Abstract
Kaposi's sarcoma-associated herpesvirus (KSHV) is a human gammaherpesvirus that has been implicated in the pathogenesis of Kaposi's sarcoma and B-cell neoplasms. The genomic organization of KSHV is similar to that of Epstein-Barr virus (EBV). EBV encodes two transcriptional factors, Rta and Zta, which functionally interact to transactivate EBV genes during replication and reactivation from latency. KSHV encodes a basic leucine zipper protein (K-bZIP), a homologue of EBV Zta, and K-Rta, the homologue of EBV Rta. EBV Rta and Zta are strong transcriptional transactivators. Although there is ample evidence that K-Rta is a potent transactivator, the role of K-bZIP as a transcriptional factor is much less clear. In this study, we report that K-bZIP modulates K-Rta function. We show that K-bZIP directly interacts with K-Rta in vivo and in vitro. This association is specific, requiring the basic domain (amino acids 122 to 189) of K-bZIP and a specific region (amino acids 499 to 550) of K-Rta, and can be detected with K-bZIP and K-Rta endogenously expressed in BCBL-1 cells treated with tetradecanoyl phorbol acetate. The functional relevance of this association was revealed by the observation that K-bZIP represses the transactivation of the ORF57 promoter by K-Rta in a dose-dependent manner. K-bZIP lacking the interaction domain fails to repress K-Rta-mediated transactivation; this finding attests to the specificity of the repression. Interestingly, this repression is not observed for the promoter of polyadenylated nuclear (PAN) RNA, another target of K-Rta; thus, repression is promoter dependent. Finally, we provide evidence that the modulation of K-Rta by K-bZIP also occurs in vivo during reactivation of the viral genome in BCBL-1 cells. When K-bZIP is overexpressed in BCBL-1 cells, the level of expression of ORF57 but not PAN RNA is repressed. These data support the model that one function of K-bZIP is to modulate the activity of the transcriptional transactivator K-Rta.
Kaposi's sarcoma (KS)-associated herpesvirus (KSHV), also known as human herpesvirus 8, is a member of the gammaherpesvirus family, which includes Epstein-Barr virus (EBV) and herpesvirus saimiri. KSHV infection is associated with all types of KS, including AIDS-associated KS, endemic forms of KS, and renal transplant-related KS (1, 2, 13, 24, 25, 35). It has also been implicated in B-cell lymphoproliferative diseases, such as primary effusion lymphoma (PEL) and multicentric Castleman's disease (3, 34). The KSHV genome exists in virtually all tumor samples from KS cases (6) as well as in peripheral blood mononuclear cells in up to 50% of homosexual AIDS patients with KS (38). Most of the spindle cells in KS tumors are latently infected with KSHV, implicating the direct involvement of latent genes in the transformation of KS cells. However, some of the cells in KS lesions also express markers for lytic replication (35), and several lines of evidence (e.g., reference 23) suggest that KSHV lytic replication also contributes significantly to the formation of KS lesions by either facilitating viral spread to the target site or releasing paracrine factors to support the growth of neighboring KS tumor cells. Similarly, the KSHV genome is frequently detected in human immunodeficiency virus-associated PEL biopsies, and all cell lines derived from PEL harbor KSHV with various degrees of expression of latent and lytic genes (15, 35). It thus appears that both classes of genes contribute to oncogenic processes and that the transition from latent to lytic infection is a dynamic process controlled by both exogenous factors and viral genes.
The switch from latent to lytic infection has been studied for several gammaherpesviruses; the viral gene(s) responsible for reactivation has been identified in some of these viruses (8, 9, 26). Notable examples are the Zta (also known as BZLF1 or ZEBRA) and Rta (also known as BRLF1) genes of EBV (9, 19). The expression of either the Zta or the Rta gene is sufficient to reactivate the EBV genome in latently infected cells (9, 29, 40). These two genes, both encoding transcriptional factors, have the ability to individually or synergistically regulate the expression of a host of other viral genes, leading to the lytic-cycle cascade. The Rta and Zta loci are linked in the EBV genome, and both are expressed as immediate-early genes following tetradecanoyl phorbol acetate (TPA)-induced reactivation of latently infected cell lines.
Structural and positional analogues of the EBV Rta and Zta genes have been identified in the KSHV genome. ORF50 (or K-Rta) has been shown to be the homologue of EBV Rta (22, 31, 36). Various investigators (20, 41) have independently identified a basic leucine zipper protein of KSHV (K-bZIP, or K8) as a homologue of EBV Zta. Studies by a number of laboratories have shown that K-Rta is sufficient to induce lytic reactivation of the latent KSHV genome in the BCBL-1 cell line model for PEL (22, 36). K-Rta is a potent transcriptional factor with an N-terminal putative DNA-binding domain and a carboxyl-terminal transactivation domain (21). A number of KSHV promoters activated by K-Rta have been identified. These include promoters for ORF57, K-bZIP, MIP, polyadenylated nuclear (PAN) RNA, thymidine kinase (TK), kaposin, and viral interferon regulatory factor (7, 21, 33, 37). K-Rta appears to bind DNA directly; at least two Rta response elements (RRE) have been identified, respectively, on the ORF57 (21) and PAN RNA (5, 33) promoters. Considering the large number of target genes of K-Rta, it is likely that the binding repertoire of K-Rta increases to include sites anchored by transcriptional factors associated with K-Rta. The K-Rta gene is among the first genes to be induced after treatment with stimuli that disrupt latency (TPA, butyrate, and so forth); its expression follows the kinetics of an immediate-early gene, with transcripts emerging within 2 h after induction, accumulating to a peak level at about 48 h, and declining thereafter. This regulation presumably involves a number of viral and cellular factors, among which is K-Rta itself (32).
While the role of K-Rta in reactivation and lytic replication has been generally established, the function of K-bZIP remains elusive. It was shown previously that the K-bZIP gene is expressed as an early lytic gene (20), and Katano et al. (15) demonstrated its localization in both the nucleus and the cytoplasm. K-bZIP contains at its C terminus a basic leucine zipper domain, a motif known to be involved in dimerization and DNA binding. At present, however, neither its DNA-binding nor its transactivation activities have been confirmed. With a transient overexpression system, K-bZIP was found to associate with the cellular factors p53 and CBP and to repress the transactivation potential of these molecules (14, 27). Attempts to overexpress K-bZIP in the PEL cell line to disrupt KSHV latency have thus far met with negative results (28). Thus, the role of K-bZIP in viral replication and reactivation remains largely unknown.
In this study, we report that one of the potential functions of K-bZIP is to modulate the transactivation potential of K-Rta. We show here that K-bZIP directly associates with and colocalizes with K-Rta in naturally infected BCBL-1 cells. K-bZIP represses the transactivation potential of K-Rta, interestingly, in a promoter-dependent manner. This finding was demonstrated not only in a cotransfection setting by using promoter-reporter constructs but also by overexpressing K-bZIP in the BCBL-1 cell line reactivation model. To our knowledge, this is the first example of K-bZIP participation in the regulation of KSHV genes; this finding provides a framework for understanding the role of K-bZIP in KSHV replication and reactivation.
MATERIALS AND METHODS
Cell cultures.
BCBL-1 cells (30) and GA-10 cells (ATCC CRL-2392) were grown at 37°C in RPMI 1640 supplemented with 15% fetal calf serum in the presence of 5% CO2. To establish stable BCBL-1 cells expressing hemagglutinin (HA)-K-bZIP, cells were transfected with 20 μg of HA-K-bZIP-expressing plasmid pcDNA-HA-K-bZIP, which contains a neomycin resistance gene. Stable HA-K-bZIP-expressing cells were selected in RPMI 1640 supplemented with 15% fetal calf serum and containing 400 μg of G418 (Gibco BRL)/ml in the presence of 5% CO2. When necessary, nonviable cells were removed with Ficoll (Amersham Pharmacia) by centrifugation at 400 × g for 15 min at room temperature (RT), and cells were seeded at a small scale to maintain cell density (>5 × 104 cells/ml). Virus lytic replication was induced by treatment of log-phase cells with TPA (20 ng/ml). 293 or 293T cells were maintained in Dulbecco's modified Eagle medium supplemented with 10% fetal calf serum in the presence of 5% CO2. Transfection of plasmids into 293 or 293T cells was performed by using FuGene 6 according to the recommendations of the supplier (Roche Molecular Biochemicals). At 48 h after transfection of or TPA-induced virus replication in BCBL-1 cells, protein lysates were prepared in lysis buffer (EBC buffer; 50 mM Tris-HCl [pH 7.5], 120 mM NaCl, 0.5% NP-40, 50 mM NaF, 200 μM Na2VO4, 1 mM phenylmethylsulfonyl fluoride).
