Abstract
Herpesviruses start their transcriptional cascade at nuclear domain 10 (ND10). The deposition of virus genomes at these nuclear sites occurs due to the binding of the interferon-inducible repressor protein promyelocytic leukemia protein (PML) and/or Daxx to a viral DNA-protein complex. However, the presence of repressive proteins at the nuclear site of virus transcription has remained unexplained. We investigated the mouse cytomegalovirus (MCMV) immediate-early 1 protein (IE1), which is necessary for productive infection at low multiplicities of infection and therefore likely to be involved in overcoming cellular repression. Temporal analysis of IE1 distribution revealed its initial segregation into ND10 by binding to PML and/or Daxx and IE1-dependent recruitment of the transcriptional repressor histone deacetylase-2 (HDAC-2) to this site. However, these protein aggregates are dissociated in cells producing sufficient IE1 through titration of PML, Daxx, and HDAC-2. Importantly, binding of IE1 to HDAC-2 decreased deacetylation activity. Moreover, inhibition of HDAC by trichostatin-A resulted in an increase in viral protein synthesis, an increase in cells starting the formation of prereplication compartments, and an increase in the total infectious viruses produced. Thus, IE1, like trichostatin-A, reverses the repressive effect of HDAC evident in the presence of acetylated histones in the immediate-early promoter region. Since HDAC also binds to the promoter region of IE1, as determined by the chromatin immunoprecipitation assay, these combined results suggest that IE1 inhibits or reverses HDAC-mediated repression of the infecting viral genomes, possibly by a process akin to activation of heterochromatin. We propose that even permissive cells can repress transcription of infecting viral genomes through repressors, including HDAC, Daxx, and PML, and the segregation of IE1 to ND10 that would inactivate those repressors. The virus can counter this repression by overexpressing IE1 when present in sufficient copy number, thus reducing the availability and effectiveness of these repressors.
The biological features of murine cytomegalovirus (MCMV) and human cytomegalovirus (HCMV) infection are similar. Both MCMV and HCMV genes are expressed in a sequential and coordinated manner. The genomes are of similar large size and organization and display analogous gene products, replication patterns, and pathogenesis (31, 32, 36). During infection, MCMV brings a number of tegument proteins into the cells, which transactivate the immediate-early transcription unit. Immediate-early proteins are produced first and regulate the subsequent early protein synthesis, DNA replication, and late protein synthesis. Immediate-early protein 1 (IE1) shares exons 1, 2, and 3 with IE3.
IE1 of MCMV is phosphorylated, with an apparent molecular weight of 89,000 (31), and is the most abundant viral protein of the immediate-early phase. It has been characterized to have the same general molecular structure as IE1 of HCMV, but IE1 of MCMV has no amino acid similarity with IE1 of HCMV except for a stretch in the Glu-rich region (19, 20). Although IE1 proteins are necessary for the production of early proteins, in transfected cells, IE1 cannot activate the MCMV early gene promoter e1. It requires IE3, the equivalent of the HCMV ie2 gene product (6). In the absence of IE1, MCMV replication is markedly restricted.
To effect productive infection with virus devoid of IE1, multiple hits appear to be necessary (15), suggesting that IE1 is not essential but enhances early protein synthesis through its effect on the host cell. IE1 of MCMV has a domain homologous to one in histone 2B, but a different binding region interacts with cellular histones (33). Binding to cellular histones is quite avid, and no association of IE1 with cellular DNA in dividing cells could be detected. However, in vitro, a domain different from the H2B-like domain has been identified that binds without sequence specificity to DNA (34).
Expression of IE1 is under the control of the major immediate-early promoter, a large and complex enhancer sequence that can bind to a variety of activators and repressors, all of which are nuclear transcription factors. The role of these transcription factors in viral infection remains unclear, although recent studies point to the importance of the interaction between repressors and the major immediate-early promoter in CMV infection. For example, it was found that all the repressors capable of binding with the major immediate-early promoter are preferentially expressed in undifferentiated cells which are resistant to HCMV replication (30). Also, retinoic acid treatment of undifferentiated cells before but not after infection renders these cells permissive (30), suggesting that once the genome is repressed, removal of the repressors does not reactivate it. In addition, trichostatin-A treatment to inhibit the nuclear repressor histone deacetylase (HDAC) results in the release of infectious viruses from latently infected NT2 cells, suggesting a role for HDAC in maintaining suppression of competent viral genomes (35).
Herpesviruses begin their reproductive cycle at nuclear domain 10 (ND10) (18, 27), supramolecular aggregates of various proteins that appear as 0.3- to 1-μm nuclear structures upon immunostaining of its constitutive proteins promyelocytic leukemia protein (PML), Sp100, and Daxx, all of which are repressive and can be upregulated by interferon (8, 9, 14, 16, 24, 38). PML is considered to have suppressive activity on transcription (21, 25, 40, 46, 47). The transcriptional suppression by Daxx is supported by its interaction with Pax3 and HDAC (25, 26). The herpesviruses have evolved genes that destroy or disperse this nuclear structure, and in the absence of such genes, the reproductive success of the virus is dramatically reduced.
Immediate-early proteins that disperse ND10, such as ICP0 for herpes simplex virus type 1 (13, 28) and IE1 for HCMV (1, 22, 45), are considered transactivators because they augment or enhance the transcription and/or viral protein synthesis of the early genes. Such transactivation might reflect enhancement of viral and host transcription to benefit the viral replicative program or to inhibit any host defenses (27). In the case of herpes simplex virus type 1, ICP0 appears to induce the hydrolysis of PML and other ND10 proteins by a ubiquitination process based on the E3-like properties of the ICP0 RING finger domain (12). In the case of HCMV IE1, ND10 destruction has been suggested to come about by binding of IE1 to PML (1).
HCMV IE1 is produced in substantially larger quantities than IE2 from the same transcript through differential splicing. Because high concentrations of IE1 do not appear to be required for transactivation of a few viral genomes and genes or for the potential activation of host genes, we asked whether one mechanism for IE1-based success in viral replication might be the repression of a host defense. The possibility for a nucleus-based innate defense might lie in the ND10-associated proteins because PML, Daxx, and Sp100 are thought to represent repressors and they can be upregulated by interferon (8, 9, 16, 24, 38). The basic hypothesis to be tested, therefore, is whether IE1 functions to antagonize cellular defenses by rendering ineffective certain host-repressive proteins.
In this study, we investigated nuclear localization and molecular interaction between MCMV IE1 and host nuclear repressors, including ND10-associated proteins Daxx and PML and the Daxx-interacting histone deacetylase HDAC-2, during acute infection to determine whether the cell has an innate mechanism to suppress productive infection. We found evidence for a single mechanism, binding of host repressor proteins with IE1, that results in two competing processes, viral repression and viral activation, depending on the amount of IE1 produced in a given infected cell.
MATERIALS AND METHODS
Tissue culture and viruses.
Primary mouse embryo fibroblasts (MEF), PML-negative MEF (PN2) (44), and Daxx-negative MEF (Daxx−/− MEF; Ishov et al., unpublished data) were maintained in Dulbecco's modified Eagle's medium supplemented with 10% fetal calf serum, and 1% penicillin-streptomycin. For immunohistochemical staining, cells were grown on round coverslips (Corning Glass Inc., Corning, N.Y.) in 24-well plates (Falcon; Becton Dickinson Labware, Lincoln Park, N.J.). Wild-type MCMV (42, 43) and its IE1 deletion mutant (ΔIE1 MCMV) were kindly provided by M. Messerle; in the ΔIE1 genome, exon 4 of ie1 was completely removed, and exon 3 was fused to exon 5 (M. Messerle, personal communication). Cells were infected when 80% confluent with virus at a multiplicity of infection of 1 PFU/cell. In experiments to assess the effect of trichostatin-A on viral infection, cells were treated with the HDAC inhibitor for 2 h before infection.
Effect of trichostatin-A on MCMV infection in MEF.
