Abstract
Human T-cell leukemia virus (HTLV) envelope (Env) glycoproteins induce fusion, leading to rampant syncytium formation in a broad range of cell lines. Here, we identified murine, hamster, canine, and porcine cell lines that are resistant to HTLV-1 Env-induced syncytium formation. This resistance was not due to the absence of functional receptors for HTLV Env, as these cells were susceptible to infection with HTLV Env-pseudotyped virions. As murine leukemia virus (MLV) Env and HTLV Env present close structural homologies (F. J. Kim, I. Seiliez, C. Denesvre, D. Lavillette, F. L. Cosset, and M. Sitbon, J. Biol. Chem. 275:23417-23420, 2000), and because activation of syncytium formation by MLV Env generally requires cleavage of the R peptide in the cytoplasmic domain of the Env transmembrane (TM) component, we assessed whether truncation of the cytoplasmic domain of HTLV Env would alleviate this resistance. Indeed, in all resistant cell lines, truncation of the last 8 amino acids of the HTLV Env cytoplasmic domain (HdC8) was sufficient to overcome resistance to HTLV Env-induced syncytium formation. Furthermore, HdC8-mediated cell-to-cell infection titers varied according to the target cell lines and could be significantly higher than that observed with HTLV Env on HeLa cells. These data indicate that a determinant located within the 8 carboxy-terminal cytoplasmic amino acids of TM plays a distinct role in HTLV Env-mediated cell-to-cell infection and syncytium formation.
Current understanding of fusion mediated by retroviral envelope (Env) glycoproteins suggests that interaction of the Env surface (SU) component with a receptor(s) leads to conformational changes that unmask fusion determinants of the transmembrane (TM) subunit (29). The TM ectodomain is highly conserved among retroviral Envs, whereas the carboxy-terminal cytoplasmic domain is significantly more variable and has been shown to modulate Env-mediated cell fusion and infection in various retroviruses (2, 10, 11, 13, 20-22, 25, 30), including human T-cell leukemia virus (HTLV) (19).
HTLV infects and induces fusion with rampant syncytium formation in a broad range of cell lines in vitro. However, cell-free virus titers are generally poor, and infection by HTLV occurs mostly by transmission through cell-to-cell contact (7, 18, 26). The factors and mechanisms underlying the apparently paradoxical characteristics of highly fusogenic Env and poorly infectious virions remain unclear. We describe several mammalian cell lines that are resistant to HTLV Env-mediated syncytium formation, including the murine NIH 3T3 cell line lacking thymidine kinase (TK−) originally derived from NIH 3T3 cells (28). Despite the lack of syncytium formation, these syncytium formation-resistant cells were susceptible to HTLV Env-mediated infection. Also, partial truncations in the HTLV Env TM cytoplasmic domain overcame resistance to HTLV Env-mediated syncytium formation in all of the resistant cell lines tested.
MATERIALS AND METHODS
Construction of HTLV Env cytoplasmic tail truncation mutants.
The parental HTLV-1 and Friend murine leukemia virus (MLV) Env expression vectors pCEL/H and pCEL/F were based on the MT2 strain of HTLV-1 (GenBank M37747 and AAA46185) and the 57 strain of Friend MLV (GenBank X02794), respectively. Three allelic restriction sites have been introduced in the two Envs, at the SU/TM cleavage site, the amino terminus of the membrane-spanning domain of the TM, and immediately after the Env stop codon, to facilitate domain swapping (8, 9, 14). HTLV Env lacking the entire cytoplasmic domain, HΔC, and the Friend MLV R peptide truncation mutant pCEL/FΔR have been described previously (9, 14).
HTLV-1 TM-truncated mutants were obtained in pCEL/H after insertion of a TAATATT sequence that combined an in-frame TAA stop codon with an overlapping AATATT SspI site. Stop codons were located immediately after codon 480 (HdC8) and 472 (HdC16). The 167-bp NsiI-PstI cassettes containing the mutation were reintroduced into the parental pCEL/H Env vector and verified by using ABI Prism sequencers.
Cell lines.
We used the following cell lines: NIH 3T3 murine fibroblasts, NIH 3T3(TK−) murine fibroblasts, derived from NIH 3T3 murine fibroblasts after selection with bromodeoxyuridine (28), A23 hamster fibroblasts, XC rat fibroblasts, D17 canine osteosarcoma cells, PK15 porcine kidney fibroblasts, COS African green monkey kidney fibroblasts, HeLa human cervical carcinoma cells, and 293T human fetal kidney cells expressing the simian virus 40 T antigen. The NIH 3T3(Hygr), NIH 3T3(TK−)(Hygr), and HeLa(Hygr) hygromycin B-resistant cell lines used as target cells in the coculture infection assay were generated after stable transfection of the pSV/hygro plasmid and selection in 0.3 mg of hygromycin B (InVitrogen) per ml. All cells were cultured in Dulbecco's modified Eagle's medium (InVitrogen) supplemented with 10% fetal calf serum (Gibco), 2 mM l-glutamine, and penicillin-streptomycin at 37°C and 5% CO2 in a humidified incubator.
