Abstract
A simple method is described for determining the valency of binding of immunoglobulin G to immobilized influenza A virus. Where there is a free Fab arm (monovalent binding), a second virus particle is captured. This is detected by surface plasmon resonance. The methodology should be applicable to all enveloped and nonenveloped viruses.
Antibodies are proteins built on a basic pattern of two pairs of identical light and heavy polypeptide chains. The C-terminal (or Fc) part of each polypeptide has a relatively invariant sequence (constant region) and has various receptor functions for complement and Fc receptors. At the N terminus of each polypeptide is a variable region amounting to approximately 50% of the light chain and 25% of most heavy chains. These carry hypervariable or complementarity-determining regions that fold together to form the paratope that recognizes and binds to the cognate epitope, often with an affinity that is nanomolar or better (reviewed in reference 1).
Antibodies aid recovery from many virus infections and provide protection against most reinfections. The majority of antibodies act by binding to virus particles and neutralizing virus infectivity directly, although the Fc regions of some antibody isotypes can bind and activate complement proteins or mediate the uptake and destruction of virus by phagocytic cells through binding to Fc receptors on the cell surface (4).
Since antibodies are homobivalent and since viruses comprise a mosaic of repeating epitopes on their surface, antibodies have the potential to bind bivalently, providing that the epitopes fall within the effective span of the antibody, which is at least 6 nm but not more than 9 to 15 nm apart (10). However, about half of the small number of antibodies that have been examined bind only monovalently, because the angle formed between the epitope and paratope directs the other Fab arm of the antibody away from the virus particle and out into solution. (Fab is the fragment of antibody formed by the light chain and an equivalent portion of the heavy chain that has been cleaved by protease digestion.) Monovalency is not optional but is determined by the precise interaction of the amino acid residues that make up the interacting paratope and epitope surfaces. The mode of binding has a number of functional implications. For instance, monovalent binding is inherently less stable than bivalent binding, as both paratopes have to dissociate simultaneously for a bivalently bound antibody to detach from the surface of a virion. Thus, the difference between monovalent and bivalent binding can determine whether or not the antibody has sufficient functional affinity to enact neutralization or other immune effector functions. In addition, all antibodies that bind monovalently have the potential to bind a second virus particle and to form aggregates. This reduces the number of available infectious units and results in virus neutralization.
Presently there is no easy way to determine the valency of antibody binding. The favored method is cryoelectron microscopy (cryo-EM), which has high capital cost and requires considerable training and expertise. Further, the technique is restricted to regular geometric viruses, as it deduces the valency of binding from the angle subtended by a monovalent Fab and the virus surface (7, 12). The cryo-EM process shows that some immunoglobulins G (IgGs) bind bivalently (3, 6, 13, 14), while others bind monovalently (3, 8, 9, 11, 19, 20). So far the method has been restricted to canine parvovirus (20), cowpea mosaic virus (11), rhinoviruses (3, 6, 8, 13, 14), and foot-and-mouth disease virus (9, 19). The cryo-EM process is not suitable for determining the valency of antibody binding to enveloped viruses. Other techniques for determining monovalency include analysis of virus aggregation by neutralization, electron microscopy, or centrifugation. Such antibodies may have a U-shaped neutralization curve when infectivity loss is plotted against antibody concentration (10), as aggregation is lost when epitopes are saturated at high applied antibody concentrations. Virus-antibody complexes can also be analyzed qualitatively by transmission electron microscopy (16) or semiquantitatively by sedimentation analysis (17, 18).
For the work described here, we used the Mount Sinai strain of human influenza virus A/Puerto Rico/8/34 (H1N1) (PR8), an enveloped virus approximately 100 nm in diameter. The virus has three envelope proteins, the hemagglutinin (HA), the neuraminidase, and the M2 ion channel protein. The HA is a homotrimer and is the major attachment, fusion, and neutralization protein; there are around 700 trimers per virion. Virus was grown in embryonated eggs and purified by sucrose gradient centrifugation. The hemagglutination titer and protein concentration of the virus were determined as previously described (approximately 1 hemagglutination unit [HAU]/ng with 0.15% chicken red blood cells [5]). The monoclonal antibodies (MAbs) used (H36-4.5-2 [IgG2a and specific for site Sb/B] and H37-45-5R3 [IgG3, site Ca2/A]) were specific for the HA1 subunit of PR8 (2, 15) and were kindly provided by W. Gerhard (Wistar Institute, Philadelphia, Pa.). Both MAbs bind to epitopes at the distal end of the HA trimeric spike (5). They were chosen for valency investigation, as the functional affinities (Kd) of the H36 IgG and Fab for immobilized whole-virus particles as determined by surface plasmon resonance were both 0.56 nM while that of H37 IgG (0.39 nM) was 23-fold higher that that of the H37 Fab (9.0 nM) (5). These data suggested that H36 IgG bound monovalently to virus and that H37 IgG bound bivalently. The approach used here was to bind biotinylated virus to a streptavidin (SA) sensor chip (Biacore AB, Uppsala, Sweden), bind MAb to the biotinylated virus, and then determine the valency of binding of that MAb by its ability to capture detector virus (Fig. 1).