MEF cells were seeded in 12-well plates. When the cells reached confluence, one plate of MEF cells was treated with 50 ng of trichostatin-A per ml for 2 h, and another remained untreated as control. Cells were infected with wild-type MCMV at multiplicity of infection of 1. After 2 h of adsorption, unadsorbed virus was removed by washing twice with Dulbecco's modified Eagle's medium and adding new Dulbecco's modified Eagle's medium containing 5% fetal calf serum. For the trichostatin-A treatment plate, the medium contained 50 ng of trichostatin-A per ml. Cells and supernatant were collected after washing and at days 1, 2, 3, 4, and 5. After three freeze-thaw cycles, the cellular debris was removed by centrifugation, and the supernatant was kept at −70°C for detection of virus titers by plaque assay in MEF.
Antibodies and plasmids.
Mouse cell ND10-associated proteins were visualized with the following antibodies: rabbit serum R14 produced against the N-terminal half of PML, monoclonal antibody 116 against PML (S. Lowe; Cold Spring Harbor), rabbit antibodies against Daxx (Santa Cruz Biotechnology, Inc., Santa Cruz, Calif.). Rabbit antibody against HDAC-2 was purchased from Zymed Laboratory Inc. (South San Francisco, Calif.), and the acetylhistone H3(Lys23) antibodies were from Cell Signaling Technologies (Beverly, Mass.). The monoclonal antibody against tubulin was from Sigma Co. (St. Louis, Mo.). The monoclonal antibodies against MCMV IE1 and E1 and plasmid pp89UC, containing all introns and exons of ie1 under the control of the major immediate-early promoter, were generous gifts from M. Messerle.
Immunohistochemistry.
The localization of ND10 by immunohistochemistry has been described (39). Briefly, 24 h after seeding cells, coverslips were washed twice with phosphate-buffered saline, fixed in 1% paraformaldehyde for 10 min at room temperature, washed again twice with phosphate-buffered saline and permeabilized with 0.2% Triton X-100 on ice for 20 min. Primary antibody was added for 30 min at room temperature, and cells were washed twice with phosphate-buffered saline before addition of secondary antibody labeled with Texas Red, indodicarbocyanine (blue), or fluorescein isothiocyanate (green) of either anti-rabbit or anti-mouse immunoglobulin G for another 30 min at room temperature. After washing with phosphate-buffered saline, cells were stained with Hoechst 33258.
Immunoblot analysis.
Immunoblot analysis was performed to detect proteins by loading 10 to 20 μg in each lane for sodium dodecyl sulfate-7.5% polyacrylamide gel electrophoresis (SDS-PAGE). After electrophoresis, proteins were transferred to nitrocellulose membranes (Amersham Inc., Piscataway, N.J.) and blocked with 5% nonfat milk for 60 min at room temperature. Membranes were incubated at 4°C overnight with primary antibody, followed by incubation with a horseradish peroxidase-coupled secondary antibody (Amersham Inc.) and detection with enhanced chemiluminescence (Pierce, Rockford, Ill.) according to standard methods. Membranes were stripped with stripping buffer (100 mM β-mercaptoethanol, 2% SDS, 62.5 mM Tris-HCl [pH 6.8]), washed with phosphate-buffered saline-0.1% Tween 20, and used to assay for additional proteins.
Preparation of nuclear extracts.
Nuclear extracts were obtained essentially as described before (3). Briefly, monolayer cells were washed with phosphate-buffered saline once and scraped into fresh Eppendorf tubes. Cell pellets were resuspended in cold buffer A (10 mM HEPES-KOH [pH 7.9] at 4°C, 1.5 mM MgCl2, 10 mM KCl, 0.5 mM dithiothreitol, 0.2 mM phenylmethylsulfonyl fluoride) and incubated at 4°C for 10 min. After centrifugation, pellets were resuspended in cold buffer C (20 mM HEPES-KOH [pH 7.9], 25% glycerol, 420 mM NaCl, 1.5 mM MgCl2, 0.2 mM EDTA, 0.5 mM dithiothreitol, 0.2 mM phenylmethylsulfonyl fluoride) by vortexing and incubated at 4°C for 30 min. Clarified extracts were transferred to fresh tubes and stored at −70°C until use.
Coimmunoprecipitation.
Antibodies were coupled to Dynabead M-450 paramagnetic beads (Dynal, Oslo, Norway) coated with anti-rabbit or anti-mouse IgG as described by the manufacturer. After washing with phosphate-buffered saline-0.1% bovine serum albumin, beads were incubated overnight at 4°C with clarified extracts, washed again in phosphate-buffered saline-0.1% bovine serum albumin, and resuspended in a mixture of phosphate-buffered saline and 2× Laemmli buffer (20 μl of each). After heating at 95°C for 5 min, beads were removed by centrifugation, and supernatants were analyzed by SDS-PAGE and immunoblotting.
Chromatin immunoprecipitation assay.
Adherent cells were washed twice with phosphate-buffered saline and incubated in 1% formaldehyde for 30 min at room temperature for DNA-protein cross-linking. After addition of glycine (0.125 M final concentration), cells were incubated for 5 min, washed twice in phosphate-buffered saline, scraped off, and resuspended in immunoprecipitation buffer (20 mM Tris [pH 8.0], 0.1% deoxycholate, 0.5% Triton X-100, 2 mM EDTA, 150 mM NaCl, 1 mM phenylmethylsulfonyl fluoride). Cells were sonicated six times at 30% output for 10 s to yield 200- to 1,000-bp DNA fragments. After centrifugation in a Sorvall RT7 centrifuge (4,000 rpm for 20 min), antibodies against Daxx (Santa Cruz; 5 μg/ml) along with bovine serum albumin (100 μg/ml) were added to the supernatant and incubated overnight at 4°C. An untreated aliquot of each supernatant served as a control for input DNA.
In parallel, protein G beads (Amersham Pharmacia Biotech, N.J.) were washed twice with immunoprecipitation buffer and incubated overnight at 4°C in immunoprecipitation buffer containing 0.5 mg of bovine serum albumin and 0.5 mg of heat-denatured salmon sperm DNA per ml. Beads were washed twice with immunoprecipitation buffer, incubated with the antibody-containing supernatants for 2 h at 4°C, and retrieved by centrifugation, followed by sequential washing with immunoprecipitation buffer which contained 600 mM NaCl, LiCl buffer (10 mM Tris [pH 8.0], 0.1% deoxycholate, 0.5% Triton X-100, 5 mM EDTA, 0.5% Nonidet P40, 250 mM LiCl, 1 mM phenylmethylsulfonyl fluoride), and TE (Tris-EDTA) buffer.
To purify DNA from the beads and from untreated control supernatants, samples were supplemented with 1% SDS, incubated overnight at 65°C, extracted with phenol-chloroform, and ethanol, precipitated. Samples were analyzed by PCR with 30 cycles of 94°C, 56°C, and 72°C for 1 min each. Primers used for the MCMV ie2 promoter were 5′-GGC TCC GTT CAC CCG CTC GT-3′ (sense) and 5′-TAA AGG CCA TTG AGT CAC CA-3′ (antisense); for the MCMV ie1/ie3 promoter, the primers were 5′-GTA CAA AAG GTC AAT AGG GG-3′ (sense) and 5′-GTA CCG ACG CTG GTC GCG CC-3′ (antisense). PCR products were visualized on 2% agarose gels.
Preparation of HDAC complex.
HDAC complexes were isolated by using protein G (Amersham Pharmacia Biotech) according to the manufacturer's instructions. Briefly, protein G-coupled beads were washed twice with phosphate-buffered saline-0.1% bovine serum albumin and incubated with primary antibody (anti-IE1 or anti-HDAC-2) overnight at 4°C with continuous rolling. After extensive washing with phosphate-buffered saline-0.1% bovine serum albumin, beads were incubated with nuclear extracts overnight at 4°C on a roller. Beads were washed at least three times, and an aliquot was used for Western blotting to determine the concentration of HDAC-2; the remainder was stored at −70°C. Blots were scanned, and the amount of HDAC-2 was determined with ImageQuant Systems software. The results were used to normalize the input in the HDAC deacetylation assay.
