Abstract
The expression levels of ∼4,600 cellular RNA transcripts were assessed in CD4+-T-cell lines at different times after infection with human immunodeficiency virus type 1 strain BRU (HIV-1BRU) using DNA microarrays. We found that several classes of genes were inhibited by HIV-1BRU infection, consistent with the G2 arrest of HIV-1-infected cells induced by Vpr. These included genes involved in cell division and transcription, a family of DEAD-box proteins (RNA helicases), and all genes involved in translation and splicing. However, the overall level of cell activation and signaling was increased in infected cells, consistent with strong virus production. These included a subgroup of transcription factors, including EGR1 and JUN, suggesting they play a specific role in the HIV-1 life cycle. Some regulatory changes were cell line specific; however, the majority, including enzymes involved in cholesterol biosynthesis, of changes were regulated in most infected cell lines. Compendium analysis comparing gene expression profiles of our HIV-1 infection experiments to those of cells exposed to heat shock, interferon, or influenza A virus indicated that HIV-1 infection largely induced specific changes rather than simply activating stress response or cytokine response pathways. Thus, microarray analysis confirmed several known HIV-1 host cell interactions and permitted identification of specific cellular pathways not previously implicated in HIV-1 infection. Continuing analyses are expected to suggest strategies for impacting HIV-1 replication in vivo by targeting these pathways.
Human immunodeficiency virus type 1 (HIV-1) is an enveloped retrovirus that causes severe depletion of the immune system in humans, leaving them susceptible to infection with other pathogens. Understanding the biochemical changes that occur in HIV-1 infection is important to continuing efforts aimed at countering its effects. To optimize virus production, HIV-1 is expected to redirect cellular machinery for its replication and interfere with cellular pathways that inhibit virus replication. Indeed, many of the virus-encoded proteins have been shown to interact with cellular proteins, e.g., Gag and Tsg101 (9, 13, 28, 46), Gag and cyclophilins (26), integrase and Ini1 (22), Vpr and PP2A (20), Vpr and HHR23A (48), Vpr and UNG (40), Vif and antiviral proteins (27, 42), Vif and HP68 (52), and Tat and cyclin T1 (47, 50). These interactions between viral and cellular proteins are essential for the viral life cycle and have been found to determine the species specificity of virus replication in some instances (44, 47). HIV-1 infection also has more general effects on cell morphology, gene expression, and metabolism, most clearly illustrated by the G2 arrest induced by the viral Vpr protein (19, 21, 34, 36). However, the global effects of viral infection on host cell gene expression patterns are largely unknown.
Recently, the study of pathogen-host interactions has been greatly aided by gene expression analysis using high-density microarrays. Microarrays consist of ordered sets of cDNAs or oligonucleotides, each representing a single gene, immobilized as spots on solid substrates. The level of expression of each gene is then investigated by hybridization with probes derived from mRNA isolated from cell populations of interest. In this way, expression levels of thousands of genes can be measured simultaneously, generating a global picture of the transcriptional changes effected by pathogens such as bacteria, viruses, and parasites (reviewed in references 2, 23, and 32).
Previous microarray studies on HIV-1 have used infection of cell lines at low multiplicity of infection (MOI) (MOI, 0.1 to 0.5) to study changes up to 3 days after infection (8, 15, 39) or heterogeneous target cell populations (45). These experimental protocols result in nonsynchronous infection and in selection of certain cells resistant to infection, as several rounds of infection are needed to derive a completely infected population. This reduces sensitivity and specificity for the detection of gene expression changes in infected cells. The goal of our study was to analyze global changes that occur in a near-synchronous infection cycle of HIV-1 to increase our chances of finding relevant changes induced by HIV-1 infection. To this end, we used (i) infection at a high MOI with sampling within the first 24 h postinfection, in an effort to study a single replication cycle within synchronously infected cells; (ii) cell lines, to minimize heterogeneous host cell responses to infection and to allow precise enumeration of infected cells; and (iii) several different T-cell lines, to study variability and consistency of changes induced by viral infection. This approach has allowed us to identify cellular pathways not previously implicated in HIV-1 infection.