Syncytium assay.
Twenty-four hours prior to transfection, 2 × 105 NIH 3T3, NIH 3T3(TK−), and other cells, when indicated, were seeded onto six-well tissue culture plates (Nunc). Approximately 75% confluent monolayers were transfected with 0.1 to 6 μg of pCEL/Env by using Lipofectamine Plus (InVitrogen). Immediately after transfection, cells were detached, reseeded in 12-well or 6-well tissue culture plates, and cultured for an additional 24 to 48 h. Separate duplicate wells were used for immunoblot analyses and for methanol fixation before May-Grunwald-Giemsa staining. When indicated, cocultures were performed as described previously (14), except May-Grunwald-Giemsa staining was performed. Syncytia were quantified on a stereomicroscope from nine randomly selected fields on a premarked grid and are represented as the mean number of syncytia per field (± standard deviation). Mean number of nuclei per syncytium (± standard deviation) was calculated by counting nuclei in five random syncytia in each of the nine fields.
Cell-to-cell infection assay by coculture and hygromycin selection.
Virions consisting of MLV cores pseudotyped with HTLV-1 Env were produced after transfection of 106 293T cells by a modified version of the calcium phosphate method, with 1.5 μg of pCEL/Env, 1.5 μg of pCL/Gag-Pol, and 3 μg of pCLMFG-LacZ, providing a packageable RNA coding for the lacZ gene marker (16). The Moloney MLV-based pCL/Gag-Pol expression vector was derived from pCL-Eco (16) by excising an env gene fragment between the ScaI and ClaI sites.
Following overnight transfection, 2 × 105 293T virion-producing cells were used as infectious centers and cocultivated for 24 to 48 h with either 2 × 105 HeLa(Hygr), NIH 3T3(Hygr), or NIH 3T3(TK−)(Hygr) target cells in six-well tissue culture plates. Most of the 293T virion-producing cells were then detached by several cold washes with a phosphate-buffered saline solution, and the cocultivated cells were subsequently trypsinized and reseeded at 1:4, 1:8, and 1:12 dilutions on new 35-mm culture dishes (Nunc) in medium containing 0.3 mg of hygromycin B (InVitrogen) per ml to eliminate 293T producer cells. Cells were washed, and hygromycin-containing selection medium was changed every 2 days for approximately another 5 to 7 days, until the indicator target cells reached 90 to 100% confluence.
Target cells were fixed with 0.5% (wt/vol) glutaraldehyde in phosphate-buffered saline and stained with a 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) solution to reveal β-galactosidase activity. Blue CFU, resulting from the infection of hygromycin B-resistant target cells with MLV(HTLV-1) virions produced by the 293T cells, were counted, and titers are expressed as CFU per 2 × 105 virion-producing cells, taking into account the respective dilutions of the producer and target cells. Statistical significance of differences was determined by using a pairwise Student's t test. Data are represented as mean interference ± standard deviation.
Envelope expression and incorporation into virions.
Twenty-four hours prior to transfection, 2 × 106 to 5 × 106 293T cells were seeded in 100-mm culture dishes. Transfections were performed as described above with 4 μg of pCL/Gag-Pol, 8 μg of pCLMFG-LacZ, and 4 μg of the following pCEL/Env constructs: the parental HTLV Env (pCEL/H), the HdC8, HdC16 and HΔC TM truncation mutants, or the pcDNA3.1 (InVitrogen) control vector with no env gene. Virion-producing cell extracts were collected 48 h posttransfection in 1 ml of cell lysis buffer (50 mM Tris-HCl [pH 8.0], 150 mM NaCl, 0.1% sodium dodecyl sulfate [SDS], 1.0% Nonidet P-40, 0.5% deoxycholate, and a cocktail of mammalian protease inhibitors [Sigma]) and clarified by two successive centrifugations at 13,000 rpm for 10 min at 4°C in a microcentrifuge.
Approximately 20 μl of each extract, adjusted after normalization for protein concentration by using the Bradford assay (Sigma), were subjected to electrophoresis on SDS-15% acrylamide gels, followed by transfer onto nitrocellulose (Protran; Schleicher & Schuell). Membranes were blocked in phosphate-buffered saline containing 5% powdered milk and 0.5% Tween 20, probed with a 1:100 dilution of the 5a rat anti-HTLV-1 TM monoclonal antibody (3) (a kind gift of C. Carrington and T. Schulz), washed three times with phosphate-buffered saline-0.1% Tween 20, and probed with the corresponding horseradish peroxidase-conjugated anti-rat immunoglobulin before three additional washes and detection by chemiluminescence were performed.