HDAC activity assay.
HDAC activity was assessed with the HDAC activity assay kit (Upstate Biotech, Lake Placid, N.Y.) according to the manufacturer's instructions. Immune complexes were incubated with 20,000 cpm of [3H]acetyl-labeled histone H4 peptide (Upstate Biotech) in 1× HDAC buffer at room temperature for 24 h with rolling. Reactions were stopped by adding 50 μl of 0.12 N acetic acid-0.72 N HCl. The released acetate was extracted in 0.5 ml of ethyl acetate and mixed in 5 ml of scintillation solution, and radioactivity was measured in a scintillation counter. All assays were performed in duplicate.
Confocal microscopy.
Cells were examined with a Leica TCS SPII confocal laser scanning system. Two or three channels were recorded simultaneously and/or sequentially and controlled for possible breakthrough between the fluorescein isothiocyanate and Texas Red signals and between the blue and red channels.
RESULTS
IE1 of MCMV colocalizes with PML and Daxx and recruits HDAC.
To determine where IE1 is positioned in the nucleus, MEF were infected with MCMV at a multiplicity of infection of 1, fixed at different times after infection, and immunostained for IE1 and PML. At early times (2 h postinfection) and at low IE1 expression levels at later times (3 to 5 h postinfection), IE1 colocalized almost completely with PML (Fig. 1A, left nucleus), indicating that IE1 at that stage is segregated into ND10. The same observations were made for Daxx (Fig. 1B), as expected because of the association of Daxx with SUMO-modified PML in ND10 (17). The apparently uninfected cells to the right in both Fig. 1A and B are not labeled with IE1 antibody, providing an internal control that the IE1 antibodies do not recognize ND10 antigens. Unexpected was the finding that already at 2 h postinfection, HDAC-2 colocalizes with PML and IE1 (Fig. 1C to F) and therefore at ND10.
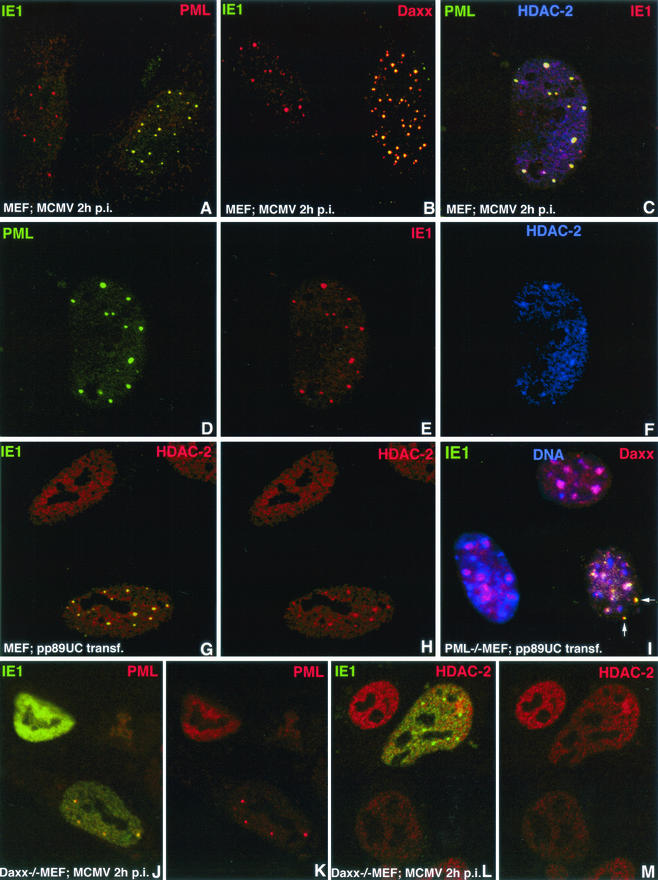
FIG.1.
Immunohistochemical localization of IE1 in infected and transfected cells. Proteins recognized by the respective antibodies are indicated by the colors used in the upper corners of each panel. The cell types used were MEF, PML-knockout MEF (PML−/− MEF), and Daxx-knockout MEF (Daxx−/− MEF). Infection with MCMV or transfection with IE1-producing plasmid pp89UC is indicated at the bottom of each panel. (A) The cell on the right was infected with MCMV and produced IE1 colocalizing with PML-positive sites (ND10). (B) The cell on the right was MCMV infected, and IE1 colocalized with Daxx, i.e., Daxx was not removed from ND10 by IE1. (C) MCMV-infected green fluorescent protein-PML fusion-producing MEF were double labeled for HDAC and IE1, revealing enrichment of HDAC-2 at PML-positive sites to which IE1 has been recruited. (D to F) Color separation demonstrating the location of IE1, PML, and HDAC-2, as shown combined in C. (G) The lower transfected cell produced small amounts of IE1 aggregated into specific domains, which also contained increased amounts of HDAC-2, proving that no other viral protein is necessary for HDAC-2 accumulation. (H) Same as G, showing HDAC-2 only. The upper cell has the normal diffuse distribution of HDAC-2 except for exclusion from some spaces representing nucleoli. (I) PML−/− cells revealing Daxx enrichment in regions of condensed chromatin (solid blue from Hoechst 33258 staining). In the IE1-producing cell (lower right), most Daxx was removed from condensed chromatin and colocalized with IE1 at sites without chromatin (arrow). (J) Daxx−/− cell in lower right shows that in the presence of a small amount of IE1, PML was still in aggregates, whereas the cell in the upper left had PML dispersed in the presence of large amounts of IE1. (K) Same as J but stained only for PML, revealing dispersion of PML in the upper left cell. (L) Infected Daxx−/− cell in which Daxx HDAC-2 did not cosegregate with IE1 into specific domains. (M) Same as L, showing the distribution of HDAC-2 alone. p.i., postinfection.
Figure 1D to F depict the infected nucleus in Fig. C with three colors used for separated IE1, PML, and HDAC-2, showing the enrichment of HDAC-2 at ND10. To establish whether HDAC's recruitment to ND10 was due to IE1 or some other viral protein, we transfected MEF with pp89UC and tested for IE1 and HDAC-2. As shown in Fig. 1G and H, the transfected lower cell has HDAC-2 enriched at sites of IE1 aggregation, whereas in the untransfected cell HDAC-2 was evenly distributed throughout the nucleus except in the nucleolus (upper cell in Fig. 1G and H). Thus, IE1 and not some other viral protein is responsible for the presence of HDAC-2 at ND10. The presence of Daxx, with which HDAC reportedly interacts (26), at ND10 is not sufficient for this recruitment in untreated mouse fibroblasts.
PML has been shown to interact with IE1 of HCMV (1). We asked whether PML was necessary for IE1 of MCMV to be segregated. Both by infection and by transfection of PML−/− cells, we found that IE1 and Daxx colocalized in individual dots (shown in Fig. 1I for transfection). In most uninfected or untransfected PML−/− cells, Daxx was highly enriched in condensed chromatin (17). During IE1 accumulation, Daxx appeared to be enriched together with IE1 in certain aggregates and possibly diffuse in the nucleoplasm, while the condensed chromatin accumulations of Daxx were depleted. Therefore, it remains unclear whether the known interaction of Daxx with PML is essential for the recruitment of IE1 to ND10.
We used Daxx−/− cells to test whether Daxx is necessary for the deposition of IE1 at ND10. At 2 h postinfection, Daxx−/− cells showed colocalization of IE1 and PML (lower right nucleus in Fig. 1J and K), demonstrating that IE1 can be recruited to ND10 in the absence of Daxx. However, any nucleus with high IE1 content showed IE1 dispersed throughout the nucleus as well as the PML dispersal (upper left nucleus in Fig. 1J and K). These images were prevalent at 3 h postinfection and later. Normal mouse cells infected with MCMV or transfected with pp89UC also revealed dispersal of ND10 (data not shown) and thus showed an effect similar to that of IE1 of HCMV on human cells. The dispersal of ND10 by transient expression of IE1 takes many hours, as previously reported for human IE1 (data not shown) (18). Thus, IE1 recruitment to ND10 can be affected by either PML or Daxx, and the dispersion of ND10 by the latter was due to high concentrations of IE1.
