Abstract
Human immunodeficiency virus type 1 (HIV-1) containing mutations in the nucleocapsid (NC) Zn2+ finger domains have greatly reduced infectivity, even though genome packaging is largely unaffected in certain cases. To examine replication defects, viral DNA (vDNA) was isolated from cells infected with viruses containing His-to-Cys changes in their Zn2+ fingers (NCH23C and NCH44C), an integrase mutant (IND116N), a double mutant (NCH23C/IND116N), or wild-type HIV-1. In vitro assays have established potential roles for NC in reverse transcription and integration. In vivo results for these processes were obtained by quantitative PCR, cloning of PCR products, and comparison of the quantity and composition of vDNA generated at discrete points during reverse transcription. Quantitative analysis of the reverse transcription intermediates for these species strongly suggests decreased stability of the DNA produced. Both Zn2+ finger mutants appear to be defective in DNA synthesis, with the minus- and plus-strand transfer processes being affected while interior portions of the vDNA remain more intact. Sequences obtained from PCR amplification and cloning of 2-LTR circle junction fragments revealed that the NC mutants had a phenotype similar to the IN mutant; removal of the terminal CA dinucleotides necessary for integration of the vDNA is disabled by the NC mutations. Thus, the loss of infectivity in these NC mutants in vivo appears to result from defective reverse transcription and integration processes stemming from decreased protection of the full-length vDNA. Finally, these results indicate that the chaperone activity of NC extends from the management of viral RNA through to the full-length vDNA.
The nucleocapsid (NC) protein is a small, highly basic protein generated during protease processing of the Gag polyprotein that occurs during retrovirus maturation (20, 21). In all orthoretroviruses, these proteins are characterized by the presence of one or two Zn2+ finger domains with a common sequence motif, -Cys-X2-Cys-X4-His-X4-Cys- (CCHC) (4, 14, 31), in which the Cys and His residues coordinate a Zn2+ ion (11, 51). As a domain of the Gag polyprotein, its role in recognition, packaging, and stabilization of the viral RNA genome has long been documented.
In addition to its RNA encapsidation function, we and others have presented data indicating that NC may play a role in events that occur early in the infection process after viral entry (5, 26, 53). The importance of the native conformation of the Zn2+ finger moieties has been demonstrated by the loss of infectivity in viruses containing mutations within these domains. For instance, mutants of Moloney murine leukemia virus (Mo-MuLV) were studied that contained substitutions within the single Zn2+ finger of its NC (25). The His residue was replaced with Cys (CCCC) in one such mutant so that the Zn2+-coordinating property was retained; the virions packaged wild-type levels of viral RNA but were replication defective. This mutant also exhibited marked differences in the 2-LTR circular viral DNA (vDNA) isolated from infected cells relative to wild type. Unlike wild-type virus, this mutant produced 2-LTR circles that appeared to arise from vDNA transcripts with incomplete termini, suggesting either a deficiency in reverse transcription, the process by which a vDNA copy of the genomic RNA is produced, or a deficiency in the ability of NC to protect the newly synthesized vDNA from exonucleases.
Similarly, mutations were generated in human immunodeficiency virus type 1 (HIV-1) in either or both of its Zn2+ fingers with comparable results: substitution of Cys for His in either or both fingers (NCH23C and/or NCH44C) also resulted in viruses in which the quantity of RNA packaged relative to that for the wild type was much larger than their infectivity in short- and long-term assays (26). Thus, in the Mo-MuLV and HIV-1 mutants, the RNA-packaging role of NC was detached from functions it apparently serves during infection.
Several groups have examined in vitro systems that represent early, postentry events that occur during retroviral infection. Models have been developed to study the process of reverse transcription (3, 28-30, 34, 35, 38, 39, 46, 47, 57, 58) and subsequent integration processes (9, 10, 22a). A number of these studies demonstrate the nucleic acid chaperone properties of NC, in which it acts to melt secondary structures and anneal complementary sequences (44, 55, 56). In the HIV-1 system, this property directly supports reverse transcription at various points, including the formation of a productive initiation complex by placement of tRNA3Lys at the primer binding site (PBS) on the genome, allowing elongation to proceed through otherwise stable stem-loop structures present in the genome and aiding strand transfer events and strand displacement syntheses that are required for completion of vDNA synthesis. It has also been proposed that specific protein-protein interactions between HIV-1 NC and reverse transcriptase (RT) may be the basis for the enhancement of the polymerization rate and RNase H activity observed in many different assays (8). In fact, Cameron et al. (8) and others (16) suggest that there is an interaction between the two proteins in vitro that is dependent on intact Zn2+ finger structures. In addition, we and others have shown that NC is able to enhance the stimulation of concerted end joining in in vitro integration reactions (9, 10, 22a). In this in vitro system, the integration reaction proceeds as observed during a productive integration event in vivo.
The stringent requirement of the wild-type Zn2+ finger domains of HIV-1 NC for infectivity was demonstrated previously (26). In this work, we have further examined the effects in vivo of mutating the Zn2+ coordinating His residues to Cys in either of the NC Zn2+ fingers, generating the NCH23C and NCH44C mutants (26). A virus containing a mutation in integrase (IN), IND116N, described previously (17, 18), and the double mutant NCH23C/IND116N were also examined to investigate possible integration defects resulting from mutations of the NC Zn2+ fingers. This work presents in vivo requirements for the wild-type HIV-1 NC Zn2+ fingers. In addition to efficient reverse transcription processes and vDNA protection, as proposed previously (53), they are required for initial integration events. Presented in this study is a mechanistic basis for the loss of infectivity in these HIV-1 NC Zn2+ finger mutants.
MATERIALS AND METHODS
Plasmid constructs.
The mutant and wild-type plasmids examined in this study use the HIV-1 proviral clone pNL4-3 (GenBank accession numbers AF324493 and M19921 [1]). The NCH23C, NCH44C, and RTD185K/D186L plasmids were described previously (26). The IN mutant in pNL4-3 (IND116N) has been described previously (18), and the combination NCH23C/IND116N mutant was generated by inserting the 1,818-bp PinAI- PflMI fragment from the IND116N plasmid into the corresponding sites of the NCH23C mutant plasmid. A frameshift mutation was also introduced in the env region (described previously [41]) of all of these plasmids so that when the plasmid was cotransfected with the vesicular stomatitis virus G (VSV-G) Env-expressing plasmid, pHCMV-g (6), the viruses produced were capable of only a single round of infection (see below). DNA manipulations were performed using standard molecular biological reagents and procedures. All mutations were verified by sequencing.
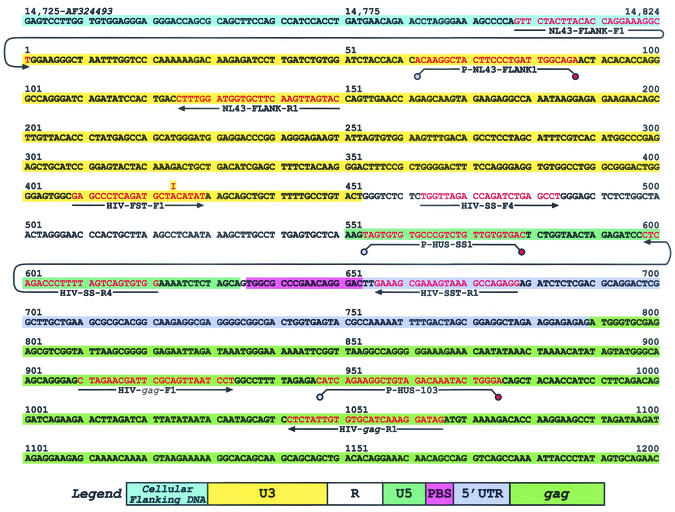
The standard template used in the quantitation of transcription intermediates (denoted pRB1008) was generated by insertion of the 1,789-bp StuI-SpeI fragment from pNL4-3 into the EcoRV-SpeI sites of pZeRO-1 (Invitrogen, Carlsbad, Calif.). This fragment contained the cellular DNA sequence flanking the 5′ end of the HIV-1 coding sequence through the SpeI site within the gag coding region. Figure 1 shows the target sequence, primers, and probes for quantitative PCR (see below). Since this plasmid was used as a standard in the quantitation of linear transcripts, it was digested with SpeI, which recognizes a single site within pRB1008.
FIG. 1.
