Abstract
Human cytomegalovirus encodes an unusual protein kinase, UL97, that activates the established antiviral drug ganciclovir and is specifically inhibited by a new antiviral drug, maribavir. We used maribavir and a UL97 null mutant, which is severely deficient in viral replication, to determine what stage of virus infection critically requires UL97. Compared with wild-type virus, there was little or no decrease in immediate-early gene expression, viral DNA synthesis, late gene expression, or packaging of viral DNA into nuclease-resistant structures in mutant-infected or maribavir-treated cells under conditions where the virus yield was severely impaired. Electron microscopy studies revealed similar proportions of various capsid forms, including DNA-containing capsids, in the nuclei of wild-type- and mutant-infected cells. However, capsids were rare in the cytoplasm of mutant-infected or maribavir-treated cells; the magnitudes of these decreases in cytoplasmic capsids were similar to those for virus yield. Thus, genetic and pharmacological evidence indicates that UL97 is required at the stage of infection when nucleocapsids exit from the nucleus (nuclear egress), and this poorly understood stage of virus infection can be targeted by antiviral drugs. Understanding UL97 function and maribavir action should help elucidate this interesting biological process and help identify new antiviral drug targets for an important pathogen in immunocompromised patients.
Protein kinases are crucial for specific stages in the replication of many different viruses. Human cytomegalovirus (HCMV), a herpesvirus that causes severe disease in newborns and in immunocompromised adults (reviewed in reference 27), encodes an unusual protein kinase known as UL97 (so designated by the open reading frame in the HCMV genome that encodes it [5, 12]). This enzyme, remarkably, phosphorylates and thereby activates the antiviral nucleoside analogs ganciclovir and acyclovir (17, 37, 38). On peptide substrates, it exhibits an unusual dependence on the identity of the residue five positions C terminal to the site of phosphorylation (1). Although not absolutely essential, UL97 is critical for viral replication in cell culture inasmuch as a UL97 deletion mutation causes a ∼100-fold decrease in viral yield (28).
The stages of HCMV infection resemble those of other herpesviruses. Following attachment to cells and entry, the internalized nucleocapsid travels to the nuclear membrane and deposits its DNA in the nucleus, wherein gene expression and DNA replication ensue (reviewed in reference 23). Viral genes are expressed in three temporally regulated classes: immediate early, early, and late. Viral DNA replication proceeds via high-molecular-weight, concatemeric intermediates, which are cleaved to genome-length units during packaging into capsids. Three capsid forms are observed in the nuclei of infected cells: nonproductive forms thought to result from failed packaging attempts (A capsids), productive intermediates that contain a scaffolding protein (B capsids), and assembled forms in which the scaffolding protein has been removed and replaced with viral DNA (C capsids). Once assembled, these nucleocapsids make their way to the inner nuclear membrane and bud through it by mechanisms of nuclear egress that remain very poorly understood (reviewed in reference 22). The particles then traverse the cytoplasm and are released from the cell as infectious, enveloped virions. Thus, there are multiple stages of virus infection that could require UL97 function.
Although several drugs have been approved for the treatment of HCMV infection, their utility is limited by problems with pharmacokinetics, toxicity, and resistance (11). Because of the unusual properties of UL97 and its critical role in viral replication, it is a promising target for the development of new drugs. Indeed, a new drug, maribavir (also known as 1263W94), specifically inhibits UL97 protein kinase activity and potently inhibits HCMV replication in cell culture via effects on UL97, as a maribavir resistance mutation maps to UL97 (3). Additionally, the drug is orally available and reasonably well tolerated and exhibits anti-HCMV activity in humans (6, 16a).
To investigate which stage of HCMV replication critically requires UL97 activity, we made use of both a UL97 deletion mutant and maribavir. Our results show that the major role for UL97 during viral replication manifests itself after packaging of viral DNA into capsids and prior to the accumulation of particles in the cytoplasm of infected cells, i.e., nuclear egress, the process by which nucleocapsids exit the nucleus.
MATERIALS AND METHODS
Cells and viruses.
Human foreskin fibroblast cells were obtained from the American Type Culture Collection and grown in Dulbecco's modification of Eagle's medium (DMEM) (Clontech) supplemented with 10% fetal bovine serum (FBS) (Sigma). Cells were subcultured every 2 to 3 days and screened for mycoplasma once a month by PCR. HCMV wild-type (wt) strain AD169 was obtained from the American Type Culture Collection. Two independent UL97 mutant isolates derived from AD169 (RCΔ97.08 and RCΔ97.19), which contain an insertion including the Escherichia coli lacZ and gpt genes replacing most of UL97 (28), were generously provided by Mark Prichard. Viral stocks were prepared and titers were determined as described previously (39). Stocks of RCΔ97 mutants were plaque purified and screened for the presence of plaques that failed to express β-galactosidase. Unless otherwise noted, isolate RCΔ97.08 was used. Prior to virus infections, cells were seeded and incubated overnight at 37°C. Cells were infected at a multiplicity of infection (MOI) of 1 PFU/cell (inoculum titers were confirmed by back titration) in DMEM with 5% FBS for 2 h. Inocula were removed, and the cells were washed two times with DMEM plus 5% FBS. DMEM plus 5% FBS, either with the indicated concentrations of drugs or without drugs, was then added, and incubation at 37°C was continued for the indicated times postinfection (p.i.).
Analysis of gene expression.
RNA was prepared at either 12 or 72 h p.i. from 106 cells/100-mm-diameter dish by using QIAgen RNeasy columns, according to the manufacturer's instructions. RNA was quantified by A260, and threefold serial dilutions were prepared. Samples were resolved by gel electrophoresis and transferred to membranes as described previously (10). The membranes were hybridized in QuikHyb (Stratagene) solution with restriction fragments or PCR products complementary to a given transcript that were radiolabeled by using a random priming kit (BMB/Roche). Restriction fragments used were a BamHI fragment from pRSET44 (kindly provided by P. Ertl) for UL44 and a commercially available DNA (Ambion) for the actin gene. PCR products were generated as described previously (15) by using the following primers: UL122, ACAATAGCCTCTTCCTCATC and CGAGATAGCGATAAATGAG; UL123, AGAAGATGCGCACCTGACC and ACAGGAAGACATCAAGCCCG; UL83, AAAAGAAGAGCGCAGCCAC and AAGAGGACCTGACGATGACC; and UL32, ACGACGACGATGATGATGAC and TGACTGGGAGGCGAAATGAC. Images were collected either on X-ray film or with a phosphorimager (Bio-Rad) and, when relevant, were quantified with a phosphorimager. Following acquisition of data by using one probe, the labeled probe was stripped from the filter, and the filter was hybridized with a subsequent probe. Conditions for hybridization, washing, and stripping were those recommended by the QuikHyb manufacturer.
Analysis of viral DNA synthesis.
