Abstract
All currently licensed yellow fever (YF) vaccines are propagated in chicken embryos. Recent studies of chick cell-derived measles and mumps vaccines show evidence of two types of retrovirus particles, the endogenous avian retrovirus (EAV) and the endogenous avian leukosis virus (ALV-E), which originate from the chicken embryonic fibroblast substrates. In this study, we investigated substrate-derived avian retrovirus contamination in YF vaccines currently produced by three manufacturers (YF-vax [Connaught Laboratories], Stamaril [Aventis], and YF-FIOCRUZ [FIOCRUZ-Bio-Manguinhos]). Testing for reverse transcriptase (RT) activity was not possible because of assay inhibition. However, Western blot analysis of virus pellets with anti-ALV RT antiserum detected three distinct RT proteins in all vaccines, indicating that more than one source is responsible for the RTs present in the vaccines. PCR analysis of both chicken substrate DNA and particle-associated RNA from the YF vaccines showed no evidence of the long terminal repeat sequences of exogenous ALV subgroups A to D in any of the vaccines. In contrast, both ALV-E and EAV particle-associated RNA were detected at equivalent titers in each vaccine by RT-PCR. Quantitative real-time RT-PCR revealed 61,600, 348,000, and 1,665,000 ALV-E RNA copies per dose of Stamaril, YF-FIOCRUZ, and YF-vax vaccines, respectively. ev locus-specific PCR testing of the vaccine-associated chicken substrate DNA was positive both for the nondefective ev-12 locus in two vaccines and for the defective ev-1 locus in all three vaccines. Both intact and ev-1 pol sequences were also identified in the particle-associated RNA. To investigate the risks of transmission, serum samples from 43 YF vaccine recipients were studied. None of the samples were seropositive by an ALV-E-based Western blot assay or had detectable EAV or ALV-E RNA sequences by RT-PCR. YF vaccines produced by the three manufacturers all have particles containing EAV genomes and various levels of defective or nondefective ALV-E sequences. The absence of evidence of infection with ALV-E or EAV in 43 YF vaccine recipients suggests low risks for transmission of these viruses, further supporting the safety of these vaccines.
Yellow fever (YF) vaccines are highly efficacious in preventing mortality and morbidity in humans living in or traveling to regions of tropical Africa and South America where YF is endemic (3, 20). To date, more than 400 million doses of YF vaccines have been administered (20). Currently licensed YF vaccines include the attenuated 17D strain, which is propagated in chicken embryos. Vaccine preparation typically includes inoculation of 7- to 9-day-old embryonated eggs with the vaccine strain. Infected embryos are harvested, pooled, and then homogenized. Homogenates are clarified by low-speed centrifugation, and the supernatant fluid containing the vaccine harvest is titrated and lyophilized under standardized conditions (33).
Because of the risk of contamination with substrate-derived avian pathogens, regulations for vaccine manufacture require all avian embryos used for propagation of vaccine virus to be derived from a closed, specific pathogen-free (SPF) flock devoid of known avian pathogens including exogenous retroviruses of the avian leukosis virus (ALV) and the reticuloendotheliosis virus groups (32). Despite the use of SPF chicken flocks, low-level reverse transcriptase (RT) activity, an indication of retroviruses, was recently detected in YF vaccines and other chick cell-derived measles and mumps vaccines produced by several manufacturers in Europe and the United States (5, 19, 28). The origin of RT activity has thus far been examined in measles vaccines produced in chicken embryonic fibroblasts (CEF). These studies have shown that this RT activity was associated with particles containing RNA from endogenous avian virus (EAV) and endogenous ALV (ALV-E) (17, 28, 30). These findings were consistent with current vaccine manufacturing regulations that require the elimination of exogenous retroviral infections from source chickens. These manufacturing regulations do not address the presence of endogenous retroviruses because such particles have not previously been known to be associated with chick cell-derived vaccines.
Both EAV and ALV-E are members of endogenous retroviral families present in the chicken germ line. Little is known about the EAV family, which is distinct from but related to the ALV family. EAV elements are present in at least 50 copies per chicken genome (23). The majority of EAV loci remain uncharacterized, and the nucleotide sequences of full-length EAV genomes are not available. Partial EAV sequences show numerous frameshift mutations or sizable deletions in the env region (6). The genome EAV-0, the most complete EAV derived from ALV-E-free (line 0) chickens, is approximately 5.8 kbp in length (23). None of the known EAV sequences represent full-length and intact retroviral genomes, and no infectious EAV isolates have yet been identified (6).
ALV-E particles are expressed from ev loci that are inheritable proviral elements (21). On the basis of its envelope sequence, ALV-E is differentiated from ALV subgroups A to D and J, which are all exogenously acquired infectious agents (11, 22). While exogenous ALVs have been shown to cause several neoplastic diseases in infected chickens (8, 9) and non-neoplastic diseases such as myocarditis (13) and osteopetrosis (26), ALV-E is not known to be pathogenic to chickens (10, 18). At least 22 different ev loci, designated ev-1 to ev-22, have been identified in White Leghorn chickens (21). ev loci are grouped on the basis of the viral phenotypes they confer (7, 21). These phenotypes range from structurally and enzymatically complete infectious particles (e.g., ev-2, ev-11, ev-12, ev-18, and ev-21) to the absence of detectable viral protein expression (e.g., ev-4, ev-5, and ev-8) to structurally or enzymatically defective particles (e.g., ev-1, ev-7, and ev-9). The defective nature of ev-1 is a result of a +1 frameshift mutation at nucleotide position 5026 within the pol region. This signature mutation truncates the α subunit of RT (RT-α) and inactivates integrase (17). ev-1 is ubiquitous in White Leghorns, and the other loci are less prevalent and vary in number depending on the chicken line (14). Since any given chicken may contain several different loci, the phenotypes of endogenous particles expressed by vaccine substrates may vary with the embryo pool used.
