI fragment of pRB3710/α0:165-167A (both sites were blunt ended by treatment with T4 DNA polymerase) into the blunt-ended SalI site of pKO5M. pRB442, pRB4867, pRB4986, and pRB5160 were obtained from B. Roizman.
Construction of the recombinant HSVs and E. coli strains harboring HSV-BACs.
Recombinant virus YK301 was constructed by cotransfection of rabbit skin cells with intact R7205 viral DNA and pB2. Viral DNAs were extracted from infected cells and purified on a 5 to 20% potassium acetate gradient as described elsewhere (18). TK-negative recombinant viruses were selected on 143TK− cells overlaid with DMEM containing 5% FCS and 40 μg of bromodeoxyuridine (BUdR) per ml as described previously (23). The recombinants were screened by PCR with primers that correspond to pBeloBAC11 sequences and then verified by Southern blotting.
Circular viral DNA of YK301 was isolated from infected Vero or rabbit skin cells by the Hirt method (16). Briefly, nearly confluent Vero or rabbit skin cells were infected with 3 PFU of YK301 per cell for 6 h, harvested, washed once with phosphate-buffered saline (PBS), and lysed in 0.4 ml of buffer A (0.6% sodium dodecyl sulfate and 10 mM EDTA) for 30 min at room temperature. Next, 0.1 ml of 5 M NaCl was added, and after incubation of the cell lysate at 4°C for 24 h, cell debris was cleared by centrifugation. The supernatant was extracted with phenol-chloroform twice, and DNA was precipitated with ethanol. To construct E. coli YEbac101, which harbors circular viral DNA of YK301, the DNA from infected cells was electroporated into E. coli DH10B (Invitrogen), using a Bio-Rad Gene Pulser II with 0.1-cm cuvettes at 1.8 kV, 25 μF, and 200 Ω. The transformed bacteria were grown on Luria-Bertani (LB) agar plates containing 12.5 μg of chloramphenicol per ml. HSV-BAC DNAs were isolated from the E. coli cultures of chloramphenicol-resistant colonies by standard alkaline lysis procedures (37) and screened by PCR with appropriate primer sets or restriction enzyme digestion. For transfection experiments, HSV-BAC DNAs were isolated by the alkaline lysis procedures and further purified by centrifugation in cesium chloride gradients (37).
To generate YK302, 1 μg of the HSV-BAC DNA (pYEbac101) from YEbac101 was transfected into rabbit skin cells in 12-well plates by calcium phosphate precipitation (14) and incubated at 37°C. Viral plaques were observed 2 or 3 days after transfection. YK303 was constructed by cotransfection of rabbit skin cells with intact viral DNA of YK302 and pRB4867. TK-positive recombinant viruses were selected on 143TK− cells overlaid with DMEM supplemented with hypoxanthine, aminopterin, thymidine and 5% FCS (HAT medium). The recombinants were screened by PCR with primers that correspond to the TK gene of HSV-1 and then verified by Southern blotting.
E. coli YEbac102, which harbors circular viral DNA of YK303 was constructed by exactly the same procedure as for YEbac101 except that E. coli DH10B was electroporated with circular viral DNA extracted from YK303-infected cells. YK304 was generated by transfection of rabbit skin cells with the HSV-BAC DNA (pYEbac102) extracted from YEbac102. E. coli YEbac103 and -104 were constructed by electroporation of RecA-positive E. coli RR1 (ATCC CRL 31343) with pYEbac102. YEbac103 and YEbac104 were derived from different colonies.
BAC excision.
To excise the BAC DNA backbone, Vero cells were infected with a recombinant adenovirus, AxCANCre, at a multiplicity of 100 PFU per cell. After 2 h of viral adsorption, cells were extensively washed with DMEM, and then DMEM containing 5% FCS was added and infection proceeded. At 24 h postinfection, cells were superinfected with YK304 at a multiplicity of 0.03 PFU per cell, and the infection was allowed to proceed for another 24 h. The supernatants were then harvested, and BAC excision was verified by Southern blotting. YK311 was derived from the supernatants and plaque purified once on Vero cells.
Animal studies.
Three-week-old male mice were anesthetized with pentobarbital sodium and injected intracerebrally with 10-fold serial dilutions of each virus (10 mice per dilution). Mice were monitored daily; mortality from 2 to 14 days after infection was attributed to the inoculated viruses. The 50% lethal dose (LD50) ratios were calculated by the Behrens-Karber method.
[35S]methionine labeling of cells and preparation of labeled protein extracts.
Nearly confluent Vero cells in 60-mm-diameter dishes were mock infected or infected with 5 PFU of either YK304 or HSV-1(F) per cell. After adsorption, the inoculum was replaced with DMEM containing 1% FCS and incubated at 37°C for 14 h. The cells were then washed with the same medium without methionine and overlaid with 4 ml of the medium lacking methionine but supplemented with 100 μCi of [35S]methionine (specific activity, >1,000 Ci/mmol) for an additional 12 h. The cells were then harvested; washed with PBS; solubilized with PBS containing 1% Nonidet P-40, 1% deoxycholic acid, and 1 mM phenylmethylsulfonyl fluoride; subjected to electrophoresis in a denaturing gel; transferred to a polyvinylidene difluoride membrane; and subjected to autoradiography as previously described (7).
Southern blotting.
Viral DNAs were digested with EcoRI or BglII, subjected to electrophoresis on 0.7% agarose gels, and transferred to a Hybond-N+ nylon membranes (Amersham Pharmacia). The blots were hybridized with the appropriate DNA probe labeled with [α-32P]dCTP by using a Rediprime II labeling kit (Amersham-Pharmacia) as instructed by the manufacturer.
Allelic exchange.
Allelic exchange was carried out as described previously (17). In brief, E. coli YEbac103 was electroporated with pKO5M/α0:165-167A by using a Bio-Rad Gene Pulser with 0.1-cm cuvettes at 1.8 kV, 25 μF, and 200 Ω. After incubation for 2 h at 30°C in 1 ml of SOC medium, the bacteria were plated on prewarmed zeocin-chloramphenicol (each at 20 μg/ml)-LB plates and incubated overnight at 43°C. The next day, six colonies were picked, diluted serially in LB medium, plated on chloramphenicol-10% sucrose-LB plates, and incubated overnight at 30°C. To confirm the loss of the replacement vector, chloramphenicol-sucrose-resistant colonies were restreaked in duplicate on chloramphenicol-sucrose-LB and zeocin-LB plates and incubated at 30°C overnight. Sucrose- and chloramphenicol-resistant colonies and zeocin-sensitive colonies were screened by DNA sequence analyses. A colony (YEbac106) confirmed by sequence analyses was grown in LB-chloramphenicol medium, and the mutated HSV-BAC DNA (pYEbac106) was isolated as described above. A mutant virus (YK321) was generated by transfection of pYEbac106 into rabbit skin cells as described above.
Antibody and immunoblotting.
A monoclonal antibody to ICP0 (H1083) was purchased from the Goodwin Institute. The electrophoretically separated proteins transferred to a nitrocellulose sheet were reacted with the antibody to ICP0 as described previously (20, 23).
RESULTS
Generation of recombinant viruses and BAC plasmids.