MATERIALS AND METHODS
T-cell lines and virus stocks.
The human T-lymphoblastoid-cell lines CCRF-CEM, Jurkat clone E6-1, and SupT1 were obtained from the American Type Culture Collection (CCL-119, TIB-152, and CRL-1942, respectively). The CCRF-CEM subclone CEMss was obtained from P. L. Nara through the AIDS Research and Reference Reagent Program (ARP), Division of AIDS, National Institute of Allergy and Infectious Diseases, National Institutes of Health. CEMfh are CEMss cells that were maintained at the Fred Hutchinson Cancer Research Center for several years prior to use in this study.
HIV-1BRU was obtained from J.-M. Bechet and L. Montagnier through the ARP. Virus stocks were made by exposing CCRF-CEM cells (107/ml) to virus at an MOI of 0.01. After 1 h, cells were washed and then cultured at 106/ml. After 3 days, culture medium was replaced, and cells were cultured for an additional 24 h before harvesting the virus-containing supernatant. Infectivity was determined on the same cells and was generally ∼107 50% tissue culture infective doses/ml. Supernatants were also obtained from mock infections, with cells treated as described above but without adding infectious virus. Heat-inactivated virus was prepared by heating for 2 h at 56°C in a water bath with frequent mixing. All mock and virus preparations were kept on ice until added to cells (within 30 min).
Infections.
Infection of T-cell lines with HIV-1BRU was performed at an MOI of 2 at 37°C (Table 1). After 1 h of exposure to virus, cells were washed with phosphate-buffered saline and cultured at 106 cells per ml in RPMI with 10% fetal bovine serum. Mock cultures were treated the same way, substituting virus-containing supernatant with non-virus-containing tissue culture supernatant. Cells were harvested for RNA extraction at the indicated times by washing twice with phosphate-buffered saline and resuspending in 4 M guanidinium thiocyanate-25 mM sodium citrate-0.5% Sarkosyl-0.1 M 2-mercaptoethanol.
TABLE 1.
CD4+-T-cell line infection experimentsa with HIV-1BRU
| Cell line | Virus | Time (h) | No. of infections |
|---|---|---|---|
| CCRF-CEM | HIV-1BRU | 1 | 2 |
| HIV-1BRU | 4 | 2b | |
| HIV-1BRU | 8 | 2b | |
| HIV-1BRU | 12 | 2 | |
| HIV-1BRU | 24 | 4b | |
| Heat inactivated | 24 | 2 | |
| CEMfh | HIV-1BRU | 24 | 1 |
| CEMss | HIV-1BRU | 24 | 1 |
| SupT1 | HIV-1BRU | 24 | 1b |
| Jurkat | HIV-1BRU | 24 | 1 |
One experiment was defined as two hybridizations for each mock-HIV combination, one with the infected cells labeled with Cy3 and the mock-infected cells labeled with Cy5 and one with the color scheme reversed. As each cDNA was spotted in duplicate on each slide, this resulted in four data points per test.
Samples from one or more of the biological replicates at these time points were tested twice.
Flow cytometry.
Fluorescence-activated cell sorting (FACS) analysis of intracellular p24 expression was performed using the Fix and Perm kit (CALTAG, Burlingame, Calif.) and anti-p24 KC57-FITC antibody (Beckman-Coulter, Fullerton, Calif.) according to the manufacturer's protocol. For analysis of differentially expressed cell surface antigens CD53, CD69, and TFRC (CD71), purified specific monoclonal antibodies were purchased from BD Pharmingen (San Diego, Calif.). Staining was performed according to the manufacturer's protocol using isotype-control antibodies to assess nonspecific binding.
Hybridization probes.
Isolation of total RNA was performed using a modified guanidinium thiocyanate method (7). Oligotex (Qiagen, Valencia, Calif.) was used to isolate mRNA according to the manufacturer's protocol. Fluorescence-labeled cDNA probes were generated and hybridized to the slides as described by Geiss et al. (14).
Microarray construction and analysis.