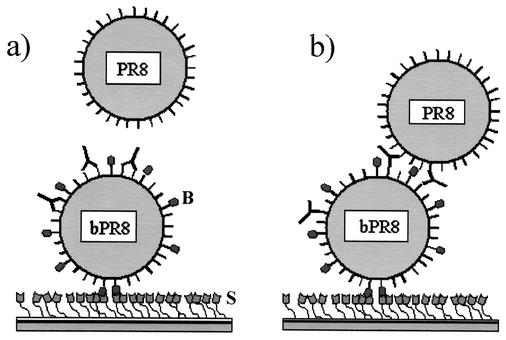
FIG. 1.
Schematic showing the rationale and hypothesis presented in this report. Shown in panels a and b, the SA (S)-coated chip surface at the bottom of the figure binds an influenza virus particle (bPR8) that has biotin residues (B) covalently linked to its surface proteins. IgG (Y) then flows over the virus. Only IgG that binds is shown. Detector virus then flows over the bPR8-IgG complexes. (a) Shown is an IgG that binds bivalently to the virus surface and is postulated not to be able to bind detector nonbiotinylated virus particle (PR8). (b) Shown is another IgG that recognizes a different epitope and binds monovalently and is predicted to bind detector particles by means of the free Fab arms. Binding of detector virus is determined by surface plasmon resonance and is quantitated in RU (see text).
PR8 virus (100 μg of protein/ml or 105 HAU) was biotinylated (bPR8) for 2 h on ice using sulfo-N-hydroxysuccinimide-long chain-long chain-biotin (with a spacer arm of 30.5 Å) (40 μl; Pierce) according to the manufacturer's guidelines. This was carried out in the presence of 10 mg of bovine serum albumin (BSA; Sigma) per ml of phosphate-buffered saline (PBS) to avoid overbiotinylation. Virus was separated from free biotin and BSA by centrifugation through 20% (wt/vol) sucrose in PBS (60,000 × g for 90 min at 4°C). BSA (10 mg) was biotinylated as a control (bBSA) by the same method, with free biotin being removed by using a microconcentrator with a cutoff at an Mr of 10,000 (Millipore).
An SA sensor chip was docked in a BIAcore 2000 instrument and treated sequentially with HBS-EP buffer (10 mM HEPES buffer [pH 7.4], 150 mM NaCl, 3 mM EDTA, and 0.005% [vol/vol] polysorbate 20; Biacore), 40% glycerol, and 50 mM NaOH in 1 M NaCl according to the manufacturer's instructions. bPR8 (30 μl; 1,500 HAU/ml in HBS-EP buffer) was then passed over track 2 of the chip at 5 μl/min. This was followed by 1 M NaCl for 1 min to remove loosely associated material and by more bPR8, until no more virus bound. Finally, 1 M NaCl was flowed through (in pulses of 30 s) until the virus baseline had stabilized, and then bBSA (200 μg/ml) was used to block any remaining free SA. Approximately 4,000 resonance units (RU) of bPR8 bound to the chip. A negative control (track 1) was coated with bBSA and was stabilized with 1 M NaCl. Next, MAbs (25 μl) at the required concentration were passed over tracks 1 and 2 at 10 μl/min, followed by detector PR8 (10 μl; 1,500 HAU/ml) at 10 μl/min. Between the applications of MAb concentrations, bound antibody and detector virus were removed with a 10-s pulse of 1 M NaCl and a 5-s pulse of 4 M MgCl2. This left the bPR8 ready to bind more antibody. Binding curves were recorded as sensorgrams using the BIAcore 2000 control software and were processed using BIAevaluation software.