For pelleted virion preparations, HTLV Env-pseudotyped virions [MLV(H)] were generated as described for the cell-to-cell infection assay. Three milliliters of cell culture medium was recovered 48 h posttransfection, filtered through a 0.45-μm-pore-size filter, and ultracentrifuged on a 2-ml 20% sucrose cushion (in Tris [pH 7.4], 100 mM NaCl, and 10 mM EDTA) at 24,000 rpm for 2 h in an SW41 rotor (Beckman). The pellet was lysed in 50 μl of the cell lysis buffer and electrophoresed in SDS-15% polyacrylamide (anti-TM immunoblots) and SDS-12.5% polyacrylamide gels (anti-SU and anti-CA immunoblots) before transfer. Immunoblot analyses were performed as described above, with the additional detection of the MLV capsid (CA) protein with a 1:200 dilution of the rat R187 anti-CA monoclonal antibody (a kind gift of B. Chesebro) as described previously (1). Detection of mature SU and Env precursor was performed with a 1:100 dilution of the 1C11 mouse anti-HTLV-1 SU monoclonal antibody (Epitope).
RESULTS
Resistance of several cell lines to HTLV Env-mediated syncytium formation.
While most vertebrate cell lines are sensitive to HTLV Env syncytium formation, we found several mammalian adherent cell lines, including hamster A23, canine D17, porcine PK15, and murine NIH 3T3(TK−) cells, to be resistant to HTLV Env-mediated syncytium formation. We compared in more detail the NIH 3T3(TK−) cell line with the parental NIH 3T3 cells. The two cell lines were equally sensitive to syncytium formation by the Friend MLV ecotropic Env lacking the R peptide (FΔR) (Fig. 1A). A fusion-active form of the ampho-MLV Env (AΔR) and the vesicular stomatitis virus G glycoprotein also induced equivalent numbers of large syncytia in NIH 3T3(TK−) and NIH 3T3 cells (not shown).
FIG. 1.
Distinct susceptibility of NIH 3T3 and NIH 3T3(TK−) cells to HTLV Env-induced syncytium formation. (A) NIH 3T3 and NIH 3T3(TK−) cells were tested for their susceptibility to HTLV-1 Env (H)-induced syncytium formation. Cells were transfected with up to 6 μg of envelope expression vector. As controls, both cell lines were transfected with vectors encoding either the parental, full-length Friend MLV Env (F) or the fusogenic Friend MLV Env lacking the R peptide (FΔR). Syncytia are observed as dark plaques. (B) NIH 3T3(TK−) cells transfected with the HTLV-1 Env, NIH 3T3(TK−)+H, were tested for their ability to trigger syncytium formation with overlaid HeLa (left panel) and NIH 3T3(TK−) target cells (right panel).
Thus, although NIH 3T3(TK−) cells were not intrinsically resistant to syncytium formation, no syncytia were observed upon transfection with as much as 6 μg of HTLV Env expression vector (Fig. 1A). In contrast, introduction of only 1 μg of the same vector in NIH 3T3 cells resulted in the formation of several hundred clearly detectable syncytia per well (Fig. 1A and Table 1). Since the NIH 3T3(TK−) as well as the nonmurine A23, D17, and PK15 resistant cell lines were sensitive to syncytium formation induced by the AΔR Env and vesicular stomatitis virus G proteins (not shown), resistance of these four cell lines to syncytium formation was specific to the HTLV Env.
TABLE 1.
HTLV Env truncation mutants overcome resistance of NIH 3T3(TK−) cells to HTLV Env-mediated syncytium formationa
| Env | Mean no. of syncytiab ± SD
|
|
|---|---|---|
| NIH 3T3 | NIH 3T3(TK−) | |
| Mock | <1 | <1 |
| H | 276 ± 15 | <1 |
| HdC8 | 457 ± 7 | 83 ± 5 |
| HdC16 | 453 ± 32 | 32 ± 5 |
| HΔC | <1 | <1 |
| F | <1 | <1 |
| FΔR | 650 ± 74 | 678 ± 35 |
NIH 3T3 and NIH 3T3(TK−) cells were transfected with 4 μg of pCEL/Env. Data are from three experiments performed in duplicate and are represented as mean ± standard deviation.
Syncytia containing at least four nuclei were counted in nine fields for each experiment.
We verified that lack of syncytium formation in NIH 3T3(TK−) cells was not due to lower levels of HTLV Env expression (not shown) or to expression of nonfunctional Env, since NIH 3T3(TK−) cells transfected with HTLV Env induced significant syncytium formation with cocultured HeLa cells (Fig. 1B). This coculture led to smaller fusion foci compared to the rampant formation of large syncytia observed with homogeneously susceptible cell cocultures (14). Furthermore, the lack of syncytium formation in NIH 3T3(TK−) cells was not due to the absence of functional HTLV receptors at the surface of these cells, since a soluble form of HTLV Env receptor-binding domain bound NIH 3T3(TK−) cells at least as efficiently as NIH 3T3 cells (F. J. Kim, E. N. Garrido, N. Manel, M. Sitbon, and J.-L. Battini, unpublished data). This was further confirmed by the finding that both cell-free virion titers (not shown) and cell-to-cell infection titers (Table 2) of MLV(H) virion pseudotypes were similar for NIH 3T3 and NIH 3T3(TK−) cells. Similarly, the A23, D17, and PK15 syncytium-resistant cell lines were readily susceptible to infection by MLV(H) virion pseudotypes (not shown).