The cDNA microarrays used in this study were generated in-house, and a description of the cDNA clones and construction has been published (14). Each slide contained 9,216 spots, corresponding to 4,608 genes spotted in duplicate. Included in this number was a set of 384 selected control genes spotted on each slide. These included HIV-1 genes used as positive controls, nonhuman genes used as negative controls, and a variety of genes selected from the Research Genetics 15K human gene set (Invitrogen, Carlsbad, Calif.).
Data analysis and selection of differentially expressed clones.
For each of the infection conditions (Table 1), duplicate slides were hybridized with probes generated from the same RNAs but with the fluorescent labels reversed to control for dye-specific effects (15). Intensity values in Cy3 and Cy5 channels were extracted from each image with customized software (Spot-On Image, developed by R. E. Bumgarner and Erick Hammersmark). The resulting data files were entered into the Expression Array Manager (EAM) (developed by Jeff Furlong). EAM includes a database for storing microarray data, software tools for uploading data to the database, and software for uploading data to the Rosetta Resolver gene expression data analysis system (purchased from Rosetta Inpharmatics, Kirkland, Wash.). Spot-On data files were combined with information regarding slide and sample preparation and hybridization images. Resolver was then used to analyze hybridization intensity for each spot, to calculate ratios and standard deviations of replicate experiments after elimination of uninformative data, and to select statistically significant, differentially expressed genes. This included normalization for nonlinearities in the Cy3/Cy5 ratio as a function of intensity. The lists of genes selected in all experiments were combined to allow clustering using agglomerative clustering with complete linkage as the heuristic criterion to compute the between-cluster similarity measures (18). In complete linkage, the updated distance (or dissimilarity) of the merged cluster (A,B) is equal to the longest distance over all pairs of objects between clusters A and B.
RT-PCR quantification.
Differential gene expression for a sample of eight genes was verified using reverse transcription (RT)-PCR with gene-specific primer pairs (Table 2). These genes were chosen based on either a substantial change in their expression levels by microarray analysis or their being regulated by the same transcription factor (SREBP [5]). Total RNA from a 24-h HIV-1BRU infection of CEMfh cells was used for quantification by multiplex RT-PCR using QuantumRNA Classic 18S internal standards (Ambion, Austin, Tex.) according to the manufacturer's protocol. PCR products were quantified on an Agilent (Palo Alto, Calif.) model 2100 Bioanalyzer DNA chip with internal standards as controls for variable amplification and loading of PCR products.
TABLE 2.
Gene-specific primers for RT-PCR
| Gene | Image clone | Sequence
|
Product size (bp) | Annealing temp (°C) | |
|---|---|---|---|---|---|
| Forward primer | Reverse primer | ||||
| FDPS | 80410 | 5′-GTGCTGACTGAGGATGAGATG-3′ | 5′-AAGAGGTCAAGGTAATCATCCTG-3′ | 668 | 56.3 |
| LSS | 770355 | 5′-CTGGCTGGCTGTCCTGAATG-3′ | 5′-GCATCTGGCGGTAGTACTTCTG-3′ | 728 | 59.8 |
| SCD | 123474 | 5′-CCTACCTGCAAGTTCTACACCTG-3′ | 5′-CAGCGGTACTCACTGGCAGAG-3′ | 665 | 56.8 |
| IDI1 | 44975 | 5′-CGTGTTGTAGTCATCCATTAAG-3′ | 5′-CAAGTCCTCTGCAAGTGCTC-3′ | 795 | 50.3 |
| MYC | 812965 | 5′-GCTTCACCAACAGGAACTATGAC-3′ | 5′-AAGCCGCTCCACATACAGTC-3′ | 397 | 58.9 |
| CD53 | 504226 | 5′-CAAGAATATCACGGCATGG-3′ | 5′-CCACAGAACTACTGCAGATCATAG-3′ | 693 | 54.9 |
| NAB1 | 843249 | 5′-TGGTGATGATGTCCAGCAACTCTG-3′ | 5′-TTGAGCAGCCGCTTCATTAACAGTG-3′ | 745 | 58.6 |
| NAB2 | 770868 | 5′-TACTATGAGACCTTCATCCAGCAG-3′ | 5′-TACCATCTCTGGCTCCAGTCG-3′ | 558 | 60.2 |
Detailed protocols are available at http://ubik.microbiol.washington.edu/protocols/protocols.htm.