In contrast to the information available on avian retroviruses in CEF-derived measles vaccines, little is known about the presence and origin of adventitious retroviruses in currently used YF vaccines. The study of retroviral contamination in YF vaccines and the assessment of any associated risk for vaccine recipients are needed to fully define the safety of these vaccines. These studies are also important for determining whether propagation of YF vaccines in chicken embryos may be associated with increased contamination with endogenous retroviruses. Here, we (i) examined YF vaccines from three different manufacturers for RT activity and the presence of ALV and EAV, (ii) characterized ALV-E sequences and substrate-derived ev loci, and (iii) tested YF vaccine recipients for evidence of infection with ALV and EAV.
MATERIALS AND METHODS
YF and MMRII vaccines.
Three currently licensed YF vaccines, YF-vax (Connaught Laboratories), Stamaril (Aventis [formerly Pasteur Merieux]), and YF-FIOCRUZ (FIOCRUZ-Bio-Manguinhos, Rio de Janeiro, Brazil), were studied. The YF vaccine lots tested were 7A81464 and UA037AA for YF-vax, 00A13 for Stamaril, and 968FB026Z and 102/84 for YF-FIOCRUZ. The YF-vax and Stamaril vaccines were purchased, whereas YF-FIOCRUZ was donated by the manufacturer. MMRII vaccine (lot 0132J; Merck Inc.) was also purchased. YF-vax and Stamaril were obtained in single-dose vaccine vials; YF-FIOCRUZ was provided as 50 vaccine doses per vial.
Detection of RT activity.
The PCR-based Amp-RT assay was used to detect RT activity (15). Single-dose YF vaccines were serially diluted 10-fold. Five microliters each of undiluted and diluted vaccine was used for detection of RT activity. Rous-associated virus-0 (RAV-0), a prototype ALV-E isolate, was used as a positive control. Briefly, samples were suspended in RT buffer containing a 350-bp encephalomyocarditis virus (EMCV) RNA template and 200 ng of a complementary reverse primer. The mixture was incubated at 37°C for 2 h and then inactivated by heating at 95°C. The reaction-generated EMCV cDNA was amplified by PCR using 2.5 U of Taq polymerase and a forward primer for the EMCV sequence. Amplified Amp-RT products were detected by Southern blot hybridization using a 32P-labeled internal probe (15). RAV-0 was used to spike YF vaccine to ascertain any inhibition of the testing.
Detection of RT proteins by Western blotting.
Reconstituted preparations of YF-VAX, Stamaril, and YF-Brazil were clarified by centrifugation at 12,000 × g for 10 min. The supernatants were ultracentrifuged at 100,000 × g for 1 h to pellet virus particles. Virus particles were lysed by resuspending each vaccine pellet in 50 μl of buffer (125 mM Tris-HCl [pH 8.3], 125 mM KCl, 25 mM MgCl2, 1 mM EGTA, 0.15% NP-40, 5 mM dithiothreitol) and vortexing for 30 s. Lysates of the YF vaccines were boiled in loading dye for 8 min, and then one-fourth-dose equivalents of YF-Vax and one-dose equivalents of Stamaril and YF-FIOCRUZ were loaded, along with RT from an exogenous avian myeloblastosis virus (AMV) isolate, into two 10% acrylamide Ready Gels (Bio-Rad, Hercules, Calif.). The lysates were fractionated at 95 V for 2 h and then transferred onto natural nitrocellulose (0.2 μm; Bio-Rad) for 1.5 h at 90 V. The membranes were blocked overnight at 4°C in blocking buffer (Tris-buffered saline-5% milk-0.1% Tween 20). One blocked membrane was incubated overnight at 4°C with goat anti-AMV RT (Quality Biotech Incorporated-Resource Laboratory, Camden, N.J.) at a 1:1,750 dilution in blocking buffer, and the other membrane was incubated with goat preimmune serum. The membranes were then washed four times for 30 min each with wash buffer (Tris-buffered saline-1% Tween 20) and incubated for 1.5 h with horseradish peroxidase (HRP)-conjugated donkey anti-goat immunoglobulin G. After the secondary incubation, the membranes were again washed four times. The blots were developed by addition of ECL substrate (Amersham, Piscataway, N.J.) and exposure to Hyperfilm (Amersham) for 3 min (16). The sizes of reactive bands were determined from a standard curve made with Kaleidoscope prestained 202-, 121-, 79-, 41-, and 31.6-kDa markers (Bio-Rad) and the 92- and 68.4-kDa AMV RT proteins.
Extraction of vaccine-associated substrate DNA and particle-associated RNA from YF vaccines.
YF vaccines were reconstituted and were then clarified by centrifugation at 10,000 × g for 5 min. Substrate DNA from single-dose YF vaccines was extracted using a Promega Wizard genomic DNA isolation kit. Particle-associated RNA was extracted as previously described (17). Briefly, supernatants were ultracentrifuged at 100,000 × g for 1 h and pellets were resuspended in 100 μl of Dulbecco's phosphate-buffered saline. Free RNA and DNA were digested with RNase (2 U) and twice with DNase (5 U) for 1 h at 37°C, respectively. Particle-associated RNA in digested pellets was then extracted using a QIAamp viral RNA kit (QIAGEN Inc., Valencia, Calif.). Extracted RNA was resuspended in 60 μl of elution buffer provided in the QIAamp viral RNA kit. Culture supernatant containing RAV-0 was used as the positive control to validate RNA extraction.
Detection of exogenous ALV DNA and RNA sequences in YF vaccines.
The presence of exogenous ALV DNA in single-dose YF vaccines was determined by a newly developed PCR assay. The assay uses primers that are conserved among the U3-U5 regions of the long terminal repeats (LTRs) of exogenous ALV subgroups A to D. DNA equivalent to one-fifth of a dose was amplified with the forward primer exoALV LTRF1 (5′-ACG ATG AGT TAG CAA CAT GCC TTA CAA GG-3′) and the reverse primer exoALV LTRR1 (5′-GTC CGG CCA TCA ACC CAG GTG-3′). The PCR conditions were 94°C for 1 min, 57°C for 1 min, and 72°C for 1 min for 39 cycles. Plasmid pATV-8 containing the full-length genome of exogenous ALV subgroup C was used as a positive control (ATCC 45033; American Type Culture Collection, Manassas, Va.). PCR products were detected by Southern blot hybridization using the 32P-labeled exoALV LTR.1P probe (5′-AGA GAA AAA GCA CCG TGC ATG-3′). The detection threshold of the PCR assay was measured using known copy numbers of the pATV-8 plasmid. Particle-associated RNA from each YF vaccine was analyzed by RT-PCR for exogenous ALV sequences by using primers derived from the U3-R region of the LTR. The forward primer was exoALV LTRF1 and the reverse primer was exoALV LTRR2 (5′-ATG CGG AAT TCA GTG GTT CGT CCA ATC-3′). The RT reactions were primed with exoALV LTRR2 at 37°C for 60 min followed by 95°C for 5 min. PCR amplification and detection of the amplified products with exoALV LTR.1P were performed as described for the proviral LTR sequences.