TABLE 2.
Cell-to-cell transmission of HTLV-1 Env-pseudotyped virions into NIH 3T3, NIH 3T3(TK−), and HeLa target cellsa
| Pseudotyping Envb | No. of CFU in hygromycin-resistant target cells
|
||
|---|---|---|---|
| NIH 3T3 | NIH 3T3(TK−) | HeLa | |
| None | <10 | <10 | <10 |
| H | 784-1,082 | 608-888 | 2,720-7,080 |
| HdC8 | 353-576 | 292-384 | 6,752-13,128 |
| HdC16 | 304-360 | 128-196 | 2,368-7,680 |
| HΔC | <10 | <10 | <10 |
Cell-to-cell transmission was evaluated by the number of LacZ-positive blue CFU of the specified hygromycin-resistant target cells obtained after coculture with 2 × 105 293T cells producing Env-pseudotyped lacZ virions. Data are presented as the range of titers obtained in three independent experiments.
Pseudotyped MLV cores were produced by 293T cells transiently transfected with control plasmid DNA (none) or with parental (H) or HdC8, HdC16, or HΔC HTLV-derived Env truncation mutants (diagrammed in Fig. 2A).
Partial truncation of the HTLV Env cytoplasmic tail allows syncytium formation in resistant cell lines. Since HTLV and MLV Env have a common SU modular organization (14; F. J. Kim, E. N. Garrido, N. Manel, M. Sitbon, and J.-L. Battini, unpublished data), and since the carboxy-terminal R peptide of the MLV TM cytoplasmic tail inhibits syncytium formation (20, 21), we evaluated whether partial cleavage of the HTLV Env cytoplasmic domain would reveal an effect similar to that observed after cleavage of the MLV R peptide. Because no consensus R peptide cleavage sequence could be identified in the HTLV TM, the HdC8 and HdC16 cytoplasmic domain truncation mutants were empirically generated by eliminating the carboxy-terminal 8 and 16 amino acids, respectively, of the cytoplasmic domain, immediately carboxy terminal of serines 480 and 472 of the parental HTLV Env (Fig. 2A).
FIG. 2.
Truncation of HTLV-1 Env TM induces syncytium formation in NIH 3T3(TK−) cells. (A) Schematic representation of HTLV Env. The SU and TM subunits are indicated. The HTLV-1 Env signal peptide, fusion peptide, and TM membrane anchor are shown as dotted boxes, from the amino terminus to the carboxy terminus, respectively. Amino acid residues of the membrane anchor (anchor) and cytoplasmic domain (CD) are shown, and the position of the last carboxy-terminal residue of each mutant is indicated with an arrowhead. Amino acid residue numbering starts from the first signal peptide methionine of the HTLV-1 Env precursor. (B) Parental HTLV-1 Env (H) and truncation mutants lacking the 8 (HdC8) or 16 (HdC16) carboxy-terminal amino acids of the TM were tested for their ability to form syncytia after transfection in NIH 3T3 or NIH 3T3(TK−) cells. Open arrows in the panels on the right point to syncytia.
Both TM truncations led to significantly increased numbers of syncytia in NIH 3T3 cells. Furthermore, transfection of HdC8 and HdC16 resulted in the formation of syncytia in the initially resistant NIH 3T3(TK−) cells (Fig. 2B), albeit to a lesser extent than observed in parental NIH 3T3 cells (Table 1). Despite the more compact appearance of the NIH 3T3(TK−) syncytia (Fig. 2B), no significant differences were observed in the number of nuclei per NIH 3T3 and NIH 3T3(TK−) syncytium formed by the parental, HdC8, and HdC16 HTLV Envs. In all cases, the mean number of nuclei per syncytium ranged from 11 to 18 (± 6 to 10). As expected from a previous study using NIH 3T3 cells (9), the HΔC mutant, which lacked the entire cytoplasmic domain of the TM, was also unable to induce syncytium formation in NIH 3T3(TK−) cells. The ability of the HdC8 and HdC16 truncation mutants to induce syncytium formation was also observed in the resistant A23, D17, and PK15 cell lines (not shown).
Cell-to-cell transmission of MLV virions pseudotyped with parental and truncated HTLV Env.
Infection by HTLV or HTLV Env-pseudotyped virions has been shown to be significantly more efficient upon cell-to-cell contact than via cell-free viral supernatants (7, 26). We therefore assayed transmission of MLV virions pseudotyped with either the parental HTLV-1 Env, MLV(H), the HdC8 mutant MLV(HdC8), or the HdC16 mutant MLV(HdC16), using a cell-to-cell transmission assay. Infection of hygromycin B-resistant target cells was assessed following coculture with pseudotyped virion-producing 293T cells. Input 293T cells were progressively eliminated by hygromycin treatment, and CFU were quantified in the resistant target cells. Although slightly more efficient on NIH 3T3(Hygr) cells than on NIH 3T3(TK−)(Hygr) target cells, cell-to-cell transmission of MLV(H) virions was similar (P > 0.15) in the two cell types, with titers ranging from 784 to 1,082 CFU (mean ± standard deviation, 947 ± 151 CFU) and 608 to 888 CFU (mean, 712 ± 153 CFU) per 2 × 105 virion-producing cells, respectively (Table 2). This similar susceptibility to infection was in striking contrast to the aforementioned distinct susceptibilities of the two cell lines to HTLV Env-mediated syncytium formation (Fig. 1 and Table 1).