RESULTS
HIV-1 test system.
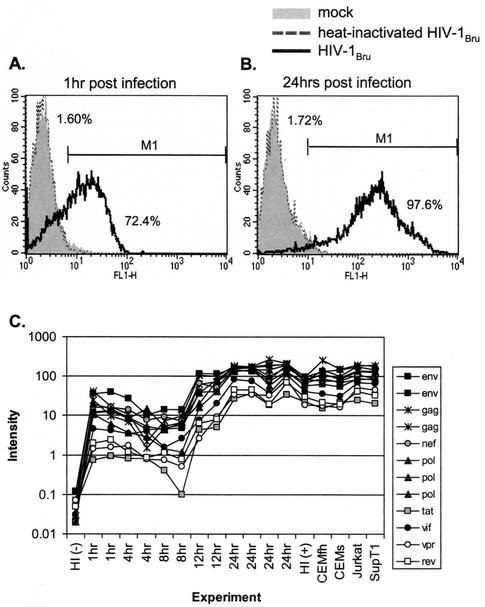
To accurately analyze changes in cellular gene expression induced by HIV-1 infection, uninfected cells need to be compared with a population of homogenously HIV-1-infected cells. For this purpose, the CD4+-T-cell line CCRF-CEM was inoculated with HIV-1BRU at an MOI of 2, an inoculum sufficient to ensure that every cell is contacted by infectious virus particles. Mock and infected cells were harvested at 1, 4, 8, 12, or 24 h after infection and subjected to FACS analysis of intracellular p24 expression to evaluate the synchrony of infection. The result of one such experiment is shown in Fig. 1A and B. A low level of p24 was detected on the majority of cells 1 h after infection (Fig. 1A), reflecting the high level of input virus attached to the cells. However, by 24 h most if not all cells showed high-level p24 expression, reflecting productive infection (Fig. 1B). The shift between low- and high-level p24 detection occurred between 12 and 16 h postinfection (data not shown), consistent with previous studies of HIV life cycle kinetics (17, 24, 35). Infection with heat-inactivated virus did not result in any detectable p24 expression, indicating that both the early (1 h) and late (24 h) p24 expression detected in the time course experiments was not the input virus nonspecifically sticking to the cells. We therefore concluded that within 24 h, nearly all of the cells were productively infected.
FIG. 1.
Infection of CD4+-T-cell line CCRF-CEM with HIV-1BRU. (A and B) Cells were exposed to infectious virus at a multiplicity of infection of 2, heat-inactivated virus, or mock-infected cell supernatant. At 1 and 24 h postexposure, cells were fixed and permeabilized for intracellular p24 staining. Only cells exposed to the infectious virus stock showed detectable levels of p24 at 1 h postinfection (A) and increased p24 expression after 24 h (B). (C) Expression of HIV-1 RNA transcripts during infection. The graph depicts results for 17 HIV-1BRU infection experiments: a time course using CCRF-CEM cells and after 24 h of infection of CEMfh, CEMss, Jurkat, and SupT1 cells. Twelve PCR products representing different regions of the HIV-1BRU genome were spotted on each slide; nucleotide positions relative to the HIV-1BRU genome were 317 to 1281 (gag), 853 to 2471 (gag), 2179 to 3097 (pol), 3015 to 4363 (pol), 4337 to 5598 (pol), 5538 to 6974 (env), 6583 to 7404 (env), 7115 to 8631 (env), 8276 to 9225 (nef), 5402 to 5666 (tat), 7951 to 8272 (rev), 4619 to 5257 (vif), and 5124 to 5446 (vpr). Plotted are hybridization intensities of each of the 12 spots in the treated cells. HI (-) refers to mock infection versus heat-inactivated BRU, and HI (+) refers to heat-inactivated HIV-1BRU versus infectious HIV-1BRU.