Antibodies.
Anti-K-bZIP antibody was a generous gift from M.-T. Li (Harvard Medical School). Anti-HA antibody (Babco), anti-T7 tag antibody (Novagen), anti-Flag antibody (Upstate), anti-promyelocytic leukemia protein (PML) mouse monoclonal antibody (PG-M3; Santa Cruz), and antiactin goat polyclonal antibody (sc-1615; Santa Cruz) were commercially prepared. Anti-K-Rta chicken immunoglobulin Y (IgY) was raised against a peptide spanning amino acids 516 to 530 (KRKQRSKERSSKKRK) and used for immunoprecipitation, immunoblotting, and immunofluorescence analyses. Anti-Flag M2 affinity gel (Sigma) was used for the immunoprecipitation of Flag-tagged proteins.
Plasmids.
Plasmids encoding full-length K-bZIP (K-bZIP/wt; residues 1 to 273) were constructed by using pcDNA3.1 (Invitrogen). Cloning introduced a CpoI site and either an HA tag, a Flag tag, or a T7 tag at the N terminus as described previously (20). The resulting plasmids were denoted pHA-K-bZIP, pFlag-K-bZIP, and pT7-K-bZIP, respectively. Deletion fragments of both K-bZIP and K-Rta were amplified by PCR with Pfu-Turbo (Strategene) and with the primers listed in Table 1 and then were cloned into pcDNA3.1 or pGEX4T-2 (Amersham Pharmacia) to produce glutathione S-transferase (GST) fusion proteins. An expression plasmid with a deletion of the K-bZIP basic region (positions 123 to 189) (K-bZIPΔBR) was constructed as follows. First, the K-bZIP transactivation domain and leucine zipper domain were independently amplified by PCR with primer pairs (K-bZIP NF-K-bZIP TA-R and K-bZIP LZ-F-K-bZIP CR) (Table 1). These primers introduced an overlapping region that allowed the resulting products to be mixed together and then amplified with primers K-bZIP NF and K-bZIP CR to produce a K-bZIP coding region lacking the basic region. Amplified fragments were digested with CpoI (Fermentas) and then inserted in frame 3′ of the Flag tag sequence of pcDNA-Flag after purification of both the fragments and pcDNA-Flag from an agarose gel. The resulting plasmid was denoted pFlag-K-bZIPΔBR.
TABLE 1.
Primers used for PCR amplification
| Primer | Sequence (5′→3′)a |
|---|---|
| K-bZIP NF | TAcggtccgaccATGCCCAGAATGAAGGA CATACC |
| K-bZIP 120F | CGggatccCTTCCAACTCGCAGATCCAA GAGGCGA |
| K-bZIP 122F | AAcggtccgACTCGCAGATCCAAGAGG CGA |
| K-bZIP 190F | AAcggtccgGCATTAGAAGAAAAGGATG CACAA |
| K-bZIP 122R | ATcggaccgAAGCTGTTGCGAAATGTGT GGTCC |
| K-bZIP 189R | AAcggaccgCTGCTGCAGCTGTCTTGTGTA |
| K-bZIP CR | ATcggaccgCAATAAACCCACAGCCCAT AG |
| K-bZIP TA-R | ctaatgcAAGCTGTTGCGAAATGTGTGG |
| K-bZIP LZ-F | acagcttGCATTAGAAGAAAAGGATGC ACAAC |
| K-Rta N-Cpo | TAcggtccgACCATGGCGCAAGATGACAA |
| K-Rta 110F | TAcggtccgATTATTCGGATCCTCACGG AG |
| K-Rta 239F | TAcggtccgATTACCACCGGCAAGGTCAC |
| K-Rta 299F | TAcggtccgAAACCCCATCCCAACATG |
| K-Rta 499F | TAcggtccgTGTAGAGATTCAACGGC |
| K-Rta 550F | TAcggtccgTTGGGATCAATTACCACCC |
| K-Rta 111R | TAcggaccgAATAATGCCTTGGGATGCCTC |
| K-Rta 240R | TAcggaccgGTAATTGGCCGGCGTTTCT |
| K-Rta 297R | TAcggaccgCTAATGACAAACTGGCTCA GG |
| K-Rta 503R | TAcggaccgTTGAATCTCTACACGGCAC AC |
| K-Rta 550R | TAcggaccgCAAAGAGGTACCAGGTGTC GT |
| K-Rta CR | TAcggaccgTCAGTCTCGGAAGTAATTAC GC |
| ORF57 promoter F | GAAgagctcCAAGACCATTAGCTATCTG CC |
| ORF57 promoter R | GAtagctaGCCTATTTTGGGAACCTGGC AG |
| PAN RNA promoter F | GAAgagctcGGACCGTGGGCGAGCCGA AATA |
| PAN RNA promoter R | GAtagctaGCTGGGCAGTCCCAGTGCTA AAC |
| K-Rta probe F | GTGCACGCCACGGATGTCGCCACG |
| K-Rta probe R | TCAGTCTCGGAAGTAATTACGCC |
| ORF57 probe F | GTTACCAGATTTAGATTACTTCATC |
| ORF57 probe R | CTTAAGAAAGTGGATAAAAGAATAA ACCC |
| PAN RNA probe F | GACTAAAGTGGTGTGCGGCAG |
| PAN RNA probe R | CAATCGACGCAAGTCAAGACAC |
| Probe S.B(unique).-F | CAAGAGTGCCATGTTTATATCTGC |
| Probe S.B(unique).-R | CTAATAAAGGATGGAAAACAGTCTG |
| Probe S.B(TR).-F | CCTGGTAGCCAGTACTTACCAATAAT TCC |
| Probe S.B(TR).-R | GGTGTTCACGTAGTGTCCAGGGCTC CAC |
In all sequences, the italic lowercase nucleotides represent restriction enzyme sites used for cloning the PCR products. The lower case nucleotides of primers K-bZIP TA-R and K-bZIP LZ-F show the overlapping region allowing the PCR products to anneal to each other for the K-bZIPΔBR basic region deletion mutant.
Immunofluorescence assay.
TPA-treated BCBL-1 cells spotted on slides were fixed with methanol-acetone (1:1) for 15 min at RT and then washed three times with phosphate-buffered saline (PBS). Cells were incubated with anti-K-bZIP rabbit serum (1:500) and anti-K-Rta IgY (1:500) or anti-PML mouse monoclonal antibody in PBS-2% bovine serum albumin (BSA) for 1 h at 37°C. After four washes with PBS, rhodamine-conjugated anti-rabbit goat IgG F(ab′)2 (1:1,000) (ICN Biomedicals, Inc.) and fluorescein isothiocyanate (FITC)-conjugated anti-chicken IgY donkey IgG F(ab′)2 (1:1,000) (Jackson ImmunoResearch) or FITC-conjugated anti-mouse sheep IgG F(ab′)2 (1:1,000) (ICN) in PBS-2% BSA were applied as secondary antibodies and allowed to react at 37°C for 1 h. DNA staining was performed with 2.5 μM TO-PRO-3 (Molecular Probes) in PBS for 2 min. Imaging was performed by using a confocal microscope equipped with an argon-krypton laser (LSM510-MicroSystem; Carl Zeiss Co., Ltd.).
Preparation and purification of GST fusion proteins.