Cell-to-cell transmission was undetectable with the HΔC pseudotypes, and despite their increased fusogenic properties, the partially truncated HdC8 and HdC16 TM mutants demonstrated decreased cell-to-cell transmission compared to parental HTLV Env-pseudotyped virions (Table 2). Lower infectious titers of the HdC8 and HdC16 Env mutants were observed on both NIH 3T3 and NIH 3T3(TK−) target cells, with titers ranging between 353 and 576 CFU (mean, 448 ± 115 CFU) and 304 and 360 CFU (mean, 326 ± 30 CFU) in NIH 3T3 cells and between 292 and 384 CFU (mean, 345 ± 48 CFU) and 128 and 196 CFU (mean, 164 ± 34 CFU) in NIH 3T3(TK−) cells, respectively (P < 0.03 in all cases, compared to parental HTLV) (Table 2).
Coculture controls, as described elsewhere (F. J. Kim, E. N. Garrido, N. Manel, M. Sitbon, and J.-L. Battini, unpublished data), were performed in parallel and confirmed that LacZ-positive blue colonies were indeed due to infection by MLV(Env) virions and not to either Env-induced cell-to-cell fusion or spontaneous transduction of target cells with the pCLMFG-LacZ vector. Blue colonies above background were not observed in the absence of virions or Env. Furthermore, no blue colonies were observed in control experiments where human HeLa target cells were cocultured with virions pseudotyped with ecotropic MLV Env (not shown).
Interestingly, when HeLa(Hygr) cells were used as target cells in the cell-to-cell virion transmission assay, the highest titers were reproducibly obtained with MLV(HdC8) virion pseudotypes, with titers (per 2 × 105 virion-producing cells) ranging from 6,752 to 13,128 CFU, versus 2,720 to 7,080 and 2,368 to 7,680 CFU with MLV(H) and MLV(HdC16) virion pseudotypes, respectively (P < 0.02 compared to parental HTLV and HdC16) (Table 2). As these data are in contrast to those observed with NIH 3T3 and NIH 3T3(TK−) cells, it appears that distinct properties of target cells modulate the impact of the TM cytoplasmic domain truncation on cell-to-cell infection.
Expression of parental and truncated HTLV Env and their incorporation into viral particles.
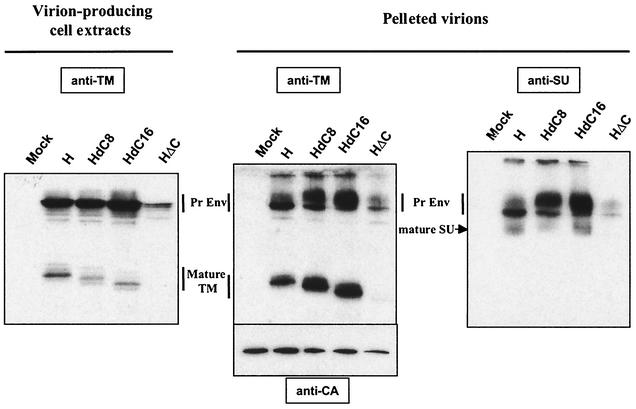
To assess whether infection with the various HTLV Env-pseudotyped virions was modulated by distinct incorporation of the mutant Env into virions, the amount of H, HdC8, and HdC16 present in the MLV particles was monitored by immunoblotting with an anti-TM monoclonal antibody. This allowed detection of incorporated Env irrespective of the level of SU shedding. Under these conditions, we detected significant amounts of the mature parental and truncated TM incorporated in all virion preparations with the exception of the HΔC TM mutant (Fig. 3, center panel), which appeared to be expressed at a lower level than the other Env constructs (Fig. 3, left panel). Accumulation of slower-migrating uncleaved precursors in all virion preparations was assessed by dual anti-TM and anti-SU recognition. Whether these forms were cell membrane- or virion-associated Env precursor intermediates remains unclear. It is noteworthy that low levels of cleaved SU were detected in all virion preparations (Fig. 3, right panel), indicative of a labile SU/TM association which may account for the low cell-free infectivity of HTLV Env-harboring particles.
FIG. 3.