Sequence used for real-time PCR detection of reverse transcription intermediates. A portion of the sequence contained in pRB1008 that is used as a standard template for real-time PCR is presented. The sequence and positions in the pNL4-3 sequence of the 5′ cellular flanking region, the 5′ LTR, and a portion of the gag region are shown. The location in the pNL4-3 sequence is denoted by the numbers at the top of the sequence. Positions of primers and probes used are shown, with the sequences depicted in red text (probes are designated with a P- prefix). Primers and probes used for the detection of cellular flanking sequences are NL43-FLANK-F1, NL43-FLANK- R1, and P-NL43-FLANK1, respectively. The R-U5 region, representative of minus-strand strong-stop vDNA, is detected using HIV-SS-F4, HIV-SS-R4, and P-HUS-SS1. Detection of the U3-U5 region generated after minus-strand transfer is performed using HIV-FST-F1, HIV-SS-R4, and P-HUS-SS1. HIV-FST-F1 has an inosine at position 16 (from the 5′ end) instead of an adenosine. The inosine forms a base pair with thymidine in the standard template pRB1008 (the U3 region was derived from the 5′ LTR of pNL4-3), and in the vDNA target the inosine forms a base pair with cytosine since the source of the U3 region is derived from the 3′ end of the genome (i.e., the 3′ LTR of pNL4-3). The inosine-thymidine and inosine-cytosine interactions involve the same number of hydrogen bonds, and thus the HIV-FST-F1 will have roughly the same Tm with either target. Sequences in gag are quantitated using HIV-gag-F1, HIV-gag-R1, and P-HUS-103, and this region represents sequences generated after late-minus-strand synthesis. R-5′UTR targets, representative of sequences generated after plus-strand transfer, are detected using HIV-SS-F4, HIV-SST-R1, and P-HUS-SS1.
The standard template used in the quantitation of cellular DNA equivalents (designated pJB1057) was generated by first amplifying a 1,301-bp portion of the region surrounding introns 4 to 6 of the gene encoding porphobilinogen deaminase (PBGD) (13, 27) with sense and antisense primers, PB-HindIII (5′-ATC CAA GCT TAG CCC AAA GAT GAG AGT G-3′) and PB-BglII (5′-CCT TCT CAA GAT CTC AGG AGC ATG A-3′), respectively (restriction sites are denoted in boldface type). The PCR product was digested with HindIII and BglII and ligated into the corresponding sites of the pTRI-18 vector (Ambion, Austin, Tex.).
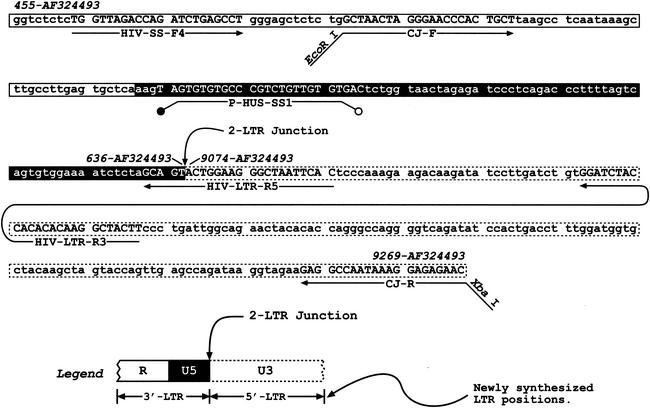
The template used as a standard in the quantitation of 2-LTR circular vDNA (denoted pJB1041) was generated by first PCR amplifying a region spanning the 2-LTR junction within circularized vDNA that was isolated from cells infected with wild-type virus. The contiguous sequence of this junction fragment corresponds to nucleotides (nt) 455 to 636 joined to nt 9074 to 9269 in the pNL4-3 sequence. Figure 2 shows a portion of the sequence along with primers and the probe used for real-time PCR (see below). The PCR products were ligated into pGEM-T (Promega, Madison, Wis.).
FIG. 2.
HIV-1 2-LTR circle junction fragment sequence, with PCR and cloning primers, and the probe. A portion of the full-length 2-LTR junction sequence is presented, along with the locations of various primers and the probe. Nucleotide positions in the pNL4-3 sequence are denoted at the top in italics. The position of the 2-LTR junction is also shown. This sequence region contained in the pJB1041 plasmid was used as a standard template for real-time PCR. The legend depicting the various regions (R, U5, and U3) is shown at the bottom of the figure. The primers and probe used to quantify full-junction 2-LTR circles are HIV-SS-F4, HIV-LTR-R5, and P-HUS, SS1, respectively. Total 2-LTR circles are detected using HIV-SS-F4, HIV-LTR-R3, and P-HUS-SS1. The CJ-F and CJ-R primers have been described as the upstream and downstream primers, respectively, by Julias et al. (37).
Cell lines, transfections, infections, and total-cell DNA isolation.
The 293T cell line (293 cell line containing the simian virus 40 large T antigen) was maintained as described previously (26). A human osteosarcoma (HOS) cell line was the generous gift of John G. Julias and was maintained as described previously (36). All lines were maintained under 7% CO2 at 37°C.
Pseudotyped viruses, capable of only a single round of infection, were generated from 293T cells by transfection as described previously (36). Infection of HOS cells and isolation of total cellular DNA were performed as described previously (36), except that 2 μg of hexadimethrine bromide (Polybrene [Sigma-Aldrich Corp., St. Louis, Mo.]) per ml was included in culture fluids applied to the HOS cells.
Quantitative PCR.
Quantitative, real-time PCR was performed using an ABI Prism 7700 sequence detection system (Applied Biosystems, Foster City, Calif.). Appropriate sense and antisense primers were used, along with oligonucleotide probes as described in the ABI-7700 Users' Manual and elsewhere (Applied Biosystems part no. 402823 [52]). Plasmid DNA for each template described above was prepared from a plasmid with the expected insert identified by sequence analysis, quantitated by measuring the absorbance at 260 nm, and stored in aliquots at concentrations of 106 copies/μl at −20°C. Primer and probe combinations used to detect reverse transcription intermediates are presented in the legend to Fig. 1. To verify that the vDNA isolated from the infection of the HOS cells was not contaminated with transfected plasmid DNA and that the detected targets were generated from a reverse transcription event, an additional primer-probe combination was designed to detect “cellular flanking” DNA, which is presented in Fig. 1. By using this in combination with the HIV-1 RT-defective clone (RTD185K/D186L), we were able to confirm that no carryover DNA was present in the vDNA preparations.
The 2-LTR circular vDNA was quantitated in a similar manner by using two different reactions for each sample. They discriminate between circular vDNA containing the expected sequence for full-length linear vDNA, prior to ligation, and other species that contain both the full-length species and species with a range of deletions at either the 5′ or 3′ long terminal repeat (LTR) ends, up to ∼50 bp. Both reactions utilize the same sense primer (HIV-SS-F4) and fluorogenic probe (P-HUS-SS1 [see Fig. 2]) shown in Fig. 1. The positions of the antisense primers were varied to detect the expected sequence at the junction of either full-length vDNA or end-to-end ligated vDNAs that either are full length or may have been truncated prior to ligation. For primer-probe combinations, see Fig. 2. Quantitation was performed by comparing amplification profiles for unknown samples with those for a dilution series of a stock solution of known copy number of the standard pJB1041 (see above).
Typically DNA from ∼105 cells (determined by measuring the PBGD content, a single-copy/cell assay, of the vDNA sample isolated) was analyzed by real-time PCR. Copy numbers were adjusted on a per cpm of RT activity basis as well as on a per cell basis and arbitrarily reported as corrected copies present for 4 × 1011 cells. TaqMan quantitation of PBGD targets in DNA samples was carried out with the primers PBGD-DF3 (5′-AGG GCA GGA ACC AGG GAT TAT G-3′) and PBGD-DR1 (5′-GGG CAC CAC ACT CTC CTA TCT TT-3′) and P-PBGD-01 probe (5′-FAM-ATG TCC ACC ACA GGG GAC AAG ATT C-TAMRA-3′). This gene is present in two copies in the diploid human genome (13, 27) and is thus used as a measure of cell number (48). The primer-probe combination is designed to amplify a 334-bp region spanning introns 4 to 6 of the PBGD gene (48). The cell equivalent DNA present in a given sample was determined by comparison with a dilution series of a standardized stock of pJB1057 described above.
The primers used in this study were chosen empirically according to which functioned best within the constraints of the TaqMan assays. These PCR amplifications were carried out in 96-well optical plates (Marsh Bio Products, Rochester, N.Y.) in a total volume of 50 μl. A sample volume of 10 μl was added to 40 μl of a master mix consisting of 3 μl of TaqMan buffer A, 2 μl of AmpliTaq Gold buffer, 300 μM each dATP, dGTP, and dCTP, 600 μM dUTP, 4 mM MgCl2, primers as described above [1 μM each], 100 nM probe, and 1.25 U of AmpliTaq Gold. Buffers, MgCl2, and enzyme were obtained from Applied Biosystems, deoxynucleoside triphosphates were obtained from Promega, primers were obtained from Invitrogen, and probes were obtained from Biosource International (Camarillo, Calif.). The reaction mixtures were initially incubated for 10 min at 94°C, followed by 45 cycles of two-step amplification regimens (95° C for 15 s and 60°C for 1 min) for all reactions except the full-length and total 2-LTR assays and the cellular flanking DNA assay, which were amplified by using 95°C for 15 s and 66°C for 1 min.
PCR amplification, product cloning, and sequence determination of 2-LTR junction fragments.