GST fusion proteins were expressed in Escherichia coli strain BL21 transformed with the following plasmids encoding distinct domains of K-bZIP and K-Rta: pGEX-K-bZIP F.L. (residues 1 to 237), pGEX-K-bZIP I (residues 1 to 121), pGEX-K-bZIP II (residues 1 to 189), pGEX-K-bZIP III (residues 120 to 189), pGEX K-bZIP IV (residues 122 to 237), pGEX K-bZIP V (residues 190 to 237), pGEX-K-Rta F.L. (residues 1 to 691), pGEX-K-Rta I (residues 1 to 109), pGEX-K-Rta II (residues 110 to 295), pGEX-K-Rta III (residues 239 to 503), pGEX-K-Rta IV (residues 299 to 550), pGEX-K-Rta V (residues 499 to 550), pGEX-K-Rta VI (residues 550 to 691), and pGEX4T-2. GST fusion proteins were then purified with glutathione-agarose beads (Amersham Pharmacia) by standard procedures. Bacterial cells (500 ml) were cultured in Luria broth for each construct. Protein expression was induced with 1 mM (final concentration) isopropyl-β-d-thiogalactopyranoside. Bacterial cells were washed once in PBS and then lysed in PBS containing 1% Triton X-100 and 1% sarcosyl by sonication. After clearing by centrifugation at 7,000 × g for 10 min at 4°C, glutathione-agarose beads (500 μl of a 1:1 slurry in PBS) were added to the lysates for affinity purification. After incubation overnight at 4°C with rotation, the beads were washed four times in PBS containing 1% Triton X-100 and 1% sarcosyl. The proteins immobilized on the glutathione-agarose beads were quantified by Coomassie blue staining with BSA as a protein standard.
Immunoprecipitation and immunoblot analyses.
Briefly, BCBL-1 cells were rinsed in ice-cold PBS and lysed at 2 × 107 cells/ml in EBC buffer with protease inhibitor cocktails (Roche). After clearing of debris by centrifugation at 15,000 × g for 10 min at 4°C, 20 μl of anti-chicken IgY immobilized on agarose beads (Promega, Madison, Wis.) was added to the lysates and incubated overnight at 4°C to reduce nonspecific binding. Each cell lysate (1 mg/ml) was reacted with anti-K-Rta chicken IgY (1:100) for 3 h at 4°C with gentle rotation. The immunocomplex was captured by the addition of 50 μl of anti-chicken IgY immobilized on agarose beads and rocking for an additional 2 h at 4°C. Beads were washed four times with EBC buffer and then boiled for 5 min in 20 μl of 2× sodium dodecyl sulfate (SDS) sample buffer (125 mM Tris-HCl [pH 6.8], 4% SDS, 10% 2-mercaptoethanol, 20% glycerol, 0.6% bromphenol blue). 293T cells were cotransfected with 2 μg of K-bZIP expression plasmid and 8 μg of K-Rta expression plasmid by using FuGene 6 (Roche). The total amount of plasmid was maintained with an empty expression plasmid. Cells were harvested 48 h after transfection and lysed in EBC buffer. Cell lysates (500 μg) were immunoprecipitated with 25 μl of Flag antibody-conjugated agarose (Sigma). Beads were washed four times with EBC buffer and then boiled for 5 min in 20 μl of 2× SDS sample buffer. Protein samples from total cell lysates (100 μg/lane) or immunoprecipitates were subjected to SDS-10% polyacrylamide gel electrophoresis and then transferred to polyvinyldene difluoride membranes (Biotechnology Systems, Boston, Mass) by using a semidry apparatus (Amersham Pharmacia). After being blocked for 1 h at RT in TBST (20 mM Tris-HCl [pH 7.5], 137 mM NaCl, 0.05% Tween 20)-5% skim milk, the membranes were incubated with primary antibodies for 2 h at RT. The membranes were subsequently washed with TBST three times for 10 min each time at RT. The membranes were incubated with horseradish peroxidase-conjugated antibodies for 1 h at RT. After incubation with the secondary antibodies, the membranes were washed three times with TBST and visualized with enhanced chemiluminescence reagents (Amersham Pharmacia). Final dilutions of the primary antibodies were 1:3,000 for the anti-K-bZIP rabbit antibody, 1:2,500 for the anti-T7 tag monoclonal antibody, 1 μg of antiactin goat antibody/ml, and 1:2,000 for anti-K-Rta chicken IgY in TBST-5% skim milk.
In vitro interaction assay.
GST fusion protein beads containing 2.0 μg of proteins were resuspended in binding buffer (20 mM HEPES [pH 7.9], 150 mM NaCl, 1 mM EDTA, 4 mM MgCl2, 1 mM dithiothreitol, 0.02% NP-40, 10% glycerol supplemented before use with 1 mg of BSA/ml, 0.5 mM phenylmethylsulfonyl fluoride, protease inhibitor cocktails) and then incubated with 10 μl of in vitro-translated proteins labeled with [35S]methionine by using a TNT coupled transcription-translation system (Promega) for 30 min at 4°C. Beads were washed four times with binding buffer and resuspended and boiled in 2× SDS sample buffer. After proteins were separated by SDS-polyacrylamide gel electrophoresis, radiolabeled polypeptides retained on the beads were visualized by autoradiography.
Reporter assays.
As described in previous reports (33, 37), we cloned the ORF57 promoter (nucleotides [nt] 81556 to 82008) and the PAN RNA promoter (nt 27996 to 28680) into the pGL3-Basic vector (Promega). Promoter regions were amplified with primer pairs (PAN RNA promoter F-PAN promoter R and ORF57 promoter F-ORF57 promoter R) (Table 1) from total genomic DNA of BCBL-1 cells. The PCR products were digested with restriction enzymes (SacII and NheI) and cloned into the pGL3-Basic vector.
A dual luciferase reporter system (Promega) was used to quantify promoter activity. 293 cells were transfected in 12-well plates, washed with PBS, and incubated with 250 μl of passive lysis buffer provided by the manufacturer. A Renilla TK expression vector (10 ng/well) served as an internal control. Ten microliters of cell lysate per well was assayed by using an Optocomp I Luminometer (MGM Instruments, Hamden, Calif.) according to the manufacturer's recommendations. At least three independent assays were carried out for each experiment. DNA amounts used are indicated in the figure legends.
Southern and Northern blotting.
Total DNA was extracted from BCBL-1 or K-bZIP-overexpressing BCBL-1 cells by using a QIAamp blood kit (Qiagen GmbH, Hilden, Germany) at various times after induction by TPA. Five micrograms of total DNA was digested with restriction enzymes (EcoRI or EcoRI and NcoI) and then separated on an 0.8% agarose gel. The gel was depurinated by incubation in depurination buffer (0.25 M HCl) and sequentially denaturated in denaturation buffer (1.5 M NaOH, 0.5 M NaCl) for 20 min each. After denaturation, the restriction fragments were transferred to a nylon membrane (Biodyne; Pall Gelman Laboratory, Ann Arbor, Mich.) by standard procedures. The DNA was immobilized on the membrane by drying at RT for 1 h and UV cross-linking.
Total RNA was prepared with Isogen (Nippon Gene, Tokyo, Japan) as recommended by the manufacturer. Total RNA (10 μg/lane) was separated on a 1.2% agarose-formaldehyde gel and then transferred to a nylon membrane. The RNA was immobilized on the membrane by drying at RT for 1 h and UV cross-linking. DNA probes were prepared from total genomic DNA of BCBL-1 cells by PCR with primer sets (Table 1). DNA fragments purified from the agarose gel were radiolabeled with [α32-P]dATP by using a Strip-EZ DNA kit (Ambion) as recommended by the supplier. The glyceraldehyde-3-phosphate-dehydrogenase (GAPDH) probe, a generous gift from Dan Robinson (UC Davis Cancer Center), served as the control. Hybridization was performed with ULTRAhyb buffer (Ambion) as recommended by the supplier.
RESULTS
K-bZIP associates with K-Rta in BCBL-1 cells.