At selected time points following infection of 104 cells per well of 96-well dishes, the medium was removed from wells and stored at −80°C for subsequent virus titration, the cells were lysed in 10 mM Tris (pH 7.5)-1 mM EDTA with 0.5% sodium dodecyl sulfate (SDS), and the lysates were stored at −80°C. After all samples were collected, they were thawed and treated with 0.5 mg of proteinase K (Sigma) per ml at 55°C for 2 h, and triplicate aliquots were blotted onto Nytran membranes by using a multiwell blot apparatus (Schleicher & Schuell). For some experiments, the cells were treated with 0.4 M NaOH and 10 mM EDTA for 30 min at 37°C, and dilutions of aliquots of these solutions were blotted. The membranes were hybridized for virus-specific sequences as described above, using cosmid L11 (kindly provided by Gilbert Mulamba), which encodes HCMV HindIII fragments E, M, F, D, and L.
Analyses of DNA cleavage and packaging.
Infected cells were rinsed with phosphate-buffered saline (PBS), harvested with 0.05% trypsin and 0.5 mM EDTA in PBS, collected by centrifugation (5 min at 1,000 × g and 4°C), and washed once with PBS. For analysis of protection from digestion by staphylococcal nuclease, the cells were gently lysed on ice, and nuclei were collected and embedded in agarose plugs as described previously (19). One set was treated with proteinase K (40), and a second set was treated with staphylococcal nuclease as described previously (19, 36) except that 7.5 U of nuclease was used per 106 nuclei and the 37°C incubation step was for 2 to 3 h. To inactivate nucleases, the nuclease-treated plugs were subsequently incubated in proteinase K digestion buffer (0.5 M EDTA [pH 9], 1% lauryl sarcosine) at room temperature with shaking for 15 to 30 min prior to proteinase K digestion. For analysis of protection from digestion by DNase, the cells were gently lysed and the nuclei were collected and embedded in agarose plugs as described previously (19). One set of plugs was treated with proteinase K as described previously (40). A second set was treated with DNase I (Roche) for 1 h, as described previously (19), and then, unless otherwise specified, equilibrated with 0.5 M EDTA (pH 9) for 15 min to inactivate the DNase prior to proteinase K digestion. The DNA was resolved alongside molecular weight markers by pulsed-field gel electrophoresis with a Bio-Rad CHEF-DR II as described previously (40), and the DNA was visualized with ethidium bromide, denatured, and transferred to Hybond-N+ (Amersham) as described previously (20). Membranes were probed for virus-specific sequences as described above, using the L11 cosmid.
Electron microscopy (EM) of infected cells.
Cells in 24-well dishes were infected as described above. At 72 or 96 h p.i., an aliquot of the medium was removed for titration of infectious virus. The cells were washed with PBS and fixed for 2 h in 1.2% formaldehyde-2.5% glutaraldehyde-0.03% picric acid in 100 mM Cacodylate buffer. They were postfixed with 1% osmium tetroxide and 1.5% potassium ferrocyanide, stained with 1% uranyl acetate, dehydrated though a series of increasing concentrations of ethanol, equilibrated in propyleneoxide, and infiltrated with Epon-Araldite mixed 1:1 with propylene oxide. After 2 h, the cells in propylene oxide were removed from the dishes and pelleted by low-speed centrifugation. They were then transferred to freshly mixed Epon-Araldite, and the samples were left to polymerize at 60°C for 24 to 48 h. Thin sections were prepared by using a Diatome diamond knife on a Rechert Ultracut-S microtome and viewed with a JEOL 1200EX transmission electron microscope. Nuclei and cytoplasm were randomly selected by sequentially observing each grid square and photographing images from infected cells that, at a magnification of ×10,000, filled at least half of the photographic frame. The resulting negatives were printed, and nuclear capsids and cytoplasmic particles were counted from the photographs. Differences between wt and mutant or between wt and drug-treated samples were evaluated by using the F test for two counts (13).
RESULTS
UL97 is not required for very early events in HCMV infection.
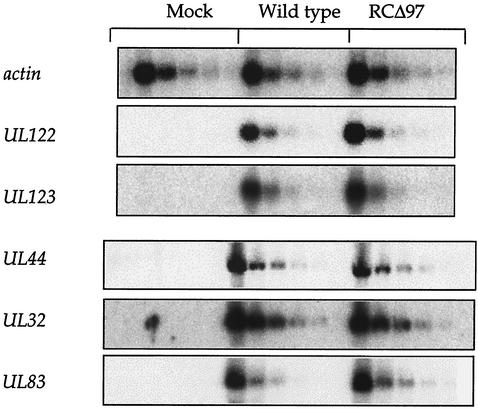
A possible role for UL97 very early in infection was suggested by studies of the homologous UL13 protein of herpes simplex virus (HSV), whose functions can be partially replaced by HCMV UL97 (26). HSV UL13 has been suggested to be involved in virus entry (24) and, in certain cells, is important for expression of particular immediate-early genes (30). To examine events up to and including immediate-early gene expression, RNA was harvested at 12 h p.i. from cells infected with either wt HCMV or a mutant lacking the UL97 gene (RCΔ97), and serial dilutions of each RNA (to aid quantification and verify linearity of the assay) were analyzed by Northern blot hybridization (Fig. 1). Little or no difference in expression of immediate-early transcripts UL122 (IE1) and UL123 (IE2) was observed when RNA from cells infected with wt virus was compared to RNA from cells infected with RCΔ97. Thus, HCMV UL97 is not critically required for the expression of these genes or for prior events in HCMV infection.
FIG. 1.
Northern blot hybridization analysis. RNA was isolated from mock-infected (left five lanes), wt-infected (middle five lanes), or RCΔ97-infected (right five lanes) cells at 12 h p.i. (top three panels) or 72 h p.i. (bottom three panels), and threefold serial dilutions were prepared. The RNAs were resolved by formaldehyde-agarose gel electrophoresis, transferred to a membrane, and probed sequentially with radiolabeled DNAs corresponding to the genes indicated on the left.
UL97 is not critically required for viral DNA synthesis.
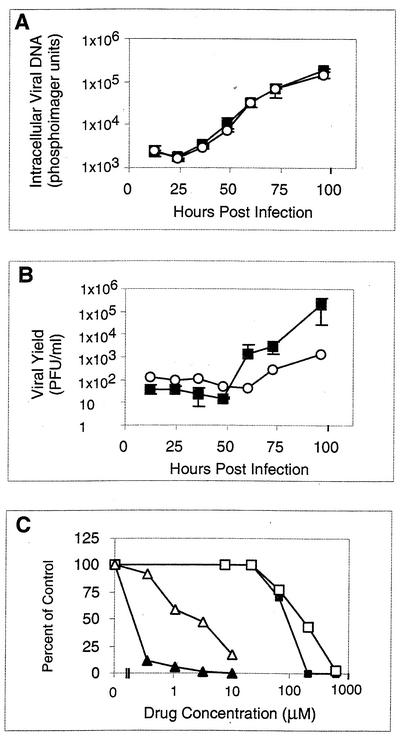
Following immediate-early gene expression, early gene expression and viral DNA synthesis ensue. To examine viral DNA synthesis, DNA was harvested from wt- or mutant-infected cells at various times p.i. and blotted onto a membrane, and the relative amounts of intracellular viral DNA were quantified by hybridization analysis. Simultaneously, infectious virus released into the medium from the same cells was titrated. In the experiment shown, equivalent amounts of viral DNA accumulated in cells infected with wt or RCΔ97 throughout the infectious cycle (Fig. 2A), despite ∼10- and 100-fold differences in viral yields at 72 and 96 h p.i., respectively (Fig. 2B). In other experiments, RCΔ97 was more highly impaired for viral yield (30- to 100-fold reduced at 72 h p.i.) but never exhibited decreases in viral DNA accumulation that were more than 3-fold (not shown).
