Abstract
Memory CD4 T-cell responses against respiratory syncytial virus (RSV) were evaluated in peripheral blood mononuclear cells of healthy blood donors with gamma interferon enzyme-linked immunospot (Elispot) assays. RSV-specific responses were detected in every donor at levels varying between 0.05 and 0.3% of CD4 T cells. For all donors tested, a considerable component of the CD4 T-cell response was directed against the fusion (F) protein of RSV. We characterized a set of 31 immunodominant antigenic peptides targeted by CD4 T cells in the context of the most prevalent HLA class II molecules within the Caucasian population. Most antigenic peptides were HLA-DR restricted, whereas two dominant DQ peptides were also identified. The antigenic peptides identified were located across the entire sequence of the F protein. Several peptides were presented by more than one major histocompatibility complex class II molecule. Furthermore, most donors recognized several F peptides. Detailed knowledge about immunodominant antigenic peptides will facilitate the ability to monitor CD4 T-cell responses in patients and the measurement of correlates of protection in vaccinated subjects.
Human respiratory syncytial virus (RSV), classified in the Pneumovirus genus of the family Paramyxoviridae, is a major cause of lower respiratory disease in young infants, immunocompromised individuals, and elderly people (15, 16, 17, 21, 26, 35). A vaccine is currently not available. In an early vaccine trial with a formalin-inactivated alum-precipitated human RSV vaccine given intramuscularly, enhancement of disease occurred in vaccinees upon subsequent exposure to the natural virus (12, 30). This enhanced illness was characterized by bronchiolitis, hypoxemia, and pneumonia and infiltration into the lungs of lymphocytes, neutrophils, and eosinophils (12, 30, 41). This cellular infiltration suggested that immune-mediated injury can contribute to the pathogenicity of RSV disease.
Primary infection with RSV can cause lower respiratory tract disease in young infants, manifesting as pneumonia or bronchiolitis (43). The disease is associated with an inflammatory response to infection, likely involving the production of cytokines and chemokines by lung epithelial cells and the recruitment of immune cells into the lungs. The T-cell response is an essential component of the immune response needed for viral clearance from the lungs (1, 18, 24). Antibodies against the fusion protein (F) and the attachment protein (G) are generated during RSV infection, but even in the presence of high levels of virus-neutralizing antibodies, reinfections occur (27), and antibodies are not needed for viral clearance (25). Also, vaccination with the formalin-inactivated RSV vaccine induced high titers of RSV-specific antibodies in vaccinees, yet caused more severe clinical disease upon natural infection (12, 36, 29).
While T-cell responses are required in RSV infections for clearance of RSV from the lung, it has been shown in murine studies that CD4 as well as CD8 T cells can be responsible for enhanced lung pathology (4, 9). Severe pneumonia with extensive influx of eosinophils into the lungs can be elicited in mice by type 2 helper T (Th2) cells specific for RSV G, which are primed during vaccination with formalin-inactivated RSV before intranasal challenge with live RSV (13, 49). This strong eosinophilic inflammation is also observed after RSV challenge of mice vaccinated with a vaccinia virus recombinant expressing solely the G protein of RSV (39).
A peptide corresponding to residues 183 to 195 in the G protein of RSV is recognized by Th2 cells in BALB/c mice (46). CD8 T-cell responses against epitopes derived from the G protein have not been observed in either BALB/c mice or humans (2, 6, 11, 38). CD4 T cells in mice that are vaccinated with a vaccinia virus recombinant expressing the F protein of RSV that are subsequently challenged with live virus produce less interleukin-4 and interleukin-5 than G-primed mice, and there is no eosinophil influx into the lungs (45).
CD8 T-cell responses against F are common in mice of different major histocompatibility complex (MHC) types (10; G. van Bleek, unpublished results). In the BALB/c model, a CD8 T-cell response has been shown to regulate the outcome of CD4 T-cell responses preventing enhanced disease (44). Whether these insights obtained from the murine model are relevant for the human situation has to be evaluated.
In peripheral blood mononuclear cells (PBMC) from healthy adults as well as from diseased infants, CD8 T-cell responses and CD4 T-cell responses can be detected. Although some data on the molecular targets of the human antiviral CD4 and CD8 T-cell responses have been published, epitopes have only been described for a limited number of MHC molecules and not in much molecular detail (8, 23, 34).
In the present study, we describe the characterization of the immunodominant epitopes derived from the RSV F protein that are recognized by human CD4 T cells in the context of those HLA class II molecules that are expressed most frequently within the Caucasian population. Antigenic peptides are found along the entire length of the F protein. In most donors, CD4 T cells recognize more than one peptide within the F protein. Several peptides are productively presented in the context of more than one HLA class II molecule, while at the level of detection of the direct gamma interferon (IFN-γ) enzyme-linked immunospot (Elispot) assays performed, there are also peptides that are haplotype specific. Most peptides are presented by HLA-DR molecules. In general, donors that share HLA class II molecules recognize the same pattern of peptides. In all donors, the CD4 T-cell response against the F peptides constituted a considerable part of the total response against RSV. Thus, RSV F is a major target structure for human memory CD4 T cells.
MATERIALS AND METHODS
PBMC.
Buffy coats were obtained from healthy adult blood donors with informed consent. Since every individual has encountered RSV by the age of 2, all the adult donors used in the present study should have been exposed to RSV regularly. PBMC were isolated by density centrifugation with Lymphoprep (Nycomed Pharma). PBMC were either thawed from cryopreserved samples (−135°C) in RPMI 1640 (Gibco)-10% dimethyl sulfoxide-30% fetal bovine serum (HyClone), or used fresh.
Virus and peptides.
Human RSV strain A2 was propagated in HEp-2 cells, and the viral titer was determined by plaque assay. The virus was routinely used for infection of PBMC at a multiplicity of infection of 1. A series of 94 peptide amides, 18 amino acid residues long, were synthesized by standard solid-phase Fmoc chemistry. The peptides synthesized were based on the sequence of fusion protein F of RSV strain A2. They overlapped by 12 amino acid residues. The purity of the peptides varied between 50 and 90%, as determined by analytical reverse-phase high-performance liquid chromatography. Peptides 11, 12, and 85 were missing from the set.
Elispot assay.
Filtration plates (96-well; MAIPS4510; Millipore) were coated overnight with anti-IFN-γ coating antibody 1-D1K (100 μl, 15 μg/ml; Mabtech) in 0.1 M carbonate-bicarbonate buffer pH 9.6, at 4οC. Before adding the cells, the plates were washed thoroughly with phosphate-buffered saline and blocked for 1 h at 37°C with RPMI 1640 containing 10% fetal bovine serum. Cells and either virus or peptide at the indicated amount were added to the well in a final volume of 200 μl of RPMI 1640 and 10% fetal bovine serum and penicillin and streptomycin. Cells were incubated at 37°C for 24 h in a humidified incubator. Then cells were removed by thoroughly washing in phosphate-buffered saline, and 100 μl of detecting monoclonal antibody 7-B6-1-biotin (Mabtech), diluted to 1 μg/ml in phosphate-buffered saline-0.5% fetal bovine serum, was added to the wells.