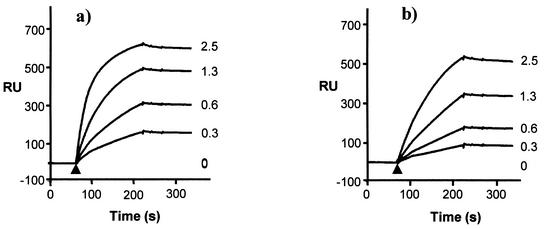
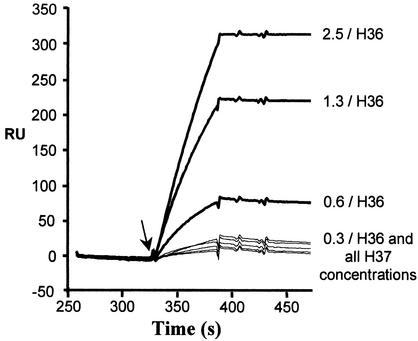
Figure 2a and b show that MAbs H36 and H37 bound to bPR8 attached to the SA chip in proportion to the applied concentration (0.3 to 2.5 μg/ml) and in similar amounts. No detector PR8 bound to the H36 IgG-bPR8 complexes at the lowest antibody concentration (Fig. 3), but, as the amount of attached antibody increased, binding was clearly seen, and this increased in proportion to the amount of bound IgG. Clearly this could only occur if the IgG had attached monovalently and had a free Fab arm. At the lowest applied antibody concentration, there was probably insufficient IgG attached to bPR8 to successfully tether the detector virus. Figure 3 also shows the data obtained with MAb H37, an IgG of similar functional affinity. Here, when the same IgG concentrations were used, there was little, if any, significant attachment of detector PR8. These latter data are consistent with H37 binding bivalently with no free Fab arm and with the fact that H37 IgG has a 23-fold higher Kd than the H37 Fab. Quantitation of antibody bound to bPR8 and detector virus captured in RU is shown in Table 1. At the maximum amount of applied antibody, there was an 11-fold difference in the ratio of captured virus to antibody bound, in favor of MAb H36.
FIG. 2.
Binding of IgG to immobilized influenza A virus. Initially approximately 4,000 resonance units (RU) of biotinylated virus (bPR8) were bound to SA sensor chips coated with SA in a series of injections (not shown). This was taken as the baseline position. Various concentrations of MAb H36 (a) and MAb H37 (b) (as indicated, in micrograms per milliliter) were injected (arrowhead) and were allowed to flow over and attach to virus. Sensorgrams of the binding of MAbs in real time are shown.
FIG. 3.
Determination of the valency of H36 and H37 IgGs bound to the immobilized PR8 shown in Fig. 2 by the capture of detector PR8 virus. Detector (nonbiotinylated) virus was injected (arrow) and was allowed to flow over the bPR8-IgG complexes. Sensorgrams record the binding of detector virus in real time. The figure is zeroed to the plateau of each IgG concentration in Fig. 2 to allow comparison of the amounts of detector virus bound. The time scale is continued from Fig. 2. H36 and H37 IgG concentrations are indicated in micrograms per milliliter. RU, resonance units.
TABLE 1.
Quantitation of antibody binding to immobilized bPR8 and of detector virus captured by that antibodya
| MAb (Concn in μg/ml) | Binding of MAb to immobilized bPR8 (a) (RU) | Capture of detector PR8 by bPR8-IgG complexes (b) (RU) | Ratio of b/a |
|---|---|---|---|
| H36 (2.5) | 581 | 319 | 0.55 |
| H37 (2.5) | 503 | 24 | 0.05 |
| H36 (1.3) | 466 | 225 | 0.48 |
| H37 (1.3) | 324 | 20 | 0.06 |
| H36 (0.6) | 296 | 80 | 0.27 |
| H37 (0.6) | 166 | 15 | 0.09 |
| H36 (0.3) | 153 | 27 | 0.18 |
| H37 (0.3) | 81 | 13 | 0.16 |
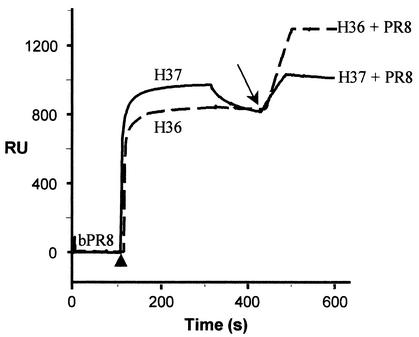
Although H37 bound to virus bivalently, it was possible that at high applied antibody concentrations it might bind monovalently, since a second antibody molecule might then be able to compete successfully with the free Fab arm of the first IgG. To test this, we repeated the above experiments with 160 μg of both MAbs H36 and H37 per ml—a 64-fold increase in antibody concentration. Figure 4 shows that there was an almost instant increase in bound H36 and H37, which then reached a plateau. After some dissociation of H37, both IgGs stabilized to approximately 820 RU. H36 IgG then captured 465 RU of detector virus, and H37 IgG captured 220 RU. Thus, it appears that under the conditions used here, some of the bivalently binding H37 antibodies are forced to bind monovalently. It will be necessary to titrate antibodies when determining valency of binding by this method. It is also of interest that bPR8 could be saturated by H37 IgG when a relatively low concentration (20 μg/ml) was passed repeatedly over virus. However, under these conditions, no detector virus was captured, presumably because there was time for each Fab arm to find an epitope and hence for the IgG to bind bivalently (data not shown).