FIG. 2.
Effects of inhibition of UL97 on viral DNA synthesis and yield. (A and B) Cells were infected with wt HCMV (closed squares) or RCΔ97 (open circles) at an MOI of 1, and at the indicated times, intracellular viral DNA was quantified by hybridization with a radiolabeled cosmid containing viral sequences (A), and infectious virus released into the medium was titrated (B). (C) Cells were infected with wt HCMV at an MOI of 1 at various concentrations of maribavir (triangles) or foscarnet (squares). At 72 h p.i., intracellular viral DNA (open symbols) and infectious virus released into the medium (closed symbols) were quantified. The amounts obtained were normalized to the amounts in the absence of drug, which was set at 100%. Error bars indicate standard deviations.
To investigate the role of UL97 in DNA synthesis further, the effect of the UL97 inhibitor maribavir was compared to that of foscarnet, which inhibits HCMV replication via viral DNA polymerase (2, 7). Infected cells were treated with various concentrations of maribavir or foscarnet, and intracellular viral DNA and viral yields were assayed (Fig. 2C). The 50% inhibitory concentration (IC50) of foscarnet for inhibition of viral DNA synthesis (160 μM) was <2-fold higher than that for viral yield (90 μM), as expected. In contrast, the IC50 of maribavir for inhibition of DNA synthesis (1.1 μM) was >10-fold higher than that for viral yield (<0.1 μM). Indeed, the yield was reduced >10-fold at the maribavir IC50 for DNA synthesis. (In a separate experiment, the IC50 of maribavir for inhibition of the [already-reduced] yield of RCΔ97 was ∼20 μM, much higher than that for wt virus, confirming studies [3] showing that UL97 mutations can confer resistance to maribavir.) Thus, the results with both the UL97 null mutant and the UL97 inhibitor suggest that the critical stage for UL97 function follows viral DNA synthesis.
UL97 is not required for late gene expression.
After viral DNA synthesis, late gene expression becomes maximal. Especially since the UL97 homolog HSV UL13 is required for efficient expression of certain late genes (30), we investigated the expression of three late genes by Northern blot hybridization of RNA harvested at 72 h p.i. from cells infected with either the wt or RCΔ97 (Fig. 1). Three transcripts were selected: UL32, UL83, and UL44. UL32 is a true late transcript whose expression depends critically on DNA synthesis, UL83 is a so-called leaky late transcript, and UL44 has both early and late characteristics (reviewed in reference 23). In the experiment shown, there was no more than a 5% decrease in levels of UL44 transcripts and, if anything, a slight increase in UL32 and UL83 transcripts in the absence of UL97. In other experiments, there was at most a threefold decrease in certain late transcripts (data not shown). Thus, UL97 is not critically required for the expression of late genes.
UL97 is not required for DNA cleavage and packaging.
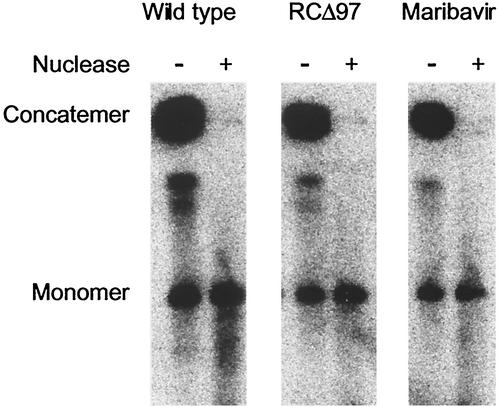
After DNA synthesis, concatemeric HCMV DNA is cleaved to genome-length units and packaged within preformed capsids, where it becomes resistant to nuclease digestion (21). To investigate this stage of infection, we assayed cleavage of viral DNA and its resistance or sensitivity to staphylococcal nuclease at 96 h p.i. Cells infected with wt virus, in the presence or absence of maribavir, or with RCΔ97 were suspended in agarose plugs and digested with proteinase K in the presence of SDS to gently release the DNA. Selected samples were digested with staphylococcal nuclease prior to proteinase K treatment and analyzed by pulsed-field gel electrophoresis and Southern blot hybridization with radiolabeled viral sequences (Fig. 3). As previously observed (21), in the absence of nuclease treatment, most of the hybridization was to viral DNA that remained in the wells of the pulsed-field gel (concatemer). This material comprises high-molecular-weight, concatemeric replication intermediates (21). This material and species that migrated a relatively short distance into the gel were highly susceptible to nuclease treatment (compare left and right lanes in Fig. 3, left panel). Monomeric, genome-length viral DNA (230 kbp) was also observed. This material results from viral DNA replication rather than from input virus, as none was observed in infected cells treated with ganciclovir (data not shown). Unlike the higher-molecular-weight material, the monomeric species in samples prepared from wt-infected cells were highly resistant to nuclease digestion (left panel). Equivalent results were obtained with samples from UL97 mutant-infected cells (middle panel) and with samples from maribavir-treated wt-infected cells (right panel); thus, UL97 was not required for cleavage and packaging in this assay. Similar results were obtained with infected cells harvested at 72 h p.i. (data not shown).
FIG. 3.
Analysis of viral DNA cleavage and packaging by using staphylococcal nuclease. Cells were infected at an MOI of 1 with wt HCMV in the absence of drug (left panel) or in the presence of 1 μM maribavir (right panel) or with RCΔ97 in the absence of drug (middle panel). At 96 h p.i., aliquots of infected cells were embedded in agarose and either lysed with SDS and proteinase K immediately (lanes −) or treated with staphylococcal nuclease prior to equilibration with 0.5 M EDTA-containing buffer and lysis (lanes +). The DNA was resolved by pulsed-field gel electrophoresis, transferred to a nylon membrane, and hybridized for virus-specific sequences with a radiolabeled probe. The positions of DNA concatemers that remain in the well of the gel and monomer (genome-length) HCMV DNA are indicated.