We had observed that HDAC-2 needed IE1 to be deposited at ND10 in wild-type MEF. When we tested whether MCMV-infected Daxx−/− MEF had HDAC-2 at ND10 at very early infection times and before ND10 dispersal, we found no enrichment for HDAC-2 at PML aggregates (Fig. 1L and M). Recruitment of HDAC-2 therefore appears to depend on the presence of both IE1 and Daxx.
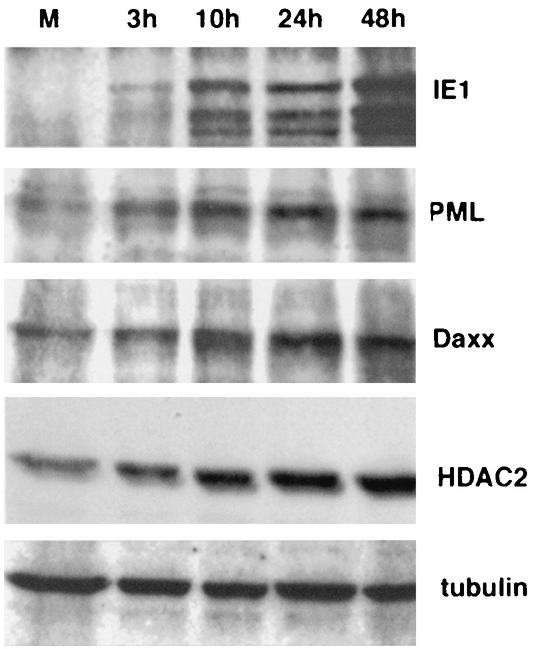
Dispersion of ND10 in herpes simplex virus type 1 is due to the ubiquitination activity of ICP0, followed by the proteosome-dependent hydrolysis of PML, Daxx, and Sp100 (11). To determine whether the MCMV IE1-dependent dispersion of ND10 might involve a similar process of protein degradation, we monitored the abundance of PML, Daxx, and HDAC-2 at different times of infection by Western blotting as shown in Fig. 2. IE1 synthesis was evident at 3 h postinfection, and the production increased over time. PML was present at low concentrations in uninfected cells, but instead of a reduction, an increase was already apparent after 3 h of infection, and it became even more abundant at 10 h postinfection The same results were observed for Daxx and HDAC-2. Tubulin served as a loading control. The increase in ND10-associated proteins, which usually results in larger ND10 (32), shows that MCMV IE1 disperses ND10 by a mechanism other than degradation of the constituent proteins.
FIG. 2.
Western blot analysis of various proteins from MEF infected with MCMV at increasing times postinfection There was an increase in PML, Daxx, and HDAC-2 beginning at 3 h postinfection which was quite obvious at 10 h postinfection. Tubulin was used as a loading control. Lane M, mock-infected MEF.
IE1 binds PML, Daxx, and HDAC-2 independently.
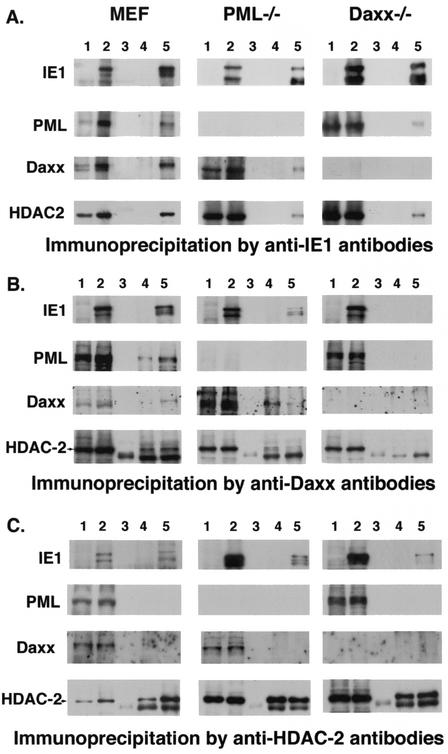
Because single-cell analysis by immunohistochemical methods provides spatial data but cannot address whether IE1 actually binds to the proteins colocalizing with it, a coimmunoprecipitation assay was used to test for direct or indirect binding of IE1 to PML, Daxx, and HDAC-2. Since PML can bind to Daxx (17) and Daxx can bind to HDAC (26), we used PML−/− and Daxx−/− cells in addition to wild-type MEF to determine whether any of the interactions are mediated by PML or Daxx. MEF and PML−/− and Daxx−/− MEF were infected with MCMV, and nuclear extracts were prepared for immunoprecipitation at 16 h postinfection.
In Fig. 3A, we show in each row the input protein from uninfected cells (lane 1) and infected cells (lane 2). Lane 3 shows the control for beads with nonimmune serum or beads with no serum from infected cells, whereas lane 4 shows the immunoprecipitate from anti-IE1-charged beads used on the uninfected cell extract and lane 5 shows the experimental sample where anti-IE1-charged beads were used on extracts from infected cells. The anti-IE1-charged beads immunoprecipitated IE1 in all three cell types. When the gels were analyzed for coimmunoprecipitated proteins, we found that IE1 brought down PML, Daxx, and HDAC-2 from MCMV-infected MEF extract. In PML−/− cells, Daxx and HDAC-2 were coimmunoprecipitated, and in Daxx−/− cells, PML and HDAC-2 were immunoprecipitated. These results suggest that PML and HDAC-2 are not dependent on Daxx and that Daxx and HDAC-2 are not dependent on PML for binding to IE1.
FIG. 3.
Coimmunoprecipitation analysis with antibodies against IE1, Daxx, and HDAC-2 in MCMV-infected wild-type cells. PML−/− and Daxx−/− cells were used as controls. Lane 1 shows the uninfected and lane 2 the infected nuclear extract; lane 3 contains the proteins precipitated by bare beads (when used with monoclonal antibodies) (A) or with beads charged with nonimmune serum (when used with rabbit serum) (B and C); lane 4 contains the proteins immunoprecipitated from uninfected cells. and lane 5 contains those from infected cells. In B and C, lanes 3 to 5 of the HDAC-2 row, the lower band represents the IgG heavy chain.
The reverse immunoprecipitation, to confirm the binding of IE1 to Daxx and HDAC-2, revealed that IE1 and, as expected, PML and HDAC-2 precipitated with anti-Daxx antibodies (Fig. 3B). The specificity of the coimmunoprecipitation was confirmed by the absence of IE1, PML, and HDAC-2 in Daxx−/− cells (Fig. 3B, lanes 4 and 5). The lower band labeled in the anti-HDAC-2-probed samples represents the heavy chain because we used rabbit serum. When anti-HDAC-2 antibodies were used, IE1 but not Daxx or PML immunoprecipitated (Fig. 3C). At present it is not clear why PML and Daxx were not precipitated by HDAC-2 in infected cells, although HDAC-2 did also not precipitate Daxx in the uninfected cells, contrary to expectations based on a previous study (26). Immunoprecipitation was not successful with the available antibodies against mouse PML.
Figure 4 summarizes the results from the coimmunoprecipitation analyses in schematic form. The arrowheads point to the protein used to “capture” the protein from which the arrow originates. The figure indicates that IE1 can bind to PML, Daxx, and HDAC-2 and that neither Daxx nor PML is necessary for the other proteins to interact with IE1, validating the recruitment data derived by in situ staining of cells at very early times after infection.
FIG. 4.
Schematic diagram of coimmunoprecipitation results. The arrowhead points to the protein used to capture the protein from which the arrow originates.