After incubation for 2 h at room temperature and washing (phosphate-buffered saline), 100 μl (diluted 1:1,000 in phosphate-buffered saline-0.5% fetal bovine serum), ExtraAvidine alkaline phosphatase conjugate (Sigma) was added and incubated for 1 to 2 h at room temperature. Then the plates were washed in phosphate-buffered saline, and 5-bromo-4-chloro-3-indolylphosphate-nitroblue tetrazolium substrate was added at 100 μl/well (Sigma; one tablet dissolved in 10 ml of H2O). Spots were counted by two investigators. Data are represented as the number of spots per 106 PBMC minus background of unstimulated samples. In Table 1, the uncorrected number of spots is shown. In the monoclonal antibody blocking experiments, the cells and monoclonal antibody were incubated for 30 min at 37°C, after which the peptides were added to the cultures. The monoclonal antibodies used are culture supernatants of hybridomas B8.11.2, producing anti HLA-DR, and SPVL3, producing anti-HLA-DQ.
TABLE 1.
Characterization of immunodominant domains within the RSV-A2 fusion proteina
| Donor HLA type or pool | Peptides in pool | No. of spots/106 PBMC
|
||||||
|---|---|---|---|---|---|---|---|---|
| CE-7 | CE-4 | CE-3 | VB-5 | VB-2 | VP-1 | CH-1 | ||
| HLA-A | 3, 28 | 2 | 2 | 2 | 1, 29 | 2 | 2 | |
| HLA-B | 7, 35 | 8, 18 | 40 | 35, 62 | 44, 57 | 44 | 27, 6 | |
| HLA-C | 7 | 4 | 5 | 2, 3 | ||||
| HLA-DR | DRB1*01 | DRB1*01 | DRB1*0401 | DRB1*0401 | DRB1*0701 | DRB1*1302 | DRB1*15 | |
| DRB1*03 | DRB1*03 | DRB1*0403 | DRB1*1101 | DRB1*15 | DRB1*16 | |||
| HLA-DQ | DQB1*05 | DQB1*02 | DQB1*02 | DQB1*03 | DQB1*03 | DQB1*06 | DQB1*0502 | |
| DQB1*05 | DQB1*03 | DQB1*0602 | ||||||
| Untreated | 4 | 12 | 4 | 46 | 10 | 14 | 32 | |
| RSV | 92b | 66 | 108 | >350 | 162 | 156 | >250 | |
| Pool 1 | 1-6 | 22 | 12 | 42 | 240 | 34 | 94 | 42 |
| Pool 2 | 7-10, 13, 14 | 6 | 12 | 40 | 220 | 38 | 36 | 68 |
| Pool 3 | 15-20 | 10 | 12 | 28 | 152 | 38 | 12 | 48 |
| Pool 4 | 21-26 | 0 | 4 | 0 | 62 | 18 | 10 | 20 |
| Pool 5 | 27-32 | 12 | 12 | 12 | 100 | 48 | 22 | 50 |
| Pool 6 | 33-38 | 4 | 16 | 24 | 114 | 28 | 26 | 66 |
| Pool 7 | 39-44 | 20 | 32 | 26 | 98 | 100 | 32 | 28 |
| Pool 8 | 45-50 | 28 | 22 | 26 | 142 | 28 | 140 | 38 |
| Pool 9 | 51-56 | 28 | 24 | 24 | 176 | 26 | 88 | 26 |
| Pool 10 | 57-62 | 8 | 24 | 40 | 106 | 40 | 42 | 28 |
| Pool 11 | 63-68 | 10 | 12 | 18 | 150 | 12 | 12 | 28 |
| Pool 12 | 71-76 | 8 | 14 | 14 | 176 | 28 | 24 | 26 |
| Pool 13 | 77-82 | 18 | 10 | 14 | 165 | 18 | 46 | 138 |
| Pool 14 | 83, 84, 86-88, 90 | 10 | 14 | 36 | 144 | 24 | 34 | 78 |
| Pool 15 | 69, 70, 91-94 | 4 | 4 | 8 | 42 | 84 | 26 | 72 |
Pools of six consecutive peptides (20 μM) were tested in a direct 24-h Elispot assay (triplicate) with PBMC from healthy adult blood donors. No. of spots per 106 PBMC are depicted.
The number of background spots (no peptide) was not subtracted.
Short-term T-cell lines.
PBMC were cultured in 96-well round-bottomed plates (2 × 105/well) in AIM-V medium (Gibco) supplemented with 1% human pooled AB serum, penicillin-streptomycin, and 20 μM peptide. Ten days after the initiation of the cultures, 10 U of recombinant human interleukin-2 per ml was added. Lines were tested on day 14 or restimulated on day 14 with 2 × 105 irradiated autologous PBMC and peptide. After the second stimulation, cells were expanded in AIM-V and 10 U of interleukin-2 per ml until day 24, when a second series of functional tests was performed.
Preparation of responder T-cell and antigen presenting cell fractions.
T-cell depletions of antigen-presenting cell populations and antigen-presenting cells plus CD8 depletions from responder T-cell populations were performed by negative selections on midiMACS columns (Miltenyi Biotec, Bergisch-Gladbach, Germany) according to the manufacturer's instructions. In brief, cells were incubated with phycoerythrin-labeled anti-CD8 and anti-HLA-DR (T-cell responder population) or anti-CD3 (antigen-presenting cell population) antibodies for 20 min on ice in phosphate-buffered saline supplemented with 2% fetal calf serum, washed once in 50 ml of the same buffer, and then incubated for 15 min at 4 to 7°C with antiphycoerythrin microbeads. The magnetically labeled fraction was retained on a midiMACS column, and the unbound cell fractions were further used in experiments. After depletions, less than 2% of the cell fraction that had been depleted remained.
Proliferation assay.
T-cell lines at day 24 of culture (2 × 104) were stimulated with 105 irradiated PBMC in the presence of 5 μM peptide in AIM-V medium supplemented with 1% human AB serum and penicillin-streptomycin. After 72 h, cultures were pulsed with [3H]thymidine for 20 h.
IFN-γ production.
T-cell lines at day 14 after a single in vitro stimulation with peptide (0.4 × 105/well) or at day 24 after two peptide stimulations (105/well) were used as responders to test reactivity (IFN-γ production) against heterozygous and homozygous antigen-presenting cell populations (PBMC depleted of CD3+ cells, 3 × 105/well) pulsed with the peptides (at 5 μM) that were used to make the lines. Cells were cultured in 96-well round-bottomed plates in 200 μl of AIM-V medium plus 1% human AB serum. After 72 h, supernatants were harvested and the IFN-γ concentration determined by standard enzyme-linked immunosorbent assay. As a control, responder cells alone and antigen-presenting cells alone did not produce IFN-γ. In some wells, antigen presentation was blocked with 1:10 diluted culture supernatant of B8.11.2, a hybridoma producing an anti-DR blocking antibody. This concentration of antibody was not toxic and specific for the antigen presentation of peptides in the context of HLA-DR. The antibody was incubated for 30 min at 37°C before peptides and responder cells were added to the cultures.