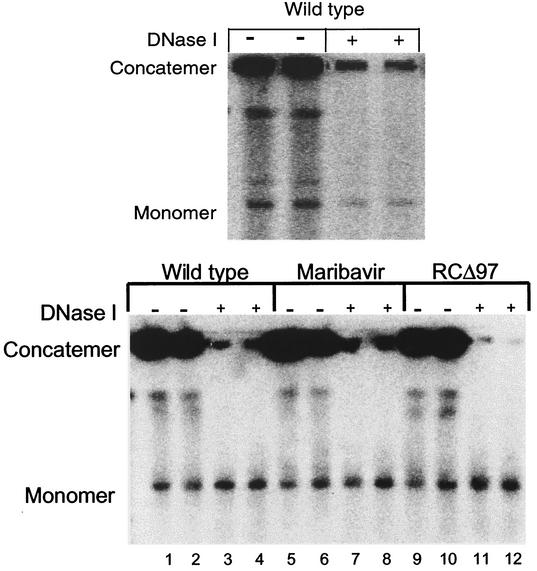
As we initially prepared this work for publication, we became aware of another study that reported substantial decreases in packaging of DNA into DNase I-resistant structures by RCΔ97 (44). We therefore assayed the effects of DNase I on concatemeric and monomeric HCMV DNAs by using pulsed-field gel electrophoresis. Our initial experiments followed a published protocol developed for studies of HSV encapsidation (20). Using this protocol, we found that highly variable and often rather small amounts of monomeric, wt HCMV DNA were protected (Fig. 4, top panel), even when a substantial fraction of concatemeric viral DNA remained undigested (data not shown). As our staphylococcal nuclease protocol included a step to inactivate nuclease in which the agarose plugs were equilibrated with a solution containing 0.5 M EDTA prior to proteinase K digestion, we included such a step in the DNase protocol. With this added step, the monomeric, genome-length viral DNA from wt-infected cells harvested at 72 h p.i. was quite resistant to DNase digestion, while concatemeric DNA was highly susceptible (Fig. 4, bottom panel, compare lanes 1 and 2 to lanes 3 and 4). Again, equivalent results were obtained with samples from maribavir-treated wt-infected cells (Fig. 4, bottom panel, lanes 5 to 8) and with samples from UL97 mutant-infected cells (lanes 9 to 12). In drug-treated and mutant-infected cells assayed in parallel, yields of infectious virus were severely diminished (25- to 100-fold) at 72 h p.i. Thus, at two different times p.i., neither the UL97 deletion mutation nor maribavir treatment meaningfully inhibited either cleavage of concatemeric DNA into genome-length species or packaging of genome-length DNA into structures resistant to staphylococcal nuclease or DNase (i.e., capsids).
FIG. 4.
Analysis of viral DNA cleavage and packaging by using DNase I. (Top panel) Cells were infected at an MOI of 1 with wt HCMV in the absence of drug. At 72 h p.i., aliquots of infected cells were embedded in agarose and either lysed with SDS and proteinase K immediately (lanes −) or treated with DNase I before lysis (lanes +). (Bottom panel) Cells were infected at an MOI of 1 with wt HCMV in the absence of drug (lanes 1 to 4) or in the presence of 1 μM maribavir (lanes 5 to 8) or with RCΔ97 in the absence of drug (lanes 9 to 12). At 72 h p.i., aliquots of infected cells were embedded in agarose and either lysed with SDS and proteinase K immediately (lanes 1, 2, 5, 6, 9, and 10) or, following equilibration with 0.5 M EDTA (pH 9), treated with DNase I and then lysed (lanes 3, 4, 7, 8, 11, and 12). In both panels, the DNA was resolved by pulsed-field gel electrophoresis. In the top panel, the gel was stained with ethidium bromide and photographed. Subsequent blotting and hybridization confirmed the locations of monomer and concatemer (data not shown). In the bottom panel, the DNA was transferred to a nylon membrane and hybridized for virus-specific sequences with a radiolabeled probe. The positions of DNA concatemers that remain in the well of the gel and monomer (genome-length) HCMV DNA are indicated.
Inhibition of UL97 specifically decreases accumulation of cytoplasmic capsids.
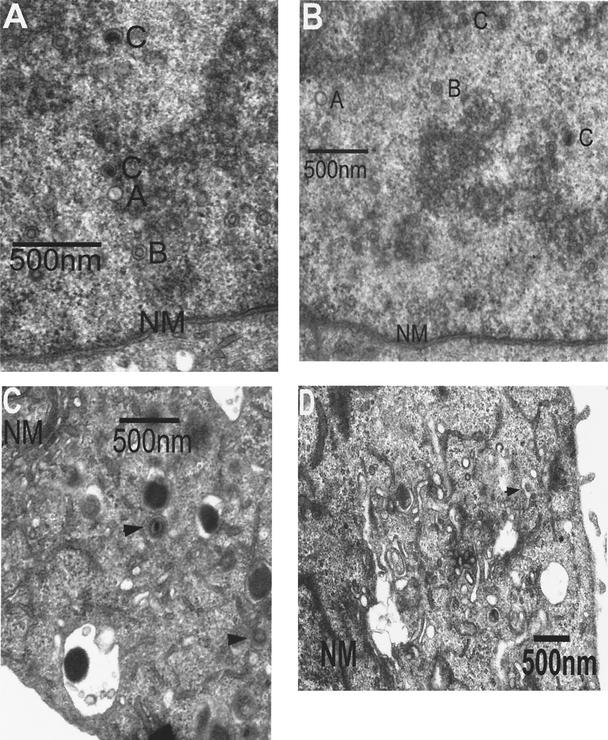
To confirm and extend these observations, cells were infected with wt virus or either of two independent isolates of the UL97 null mutant virus, and at 3 days p.i. the cells were analyzed by EM. All three intranuclear capsid forms (A, B, and C) were observed in wt- and mutant-infected cells (Fig. 5A and B). To quantify the proportions of the various capsid forms, they were counted in micrographs of randomly selected nuclei. Consistent with the nuclease resistance of genome-length DNA (Fig. 3 and 4, bottom panel), there was no decrease in DNA-containing C capsids in nuclei of mutant-infected cells (Table 1). In fact, there was a slight increase. There was a slight increase in the proportion of nonproductive A capsid forms and a slight decrease in the proportion of B capsid forms. Similar results were obtained at 4 days p.i. (not shown) Thus, UL97 was not required for formation of DNA-containing intranuclear capsids.
FIG. 5.
EM analysis of capsid formation. Cells were infected at an MOI of 1 with wt HCMV (A and C) or RCΔ97 (B and D) and analyzed by EM at 72 h p.i. (A and B) Examples of nuclei of infected cells. NM, nuclear membrane; A, B, and C, examples of capsid forms. (C and D) Examples of cytoplasms of infected cells. NM, nuclear membrane; arrowheads, examples of capsids. The capsid in mutant-infected cytoplasm in panel D is one of only four observed in a total of 64 fields. Magnifications: ×43,000 (A), ×30,000 (B), ×32,000 (C), and ×18,000 (D).