2-LTR junction fragments were PCR amplified, cloned, and sequenced as described by Julias et al. (37); the positions of the primers used for the PCR amplification are presented in Fig. 2. Sequences of the DNA inserts were determined using ABI Big Dye terminator version 2 chemistry (Applied Biosystems) and M13 reverse primer (Invitrogen) on an ABI-373XL DNA sequencer (Applied Biosystems) as specified by the manufacturer. Sequences were aligned with the aid of the ClustalW alignment protocol, used through the following URL and managed by the Baylor College of Medicine: http://searchlauncher.bcm.tmc.edu/multi-align/multi-align.html.
RESULTS
NC functions during early infection events. To more fully define the role performed by NC in these events, the course of reverse transcription (54) for wild-type and mutant viruses was examined using primers and probes complementary to sequences that are generated after discrete, critical points in the process. The quantity of transcripts containing the R-U5, U3-U5, gag, and R-5′ untranslated region (UTR) regions were measured to monitor reverse transcription from minus-strand strong-stop DNA synthesis through the minus-strand transfer event, late minus-strand synthesis, and finally the plus-strand transfer event, respectively (Fig. 1).
Reverse transcription in wild-type and IN mutant viruses.
Typical results obtained with each mutant are presented in Table 1. Percent copy numbers relative to wild type for each intermediate are also presented. In addition, values for the percent copy numbers for each successive intermediate, relative to the R-U5 sequence for each virus, are presented. We observed interassay variation in the absolute copy number derived from a particular mutant among different experiments. In contrast, the intra-assay variation was surprisingly low once copy numbers were normalized for RT activity and infected-cell number. This variability between experiments presumably reflects infection efficiency and/or other factors other than the controls and does not affect the relative results between viruses. Transfections with sheared salmon sperm DNA and plasmid containing a nonfunctional RT (RTD185K/D186L) were included as negative controls for each panel of mutants tested. The RTD185K/D186L mutant was also included to ensure that plasmid DNA was not being carried over from the initial transfection step into the infection, since this plasmid contains the same mammalian flanking DNA sequences as all of the other plasmids (Fig. 1).
TABLE 1.
Quantitation of transcription intermediates
| Virus | Targeta copy no.b
|
% of target copies relative to wild typee
|
% of target copies relative to R-U5f
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| R-U5 | U3-U5 | Gag | R-5′UTR | R-U5 | U3-U5 | Gag | R-5′UTR | R-U5 | U3-U5 | Gag | R-5′UTR | |
| (−)-Controlc,d | 0 | 0 | 0 | 0 | ||||||||
| RTD185K/D186Ld | 2 | 0 | 0 | 0 | ||||||||
| Wild type | (1.4 ± 0.1) × 106 | (1.8 ± 0.1) × 106 | (7.8 ± 1.9) × 105 | (5.5 ± 1.4) × 105 | 100 | 100 | 100 | 100 | 100 | 120 ± 12 | 56 ± 12 | 37 ± 7 |
| IND116N | (1.7 ± 0.1) × 106 | (1.6 ± 0.2) × 106 | (6.4 ± 0.4) × 105 | (4.4 ± 0.5) × 105 | 120 ± 17 | 89 ± 15 | 88 ± 26 | 88 ± 31 | 100 | 100 ± 23 | 41 ± 10 | 27 ± 10 |
| NCH23C | (3.6 ± 0.5) × 104 | (1.4 ± 0.1) × 104 | (4.8 ± 0.2) × 104 | (7.9 ± 3.8) × 101 | 2.6 ± 0.6 | 0.78 ± 0.06 | 6.6 ± 1.9 | 0.017 ± 0.011 | 100 | 32 ± 20 | 110 ± 51 | 4 ± 5 |
| NCH23C/IND116N | (2.2 ± 0.3) × 104 | (5.2 ± 0.1) × 103 | (1.0 ± 0.1) × 104 | (6.2 ± 0.8) × 102 | 1.6 ± 0.3 | 0.29 ± 0.01 | 1.3 ± 0.4 | 0.12 ± 0.05 | 100 | 31 ± 20 | 110 ± 77 | 4 ± 5 |
| NCH44C | (1.7 ± 0.2) × 105 | (1.4 ± 0.2) × 105 | (3.6 ± 0.2) × 105 | (4.3 ± 0.1) × 104 | 12 ± 3 | 7.8 ± 1.1 | 50 ± 14 | 8.4 ± 2.3 | 100 | 57 ± 23 | 230 ± 31 | 19 ± 6 |
Target sequences (R-U5, U3-U5, etc.) are depicted in Fig. 1.
Copy numbers are expressed in units of corrected mean copy number ± standard deviation. Representative values taken from a typical experiment with samples run in duplicate. All values are normalized with respect to the virus stock used in the infection and cell number, as described in Materials and Methods.
Transfection and infection using sheared salmon sperm DNA.
Copy numbers are the actual values obtained from the total DNA from ∼105 cells. Adjustments for RT activities with these samples are not possible since there is no active RT in these samples.
Refers to the percent target copy number for a mutant relative to the copy number for same target in a wild-type infection, using the values in the Copy number column. Values are expressed as mean ± standard deviation.
Percentages expressed as ratio (mean ± standard deviation) of copy number for designated species to copy number obtained for R-U5 DNA, from at least three individual transfection-infection experiments.
The wild-type and IND116N viruses produced approximately equal amounts of each reverse transcription intermediate, as expected for a mutation that has little effect on reverse transcription (Table 1). In addition, the number of R-U5 and U3-U5 DNA copies was essentially the same for both viruses, implying that the minus-strand transfer process is efficient. Finally, about one-third of the transcripts containing the R-U5 sequence were also detected with the primers complementary to R-5′UTR, suggesting that many transcripts isolated at the time of harvest were nearly complete (Table 1). The IND116N mutants were included in attempts to determine the integration efficiency of the NC mutants relative to wild type (see below).
Reverse transcription in NC mutant viruses.
Viruses containing mutations in either NC Zn2+ finger, however, exhibited a very different phenotype from the wild type. Viruses containing the Cys substitution in the amino-terminal finger (NCH23C) were particularly affected, yielding <3% of the R-U5 transcript relative to wild type. In addition, progression through both strand transfer events appeared to be enormously reduced relative to wild type, with the most severe defect occurring after plus-strand transfer (Table 1). However, simply comparing the value of the R-U5 copy number with that of U3-U5 to determine the amount of minus-strand transfer may be misleading: it has been suggested previously that this particular mutation may lead to increased degradation (decreased stability) of the vDNA by host exonucleases (53). Therefore, decreased stability of the vDNA molecule could lead to an underestimation of U3 and U5 quantities relative to more interior and possibly more protected sequences. This probably explains why we observed a much larger quantity of gag copies relative to R-U5 in all Zn2+ finger mutants than we did in the wild type. Indeed, there was frequently a higher level of gag than U3-U5 targets in these mutant vDNAs (Table 1).
Virus with the mutation in the carboxy-terminal Zn2+ finger (NCH44C) exhibited a slightly different phenotype. While the copy number of all intermediates was substantially reduced relative to that of the wild type, the ratio of the R-5′UTR to R-U5 sequence was not as dramatically lowered; it was only about one-half that observed for the wild type (Table 1). Thus, it appears that the strand transfer events in this mutant are not as critically affected as in the NCH23C mutant. The gag copy number obtained was approximately twice that for R-U5 in this mutant, which we again interpret as evidence for substantially decreased stability of the vDNA transcript with this mutation.
Quantitation of 2-LTR circular vDNA.
To obtain an idea of how many vDNAs were actually completed, vDNA termini were examined by studying 2-LTR circles, which are found in infected cells (19, 32, 50). The 2-LTR circular vDNA species are actually dead-end products of reverse transcription, but they are experimentally useful. Two-LTR circles appear to be formed through the action of a host cell ligation activity, composed of DNA ligase IV and other factors (40), that joins the free ends of the linear vDNA, and it has recently been shown that the circular form is relatively long lived (7, 42). Real-time PCR was used to determine the 2-LTR copy number in DNA samples generated by HIV-1 infection. The TaqMan quantitative method was performed with the primers and probe shown in Fig. 2. The same samples used for analysis of reverse transcription intermediates were examined here.
To distinguish between full-length sequences and those containing deletions, different primer-probe systems were designed. Both used the same sense primer (HIV-SS-F4) and probe (P-HUS-SS1) located in the R and U5 regions, respectively. To detect circle junctions formed by ligation of full-length linear vDNA (i.e., full-junction 2-LTR circles), the antisense primer HIV-LTR-R5 was used, which is complementary to the precise sequence expected across the junction. In another detection system, the antisense primer (HIV-LTR-R3) used was designed to be complementary to a sequence located further downstream (75 nt into the U3 region [Fig. 2]). This second primer-probe combination allowed the detection of both full-length sequence and sequences that contained deletions between the 3′ ends of the probe and antisense primer sequences. Copy numbers determined with this second primer-probe set were therefore termed total 2-LTR circles.