RESULTS
Identification of RSV-specific T-cell epitopes.
A set of overlapping 18-mer peptides (12-amino-acid overlap), spanning the entire sequence of the F protein of RSV strain A2, were synthesized. In a preliminary search for antigenic epitopes, 15 pools of six consecutive peptides were made. These peptide pools were analyzed in an Elispot assay for their ability to induce IFN-γ production in PBMC from healthy donors. To obtain an estimate of the total antiviral response, we also stimulated T cells with the entire spectrum of viral antigens, by infecting PBMC with RSV at a multiplicity of infection of 1. In Table 1, the data are summarized for a group of donors in which the most frequently occurring HLA class II molecules within the Caucasian population are represented.
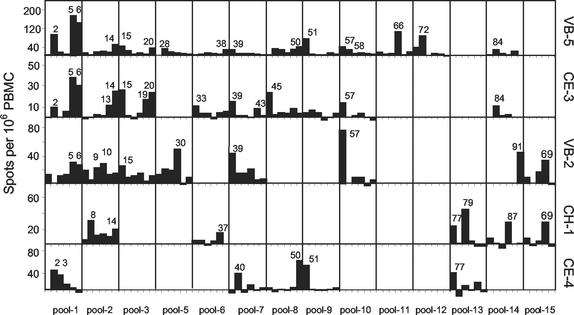
We were able to detect IFN-γ production in response to stimulation of PBMC with the peptide pools as well as with live RSV at a multiplicity of infection of 1. The magnitude of the response varied from donor to donor, but was generally on the order of 100 to 500 spots per 106 PBMC (Table 1 and data not shown). Most donors responded to more than one peptide pool. Next, we mapped the individual peptides from the positive pools for five individual donors (Fig. 1). For most pools, a clear response identified one or two positive peptides. In several cases, two adjacent peptides were found positive, possibly containing an overlapping epitope.
FIG. 1.
Determination of antigenic peptides within positive peptide pools. Direct IFN-γ Elispot assays were performed with single 18-mer F peptides at 20 μM. Only peptides of pools that were positive in previous assays were tested. The values are number of spots per 106 PBMC minus the number of spots in unstimulated PBMC. Donor VB-5, HLA-A2, B35,62, C4, DRB1*0401, DRB1*0403, DQB1*03; donor CE-3, HLA-A2, B40, DRB1*03,*0401, DQB1*02,03; donor VB-2, HLA-A1,29, B44,57, DRB1*0701, DRB1*1101, DQB1*03; donor CH-1, HLA-A2, B27,60, C2,3, DRB1*15,*16, DQB1*0502, DQB1*0602; and donor CE-4, HLA-A3,28, B7,35, DRB1*01, DQB1*05.
RSV T-cell epitopes are presented by MHC class II to antiviral CD4 T cells.
We then sought to determine the T-cell subset responsible for the antiviral T-cell responses that we measured. The peptide length of 18 amino acid residues is sufficiently long to encompass MHC class II peptides, but is less suitable for MHC class I binding. For presentation by MHC class I molecules, the 18-mer peptides require cleavage to 9 to 11 amino acid residues in order to bind to the class I binding groove. However, in our initial screens, we used fairly high concentrations of crude peptides (i.e., non-HPLC-purified). Thus, we could be measuring either CD4 or CD8 T-cell responses.
To determine the T-cell subset responding to the peptides, we performed IFN-γ Elispot assays with PBMC depleted for either CD8 T cells or CD4 T cells. For all the positive peptides that we found, the responses were completely abrogated after CD4 depletion (data not shown). The involvement of CD4 T cells in the peptide-specific responses was further confirmed by peptide stimulations of PBMC in the presence of HLA-DR blocking antibody B8.11.2 or HLA-DQ blocking antibody SPVL3 (Fig. 2). For most peptides, IFN-γ production was completely abrogated when the peptide concentration was titrated in the presence of HLA-DR blockade (Fig. 2A, C, D, and F). However, in two donors with HLA-DRB1*1302,-DRB1*15,-DQB1*06 and HLA-DRB1*1301,-DQB1*06, the response against peptides 50 and 51 was HLA-DQ restricted (Fig. 2B). These peptides were also recognized in a HLA-DQ-restricted fashion by a donor expressing HLA alleles DRB1*01 and DQB1*05 (Fig. 2D). A third donor (HLA-DRB1*15,*09, -DQB1*03,*06) recognized two adjacent peptides (peptide numbers 45 and 46) also in the context of HLA-DQ (Fig. 2E).
FIG. 2.
MHC restriction of F peptide-specific CD4 T-cell responses. Inhibition of IFN-γ production by MHC class II-specific monoclonal antibodies B8.11.2 (anti-HLA-DR) and SPVL3 (anti-HLA-DQ). (A, C, and F) IFN-γ Elispot assay with PBMC from donors VB-5(A), VB-2 (C), and VB-7 (F). F peptide 18-mers (peptide numbers are given in the figure) were titrated, and B8.11.2 culture supernatant was used diluted 1:100 (white bars) and 1:4 (grey bars). The number of IFN-γ spots in unblocked cultures is shown (black bars). As a control for the specificity of MHC class II blocking, MHC class I-mediated stimulation by the influenza virus (Flu) matrix peptide M57-65 was also performed in the presence of B8.11.2. (B, D, and E) IFN-γ Elispots of donors VP-1 (B, left panel) and MP-4 (B, right panel), donor CE-7 (D), and donor JB-1 (E). Blocking performed with culture supernatant of B8.11.2 diluted 1:50 (white bars) and culture supernatant of SPVL3 diluted 1:100 (grey bar). Unblocked cultures are shown as black bars.
Another observation made from these experiments was that there were large differences in the minimal peptide concentration (40 nM to 5 μM) necessary for optimal T-cell activation for the different individual peptides. Nevertheless, the fact that we detected responses in the direct IFN-γ Elispot assay without preexpansion of T cells indicates that during a natural infection, the total array of peptides that we have characterized can be functionally presented to the immune system.
Assigning restriction elements to RSV F CD4 T-cell epitopes: HLA-DR and -DQ restriction.
We then set out to assign specific restriction elements to the epitopes that we found. The set of 30 antigenic peptides that were characterized in Fig. 1 were used to test a larger panel of donors with HLA-DR types most frequently occurring within the Caucasian population (32) (Table 2). Most donors sharing a certain HLA class II allele recognized the same set of peptides. Thus, the HLA allele that is involved in the presentation of a certain peptide can be predicted from this shared response.
TABLE 2.