TABLE 1.
Nuclear capsids and cytoplasmic particles in wt- and mutant-infected cells: relationship to virus yieldsa
| Virus | No. of nuclei counted | No. of the following capsid form per nucleus (% of total capsids)
|
Cytoplasmic particles
|
% of wt yield | |||||
|---|---|---|---|---|---|---|---|---|---|
| A | B | C | No. of fields | No. of particles | No. of particles/field | % wt | |||
| AD169 (wt) | 31 | 2.2 (6) | 31 (76) | 7.2 (18) | 39 | 80b | 2.1 | 100 | 100 |
| RCΔ97.08 | 26 | 3.8 (9) | 29 (63) | 13 (28) | 38 | 4c | 0.11 | 5.2 | 2.2 |
| RCΔ97.19 | 13 | 5.8 (13) | 28 (63) | 11 (23) | 16 | 0 | <0.06 | <2.9 | 8.6 |
Electron micrographs of nuclei and cytoplasms of infected cells were prepared at 72 h. p.i.; infectious virus in the media of these infected cells was titrated. Intranuclear capsid forms and cytoplasmic particles (Fig. 5) were counted from micrographs. Data for AD169 and RCΔ97.08 are pooled from two independent experiments; the other data are from one representative experiment.
Of the 80 particles, 3 were A form, 60 were B form, and 17 were C form.
Of the four particles, three were B form and one was C form.
However, major differences were observed in the numbers of cytoplasmic particles in wt-infected versus mutant-infected cells (Fig. 5C and D; Table 1). In wt-infected cells, an average of two particles was found per field of cytoplasm, while cytoplasmic particles were very rare in cells infected with either independent RCΔ97 isolate. This reduction in cytoplasmic particles was highly significant (P < 0.01 by the F test for two counts) and was similar in magnitude to the reduction in yield of infectious virus in the medium of the same cells (Table 1). The ratios of B to C capsid forms in the nucleus were similar to the ratios of the corresponding forms in the cytoplasm, in both mutant- and wt-infected cells (Table 1, footnotes b and c; cytoplasmic particles included virions, noninfectious enveloped particles, and an occasional nonenveloped capsid). Strong reductions in cytoplasmic particles were also observed at 4 days p.i. (not shown).
Similarly, we infected cells with wt virus in either the presence or absence of maribavir and analyzed the numbers of cytoplasmic particles by EM. Cytoplasmic particles were significantly (P < 0.01) reduced in maribavir-treated, wt-infected cells at 3 days p.i. (one capsid in 17 fields; 3% of the wt value). This reduction in cytoplasmic capsids equaled the reduction in virus yield (3% of the wt value). Strong reductions in cytoplasmic particles were also observed at 4 days p.i. (not shown). Taken together, these data indicate that the effects of the deletion mutation and the drug manifest themselves after the packaging of viral DNA and prior to or during the cytoplasmic accumulation of virions, i.e., the stage of nuclear egress.
DISCUSSION
HCMV UL97 is an unusual protein kinase and drug target whose role during viral replication is poorly understood. In this study, we studied the effects of genetic or pharmacological inhibition of UL97 on different stages of viral replication. We found that UL97 is not critically required for very early events in viral replication, for expression of immediate-early or late genes, for replication of viral DNA, or for the cleavage and packaging of this DNA into intranuclear capsids. However, cells infected with a UL97 null mutant or cells infected with wt virus and treated with maribavir exhibited severe defects in accumulation of cytoplasmic capsids that can account for the decrease in production of infectious virus observed.
We infer that this defect at the stage of nuclear egress is due to loss of UL97 function: (i) two independent isolates containing the UL97 deletion mutation exhibited this defect, making it extremely unlikely that the effect could be due to other mutations; (ii) the deletion has little effect on the expression of neighboring genes (unpublished data); and (iii) the same defect was observed upon treatment with maribavir, which specifically inhibits UL97 activity in vitro and which inhibits HCMV replication via UL97 (1, 3; this study).
Most antiviral drugs have targeted genome replication (e.g., DNA polymerase or reverse transcriptase inhibitors) or assembly and proteolytic maturation of viral particles (e.g., DNA-packaging or protease inhibitors) (8, 11, 32). To our knowledge, maribavir is the first drug to target the stage of nuclear egress.
Viral gene expression.
Our failure to detect effects on viral gene expression contrasts with results obtained in studies of null mutants of HSV UL13, which is a homolog of HCMV UL97. In the absence of HSV UL13, there is reduced or altered phosphorylation of the HSV regulatory proteins ICP22 and ICP0 and of the large subunit of RNA polymerase II. These changes have been correlated with reduced accumulation of a subset of viral mRNAs and proteins (18, 30, 31). This apparent contrast between the two protein kinases is interesting in that the modest (four- to fivefold) replication defect of an HSV UL13 mutant can be repaired by expression of HCMV UL97, but this does not restore completely the modification of ICP22 (26). It is possible, however, that UL97 is crucial for the expression of some specific HCMV transcripts whose expression we did not assay.
Relation to other studies of UL97 function.
While this work was being prepared for publication, two other reports regarding inhibition of UL97 came to our attention. One, although not excluding a role for UL97 in late stages of viral replication, emphasized inhibition of HCMV DNA synthesis by maribavir. In this study (3), maribavir exhibited an IC50 of 0.06 μM for virus yield and an IC50 of 0.28 μM for viral DNA synthesis, a ∼5-fold difference, while we observed a ∼10-fold difference (Fig. 2C). In our view, the spread between the maribavir IC50s in the two studies, along with the relatively small DNA synthesis defect of the UL97 null mutant, is consistent with a minor role for UL97 in viral DNA synthesis that does not suffice to explain the effects of maribavir or the UL97 deletion on viral yield. However, our results certainly do not exclude the possibility that maribavir's antiviral activity is due in part to inhibition of viral DNA synthesis. At 1 μM maribavir, Biron et al. observed a >10-fold reduction in viral DNA synthesis (3). Although we observed only a ∼2-fold reduction in DNA synthesis at this drug concentration, we did observe reductions at higher concentrations (Fig. 2C), which may be due to effects on targets other than UL97. However, other explanations for the differences in results are possible (see below).