HDAC activity is suppressed by binding to IE1.
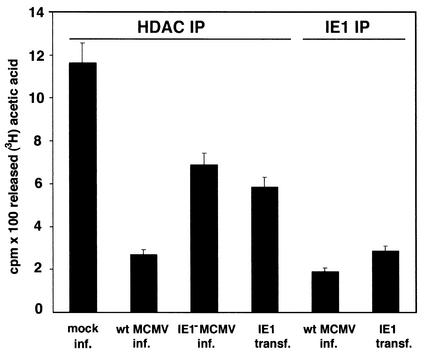
If IE1 binds PML, Daxx, and HDAC-2, we wondered what effect this binding has on these repressors. For PML and Daxx, indirect evidence from transient overexpression studies exists for repressive activities, but no direct in vitro assay is known. For HDAC, however, an in vitro assay exists with which to assess the deacetylation activity of HDAC. We used HDAC-2 immunoprecipitated from uninfected control cells and compared the deacetylation activity with that of HDAC-2 from MCMV-infected cells. The conditions of the in vitro assay were normalized by determining the amount of immunoprecipitated HDAC-2 with a Western blot as in Fig. 3. The immunoprecipitated HDAC was within a twofold range, and its concentration was adjusted accordingly before addition to the assay mixture.
Deacetylation activity was substantially reduced in infected cells (Fig. 5, lane 2). Also in IE1-transfected cells (Fig. 5, lane 4), HDAC activity was reduced despite the fact that only about 30% of the cells were transfected and therefore much of the HDAC was not exposed to IE1 in the cell. Potentially soluble IE1 and HDAC interacted after the opening of the cell. A somewhat stronger repression of HDAC-2 activity was observed when anti-IE1 antibodies were used to coimmunoprecipitate HDAC from MCMV-infected cells and thus all of the HDAC used for the activity assay was bound at the time of immunoprecipitation. A strong reduction in HDAC-2 activity was also found when anti-IE1 antibodies were used to coimmunoprecipitate HDAC-2 from IE1-transfected cells (Fig. 5, lane 5). However, the decrease in HDAC activity in infected cells does not appear to be due solely to IE1, because when IE1− MCMV was used for infection, we found a repeatable lowering of HDAC activity (Fig. 5, lane 3). The HDAC activity assay after infection or transfection strongly suggests that binding of HDAC-2 to IE1 decreases the deacetylation activity and that binding is not transitory.
FIG. 5.
Deacetylation activity with 3H-acetylated histone 4 as the substrate and HDAC immunoprecipitated (IP) by anti-HDAC-2 antibodies or anti-IE1 antibodies. Infection substantially reduced HDAC activity (second bar). IE1 bound HDAC from infected (inf.) and transfected (transf.) cells showed low activity (fifth and sixth bars, respectively). Results are mean 102 cpm from two independent experiments.wt, wild type.
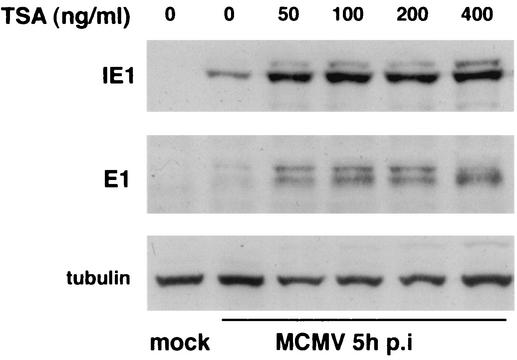
If deacetylation suppresses transcriptional activity, preventing this deacetylation activity should have the opposite effect. We tested this contention on MCMV-infected cells by inhibiting HDAC activity with various concentrations of the HDAC inhibitor trichostatin-A and assaying for IE1 and E1 protein accumulation. Western blot analysis at 5 h postinfection revealed a substantial increase in IE1 at the lowest concentration of trichostatin-A used (50 ng/ml) (Fig. 6). E1 accumulation tested at 5 h postinfection with 50 ng of trichostatin-A per ml showed a similar increase. Together, the results of the in vitro deacetylation assay and the in vivo HDAC inhibition assay show independently that HDAC is inhibited by IE1.
FIG. 6.
Comparison of IE1 and E1 protein production at 5 h postinfection in trichostatin-A-treated and untreated MCMV-infected MEF. Infection of MEF with wild-type virus had been predetermined to produce approximately equal numbers of E1-positive cells in the absence of trichostatin-A. Cells were exposed to various concentrations of trichostatin-A (TSA) 2 h before and during infection. Both IE1 and E1 production increased in wild-type-infected cells at the lowest trichostatin-A concentration tested. Tubulin was used as a loading control. p.i., postinfection.
HDAC and acetylated histone 3 bind to the major immediate-early promoter.
If trichostatin-A leads to an increase in IE1 protein expression, HDAC may, under normal conditions, repress immediate-early transcription. It may deacetylate the major immediate-early promoter or keep it deacetylated. To test whether HDAC is associated with the immediate-early promoter, we used chromatin immunoprecipitation analysis. Chromatin was precipitated with anti-HDAC-2 antibodies, followed by PCR with primers against two parts of the immediate-early promoter region. Figure 7 shows that both regions tested were precipitated by anti-HDAC-2 antibodies. Rabbit serum-coated beads were used as a control. If the major immediate-early promoter exists in a nucleosomal form, the histones of the active promoter should be acetylated. Chromatin precipitated with the antibodies against histone 3 acetylated at K23 contained the expected PCR products with the two primers of the IE1 promoters. These results demonstrate that MCMV genomes are present as chromatin and that some histone associated with those viral genomes is acetylated, apparently representing the active histones. Other nucleosome-bound histones of viral genomes are deacetylated and may represent inactive genomes.
FIG. 7.
Chromosome immunoprecipitation assay of MCMV-infected MEF. Cells were fixed 24 h postinfection, and the sonicated chromatin was immunoprecipitated with anti-HDAC2 and anti-histone 3 K23 antibodies. PCR products are shown for the primers at the indicated sites of the immediate-early promoter. Beads coated with preimmune serum were used as controls. Both the HDAC-2 and histone 3 K23 antibodies precipitated both immediate-early promoter domains.
In vivo effect of HDAC inhibition on infectivity and prereplication domain formation.
If there are competent inactive genomes in the nucleus and if they are held inactive by the deacetylation of histones bound to the immediate-early promoter, inhibition of deacetylation should relieve the repression. Also, the possibility existed that a 2-h pretreatment with trichostatin-A might change the infectivity of the cell. This could also result in changes in IE1 and E1 production in cultures of cells incubated with the same number of particles. We quantitated the number of infected cells in cultures exposed or not exposed to 100 ng of trichostatin-A per ml by labeling with anti-IE1 and anti-E1 antibodies. Lengthy trichostatin-A treatment was avoided to guard against potential trichostatin-A-dependent detrimental effects on the host cell (2, 41). Cells producing IE1 at 3 h postinfection were counted as infected.
IE1 was found in ND10 only, in ND10 and diffuse, or completely diffuse (where ND10 is already dispersed) (Fig. 8A). The different images provide a rough measure of the progression of infection, with high concentrations and diffuse IE1 distribution representing the more advanced stage (18). The different images in the same culture at the same time after infection are thought to depend on the number of competent particles infecting individual cells (18). We found that the total number of cells displaying IE1 was the same independent of trichostatin-A treatment (Table 1), indicating that the rate of infection was not influenced by trichostatin-A. However, the number of cells with IE1 diffusely distributed throughout the nucleus was increased after trichostatin-A treatment versus untreated control cells. This suggests either that an increased number of viral genomes actively transcribe in the presence of trichostatin-A or that a constant number of viral genomes have increased transcription.
FIG. 8.