The number of full-junction and total 2-LTR circles was determined in duplicate assays for each sample; typical results are presented in Table 2. The 6- to 45-fold decrease in total 2-LTR copy numbers for all NC Zn2+ finger mutants relative to wild type suggests that vDNA formation is hampered by these mutations, as has been proposed previously (53). Notably, virus containing the mutation in the proximal Zn2+ finger clearly produces drastically reduced numbers of full-length reverse transcripts and a major defect occurs at the point of plus-strand transfer, as evidenced by both the extremely small number of R-5′UTR copies (Table 1) and 2-LTR circular DNA sequence targets (Table 2). A similar but less severe reduction was observed with the NCH44C mutant. The mutation in IN resulted in an increase of 16 times the total 2-LTR copies and 40 times the full-length 2-LTR copies over wild type. These values are consistent with the hypothesis that at least one fate of unintegrated vDNA is ligation-circularization (Fig. 3A). The similarity between the values obtained from the IND116N mutant for R-5′UTR (Table 1) and total 2-LTR circle copies (Table 2) again supports the conclusion that the unintegrated vDNA in this mutant becomes ligated. Furthermore, the increase in the number of 2-LTR circles observed with the IND116N mutant did not result from increased vDNA synthesis, since Table 1 shows that the levels of reverse transcription intermediates were roughly equivalent between wild-type and IND116N infections. These data also indicate that >95% of the full-junction vDNAs synthesized became integrated in a wild-type infection after 24 h, which is consistent with previous results from this laboratory (26).
TABLE 2.
Quantitation of 2-LTR circular vDNA
| Virus | 2-LTR circle copy no.a
|
|
|---|---|---|
| Full junction | Total | |
| (−)-Controlb,c | 0 | 0 |
| RTD185K/D186Lc | 1 | 0 |
| Wild type | (1.4 ± 0.2) × 104 | (3.6 ± 0.8) × 104 |
| IND116N | (5.6 ± 0.2) × 105 | (6.0 ± 0.1) × 105 |
| NCH23C | (4.5 ± 1.6) × 102 | (8.0 ± 4.0) × 102 |
| NCH23C/IND116N | (7.0 ± 0.5) × 102 | (1.3 ± 0.4) × 103 |
| NCH44C | (3.6 ± 0.3) × 103 | (6.0 ± 0.4) × 103 |
2-LTR circle copy numbers are expressed in units of corrected mean copy number ± standard deviation. Representative values are taken from a typical experiment with samples run in duplicate using the primers and probe depicted in Fig. 2. All values are normalized with respect to the virus stock and cell number used in infection, as described in Materials and Methods.
Transfection and infection using sheared salmon sperm DNA.
Copy numbers are the actual values obtained from the total DNA from ≈105 cells. Adjustments for RT activities with these samples are not possible since there was no activity.
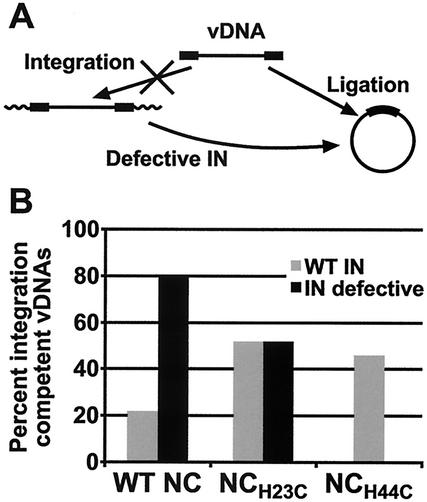
FIG. 3.
Fates of vDNA with and without a competent IN protein. (A) Schematic representation of the fate of integrated vDNA when the IN protein is knocked out. Viral DNA that could have been integrated now becomes ligated when IN is defective. The vDNA is represented by the black boxes (LTR regions) and the black line, and the cellular flanking DNA into which the provirus in inserted is represented by a thick wavy line. (B) Graph showing the percentage of integration-competent vDNAs isolated from cells infected with mutant and wild-type viruses. Values were calculated by assuming that the four types of linear vDNA sequences in Fig. 5 (U/U, U/P, P/U, and P/P) would have been integration competent if not for some defect other than vDNA end truncations or insertions. Values are reported as the percentage of integration-competent sequences of the total sequences obtained in Fig. 5.
Sequence analysis of 2-LTR circle junction fragments.
We PCR amplified, cloned, and sequenced the PCR products to compare 2-LTR circular vDNAs formed after infection with NC Zn2+ finger mutant and wild-type HIV-1. These sequenced junction fragments presented us with a snapshot of the state of the linear vDNA termini just prior to ligation. The sequence integrity at the junction of the ligated ends was examined as an indicator of three processes: (i) the progression of reverse transcription to its completion, (ii) the productivity of the integration process (using the IND116N-containing mutants), and (iii) the ability of NC to protect vDNA from degradation by host exonuclease and/or ligase activity.
Oligonucleotides complementary to the expected sequence surrounding the circle junction were used to PCR amplify 2-LTR circular vDNA (Fig. 2) (37) arising from infections with either wild-type or mutant viruses. Each primer contained a restriction site to facilitate cloning of the resulting products. Using the R-U3 primers with the probe for quantifying 2-LTR circle species (Table 2) allowed the detection of a slightly different array of sequences from those detected by using the primers in these cloning experiments (Fig. 2). Obviously, the quantification does not account for all 2-LTR circles formed, but it does detect most of those revealed with the primer set used in these cloning experiments.
PCR products from infected-cell lysates were cloned, and the inserts were analyzed for size. Distributions of the lengths of the inserts cloned for each of the mutants are presented in Fig. 4. Infections are arranged in decreasing order of percentage of expected full-length sizes as follows: IND116N > NCH44C > wild type > NCH23C = NCH23C/IND116N. The clones obtained with the wild type had a higher level of aberrantly small junction fragments than did those obtained with all of the mutants. In contrast, the IND116N mutant infection gave almost exclusively the proper sizes of 2-LTR junction fragments.
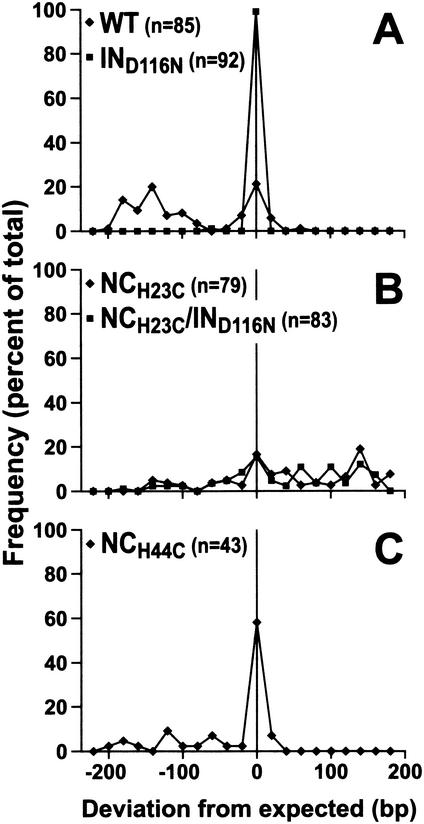
FIG. 4.
Frequency distribution of the sizes of cloned 2-LTR circular vDNA PCR products from cells incubated with mutant and wild-type viruses. Inserts of the PCR product clones were analyzed for their sizes by either agarose gel electrophoresis or sequence analysis. Results are reported as the deviation from the expected size of 346 bp (Fig. 2). The frequencies were compiled in 20-nt windows from the expected size. Frequencies of the variance of insert sizes are reported as a percentage of the total number of samples examined. Distributions for wild-type (⧫) and IND116N (▪) (A), NCH23C (⧫) and NCH23C/IND116N (▪) (B), and NCH44C (⧫) (C) virus infections are shown.
The nucleotide sequences were determined for a number of these clones, which were selected to produce an array of sequences that represented the size distribution proportionately. Viral sequences obtained from each mutant are presented schematically in Fig. 5, and properties of the termini are summarized in Table 3. The results in Fig. 4 parallel those in Fig. 5. There is a large degree of heterogeneity in the wild-type samples and less so with the mutants. The IND116N mutant is the most homogeneous, containing mostly full-length 2-LTR junctions.
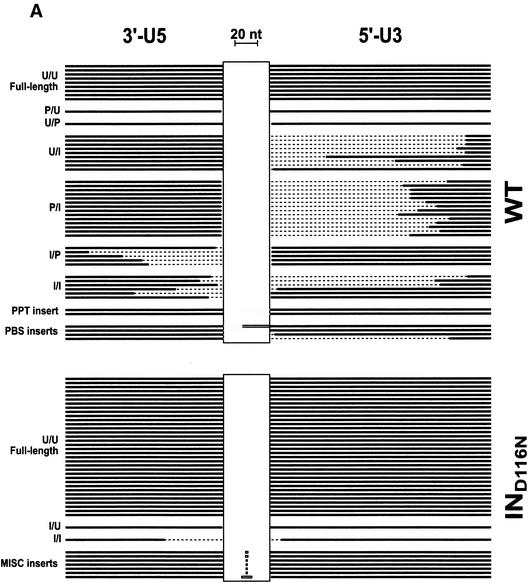
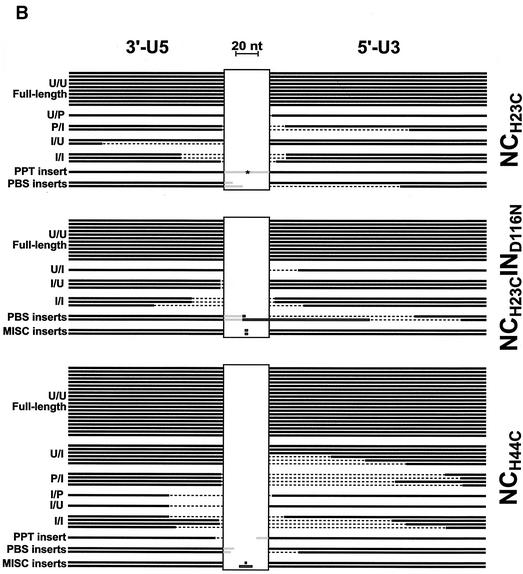
FIG. 5.