Summary of F-derived peptides that are involved in CD4 memory T-cell responses in PBMC from healthy donorsa
| Peptide | No. of spots/106 PBMC
|
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CE-8 (DRB1*03, DQB1*02) | CE-4 (DRB1*01, DRB1*03, DQB1*05, DQB1*02) | CE-7 (DRB1*01, DQB1*05) | VB-6 (DRB1*0401, DRB4*01, DQB1*03) | MP-7 (DRB1*0401, DRB1*0407, DQB1*03) | VB-5 (DRB1*0401, DRB1*0403, DQB1*03) | CE-3 (DRB1*0401, DRB1*03, DQB1*03, DQB1*02) | MP-1 (DRB1*0401, DRB1*1301, DQB1*03, DQB1*06) | MP-4 (DRB1*1301, DQB1*06, DQB1*02) | VP-1 (DRB1*15, DRB1*1302, DQB1*06) | VB-2 (DRB1*1101, DRB1*0701, DQB1*03) | VB-7 (DRB1*1101, DRB1*15, DQB1*03, DQB1*06) | CH-1 (DRB1*16, DRB1*15, DQB1*0502, DQB1*0602) | JB-1 (DRB1*15, DRB1*09, DQB1*0602, DQB1*03) | |
| 2 | 4 | 13 | 46 DR | 21 | 103 | 94 DR | 9 | 47 | 10 | 6 | 0 | 15 | 13 | |
| 3 | 35 DR | 14 | 0 | 2 | 10 | NT | ||||||||
| 5 | 5 | 5 | 79 | 104 | 176 | 37 | 55 | 1 | −2 | 40 | −4 | 8 | ||
| 6 | 9 | 5 | −6 | 70 | 104 | 143 DR | 29 | 43 | 26 DR | −4 | 3 | |||
| 8 | 8 | 3 | 8 | 9 | 2 | −1 | 4 | 1 | 30 | −1 | ||||
| 9 | 0 | 8 | 2 | 45 | 15 | 2 | 22 | −1 | 11 | −5 | ||||
| 10 | −1 | 10 | 4 | 39 | 20 | 1 | 0 | 30 DR | −3 | 13 | −3 | |||
| 13 | 9 | 1 | 9 | 28 | 13 | 11 | 20 | −1 | 12 | 21 DR | 10 | 5 | ||
| 14 | 0 | 5 | 60 | 72 | 50 DR | 24 | 55 | 2 | 14 | 1 | 19 | 21 | ||
| 15 | 3 | 4 | 49 | 72 | 41 | 25 | 39 | 24 | 3 | 12 | ||||
| 19 | 3 | 1 | −1 | 35 | 5 | 16 | 2 | −7 | 5 | |||||
| 20 | 3 | 3 | −3 | −1 | 31 | 23 | 10 | 5 | 5 | |||||
| 30 | 9 | 2 | 0 | 9 | 12 | 0 | 0 | 48 DR | 5 | −1 | ||||
| 33 | 1 | 1 | 8 | 7 | 4 | 10 | 1 | 6 | 37 DR | 5 | 1 | |||
| 39 | −1 | 10 | −7 | −1 | 4 | 24 | 14 | 0 | 0 | 42 | 47 DR | −7 | ||
| 40 | 4 | 18 | 38 DR | −1 | 9 | 7 | 1 | −3 | 0 | 8 | −4 | |||
| 45 | 25 | 7 | 4 | −3 | 4 | 23 | 8 | −3 | 44 DQ | |||||
| 46 | 13 | 9 | −4 | 6 | 30 | 2 | 10 | 8 | 68 DQ | |||||
| 50 | 5 | 1 | 68 DQ | −3 | 11 | 37 DR | 3 | 4 | 23 DQ | 76 DQ | 4 | −4 | ||
| 51 | 3 | 3 | 57 DQ | 3 | 21 | 73 DR | 4 | 41 | 22 DQ | 70 DQ | −1 | 0 | ||
| 57 | 44 | 18 | −3 | 11 | 37 | 13 | 43 | 51 | 4 | 58 DR | 103 DR | −4 | ||
| 58 | 7 | 2 | −5 | 15 | 25 | 0 | 10 | 20 | 13 | −7 | −1 | |||
| 66 | 3 | 2 | 23 | 79 | 105 DR | 31 | 2 | −3 | 8 | |||||
| 69 | −4 | −5 | 4 | 32 DR | 15 | 28 DR | 23 | |||||||
| 72 | 4 | −1 | 17 | 63 | 85 DR | 25 | 0 | −5 | 1 | |||||
| 77 | 3 | 11 | 40 DR | −3 | 13 | 2 | 1 | −5 | 23 | 5 | ||||
| 79 | 4 | 10 | −1 | 5 | −3 | 14 | 5 | 44 DR | 12 | |||||
| 83 | 3 | −3 | 21 | 0 | 4 | 7 | −7 | |||||||
| 84 | 9 | 2 | 23 | 24 | 10 | 3 | 4 | −7 | ||||||
| 87 | 15 | −3 | 3 | 7 | 28 | 5 | ||||||||
| 91 | 11 | −3 | 1 | 0 | −3 | 44 DR | −4 | 1 | 15 | |||||
Individual peptides were tested in a direct 24-h Elispot assay (triplicate) with PBMC of healthy blood donors. Numbers of spots per 106 PBMC are depicted. Peptides found positive in more than one experiment are boldfaced. For donors CE-3, 4, and 7, MP-1 and 7, VP-1, VB-2, and CH-1, only single peptides of positive pools were tested. When antibody blocking was performed to distinguish between HLA-DR and -DQ responses, this is indicated. NT, not tested.
We confirmed the validity of these predictions by testing T-cell responder populations from several heterozygous donors on peptide-loaded homozygous antigen-presenting cells. First, we determined the HLA class II restriction element for two peptides recognized by donor VB-7 (HLA-DRB1*1101, -DRB1*1501). Antigen-presenting cells and CD8-positive cells were depleted from the PBMC of this heterozygous donor. Elispot assays were performed with these antigen-presenting cell-depleted PBMC as responder cells and homozygous PBMC that were depleted of T cells as antigen-presenting cells.
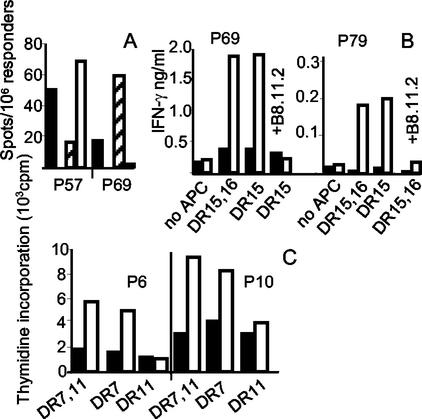
Figure 3A shows that peptide 57 was presented by HLA-DRB1*1101 and peptide 69 by HLA-DRB1*1501/B4*01 to the T cells of donor VB-7. In Fig. 3B and 3C, results are shown from T-cell stimulation assays with short-term T-cell lines from donors CH-1 (DRB1*1501, -*16) and VB-2 (DRB1*0701, -*1101) as responder cells and homozygous antigen-presenting cell populations as stimulator cells. From these experiments, we concluded that peptides 69 and 79 are presented by HLA-DRB1*1501 (Fig. 3B). Peptides 6 and 10 are both restricted by HLA-DRB1*0701 (Fig. 3C).
FIG. 3.