MCMV-infected MEF used for quantitative assessment of infection and prereplication compartment formation. (A) Cells used for quantitative assessment of infection were tested after staining for IE1 at 3 h postinfection (p.i.). The image presented shows the three IE1 staining categories distinguished, although infected cells were usually not as closely spaced. (B) Location of prereplication domains, with most cells showing several small ones and about two large ones, with substantial amounts of E1 in the nucleoplasm. Large and small prereplication compartments were counted separately.
TABLE 1.
Effect of trichostatin-A (TSA) on infection and prereplication domain formation
| MCMV infection and treatment | Time postinfection (h) | Total no. of cells counted | % IE positive (no.) | % E1 positive (no.) | Ratio, no TSA/TSA |
|---|---|---|---|---|---|
| Wild type | 3 | 1:1.04 | |||
| No TSA | 1,038 | 14.3 (147) | |||
| TSA | 1,089 | 14.9 (162) | |||
| Wild type | 5 | 1:1.47 | |||
| No TSA | 1,056 | 6.6 (69) | |||
| TSA | 1,065 | 9.7 (103) | |||
| ΔIE1 | 5 | 1:1.81 | |||
| No TSA | 1,529 | 6.5 (100) | |||
| TSA | 1,448 | 11.8 (170) |
One estimate of the number of active viral particles proceeding to the replicative state is the formation of prereplication domains, visualized as aggregation of the DNA-binding protein E1, which, like ICP8 for herpes simplex virus type 1, indicates the location where replication domains are forming (27, 29). E1 aggregates can be found as early as 2 h postinfection. Analysis of E1 distribution in wild-type MCMV-infected MEF at 5 h postinfection revealed large and small aggregates (Fig. 8B) in which progeny viral DNA will form (Tang et al., unpublished data). When they were independently assessed, we found that the number of small or large aggregates did not differ in trichostatin-A-treated versus untreated cells (10.7 versus 10.6), suggesting that it takes a certain number of active genomes to reach productive infection. However, the total number of cells with E1 accumulations was nearly 50% higher when trichostatin-A was present. So, in contrast to the infection rate, the number of cells with E1 accumulations or prereplication domains was lower when deacetylation activity was not inhibited.
If IE1 inhibits HDAC activity, HDAC activity should be higher in the absence of IE1 and more viral genomes should be suppressed. Trichostatin-A exposure of these infected cells should prevent HDAC-dependent suppression. An apparently higher productive infectivity should then be noticed in the presence of trichostatin-A. When IE1-deleted MCMV was used to infect MEF, we found that nearly twice as many cells formed prereplication sites as in the absence of HDAC inhibition with trichostatin-A (Table 1). Most of the additional cells had small prereplication sites. These differences suggest that inhibition of HDAC activity can bring additional cells into the productive replicative cycle and that IE1 is involved in the suppression of HDAC. They also suggest that infected fibroblasts with competent virus genomes exist that apparently do not proceed to the replicative state.
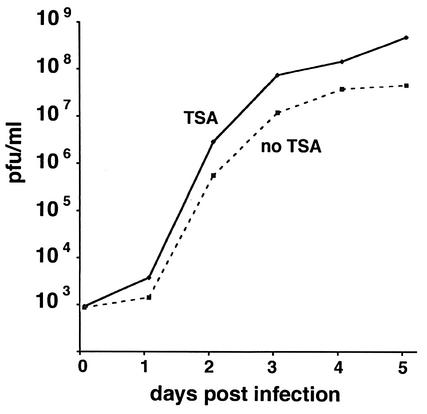
The increase in cells with prereplication domains after exposure to trichostatin-A was about 50% in wild-type MCMV-infected cells, but this does not prove that an increase in productivity can be achieved by the suppression of HDAC. An increase in productively infected cells should be reflected in an increase in competent viral particle formation. We therefore determined the PFU produced in MEF after infection with about 0.1 PFU in the presence and absence of trichostatin-A. Figure 9 shows that there was already an early increase in PFU at 24 h postinfection, which increased over time to a 10-fold-higher number of PFU in trichostatin-A-treated cells.
FIG. 9.
Virus produced from equally infected MEF cultures treated or not with 50 ng of trichostatin-A per ml were collected over time and used to determine the number of PFU. Trichostatin-A (TSA)-treated cultures produced about 8- to 10-fold more PFU than untreated cells.
DISCUSSION
Our analysis of MCMV-infected mouse cells showed that ND10 are dispersed during immediate-early protein synthesis, analogous to the findings in HCMV infection (18, 27, 29). Use of an IE1-deleted virus and a plasmid transiently expressing IE1 demonstrated that MCMV IE1, as in the human system, disperses PML and, in turn, the other ND10-associated proteins. However, unlike the ND10-dispersing ICP0 of herpes simplex virus type 1, IE1 of MCMV did not lead to hydrolysis of PML but instead to increased amounts of PML, consistent with the increased transcripts of interferon-induced proteins in HCMV infection (5), but an increase in Daxx and HDAC-2 was also observed. If these proteins are repressors and aredestroyed by ICP0 to advance the replicative success of herpes simplex virus type 1, MCMV must use a different mechanism to inactivate the increasing amounts of these repressor proteins.
IE1 of MCMV, like its HCMV counterpart, is first segregated into ND10 and only later disperses these structures when present in larger quantities throughout the nucleus. IE1 expressed after transfection also remains segregated in ND10 for prolonged times or until substantial amounts of IE1 have accumulated in the nucleus. Thus, rather than an enzymatically dependent dispersion, ND10 might undergo a gradual decline as IE1 binds PML and titrates it from ND10, as suggested previously (1). Interestingly, HDAC-2 was recruited into ND10 by IE1 and later released upon the IE1-dependent dispersal of ND10. From this observation, we contemplate the possibility of two competing processes mediated through the same IE1 binding properties.
The first is the segregation of IE1 by the cell into ND10, which, if totally effective, would result in a low productive infection, as seen with competent IE1-deleted viral genomes (15). The second process is overwhelming the cellular defense by the abundant production of IE1, thus titrating the repressive proteins of the cell through IE1-binding induced inactivation. The former may more likely happen in low-particle infection, and the latter with higher particle infection. The higher ratio of cells forming prereplication domains in trichostatin-A-treated IE1-deleted virus infection than in trichostatin-A-treated IE1-producing cells suggests that many competent viruses are repressed by the cell in the absence of IE1 (15). IE1 then should function by counteracting the repressive activities of the cell.
The ability of IE1 to bind to certain repressive host proteins and their functional inactivation through this binding would be the crucial property of IE1 that allows it to advance productive infection. Our coimmunoprecipitation experiments demonstrated the binding of IE1 to PML, Daxx, and HDAC-2. In those analyses, mouse PML−/− and Daxx−/− cells provided crucial controls and the additional information that PML can recruit and bind to IE1 without the adapter function of Daxx. Conversely, Daxx can bind to IE1 in the absence of PML. Thus, these proteins can interact with IE1 independently. It even appears that Daxx can aggregate or precipitate small amounts of IE1 in the absence of PML. Binding of the repressor proteins Daxx and PML directly or indirectly is therefore established for the multifunctional IE1 of MCMV. Unfortunately, there are no direct assays for the functions of PML and Daxx as an independent means to test if IE1 binding changes the functions of the interferon-upregulated PML and Daxx proteins that could change the replicative success of the virus.
Daxx had been reported to bind to HDAC (26), although we could not detect any HDAC at ND10 in uninfected cells. HDAC might bind to the same C-terminal domain on Daxx that is used to bind to PML (17) and therefore not bind to PML at ND10. However, HDAC-2 and presumably HDAC-1, which is often found in the same complex (4), are segregated to ND10 by IE1, and there Daxx appears to exert essential adapter functions, as Daxx−/− cells show no enrichment of HDAC at PML-positive sites. An in vitro assay exists for HDAC, and we could show that HDAC bound to IE1 has substantially decreased activity. This assay as well as the coimmunoprecipitation analysis provided strong evidence that IE1 and HDAC-2 do not interact transiently but remain bound. Assuming that this nontransient binding also occurs in the cell, IE1 might inhibit HDAC activity by making it unavailable to its histone substrate and thus retaining the acetylated state conductive to continued transcription.