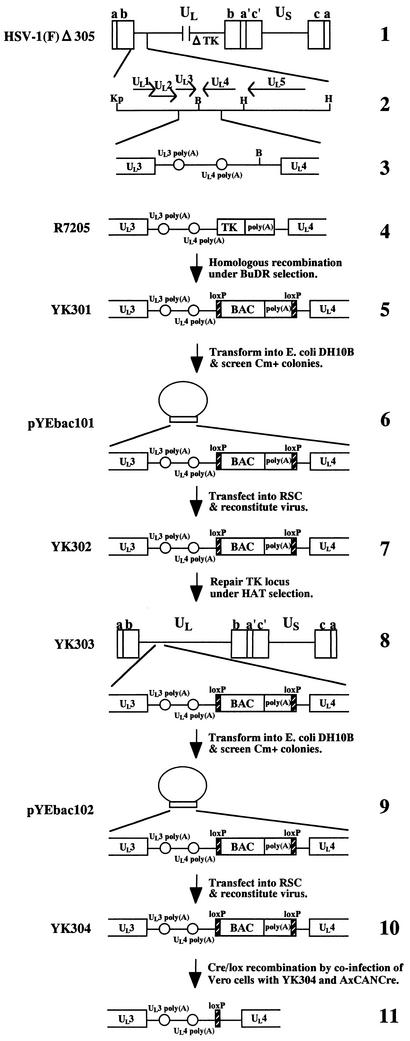
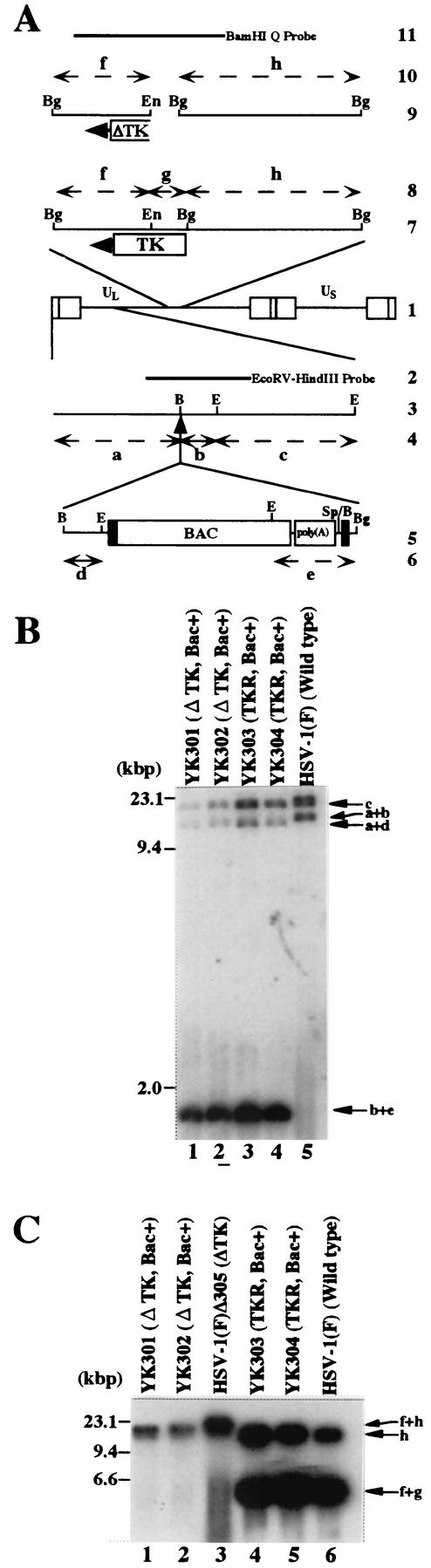
We have cloned the full-length HSV-1(F) genome into a BAC. The BAC vector was inserted into a BamHI site located in the intergenic region between the UL3 and UL4 genes without deletion of any viral sequence. We chose this site for insertion of the BAC vector since it has been reported that the insertion of foreign genes into this site was stable and had no effect on viral growth in cell culture (8, 21). The strategy for cloning is summarized in Fig. 1 . As a first step, we generated a TK-negative recombinant YK301 in which a cassette containing a BAC vector and poly(A) signals flanked by loxP sites was inserted into the intergenic region between UL3 and UL4. The loxP sites were used to make the BAC vector sequence removable by Cre/Lox site-specific recombination (42). Insertion of the poly(A) signals was necessary to properly terminate UL4 and UL5 transcription because once the BAC was inserted into the BamHI site, the poly(A) signal for UL4 and UL5 was no longer functional (Fig. 1). The transfer plasmid pB2, generated as described in Materials and Methods, was cotransfected into rabbit skin cells with intact R7205 viral DNA in which native TK was deleted and the TK expression cassette under control of the α27 promoter was inserted into the BamHI site between the UL3 and UL4 genes (Fig. 1), and the progeny of transfection were plated on 143TK− cells in the presence of BUdR. The TK-negative progeny viruses were isolated and plaque purified, their viral DNAs were screened for the presence of the BAC sequence by PCR (data not shown), and a TK-negative virus (YK301) was analyzed by Southern blotting (Fig. 2B). The DNAs of YK301 and HSV-1(F) were digested with EcoRI and subjected to Southern blotting with the EcoRV-HindIII fragment of pRB442 as a probe (Fig. 2A). As could be predicted from the construction of these viruses (Fig. 2A), the 32P-labeled probe hybridized to fragments a+b (12.6 kbp) and c (15.0 kbp) in HSV-1(F) (Fig. 2B, lane 5) and to fragments a+d (10.6 kbp), c (15.0 kbp), and b+e (1.9 kbp) in YK301 (Fig. 2B, lane 1). These results indicated that YK301 harbors the BAC vector sequence inserted into the intergenic region of the UL3 and UL4 genes. The same results were obtained for the BAC-containing recombinant viruses YK302, YK303, and YK304 described below (Fig. 2B, lanes 2 to 4).
FIG. 1.
Strategy used to clone the entire HSV-1(F) genome into a BAC. Schematic diagrams of the arrangement of the genome of HSV-1(F) and relevant domains of the recombinant viruses and HSV-BACsare shown. Line 1, a linear representation of the HSV-1(F)Δ305 genome. The unique sequences are represented as unique long (UL) and short (US), and the terminal repeats flanking them are shown as open rectangles with their designation given above. Line 2, an expanded section of the domain encoding UL1 to UL5. Arrows indicate the polarity and position of each open reading frame. Line 3, an intergenic region between UL3 and UL4 open reading frames of HSV-1(F)Δ305. Open reading frames and poly(A) signals are shown as open rectangles and circles, respectively. The BamHI site used for the insertion of the BAC is also shown. Line 4, an intergenic region between UL3 and UL4 of R7205. A chimeric α27-tk gene and upstream poly(A) signal were inserted into the BamHI site placed between the UL3 and UL4 open reading frames. Line 5, sequence of the recombinant virus, YK301, which was selected in the presence of BUdR on 143TK− cells from among the progeny of transfection of rabbit skin cells with intact R7205 DNA and plasmid pB2. The BAC sequence and the poly(A) signals are shown as open rectangles. loxP sites are represented as filled rectangles. Line 6, arrangement of plasmid pYEbac101 maintained in E. coli YEbac101, which was generated by electroporation of circular viral DNA of YK301 into E. coli DH10B. The expansion of the intergenic region between the UL3 and UL4 open reading frames of pYEbac101 is also shown. Line 7, sequence of YK302, which was reconstituted by transfection of pYEbac101 into rabbit skin cells. Line 8, arrangement of YK303 selected in the presence of HAT on 143TK− cells from among the progeny of transfection of rabbit skin cells with intact YK302 DNA and plasmid pRB4867 containing the BamHI Q fragment of HSV-1(F). The expansion of the intergenic region between open reading frames UL3 and UL4 of YK303 is also shown. Line 9, arrangement of plasmid pYEbac102 maintained in E. coli YEbac102, constructed by electroporation of circular DNA of YK303 into E. coli DH10B. Line 10, sequence of YK304, reconstituted by transfection of pYEbac102 into rabbit skin cells. Line 11, sequence of the BAC-excised recombinant virus, generated by coinfection of Vero cells with YK304 and a recombinant adenovirus AxCANCre. As a result of Cre-mediated site-specific recombination, the BAC vector backbone was excised and a single loxP site remained in the intergenic region between open reading frames UL3 and UL4. Restriction sites: B, BamHI; Kp, KpnI; H, HindIII.