Expression of HTLV-1 Env TM truncation mutants and their incorporation into virions. Equal amounts of cell extracts were obtained from 2 × 106 to 5 × 106 293T cells cotransfected with 8 μg of an MLV-based lacZ reporter gene vector, 4 μg of the MLV p/CLGag-Pol expression vector, and 4 μg of either an HTLV Env-derived expression vector (H, HdC8, HdC16, or HΔC) or a control vector (Mock). The different HTLV Env TM mutants are diagrammed in Fig. 2. Cell extracts were immunoblotted with the 5a anti-HTLV TM monoclonal antibody (3) (left panel). Pelleted virions were obtained from supernatants of 293T transfectants after concentration by ultracentrifugation on a 20% sucrose cushion. Resulting pellets were immunoblotted with either the 5a anti-HTLV TM monoclonal antibody (anti-TM, center panel) or the 1C11 anti-HTLV SU monoclonal antibody (anti-SU, right panel). Respective levels of virion expression were controlled by immunoblotting with the R187 anti-MLV capsid monoclonal antibody (a kind gift of B. Chesebro) (anti-CA, center panel). The positions of the HTLV-1 Env precursors (Pr Env), mature TM, and mature SU envelope proteins are indicated.
DISCUSSION
The HTLV Env induces the formation of syncytia and mediates infection of a broad range of vertebrate cell lines (26, 27), including murine NIH 3T3 cells (9, 26). Here, we report several mammalian adherent cell lines that are specifically resistant to HTLV Env-mediated syncytium formation. This resistance was overcome by partial truncation of the TM cytoplasmic domain carboxy terminus (Fig. 2). Cytoplasmic domain truncation did not lead to increased production of mature Env in the resistant NIH 3T3(TK−) cell line, as monitored with anti-TM antibody (not shown). Among other mechanisms, the TM carboxy-terminal amino acids may influence the kinetics of Env maturation, cell membrane interactions, and stability of the SU-TM association.
We have previously proposed that lability of the SU-TM association might explain the increased fusion ability of proline-rich region mutants of the MLV Env SU (15). A weaker SU-TM association would also favor infection by cell-to-cell transmission due to the continuous production of enveloped virions in close proximity to target cells, whereas infection by cell-free virions lacking SU would be considerably reduced. Truncation of the MLV R peptide has been shown to be essential for MLV Env function (20, 21). However, it remains to be determined whether equivalent cleavage occurs in bona fide HTLV virions. Indeed, in the absence of a defined R peptide consensus cleavage site, our truncation mutations were introduced at arbitrary positions in the HTLV Env cytoplasmic domain.
Among the syncytium-resistant cell lines, we extensively studied the NIH 3T3(TK−) cell line, which was selected for resistance to bromodeoxyuridine and lack of thymidine kinase activity (28). However, neither bromodeoxyuridine resistance nor lack of TK activity appeared to directly correlate with resistance to HTLV Env-mediated syncytium formation. Indeed, other bromodeoxyuridine-resistant cell lines recently derived in our laboratory to address this point were readily susceptible to HTLV Env-induced syncytium formation (not shown). Furthermore, introduction of the herpes simplex virus type 1 tk gene in NIH 3T3(TK−) cells did not lead to HTLV Env syncytium susceptibility (not shown).
Resistance to syncytium formation appeared to be limited to HTLV Env, since NIH 3T3(TK−) (Fig. 1) and the other resistant cell lines were fully susceptible to several other fusogenic Envs, such as FΔR (Fig. 1), AΔR, and the vesicular stomatitis virus G glycoprotein (not shown). Also, resistance was due neither to lower expression levels of mature Env by NIH 3T3(TK−) cells (not shown) nor to a lack of functional HTLV Env expression in these cells, as coculture with HeLa or NIH 3T3 target cells resulted in readily detectable syncytium formation (Fig. 1B and data not shown). Furthermore, resistance to syncytium formation was not due to the absence of HTLV receptors, as NIH 3T3(TK−) cells bound a soluble form of the HTLV Env receptor-binding domain (F. J. Kim, E. N. Garrido, N. Manel, M. Sitbon, and J.-L. Battini, unpublished data) and were infected by MLV(H) virions to levels similar to those observed with NIH 3T3 cells (Table 2).
The two HTLV Env truncation mutants that overcame resistance to syncytium formation were infectious for all cell lines, as determined by our cell-to-cell transmission assay with MLV-pseudotyped virions (Table 2 and data not shown). It is noteworthy that both mutations resulted in either partial (HdC8) or total (HdC16) deletion of the YSLI motif, which has been described previously as being required for cell-to-cell transmission of HTLV virions (6). This apparent discrepancy between our results and the reported importance of the YSLI motif in cell-to-cell transmission might be due to different experimental parameters, including assay read-outs, use of MLV versus HTLV cores, and the precise location of the TM truncations.
The HTLV TM cytoplasmic domain truncation mutants described here had distinct effects on cell-to-cell infection of NIH and HeLa target cells. In HeLa target cells, MLV(HdC8) but not MLV(HdC16) virion pseudotypes demonstrated consistently increased coculture infection titers compared to parental HTLV Env (Table 2). In contrast, virions that were pseudotyped with either mutant had decreased titers in NIH 3T3 cells. These results most likely illustrate the fact that cell membrane components, including adhesion molecules (4, 5, 12), cytoskeleton-associated proteins (24), lipids (23), and lipid raft components (17), have distinct influences on HTLV Env-mediated cell-to-cell fusion and virus transmission.