HLA allele-specific presentation of peptides. (A) IFN-γ Elispot assay with PBMC of donor VB-7 (DRB1*1101,*1501) depleted of CD8- and HLA class II-positive cells. As antigen-presenting cells, we used PBMC from homozygous donors (VB-10, DRB1*1101, and VB-12, DRB1*1501) that were depleted of CD3-positive cells. Black bars, VB-7 PBMC not depleted; white bars, DRB1*1101 antigen-presenting cells; hatched bars, DRB1*1501 antigen-presenting cells. From the number of spots depicted, the background in the absence of peptide has been subtracted. (B) IFN-γ production of short-term T-cell lines. The responder populations were incubated for 3 days with heterozygous (CH-1, DRB1*1501, -*16) antigen-presenting cells or homozygous (VB-12, DRB1*1501) antigen-presenting cells. Both antigen-presenting cell populations were CD3 depleted. Black bars, no peptide; white bars, 20 μM peptide 69 (left side) or 20 μM peptide 79 (right side). B8.11.2, anti-HLA-DR antibody. (C) Proliferation assay with short-term T-cell lines of donor VB-2 (DRB1*0701, *1101). Antigen-presenting cells were from VB-8 (DRB1*0701) or VB-10 (DRB1*1101). Black bars, no peptide; white bars, 20 μM peptide 6 (left side) or 20 μM peptide 10 (right side).
In Table 3, the amino acid sequences of the antigenic peptides are compiled. For most peptides, the restricting MHC alleles are indicated. Furthermore, the level of conservation of the amino acid sequences representing the antigenic peptides in 10 different RSV strains is indicated (see Discussion).
TABLE 3.
Sequences of dominant F peptides that are presented by HLA class II moleculesa
| Peptide | Residues | Amino acid sequence | HLA restriction | Variability between virus strains |
|---|---|---|---|---|
| 2 | 7-24 | KANAITTILTAVTFCFAS | DRB1*0101 / DRB1*0401 | Variable |
| 3 | 13-30 | TILTAVTFCFASGQNITE | DRB1*0101 | Variable |
| 5 | 25-42 | GQNITEEFYQSTCSAVSK | DRB1*0401 / DRB1*0701 | U31561: (A39V), B1: (K42R) |
| 6 | 31-48 | EFYQSTCSAVSKGYLSAL | DRB1*0401 / DRB1*0701 | U31561: (A39V), B1: (K42R) |
| 8 | 43-60 | GYLSALRTGWYTSVITIE | (<DRB1*16-DQB1*05>) | U31561: (T50I), B1 (L45F) |
| 9 | 49-66 | RTGWYTSVITIELSNIKE | (DRB1*0407) | U31561: (T50I) |
| 10 | 55-72 | SVITIELSNIKENKCNGT | DRB1*0701 | All A strains conserved; B1: (N67T) |
| 13 | 73-90 | DAKVKLIKQELDKYKNAV | Variable | |
| 14 | 79-96 | IKQELDKYKNAVTELQLL | DRB1*0401 | Variable |
| 15 | 85-102 | KYKNAVTELQLLMQSTPP | DRB1*0401 | Variable |
| 19 | 109-126 | RELPRFMNYTLNNAKKTN | Variable | |
| 20 | 115-132 | MNYTLNNAKKTNVTLSKK | Variable | |
| 30 | 175-192 | NKAVVSLSNGVSVLTSKV | (DRB1*0701) | Conserved A + B |
| 33 | 193-210 | LDLKNYIDKQLLPIVNKQ | (DRB1*1101) | U31561: (N197T), B strains several changes |
| 39 | 229-246 | RLLEITREFSVNAGVTTP | (DRB1*1101) | Conserved in all A strains |
| 40 | 235-252 | REFSVNAGVTTPVSTYML | DRB1*0101 | Conserved in all A strains |
| 45 | 265-282 | PITNDQKKLMSNNVQIVR | <DRB1*03-DQB1*02> / DQB1*03 | Conserved in all A strains, single change in B1: (N276S) |
| 46 | 271-288 | KKLMSNNVQIVRQQSYSI | <DRB1*03-DQB1*02> / DQB1*03 | Concerved in all A strains, single change in B1: (N276S) |
| 50 | 295-312 | EVLAYVVQLPLYGVIDTP | DQB1*05 / DQB1*06 | A strains conserved, single change in B1: (L305-I) |
| 51 | 301-318 | VQLPLYGVIDTPCWKLHT | DQB1*05/DQB1*06 | B1: (L305-I) |
| 57 | 337-354 | TDRGWYCDNAGSVSFFPQ | (DRB1*03-DQB1*02) / DRB1*1101 / <DRB1*1301-DQB1*06> | Single variation in strain U31559: (Q354L) |
| 58 | 343-360 | CDNAGSVSFFPQAETCKV | Variable | |
| 66 | 391-408 | YDCKIMTSKTDVSSSVIT | DRB1*0401 | Conserved in all A strains, single change in B1: (V402I) |
| 69 | 409-426 | SLGAIVSCYGKTKCTASN | DRB1*0701 / <DRB1*1501-DRB5*01> | Conserved A + B |
| 72 | 427-444 | KNRGIIKTFSNGCDYVSN | DRB1*0401 | Single change in Long strain: (V442A) |
| 77 | 457-474 | YYVNKQEGKSLYVKGEPI | DRB1*0101 | Conserved in all A strains |
| 79 | 469-486 | VKGEPIINFYDPLVFPSD | <DRB1*1501 -DRB5*01> | Conserved in all A strains, single change in B1: (F477Y) |
| 83 | 493-510 | SQVNEKINQSLAFIRKSD | Conserved A + B | |
| 84 | 499-516 | INQSLAFIRKSDELLHNV | Long: (N515H), B1: (K508R) | |
| 87 | 517-534 | NAGKSTTNIMITTIIIVI | (<DRB1*16-DQB1*0502>) | Conserved in A strains, single change in B1: (A518T) |
| 91 | 541-558 | LIAVGLLLYCKARSTPVT | (DRB1*0701) | U31558 (L547F), B1: several changes |
The sequences of antigenic peptides were checked for their variability in eight different A strain viruses and two B strain viruses: Long strain and F proteins with GenBank accession numbers U31558, U31559, U31560, U31561, U31562, Z26524-short, L25351-short, and RSV B strains D00334 and B1 (accession number AF013254). Restriction elements in parentheses are ascribed on the basis of responses found in donors sharing these HLA molecules, <>, DR and DQ contribution not determined by blocking experiments. Boldface, overlapping stretches in adjacent peptides.
DISCUSSION
The T-cell response against RSV is crucial for the efficient eradication of virus from the infected lung. However, there are also indications that the nature of the immune response during primary and secondary infections with RSV determines the severity of lower respiratory tract illness and may play a role in RSV-related childhood wheezing and asthma (14, 31). Despite the key role of the antiviral cellular immune response in protection and pathogenesis, there exists only limited knowledge on the magnitude and longevity of the response and on the precise molecular targets. Identification of T-cell epitopes is therefore of considerable importance to furthering our understanding of this infection.