The second report (44) observed that RCΔ97 exhibited a 4- to 6-fold decrease in viral DNA synthesis, which is only slightly more severe than the ∼3-fold decrease that we sometimes observed, and slight effects on accumulation of late proteins. In agreement with our conclusions, those authors inferred that effects on DNA synthesis and viral proteins do not account for the poor replication of the mutant virus. However, in contrast with our findings, those authors reported substantial decreases in packaging of DNA into nuclease-resistant structures and in intranuclear DNA-containing C capsids. Among several differences in methodology between the two studies, one that may be important is that our biochemical studies of packaging used equilibration in 0.5 M EDTA to chelate divalent cations and inactivate nucleases prior to release of viral DNA. Without this EDTA step, we obtained variable and usually incomplete protection of cleaved DNA from DNase I, even with samples from wt-infected cells (Fig. 4, top panel). With the EDTA step, complete staphylococcal nuclease or DNase digestion of concatemeric DNA and complete protection of monomer-length DNA were observed, as expected (Fig. 3 and 4, bottom panel). The other study (44) used only 1 mM EDTA to chelate the 1.5 mM MgCl2 used in the DNase step. Substantial DNase digestion of terminal fragments (markers of unit-length genomes) of wt HCMV DNA is noticeable in the other analysis (44), consistent with the incomplete protection we observed without the 0.5 M EDTA equilibration step.
A second potentially important difference in methodology is that we performed our EM studies at an MOI of 1, rather than 0.1. This made it easier to count many infected cells at the same stage of infection and document the average numbers and proportions of different capsid forms per nucleus (Table 1). These numbers were not documented in the other paper. Regardless, Wolf et al. reported only a 27-fold change in the ratios of A capsids to C capsids in wt- versus mutant-infected cells (44). Especially as this change in ratio was stated to be due in part to a significant increase in the numbers of A capsids in mutant-infected cells, the reduction in DNA-containing C capsids observed by those authors is not sufficient to account for the ∼1,000-fold reduction in virus yield that they reported with this MOI (44) or even the 100-fold reduction reported previously with this MOI at 72 h p.i. (28).
On the other hand, it is possible that differences in methodologies among the three laboratories (e.g., differences in serum or cells) might account for differences in the importance of UL97 for various stages of viral infection. Consistent with this possibility is the threefold variability we observed in viral DNA synthesis of RCΔ97. Also, in one experiment, we did observe a single RCΔ97-infected cell that contained an unusually high proportion of intranuclear A capsids, akin to what was observed by Wolf et al. (44). It may be that UL97 has multiple viral or cellular protein substrates in the infected cell and, depending on the state of the infected cell, the phosphorylation state of these proteins may be more or less important for efficient viral replication. Similarly, it may be that under certain conditions, cellular protein kinases can phosphorylate the substrates of UL97, thereby fulfilling the role of UL97 in viral DNA synthesis or viral DNA packaging, while under other conditions, there is a greater requirement for UL97 activity for these stages. Determining the physiologically relevant substrates of UL97 will be important for testing these possibilities. Regardless, our results indicate a crucial role for UL97 at the stage of nuclear egress.
Possible roles of UL97 at the stage of nuclear egress.
In principle, UL97 could be required for nuclear egress per se or for the stability of particles following egress. One might expect that the loss of UL97, which is a virion component (41), could decrease particle stability. However, maribavir, which simply inhibits UL97 kinase activity, also drastically reduced the numbers of cytoplasmic particles, suggesting that the phenotype is not due simply to particle destabilization resulting from a lack of UL97 protein. A defect in nuclear egress per se might be expected to increase the numbers of DNA-containing capsids in the nucleus. Indeed, there was a slight increase in C capsids in the nucleus in UL97 deletion mutant-infected cells (Table 1). Whether this increase is sufficient to account for the decrease in cytoplasmic capsids depends on the efficiencies and rates of capsid assembly, DNA packaging, nuclear egress, and release from the cell. As observed previously (21) and here, DNA packaging appears to be relatively inefficient. Given that release from the cell is probably 50% efficient at best (23), the paucity of cytoplasmic particles relative to nuclear particles that we observed, even in wt-infected cells (Fig. 5 and Table 1), appears to be consistent with nuclear egress ordinarily being relatively inefficient. Although more data are required, our working hypothesis is that UL97 is required for nuclear egress itself rather than for particle stability following nuclear egress.
Studies of mutants of HSV and pseudorabies virus suggest possible substrates of UL97 that might explain its role in nuclear egress. Like RCΔ97, HSV UL12 (alkaline nuclease) mutant viruses replicate poorly in cell culture. Despite nearly wt levels of DNA synthesis and processing, these UL12 mutants are deficient in nuclear egress of nucleocapsids, which is thought to be due to abnormalities in packaged DNA (19, 36). HCMV UL98 is a phosphoprotein (reference 16 and unpublished results) that is homologous to HSV UL12, and HCMV UL98 can complement the defects of an HSV UL12 deletion mutant (9). However, the importance of alkaline nuclease for nuclear egress may differ between HSV and CMV. Whereas nuclear egress of HSV appears to involve a mechanism for sensing the state of packaged DNA (19, 36, 42), in HCMV-infected cells, enveloped particles that contain capsids lacking DNA (noninfectious enveloped particles) are efficiently produced (reviewed in reference 23).
HSV and pseudorabies virus mutants with deletions in UL31 or UL34 also replicate poorly in cell culture and exhibit defects in nuclear egress (4, 14, 22, 34). Interestingly, HSV UL34 is a phosphoprotein whose phosphorylation depends on an HSV protein kinase, US3 (29). UL31, which is also a phosphoprotein, and UL34 localize to the nuclear rim, and their normal distribution depends on US3 (33). Although HSV US3 deletion mutants replicate well in cell culture, pseudorabies virus US3 deletion mutants do not, and the latter exhibit a defect in nuclear egress (43). Perhaps one or more of the HCMV homologs of HSV UL12, UL31, and UL34 require phosphorylation by UL97 for proper functioning and this accounts for a requirement for UL97 for nuclear egress.
After submission of this paper, Muranyi et al. (25) reported that transfection of the murine CMV (MCMV) and HCMV homologs of HSV UL34 and UL31 results in colocalization of the viral proteins with protein kinase C at the nuclear rim and alterations in nuclear lamina. Additionally, MCMV and HCMV infection induced lamin phosphorylation, which was reduced by a protein kinase C inhibitor (25). Such alterations in nuclear lamina may be important for nuclear egress. These interesting results leave open the possibility that UL97 may also phosphorylate and alter nuclear lamins. Of course, other viral or cellular targets of UL97 phosphorylation could be involved in nuclear egress.
Understanding herpesvirus nuclear egress, which, at least for HSV, involves the state of packaged DNA (19, 36, 42), intranuclear movements, effects on the nuclear lamina (35), and dynamic interactions with the nuclear membrane, seems certain to illuminate previously unknown areas of both virus and cell biology. Determining the natural substrates of UL97 and the consequences of their phosphorylation, which will be abetted by maribavir, should shed considerable light on this problem but may also reveal new potential antiviral drug targets. Antiviral drugs, including maribavir, that block nuclear egress not only should aid studies of this stage of infection but also may be especially useful in treating viral diseases, as they are likely to exhibit different toxicities and patterns of resistance than do currently existing drugs.