FIG.2.
(A) Schematic diagram of genome structures of HSV-1(F) and relevant domains of the recombinant viruses. Line 1, sequence arrangement of HSV-1(F) genome. Line 2, location of the EcoRV-HindIII fragment used as a radiolabeled probe in panel B. Line 3, an enlarged portion of the EcoRI J and D fragments of HSV-1(F). Line 5, sequence of the BAC-containing recombinant virus YK301, YK302, YK303, or YK304. Lines 4 and 6, expected sizes of DNA fragments generated by cleavage of the DNA. The fragment designations shown here are identical to those described in the text and in panel B. Line 7, an enlarged portion of the BglII M and I fragments of HSV-1(F), YK303 or YK304. Line 9, Δ305, YK301, or YK302 has a deletion (fragment g) in the TK locus. Lines 8 and 10, expected sizes of DNA fragments generated by cleavage of the DNA. The fragment designations shown here are identical to those described in the text and in panel C. Line 11, location of the BamHI Q fragment used as a radiolabeled probe in panel C. Restriction sites: E, EcoRI; B, BamHI; Bg, BglII; En, EcoNI; Sp, SphI. (B and C) Autoradiographic images of electrophoretically separated EcoRI (B) or BglII (C) digests of YK301, YK302, YK303, YK304, HSV-1(F)Δ305, and HSV-1(F) DNAs, hybridized to the radiolabeled EcoRV-HindIII fragment of pRB442 or BamHI Q fragment of HSV-1(F), respectively. The letters on the right refer to the designations of the DNA fragments generated by restriction endonuclease cleavage. Molecular sizes are shown on the left.
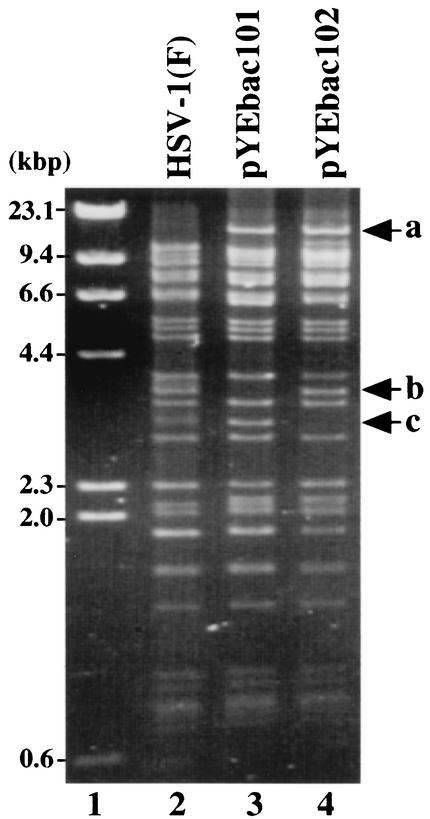
To isolate circular DNA of YK301, Vero cells were infected with YK301 for 6 h and harvested. The circular DNA of YK301, extracted as described in Materials and Methods, was electroporated into E. coli DH10B, and recombinant colonies resistant to chloramphenicol were screened for the presence of the α0 and BAC sequences by PCR. One chloramphenicol-resistant colony (YEbac101) was selected for further analysis, and the HSV-BAC DNA (pYEbac101) extracted from YEbac101 was examined by restriction digestion with BamHI. As shown in Fig. 3, the digestion of pYEbac101 yielded a complex pattern similar to that of HSV-1(F) nucleocapsid DNA, with the following exceptions (lanes 2 and 3). The digestion of pYEbac101 produced an unique 16.3-kbp fragment a due to the BAC insertion (Fig. 3, lanes 2 and 3). A 3.3-kbp fragment b observed in HSV-1(F) DNA (Fig. 3, lane 2) was missing in pYEbac101 because pYEbac101 has a deletion in the TK locus (Fig. 3, lane 3). These results indicated that the HSV-1(F) genome with a deletion in the TK locus was cloned as a BAC plasmid.
FIG. 3.
Agarose gel electrophoresis of molecular weight markers (lane 1), BamHI-digested HSV-1(F) viral DNA (lane 2), and the cloned HSV-1 BAC plasmid DNAs pYEbac101 (lane 3) and pYEbac102 (lane 4). Fragment a of approximately 16.3 kbp, detected in pYEbac101 and pYEbac102, results from the fusion of the BAC sequence and BamHI C fragment of HSV-1(F). Fragment c of approximately 2.9 kbp, detected only in pYEbac101, results from a deletion in the viral TK locus. After the TK deletion was repaired, fragment b was restored in pYEbac102. Molecular sizes are shown on the left.
To test whether pYEbac101 was infectious, it was transfected into rabbit skin cells. Plaque formation was observed within 2 days after transfection, and the cytopathic effect was expanded (data not shown), indicating that pYEbac101 was able to produce infectious virus (YK302). One-step and multistep growth curve experiments revealed that there were no significant differences in the growth rate on Vero cells between YK302 and HSV-1(F)Δ305, each of which has the same deletion in the viral TK locus (data not shown). To repair the native TK gene of YK302, rabbit skin cells were cotransfected with intact YK302 DNA and the BamHI Q fragment in pRB4867 containing the entire TK-coding sequence. TK-positive progeny viruses were selected on 143TK− cells overlaid in HAT medium, isolated, and plaque purified. Viral DNAs of the isolates were screened for the presence of the TK sequence by PCR (data not shown), and a TK-positive virus (YK303) was analyzed by Southern blotting with the BamHI Q fragment as a probe (Fig. 2C). As predicted (Fig. 2A), the BamHI Q probe hybridized with fragments h (15.5 kbp) and f+g (6.0 kbp) in TK-repaired YK303 (Fig. 2C, lane 4) and wild-type HSV-1(F) (Fig. 2C, lane 6) and with fragment f+h (21.5 kbp) in TK-negative viruses YK301 (Fig. 2C, lane 1), YK302 (Fig. 2C, lane 2), and HSV-1(F)Δ305 (Fig. 2C, lane 3). The same result was obtained with another TK-positive virus, YK304, described below (Fig. 2C, lane 5).