In the present study, we took a first step to mapping the targets of the antiviral CD4 T-cell response. We quantitated the responses directly ex vivo with IFN-γ Elispot assays. Therefore, our results shed new light not only on the specificity of the response, but also on the magnitude of the memory T-cell response. We show for a panel of healthy blood donors that CD4 T-cell memory responses against RSV can be directly detected without preexpansion of T cells. The frequency of CD4 memory T-cell responses varied between donors but was generally on the order of 0.05 to 0.3% of CD4 T cells (Table 1 and data not shown). These values are in the range of the memory T-cell responses reported for mumps, but are approximately 10-fold lower than the responses found for cytomegalovirus (48). We did not find a correlation between the higher-responding donors and the time of year that their blood was sampled, i.e., the RSV season. Moreover, in those isolated cases in which the same donor was sampled twice, the level of anti-RSV CD4 T-cell memory was similar.
We tested a panel of donors that together covered the most frequently occurring HLA class II haplotypes within the Caucasian population for their T-cell responsiveness against peptides derived from RSV F. A broad repertoire of CD4 memory T cells are primed in vivo, because most of the donors tested responded against multiple epitopes within the F protein of RSV. The F protein appears to be one of the dominant antigenic proteins within RSV, because in most donors the numbers of T cells responding to F peptides constituted a considerable fraction of the T cells that responded to the entire virus.
In one earlier study by Levely et al., a dominant CD4 T-cell epitope was characterized, encompassing amino acid residues 338 to 355 of RSV strain A2-F (34). The response against this peptide was HLA-DR restricted, but the presenting HLA molecule was not further identified. The peptide was recognized by different donors that did not share a single HLA class II molecule. Our data indicate that peptide 57, corresponding to residues 337 to 354 of the F protein, was most strongly recognized in the context of HLA-DRB1*1101 and -DRB1*1301 and further in the context of HLA-DRB1*03 and -DRB1*0403. These data are in good agreement with the Levely study, in which responding donors carried HLA-DR4, -DR5, or -DR6.
Levely and coworkers also identified other regions that induced weak proliferative responses which were not further characterized. However, we noticed that some of these regions overlap the peptides that we characterized in our Elispot assays. For instance, responses against sequences 31 to 45, 81 to 95, and 111 to 125 were found by Levely and coworkers in a donor carrying HLA-DR4 and -DR5. These peptides correspond to our peptides 6 (amino acid residues 31 to 48), 14 (amino acid residues 79 to 96) and 19 (amino acid residues 109 to 126), which we identified as being presented by HLA-DRB1*04. Furthermore, a weak proliferative response found by Levely et al. against peptide 231 to 251, corresponding to our peptide 39 (residues 229 to 246), may have been a response in the context of HLA-DR5 expressed by the responding donor, because we found a response to peptide 39 in the context of HLA-DRB1*1101. The very consistent finding of T-cell responses against the same peptides in our experiments and those performed 10 years ago by Levely et al. with a different functional readout (proliferation) underscores the usefulness of these peptides to monitor F-specific memory CD4 T-cell responses.
Bovine RSV F protein is 81% homologous to the human F protein (33). In cattle, the CD4 T-cell responses against F were predominantly mapped in the F1 part of the protein. In contrast, epitopes in our human volunteers were distributed along the entire length of the F protein. Only a single stretch of 46 amino acid residues (residues 121 to 168) was not recognized. This part of the protein overlaps the 27-residue peptide (110 to 136) that is removed from the fusion protein after furin cleavage in the Golgi compartment (22, 50) and extended into the hydrophobic N-terminal region of F1. However, antigenic peptides 19 and 20 also overlap the region between F1 and F2, implying that furin cleavage in the Golgi does not interfere with MHC class II presentation. Interestingly, a peptide corresponding to bovine F residues 301 to 316, differing at only one position from human RSV (305Ile →Leu), is recognized by human CD4 T cells as well as bovine T cells (19). Moreover, this peptide is presented by DQ class II molecules in both humans and cattle.
Many immunodominant regions that were identified in the present study are highly conserved between different A strain viruses (Table 3). Peptides 30 and 69 are completely conserved in A and B strains, and peptides 39, 40, 45/46, 50/51, 66, 77, 79, and 87 are conserved in all A strains. For other peptides, single substitutions were found in a single A strain (peptides 5, 6, 8, 33, 57,72, 84, and 91). Some amino acid stretches are more variable, such as the amino-terminal part of F2 (peptides 2 and 3) and the region overlapping peptides 19 and 20. It is possible that the substitutions in different virus strains affect the ability of peptides to bind to the HLA class II binding groove or influence contact with the T-cell receptor. Moreover, variations outside the epitopes may also affect antigen processing and presentation.
In vaccination studies performed in animal models, it has been shown that an exuberant T-cell response against components of RSV can cause enhanced disease characterized by inflammatory infiltrates in the lung. The nature of these infiltrates is determined by the cytokine and chemokine patterns produced by the activated T-cell population (3, 13, 24). In BALB/c mice vaccinated with formalin-inactivated RSV, a Th2 response is responsible for disease enhancement resulting from an inflammatory response in the lung that is associated with a strong eosinophilia (42).
Studies in the BALB/c mouse model have mostly focused on the Th2 type T-cell response. However, in BALB/c mice it is also possible to establish lung disease that is caused by a more Th1-dominated response that correlates with a predominant influx of neutrophils into the lung. This type 1 response can be induced by either a strong CD4 or an excessive CD8 T-cell response (9, 42). In the autopsy reports of formalin-inactivated RSV-vaccinated children that died of severe lower respiratory tract disease, eosinophils were found in the lungs (30). Also, enhanced eosinophil counts in the peripheral blood were reported for infants vaccinated with formalin-inactivated RSV who survived a subsequent infection with the natural virus (12). In contrast, eosinophils have not been observed in lung tissue of children with fatal primary RSV infection (37). Moreover, blood eosinophil counts are not generally enhanced in bronchiolitis patients (20). It is unclear whether Th1 or Th2 cells are involved in severe disease in primary RSV infections in infants.
The nature of a T-cell response can be influenced by the specific antigenic complex of MHC and peptide (7, 40). In fact, in the BALB/c model of vaccinia virus G vaccination, a Vβ14 T-cell subset recognizing a single MHC-peptide combination dominates the CD4 T-cell response and is responsible for the Th2 shift of the RSV-specific CD4 T cells. It is this Th2-biased CD4 T-cell response that triggers the cascade of events leading to severe lower respiratory tract disease (47). Therefore, knowledge of antigenic peptides recognized by human RSV-specific CD4 T cells, in combination with the MHC molecules that present them to the T cells, is an essential first step towards our understanding of the role of T cells in human immune pathology.
Acknowledgments
We thank Ger Rijkers for carefully reading the manuscript and Rein Zaat from the Bloodbank Utrecht for providing us with buffy coats (research project B95.004x).
This project was supported by the Breedtestrategie, a research initiative from Utrecht University, in conjunction with the RIVM and ID-Lelystad.