FIG. 4.
Valency of binding of H37 IgG applied at high concentration: a proportion of this IgG that bound bivalently at low concentration now binds monovalently. The baseline represents bound bPR8 (Fig. 2). The first part of each curve shows the injection of 160 μg of H37 and H36 IgGs per ml (arrowhead) and their binding to immobilized bPR8, under the conditions described in the Fig. 2 legend. After the plateau, there is some dissociation of H37 IgG. After the injection of detector nonbiotinylated PR8 (arrow), there is an increase in RU due to its capture by antibody. H37, solid line; H36, dashed line.
The above data provide proof of principle for the elucidation of the binding valency of antibodies to the surface of a virus particle using surface plasmon resonance. The methodology should be able to demonstrate the valency of binding of most monomeric Igs (see below for possible exceptions) and should reveal any free Fab arms of bound polymeric IgA and IgM. The method should be applicable to all types of virus particle.
Notwithstanding the data above, there may be circumstances where an antibody that has the potential to bind bivalently to a virus particle does not do so. Included among these would be the situation where the relevant epitopes are too close together or too far apart to permit bivalent binding (10). Such an IgG would bind monovalently, but it is a moot point whether or not the antibody would capture another virus particle, since the free Fab arms would still be directed towards the first particle. Capture ability might then depend on the degree of IgG hinge and elbow flexibility. Another possible exception might arise with a monovalently binding IgG that recognizes an epitope in a depression on the virus capsid or partially down a spike protein. This situation would effectively foreshorten the IgG so that it might not be long enough to bind to the corresponding epitope on a second virus particle and capture it. However, such epitopes are in general not well presented to the immune system and their cognate IgGs are probably rare. In any case capture data for monomeric Ig should be consistent with the Kd values of the Ig and its Fab, and these should be determined before drawing a final conclusion as to antibody valency.
Acknowledgments
We thank Walter Gerhard (Wistar Institute) for generously providing antibodies and Matt Edwards (Edward Jenner Institute for Vaccine Research, Compton, United Kingdom) and Peter Critchley (University of Warwick, Coventry, United Kingdom) for helpful discussions.
S.A.H. was supported by the Biotechnology and Biological Sciences Research Council.