Detection of ALV-E and EAV RNA sequences in YF vaccines.
RT-PCRs to test for the presence of ALV-E and EAV RNA were performed as described previously (28). RT-PCRs for ALV-E env and EAV env-like sequences were set up with 4 μl of particle-associated RNA (equivalent to ∼6% of the total RNA from one dose of vaccine). The primers used for ALV-E RT-PCR were ALVENVF2 (5′-CGG TGC ATA TGG CTA CAG ATT TTG-3′) and ALVENVR2 (5′-TTT CCA CAA CAT CCG CTG ACA TTA-3′); the specific probe was ALVENVP1 (5′-AAG GAA ATT AAT GAG ACA GAG CCG-3′). The primers used for EAV RT-PCR were EAVENVF10 (5′-ACA GAA GAT CAA GAT GCA GGC CGA-3′) and EAVENVR10 (5′-GCT CCT GAA TGC GCT GAT ACA CGT-3′); the specific probe was EAVENVP1 (5′-CCG TGG CTA AAA CAA ATG CTT-3′). PCR products were detected by Southern blot hybridization using the ALV-E- and EAV-specific 32P-labeled probes.
ev loci typing by locus-specific PCR analysis.
Locus-specific PCRs were used to identify several ev loci in YF vaccines (4, 17). The YF vaccine lots tested were UA037AA for YF-vax, 00A13 for Stamaril, and 968FB026Z for YF-FIOCRUZ. PCR primers directed against the ev LTRs and the sequences flanking known integration sites for each locus were used for amplification (17). DNA equivalent to 1/20 of a single dose of YF vaccine was used in each PCR. Amplification of the targeted sites produced fragments of defined lengths, which indicated the presence or absence of each locus (4). ev loci testing included assays for defective ev-1 and ev-3 and nondefective ev-2, ev-7, ev-9, ev-12, and ev-21 by using previously published primers (17), except in the case of ev-1, ev-3, and ev-9, for which the LTR primer was ev1LTR.1F (5′GCG TAG AC GAA GC ATG TAC GAT3′). PCR products for ev-3, ev-9, and ev-21 were detected by Southern blot hybridization using the 32P-labeled evLTRA.1P probe (5′-CCT GAA TGA AGC AGA AGG CTT CG-3′), and those for ev-2, ev-7, and ev-12 were detected with the ev1LTR.1P probe (5′-TGA AAA GTC TAA AGA CCA AAT AAG-3′). The ev-1 locus was used as a positive control, and the PCR product was probed with evLTRA.2P (5′GAT ATT AAA GTC AAT TTC TAC TAA G-3′), which allowed the determination of the heterozygous and/or homozygous presence of the locus (17).
Quantitation of particle-associated ALV-E RNA by real-time RT-PCR.
The levels of the ALV-E RNA sequences in particle-associated RNA extracts from the YF vaccines were determined by a quantitative RT-PCR assay that targeted the R-U5 region in the LTR. RNA extracts were reverse transcribed in triplicate reactions for 1 h at 37°C by using the evLTRqB.R3 reverse primer (5′-ACG ATT GCG AAC ACC TGA AT-3′) and were then amplified by PCR. RNA extracted from RAV-0 was utilized as the positive control. The PCR amplifications were performed on an iCycler unit with optical module (Bio-Rad) by using the evLTRq.F3 (5′-CAT TTT ACC ATC CAC CAC ATT GG-3′) and evLTRqB.R3 primers and the 6-carboxyfluorescein (FAM)-labeled evLTRq.3P probe (5′FAM-TGC ACC TGG GTA GAT GGA CAG ACC G-QSY7-3′). PCRs were initiated by an 11-min hot start with ampliTaq Gold polymerase (Perkin-Elmer) followed by 50 cycles of 95°C for 30 s, 50°C for 30 s, and 60°C for 30 s. Threshold (10 times the mean of the standard deviation of the background fluorescence) was calculated by using amplification cycles 2 to 20 as the baseline. A sample with a relative fluorescence threshold crossing (TC) of 40 cycles or less and with a consistent increase in relative fluorescence was considered positive. Two micrograms of DNA from quail fibroblasts and line 0 CEF, which do not contain ALV-E sequences, were used as negative controls. To generate the ALV-E standards for quantitation, an ev-1 U3-R-U5 LTR fragment was amplified from CEF DNA by using the primers evLTRdn.F2 (5′-GAC ATA TGG GCG TAG ACG AAG CTA TGT-3′) and evLTRdn.R2 (5′-CTG CTT CAT TCA GGT GTT CGC AAT CGT-3′). The amplified LTR product was ligated into the pCR2.1 vector (Invitrogen) and then used in a 42°C heat-shock transformation of Escherichia coli. One recombinant colony was selected, the plasmid clone was extracted, and the copy numbers were determined spectrophotometrically.
Serial dilutions of 104 to 101 copies/10 μl of the evLTRdn2 plasmid were amplified in each experiment, and their resultant TC values were used for the generation of the standard curve. ALV-E copy numbers in each reaction were extrapolated from the plotting of the TC values of the samples against the standard curve, provided that the amplification efficiency of each copy number fell within a 95% linear correlation (correlation coefficients, >0.95). Final ALV-E copy numbers for each vaccine were computed from the mean result of six evaluations.
Sequence analysis of ALV-E pol RNA sequences in the vaccines.