Acknowledgments
We thank the following for their generosity: C. Talarico (GlaxoSmithKline) for maribavir, M. Prichard (Aviron) for the independent RCΔ97 isolates, G. Mulamba (Coen lab) for cosmid L11, P. Ertl (GlaxoSmithKline) for pRSET44, M. Ericsson and L. Trakimas (Harvard Medical School EM facility) for assistance with EM, K. Biron for communicating unpublished results, A. Griffiths for invaluable help with preparation of figures, anonymous referees for useful comments, and K. Biron, L. Enquist, R. Harvey, D. Knipe, E. Mocarski, M. Prichard, F. Roth, and C. Talarico for helpful discussions.
This work was supported by grant RO1 AI26077 from NIH. P.M.K. was also supported by NIH training grant AI07245. The Coen lab has previously received gifts and research support from entities that are now subsumed into GlaxoSmithKline.
Footnotes
We dedicate this paper to the memory of Bill Burns, a valued colleague and friend, who dedicated himself to treating disease and understanding biology.
REFERENCES
- 1.Baek, M.-C., P. M. Krosky, Z. He, and D. M. Coen. 2002. Specific phosphorylation of exogenous protein and peptide substrates by the human cytomegalovirus UL97 protein kinase: importance of the P+5 position. J. Biol. Chem. 277:29593-29599. [DOI] [PubMed] [Google Scholar]
- 2.Baldanti, F., M. R. Underwood, S. C. Stanat, K. K. Biron, S. Chou, A. Sarasini, E. Silini, and G. Gerna. 1996. Single amino acid changes in the DNA polymerase confer foscarnet resistance and slow-growth phenotype, while mutations in the UL97-encoded phosphotransferase confer ganciclovir resistance in three double resistant human cytomegalovirus strains recovered from patients with AIDS. J. Virol. 70:1390-1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Biron, K. K., R. J. Harvey, S. C. Chamberlain, S. S. Good, A. A. Smith III, M. G. Davis, C. L. Talarico, W. H. Miller, R. Ferris, R. E. Dornsife, S. C. Stanat, J. C. Drach, L. B. Townsend, and G. W. Koszalka. 2002. Potent and selective inhibition of human cytomegalovirus replication by 1263W94, a benzimidazole l-riboside with a unique mode of action. Antimicrob. Agents Chemother. 46:2365-2372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chang, Y. E., C. Van Sant, P. W. Krug, A. E. Sears, and B. Roizman. 1997. The null mutant of the UL31 gene of herpes simplex virus 1: construction and phenotype in infected cells. J. Virol. 71:8307-8315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chee, M. S., G. L. Lawrence, and B. G. Barrell. 1989. Alpha-, beta- and gammaherpesviruses encode a putative phosphotransferase. J. Gen. Virol. 70:1151-1160. [DOI] [PubMed] [Google Scholar]
- 6.Chulay, J., K. Biron, L. Wang, M. Underwood, S. Chamberlain, L. Frick, S. Good, M. Davis, B. Harvey, L. Townsend, J. Drach, and G. W. Koszalka. 1999. Development of novel benzimidazole riboside compounds for treatment of cytomegalovirus disease. Adv. Exp. Med. Biol. 458:129-134. [DOI] [PubMed] [Google Scholar]
- 7.Erriksson, B., B. Öberg, and B. Wahren. 1982. Pyrophosphate analogs as inhibitors of DNA polymerases of cytomegalovirus, herpes simplex virus and cellular origin. Biochim. Biophys. Acta 696:115-123. [DOI] [PubMed] [Google Scholar]
- 8.Flint, S. J., L. W. Enquist, R. M. Krug, V. R. Racaniello, and A. M. Skalka. 2000. Principles of virology: molecular biology, pathogenesis, and control. ASM Press, Washington, D.C.
- 9.Gao, M., B. J. Robertson, P. J. McCann, D. R. O'Boyle, S. K. Weller, W. W. Newcomb, J. C. Brown, and S. P. Weinheimer. 1998. Functional conservation of the alkaline nuclease of herpes simplex type 1 and human cytomegalovirus. Virology 249:460-470. [DOI] [PubMed] [Google Scholar]
- 10.Hann, L. E., W. J. Cook, S. E. Uprichard, D. M. Knipe, and D. M. Coen. 1998. The role of herpes simplex virus ICP27 in the regulation of UL24 gene expression by differential polyadenylation. J. Virol. 72:7709-7714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hayden, F. G. 2001. Antiviral agents (nonretroviral), p. 1313-1347. In J. G. Hardman, L. E. Limbird, and A. G. Gilman (ed.), Goodman & Gilman's The pharmacological basis of therapeutics, 10th ed. McGraw-Hill, New York, N.Y.
- 12.He, Z., Y.-S. He, Y. Kim, L. Chu, C. Ohmstede, K. K. Biron, and D. M. Coen. 1997. The human cytomegalovirus UL97 protein is a protein kinase that autophosphorylates on serines and threonines. J. Virol. 71:405-411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kanji, G. K. 1999. 100 statistical tests, new edition. SAGE Publications, London, United Kingdom.
- 14.Klupp, B. G., H. Granzow, and T. C. Mettenleiter. 2000. Primary envelopment of pseudorabies virus at the nuclear membrane requires the UL34 gene product. J. Virol. 74:10063-10073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Krosky, P. M., M. R. Underwood, S. R. Turk, K. W.-H. Feng, R. K. Jain, R. G. Ptak, A. C. Westerman, K. K. Biron, L. B. Townsend, and J. C. Drach. 1998. Resistance of human cytomegalovirus to benzimidazole nucleoside analogs maps to two open reading frames, UL89 and UL56. J. Virol. 72:4721-4728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lahijani, R. S., E. W. Otteson, and S. C. St. Jeor. 1992. A possible role for nonsense suppression in the synthesis of a human cytomegalovirus 58-kDa virion protein. Virology 186:309-312. [DOI] [PubMed] [Google Scholar]
- 16a.Lalezari, J. P., J. A. Aberg, L. H. Wang, M. B. Wire, R. Miner, W. Snowden, C. L. Talarico, S. Shaw, M. A. Jacobson, and W. L. Drew. 2002. Phase I dose escalation trial evaluating the pharmacokinetics, anti-human cytomegalovirus (HCMV) activity, and safety of 1263W94 in human immunodeficiency virus-infected men with asymptomatic HCMV shedding. Antimicrob. Agents Chemother. 46:2969-2976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Littler, E., A. D. Stuart, and M. S. Chee. 1992. Human cytomegalovirus UL97 open reading frame encodes a protein that phosphorylates the antiviral nucleoside analogue ganciclovir. Nature 358:160-162. [DOI] [PubMed] [Google Scholar]
- 18.Long, M. C., V. Leong, P. A. Schaffer, C. A. Spencer, and S. A. Rice. 1999. ICP22 and the UL13 protein kinase are both required for herpes simplex virus-induced modification of the large subunit of RNA polymerase II. J. Virol. 73:5593-5604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Martinez, R., R. T. Sarisky, P. C. Weber, and S. K. Weller. 1996. Herpes simplex virus type 1 alkaline nuclease is required for efficient processing of viral DNA replication intermediates. J. Virol. 70:2075-2085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McNab, A. R., P. Desai, S. Person, L. L. Roof, D. R. Thomsen, W. N. Newcomb, J. C. Brown, and F. L. Homa. 1998. The product of the herpes simplex virus type 1 UL25 gene is required for encapsidation but not for cleavage of replicated viral DNA. J. Virol. 72:1060-1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McVoy, M. A., and S. P. Adler. 1994. Human cytomegalovirus DNA replicates after early circularization by concatemer formation, and inversion occurs within the concatemer. J. Virol. 68:1040-1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mettenleiter, T. C. 2002. Herpesvirus assembly and egress. J. Virol. 76:1537-1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mocarski, E. S., and C. T. Courcelle. 2001. Cytomegaloviruses and their replication, p. 2629-2673. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology, 4th ed., vol. 2. Lippincott Williams & Wilkins, Philadelphia, Pa.