The circular DNA of YK303 extracted as before was transformed into E. coli DH10B, and a chloramphenicol-resistant colony (YEbac102) which harbors the TK-repaired YK303 DNA was isolated. An HSV-BAC DNA (pYEbac102) extracted from YEbac102 was subjected to restriction enzyme analysis with BamHI. As shown in Fig. 3, the pattern of BamHI digestion of pYEbac102 was almost identical to that of HSV-1(F) viral DNA (Fig. 3, lanes 2 and 4), with the exception that the digestion of pYEbac102 produced an unique 16.3-kbp fragment a due to the BAC insertion (Fig. 3, lanes 2 and 4). Compared to the pattern of BamHI digestion of pYEbac101, a 3.3-kbp fragment b appeared in pYEbac102 instead of a 2.9-kbp fragment c as a result of TK repair (Fig. 3, lanes 3 and 4). The pYEbac102 was infectious, based on the observation that transfection of pYEbac102 into rabbit skin cells resulted in production of infectious virus (YK304) (data not shown).
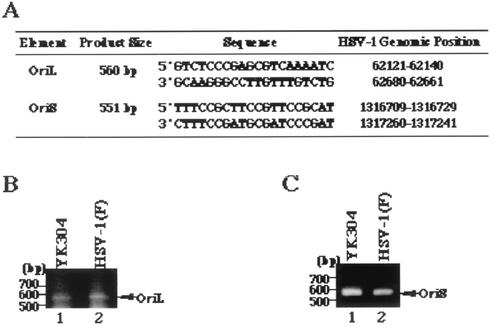
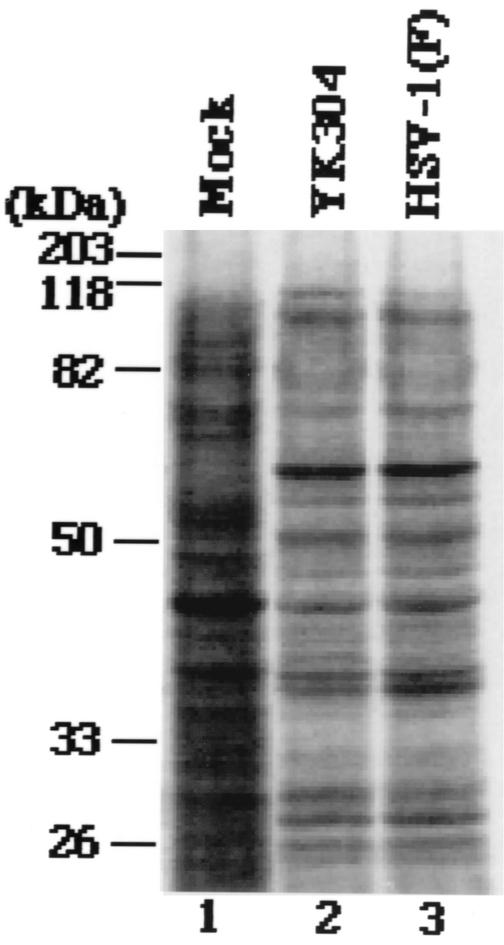
Next, we examined the intactness of viral gene elements that have been reported to be unstable in bacteria (10). To this end, a fragment containing OriS or OriL was amplified from the viral genome of HSV-1(F) or YK304 by PCR with the primer pair shown in Fig. 4A. As shown in Fig. 4B, the electrophoretic mobility of the PCR product containing OriL amplified from YK304 was identical to that of the product amplified from HSV-1(F). The same results were obtained with the PCR product containing OriS (Fig. 4C), indicating that these origin sequences are stable in YEbac102. To emphasize the similarities between YK304 and parental HSV-1(F), a profile of viral polypeptides of YK304 was compared to that of HSV-1(F). For this purpose, Vero cells were mock infected or infected with YK304 or HSV-1(F), labeled with [35S]methionine, harvested, solubilized, and subjected to electrophoresis in a denaturing gel. As shown in Fig. 5, the pattern of viral polypeptide synthesis in YK304-infected Vero cells was almost identical to that in HSV-1(F)-infected Vero cells. Furthermore, YK304 appears to maintain a temperature-sensitive mutation in the ICP4 gene, based on the observation that YK304 was not able to exhibit a cytopathic effect on Vero cells at a nonpermissive temperature (39.5°C) at 24 h postinfection (data not shown), as previously reported for wild-type HSV-1(F) (24). These results support our conclusion that the full-length infectious HSV-1(F) genome was cloned as a BAC plasmid.
FIG. 4.
(A) Primer pairs and sizes of predicted PCR products containing HSV-1 OriL or OriS obtained by using the primer pairs. (B) Agarose gel electrophoresis of PCR products containing OriL of YK304 (lane 1) and HSV-1(F) (lane 2). PCR products are indicated by arrows on the right, and molecular sizes are shown on the left. (C) Agarose gel electrophoresis of PCR products containing OriS of YK304 (lane 1) and HSV-1(F) (lane 2). PCR products are indicated by arrows on the right, and molecular sizes are shown on the left.
FIG. 5.
Autoradiographic images of [35S]methionine-labeled lysates extracted from cells mock infected (lane 1) or infected with YK304 (lane 2) or HSV-1(F) (lane 3) that were separated electrophoretically in a denaturing gel, transferred electrically to a polyvinylidene difluoride membrane, and subjected to autoradiography. Molecular masses are shown on the left.
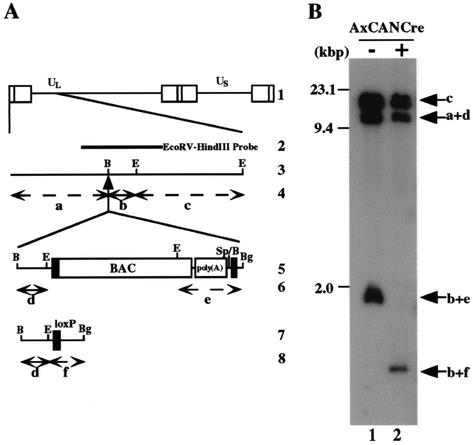
To test whether the BAC DNA backbone which is flanked by two loxP sites is in fact able to be excised from the viral genome by Cre recombinase, Vero cells were mock infected or infected with a recombinant adenovirus, AxCANCre, expressing Cre recombinase and then superinfected with YK304. At 24 h postinfection with YK304, the supernatant was harvested, and the viral genome was digested with EcoRI and analyzed by Southern blotting with the EcoRV-HindIII fragment of pRB442 as a probe (Fig. 6B). Consistent with the results in Fig. 2B, the probe hybridized with fragments c (15.0 kbp), a+d (10.6 kbp), and b+e (1.9-kbp) in the absence of AxCANCre (Fig. 6B, lane 1), whereas fragment b+e was shifted to fragment b+f (1.0 kbp) as a result of BAC excision with the aid of AxCANCre (Fig. 6A and B, lanes 1 and 2). It should be noted that the probe barely hybridized with fragment b+e after the treatment with AxCANCre (Fig. 6B, lane 2). Furthermore, sequence analysis of the intergenic region between UL3 and UL4 of the BAC-excised recombinant virus revealed that the expected Cre-mediated excision occurred and only one loxP site remained in the region (data not shown). These results indicated that the BAC vector sequence can be removed quite efficiently from the viral genome in Vero cells by treatment with AxCANCre expressing Cre recombinase.
FIG. 6.