Schematic of PCR product insert alignments. The cloned inserts of products from the PCR of vDNA isolated from cells incubated with mutant and wild-type viruses were sequenced. The virus examined is indicated on the right of each collection, and the classification of each sequence type is indicated on the left as follows: U, unprocessed end (full length); P, processed end (the CA dinucleotides were removed from the end, presumably by the viral IN protein); I, incomplete end (truncated compared to the full-length sequence). The letter to the left of the slash represents the state of the 3′-U5 end, and the letter to the right represents the state of the 5′-U3 end of the vDNA prior to ligation. The thick black lines in each panel represent nucleotides that are present in each of the clones. The dashed regions depict the nucleotides that are missing from the vDNA product insert. The grey regions depict DNA insertions of viral origin (PPT or PBS), and the horizontal boxes represent insertions of nonviral origin. The vertical boxes are spacer regions to accommodate the insertions. Black lines that are butted against the vertical boxes are full-length 2-LTR viral sequences. The lengths of lines and horizontal boxes are proportional to the length of the DNA sequence, with a 20-nt reference shown at the top of the figure. The PPT insert in the NCH23C collection of alignments, denoted by *, is 229 bp and is contiguous with the 5′-U3 end. A summary of the types of sequences for each virus is presented in Table 3. (A) Alignment of sequences from cloned PCR products of vDNA from infections with virus containing wild-type NC protein. (B) Alignment of sequences from cloned PCR products of vDNA from infections with virus containing mutant NC proteins.
TABLE 3.
Classification of LTR termini from 2-LTR junction fragments
| Virus | Total no. of sequences obtaineda | Total no. of 3′ U5 endsb
|
Total no. of 5′ U5 endsb
|
||||
|---|---|---|---|---|---|---|---|
| Uc | Pc | Ic | U | P | I | ||
| Wild type | 51 | 21 | 15 | 11 | 12 | 6 | 29 |
| IND116N | 43 | 41 | 0 | 2 | 41 | 0 | 2 |
| NCH23C | 21 | 12 | 2 | 5 | 14 | 1 | 6 |
| NCH23C/IND116N | 23 | 15 | 0 | 6 | 17 | 0 | 6 |
| NCH44C | 43 | 28 | 4 | 7 | 24 | 1 | 15 |
Sequence categories summarized are from virally derived DNAs and are tallied from sequences depicted in Fig. 5.
LTR end of linear vDNA prior to ligation.
U, unprocessed LTR end; P, processed LTR end defined as the removal of the CA dinucleotide from the 3′ end via the action of IN; I, incomplete end, defined as an end which was either not completely synthesized or was degraded once synthesized.
The Asp116-to-Asn mutation of HIV-1 IN results in almost complete inhibition of its end-processing function and DNA strand transfer activities in vitro (18). As expected, none of the sequences obtained for the IND116N mutant contained processed ends (Fig. 5A; Table 3). This mutation also resulted in a greater absolute number of 2-LTR circles than did the wild type (Table 2). In the absence of integration activity, equivalent numbers of total vDNA transcripts (linear, circular, and proviral) were expected and observed for both wild-type and IND116N viruses (Table 1). It is apparent that the vDNAs that are unavailable for integration (as in the IND116N case) are acted on by nuclear ligases in the infected cell (Fig. 3A). The His-to-Cys NC Zn2+ finger mutation in either Zn2+ finger of HIV-1 yielded proportionally more full-length 2-LTR circles in the mutants than in the wild type (Fig. 5; Table 2), although ∼10- to 50-fold fewer total circles were produced overall (Table 2). In addition to simple end joining, all viruses except the IND116N mutant yielded a small proportion of circle junctions that had an insertion of virally derived sequence, either the PBS or the 3′ polypurine tract (PPT), where minus- and plus-strand DNA synthesis is initiated, respectively (Fig. 5). These types of insertions were also detected in the analysis of 2-LTR circles formed by Mo-MuLV and are probably either a mispriming event or a defect in RNase H processing during reverse transcription (25, 37). Some nonviral insertions were also detected between the termini. It appears that the HIV-1 NC mutation does not significantly alter the frequency of these events, since they occur almost equivalently in both the NC mutants and wild type. While none of these types of inserts were observed with the IND116N mutant, the larger number of circle junctions obtained with this mutant (Fig. 5A; Table 2) probably masks these relatively rare occurrences.
The ratios of full-junction to total 2-LTR circles from (i) the real-time PCR (TaqMan) assay (Table 2) and (ii) the cloning and sequencing protocol are presented in Table 4. The effectiveness and reliability of the real-time PCR assay are demonstrated by the excellent agreement between the values obtained by the two methods. In addition to providing a measure of the integrity of the ligated termini, the normalized copy numbers provide more quantitative information than can be obtained from the cloning experiments alone. To our knowledge, this is the first reported assay of this type that can reliably distinguish between full-length and truncated 2-LTR junction fragments.
TABLE 4.
Quantitation of 2-LTR circular vDNA
| Virus | % of full junction with respect to total no. by:
|
|
|---|---|---|
| Quantitative PCRa | Cloning and sequencingb | |
| Wild type | 37 ± 5 | 43 |
| IND116N | 87 ± 9 | 91 |
| NCH23C | 54 ± 7 | 55 |
| NCH23C/IND116N | 58 ± 6 | 63 |
| NCH44C | 58 ± 3 | 61 |
Values expressed as percent ratio (mean ± standard deviation) of full junction/total 2-LTR circles from at least three individual transfection-infection experiments.
Percentage of full junctions/total 2-LTR junctions obtained on analysis of sequence data (Fig. 5), with correction for differential detection resulting from the use of primers that target slightly different regions of the expected circle junction sequence.
Effects of NC mutations on integration.
IN performs the obligate removal of two nucleotides from the 3′ ends of the linear vDNA prior to its integration (15, 49), and it appears that mutations in NC can affect this reaction in vivo. A greater proportion of sequences obtained from infections with the HIV-1 NC mutants appear to exhibit deficiencies in this end-processing function of both LTR termini relative to the wild type, particularly with respect to the 5′ U3 end. For example, 26 and 44% of the sequences examined from the wild type were not processed (Fig. 5) at the 5′ U3 and 3′ U5 ends (Fig. 5A), respectively, whereas 67 and 63%, respectively, of the sequences from the NCH23C mutant were not processed (Fig. 5B). Furthermore, adding the IND116N mutation to the NCH23C mutant virus resulted in the production of nearly equivalent levels of 2-LTR circles (Table 2), suggesting a complete lack of integration with the NCH23C mutant. In addition, the types of sequences obtained (processed versus unprocessed) were minimally affected among the two NCH23C mutants (Table 3). The NCH23C mutation somehow caused end-processing defects in the vDNA (Table 3), since the termini were almost exclusively unprocessed (as observed with the IND116N mutant), and the intact NC Zn2+ finger was required for this activity in conjunction with IN.
As a consequence of the defect in end processing, the level of integration in the NCH23C mutant appears quite defective, as mentioned above. This can be confirmed further by examining the 2-LTR junction fragments in Fig. 5. Theoretically, full-length linear vDNAs with either unprocessed or processed termini at both ends (i.e., the U/U, U/P, P/U, or P/P classes of vDNAs [Fig. 5]) would be competent for integration in virions if there were no other defects. Of course, only the vDNAs of the P/P type would be integrated. IN in these “ideal” virions would process the unprocessed ends, and integration would occur. By comparing the percentages of vDNAs that are integration competent for virus pairs with and without a functional IN, one can observe the integration competency of a particular mutant or wild-type virus. The fact that the 2-LTR circles are stable in culture (7, 42) makes this analysis attractive. For the wild-type and IND116N pair, where both contain wild-type NC, 22% of the vDNAs were integration competent in the wild type whereas the level increased to 80% for the IND116N mutant (Fig. 3B and 5A). In contrast, comparing NCH23C with and without a functional IN showed absolutely no change in the levels of integration-competent vDNAs (52%), again indicating a gross defect in the integration process. The level of integration-competent vDNAs in the NCH44C mutant was similar to that for the NCH23C (Fig. 3B and 5B).