REFERENCES
- 1.Alwan, W. H., F. M. Record, and P. J. Openshaw. 1992. CD4+ T cells clear virus but augment disease in mice infected with respiratory syncytial virus. Comparison with the effects of CD8+ T cells. Clin. Exp. Immunol. 8:527-536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alwan, W. H., F. M. Record, and P. J. M. Openshaw. 1993. Phenotypic and functional characterization of T-cell lines specific for individual respiratory syncytial virus proteins. J. Immunol. 150:5211-5218. [PubMed] [Google Scholar]
- 3.Alwan, W. H., and P. J. M. Openshaw. 1993. Distinct patterns of T and B cell immunity to respiratory syncytial virus induced by individual proteins. Vaccine 11:431-437. [DOI] [PubMed] [Google Scholar]
- 4.Alwan, W. H., W. J. Kozlowska, and P. J. M. Openshaw. 1994. Distinct types of lung disease caused by functional subsets of antiviral T cells. J. Exp. Med. 179:81-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Anderson, J. J., J. A. Harrop, H. Peers, H. Briggs, G. L. Toms, and R. Scott. 1991. Recognition of respiratory syncytial virus proteins by human and BALB/c lymphocytes. J. Med. Virol. 35:165-173. [DOI] [PubMed] [Google Scholar]
- 6.Bangham, C. R. M., P. J. M. Openshaw, L. A. Ball, A. M. Q. King, G. W. Wertz, and B. A. Askonas. 1986. Human and murine cytotoxic T cells specific to respiratory syncytial virus recognize the viral nucleoprotein (N), but not the major glycoprotein (G), expressed by vaccinia virus virus recombinants. J. Immunol. 137:3973-3977. [PubMed] [Google Scholar]
- 7.Blander, J. M., D. B. Sant'Angelo, K. Bottomly, and C. A. Janeway, Jr. 2000. Alteration at a single amino acid residue in the T-cell receptor α chain complementary determining region 2 changes the differentiation of naive CD4 T cells in response to antigen from T helper cell type 1 (Th1) to Th2. J. Exp. Med. 191:2065-2074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brandenburg, A. H., L. de Waal, H. H. Timmerman, P. Hoogerhout, R. L. de Swart, and A. D. M. E. Osterhaus. 2000. HLA class I-restricted cytotoxic T-cell epitopes of the respiratory syncytial virus fusion protein. J. Virol. 74:10240-10244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cannon, M. J., P. J. Openshaw, and B. A. Askonas. 1988. Cytotoxic T cells clear virus but augment lung pathology in mice infected with respiratory syncytial virus. J. Exp. Med. 168:1163-1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chang, J., A. Srikiatkhachorn, and T. J. Braciale. 2001. Visualization and characterization of respiratory syncytial virus F-specific CD8+ T cells during experimental virus infection. J. Immunol. 167:4254-4260. [DOI] [PubMed] [Google Scholar]
- 11.Cherrie, A. H., K. Anderson, G. W. Wertz, and P. J. M. Openshaw. 1992. Human cytotoxic T cells stimulated by antigen on dendritic cells recognize the N, SH, F, M, 22K, and 1b proteins of respiratory syncytial virus. J. Virol. 66:2102-2110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chin, J., R. L. Magoffin, L. A. Shearer, J. H. Schieble, and E. H. Lennette. 1969. Field evaluation of a respiratory syncytial virus vaccine and a trivalent parainfluenza virus vaccine in a pediatric population. Am. J. Epidemiol. 89:449-463. [DOI] [PubMed] [Google Scholar]
- 13.Connors, M., A. B. Kulkarni, C.-Y. Firestone, K. L. Holmes, H. C. Morse, A. V. Sotnikov, and B. R. Murphy. 1992. Pulmonary histopathology induced by respiratory syncytial virus (RSV) challenge of formalin-inactivated RSV-immunized BALB/c mice is abrogated by depletion of CD4+ T cells. J. Virol. 66:7444-7451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ehlenfield, D. R., K. Cameron, and R. C. Welliver. 2000. Eosinophilia at the time of respiratory syncytial virus bronchiolitis predicts childhood reactive airway disease. Pediatrics 105:79-83. [DOI] [PubMed] [Google Scholar]
- 15.Englund, J. A., C. J. Sullivan, M. C. Jordan, L. P. Dehner, G. M. Vercellotti, and H. H. Balfour. 1988. Respiratory syncytial virus infection in immunocompromised adults. Ann. Intern. Med. 109:203-208. [DOI] [PubMed] [Google Scholar]
- 16.Falsey, A. R., and E. E. Walsh. 1998. Relationship of serum antibody to risk of respiratory syncytial virus infection in elderly adults. J. Infect. Dis. 177:463-467. [DOI] [PubMed] [Google Scholar]
- 17.Falsey, A. R., and E. E. Walsh. 2000. Respiratory syncytial virus infection in adults. Clin. Microbiol. Rev. 13:371-384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fishaut M., D. Tubergen, and K. Mcintosh. 1980. Cellular response to respiratory viruses with particular reference to children with disorders of cell-mediated immunity. J. Pediatr. 96:179-186. [DOI] [PubMed] [Google Scholar]
- 19.Fogg, M. H., K. R. Parsons, L. H. Thomas, and G. Taylor. 2001. Identification of CD4+ T-cell epitopes on the fusion (F) and attachment (G) proteins of bovine respiratory syncytial virus (BRSV). Vaccine 19:3226-3240. [DOI] [PubMed] [Google Scholar]
- 20.Garofalo, R., A. Dorris, S. Ahlstedt, and R. C. Welliver. 1994. Peripheral blood eosinophil counts and eosinophil cationic protein content of respiratory secretions in bronchiolitis: relationship to severity of disease. Pediatr. Allergy Immunol. 5:111-117. [DOI] [PubMed] [Google Scholar]
- 21.Glezen, P., and F. W. Denny. 1973. Epidemiology of acute lower respiratory disease in children. N. Engl. J. Med. 288:498-505. [DOI] [PubMed] [Google Scholar]
- 22.Gonzalez-Reyes, L., M. B. Ruiz-Arguello, B. Garcia-Barreno, L. Calder, J. A. Lopez, J. P. Albar, J. J. Skehel, D. C. Wiley, and J. A. Melero. 2001. Cleavage of the human respiratory syncytial virus fusion protein at two distinct sites is required for activation of membrane fusion. Proc. Natl. Acad. Sci. USA 98:9859-9864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Goulder, P. J. R., F. Lechner, P. Klenerman, K. McIntosh, and B. A. Walker. 2000. Characterization of a novel respiratory syncytial virus-specific human cytotoxic T-lymphocyte epitope. J. Virol. 74:7694-7697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Graham, B. S., L. A. Bunton, P. F. Wright, and D. T. Karzon. 1991. Role of T lymphocyte subsets in the pathogenesis of primary infection and rechallenge with respiratory syncytial virus in mice. J. Clin. Investig. 88:1026-1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Graham, B. S., L. A. Bunton, P. F. Wright, and D. T. Karzon. 1991. Respiratory syncytial virus infection in anti-mu treated mice. J. Virol. 65:4936-4942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hall, C. B., K. R. Powell, N. E. MacDonald, et al. 1986. Respiratory syncytial virus infection in children with compromised immune function. N. Engl. J. Med. 315:77-81. [DOI] [PubMed] [Google Scholar]