(A) Schematic diagram of the sequence of HSV-1(F) and of the recombinant virus YK304 and the BAC-excised recombinant virus derived from it. Line 1, sequence arrangement of the HSV-1(F) DNA. Line 2, location of the EcoRV-HindIII fragment used as a radiolabeled probe in panel B. Line 3, an enlarged portion of the domain of HSV-1(F) encoded by EcoRI J and D fragments. Line 5, sequence of the recombinant virus YK304. Line 7, sequence of the BAC-excised recombinant virus, which is generated by coinfection of Vero cells with YK304 and AxCANCre. Lines 4, 6, and 8, expected sizes of DNA fragments generated by cleavage of the DNA. The fragment designations shown here are identical to those used in the text and panel B. (B) Autoradiographic image of electrophoretically separated EcoRI digests of DNAs of YK304 (lane 1) or the BAC-excised recombinant virus (lane 2) hybridized to the radiolabeled EcoRV-HindIII fragment of pRB442. The letters on the right refer to the designations of the DNA fragments generated by restriction endonuclease cleavage. Molecular sizes are shown on the left.
Characterization of recombinant viruses reconstituted from the infectious HSV-BAC pYEbac102.
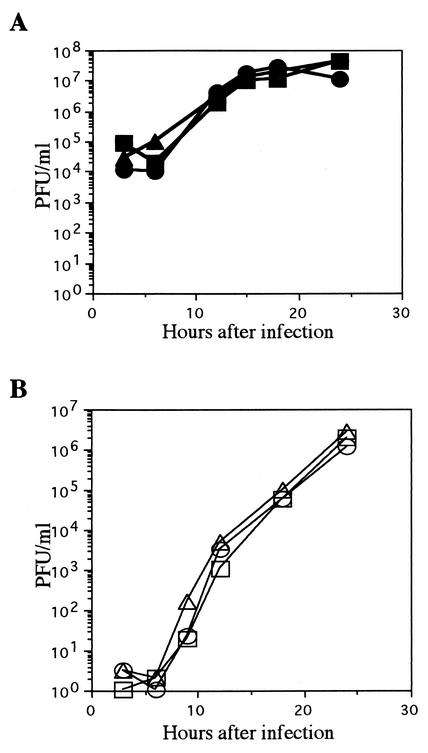
To characterize the reconstituted viruses (YK304 and YK311) from the full-length infectious HSV-1(F) genome cloned as the BAC plasmid (pYEbac102), we performed two series of experiments. YK311, in which the BAC vector sequence was removed by Cre-LoxP recombination, was derived from YK304 as described in Materials and Methods. In the first series of experiments, Vero cells were infected with YK304, YK311, or HSV-1(F) at a multiplicity of 10 or 0.01 PFU per cell and harvested at the indicated time points (Fig. 7). The titers of each sample were then determined by standard plaque assay on Vero cells, and growth curves of HSV-1(F), YK304, and YK311 were plotted. As shown in Fig. 7, YK304 and YK311 showed growth kinetics almost identical to those of wild-type HSV-1(F) on Vero cells at low and high multiplicities of infection. The experiment was repeated, and similar results were obtained (data not shown).
FIG. 7.
Comparison of the in vitro growth properties of wild-type HSV-1(F) and the recombinant viruses YK304 and YK311. Vero cells were infected with HSV-1(F) (circles), YK304 (squares), or YK311 (triangles) at a multiplicity of 10 (A) or 0.01 (B) PFU per cell. Supernatant and cells were harvested at the indicated time points, and titrations of cell lysates were performed on Vero cells.
In the second series of experiments, mice were injected intracerebrally with HSV-1(F), YK304, or YK311, and mortality was monitored from day 2 to 14. HSV-1(F), YK304, and YK311 showed similar PFU/LD50 values of 10−0.5, 10−0.4, and 10−0.5, respectively. The experiment was repeated, and similar results were obtained (data not shown). Taken together, these results indicated that as far as was tested, the reconstituted viruses (YK304 and YK311) from pYEbac102 maintain wild-type properties such as ability to grow in cell culture and virulence in mice.
Allelic exchange of HSV-BAC in E. coli.
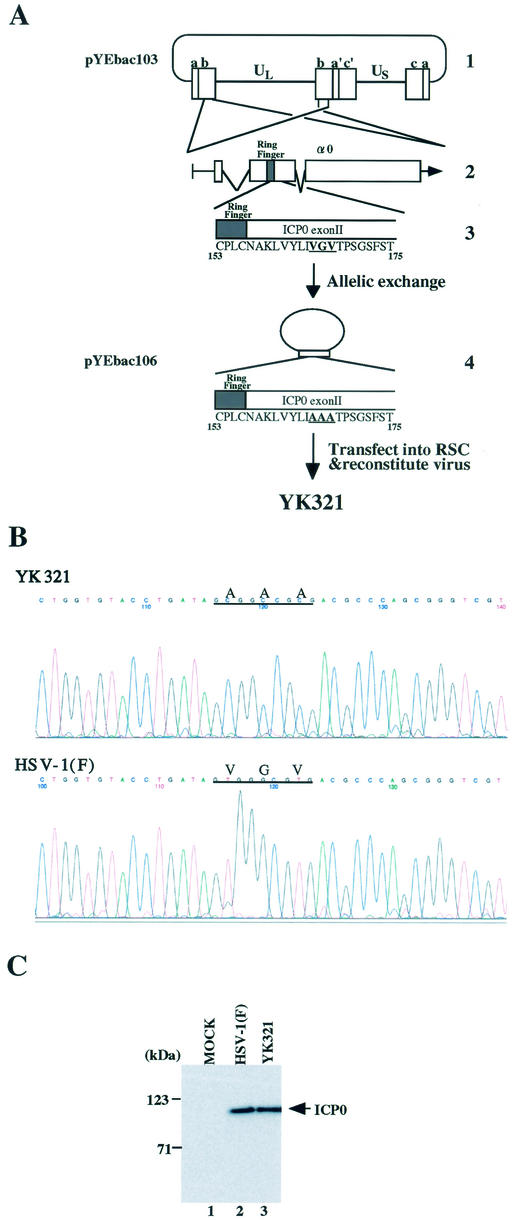
To examine whether the BAC-cloned HSV-1 infectious genome (pYEbac102) constructed in this study in fact can be manipulated by mutagenesis in E. coli, we decided to introduce amino acid substitutions into the α0 gene within the HSV-1 genome by using a two-step homologous recombination approach which is described elsewhere (17, 25). Since the HSV-1 genome contains two copies of the α0 gene located in the inverted repeats (Fig. 8A), the introduction of this kind of mutation into the HSV-1 genome is considered one of the most complicated manipulations. As a first step, pYEbac102 was transferred to RecA-positive E. coli strain RR1 cells, and chloramphenicol-resistant colonies (YEbac103 and YEbac104) which harbor the HSV-1(F) genome were selected for further analysis. Reconstituted viruses from YEbac103 and YEbac104 showed similar growth properties in Vero cells (data not shown). For allelic exchange in E. coli, YEbac103 was transformed with pKO5M/α0:165-167A, in which codons 165 to 167 of the α0 gene were replaced with alanine codons. After a series of selections (described in Materials and Methods), the clones meeting selection criteria were further analyzed by sequence analyses. An E. coli clone that harbors HSV-BAC with the amino acid substitutions in both copies of the α0 gene was isolated and designated YEbac106. Transfection of the HSV-BAC plasmid (pYEbac106) extracted from YEbac106 into rabbit skin cells resulted in the production of viral plaques within 3 days posttransfection, and the recombinant virus (YK321) was able to replicate in the cells (data not shown). The presence of desired mutations in the viral genome of YK321 was verified by sequencing the PCR-amplified fragment of α0 exon II (Fig. 8B), and YK321 produced the mutant ICP0 with an apparent Mr of 110,000, which is the same size as that of wild-type ICP0 in infected Vero cells (Fig. 8C). These results confirmed that the intended alteration can be introduced into the full-length HSV-BAC generated in this study.