Particle-associated RNA equivalent to one-half dose of each vaccine was used in RT-PCR amplifications of a 2,064-bp region in the pol gene. Reverse transcription reactions were carried out for 1 h at 39°C by using the evpolnesREV primer (5′-ACC TTC GGA AAT AGG GGA CGG GAT-3′). The cDNA product was PCR amplified for 40 cycles (melting at 95°C for 1 min, annealing at 55°C for 1 min, and extension at 72°C for 2 min) by using the forward primer polUF2 (5′-TTC TAT GAG CAG TTA CGA GGG TC-3′). This region was further amplified for 30 cycles by using the nested primers polF6 (5′-CCA CAC ACC CAA GGC CAT GTT TG-3′)-polR4 (5′-GAC GTG AAG CAG GAC CCA TTA TCT G-3′) and polF8 (5′-GCG TAT CCC TTG AGA GAG GCT AAA G-3′)-polR3 (5′-CCT TTT TCC CAC TCC CCT GTC TCT-3′). For each RNA extract in which RT was omitted prior to nested PCR amplification, additional reactions were performed to rule out the presence of residual DNA. Double-stranded sequence analysis was performed on the overlapping nested fragments by the chain termination method with Big Dye Terminator reagents (Perkin-Elmer ABI).
YF vaccine recipients.
Serum samples obtained after YF vaccination were available from 43 Brazilian subjects who have documented vaccinations with YF-FIOCRUZ. Of the 43 vaccine recipients, 34 received one dose of YF vaccine, 2 received two doses, 3 received three doses, and 4 received four or more doses. The mean times of serum sample collection following the last YF vaccination were 4.37 years (range, 0.25 to 25 years), 6.0 years (range, 5 to 7 years), 3.3 years (range, 1 to 8 years), and 2 years (range, 1 to 4 years) for recipients receiving one, two, three, and four or more doses of vaccine, respectively. Baseline serum samples obtained before YF vaccination were also available for all 43 subjects.
Detection of antibodies to ALV-E gag p27 in recipients of YF vaccine.
A recently developed and validated Western blot assay was used to detect antibodies to ALV-E in YF vaccine recipients (16). The source of the antigen for the Western blot assay was RAV-0-infected 15B1 CEF. Uninfected 15B1 CEF cell lysates were used as the negative control antigen to test reactive samples for specificity. Serum samples were diluted 1:50 in blocking buffer. Rabbit anti-AMV p27 antibody (SPAFAS, Norwich, Conn.) was used as a positive control. Anti-rabbit and anti-human antibodies conjugated to HRP were used as secondary antibodies for rabbit and human plasma samples, respectively, at dilutions of 1:6,000. Control human immunoglobulin G was used as an assay control for anti-human HRP-conjugated secondary antibody. Seroreactivity to ALV p27 Gag protein was visualized by chemiluminescence with ECL (Amersham, Rockford, Ill.) Western blotting reagents (16).
Detection of ALV and EAV RNA sequences in sera of YF vaccine recipients by RT-PCR.
RNA was extracted from 200 μl of serum by using a QIAGEN viral RNA mini kit. RNA extract from 34 μl of serum was used for each RT-PCR as previously described (16). The primers used for ALV RT-PCR were ALVENVF2 and ALVENVR2, and the specific probe was ALVENVP1. The primers used for EAV RT-PCR were EAVENVF10 and EAVENVR10, and the specific probe was EAVENVP1 (16). The RT reaction was carried out at 37°C for 2 h followed by 95°C for 5 min. The PCR conditions included 35 cycles of 95°C for 1 min, 55°C for 1 min, and 72°C for 1 min. PCR products were detected by Southern blot hybridization using specific 32P-labeled internal probes. RNA extracts from culture supernatants containing RAV-0 and EAV were used as positive controls.
Nucleotide sequence accession number.
The ALV-E sequence obtained from the YF-FIOCRUZ vaccine (see below) has been deposited in GenBank under accession no. AY127567.
RESULTS
Screening for RT activity.
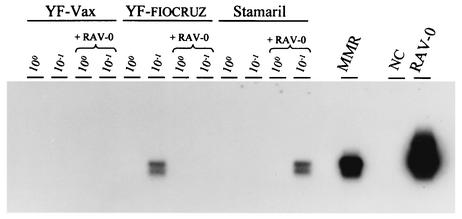
The results of the RT testing with the Amp-RT assay are shown in Fig. 1. No RT activity was detected in any of the three undiluted YF vaccines (YF-FIOCRUZ, lot 968FB026Z; Stamaril, lot 00A13; and YF-vax, lot UA037AA). Similarly, the results of the testing of 10-fold dilutions were also negative, with only a weak signal detected for the YF-FIOCRUZ vaccine. In contrast, the measles, mumps, and rubella (MMR) vaccine had detectable RT activity. Both undiluted and diluted YF vaccines spiked with RAV-0 tested negative, with the exception of a weak signal observed in only one reaction. These data confirm the presence of assay inhibitors and suggest that the observed negative results do not indicate the absence of RT activity.
FIG. 1.
Testing for RT activity in three YF vaccines by the Amp-RT assay and demonstration of assay inhibition. 100, undiluted; 10−1, 1/10 dilution; +RAV-0, spiked with RT-positive RAV-0; MMR, measles, mumps, and rubella vaccine; NC, negative control (water).
Detection of RT proteins by Western blotting.
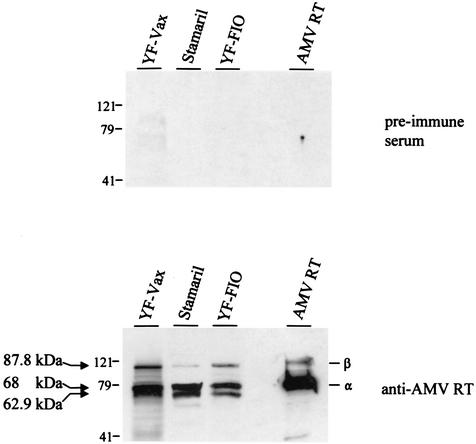
Since enzymatic detection of RT activity was not possible, we used Western blotting to look for evidence of RT proteins that are reactive to an anti-AMV RT antiserum. Figure 2 shows representative results from the Western blot analysis. All three vaccines tested had detectable bands. The predominant band migrated at 68 kDa, similar in size to the AMV RT-α. The second most predominant band migrated at ∼62.9 kDa and is close in size to the expected 61.6-kDa full-length EAV-0 RT-α. A third band of ∼87.8 kDa was also visible and may represent the RT-β subunit of ev-1, which is known to prematurely terminate in pol and is expected to yield an RT-β of 87.5 kDa. No reactive bands were seen with the control preimmune serum, confirming the specificity of the observed reactivity.