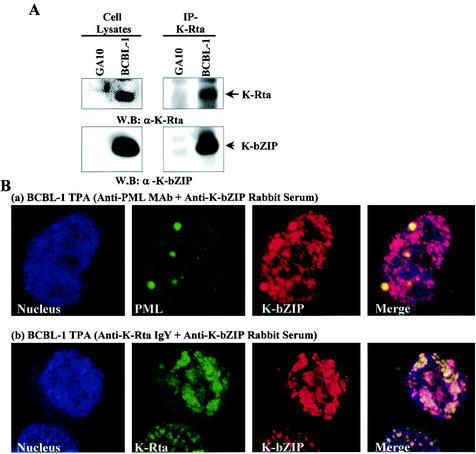
K-bZIP is the positional and structural analogue of EBV Zta, a strong transcriptional factor and a potent trigger of EBV lytic replication. Parallel studies with K-bZIP thus far, however, have failed to show its ability to reactivate the latent KSHV genome harbored in the BCBL-1 cell line (reference 28 and data not shown). Thus, the role of K-bZIP in viral replication remains elusive. To explore the possible function(s) of K-bZIP, we studied viral and cellular proteins which might interact with K-bZIP under physiologyical conditions; this was done with a cell system in which the interactions between endogenous rather than ectopically overexpressed proteins can be studied. The BCBL-1 cell line was chosen because it expresses K-bZIP and other viral lytic gene products upon treatment with TPA. The expression of K-bZIP peaks at about 48 h after induction (27; also see below). Extracts were isolated from BCBL-1 cells at 48 h after TPA treatment and immunoprecipitated with antibodies against various KSHV proteins. Among the viral genes analyzed, anti-K-Rta coprecipitated K-bZIP (Fig. 1A). Specificity was shown by the lack of K-bZIP in the anti-HA immunoprecipitate of BCBL-1 cells (data not shown) or the anti-K-Rta immunoprecipitate of GA-10, a B-cell line negative for KSHV infection (Fig. 1A). These data provide the first evidence that K-bZIP associates with K-Rta.
FIG. 1.
Coimmunoprecipitation assay. (A) Coimmunoprecipitation assay with the KSHV-positive BCBL-1 cell line. The GA-10 cell line was used as a KSHV-negative B-cell control. BCBL-1 cell lines induced with TPA (48 h) were harvested, and the same amounts of lysates (1.0 mg) were precipitated with anti-K-Rta chicken IgY and then immunoblotted with anti-K-bZIP rabbit antibody. IP, immunoprecipitation; W.B, Western blotting; α, anti. (B) Colocalization of K-bZIP with PML (a) and K-Rta (b) in BCBL-1 cells. Confocal analyses were performed by using anti-K-bZIP rabbit serum and anti-PML mouse monoclonal antibody (MAb) or anti-K-Rta chicken IgY. K-bZIP (red), PML (green), and K-Rta (green) were detected with rhodamine-conjugated anti-rabbit IgG and FITC-conjugated anti-mouse IgG or anti-chicken IgY. The nucleus was counterstained with TO-PRO-3 (blue). These panels are representative of 10 different fields.
K-bZIP colocalizes with K-Rta in BCBL-1 cells.
If a physical interaction between K-bZIP and K-Rta exists in intact cells, we would expect these two molecules to be colocalized in TPA-treated BCBL-1 cells. For this experiment, we raised chicken antibody against K-Rta (IgY isotype), so that it could be distinguished from rabbit IgG antibody against K-bZIP. Rhodamine-conjugated anti-rabbit IgG antiserum and FITC-conjugated anti-chicken IgY antiserum were used, respectively, to detect K-bZIP (red) and K-Rta (green). Previously, Katano et al. (16) and Wu et al. (39) showed that K-bZIP colocalized with PML. Therefore, we first performed a similar experiment to validate the fidelity of the K-bZIP antibody. As shown in Fig. 1B, panel a, K-bZIP colocalized with PML in PML bodies, as previously reported (16, 39). K-bZIP, however, had a much wider distribution in the nucleus, occupying both the nucleoplasm and other subnuclear structures. We next performed a colocalization assay for K-bZIP and K-Rta. As shown in Fig. 1B, panel b, both K-bZIP and K-Rta were found to be localized primarily in the nucleus, consistent with previous reports (15, 16). Merging the images of K-bZIP and K-Rta yielded a yellow color, indicating colocalization of these proteins in the nucleus. These results suggest that K-bZIP and K-Rta physically associate with each other in BCBL-1 cells.
Association of coexpressed K-bZIP and K-Rta in 293T cells.
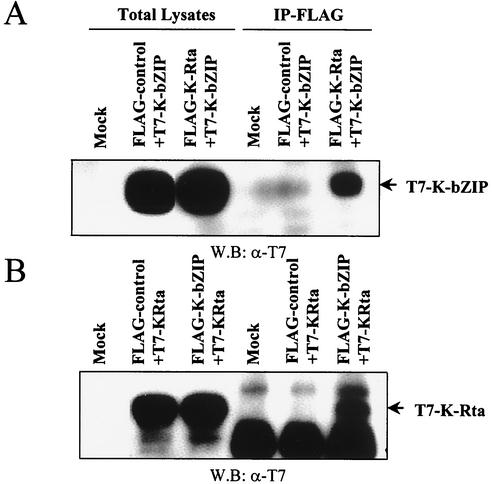
As an independent test of the association between K-bZIP and K-Rta and to rule out the possibility that the observed association in BCBL-1 cells was due to artifacts from the use of polyclonal antibodies, we constructed epitope-tagged K-Rta and K-bZIP expression vectors with Flag and T7, respectively. These constructs were linked to a cytomegalovirus (CMV) promoter and cotransfected into 293T cells. At 48 h after transfection, the cells were harvested and lysed under mild conditions (0.5% NP-40) to prevent destruction of the protein-protein complex. Coprecipitation analyses were carried out with Flag antibody-conjugated agarose. As shown in Fig. 2A, Flag-K-Rta coprecipitated T7-K-bZIP in cotransfected cells only and not in cells cotransfected with T7-K-bZIP and a Flag control vector. Reciprocal experiments with Flag-K-bZIP and T7-K-Rta expression vectors confirmed this result (Fig. 2B).
FIG. 2.
Association between K-bZIP and K-Rta in cotransfected 293T cells. 293T cells were cotransfected with the indicated plasmids. Cell lysates were precipitated with Flag antibody-conjugated agarose, and coimmunoprecipitation of K-bZIP (A) or K-Rta (B) was detected by using anti-T7 antibody. The expression of T7-tagged K-bZIP or T7-tagged K-Rta in total cell lysates is shown in the same blots as a control. IP, immunoprecipitation; W.B, Western blotting; α, anti.
The results presented above, based on experiments with two different cell lines and with two different approaches (coprecipitation and colocalization), provide evidence that K-bZIP associates with K-Rta in vivo.
The basic region of K-bZIP interacts with K-Rta.
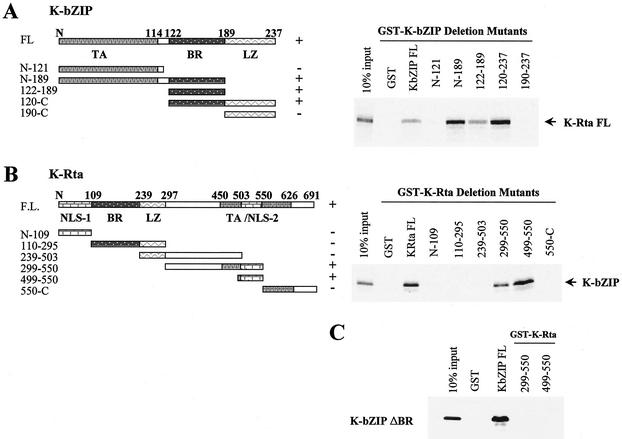
Our next task was to study whether K-bZIP directly interacts with K-Rta and, if so, to map the region(s) of K-bZIP that is responsible for the association. In vitro-translated, radiolabeled full-length K-Rta was incubated with a series of fusions of GST with different domains of K-bZIP (Fig. 3A, left panel). As shown in Fig. 3A, right panel, the K-bZIP basic region (amino acids 122 to 189) is required for the interaction with K-Rta, as only K-bZIP mutants encompassing the basic region (e.g., constructs FL, N-189, 122-189, and 120-C) could pull down in vitro-translated K-Rta. Constructs N-189 and 120-237 were most telling: the former, with a deletion precisely in the basic region, failed to bind K-Rta, and the latter, containing only the basic region, bound K-Rta. Interestingly, the level of binding by the construct containing only the basic region was lower than that of mutant N-189 or 120-237; this finding indicates that adjacent regions are also involved in the tight binding of K-Rta. GST alone and in vitro-translated luciferase were used as negative controls; both failed to bind either K-Rta or GST-K-bZIP mutants (Fig. 3A and data not shown).