- 27.Hall, C. B., E. E. Walsh, C. E. Long, and K. C. Schnabel. 1991. Immunity to and frequency of reinfection with respiratory syncytial virus. J. Infect. Dis. 163:693-698. [DOI] [PubMed] [Google Scholar]
- 28.Jackson, M., and R. Scott. 1996. Different patterns of cytokine induction in cultures of respiratory syncytial virus-specific human Th cell lines following stimulation with RS virus and RS virus proteins. J. Med. Virol. 49:161-169. [DOI] [PubMed] [Google Scholar]
- 29.Kapikian, A. Z., R. H. Mitchell, R. M. Chanock, R. A. Shvedoff, and C. E. Stewart. 1969. An epidemiological study of altered clinical reactivity to respiratory syncytial (RS) virus infection in children previously vaccinated with an inactivated RS virus vaccine. Am. J. Epidemiol. 89:405-421. [DOI] [PubMed] [Google Scholar]
- 30.Kim, H. W., J. G. Canchola, C. D. Brandt, G. Pyles, R. M. Chanock, M. Jensen, and R. H. Parrott. 1969. Respiratory syncytial virus disease in infants despite prior administration of antigenic inactivated vaccine. Am. J. Epidemiol. 89:422-434. [DOI] [PubMed] [Google Scholar]
- 31.Kimpen, J. L. L., and E. A. F. Simoes. 2001. Respiratory syncytial virus and reactive airway disease: new developments prompt a new review. Am. J. Respir. Crit. Care Med. 163:S1-S6. [DOI] [PubMed] [Google Scholar]
- 32.Knipper, A. J., P. Hakenberg, J. Enczmann, A. Kuhroeber, U. Kiesel, G. Koegler, and P. Wernet. 2000. HLA-DRB1,3,4,5 and -DQB1 allele frequencies and HLA-DR/DQ linkage disequilibrium of 321 German caucasoid patients and their corresponding 821 potential unrelated stem cell transplants. Hum. Immunol. 61:605-614. [DOI] [PubMed] [Google Scholar]
- 33.Lerch, R. A., K. Anderson, V. L. Amann, and G. W. Wertz. 1991. Nucleotide sequence analysis of the bovine respiratory syncytial virus fusion protein mRNA and expression from a recombinant vaccinia virus. Virology 181:118-131. [DOI] [PubMed] [Google Scholar]
- 34.Levely, M. E., C. A. Bannow, C. W. Smith, and J. A. Nicholas. 1991. Immunodominant T-cell epitope on the F protein of respiratory syncytial virus recognized by human lymphocytes. J. Virol. 65:3789-3796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Martin, A. J., P. S. Gardner, and J. McQuillin. 1978. Epidemiology of respiratory viral infection among pediatric inpatients over a 6-year period in north-east England. Lancet ii:1035-1038. [DOI] [PubMed]
- 36.Murphy, B. R., G. A. Prince, E. E. Walsh, et al. 1986. Dissociation between serum neutralizing and glycoprotein antibody responses of infants and children who received inactivated respiratory syncytial virus vaccine. J. Clin. Microbiol. 24:197-202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Neilson, K. A., and E. J. Yunis. 1990. Demonstration of respiratory syncytial virus in an autopsy series. Pediatr. Pathol. 10:491-502. [DOI] [PubMed] [Google Scholar]
- 38.Nicholas, J. A., K. L. Rubino, M. E. Levely, E. G. Adams, and P. L. Collins. 1990. Cytolytic T-lymphocyte responses to respiratory syncytial virus: effector cell phenotype and target proteins. J. Virol. 64:4232-4241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Openshaw, P. J. M., S. L. Clarke, and F. M. Record. 1992. Pulmonary eosinophilic response to respiratory syncytial virus infection in mice sensitized to the major surface glycoprotein G. Int. Immunol. 4:493-500. [DOI] [PubMed] [Google Scholar]
- 40.Pfeiffer, C., J. Stein, S. Southwood, H. Ketelaar, A. Sette, and K. Bottomly. 1995. Altered peptide ligands can control CD4 T lymphocyte differentiation in vivo. J. Exp. Med. 182:1591-1596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Prince, G. A., S. J. Curtis, K. C. Yim, and D. D. Porter. 2001. Vaccine-enhanced respiratory syncytial virus disease in cotton rats following immunization with lot 100 or a newly prepared reference vaccine. J. Gen. Virol. 82:2881-2888. [DOI] [PubMed] [Google Scholar]
- 42.Simmons, C. P., T. Hussell, T. Sparer, G. Walzl, P. Openshaw, and G. Dougan. 2001. Mucosal delivery of a respiratory syncytial virus CTL peptide with enterotoxin-based adjuvants elicits protective immunopathogenic, and immunoregulatory antiviral CD8+ T-cell responses. J. Immunol. 166:1106-1113. [DOI] [PubMed] [Google Scholar]
- 43.Simoes, E. A. F. 1999. Respiratory syncytial virus infection. Lancet 354:847-852. [DOI] [PubMed] [Google Scholar]
- 44.Srikiatkhachorn, A., and T. J. Braciale. 1997. Virus-specific CD8+ T lymphocytes downregulate T helper cell type 2 cytokine secretion and pulmonary eosinophilia during experimental murine respiratory syncytial virus infection. J. Exp. Med. 186:421-432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Srikiatkhachorn, A., and T. J. Braciale. 1997. Virus-specific memory and effector cell T lymphocytes exhibit different cytokine responses to antigens during experimental murine respiratory syncytial virus infection. J. Virol. 71:678-685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Varga, S. M., E. L. Wissinger, and T. J. Braciale. 2000. The attachment (G) glycoprotein of respiratory syncytial virus contains a single immunodominant epitope that elicits both Th1 and Th2 CD4+ T-cell responses. J. Immunol. 165:6487-6495. [DOI] [PubMed] [Google Scholar]
- 47.Varga, S. M., X. Wang, R. M. Welsh, and T. J. Braciale. 2001. Immunopathology in RSV infection is mediated by a discrete oligoclonal subset of antigen-specific CD4+ T cells. Immunity 15:637-646. [DOI] [PubMed] [Google Scholar]
- 48.Waldrop, S. L., C. J. Pitcher, D. M. Peterson, V. C. Maino, and L. J. Picker. 1997. Determination of antigen-specific memory/effector CD4+ T-cell frequencies by flow cytometry. J. Clin. Investig. 99:1739-1750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Waris, M. E., C. Tsou, D. D. Erdman, S. R. Zaki, and L. J. Anderson. 1996. Respiratory syncytial virus infection in BALB/c mice previously immunized with formalin-inactivated virus induces enhanced pulmonary inflammatory response with a predominant Th2-like cytokine pattern. J. Virol. 70:2852-2860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zimmer, G., L. Budz, and G. Herrler. 2001. Proteolytic activation of respiratory syncytial virus fusion protein. J. Biol. Chem. 276:31642-31650. [DOI] [PubMed] [Google Scholar]