FIG. 2.
Western blot reactivity of viral proteins in YF vaccines to an AMV RT antiserum. α, AMV RT-α; β, AMV RT-β; YF-FIO, YF FIOCRUZ vaccine. Arrows indicate the sizes of three reactive bands.
Detection of particle-associated ALV-E and EAV RNA sequences in YF vaccines.
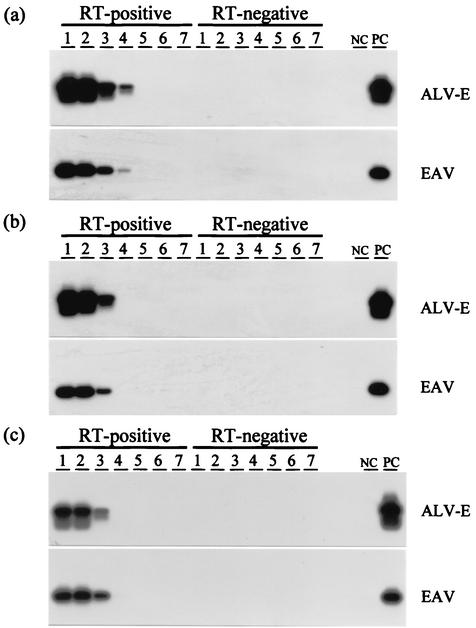
Both ALV-E and EAV RNA sequences were detectable by RT-PCR analysis of particle-associated RNA extracts from all three YF vaccines (Fig. 3). Testing of end-point 10-fold serial dilutions revealed equivalent titers of EAV and ALV-E for all three vaccines, as well as higher titers of both sequences in the YF-vax vaccine. No sequences were detected by any of the PCRs without RT, confirming that the observed positive amplifications were all of RNA origin.
FIG. 3.
Titration of ALV-E and EAV particle-associated RNA sequences in the YF vaccines YF-vax (a), YF-FIOCRUZ (b), and Stamaril (c) by RT-PCR. Lane 1, undiluted vaccine; lanes 2 to 7, vaccine at dilutions of 10−1, 10−2, 10−3, 10−4, 10−5, and 10−6, respectively. RT-negative, control amplification reactions without RT; NC, negative control; PC, positive control.
Absence of exogenous ALV DNA and RNA in YF vaccines.
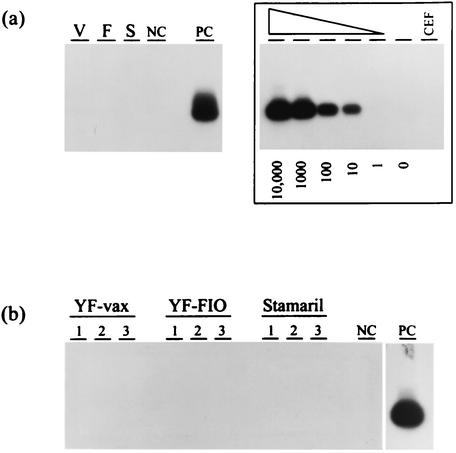
Both the vaccine-associated chicken substrate DNA and the particle-associated RNA from all three vaccines were found to be negative for exogenous ALV LTR sequences by PCR and RT-PCR analysis, respectively. Representative RT-PCR results are shown in Fig. 4. These data do not support the presence of exogenous ALV in the tested vaccines. A titration of plasmid copies showing a detection threshold for the PCR assay of 10 copies is also shown in Fig. 4.
FIG. 4.
Analysis of YF vaccines for the presence of exogenous ALV (subgroups A to D). (a) Proviral detection in vaccine-associated DNA. V, YF-vax; F, YF-FIOCRUZ; S, Stamaril. The assay detection threshold of known copy numbers (10,000 to 0) is shown. (b) RT-PCR results of 3 log dilutions of particle-associated RNA. 1, undiluted; 2, 10-fold dilution; 3, 100-fold dilution; NC, negative control; PC, positive control; CEF, PCR-negative CEF DNA.
ALV-E particle-associated RNA levels in YF vaccines.
To better quantitate the ALV-E RNA copy numbers in the vaccines, a quantitative fluorochrome-based real-time RT-PCR assay was developed and used to amplify particle-associated RNA extracts. Duplicate samplings of serial dilutions (10−1 to 10−3) of each vaccine were made and tested in two different evaluations. Based on these data, the ALV-E LTR copy numbers for each vaccine were computed. YF-vax lot UA037AA had 1,665,000 RNA copies/dose, while YF-FIOCRUZ lot 963FB026Z had 348,000 copies/dose and Stamaril lot 00A13 had 61,600 copies/dose. The MMRII lot 0132J had 18,900 copies/dose.
Identification of ev loci in YF vaccines.
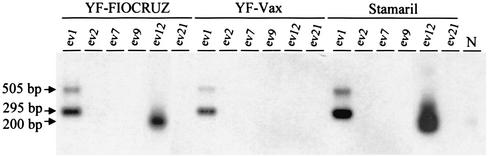
The results of ev locus-specific PCR analysis of YF-vax, Stamaril, and YF-FIOCRUZ vaccines for ev-1, ev-2, ev-7, ev-9, ev-12, and ev-21 are shown in Fig. 5. All three vaccines were positive for ev-1. YF-FIOCRUZ and Stamaril were positive for ev-12, while YF-vax tested negative for this locus. The PCR signals for ev-12 were generally weaker than those for ev-1 and required longer exposures for the Southern blot hybridizations, suggesting a lower copy number in the tested DNA. All three vaccines were negative for ev-2, ev-7, ev-9, and ev-21.
FIG. 5.
ev locus-specific PCR analysis of YF vaccine-associated DNA. Arrows show the sizes of detected Southern blot signals indicative of the presence of ev-1 (295 bp) and ev-12 (200 bp) and the absence of ev-1 (505 bp). N, CEF ev-12-negative control.
Sequence analysis of ALV-E pol in the vaccines.