FIG. 3.
In vitro interactions and interacting domains of both K-bZIP and K-Rta. (A) The domains of K-bZIP and the GST-K-bZIP mutants are indicated in the left panel. FL, full length; TA, transactivation domain; BR, basic region; LZ, leucine zipper domain. The results of GST pull-down assays for in vitro-translated, 35S-labeled full-length K-Rta and deletion mutants of K-bZIP are shown in the right panel. Pull-down-positive clones are indicated by plus signs. (B) The domains of K-Rta and the GST-K-Rta mutants are indicated in the left panel. NLS, putative nuclear localization signal. The results of GST pull-down assays for in vitro-translated, 35S-labeled full-length K-bZIP and deletion mutants of K-Rta are shown in the right panel. (C) Results of GST pull-down assays for in vitro-translated, 35S-labeled K-bZIPΔBR and K-Rta mutants. Full-length GST-K-bZIP was used as a positive control.
A C-terminal domain of K-Rta interacts with K-bZIP.
A reciprocal experiment was carried out to map the domain of K-Rta that interacts with K-bZIP. A series of GST fusion proteins that contain distinct domains of K-Rta were constructed (Fig. 3B, left panel) and incubated with in vitro-translated, radiolabeled full-length K-bZIP (Fig. 3B, right panel). This experiment helped to define a region from amino acids 499 to 550 of K-Rta as necessary for binding to K-bZIP. If this region indeed interacts with the basic region of K-bZIP, then we would predict that constructs GST-299-550 (GST fused to amino acids 499 to 550 of K-Rta) and GST-499-550 should not bind K-bZIPΔBR, which has a deletion of the basic region. As shown in Fig. 3C, this was indeed the case: K-bZIPΔBR failed to bind both construct GST-299-550 and construct GST-499-550. However, it retained the ability to bind full-length GST-K-bZIP, presumably because the dimerization domain (i.e., the leucine zipper region) remains intact in the K-bZIPΔBR construct. Based on these results, we conclude that K-bZIP and K-Rta directly interact with each other and that amino acids 120 to 189 (the basic region) of K-bZIP and amino acids 499 to 550 of K-Rta are the interacting domains.
Wild-type K-bZIP represses K-Rta-mediated transactivation in a promoter-dependent manner.
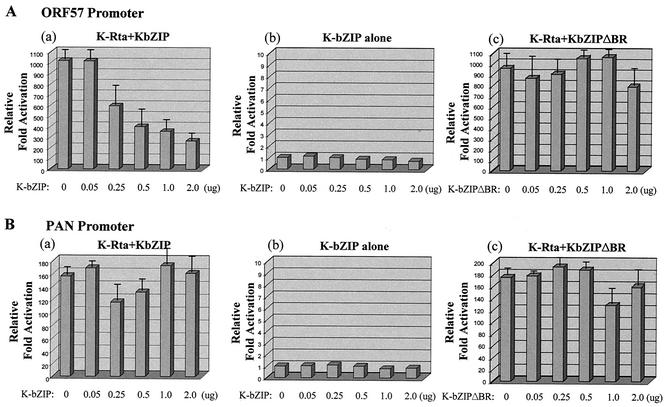
The direct interaction and colocalization of K-bZIP and K-Rta suggest that these proteins may modulate each other's activities. Little is known about the K-bZIP target genes; in contrast, several KSHV promoters, such as those of ORF57 (450 bp) and PAN RNA (685 bp), have been shown to be transcriptionally activated by K-Rta (21, 33, 37). To address the question of whether K-bZIP modulates K-Rta transactivation potential, we constructed luciferase reporter plasmids that encode the promoters of ORF57 and PAN RNA. As in previous studies, we were able to show that K-Rta is capable of activating the ORF57 promoter (1,057-fold) (Fig. 4A, panel a) and the PAN RNA promoter (155-fold) (Fig. 4B, panel a) in 293 cells. However, K-bZIP alone does not have any capacity for transactivating either of these promoters (Fig. 4A and B, panel b), nor does it appear to bind these promoters (unpublished data).
FIG. 4.
K-bZIP represses K-Rta-mediated transactivation in a sequence-specific manner. Transient reporter assays were performed with 293 cells transiently cotransfected with a Renilla control reporter and another reporter containing the ORF57 promoter or the PAN RNA promoter. The amount of the K-Rta expression vector was kept at 0.25 μg. The amounts of the K-bZIP and K-bZIPΔBR expression vectors are shown in the panels. In all of these assays, the fold activation was determined by measuring the luciferase activity derived from the reporter plasmid after normalization to the Renilla luciferase activity derived from a cotransfected Renilla TK control plasmid. All experiments were performed in triplicate, and the total amount of DNA was kept constant by using an empty expression vector. The luciferase activity of the reporter plasmid alone is normalized to a value of 1. K-bZIP alone has no transactivation or repression activity.
When a K-bZIP expression plasmid was cotransfected with a K-Rta expression vector, the K-Rta-mediated transactivation of ORF57 was largely inhibited by K-bZIP in a dose-dependent manner (Fig. 4A, panel a). K-bZIPΔBR, which has a deletion of the basic region and which failed to bind K-Rta, did demonstrate inhibition (Fig. 4A and B, panel c); this finding suggests that a direct interaction between these proteins may be important.
Interestingly, when this analysis was extended to the PAN RNA promoter, different results were obtained. Although we could reproduce the transactivation of the PAN RNA promoter by K-Rta (Fig. 4B, panel a), the addition of the wild-type K-bZIP construct or the K-bZIPΔBR construct had little effect on the transactivation potential of K-Rta (Fig. 4B, panels a and c). These results suggest that the K-bZIP modulation of K-Rta may be influenced by the precise context of the promoter.
K-bZIP represses K-Rta-mediated transactivation in the reactivation model.
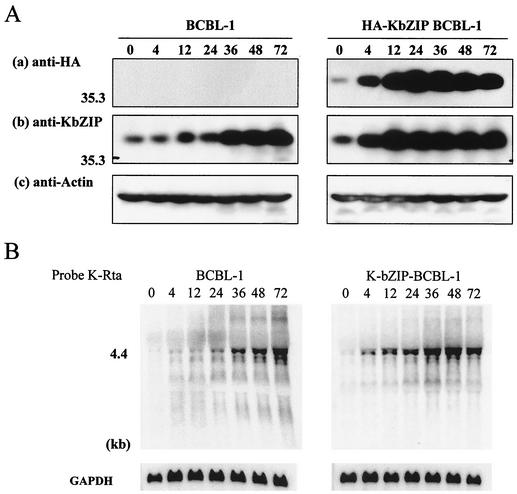
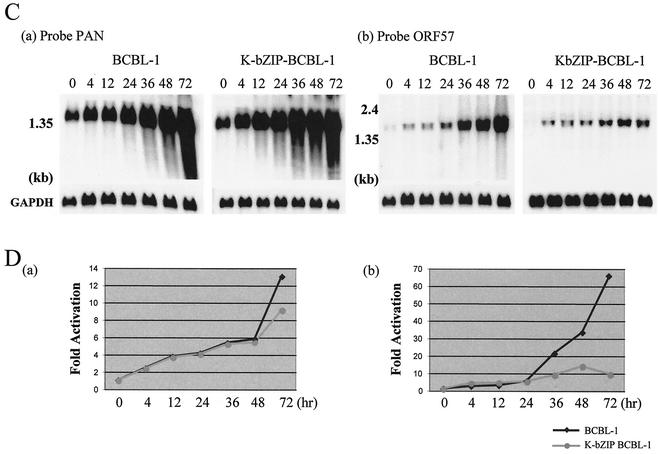
After showing that K-bZIP can repress the K-Rta-mediated activation of an isolated ORF57 promoter, we examined whether this regulation could also be demonstrated in the context of the entire KSHV genome. Accordingly, we developed a BCBL-1 cell line stably expressing K-bZIP. Because the CMV promoter can be induced by TPA (4), we expected that upon TPA treatment (to reactivate the viral genome), ectopically expressed K-bZIP would also be concurrently induced to a much higher level. To distinguish exogenous K-bZIP from endogenous K-bZIP, the CMV-K-bZIP construct was tagged with an HA epitope. Expression was confirmed by immunoblotting with anti-HA and anti-K-bZIP rabbit antibodies (Fig. 5A). The TPA treatment indeed significantly and rapidly increased the level of expression of HA-CMV-K-bZIP, such that at early time points (12 and 24 h), the total amount of K-bZIP was about five times that of endogenous K-bZIP (Fig. 5A, panel b, compare BCBL-1 and HA-K-bZIP BCBL-1). As expected, endogenous K-bZIP was also induced and reached a significant level at 48 h after induction. The purpose of this experiment was to overexpress K-bZIP early to inhibit K-Rta activity and study its effect on the activation of K-Rta target genes, such as ORF57 and PAN RNA. We extracted total cellular RNA at seven different times after TPA stimulation from K-bZIP-overexpressing BCBL-1 cells and from parental BCBL-1 cells and then performed Northern blot analyses. The overexpression of K-bZIP at early times did not seem to significantly affect the expression of K-Rta (Fig. 5B); this finding is consistent with our current understanding that K-bZIP is not an activator of K-Rta. Thus, the influence of K-Rta genes induced by K-bZIP, if any, likely occurs at a stage other than the expression of K-Rta. When the expression kinetics for PAN RNA were examined, there was no notable difference between the two cell lines at any time after TPA treatment (Fig. 5C and D, panel a). In contrast, the expression of ORF57 was repressed (about 6.5-fold) in K-bZIP-overexpressing BCBL-1 cells at 72 h after TPA treatment. These observations are consistent with the differential regulation of K-Rta by K-bZIP determined by promoter analysis.