Deacetylation has been equated with inducing transcriptional repression (23). We observed an increase in viral IE1 and E1 production as well as a substantial increase in the production of competent viral particles when deacetylation was inhibited by trichostatin-A. This is consistent with the idea that IE1 suppresses HDAC. Since HDAC may not only deacetylate histones (23), the mechanism of HDAC suppression of MCMV remains obscure. However, histone deacetylation as a potential repressive mechanism implies a requirement for chromatinization of infecting viral genomes. The ability of antibodies to precipitate the various immediate-early promoter regions showed that MCMV is present as chromatin during infection of permissive cells. Acetylation-dependent activation of the lytic cycle of Epstein-Barr virus (7) and reactivation of latent HCMV from T2 cells (35) by trichostatin-A have been reported. Here we could show that both acetylated and deacetylated IE1 promoter regions exist in the same culture of permissive cells during lytic infection.
A possible consequence of histone-induced nucleosome formation of MCMV is deacetylation of the respective histones and suppression of transcription from those genomes. If such chromatization and repression occur during normal infection of permissive cells, a block of deacetylation would allow transcription of more viral genomes. Such a shift in balance would only be observed when some infected cells successfully block all viral genomes from proceeding to the early stage of the replication cascade. Our analysis of such a scenario revealed an approximately 50% increase in the number of cells supporting productive infection when deacetylation is blocked. Significantly, an additional 87% of the cells produced prereplication sites under the same conditions when IE1 was absent. This increase was not due to a potential trichostatin-A-induced change in infection rate.
Whether by deacetylation of histones or other proteins, inhibition of HDAC activity appears to drive viral genomes into the productive cycle, genomes that would otherwise remain repressed. The mechanism that represses MCMV in infected permissive fibroblasts may have consequences for the progression to virus latency. In fact, this finding suggests that some fibroblasts may harbor latent virus if the same mechanism works in vivo and the cells are not eliminated by the immune system. This should be a testable possibility.
The mechanism with which IE1 appears to effect suppression of the cellular defense mechanism, here shown directly only for HDAC, is binding and thus blocking HDAC activity on viral genomes. If this is the case, HDAC seems to prevent acetylation or reacetylation of the viral chromatin and the accompanying activation of viral transcription. IE1 binding of other proteins such as PML and Daxx might also be important to prevent inactivation of the viral genome, although we presently have no good concept of how this might be. In this context, it is interesting that IE1 has a histone-binding domain and binds avidly to histones (33), thus making unavailable histones that would tend to form nucleosomes with the replicating viral DNA. IE1 may be using the same mechanism, binding, to prevent nucleosome formation, which is detrimental in packaging in the late stage of the replicative cycle. Inactivation of cellular repressors by binding may play an important role in the apparent transactivating properties of some viral proteins.
Acknowledgments
We thank M. Messerle and U. Koszinowski for most generously providing the MCMV IE1 deletion mutant and various probes with which to start this project, P. Pandolfi for the PML−/− cells, and S. Lowe for the PML antibodies.
This study was supported by funds from NIH AI 41136 and from the Human Frontier Science Program and the G. Harold and Leila Y. Mathers Charitable Foundation. NIH core grant CA-10815 is acknowledged for support of the microscopy and sequencing facility.
REFERENCES
- 1.Ahn, J. H., E. J. Brignole 3rd, and G. S. Hayward. 1998. Disruption of PML subnuclear domains by the acidic IE1 protein of human cytomegalovirus is mediated through interaction with PML and may modulate a RING finger-dependent cryptic transactivator function of PML. Mol. Cell. Biol. 18:4899-4913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amin, H. M., S. Saeed, and S. Alkan. 2001. Histone deacetylase inhibitors induce caspase-dependent apoptosis and downregulation of daxx in acute promyelocytic leukaemia with t(15;17). Br. J. Haematol. 115:287-297. [DOI] [PubMed] [Google Scholar]
- 3.Andrews, N. C., and D. V. Faller. 1991. A rapid micropreparation technique for extraction of DNA-binding proteins from limiting numbers of mammalian cells. Nucleic Acids Res. 19:2499.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ashburner, B. P., S. D. Westerheide, and A. S. Baldwin, Jr. 2001. The p65 (RelA) subunit of NF-κB interacts with the histone deacetylase (HDAC) corepressors HDAC1 and HDAC2 to negatively regulate gene expression. Mol. Cell. Biol. 21:7065-7077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Browne, E. P., B. Wing, D. Coleman, and T. Shenk. 2001. Altered cellular mRNA levels in human cytomegalovirus-infected fibroblasts: viral block to the accumulation of antiviral mRNAs. J. Virol. 75:12319-12330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Buhler, B., G. M. Keil, F. Weiland, and U. H. Koszinowski. 1990. Characterization of the murine cytomegalovirus early transcription unit e1 that is induced by immediate-early proteins. J. Virol. 64:1907-1919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chang, L. K., and S. T. Liu. 2000. Activation of the BRLF1 promoter and lytic cycle of Epstein-Barr virus by histone acetylation. Nucleic Acids Res. 28:3918-3925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chelbi-Alix, M. K., L. Pelicano, F. Quignon, M. H. Koken, L. Venturini, M. Stadler, J. Pavlovic, L. Degos, and H. de The. 1995. Induction of the PML protein by interferons in normal and APL cells. Leukemia 9:2027-2033. [PubMed] [Google Scholar]
- 9.Chelbi-Alix, M. K., F. Quignon, L. Pelicano, M. H. Koken, and H. de The. 1998. Resistance to virus infection conferred by the interferon-induced promyelocytic leukemia protein. J. Virol. 72:1043-1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Doucas, V., A. M. Ishov, A. Romo, H. Juguilon, M. D. Weitzman, R. M. Evans, and G. G. Maul. 1996. Adenovirus replication is coupled with the dynamic properties of the PML nuclear structure. Genes Dev. 10:196-207. [DOI] [PubMed] [Google Scholar]
- 11.Everett, R. D. 2000. ICP0 induces the accumulation of colocalizing conjugated ubiquitin. J. Virol. 74:9994-10005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Everett, R. D., P. Freemont, H. Saitoh, M. Dasso, A. Orr, M. Kathoria, and J. Parkinson. 1998. The disruption of ND10 during herpes simplex virus infection correlates with the Vmw110- and proteasome-dependent loss of several promyelocytic leukemia protein isoforms. J. Virol. 72:6581-6591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Everett, R. D., and G. G. Maul. 1994. herpes simplex virus type 1 immediate-early protein Vmw110 causes redistribution of PML. EMBO J. 13:5062-5069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gongora, R., R. P. Stephan, Z. Zhang, and M. D. Cooper. 2001. An essential role for daxx in the inhibition of b lymphopoiesis by type I interferons. Immunity 14:727-737. [DOI] [PubMed] [Google Scholar]
- 15.Greaves, R. F., and E. S. Mocarski. 1998. Defective growth correlates with reduced accumulation of a viral DNA replication protein after low-multiplicity infection by a human cytomegalovirus IE1 mutant. J. Virol. 72:366-379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Grotzinger, T., T. Sternsdorf, K. Jensen, and H. Will. 1996. Interferon-modulated expression of genes encoding the nuclear-dot-associated proteins Sp100 and promyelocytic leukemia protein (PML). Eur. J. Biochem. 238:554-560. [DOI] [PubMed] [Google Scholar]