Our observations that NIH 3T3(TK−) cells are resistant to HTLV Env-mediated syncytium formation but not to infection reinforces the hypothesis that different factors are involved in these two processes. In this regard, it is interesting that, as monitored by time-lapse video microscopy, NIH 3T3(TK−) cells were readily distinguishable from standard NIH 3T3 cells in at least two aspects: (i) the NIH 3T3(TK−) cells were considerably more mobile in culture and (ii) they exhibited a relatively compact morphology, with more compact lammellipodia and increased filopodium formation (not shown). Our data suggest that syncytium formation is not strictly dependent upon receptor availability, but is likely to involve post-receptor-binding membrane dynamics. We showed that a determinant located within the 8 carboxy-terminal residues of the TM cytoplasmic domain plays a major role in these dynamics.
Acknowledgments
We are indebted to N. Taylor for helpful discussion and critical reading of the manuscript, to C. Denesvre for continued input, to S. Kinet for help with FACS analyses, to Robert Naviaux for the pCL-Eco and pMFG-LacZ plasmids, to Françoise Carbonell for technical assistance, and to all the members of our lab for insightful discussions.
F.J.K. was supported by an award from the Philippe Foundation and successive graduate student fellowships from the Agence Nationale pour la Recherche sur le SIDA (ANRS) and the Association pour la Recherche contre le Cancer (ARC). N.M. is a fellow of the Ecole Normale Supérieure (Lyon, France). Y.B. was supported by a fellowship from ANRS. J.-L.B. and M.S. are supported by the Institut National de la Santé et de la Recherche Médicale (INSERM). This work was supported by grants from ARC (ARC No. 5989) and the Association Française contre les Myopathies (AFM No. 7706) to M.S.
REFERENCES
- 1.Audit, M., J. Dejardin, B. Hohl, C. Sidobre, T. J. Hope, M. Mougel, and M. Sitbon. 1999. Introduction of a cis-acting mutation in the capsid-coding gene of Moloney murine leukemia virus extends its leukemogenic properties. J. Virol. 73:10472-10479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brody, B. A., and E. Hunter. 1994. Postassembly cleavage of a retroviral glycoprotein cytoplasmic domain removes a necessary incorporation signal and activates fusion activity. J. Virol. 68:4620-4627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Carrington, C. V., N. Paul, J. Cordell, and T. F. Schulz. 1996. Probing the conformation of the human T-lymphotropic virus I envelope protein complex with monoclonal antibodies. J. Gen. Virol. 77:2025-2029. [DOI] [PubMed] [Google Scholar]
- 4.Daenke, S., and S. Booth. 2000. Molecular mechanisms affecting HTLV type 1-dependent fusion at the cell membrane: implications for inhibiting viral transmission. AIDS Res. Hum. Retroviruses. 16:1731-1736. [DOI] [PubMed] [Google Scholar]
- 5.Daenke, S., S. A. McCracken, and S. Booth. 1999. Human T-cell leukaemia/lymphoma virus type 1 syncytium formation is regulated in a cell-specific manner by ICAM-1, ICAM-3 and VCAM-1 and can be inhibited by antibodies to integrin beta2 or beta7. J. Gen. Virol. 80:1429-1436. [DOI] [PubMed] [Google Scholar]
- 6.Delamarre, L., C. Pique, A. R. Rosenberg, V. Blot, M. P. Grange, I. Le Blanc, and M. C. Dokhelar. 1999. The Y-S-L-I tyrosine-based motif in the cytoplasmic domain of the human T-cell leukemia virus type 1 envelope is essential for cell-to-cell transmission. J. Virol. 73:9659-9663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Delamarre, L., A. R. Rosenberg, C. Pique, D. Pham, and M. C. Dokhelar. 1997. A novel human T-leukemia virus type 1 cell-to-cell transmission assay permits definition of SU glycoprotein amino acids important for infectivity. J. Virol. 71:259-266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Denesvre, C., C. Carrington, A. Corbin, Y. Takeuchi, F. L. Cosset, T. Schulz, M. Sitbon, and P. Sonigo. 1996. TM domain swapping of murine leukemia virus and human T-cell leukemia virus envelopes confers different infectious abilities despite similar incorporation into virions. J. Virol. 70:4380-4386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Denesvre, C., P. Sonigo, A. Corbin, H. Ellerbrok, and M. Sitbon. 1995. Influence of transmembrane domains on the fusogenic abilities of human and murine leukemia retrovirus envelopes. J. Virol. 69:4149-4157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dubay, J. W., S. J. Roberts, B. H. Hahn, and E. Hunter. 1992. Truncation of the human immunodeficiency virus type 1 transmembrane glycoprotein cytoplasmic domain blocks virus infectivity. J. Virol. 66:6616-6625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gabuzda, D. H., A. Lever, E. Terwilliger, and J. Sodroski. 1992. Effects of deletions in the cytoplasmic domain on biological functions of human immunodeficiency virus type 1 envelope glycoproteins. J. Virol. 66:3306-3315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hildreth, J. E., A. Subramanium, and R. A. Hampton. 