FIG. 8.
Characterization of a recombinant virus YK321 harboring amino acid substitutions in both copies of the α0 gene. (A) Strategy for allelic exchange of the HSV-BAC genome in E. coli. Line 1, schematic diagram of the sequence of plasmid pYEbac103. Line 2, an expanded section of the domain encoding the α0 gene. The transcript and coding regions are shown. Line 3, amino acid sequence of a domain of ICP0 carrying codons 153 to 175 in pYEbac103. Line 4, arrangement of theplasmid pYEbac106, which was constructed by allelic exchange as described in Materials and Methods. An enlarged section of the ICP0 domain carrying codons 153 to 175 in pYEbac106 is also shown. A reconstituted virus YK321 was generated by transfection of pYEbac106 into rabbit skin cells. (B) Sequence of ICP0 exon II from wild-type HSV-1(F) and the recombinant virus YK321. Cytoplasmic viral DNA of YK321 or HSV-1(F) was obtained from infected Vero cells, and a 283-bp fragment was PCR amplified and then sequenced. The sequences of YK321 (upper panel) and wild-type HSV-1(F) (lower panel) were identical, with the exception of the desired mutations shown by lines. Codons 165 to 167 encoding amino acid residues VGV in the HSV-1(F) genome were replaced with codons for amino acid residues AAA in the YK321 genome. (C) Photographic image of an immunoblot of electrophoretically separated lysates of mock (lane 1)-, HSV-1(F) (lane 2)-, or YK321 (lane 3)-infected Vero cells harvested 48 h postinfection. The blot was probed with the monoclonal antibody to ICP0. Molecular masses are shown on the left.
DISCUSSION
The combination of E. coli genetics and BAC technology has undoubtedly opened new avenues for the manipulation of large herpesvirus genomes, including the genomes of mouse, human, and pig cytomegaloviruses (4, 26, 29, 31, 47), Epstein-Barr virus (11), pseudorabies virus (PRV) (38, 39), mouse gammaherpesvirus 68 (1), Kaposi's sarcoma-associated herpesvirus (48), and HSV-1 (17, 36, 41). Although several laboratories have reported the cloning of HSV-1 genomes as BACs, these were either noninfectious because of deletions of packaging signals or infectious with a deletion of one or more nonessential viral genes (17, 36, 41). In the present study, we have succeeded in cloning the full-length HSV-1(F) genome as a BAC (pYEbac102) without any deletions or destruction of viral genes and have shown that as far as was examined, viruses (YK304 and YK311) reconstituted from the HSV-BAC clone (pYEbac102) exhibited wild-type phenotypes both in vitro and in vivo. The salient features of our results can be summarized as follows.
(i) To clone the entire infectious HSV-1 genome as a BAC, we chose the BamHI site in the intergenic region between UL3 and UL4 for the insertion of the BAC vector. This location has been used as a stable insertion site for various foreign genes with no viral sequence deletions (8, 21). Thus, R8102 carrying a lacZ gene inserted into the site was constructed in B. Roizman's laboratory and has been used to monitor viral infectivity by β-galactosidase assays (8). Subsequently, our group also used the site for the insertion of a foreign gene expression cassette and obtained a high enough level of expression without affecting viral growth in cell culture (21). These results prompted us to target the BAC vector sequence in the intergenic region between UL3 and UL4. Consistent with a previous report (21), the growth of YK304 and YK311, which are reconstituted from the HSV-BAC containing the full-length HSV-1(F) genome (pYEbac102), was similar to that of wild-type HSV-1(F) in Vero cells. Furthermore, these reconstituted viruses exhibited wild-type virulence in a mouse model system when inoculated intracerebrally. These results indicate that the cloned HSV-BAC pYEbac102 in this study satisfies both of the requirements that it maintain the full-length HSV-1 genome and that the reconstituted viruses from pYEbac102 are indistinguishable from wild-type virus both in vitro and in vivo.
(ii) Infectious clones in which BAC sequences were inserted into the intergenic regions or at the end of the herpesvirus genomes without destruction of any viral genes have been constructed in several herpesviruses, including mouse gammaherpesvirus 68 (1), PRV (39), and human cytomegalovirus (47). In all of these cases, however, the viruses reconstituted from the BAC-inserted genomes exhibited slower growth in cell cultures and/or a more attenuated virulence in animal model systems than wild-type viruses, possibly because the BAC insertion affects the expression of neighboring genes and/or efficiency of packaging of the oversized genome carrying the inserted BAC sequence (1, 39, 47). Supporting this, the wild-type phenotypes were restored only when the inserted BAC sequences were excised by either homologous recombination using duplicated viral sequence or a Cre/Lox site-specific recombination system (1, 39, 44, 47). So far, only one infectious PRV-BAC clone showed the wild-type phenotype both in vitro and in vivo without removal of the BAC sequence upon delivery of the PRV-BAC clone into mammalian cells; however, this PRV-BAC clone does not contain the entire PRV genome and has a defect in the viral gG locus due to the BAC vector insertion (38). Thus, our study is the first demonstrating that reconstituted virus (YK304) from a BAC clone containing the full-length herpesvirus genome with no viral gene destruction shows wild-type phenotypes in cell cultures and in mouse model systems without excision of the BAC sequence.
(iii) Smith and Enquist (38) reported that BAC sequences were unstable in the PRV genome during the infection of mammalian cells, resulting in spontaneous deletion of the BAC and surrounding viral sequences. Therefore, the excision of the BAC sequence from the HSV-1 genome is better, even though it is unknown whether the features observed in the PRV genome are true for the HSV-1 genome, and YK304 exhibited wild-type phenotypes both in vitro and in vivo without excision of the BAC sequence as described above. The same group also developed a system for the rapid and efficient excision of the BAC sequence from the PRV genome (39). This was accomplished by flanking the BAC sequence with loxP sites and adding within the BAC vector sequence an expression cassette for a Cre-encoding gene which is specifically active only in eukaryotic cells. By using this system, the BAC vector backbone was removed from the viral genome autonomously on delivery of the PRV-BAC plasmid into mammalian cells without the need for plaque purification. In the present study we demonstrated an alternative method for the efficient excision of the BAC vector backbone from the HSV-1 genome by using the Cre-Lox site-specific recombination system. For this purpose, we used a recombinant adenovirus expressing Cre recombinase (AxCANCre) (19). The use of recombinant adenoviruses has been shown to be an excellent way to deliver and express foreign genes in virtually 100% of mammalian cells derived from various tissues and species (2, 19). The recombinant adenovirus AxCANCre has two notable features. First, AxCANCre is a replication-incompetent recombinant virus in which the E1 gene, which is essential for viral replication in cell cultures, is deleted, and therefore the virus can be propagated only in complementing cell lines that provide the E1 gene product (19). Second, AxCANCre was originally developed for Cre-mediated activation of a transgene introduced into mammalian cells, and on using AxCANCre, Cre-mediated switching of gene expression occurs in nearly 100% of mammalian cells (19). These features led us to use AxCANCre for removal of the BAC sequence from the HSV-1 genome. As expected, coinfection of Vero cells with AxCANCre and YK304 harboring the BAC sequence resulted in efficient excision of the BAC vector from the YK304 genome. Notably, the BAC sequence was barely detected after infection with AxCANCre by Southern blotting, suggesting that the efficiency of the BAC excision by AxCANCre was nearly 100% and that further plaque purification is not required to obtain recombinant viruses with BAC excised. Contamination by AxCANCre in the reconstituted virus is negligible because AxCANCre cannot grow in normal mammalian cells and is therefore easily washed out after viral adsorption of AxCANCre. Furthermore, AxCANCre is able to infect mammalian cells derived from various species. Thus, AxCANCre is a novel and efficient tool for the excision of BAC vectors not only from the HSV-1 genome but also from any mammalian herpesvirus genomes.