- 17.Ishov, A. M., A. G. Sotnikov, D. Negorev, O. V. Vladimirova, N. Neff, T. Kamitani, E. T. Yeh, J. F. Strauss 3rd, and G. G. Maul. 1999. PML is critical for ND10 formation and recruits the PML-interacting protein Daxx to this nuclear structure when modified by SUMO-1. J. Cell Biol. 147:221-234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ishov, A. M., R. M. Stenberg, and G. G. Maul. 1997. Human cytomegalovirus immediate-early interaction with host nuclear structures: definition of an immediate transcript environment. J. Cell Biol. 138:5-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Keil, G. M., A. Ebeling-Keil, and U. H. Koszinowski. 1987. Immediate-early genes of murine cytomegalovirus: location, transcripts, and translation products. J. Virol. 61:526-533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Keil, G. M., A. Ebeling-Keil, and U. H. Koszinowski. 1987. Sequence and structural organization of murine cytomegalovirus immediate-early gene 1. J. Virol. 61:1901-1908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kentsis, A., E. C. Dwyer, J. M. Perez, M. Sharma, A. Chen, Z. Q. Pan, and K. L. Borden. 2001. The RING domains of the promyelocytic leukemia protein PML and the arenaviral protein Z repress translation by directly inhibiting translation initiation factor eIF4E. J. Mol. Biol. 312:609-623. [DOI] [PubMed] [Google Scholar]
- 22.Korioth, F., G. G. Maul, B. Plachter, T. Stamminger, and J. Frey. 1996. The nuclear domain 10 (ND10) is disrupted by the human cytomegalovirus gene product IE1. Exp. Cell Res. 229:155-158. [DOI] [PubMed] [Google Scholar]
- 23.Kouzarides, T. 2000. Acetylation: a regulatory modification to rival phosphorylation? EMBO J. 19:1176-1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lavau, C., A. Marchio, M. Fagioli, J. Jansen, B. Falini, P. Lebon, F. Grosveld, P. P. Pandolfi, P. G. Pelicci, and A. Dejean. 1995. The acute promyelocytic leukaemia-associated PML gene is induced by interferon. Oncogene 11:871-876. [PubMed] [Google Scholar]
- 25.Lehembre, F., S. Muller, P. P. Pandolfi, and A. Dejean. 2001. Regulation of Pax3 transcriptional activity by SUMO-1-modified PML. Oncogene 20:1-9. [DOI] [PubMed] [Google Scholar]
- 26.Li, H., C. Leo, J. Zhu, X. Wu, J. O'Neil, E. J. Park, and J. D. Chen. 2000. Sequestration and inhibition of daxx-mediated transcriptional repression by PML. Mol. Cell. Biol. 20:1784-1796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Maul, G. G. 1998. Nuclear domain 10, the site of DNA virus transcription and replication. Bioessays 20:660-667. [DOI] [PubMed] [Google Scholar]
- 28.Maul, G. G., H. H. Guldner, and J. G. Spivack. 1993. Modification of discrete nuclear domains induced by herpes simplex virus type 1 immediate early gene 1 product (ICP0). J. Gen.. Virol. 74:2679-2690. [DOI] [PubMed] [Google Scholar]
- 29.Maul, G. G., A. M. Ishov, and R. D. Everett. 1996. Nuclear domain 10 as preexisting potential replication start sites of herpes simplex virus type-1. Virology 217:67-75.8599237 [Google Scholar]
- 30.Meier, J. L. 2001. Reactivation of the human cytomegalovirus major immediate-early regulatory region and viral replication in embryonal NTera2 cells: role of trichostatin A, retinoic acid, and deletion of the 21-base-pair repeats and modulator. J. Virol. 75:1581-1593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Messerle, M., B. Buhler, G. M. Keil, and U. H. Koszinowski. 1992. Structural organization, expression, and functional characterization of the murine cytomegalovirus immediate-early gene 3. J. Virol. 66:27-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mocarski, E. S., Jr., and C. T. Courcelle. 2001. Cytomegaloviruses and their replication, p. 2629-2673. In D. M. Knipe and P. M. Howley (ed.), Fields virology. Lippincott Williams and Wilkins, Philadelphia, Pa.
- 33.Munch, K., B. Buhler, M. Messerle, and U. H. Koszinowski. 1991. The core histone-binding region of the murine cytomegalovirus 89K immediate early protein. J. Gen. Virol. 72:1967-1974. [DOI] [PubMed] [Google Scholar]
- 34.Munch, K., M. Messerle, B. Plachter, and U. H. Koszinowski. 1992. An acidic region of the 89K murine cytomegalovirus immediate early protein interacts with DNA. J. Gen. Virol. 73:499-506. [DOI] [PubMed] [Google Scholar]
- 35.Murphy, J. C., W. Fischle, E. Verdin, and J. H. Sinclair. 2002. Control of cytomegalovirus lytic gene expression by histone acetylation. EMBO J. 21:1112-1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rawlinson, W. D., H. E. Farrell, and B. G. Barrell. 1996. Analysis of the complete DNA sequence of murine cytomegalovirus. J. Virol. 70:8833-8849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Regad, T., A. Saib, V. Lallemand-Breitenbach, P. P. Pandolfi, H. de The, and M. K. Chelbi-Alix. 2001. PML mediates the interferon-induced antiviral state against a complex retrovirus via its association with the viral transactivator. EMBO J. 20:3495-3505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stadler, M., M. K. Chelbi-Alix, M. H. Koken, L. Venturini, C. Lee, A. Saib, F. Quignon, L. Pelicano, M. C. Guillemin, C. Schindler, et al. 1995. Transcriptional induction of the PML growth suppressor gene by interferons is mediated through an ISRE and a GAS element. Oncogene 11:2565-2573. [PubMed] [Google Scholar]
- 39.Tang, Q., P. Bell, P. Tegtmeyer, and G. G. Maul. 2000. Replication but not transcription of simian virus 40 DNA is dependent on nuclear domain 10. J. Virol. 74:9694-9700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vallian, S., K. V. Chin, and K. S. Chang. 1998. The promyelocytic leukemia protein interacts with Sp1 and inhibits its transactivation of the epidermal growth factor receptor promoter. Mol. Cell. Biol. 18:7147-7156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vigushin, D. M., S. Ali, P. E. Pace, N. Mirsaidi, K. Ito, I. Adcock, and R. C. Coombes. 2001. Trichostatin A is a histone deacetylase inhibitor with potent antitumor activity against breast cancer in vivo. Clin. Cancer Res. 7:971-976. [PubMed] [Google Scholar]
- 42.Wagner, M., S. Jonjic, U. H. Koszinowski, and M. Messerle. 1999. Systematic excision of vector sequences from the BAC-cloned herpesvirus genome during virus reconstitution. J. Virol. 73:7056-7060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wagner, M., D. Michel, P. Schaarschmidt, B. Vaida, S. Jonjic, M. Messerle, T. Mertens, and U. Koszinowski. 2000. Comparison between human cytomegalovirus pUL97 and murine cytomegalovirus (MCMV) pM97 expressed by MCMV and vaccinia virus: pM97 does not confer ganciclovir sensitivity. J. Virol. 74:10729-10736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang, Z. G., L. Delva, M. Gaboli, R. Rivi, M. Giorgio, C. Cordon-Cardo, F. Grosveld, and P. P. Pandolfi. 1998. Role of PML in cell growth and the retinoic acid pathway. Science 279:1547-1551. [DOI] [PubMed] [Google Scholar]
- 45.Wilkinson, G. W., C. Kelly, J. H. Sinclair, and C. Rickards. 1998. Disruption of PML-associated nuclear bodies mediated by the human cytomegalovirus major immediate early gene product. J. Gen. Virol. 79:1233-1245. [DOI] [PubMed] [Google Scholar]
- 46.Wu, W. S., S. Vallian, E. Seto, W. M. Yang, D. Edmondson, S. Roth, and K. S. Chang. 2001. The growth suppressor PML represses transcription by functionally and physically interacting with histone deacetylases. Mol. Cell. Biol. 21:2259-2268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wu, W. S., Z. X. Xu, R. Ran, F. Meng, and K. S. Chang. 2002. Promyelocytic leukemia protein PML inhibits Nur77-mediated transcription through specific functional interactions. Oncogene 21:3925-3933. [DOI] [PubMed] [Google Scholar]