- 24.Morrison, E. E., Y.-F. Wang, and D. M. Meredith. 1998. Phosphorylation of structural components promotes dissociation of the herpes simplex virus type 1 tegument. J. Virol. 72:7108-7114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Muranyi, W., J. Haas, M. Wagner, G. Krohne, and U. H. Koszinowski. 2002. Cytomegalovirus recruitment of cellular kinases to dissolve the nuclear lamina. Science 297:854-857. [DOI] [PubMed] [Google Scholar]
- 26.Ng, T. I., C. Talarico, T. C. Burnette, K. Biron, and B. Roizman. 1996. Partial substitution of the functions of the herpes simplex virus UL13 gene by the human cytomegalovirus UL97 gene. Virology 225:347-358. [DOI] [PubMed] [Google Scholar]
- 27.Pass, R. F. 2001. Cytomegalovirus, p. 2675-2705. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology, 4th ed., vol. 2. Lippincott Williams & Wilkins, Philadelphia, Pa.
- 28.Prichard, M. N., N. Gao, S. Jairath, G. Mulamba, P. Krosky, D. M. Coen, B. O. Parker, and G. S. Pari. 1999. A recombinant human cytomegalovirus with a large deletion in UL97 has a severe replication deficiency. J. Virol. 73:5663-5670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Purves, F., D. Spector, and B. Roizman. 1992. UL34, the target of the herpes simplex virus US3 protein kinase, is a membrane protein which in its unphosphorylated state associates with novel phosphoproteins. J. Virol. 66:4295-4303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Purves, F. C., W. O. Ogle, and B. Roizman. 1993. Processing of the herpes simplex virus regulatory protein α22 mediated by the UL13 protein kinase determines the accumulation of a subset of α and γ mRNAs and proteins in infected cells. Proc. Natl. Acad. Sci. USA 90:6701-6705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Purves, F. C., and B. Roizman. 1992. The UL13 gene of herpes simplex virus 1 encodes the functions for posttranslational processing associated with phosphorylation of the regulatory protein α22. Proc. Natl. Acad. Sci. USA 89:7310-7314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Raffanti, S., and D. W. Haas. 2001. Antiretroviral agents, p. 1349-1380. In J. G. Hardman, L. E. Limbird, and A. G. Gilman (ed.), Goodman & Gilman's The pharmacological basis of therapeutics, 10th ed. McGraw-Hill, New York, N.Y.
- 33.Reynolds, A. E., B. J. Ryckman, J. D. Baines, Y. Zhou, L. Liang, and R. J. Roller. 2001. UL31 and UL34 proteins of herpes simplex virus type 1 form a complex that accumulates at the nuclear rim and is required for envelopment of nucleocapsids. J. Virol. 75:8803-8817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Roller, R. J., Y. Zhou, R. Schnetzer, J. Ferguson, and D. DeSalvo. 2000. Herpes simplex virus type 1 UL34 gene product is required for viral envelopment. J. Virol. 74:117-129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Scott, E. S., and P. O'Hare. 2001. Fate of the inner nuclear membrane protein lamin B receptor and nuclear lamins in herpes simplex virus type 1 infection J. Virol. 75:8818-8830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shao, L., L. M. Rapp, and S. K. Weller. 1993. Herpes simplex virus 1 alkaline nuclease is required for efficient egress of capsids from the nucleus. Virology 196:146-162. [DOI] [PubMed] [Google Scholar]
- 37.Sullivan, V., C. L. Talarico, S. C. Stanat, M. Davis, D. M. Coen, and K. K. Biron. 1992. A protein kinase homologue controls phosphorylation of ganciclovir in human cytomegalovirus-infected cells. Nature 358:162-164. [DOI] [PubMed] [Google Scholar]
- 38.Talarico, C. L., B. T. C., W. H. Miller, S. L. Smith, M. G. Davis, S. C. Stanat, T. I. Ng, Z. He, D. M. Coen, B. Roizman, and K. K. Biron. 1999. Acyclovir is phosphorylated by the human cytomegalovirus UL97 protein Antimicrob. Agents Chemother. 43:1941-1946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Turk, S. R., C. Shipman, Jr., R. Nassiri, G. Genzlinger, S. H. Krawczyk, L. B. Townsend, and J. C. Drach. 1987. Pyrrolo[2,3-d]pyrimidine nucleosides as inhibitors of human cytomegalovirus. Antimicrob. Agents Chemother. 31:544-550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Underwood, M. R., R. J. Harvey, S. C. Stanat, M. Hemphill, T. Miller, J. C. Drach, L. B. Townsend, and K. K. Biron. 1998. Inhibition of HCMV DNA maturation by a benzimidazole ribonucleoside is mediated through the UL89 gene product. J. Virol. 72:717-725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.van Zeijl, M., J. Fairhurst, E. Z. Baum, L. Sun, and T. R. Jones. 1997. The human cytomegalovirus UL97 protein is phosphorylated and a component of virions. Virology 231:72-80. [DOI] [PubMed] [Google Scholar]
- 42.Vlazny, D. A., A. Kwong, and N. Frenkel. 1982. Site-specific cleavage/packaging of herpes simplex virus DNA and the selective maturation of nucleocapsids containing full-length viral DNA. Proc. Natl. Acad. Sci. USA 79:1423-1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wagenaar, F., J. M. Pol, B. Peeters, A. L. Gielkens, N. de Wind, and T. G. Kimman. 1995. The US3 encoded protein kinase from pseudorabies virus affects egress of virions from the nucleus. J. Gen. Virol. 76:1851-1859. [DOI] [PubMed] [Google Scholar]
- 44.Wolf, D. G., C. T. Courcelle, M. N. Prichard, and E. S. Mocarski. 2001. Distinct and separate roles for herpesvirus-conserved UL97 kinase in cytomegalovirus DNA synthesis and encapsidation. Proc. Natl. Acad. Sci. USA 98:1895-1900. [DOI] [PMC free article] [PubMed] [Google Scholar]