DISCUSSION
This work demonstrates some of the in vivo requirements for the wild-type HIV-1 NC Zn2+ fingers in infection processes. The NC mutants examined are defective for replication despite being able to package normal levels of RNA genomes. We examined reverse transcription intermediates from infected cells and found that the wild-type NC Zn2+ fingers are required for reverse transcription processes and vDNA protection, as has been proposed previously for HIV-1 (53) and by this laboratory for Mo-MuLV NC (25). Surprisingly, this is the first report demonstrating that the NC Zn2+ fingers are required for initial integration events. Mutations to the NC Zn2+ fingers prevent proper end processing via viral IN. It can also be inferred from these results that NC remains associated with not only the viral RNA throughout reverse transcription but also the full-length vDNA up to the point of integration.
The results presented in this study generally coincide with those reported previously by this laboratory (25, 26) and extend our understanding of earlier experiments considerably. The importance of the Zn2+ finger motifs in NC proteins of several retroviruses has long been recognized. Experiments in vitro that simulate events during early infection have indicated their role in critical functions, including initiation and procession of reverse transcription (28-30, 33, 47), as well as integration of model substrates (9, 10, 22a). In addition, the importance of the Zn2+ fingers in vivo for viral replication was demonstrated previously (5, 23, 26, 43, 53). Here, we have begun to link in vitro observations of the function of NC Zn2+ fingers and the broader role of NC with events that occur in vivo.
Previous studies of Mo-MuLV (25) suggested that NC has important functions in generating and/or stabilizing the vDNA produced during infection. The present study was initiated to explore these phenomena, noting how analogous mutations in one or the other NC Zn2+ fingers of HIV-1 affect vDNA production. The NC mutants tested appear deficient in vDNA synthesis since there was a substantial decrease in the R-U5 (strong-stop product) copy numbers compared to wild type for both the NCH23C and NCH44C mutants (39- and 8-fold, respectively [Table 1]). Even more drastic reductions were observed in sequences corresponding to the vDNA ends. For example the U3-U5 (minus-strand transfer product) and R-5′UTR (plus-strand transfer product) copy numbers decreased by ∼130- and ∼6,000-fold, respectively, for NCH23C compared to wild-type virus. The reductions in the termini for the NCH44C mutant compared to wild type were ∼10-fold for the first- and second-strand transfer products.
On initial inspection of strong-stop and strand transfer vDNA quantities, one gets the impression that there are severe defects in reverse transcription processes. However, the reduction in gag levels was only 16- and 2-fold for the NCH23C and NCH44C mutants, respectively, compared to wild type (Fig. 1; Table 1). The only way that gag sequences can be obtained is via successful strand transfer events (54). The fact that gag is present at fairly substantial levels in the mutants, compared to those in the wild type, indicates that the NC mutants are deficient in protecting the ends of the vDNA from exonucleases. These data show that reverse transcription is not affected very severely. The overall numbers of vDNA molecules are reduced, but not very drastically; however, the quality of the vDNA is inferior since the ends are degraded. Degradation of the ends would account for the drastic reduction in the R-U5, U3-U5, and R-5′UTR target sequences, which represent the strong-stop, minus-stand transfer, and plus-strand transfer vDNA species, respectively. In addition, a reduction in the sequences at the vDNA ends would inflate the levels of gag targets, relative to R-U5, as observed in the NCH23C mutants and especially in the NCH44C mutant.
In addition to the obvious end degradation problem, there are reverse transcription defects in NC mutants since the levels of gag in the mutants relative to those in the wild-type virus are reduced (Table 1). Defects in the chaperone activity of the NCH23C and the NCH44C mutants have been demonstrated in vitro by Williams et al. (56) and Guo et al. (30). These mutations would certainly affect minus- and plus-strand transfer processes as well as nucleic acid secondary-structure management in the mutant virus infections. However, it is difficult to obtain meaningful data regarding the effects of these mutations on reverse transcription per se, due to the overriding degradation of the vDNA termini seen in these mutant infections.
There appears to be an additional effect on integration processes in the HIV-1 NC mutants. This is apparent when the effects of the IN mutation in IND116N are compared with results obtained for both NC Zn2+ finger mutants. The NC mutant viruses increase the proportion of full-length sequences compared to that in the wild type, and these sequences have not undergone end processing (i.e., removal of the terminal dinucleotides [Table 3; Fig. 5]). The NC mutants also yield defective or incomplete vDNAs, similar to what is observed with wild-type HIV-1. This was not the case with the Mo-MuLV NC mutants, since significant end-truncations were noted in the 2-LTR circles from the Mo-MuLV mutants whereas wild-type junction fragments had a much higher proportion of full-length sequences (25). There is, however, a reduction in the quantity of 2-LTR circular DNA molecules in the HIV-1 NC mutants, similar to observations made with the Mo-MuLV mutants.
These results demonstrate that NC, with intact Zn2+ fingers, are required for efficient removal of the 3′-terminal dinucleotides by IN in vivo. The fact that mutating IN in NCH23C does not increase the proportion of full-length junction fragments certainly supports this conclusion. The relative increase in the number of full-length 2-LTR circles for the IND116N mutant compared to wild type is due to a block in integration, yet the absence of an added effect on mutation of IN in the NCH23C mutant provides evidence that the NC mutation alone leads to the end-processing defect that blocks integration of the vDNA (Fig. 5; Table 2). In addition, there is a ∼60% increase in the level of integration-competent junction fragments between the wild-type and IND116N infections, which is not seen with the NCH23C-NCH23C/IND116N pair (Fig. 3B and 5).
NC appears to be assisting viral IN with initial integration processes through direct or indirect interactions. It is possible that the end-processing defect with the NC mutants may also be linked to vDNA protection issues. In vitro experiments have shown that purified NCH23C and NCH44C mutant proteins were able to stimulate coupled joining at ∼50% of the wild-type levels (10). Since these mutant proteins can support integration in vitro, the defect with these NC mutants in vivo may be attributable to the lack of protection of the vDNA from cellular ligases, which may act to displace IN from the vDNA prior to the removal of the terminal dinucleotides by IN. Whatever the mechanism, these results provide strong in vivo evidence for an involvement for NC during the integration processes.
Reverse transcription and integration steps of the virus life cycle are affected as described above. Since pseudotyped viruses were used in this work, it is possible that differences might be observed in the effects of the NC mutations when infections are mediated through the use of HIV-1 Env. It has been reported that HIV-1 entry via VSV-G pseudotyping can affect downstream steps in the viral life cycle (2, 12). Further studies comparing reverse transcription and integration processes from NC mutant viral infections, mediated by VSV-G and HIV-1 gp120SU Env proteins, would address this possibility. One problem involves obtaining sufficient virus titers from gp120SU/CD4-mediated infections to generate sufficient levels of reverse transcription intermediates from some of the mutants.
Results of both in vitro and in vivo experiments have shown that the two Zn2+ fingers in the HIV-1 NC protein are not interchangeable (24, 30, 56) and, furthermore, that defects in replication are more strongly affected by mutations in the NH2-terminal finger. We found previously that the NCH23C mutant is completely replication defective whereas NCH44C is able to replicate at a very low level, eventually reverting to a wild-type genotype (26). Results presented here are consistent with the different behaviors observed for the two viruses, since the NCH23C mutant clearly possesses more severe defects in reverse transcription and vDNA protection than does the NCH44C mutant (Tables 1 and 2). It appears that vDNA production with the NCH23C mutant is ∼8-fold lower than with the NCH44C mutant and ∼16-fold lower than with the wild type when gag levels are compared (Table 1).
One additional conclusion can be drawn from these results. NC is probably associated with the preintegration complex up to and possibly during the integration event, which forms the provirus. In this study, we have shown that wild-type NC is required for setting the stage for initial integration events and that processing of the vDNA termini by the viral IN protein is not occurring with the NCH23C mutant. Additionally, in vitro studies from this and other laboratories have shown that wild-type recombinant NC can enhance integration and, specifically, the coupled joining reaction (9, 10, 22a). Evidence from other laboratories also suggests the presence of NC at later stages of the infection process. Gallay et al. (22) demonstrated the presence of HIV-1 NC in the nuclear fraction from infected cells after 8 h; NC is presumably associated with preintegration complexes that were imported into the nucleus. Risco et al. (45) also showed the presence of NC in nuclei by immunogold labeling of Mo-MuLV-infected cells. They speculate that NC enters the nucleus as a component of a complex containing IN and the vDNA. NC possesses nucleic acid chaperone activity, which assists in many processes during reverse transcription (44). The results of this study strongly suggest that NC is involved in viral nucleic acid management of the full-length vDNA as well as chaperone functions previously proposed.
As has been demonstrated in this work, the lack of replication in NC mutant viruses is due to a reduction in the quantity and quality of the vDNA synthesized, with defects in integration as well. It is clear that NC is a necessary component of the reactions carried out by RT and IN. These reactions are so sensitive to the nature of the NC protein, in fact, that the relatively subtle mutations investigated here that preserve Zn2+ binding and RNA packaging are catastrophic to viral replication. Therefore, the NC component of these processes obviously represents an important target in the development of antiretroviral therapies against viruses resistant to current treatments.