(iv) pYEbac102 can be manipulated in E. coli. We followed the well-established methods for HSV-1 (17) and other herpesviruses (6) for insertion of a modification into pYEbac102 in E. coli. In the present study, we succeeded in inserting amino acid substitutions into both copies of the α0 gene in pYEbac102 by a two-step allelic exchange process using YEbac103, in which pYEbac102 was transferred into RecA-positive E. coli RR1. Because the introduction of such mutations into both copies of a diploid viral gene is considered one of the most complicated manipulations in herpesvirus genomes, it is conceivable that most mutations can be introduced into pYEbac102 by using the system described here. Furthermore, pYEbac102 is applicable to other bacterial genetics, including allelic exchange using RecE and RecT (1), transposon mutagenensis (5, 38), and site-specific recombination systems using Flp recombinase (1). Thus, pYEbac102 will facilitate the genetic manipulation of HSV-1 and be a multipurpose clone for basic research into HSV-1 and the development and improvement of HSV vectors in human therapy.
Acknowledgments
We thank B. Roizman for HSV-1(F), R7205, pRB442, pRB4867, pRB4986, pRB5160, 143TK−, and rabbit skin cells and G. Church for pKO3.
This study was supported in part by grants for Scientific Research (to Y.K. and Y.Y.) and grants for Scientific Research in Priority Areas (to Y.K. and Y.Y.) from the Ministry of Education, Science, Sports and Culture of Japan and Japan Science and Technology Cooperation. Y.K. was supported by a grant from the Uehara Memorial Foundation.
REFERENCES
- 1.Adler, H., M. Messerle, M. Wagner, and U. H. Koszinowski. 2000. Cloning and mutagenesis of the murine gammaherpesvirus 68 genome as an infectious bacterial artificial chromosome. J. Virol. 74:6964-6974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ali, M., N. R. Lemoine, and C. J. Ring. 1994. The use of DNA viruses as vectors for gene therapy. Gene Ther. 1:367-384. [PubMed] [Google Scholar]
- 3.Baines, J. D., and B. Roizman. 1991. The open reading frames UL3, UL4, UL10, and UL16 are dispensable for the replication of herpes simplex virus type 1 in cell culture. J. Virol. 65:938-944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Borst, E. M., G. Hahn, U. H. Koszinowski, and M. Messerle. 1999. Cloning of the human cytomegalovirus (HCMV) genome as an infectious bacterial artificial chromosome in Escherichia coli: a new approach for construction of HCMV mutants. J. Virol. 73:8320-8329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brune, W., C. Menard, U. Hobom, S. Odenbreit, M. Messerle, and U. H. Koszinowski. 1999. Rapid identification of essential and nonessential herpesvirus genes by direct transposon mutagenesis. Nat. Biotechnol. 17:360-364. [DOI] [PubMed] [Google Scholar]
- 6.Brune, W., M. Messerle, and U. H. Koszinowski. 2000. Forward with BACs: new tools for herpesvirus genomics. Trends Genet. 16:254-259. [DOI] [PubMed] [Google Scholar]
- 7.Chou, J., and B. Roizman. 1990. The herpes simplex virus 1 gene for ICP34.5, which maps in inverted repeats, is conserved in several limited-passage isolates but not in strain 17syn+. J. Virol. 64:1014-1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cocchi, F., L. Menotti, P. Mirandola, M. Lopez, and G. Campadelli-Fiume. 1998. The ectodomain of a novel member of the immunoglobulin subfamily related to the poliovirus receptor has the attributes of a bona fide receptor for herpes simplex virus types 1 and 2 in human cells. J. Virol. 72:9992-10002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Crumpacker, C. 2001. Antiviral therapy, p. 393-434. In D. M. Knipe, P. M. Howly, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology, 4th ed. Lippincott-Williams &Wilkins, Philadelphia, Pa.
- 10.Cunningham, C., and A. J. Davison. 1993. A cosmid-based system for constructing mutants of herpes simplex virus type 1. Virology 197:116-124. [DOI] [PubMed] [Google Scholar]
- 11.Delecluse, H. J., T. Hilsendegen, D. Pich, R. Zeidler, and W. Hammerschmidt. 1998. Propagation and recovery of intact, infectious Epstein-Barr virus from prokaryotic to human cells. Proc. Natl. Acad. Sci. USA 95:8245-8250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ejercito, P. M., E. D. Kieff, and B. Roizman. 1968. Characterization of herpes simplex virus strains differing in their effects on social behaviour of infected cells. J. Gen. Virol. 2:357-364. [DOI] [PubMed] [Google Scholar]
- 13.Field, H. J., and P. Wildy. 1978. The pathogenicity of thymidine kinase-deficient mutants of herpes simplex virus in mice. J. Hyg. (London) 81:267-277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Graham, F. L., and A. J. van der Eb. 1973. A new technique for the assay of infectivity of human adenovirus 5 DNA. Virology 52:456-467. [DOI] [PubMed] [Google Scholar]
- 15.He, B., J. Chou, D. A. Liebermann, B. Hoffman, and B. Roizman. 1996. The carboxyl terminus of the murine MyD116 gene substitutes for the corresponding domain of the gamma(1)34.5 gene of herpes simplex virus to preclude the premature shutoff of total protein synthesis in infected human cells. J. Virol. 70:84-90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hirt, B. 1967. Selective extraction of polyoma DNA from infected mouse cell cultures. J. Mol. Biol. 26:365-369. [DOI] [PubMed] [Google Scholar]
- 17.Horsburgh, B. C., M. M. Hubinette, D. Qiang, M. L. MacDonald, and F. Tufaro. 1999. Allele replacement: an application that permits rapid manipulation of herpes simplex virus type 1 genomes. Gene Ther. 6:922-930. [DOI] [PubMed] [Google Scholar]
- 18.Igarashi, K., R. Fawl, R. J. Roller, and B. Roizman. 1993. Construction and properties of a recombinant herpes simplex virus 1 lacking both S-component origins of DNA synthesis. J. Virol. 67:2123-2132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kanegae, Y., G. Lee, Y. Sato, M. Tanaka, M. Nakai, T. Sakaki, S. Sugano, and I. Saito. 1995. Efficient gene activation in mammalian cells by using recombinant adenovirus expressing site-specific Cre recombinase. Nucleic Acids Res. 23:3816-3821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kawaguchi, Y., R. Bruni, and B. Roizman. 1997. Interaction of herpes simplex virus 1 alpha regulatory protein ICP0 with elongation factor 1delta: ICP0 affects translational machinery. J. Virol. 71:1019-1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kawaguchi, Y., M. Tanaka, A. Yokoymama, G. Matsuda, K. Kato, H. Kagawa, K. Hirai, and B. Roizman. 2001. Herpes simplex virus 1 alpha regulatory protein ICP0 functionally interacts with cellular transcription factor BMAL1. Proc. Natl. Acad. Sci. USA 98:1877-1882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kawaguchi, Y., C. Van Sant, and B. Roizman. 