To better understand the ev source of the particle-associated genomic ALV-E sequences, we analyzed the pol sequences in all three vaccines. The polUF2 and evpolnesREV primers successfully amplified 2,064-bp sequences of ALV-E pol, which was further amplified by nested PCR to generate sufficient template for population-based sequencing. All three vaccines were found to have ALV-E sequences that were identical to ev-1 and had the signature insertion mutation at position 5026. A mixture of ev-1 and a less prevalent species was detected in the product amplified from the YF-FIOCRUZ vaccine. The additional sequence did not possess an insertion at position 5026 and had seven other nucleotide changes in the 668 bp preceding position 5026. This sequence was intact, had a 98.9% sequence similarity to ev-1 in this region of pol, and may represent RNA from either ev-12 or another locus not tested in our study.
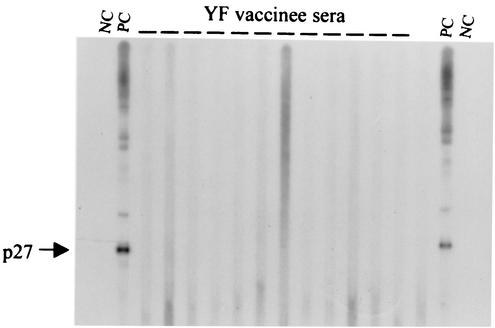
Western blot reactivity to ALV p27 in YF vaccine recipients.
Of the 43 post-YF vaccination serum samples, 42 were nonreactive to the p27 protein in the Western blot assay (Fig. 6). Only one serum sample showed weak reactivity to a band slightly larger than p27. This sample was equally reactive to the same band in the uninfected control antigen, indicating that the observed reactivity was nonspecific (data not shown). We also tested the baseline pre-YF vaccination serum sample available for this vaccine recipient and found that it was also reactive to the same band, indicating the presence of preexisting nonspecific antibodies (data not shown). No other reactivity was observed for any of the 43 serum samples tested.
FIG. 6.
Western blot reactivity of sera from YF vaccine recipients to ALV-E antigen. NC, negative control human serum; PC, positive control anti-p27 ALV Gag antiserum; —, test serum.
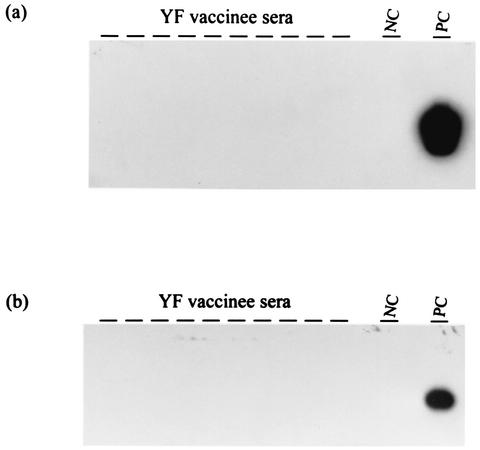
ALV and EAV RNA sequences in sera from YF vaccine recipients.
All sera from the 43 YF vaccine recipients tested negative for both ALV and EAV RNA by RT-PCR (Fig. 7). These results indicate the absence of both ALV and EAV viremia in these vaccine recipients.
FIG. 7.
RT-PCR analysis of sera from YF vaccine recipients for the presence of ALV-E (a) and EAV (b). PC, positive control; NC, negative control; —, test sample.
DISCUSSION
This study investigated the presence of ALV and EAV particles in YF vaccines produced by three vaccine manufacturers and assessed the evidence of infection with these retroviruses in YF vaccine recipients. We first showed that no exogenous ALV RNA or DNA sequences were detected in any of the three YF vaccines, a result that does not support contamination with exogenous ALV. Exogenous ALVs have previously been found in YF vaccines, and the observed absence of these viruses may be explained by the adherence of the manufacturers to current regulations that require chicken flocks used for vaccine preparation to be free of specific exogenous ALV pathogens (29, 32).
Consistent with previous reports (2, 25), we showed that assay inhibition hampered screening for enzymatic RT activity by a PCR-based RT test. However, by using a Western blot assay and an antiserum against AMV RT, we were able to document the presence of virion-derived RT proteins in all three YF vaccines. Since any given retrovirus is known to have two processed RT proteins, the detection of three reactive bands indicates that these proteins originated from more than one source. The observed sizes of these bands suggest that ev-1 and EAV are two likely sources of the RT. Since cleavage of RT subunits is known to occur late during particle budding, the detection of these RT subunits further proves the presence of viral particles in all three vaccines (11). These findings demonstrate the utility of Western blot analysis for detecting and characterizing RT contamination of biologics, particularly in products that have substances inhibitory to PCR-based RT testing.
Our data showing detectable particle-associated ALV-E and EAV RNA in all three YF vaccines indicate that genomic RNA from both ALV-E and EAV are packaged. The end-point dilution analysis showed equivalent titers of ALV-E and EAV in all three vaccines, a finding not previously seen in CEF-derived MMR vaccines in which EAV was the predominant RNA species (28, 30). The difference between the proportions of ALV-E in the MMR and YF vaccines is not well understood. It may simply reflect higher levels of ALV-E expression in embryos than in CEF, it could possibly be related to increased activation of ALV-E expression by the replication of the YF vaccine virus, or perhaps it results from variability in the copackaging of EAV or ALV-E RNA among different vaccines. The predominance of particle-producing ev loci in the substrates of the YF vaccines may also explain the higher proportion of ALV-E. However, this possibility is not supported by the fact that only ev-1 and ev-12 were found in the YF vaccines.
The quantitation of particle-associated ALV-E RNA copy numbers by real-time RT-PCR has provided new information on the estimates of particle numbers in the vaccines. We found a 27-fold variability in the ALV-E copy numbers among the three vaccines, and a substantial number of RNA copies (1,665,000) was seen in one YF-vax lot. These data suggest that while all recipients of YF vaccines are exposed to ALV-E, the degree of such exposures may vary with each vaccine. The higher ALV-E levels seen in some vaccines may be explained by increased expression of ALV-E in the substrates or may simply reflect the use of more-concentrated vaccines by some manufacturers. The latter possibility is supported by the fact that the minimum vaccine titer in YF-vax is at least 1.3 log higher than that in either Stamaril or YF-FIOCRUZ. Analysis of a larger number of vaccines of different lots will help define the range of ALV-E levels in YF vaccines.