FIG. 5.
K-bZIP represses ORF57 transcription in vivo. (A) Confirmation of exogenous K-bZIP expression in BCBL-1 cells. Protein extract (100 μg/lane) from each time point (hours) was loaded. Exogenous K-bZIP was detected with anti-HA antibody (a), and total K-bZIP was detected with anti-K-bZIP antibody (b). The actin protein served as an internal control for the amount of protein on the membrane and was detected by antiactin goat serum (c). HA-KbZIP BCBL-1, HA-tagged K-bZIP-overexpressing BCBL-1 cells. (B) K-Rta expression is not affected by the overexpression of K-bZIP. The amount of K-Rta RNA was monitored by Northern blot analysis with a K-Rta probe (nt 74228 to 74629). GAPDH mRNA served as an internal control for the amount of RNA on the membrane. (C) K-bZIP represses ORF57 transcription but not PAN RNA. The amounts of ORF57 and PAN RNAs present after TPA induction were monitored by Northern blot analyses with a PAN probe (nt 29398 to 29776) or an ORF57 probe (nt 83143 to 83545). GAPDH mRNA served as an internal control for the amount of RNA present on the membrane. (D) Quantification of transcripts. PAN RNA and ORF57 transcripts were quantified by using Quantity One (Bio-Rad). The transcript level at 0 h was normalized to a value of 1. The fold transcriptional activation is shown as a line chart.
Overexpressed K-bZIP inhibits viral DNA replication in B cells.
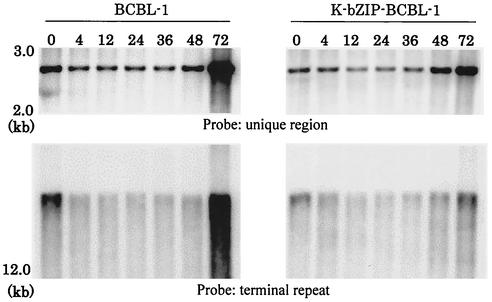
The results just presented suggest that K-bZIP is able to repress K-Rta-mediated transactivation for certain promoters, including ORF57. We next examined whether K-bZIP overexpression may compromise the ability of K-Rta to reactivate the latent genome and induce viral DNA replication. In the same experiment, we measured the increasing number of viral DNA copies after TPA treatment by Southern blotting with two different fragments (unique and terminal repeat regions of the KSHV genome) as probes. All samples were calibrated based on the total amount of cellular DNA loaded. As shown in the upper panel of Fig. 6, the total amount of KSHV DNA (at 72 h after TPA treatment) in BCBL-1 cells overexpressing K-bZIP was significantly smaller than that in parental BCBL-1 cells, as probed by an internal fragment of the KSHV genome (corresponding to the gH gene). This result indicates a lower replication rate for the KSHV genome in K-bZIP-overexpressing BCBL-1 cells. To further distinguish the lytic replication form from the latent form, we used a terminal repeat fragment as a probe (Fig. 6, lower panel). We expected that the replicating form would have more heterogeneous ends (due to the variable number of terminal repeats) and could be discerned from the latent form. A somewhat smeared pattern of the terminal repeat fragment was detected in BCBL-1 cells (at 72 h after TPA treatment) under the electrophoresis conditions used. Importantly, the hybridization intensity was greatly reduced in K-bZIP-overexpressing BCBL-1 cells, suggesting that KSHV DNA lytic replication was inhibited by the overexpression of K-bZIP. This observation is intriguing, as it suggests that the bZIP orthologues in KSHV and EBV, while both functioning to modulate Rta proteins, may have an opposite biological effect on viral replication. We recognize that this overexpression system may exaggerate the effect of K-bZIP. The natural role of K-bZIP may be to modulate K-Rta activity to ensure the proper timing of viral DNA replication.
FIG. 6.
Southern blot analyses of total DNAs from BCBL-1 cells and K-bZIP-overexpressing BCBL-1 cells (K-bZIP-BCBL-1). Five micrograms of total DNA was loaded in each lane after digestion with restriction enzymes (EcoRI for the unique probe or EcoRI and NcoI for the terminal repeat probe). A DNA fragment (nt 38900 to 39350) of the unique region (positions 3417 to 4081 of GenBank accession number U85269) of the KSHV genome was amplified by PCR and used as a probe.
DISCUSSION
The K-bZIP and K-Rta genes are among the first genes expressed during the activation of the viral lytic-cycle cascade after TPA treatment of BCBL-1 cells. These two genes likely play important roles in both viral replication and reactivation processes. Much evidence strongly implicates K-Rta as a pivotal “switch” in the reactivation process (22, 36). Our results support a new role for K-bZIP in this process; K-bZIP functions as a specific modulator of K-Rta activity. Although this may not be the sole function of K-bZIP in viral replication, it is likely to be an important one. We found that K-bZIP functions as a trans-repressor; this is a somewhat surprising finding in light of the strong transactivation potential of the EBV homolog of K-bZIP, Zta. On the other hand, the transcriptional regulation of EBV Rta by Zta and vice versa has been documented, thereby setting a precedent for a modulatory interaction between Rta and Zta (10, 17). Herpesvirus replication follows a strict temporal order, with genes in different phases being upregulated and then downregulated. Thus, trans-repression of genes may be as important as trans-activation. Given that K-Rta is an exceptionally strong transactivator and that, in our experiments, “unchecked” overexpression often leads to cell death (unpublished data), modulation of its activity in a timely fashion may be critical for maintaining the viability of the host cell to allow for the effective replication of KSHV.
Interestingly, the K-bZIP-mediated repression of K-Rta activity is promoter dependent. It has been found that K-bZIP represses the K-Rta-mediated activation of the promoters of ORF57 (this study) and K-bZIP (unpublished data and J. Giam, personal communication) but not that of PAN RNA. Recent data showed that K-Rta apparently binds different DNA sequences (RRE) on different promoters (5, 11, 21, 33) and that the RRE for the PAN RNA promoter is distinct from that for the ORF57 promoter (5, 21, 33). Notably, the K-bZIP-repressible ORF57 and K-bZIP promoters share partial sequence homology in their RRE (21). K-Rta is a versatile transactivator that appears to interact with multiple cellular transcriptional factors (e.g., sp1, Oct1, and STAT3) and target different enhancer motifs (7, 12, 32). Our data suggest that the coactivator complex assembled around K-Rta may be sequence dependent and that K-bZIP may interfere with the assembly of some, but not all, types of coactivator complexes.