Acknowledgments
This work was supported by the NCI, NIH, under contract N01-CO-12400 with SAIC-Frederick, Inc.
We thank John G. Julias and Stephen H. Hughes of the NCI at Frederick for the reagents and for valuable discussions throughout these studies. We thank David E. Ott and Louis E. Henderson of SAIC Frederick, Inc., and Frederic Bushman of the Salk Institute for many helpful discussions. We also thank Jeffrey D. Lifson, Mike Piatak, and Kalachar Suryanarayana of SAIC Frederick, Inc., for invaluable assistance with the real-time PCR technique.
REFERENCES
- 1.Adachi, A., H. E. Gendelman, S. Koenig, T. Folks, R. Willey, A. Rabson, and M. A. Martin. 1986. Production of acquired immunodeficiency syndrome- associated retrovirus in human and nonhuman cells transfected with an infectious molecular clone. J. Virol. 59:284-291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aiken, C. 1997. Pseudotyping human immunodeficiency virus type 1 (HIV-1) by the glycoprotein of vesicular stomatitis virus targets HIV-1 entry to an endocytic pathway and suppresses both the requirement for Nef and the sensitivity to cyclosporin A. J. Virol. 71:5871-5877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Allain, B., M. Lapadat-Tapolsky, C. Berlioz, and J.-L. Darlix. 1994. Transactivation of the minus-strand DNA transfer by nucleocapsid protein during reverse transcription of the retroviral genome. EMBO J. 13:973-981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Berg, J. M. 1986. Potential metal-binding domains in nucleic acid binding proteins. Science 232:485-487. [DOI] [PubMed] [Google Scholar]
- 5.Berthoux, L., C. Pechoux, M. Ottmann, G. Morel, and J. L. Darlix. 1997. Mutations in the N-terminal domain of human immunodeficiency virus type 1 nucleocapsid protein affect virion core structure and proviral DNA synthesis. J. Virol. 71:6973-6981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Burns, J. C., T. Friedmann, W. Driever, M. Burrascano, and J. K. Yee. 1993. Vesicular stomatitis virus G glycoprotein pseudotyped retroviral vectors: concentration to very high titer and efficient gene transfer into mammalian and nonmammalian cells. Proc. Natl. Acad. Sci. USA 90:8033-8037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Butler, S. L., E. P. Johnson, and F. D. Bushman. 2002. Human immunodeficiency virus cDNA metabolism: notable stability of two-long terminal repeat circles. J. Virol. 76:3739-3747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cameron, C. E., M. Ghosh, S. F. Le Grice, and S. J. Benkovic. 1997. Mutations in HIV reverse transcriptase which alter RNase H activity and decrease strand transfer efficiency are suppressed by HIV nucleocapsid protein. Proc. Natl. Acad. Sci. USA 94:6700-6705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Carteau, S., S. C. Batson, L. Poljak, J. F. Mouscadet, H. de Rocquigny, J. L. Darlix, B. P. Roques, E. Kas, and C. Auclair. 1997. Human immunodeficiency virus type 1 nucleocapsid protein specifically stimulates Mg2+-dependent DNA integration in vitro. J. Virol. 71:6225-6229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Carteau, S., R. J. Gorelick, and F. D. Bushman. 1999. Coupled integration of human immunodeficiency virus type 1 cDNA ends by purified integrase in vitro: stimulation by the viral nucleocapsid protein. J. Virol. 73:6670-6679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chance, M. R., I. Sagi, M. D. Wirt, S. M. Frisbie, E. Scheuring, E. Chen, J. W. Bess, Jr., L. E. Henderson, L. O. Arthur, T. L. South, G. Perez-Alvarado, and M. F. Summers. 1992. Extended X-ray absorption fine structure studies of a retrovirus: equine infectious anemia virus cysteine arrays are coordinated to zinc. Proc. Natl. Acad. Sci. USA 89:10041-10045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chazal, N., G. Singer, C. Aiken, M. L. Hammarskjold, and D. Rekosh. 2001. Human immunodeficiency virus type 1 particles pseudotyped with envelope proteins that fuse at low pH no longer require Nef for optimal infectivity. J. Virol. 75:4014-4018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chretien, S., A. Dubart, D. Beaupain, N. Raich, B. Grandchamp, J. Rosa, M. Goossens, and P. H. Romeo. 1988. Alternative transcription and splicing of the human porphobilinogen deaminase gene result either in tissue-specific or in housekeeping expression. Proc. Natl. Acad. Sci. USA 85:6-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Covey, S. N. 1986. Amino acid sequence homology in gag region of reverse transcribing elements and the coat protein gene of cauliflower mosaic virus. Nucleic Acids Res. 14:623-633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Craigie, R., T. Fujiwara, and F. Bushman. 1990. The IN protein of Moloney murine leukemia virus processes the viral DNA ends and accomplishes their integration in vitro. Cell 62:829-837. [DOI] [PubMed] [Google Scholar]
- 16.Druillennec, S., A. Caneparo, H. de Rocquigny, and B. P. Roques. 1999. Evidence of interactions between the nucleocapsid protein NCp7 and the reverse transcriptase of HIV-1. J. Biol. Chem. 274:11283-11288. [DOI] [PubMed] [Google Scholar]
- 17.Engelman, A., and R. Craigie. 1992. Identification of conserved amino acid residues critical for human immunodeficiency virus type 1 integrase function in vitro. J. Virol. 66:6361-6369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Engelman, A., G. Englund, J. M. Orenstein, M. A. Martin, and R. Craigie. 1995. Multiple effects of mutations in human immunodeficiency virus type 1 integrase on viral replication. J. Virol. 69:2729-2736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Farnet, C. M., and W. A. Haseltine. 1991. Circularization of human immunodeficiency virus type 1 DNA in vitro. J. Virol. 65:6942-6952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Frankel, A. D., and J. A. Young. 1998. HIV-1: fifteen proteins and an RNA. Annu. Rev. Biochem. 67:1-25. [DOI] [PubMed] [Google Scholar]
- 21.Freed, E. O. 1998. HIV-1 gag proteins: diverse functions in the virus life cycle. Virology 251:1-15. [DOI] [PubMed] [Google Scholar]
- 22.Gallay, P., T. Hope, D. Chin, and D. Trono. 1997. HIV-1 infection of nondividing cells through the recognition of integrase by the importin/karyopherin pathway. Proc. Natl. Acad. Sci. USA 94:9825-9830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23a.Gao, K., R. J. Gorelick, D. G. Johnson, and F. D. Bushman. 2003. Cofactors for human immunodeficiency virus type 1 cDNA integration in vitro. J. Virol. 77:1598-1603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gorelick, R. J., D. J. Chabot, D. E. Ott, T. D. Gagliardi, A. Rein, L. E. Henderson, and L. O. Arthur. 1996. Genetic analysis of the zinc finger in the Moloney murine leukemia virus nucleocapsid domain: replacement of zinc- coordinating residues with other zinc-coordinating residues yields noninfectious particles containing genomic RNA. J. Virol. 70:2593-2597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gorelick, R. J., D. J. Chabot, A. Rein, L. E. Henderson, and L. O. Arthur. 1993. The two zinc fingers in the human immunodeficiency virus type 1 nucleocapsid protein are not functionally equivalent. J. Virol. 67:4027-4036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gorelick, R. J., W. Fu, T. D. Gagliardi, W. J. Bosche, A. Rein, L. E. Henderson, and L. O. Arthur. 1999. Characterization of the block in replication of nucleocapsid protein zinc finger mutants from Moloney murine leukemia virus. J. Virol. 73:8185-8195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gorelick, R. J., T. D. Gagliardi, W. J. Bosche, T. A. Wiltrout, L. V. Coren, D. J. Chabot, J. D. Lifson, L. E. Henderson, and L. O. Arthur. 1999. Strict conservation of the retroviral nucleocapsid protein zinc finger is strongly influenced by its role in viral infection processes: characterization of HIV-1 particles containing mutant nucleocapsid zinc-coordinating sequences. Virology 256:92-104. [DOI] [PubMed] [Google Scholar]
- 27.Grandchamp, B., H. De Verneuil, C. Beaumont, S. Chretien, O. Walter, and Y. Nordmann. 1987. Tissue-specific expression of porphobilinogen deaminase. Two isoenzymes from a single gene. Eur. J. Biochem. 162:105-110. [DOI] [PubMed] [Google Scholar]