1997. Human T-cell lymphotropic virus type 1 (HTLV-1)-induced syncytium formation mediated by vascular cell adhesion molecule-1: evidence for involvement of cell adhesion molecules in HTLV-1 biology. J. Virol. 71:1173-1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Johnston, P. B., J. W. Dubay, and E. Hunter. 1993. Truncations of the simian immunodeficiency virus transmembrane protein confer expanded virus host range by removing a block to virus entry into cells. J. Virol. 67:3077-3086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim, F. J., I. Seiliez, C. Denesvre, D. Lavillette, F. L. Cosset, and M. Sitbon. 2000. Definition of an amino-terminal domain of the human T-cell leukemia virus type 1 envelope surface unit that extends the fusogenic range of an ecotropic murine leukemia virus. J. Biol. Chem. 275:23417-23420. [DOI] [PubMed] [Google Scholar]
- 15.Lavillette, D., M. Maurice, C. Roche, S. J. Russell, M. Sitbon, and F. L. Cosset. 1998. A proline-rich motif downstream of the receptor binding domain modulates conformation and fusogenicity of murine retroviral envelopes. J. Virol. 72:9955-9965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Naviaux, R. K., E. Costanzi, M. Haas, and I. M. Verma. 1996. The pCL vector system: rapid production of helper-free, high-titer recombinant retroviruses. J. Virol. 70:5701-5705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Niyogi, K., and J. E. Hildreth. 2001. Characterization of new syncytium-inhibiting monoclonal antibodies implicates lipid rafts in human T-cell leukemia virus type 1 syncytium formation. J. Virol. 75:7351-7361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Okochi, K., H. Sato, and Y. Hinuma. 1984. A retrospective study on transmission of adult T cell leukemia virus by blood transfusion: seroconversion in recipients. Vox Sang. 46:245-253. [DOI] [PubMed] [Google Scholar]
- 19.Pique, C., D. Pham, T. Tursz, and M. C. Dokhelar. 1993. The cytoplasmic domain of the human T-cell leukemia virus type I envelope can modulate envelope functions in a cell type-dependent manner. J. Virol. 67:557-561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ragheb, J. A., and W. F. Anderson. 1994. pH-independent murine leukemia virus ecotropic envelope-mediated cell fusion: implications for the role of the R peptide and p12E TM in viral entry. J. Virol. 68:3220-3231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rein, A., J. Mirro, J. G. Haynes, S. M. Ernst, and K. Nagashima. 1994. Function of the cytoplasmic domain of a retroviral transmembrane protein: p15E-p2E cleavage activates the membrane fusion capability of the murine leukemia virus Env protein. J. Virol. 68:1773-1781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rice, N. R., L. E. Henderson, R. C. Sowder, T. D. Copeland, S. Oroszlan, and J. F. Edwards. 1990. Synthesis and processing of the transmembrane envelope protein of equine infectious anemia virus. J. Virol. 64:3770-3778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sagara, Y., Y. Inoue, E. Kojima, C. Ishida, H. Shiraki, and Y. Maeda. 2001. Phosphatidylglycerol participates in syncytium formation induced by HTLV type 1-bearing cells. AIDS Res. Hum. Retroviruses 17:125-135. [DOI] [PubMed] [Google Scholar]
- 24.Sagara, Y., C. Ishida, Y. Inoue, H. Shiraki, and Y. Maeda. 1998. 71-kilodalton heat shock cognate protein acts as a cellular receptor for syncytium formation induced by human T-cell lymphotropic virus type 1. J. Virol. 72:535-541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Spies, C. P., G. D. Ritter, Jr., M. J. Mulligan, and R. W. Compans. 1994. Truncation of the cytoplasmic domain of the simian immunodeficiency virus envelope glycoprotein alters the conformation of the external domain. J. Virol. 68:585-591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sutton, R. E., and D. R. Littman. 1996. Broad host range of human T-cell leukemia virus type 1 demonstrated with an improved pseudotyping system. J. Virol. 70:7322-7326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Trejo, S. R., and L. Ratner. 2000. The HTLV receptor is a widely expressed protein. Virology 268:41-48. [DOI] [PubMed] [Google Scholar]
- 28.Wei, C. M., M. Gibson, P. G. Spear, and E. M. Scolnick. 1981. Construction and isolation of a transmissible retrovirus containing the src gene of Harvey murine sarcoma virus and the thymidine kinase gene of herpes simplex virus type 1. J. Virol. 39:935-944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Weissenhorn, W., A. Dessen, L. J. Calder, S. C. Harrison, J. J. Skehel, and D. C. Wiley. 1999. Structural basis for membrane fusion by enveloped viruses. Mol. Membr. Biol. 16:3-9. [DOI] [PubMed] [Google Scholar]
- 30.Zingler, K., and D. R. Littman. 1993. Truncation of the cytoplasmic domain of the simian immunodeficiency virus envelope glycoprotein increases env incorporation into particles and fusogenicity and infectivity. J. Virol. 67:2824-2831. [DOI] [PMC free article] [PubMed] [Google Scholar]