At present, we do not know the exact mechanism for the repression by K-bZIP of K-Rta-mediated transactivation. Because our work showed that K-bZIP directly interacts with K-Rta, we favor the hypothesis that K-bZIP binding to K-Rta either stably or transiently interferes with the assembly of the coactivator complex. We mapped the interacting domain of K-Rta to a stretch of 51 amino acids (499 to 550), a region previously shown to be important in transactivation function (37). The interacting domain of K-bZIP resides in the basic region (amino acids 120 to 189), although adjacent sequences in the leucine zipper region facilitate binding. A K-bZIP mutant with a deletion of the basic region failed to repress K-Rta-mediated transactivation; this finding is consistent with our hypothesis. Because the basic region is the putative DNA-binding domain of K-bZIP, we cannot rule out the possibility that DNA binding is required for this process. However, the binding of K-bZIP to the ORF57 promoter (450 bp) under conditions in which purified AP-1 was able to gel shift this fragment was not observed (T. Ellison, unpublished data).
To ensure that our data were obtained from the most relevant context, many of our studies were carried out with naturally infected BCBL-1 cells. We detected the interaction between K-Rta and K-bZIP by coprecipitation and by colocalization in TPA-induced BCBL-1 cells, in which these two molecules are made during the course of reactivation. To study the impact of K-bZIP on the lytic replication and transcriptional programming of KSHV, we isolated cells stably transfected with HA-CMV-K-bZIP. To avoid clonal variations, a pool of transfected cells was used in the analysis. We chose the CMV promoter because of its well-documented activity in human cells and its inducibility by TPA (4). The latter feature ensures that during the course of TPA treatment, exogenous HA-K-bZIP remains expressed at a very high level. This strategy worked as planned; in HA-K-bZIP-overexpressing BCBL-1 cells, the level of exogenous K-bZIP is at least fivefold higher than that in the control at early time points, allowing us to monitor its repression effect immediately after the induction of K-Rta. Initially, we were surprised that the basal level (time zero, without TPA induction) of K-bZIP was only slightly higher in the transfected cells than in the control cells. Now we know that exogenous K-bZIP expression represses that of endogenous K-bZIP (presumably through its repression of K-Rta), such that the basal level of endogenous K-bZIP in the transfected cells at time zero was virtually undetectable (unpublished data). In this setting, we were able to demonstrate that the transcription of ORF57 is significantly reduced, although that of PAN RNA is relatively unaffected. The overall replication of KSHV DNA is also blocked, presumably due to the action of this transcriptional program. This study provides the first direct evidence that K-bZIP expression has an impact on K-Rta-mediated transactivation and on viral replication.
ORF57, the target of K-bZIP-mediated repression, is involved in the posttranscriptional regulation of gene expression (18). The EBV homolog of ORF57 is BMLF1, or Mta, which performs a similar function. The transcription of EBV BMLF1 is regulated by both EBV Rta and EBV Zta (17), similar to the observations in our report. For EBV, however, Zta enhances rather than represses the transactivation mediated by Rta (17). Thus, gammaherpesviruses use evolutionarily conserved genes in different strategies to perform similar functions.
In summary, this report describes physical and functional interactions between the K-bZIP and K-Rta genes, which are immediate-early and early lytic genes involved in the transcriptional regulation of viral genes in lytic replication. These two genes are physically linked in the KSHV genome, and now we provide evidence that their products are also biochemically linked. We showed that K-bZIP acts as a transcriptional repressor, curtailing the transactivation potential of K-Rta. Our findings provide a new perspective on K-bZIP functions and suggest a different strategy for probing its role in KSHV replication and reactivation.
Acknowledgments
We appreciate the generous gifts of the modified pcDNA expression vector and the modified pGEX vector from Dan Robinson. We thank Greg Mayer for advice and assistance with confocal microscopy.
This work was supported by NIH grant CA91574 (to H.-J.K.) and Universitywide AIDS Research Program grant R00-D-034 (to P.L. and H.-J.K.).
REFERENCES
- 1.Alkan, S., D. S. Karcher, A. Ortiz, S. Khalil, M. Akhtar, and M. A. Ali. 1997. Human herpesvirus-8/Kaposi's sarcoma-associated herpesvirus in organ transplant patients with immunosuppression. Br. J. Haematol. 96:412-414. [DOI] [PubMed] [Google Scholar]
- 2.Boshoff, C., T. F. Schulz, M. M. Kennedy, A. K. Graham, C. Fisher, A. Thomas, J. O. McGee, R. A. Weiss, and J. J. O'Leary. 1995. Kaposi's sarcoma associated herpesvirus infects endothelial and spindle cells. Nat. Med. 1:1274-1278. [DOI] [PubMed] [Google Scholar]
- 3.Cesarman, E., Y. Chang, P. S. Moore, J. W. Said, and D. M. Knowles. 1995. Kaposi's sarcoma-associated herpesvirus-like DNA sequences in AIDS-related body-cavity-based lymphomas. N. Engl. J. Med. 332:1186-1191. [DOI] [PubMed] [Google Scholar]
- 4.Chan, Y.-J., C.-J. Chiou, Q. Huang, and G. S. Hayward. 1996. Synergistic interactions between overlapping binding sites for the serum response factor and ELK-1 proteins mediate both basal enhancement and phorbol ester responsiveness of primate cytomegalovirus major immediate-early promoters in monocyte and T-lymphocyte cell types. J. Virol. 70:8590-8605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chang, P.-J., D. Shedd, L. Gradoville, M.-S. Cho, L.-W. Chen, J. Chang, and G. Miller. 2002. Open reading frame 50 protein of Kaposi's sarcoma-associated herpesvirus directly activeates the viral PAN and K12 genes by binding to related response elements. J. Virol. 76:3168-3178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chang, Y., E. Cesarman, M. S. Pessin, F. Lee, J. Culpepper, D. M. Knowles, and P. S. Moore. 1994. Identification of herpesvirus-like DNA sequences in AIDS-associated Kaposi's sarcoma. Science 266:1865-1869. [DOI] [PubMed] [Google Scholar]
- 7.Chen, J., K. Ueda, S. Sakakibara, T. Okuno, and K. Yamanishi. 2000. Transcriptional regulation of the Kaposi's sarcoma-associated herpesvirus viral interferon regulatory factor gene. J. Virol. 74:8623-8634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chevallier-Greco, A., E. Manet, P. Chavrier, C. Mosnier, J. Daillie, and A. Sergeant. 1986. Both Epstein-Barr virus (EBV)-encoded trans-acting factors, EB1 and EB2, are required to activate transcription from an EBV early promoter. EMBO J. 5:3243-3249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Countryman, J., and G. Miller. 1985. Activation of expression of latent Epstein-Barr herpesvirus after gene transfer with a small cloned subfragment of heterogenous viral DNA. Proc. Natl. Acad. Sci. USA 82:4085-4089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cox, M. A., J. Leahy, and J. M. Hardwick. 1990. An enhancer within the divergent promoter of Epstein-Barr virus responds synergistically to the R and Z transactivators. J. Virol. 64:313-321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Duan, W., S. Wang, S. Liu, and C. Wood. 2001. Characterization of Kaposi's sarcoma-associated herpesvirus/human herpesvirus-8 ORF57 promoter. Arch. Virol. 146:403-413. [DOI] [PubMed] [Google Scholar]
- 12.Gwack, Y., S. Hwang, C. Lim, Y. S. Won, C. H. Lee, and J. Choe. 2002. Kaposi's sarcoma-associated herpesvirus open reading frame 50 stimulates the transcriptional activity of STAT3. J. Biol. Chem. 277:6438-6442. [DOI] [PubMed] [Google Scholar]