Gene expression in CCRF-CEM cells after infection with HIV-1BRU.
Two CCRF-CEM infection time course experiments were performed. RNA was isolated at 1, 4, 8, 12, and 24 h after HIV-1BRU infection (Table 1). In addition, to control for changes induced by non-HIV factors in the viral stock, parallel cultures of CCRF-CEM cells were treated with heat-inactivated HIV-1BRU or a mock virus preparation from uninfected cells. After conversion into dye-labeled cDNA, probes were hybridized to human cDNA arrays and spot ratios were quantified. Each mock-HIV mRNA combination was labeled twice, according to reciprocal color schemes, to control for sequence-specific dye incorporation.
Twelve control spots on each array contained PCR products spanning the length of the HIV-1 genome in segments. None of these control spots showed expression above background levels after hybridization with probes from mock-infected or heat-treated cells. The expression of HIV-1 gene segments in the HIV-1BRU-infected cells is shown in Fig. 1C. HIV-1 RNA was evident 1 h after exposure to virus, reflecting input virus bound to cells, and as expected, the abundance of these sequences decreased at 4 and 8 h postinfection as the genomic viral RNA was reverse transcribed into cDNA and then degraded. At 12 h postinfection the first increase in viral transcripts due to productive infection was observed, with maximum levels achieved by 24 h, and the intensity for each segment correlated with the length of the probe. RNA expression kinetics were consistent with the intracellular p24 expression kinetics measured by FACS (compare Fig. 1).
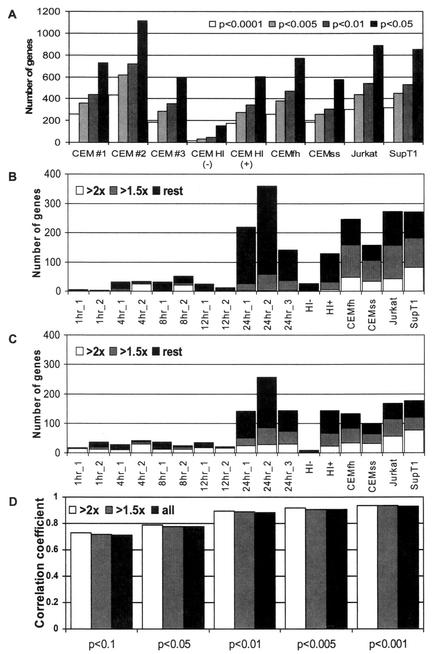
Several criteria were examined to select the set of statistically significant, differentially expressed genes (Fig. 2). P values chosen to define each experiment's set of regulated genes greatly influenced the numbers of genes selected, with close to 1,000 genes selected using a P of ≤0.05 and fewer than 300 selected using a P of ≤0.001 for the 24-h time points (Fig. 2A). The majority of changes observed at each time point were less than twofold for both down-regulation (Fig. 2B) and up-regulation (Fig. 2C). Changes were consistent across duplicate experiments for genes down- or up-regulated by both small or larger amounts, as the correlation coefficient between duplicate experiments was similar whether the experiments included all differentially regulated genes or just those regulated by at least 1.5- or 2-fold (Fig. 2D). However, decreasing the P cutoff did increase the correlation coefficient for duplicate experiments (Fig. 2D). On the basis of these analyses, we primarily used P values to select regulated genes, irrespective of the degree of change.
FIG. 2.
Genes found to be differentially regulated under different selection criteria. (A) Effect of signature P on number of genes selected. (B) Number of down-regulated genes in each experiment using a signature P of ≤0.005. Depicted are genes down-regulated by at least 2-fold (white bars) or 1.5-fold (grey bars), and all differentially regulated genes (black bars). (C) Number of up-regulated genes in each experiment using a signature P of ≤0.005 and depicted as in panel B. (D) Effect of signature P and relative change on the correlation coefficient between duplicate experiments.