To better understand the ev source of the ALV-E RNA, we analyzed the ev types in the chicken substrate DNA found in the YF vaccines. This testing can provide a direct approach to define the ev loci present in the substrate used for any given vaccine. While the amount of substrate DNA in these vaccines was not sufficient for loci typing by conventional restriction fragment length polymorphism, we show that it was adequate for ev typing by PCR-based tests. We demonstrated the presence of ev-1 in all vaccines, a finding consistent with the ubiquitous nature of this locus (27). We also found evidence of lower levels of ev-12 in two vaccines. The identification of ev-1 RNA in all three vaccines and of an additional RNA with an intact pol sequence, likely of ev-12 origin, in one vaccine further supports these findings. While both loci are known to express virus particles, only virions derived from ev-12 are infectious, since those expressed from ev-1 have a truncated RT-β and integrase (7, 17). The detection of ev-1 and ev-12 in the YF vaccines demonstrates that both defective and nondefective ev loci can contribute to the ALV-E present in YF vaccines. Our findings, however, do not exclude the presence of additional nondefective loci (e.g., ev-18 and ev-19), which could not be tested because of the absence of specific PCR assays.
The documentation of both ALV-E and EAV in all YF vaccines suggests that YF vaccine recipients may be universally exposed to these endogenous retroviral particles and thus highlights the importance of assessing the risks of transmission of these viruses to YF vaccine recipients. The possibility that YF vaccine recipients may also be exposed to nondefective ALV-E further heightens the importance of these studies. Our data on 43 recipients of YF-FIOCRUZ show no evidence of ALV-E or EAV infection. The absence of infection in nine individuals who had more than one YF vaccination further suggests that EAV and both defective and nondefective ALV-E are not easily transmissible through YF vaccination. While reassuring, our results are limited by the small number of YF vaccine recipients tested and may warrant confirmation by a study with a larger number of vaccinees. Nevertheless, the negative findings observed here are similar to those from a previous study of 206 MMR vaccine recipients (16), and collectively, these data demonstrate the low risks of ALV-E and EAV transmission through these exposures. Thus, our data provide support for current immunization policies with YF vaccines.
Several factors may explain the lack of transmission of either EAV or ALV-E to YF vaccine recipients. These include natural resistance of human cells to infection with these viruses, the presence of few or no infectious particles in the vaccines, or nonspecific virus lysis or inactivation (16, 31). While the role of each of these factors in the observed lack of infection with ALV-E or EAV is not entirely clear, the defective nature of known EAV genomes may explain the absence of EAV infection. Data presented by Adkins et al. (1) suggest that human cells may be resistant to ALV-E infection because they lack a specific surface receptor. Those authors showed that expression of the avian TVB receptor renders human 293 cells susceptible to ALV-E infection, demonstrating that ALV-E replication can be supported in these cells if virus entry is achieved. More work is necessary to define the susceptibility of different human cell types to ALV-E infection.
While our study is the first to document the presence of avian endogenous retroviruses in currently used YF vaccines, contamination of YF vaccines with exogenous ALV has been described previously (12). However, no evidence of neutralizing antibodies or increase in cancer rates has been associated with exposure to these ALV-positive vaccines (24, 29).
The detection of avian endogenous retroviruses in currently used YF vaccines raises the question of whether an alternate vaccine substrate is desirable. Obtaining YF vaccines that are free of any avian retrovirus may require a substantial change from the use of chicken embryos to that of cells from different species such as immortalized or diploid mammalian cells, which are known to be susceptible to infection with YF vaccine strains. Any change in the substrate should be evaluated for its potential to affect the safety or efficacy of the vaccine. While it may not be possible at present to obtain chick-derived substrates that do not express any ALV-E or EAV, it may be advisable to consider the use of chicken substrates that are free of nondefective ev loci and, thus, do not express replication-competent ALV-E. Since such ev loci are known, vaccine substrate providers can readily identify and eliminate them from their source SPF chickens.
Acknowledgments
We thank Thomas M. Folks for critical review of the manuscript.
This work was supported in part by a grant from the National Vaccine Program Office.
REFERENCES
- 1.Adkins, H. B., J. Brojatsch, J. Naughton, M. M. Rolls, J. M. Pesola, and J. A. T. Young. 1997. Identification of a cellular receptor for subgroup E avian leukosis virus. Proc. Natl. Acad. Sci. USA 94:11617-11622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Andre, M., S. Morgeaux, and F. Fuchs. 2000. Quantitative detection of RT activity by PERT assay: feasibility and limits to a standardized screening assay for human vaccines. Biologicals 28:67-80. [DOI] [PubMed] [Google Scholar]
- 3.Barrett, A. D. 1997. Yellow fever vaccines. Biologicals 25:17-25. [DOI] [PubMed] [Google Scholar]
- 4.Benkle, B. F. 1998. Locus-specific diagnostic test for endogenous avian leukosis-type viral loci in chickens. Poult. Sci. 77:1027-1035. [DOI] [PubMed] [Google Scholar]
- 5.Boni, J., J. Stadler, F. Reigei, and J. Shupbach. 1996. Detection of reverse transcriptase activity in live attenuated virus vaccines. Clin. Diagn. Virol. 5:43-53. [DOI] [PubMed] [Google Scholar]