- 13.Huang, Y. Q., J. J. Li, M. H. Kaplan, B. Poiesz, E. Katabira, W. C. Zhang, D. Feiner, and A. E. Friedman-Kien. 1995. Human herpesvirus-like nucleic acid in various forms of Kaposi's sarcoma. Lancet 345:759-761. [DOI] [PubMed] [Google Scholar]
- 14.Hwang, S., Y. Gwack, H. Byun, C. Lim, and J. Choe. 2001. The Kaposi's sarcoma-associated herpesvirus K8 protein interacts with CREB-binding protein (CBP) and represses CBP-mediated transcription. J. Virol. 75:9509-9516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Katano, H., Y. Sato, H. Itoh, and T. Sata. 2001. Expression of human herpesvirus 8 (HHV-8)-encoded immediate early protein, open reading frame 50, in HHV-8-associated diseases. J. Hum. Virol. 4:96-102. [PubMed] [Google Scholar]
- 16.Katano, H., K. Ogawa-Goto, H. Hasegawa, T. Kurata, and T. Sata. 2000. Human-herpesvirus-8-encoded K8 protein colocalizes with the promyelocytic leukemia protein (PML) bodies and recruites p53 to the PML bodies. Virology 286:446-455. [DOI] [PubMed] [Google Scholar]
- 17.Kenney, S., E. Holley-Guthrie, E. C. Mar, and M. Smith. 1989. The Epstein-Barr virus BMLF1 promoter contains an enhancer element that is responsive to the BZLF1 and BRLF1 transactivators. J. Virol. 63:3878-3883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kirshner, J. R., D. M. Lukac, J. Chang, and D. Ganem. 2000. Kaposi's sarcoma-associated herpesvirus open reading frame 57 encodes a posttranscriptional regulator with multiple distinct activities. J. Virol. 74:3586-3597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lieberman, P. M., J. M. Hardwick, J. Sample, G. S. Hayward, and S. D. Hayward. 1990. The Zta transactivator involved in induction of lytic cycle gene expression in Epstein-Barr virus-infected lymphocytes binds to both AP-1 and ZRE sites in target promoter and enhancer regions. J. Virol. 64:1143-1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lin, S.-F., D. R. Robinson, G. Miller, and H.-J. Kung. 1999. Kaposi's sarcoma-associated herpesvirus encodes a bZIP protein with homology to BZLF1 of Epstein-Barr virus. J. Virol. 73:1909-1917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lukac, D. M., L. Garibyan, J. R. Kirshner, D. Palmeri, and D. Ganem. 2001. DNA binding by Kaposi's sarcoma-associated herpesvirus lytic switch protein is necessary for transcriptional activeation of two viral delayed early promoters. J. Virol. 75:6786-6799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lukac, D. M., R. Renne, J. R. Kirshner, and D. Ganem. 1998. Reactivation of Kaposi's sarcoma-associated herpesvirus infection from latency by expression of the ORF 50 transactivator, a homolog of the EBV R protein. Virology 252:304-312. [DOI] [PubMed] [Google Scholar]
- 23.Martin, D. F., B. D. Kuppermann, R. A. Wolitz, A. G. Palestine, H. Li, C. A. Robinson, and the Roche Ganciclovir Study Group. 1999. Oral ganciclovir for patients with cytomegalovirus retinitis treated with a ganciclovir implant. N. Engl. J. Med. 340:1063-1070. [DOI] [PubMed] [Google Scholar]
- 24.Memar, O. M., P. L. Rady, and S. K. Tyring. 1995. Human herpesvirus-8: detection of novel herpesvirus-like DNA sequences in Kaposi's sarcoma and other lesions. J. Mol. Med. 73:603-609. [DOI] [PubMed] [Google Scholar]
- 25.Moore, P. S., and Y. Chang. 1995. Detection of herpesvirus-like DNA sequences in Kaposi's sarcoma in patients with and without HIV infection. N. Engl. J. Med. 332:1181-1185. [DOI] [PubMed] [Google Scholar]
- 26.Nicholas, J., L. S. Coles, C. Newman, and R. W. Honess. 1991. Regulation of the herpesvirus saimiri (HVS) delayed-early 110-kilodalton promoter by HVS immediate-early gene products and a homolog of the Epstein-Barr virus R trans activator. J. Virol. 65:2457-2466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Park, J., T. Seo, S. Hwang, D. Lee, Y. Gwack, and J. Choe. 2000. The K-bZIP protein from Kaposi's sarcome-associated herpesvirus interacts with p53 and represses its transcriptional activity. J. Virol. 74:11977-11982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Polson, A. G., L. Huang, D. M. Lukac, J. D. Blethrow, D. O. Morgan, A. L. Burlingame, and D. Ganem. 2001. Kaposi's sarcoma-associated herpesvirus K-bZIP protein is phosphorylated by cyclin-dependent kinases. J. Virol. 75:3175-3184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ragoczy, T., L. Heston, and G. Miller. 1998. The Epstein-Barr virus Rta protein activates lytic cycle genes and can disrupt latency in B lymphocytes. J. Virol. 72:7978-7984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Renne, R., W. Zhong, B. Herndier, M. McGrath, N. Abbey, D. Kedes, and D. Ganem. 1996. Lytic growth of Kaposi's sarcoma-associated herpesvirus (human herpesvirus 8) in culture. Nat. Med. 2:342-346. [DOI] [PubMed] [Google Scholar]
- 31.Russo, J. J., R. A. Bohenzky, M.-C. Chien, J. Chen, M. Yan., D. Maddalena, J. P. Parry, D. Peruzzi, I. S. Edelman, Y. Chang, and P. S. Moore. 1996. Nucleotide sequence of the Kaposi's sarcoma-associated herpesvirus (HHV8). Proc. Natl. Acad. Sci. USA 93:14862-14867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sakakibara, S., K. Ueda, J. Chen, T. Okuno, and K. Yamanishi. 2001. Octamer-binding sequence is a key element for the autoregulation of Kaposi's sarcoma-associated herpesvirus ORF50/Lyta gene expression. J. Virol. 75:6894-6900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Song, M. J., H. J. Brown, T.-T. Wu., and R. Sun. 2001. Transcription activation of polyadenylated nuclear RNA by Rta in human herpesvirus 8/Kaposi's sarcoma-associated herpesvirus. J. Virol. 75:3129-3140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Soulier, J., L. Grollet, E. Oksenhendler, P. Cacoub, D. Cazals-Hatem, P. Babinet, M. F. d'Agay, J. P. Clauvel, M. Raphael, L. Degos, et al. 1995. Kaposi's sarcoma-associated herpesvirus-like DNA sequences in multicentric Castleman's disease. Blood 86:1276-1280. [PubMed] [Google Scholar]
- 35.Sun, R., S.-F. Lin, K. Staskus, L. Gradoville, E. Grogan, A. Haase, and G. Miller. 1999. Kinetics of Kaposi's sarcoma-associated herpesvirus gene expression. J. Virol. 73:2232-2242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sun, R., S.-F. Lin, L. Gradoville, Y. Yuan, F. Zhu, and G. Miller. 1998. A viral gene that activeates lytic cycle expression of Kaposi's sarcoma-associated herpesvirus. Proc. Natl. Acad. Sci. USA 95:10866-10871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang, S., S. Liu, M. Wu, Y. Geng, and C. Wood. 2001. Kaposi's sarcoma-associated herpesvirus/human herpesvirus-8 ORF50 gene product contains a potent C-terminal activation domain which activates gene expression via a specific target sequence. Arch. Virol. 146:1415-1426. [DOI] [PubMed] [Google Scholar]
- 38.Whitby, D., M. R. Howard, M. Tenant-Flowers, N. S. Brink, A. Copas, C. Boshoff, T. Hatzioannou, F. E. Suggett, D. M. Aldam, A. S. Denton, R. F. Miller, I. V. D. Weller, R. A. Weiss, and T. F. Schulz. 1995. Detection of Kaposi sarcoma associated herpesvirus in peripheral blood of HIV-infected individuals and progression to Kaposi's sarcoma. Lancet 346:799-802. [DOI] [PubMed] [Google Scholar]
- 39.Wu, F. Y., J.-H. Ahn, D. J. Alecendor, W.-J. Jang, J. Xiao, S. D. Hayward, and G. S. Hayward. 2001. Origin-independent assembly of Kaposi's sarcoma-associated herpesvirus DNA replication compartments in transient cotransfection assays and associateion with the ORF-K8 protein and cellular PML. J. Virol. 75:1487-1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zalani, S., E. Holley-Guthrie, and S. Kenney. 1996. Epstein-Barr viral latency is disrupted by the immediate-early BRLF1 protein through a cell-specific mechanism. Proc. Natl. Acad. Sci. USA 93:9194-9199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhu, F. X., T. Cusano, and Y. Yuan. 1999. Identification of the immediate-early transcripts of Kaposi's sarcoma-associated herpesvirus. J. Virol. 73:5556-5567. [DOI] [PMC free article] [PubMed] [Google Scholar]