REFERENCES
- 1.Burton, D. R. 1990. Antibody: the flexible adaptor molecule. Trends Biochem. Sci. 15:64-69. [DOI] [PubMed] [Google Scholar]
- 2.Caton, A. J., G. G. Brownlee, J. W. Yewdell, and W. Gerhard. 1982. The antigenic structure of the influenza virus A/PR/8/34 hemagglutinin (H1 subtype). Cell 31:417-427. [DOI] [PubMed] [Google Scholar]
- 3.Che, Z., N. H. Olson, D. Leippe, W.-M. Lee, A. G. Mosser, R. R. Rueckert, T. S. Baker, and T. J. Smith. 1998. Antibody-mediated neutralization of human rhinovirus 14 explored by means of cryoelectron microscopy and X-ray crystallography of virus-Fab complexes. J. Virol. 72:4610-4622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dimmock, N. J. 1993. Neutralization of animal viruses. Curr. Top. Microbiol. Immunol. 183:1-149. [DOI] [PubMed] [Google Scholar]
- 5.Edwards, M. J., and N. J. Dimmock. 2000. Two influenza A virus haemagglutinin-specific Fabs neutralize by inhibiting virus attachment to target cells, while neutralization by their IgGs is complex and occurs through fusion-inhibition and attachment-inhibition simultaneously. Virology 278:423-435. [DOI] [PubMed] [Google Scholar]
- 6.Hewat, E. A., N. Verdaguer, I. Fita, W. Blakemore, S. Brookes, A. King, J. Newman, E. Domingo, M. G. Mateu, and D. Stuart. 1997. Structure of the complex of a Fab fragment of a neutralizing antibody with foot-and-mouth disease virus: positioning of a highly mobile antigenic loop. EMBO J. 16:1492-1500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hewat, E. A., and D. Blaas. 1996. Structure of a neutralizing antibody bound bivalently to human rhinovirus 2. EMBO J. 15:1515-1523. [PMC free article] [PubMed] [Google Scholar]
- 8.Hewat, E. A., and D. Blaas. 2001. Structural studies on antibody interacting with viruses. Curr. Top. Microbiol. Immunol. 260:29-44. [DOI] [PubMed] [Google Scholar]
- 9.Hewat, E. A., T. C. Marlovits, and D. Blaas. 1998. Structure of a neutralizing antibody bound monovalently to human rhinovirus. J. Virol. 72:4396-4402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mosser, A. G., D. Leippe, and R. R. Rueckert. 1989. Neutralization of picornaviruses: support for the pentamer bridging hypothesis, p. 155-167. In B. L. Semler and E. Ehrenfeld (ed.), Molecular aspects of picornavirus infection and detection. American Society for Microbiology, Washington, D.C.
- 11.Porta, C., G. Wang, H. Cheng, Z. Chen, T. S. Baker, and J. E. Johnson. 1994. Direct imaging of interactions between an icosahedral virus and conjugate fragments by cryoelectron microscopy and X-ray crystallography. Virology 204:777-778. [DOI] [PubMed] [Google Scholar]
- 12.Smith, T. J. 2001. Antibody interactions with rhinovirus: lessons for mechanism of neutralization and the role of immunity in viral evolution. Curr. Top. Microbiol. Immunol. 260:1-28. [DOI] [PubMed] [Google Scholar]
- 13.Smith, T. J., N. H. Olson, R. H. Cheng, E. S. Chase, and T. S. Baker. 1993. Structure of a human rhinovirus-bivalently bound antibody complex: implications for viral neutralization and antibody flexibility. Proc. Natl. Acad. Sci. USA 90:7015-7018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Smith, T. J., N. H. Olson, R. H. Cheng, H. Liu, E. S. Chase, W. M. Lee, D. M. Leippe, A. G. Mosser, R. R. Rueckert, and T. S. Baker. 1993. Structure of human rhinovirus complexed with Fab fragments from a neutralizing antibody. J. Virol. 67:1148-1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Staudt, L. M., and W. Gerhard. 1983. Generation of antibody diversity in the immune response of BALB/c mice to influenza virus hemagglutinin. I. Significant variation in repertoire expression between individual mice. J. Exp. Med. 157:687-704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Taniguchi, K., and S. Urasawa. 1987. Different virus-precipitating activities of neutralizing monoclonal antibodies that recognise distinct sites on poliovirus particles. Arch. Virol. 92:27-40. [DOI] [PubMed] [Google Scholar]
- 17.Thomas, A. A. M., R. Vrijsen, and A. Boeyé. 1986. Relationship between poliovirus neutralization and aggregation. J. Virol. 59:479-485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Thomas, A. A. M., P. Brioen, and A. Boeyé. 1985. A monoclonal antibody that neutralizes poliovirus by cross-linking virions. J. Virol. 54:7-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Verdaguer, N., G. Schoehn, W. F. Ochoa, I. Fita, S. Brookes, A. King, E. Domingo, M. G. Mateu, D. Stuart, and E. A. Hewat. 1999. Flexibility of the major antigenic loop of foot-and-mouth disease virus bound to a Fab fragment of a neutralizing antibody: structure and neutralisation. Virology 255:260-268. [DOI] [PubMed] [Google Scholar]
- 20.Wikoff, W. R., G. J. Wang, C. R. Parrish, R. H. Cheng, M. L. Strassheim, T. S. Baker, and M. G. Rossmann. 1994. The structure of a neutralized virus: canine parvovirus complexed with neutralizing antibody fragment. Structure 2:595-607. [DOI] [PMC free article] [PubMed] [Google Scholar]