- 6.Boyce-Jacino, M. T., K. O'Donoghue, and A. J. Faras. 1992. Multiple complex families of endogenous retroviruses are highly conserved in the genus Gallus. J. Virol. 66:4919-4929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Coffin, J. M., P. N. Tsichlis, K. F. Conklin, A. Senior, and H. L. Robinson. 1983. Genomes of exogenous and endogenous avian retroviruses. Virology 126:51-72. [DOI] [PubMed] [Google Scholar]
- 8.Cooper, M. D., L. N. Payne, P. B. Dent, B. R. Burmester, and R. A. Good. 1968. Pathogenesis of avian lymphoid leukosis. I. Histogenesis. J. Natl. Cancer Inst. 41:373-378. [PubMed] [Google Scholar]
- 9.Crittenden, L. B., W. S. Hayward, H. Hanafusa, and A. M. Fadly. 1980. Induction of neoplasms by subgroup E recombinant of exogenous and endogenous avian retroviruses (Rous-associated virus type 60). J. Virol. 33:915-919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Crittenden, L. B., R. L. Witter, and A. M. Fadly. 1979. Low incidence of lymphoid tumors in chickens continuously producing endogenous virus. Avian Dis. 23:646-653. [PubMed] [Google Scholar]
- 11.Crittenden, L. B. 1991. Retroviral elements in the genome of the chicken: implications for poultry genetics and breeding. Crit. Rev. Poult. Biol. 3:73-109. [Google Scholar]
- 12.Dougherty, R. M., and R. J. C. Harris. 1966. Contaminant viruses in two live virus vaccines produced in chicken cells. J. Hyg. Camb. 64:1-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gilka, F., and J. L. Spencer. 1990. Chronic myocarditis and circulatory syndrome in a White Leghorn strain induced by avian leukosis virus: light and electron microscope study. Avian Dis. 34:174-184. [PubMed] [Google Scholar]
- 14.Gudkov, A. V., I. B. Obukh, S. M. Serov, and B. S. Naroditsky. 1981. Variety of endogenous proviruses in the genomes of chickens of different breeds. J. Gen. Virol. 57:85-94. [DOI] [PubMed] [Google Scholar]
- 15.Heneine, W., S. Yamamoto, W. M. Switzer, T. J. Spira, and T. M. Folks. 1995. Detection of reverse transcriptase by a highly sensitive assay in sera from persons infected with human immunodeficiency virus type 1. J. Infect. Dis. 171:1210-1216. [DOI] [PubMed] [Google Scholar]
- 16.Hussain, A. I., V. Shanmugam, W. M. Switzer, S. X. Tsang, A. Fadly, D. Thea, R. Helfand, W. J. Bellini, T. M. Folks, and W. Heneine. 2001. Lack of evidence of endogenous avian leukosis virus and endogenous avian retrovirus transmission to measles, mumps and rubella vaccine recipients. Emerg. Infect. Dis. 7:66-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Johnson, J. A., and W. Heneine. 2001. Characterization of endogenous avian leukosis viruses in chicken embryonic fibroblast substrates used in production of measles and mumps vaccines. J. Virol. 75:3605-3612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Linial, M., and P. E. Neiman. 1976. Infection of chicken cells by subgroup E viruses. Virology 73:508-520. [DOI] [PubMed] [Google Scholar]
- 19.Maudru, T., and K. W. Peden. 1998. Analysis of a coded panel of licensed vaccines by polymerase chain reaction-based reverse transcriptase assays: a collaborative study. J. Clin. Virol. 11:19-28. [DOI] [PubMed] [Google Scholar]
- 20.Monath, T. P. 2001. Yellow fever: an update. Lancet Infect. Dis. 1:11-20. [DOI] [PubMed] [Google Scholar]
- 21.Payne, L. N. 1992. Biology of avian retroviruses, p. 299-403. In J. A. Levy (ed.), The Retroviridae. Plenum Press, New York, N.Y.
- 22.Payne, L. N., and H. G. Purchase. 1991. Leukosis/sarcoma group, p. 386-439. In B. W. Calnek (ed.), Diseases of Poultry, 9th ed. Iowa State University Press, Ames.
- 23.Resnick, R. M., M. T. Boyce-Jacino, Q. Fu, and A. J. Faras. 1990. Phylogenetic distribution of the novel avian endogenous provirus family EAV-0. J. Virol. 64:4640-4653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Richman, A. V., C. G. Auliso, W. G. Jahnes, and N. M. Tauraso. 1972. Avian leukosis antibody response in individuals given chicken embryo derived vaccines. Proc. Soc. Exp. Biol. Med. 139:235-237. [DOI] [PubMed] [Google Scholar]
- 25.Robertson, J. S., C. Nicolson, A. Riley, M. Bently, G. Dunn, T. Corcoran, G. C. Schild, and P. Minor. 1997. Assessing the significance of reverse transcriptase activity in chick cell-derived vaccines. Biologicals 25:403-414. [DOI] [PubMed] [Google Scholar]
- 26.Smith, R. E. 1982. Avian osteopetrosis. Curr. Top. Microbiol. Immunol. 101:75-94. [DOI] [PubMed] [Google Scholar]
- 27.Tereba, A., and S. M. Astrin. 1980. Chromosomal localization of ev-1, a frequently occurring endogenous retrovirus locus in White Leghorn chickens, by in situ hybridization. J. Virol. 35:888-894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tsang, S. X., W. M. Switzer, V. Shanmugam, J. A. Johnson, C. Goldsmith, A. Wright, A. Fadly, D. Thea, H. Jaffe, T. M. Folks, and W. Heneine. 1999. Evidence of avian leukosis virus subgroup E and endogenous avian virus in measles and mumps vaccines derived from chicken cells: investigation of transmission to vaccine recipients. J. Virol. 73:5843-5851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Waters, T. D., P. S. Anderson, G. W. Beebe, and R. W. Miller. 1972. Yellow fever vaccination, avian leukosis virus, and cancer risk in man. Science 177:76-77. [DOI] [PubMed] [Google Scholar]
- 30.Weissmahr, R. N., J. Schupbach, and J. Boni. 1997. Reverse transcriptase activity in chicken embryo fibroblast culture supernatants is associated with particles containing endogenous avian retrovirus EAV-0 RNA. J. Virol. 71:3005-3012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Welsh, R. M., N. R. Cooper, F. C. Jensen, and M. B. A. Oldstone. 1975. Human serum lyses RNA tumor viruses. Nature 257:612-614. [DOI] [PubMed] [Google Scholar]
- 32.World Health Organization Expert Committee on Biological Standardization. 1994. Requirements for measles, mumps and rubella vaccines and combined vaccines (live). Requirements for biological substances. WHO Tech. Rep. Ser. 840:100-207. [Google Scholar]
- 33.World Health Organization Expert Committee on Biological Standardization. 1998. Requirements for yellow fever vaccine. WHO Tech. Rep. Ser. 872:31-63. [Google Scholar]