- 28.Guo, J., L. E. Henderson, J. Bess, B. Kane, and J. G. Levin. 1997. Human immunodeficiency virus type 1 nucleocapsid protein promotes efficient strand transfer and specific viral DNA synthesis by inhibiting TAR-dependent self-priming from minus-strand strong-stop DNA. J. Virol. 71:5178-5188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Guo, J., T. Wu, J. Anderson, B. F. Kane, D. G. Johnson, R. J. Gorelick, L. E. Henderson, and J. G. Levin. 2000. Zinc finger structures in the human immunodeficiency virus type 1 nucleocapsid protein facilitate efficient minus- and plus-strand transfer. J. Virol. 74:8980-8988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Guo, J., T. Wu, B. F. Kane, D. G. Johnson, L. E. Henderson, R. J. Gorelick, and J. G. Levin. 2002. Subtle alterations of the native zinc finger structures have dramatic effects on the nucleic acid chaperone activity of human immunodeficiency virus type 1 nucleocapsid protein. J. Virol. 76:4370-4378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Henderson, L. E., T. D. Copeland, R. C. Sowder, G. W. Smythers, and S. Oroszlan. 1981. Primary structure of the low molecular weight nucleic acid- binding proteins of murine leukemia viruses. J. Biol. Chem. 256:8400-8406. [PubMed] [Google Scholar]
- 32.Hong, T., K. Drlica, A. Pinter, and E. Murphy. 1991. Circular DNA of human immunodeficiency virus: analysis of circle junction nucleotide sequences. J. Virol. 65:551-555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hsu, M., L. Rong, H. de Rocquigny, B. P. Roques, and M. A. Wainberg. 2000. The effect of mutations in the HIV-1 nucleocapsid protein on strand transfer in cell-free reverse transcription reactions. Nucleic Acids Res. 28:1724-1729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ji, X., G. J. Klarmann, and B. D. Preston. 1996. Effect of human immunodeficiency virus type 1 (HIV-1) nucleocapsid protein on HIV-1 reverse transcriptase activity in vitro. Biochemistry 35:132-143. [DOI] [PubMed] [Google Scholar]
- 35.Johnson, P. E., R. B. Turner, Z. R. Wu, L. Hairston, J. Guo, J. G. Levin, and M. F. Summers. 2000. A mechanism for plus-strand transfer enhancement by the HIV-1 nucleocapsid protein during reverse transcription. Biochemistry 39:9084-9091. [DOI] [PubMed] [Google Scholar]
- 36.Julias, J. G., A. L. Ferris, P. L. Boyer, and S. H. Hughes. 2001. Replication of phenotypically mixed human immunodeficiency virus type 1 virions containing catalytically active and catalytically inactive reverse transcriptase. J. Virol. 75:6537-6546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Julias, J. G., M. J. McWilliams, S. G. Sarafianos, E. Arnold, and S. H. Hughes. 2002. Mutations in the RNase H domain of HIV-1 reverse transcriptase affect the initiation of DNA synthesis and the specificity of RNase H cleavage in vivo. Proc. Natl. Acad. Sci. USA 99:9515-9520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kim, J. K., C. Palaniappan, W. Wu, P. J. Fay, and R. A. Bambara. 1997. Evidence for a unique mechanism of strand transfer from the transactivation response region of HIV-1. J. Biol. Chem. 272:16769-16777. [DOI] [PubMed] [Google Scholar]
- 39.Lapadat-Tapolsky, M., C. Gabus, M. Rau, and J. L. Darlix. 1997. Possible roles of HIV-1 nucleocapsid protein in the specificity of proviral DNA synthesis and in its variability. J. Mol. Biol. 268:250-260. [DOI] [PubMed] [Google Scholar]
- 40.Li, L., J. M. Olvera, K. E. Yoder, R. S. Mitchell, S. L. Butler, M. Lieber, S. L. Martin, and F. D. Bushman. 2001. Role of the non-homologous DNA end joining pathway in the early steps of retroviral infection. EMBO J. 20:3272-3281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ott, D. E., E. N. Chertova, L. K. Busch, L. V. Coren, T. D. Gagliardi, and D. G. Johnson. 1999. Mutational analysis of the hydrophobic tail of the human immunodeficiency virus type 1 p6Gag protein produces a mutant that fails to package its envelope protein. J. Virol. 73:19-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pierson, T. C., T. L. Kieffer, C. T. Ruff, C. Buck, S. J. Gange, and R. F. Siliciano. 2002. Intrinsic stability of episomal circles formed during human immunodeficiency virus type 1 replication. J. Virol. 76:4138-4144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ramboarina, S., N. Moreller, M. C. Fournie-Zaluski, and B. P. Roques. 1999. Structural investigation on the requirement of CCHH zinc finger type in nucleocapsid protein of human immunodeficiency virus 1. Biochemistry 38:9600-9607. [DOI] [PubMed] [Google Scholar]
- 44.Rein, A., L. E. Henderson, and J. G. Levin. 1998. Nucleic-acid-chaperone activity of retroviral nucleocapsid proteins: significance for viral replication. Trends Biochem. Sci. 23:297-301. [DOI] [PubMed] [Google Scholar]
- 45.Risco, C., L. Menendez-Arias, T. D. Copeland, P. Pinto da Silva, and S. Oroszlan. 1995. Intracellular transport of the murine leukemia virus during acute infection of NIH 3T3 cells: nuclear import of nucleocapsid protein and integrase. J. Cell Sci. 108:3039-3050. [DOI] [PubMed] [Google Scholar]
- 46.Rong, L., C. Liang, M. Hsu, X. Guo, B. P. Roques, and M. A. Wainberg. 2001. HIV-1 nucleocapsid protein and the secondary structure of the binary complex formed between tRNA(Lys. 3) and viral RNA template play different roles during initiation of (−) strand DNA reverse transcription. J. Biol. Chem. 276:47725-47732. [DOI] [PubMed] [Google Scholar]
- 47.Rong, L., C. Liang, M. Hsu, L. Kleiman, P. Petitjean, H. de Rocquigny, B. P. Roques, and M. A. Wainberg. 1998. Roles of the human immunodeficiency virus type 1 nucleocapsid protein in annealing and initiation versus elongation in reverse transcription of viral negative-strand strong-stop DNA. J. Virol. 72:9353-9358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rossio, J. L., M. T. Esser, K. Suryanarayana, D. K. Schneider, J. W. Bess, Jr., G. M. Vasquez, T. A. Wiltrout, E. Chertova, M. K. Grimes, Q. Sattentau, L. O. Arthur, L. E. Henderson, and J. D. Lifson. 1998. Inactivation of human immunodeficiency virus type 1 infectivity with preservation of conformational and functional integrity of virion surface proteins. J. Virol. 72:7992-8001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sherman, P. A., and J. A. Fyfe. 1990. Human immunodeficiency virus integration protein expressed in Escherichia coli possesses selective DNA cleaving activity. Proc. Natl. Acad. Sci. USA 87:5119-5123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Smith, J. S., S. Y. Kim, and M. J. Roth. 1990. Analysis of long terminal repeat circle junctions of human immunodeficiency virus type 1. J. Virol. 64:6286-6290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.South, T. L., P. R. Blake, R. C. Sowder, L. O. Arthur, L. E. Henderson, and M. F. Summers. 1990. The nucleocapsid protein isolated from HIV-1 particles binds zinc and forms retroviral-type zinc fingers. Biochemistry 29:7786-7789. [DOI] [PubMed] [Google Scholar]
- 52.Suryanarayana, K., T. A. Wiltrout, G. M. Vasquez, V. M. Hirsch, and J. D. Lifson. 1998. Plasma SIV RNA viral load determination by real-time quantification of product generation in reverse transcriptase-polymerase chain reaction. AIDS Res. Hum. Retroviruses 14:183-189. [DOI] [PubMed] [Google Scholar]
- 53.Tanchou, V., D. Decimo, C. Pechoux, D. Lener, V. Rogemond, L. Berthoux, M. Ottmann, and J. L. Darlix. 1998. Role of the N-terminal zinc finger of human immunodeficiency virus type 1 nucleocapsid protein in virus structure and replication. J. Virol. 72:4442-4447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Telesnitsky, A., and S. P. Goff. 1997. Reverse transcription and generation of retroviral DNA, p. 121-160. In J. M. Coffin, S. H. Hughes, and H. E. Varmus (ed.), Retroviruses. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y. [PubMed]
- 55.Tsuchihashi, Z., and P. O. Brown. 1994. DNA strand exchange and selective DNA annealing promoted by the human immunodeficiency virus type 1 nucleocapsid protein. J. Virol. 68:5863-5870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Williams, M. C., R. J. Gorelick, and K. Musier-Forsyth. 2002. Specific zinc- finger architecture required for HIV-1 nucleocapsid protein's nucleic acid chaperone function. Proc. Natl. Acad. Sci. USA 99:8614-8619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wu, T., J. Guo, J. Bess, L. E. Henderson, and J. G. Levin. 1999. Molecular requirements for HIV-1 plus-strand transfer: analysis in reconstituted and endogenous reverse transcription systems. J. Virol. 73:4794-4805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wu, W., L. E. Henderson, T. D. Copeland, R. J. Gorelick, W. J. Bosche, A. Rein, and J. G. Levin. 1996. Human immunodeficiency virus type 1 nucleocapsid protein reduces reverse transcriptase pausing at a secondary structure near the murine leukemia virus polypurine tract. J. Virol. 70:7132-7142. [DOI] [PMC free article] [PubMed] [Google Scholar]