1998. Eukaryotic elongation factor 1delta is hyperphosphorylated by the protein kinase encoded by the U(L)13 gene of herpes simplex virus 1. J. Virol. 72:1731-1736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kawaguchi, Y., C. Van Sant, and B. Roizman. 1997. Herpes simplex virus 1 alpha regulatory protein ICP0 interacts with and stabilizes the cell cycle regulator cyclin D3. J. Virol. 71:7328-7336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lagunoff, M., and B. Roizman. 1994. Expression of a herpes simplex virus 1 open reading frame antisense to the gamma(1)34.5 gene and transcribed by an RNA 3′ coterminal with the unspliced latency-associated transcript. J. Virol. 68:6021-6028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Link, A. J., D. Phillips, and G. M. Church. 1997. Methods for generating precise deletions and insertions in the genome of wild-type Escherichia coli: application to open reading frame characterization. J. Bacteriol. 179:6228-6237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Marchini, A., H. Liu, and H. Zhu. 2001. Human cytomegalovirus with IE-2 (UL122) deleted fails to express early lytic genes. J. Virol. 75:1870-1878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Markert, J. M., M. D. Medlock, S. D. Rabkin, G. Y. Gillespie, T. Todo, W. D. Hunter, C. A. Palmer, F. Feigenbaum, C. Tornatore, F. Tufaro, and R. L. Martuza. 2000. Conditionally replicating herpes simplex virus mutant G207 for the treatment of malignant glioma: results of a phase I trial. Gene Ther. 7:867-874. [DOI] [PubMed] [Google Scholar]
- 28.Markert, J. M., J. N. Parker, G. Y. Gillespie, and R. J. Whitley. 2001. Genetically engineered human herpes simplex virus in the treatment of brain tumours. Herpes 8:17-22. [PubMed] [Google Scholar]
- 29.McGregor, A., and M. R. Schleiss. 2001. Molecular cloning of the guinea pig cytomegalovirus (GPCMV) genome as an infectious bacterial artificial chromosome (BAC) in Escherichia coli. Mol. Genet. Metab. 72:15-26. [DOI] [PubMed] [Google Scholar]
- 30.McKnight, J. L., T. M. Kristie, and B. Roizman. 1987. Binding of the virion protein mediating alpha gene induction in herpes simplex virus 1-infected cells to its cis site requires cellular proteins. Proc. Natl. Acad. Sci. USA 84:7061-7065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Messerle, M., I. Crnkovic, W. Hammerschmidt, H. Ziegler, and U. H. Koszinowski. 1997. Cloning and mutagenesis of a herpesvirus genome as an infectious bacterial artificial chromosome. Proc. Natl. Acad. Sci. USA 94:14759-14763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Post, L. E., S. Mackem, and B. Roizman. 1981. Regulation of alpha genes of herpes simplex virus: expression of chimeric genes produced by fusion of thymidine kinase with alpha gene promoters. Cell 24:555-565. [DOI] [PubMed] [Google Scholar]
- 33.Post, L. E., and B. Roizman. 1981. A generalized technique for deletion of specific genes in large genomes: alpha gene 22 of herpes simplex virus 1 is not essential for growth. Cell 25:227-232. [DOI] [PubMed] [Google Scholar]
- 34.Rampling, R., G. Cruickshank, V. Papanastassiou, J. Nicoll, D. Hadley, D. Brennan, R. Petty, A. MacLean, J. Harland, E. McKie, R. Mabbs, and M. Brown. 2000. Toxicity evaluation of replication-competent herpes simplex virus (ICP 34.5 null mutant 1716) in patients with recurrent malignant glioma. Gene Ther. 7:859-866. [DOI] [PubMed] [Google Scholar]
- 35.Roizman, B., and D. M. Knipe. 2001. Herpes simplex viruses and their replication, p. 2399-2459. In D. M. Knipe, P. M. Howly, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology, 4th ed. Lippincott-Williams &Wilkins, Philadelphia, Pa.
- 36.Saeki, Y., T. Ichikawa, A. Saeki, E. A. Chiocca, K. Tobler, M. Ackermann, X. O. Breakefield, and C. Fraefel. 1998. Herpes simplex virus type 1 DNA amplified as bacterial artificial chromosome in Escherichia coli: rescue of replication-competent virus progeny and packaging of amplicon vectors. Hum. Gene Ther. 9:2787-2794. [DOI] [PubMed] [Google Scholar]
- 37.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 38.Smith, G. A., and L. W. Enquist. 1999. Construction and transposon mutagenesis in Escherichia coli of a full-length infectious clone of pseudorabies virus, an alphaherpesvirus. J. Virol. 73:6405-6414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Smith, G. A., and L. W. Enquist. 2000. A self-recombining bacterial artificial chromosome and its application for analysis of herpesvirus pathogenesis. Proc. Natl. Acad. Sci. USA 97:4873-4878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Spaete, R. R., and N. Frenkel. 1982. The herpes simplex virus amplicon: a new eucaryotic defective-virus cloning-amplifying vector. Cell 30:295-304. [DOI] [PubMed] [Google Scholar]
- 41.Stavropoulos, T. A., and C. A. Strathdee. 1998. An enhanced packaging system for helper-dependent herpes simplex virus vectors. J. Virol. 72:7137-7143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sternberg, N., and D. Hamilton. 1981. Bacteriophage P1 site-specific recombination. I. Recombination between loxP sites. J. Mol. Biol. 150:467-486. [DOI] [PubMed] [Google Scholar]
- 43.Todo, T., S. D. Rabkin, P. Sundaresan, A. Wu, K. R. Meehan, H. B. Herscowitz, and R. L. Martuza. 1999. Systemic antitumor immunity in experimental brain tumor therapy using a multimutated, replication-competent herpes simplex virus. Hum. Gene Ther. 10:2741-2755. [DOI] [PubMed] [Google Scholar]
- 44.Wagner, M., S. Jonjic, U. H. Koszinowski, and M. Messerle. 1999. Systematic excision of vector sequences from the BAC-cloned herpesvirus genome during virus reconstitution. J. Virol. 73:7056-7060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Whitley, R. J. 2001. Herpes simplex viruses, p. 2461-2509. In D. M. Knipe, P. M. Howly, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology, 4th ed. Lippincott-Williams &Wilkins, Philadelphia, Pa.
- 46.Wolfe, D., W. F. Goins, D. J. Fink, and J. C. Glorioso. 2000. Design and use of herpes simplex viral vectors for gene therapy, p. 81-108. In N. S. Templeton and D. D. Lasic (ed.), Gene therapy. Marcel Dekker, New York, N.Y.
- 47.Yu, D., G. A. Smith, L. W. Enquist, and T. Shenk. 2002. Construction of a self-excisable bacterial artificial chromosome containing the human cytomegalovirus genome and mutagenesis of the diploid TRL/IRL13 gene. J. Virol. 76:2316-2328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhou, F. C., Y. J. Zhang, J. H. Deng, X. P. Wang, H. Y. Pan, E. Hettler, and S. J. Gao. 2002. Efficient infection by a recombinant Kaposi's sarcoma-associated herpesvirus cloned in a bacterial artificial chromosome: application for genetic analysis. J. Virol. 76:6185-6196. [DOI] [PMC free article] [PubMed] [Google Scholar]