The number of genes down- and up-regulated using a signature P of ≤0.005 is shown in Fig. 2B and C, respectively. Consistent with viral gene expression, few cellular genes were found to be differentially regulated as early as 1 h postinfection (5 genes), while the majority of changes were observed by 24 h postinfection (281 genes). The lists of differentially expressed genes selected at each time point were combined to allow clustering of genes over the entire time course. The experiments with heat-inactivated virus (HI− and HI+) were included as specificity controls. Because every time point was tested at least two times, only genes differentially regulated (P ≤ 0.005) in at least 2 of the 13 CCRF-CEM experiments were included in the following analyses.
Kinetics and cell-type-specific expression of host cell genes.
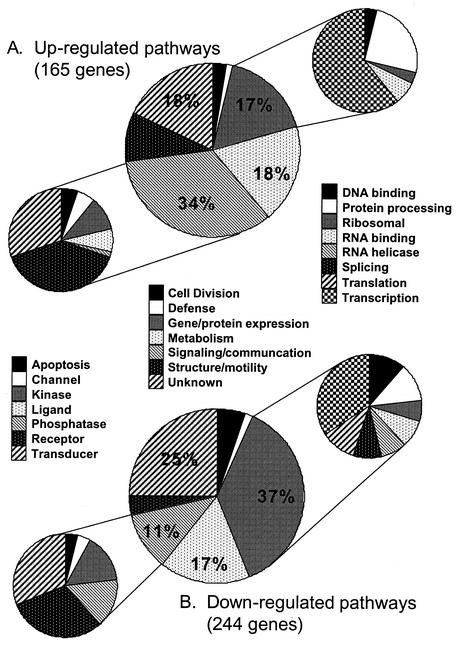
409 host cell genes fulfilled our requirements for differential expression (see supplemental data S1 and S2 for hierarchical cluster, gene names, IMAGE clone numbers and mean ratios) and were divided into groups based on their known function (Fig. 3). The majority of changes in gene expression were observed at 24 h. These included several genes previously implicated in HIV-1 infection, such as EGR1 (11), CDK4 (30), CD69 (3), CD71 (33, 37), CD87 (43), and JUN (6, 25), providing support for the validity of our other findings. Overall, genes involved in signaling and communication were stimulated, while genes involved in cell division, transcription, translation, and splicing, were inhibited by HIV-1BRU infection (Fig. 3). Many receptors and kinases were up-regulated, including CD69, T-cell receptor α, LDLR, mitogen-activated protein 2 kinase 1, and DYRK2. Positive cell cycle regulators were down-regulated and negative cell cycle regulators were up-regulated, reflecting G2 arrest of HIV-1 infected cells. Five members of the SR family of splicing factors were down-regulated, perhaps to facilitate the production of unspliced and single-spliced viral RNA messages needed at the later stages of the virus life cycle. Likewise, the expression of seven DEAD-box proteins (DDX, RNA helicases), several elongation initiation factors, and heterogeneous nuclear ribonucleoproteins was down-regulated. Though most transcription factors were down-regulated (e.g., MYC), a small group of transcription factors including EGR1, JUN, TNRC3, DRPLA, ARP, SMARCD2, SP140, GATA3, RELB, ELF1, and TAL1 were up-regulated by HIV-1 infection, implicating these factors in facilitating HIV-1 production. The majority of enzymes were down-regulated, including strong repression of MAT2A, ODC1, UNG, and ADSL. Interestingly, the genes encoding enzymes in the cholesterol biosynthesis pathway with sterol-responsive elements in their promoters (5), were all consistently up-regulated following HIV-1BRU infection (van 't Wout et al., submitted for publication).
FIG. 3.
Gene groups up- or down-regulated by HIV-1BRU infection of CCRF-CEM cells. (A) Up-regulated genes; (B) down-regulated genes. Genes were divided into seven broad categories based on their function (middle pie charts), with each category depicted by a separate pattern and numbers reflecting percentages of genes in that category compared to total number of genes regulated. Genes involved in cell signaling and communication were mostly up-regulated; this category was further divided into seven classes (pie charts at left). Genes involved in gene and protein expression were mostly down-regulated; this category was further divided into eight classes (pie charts at right).
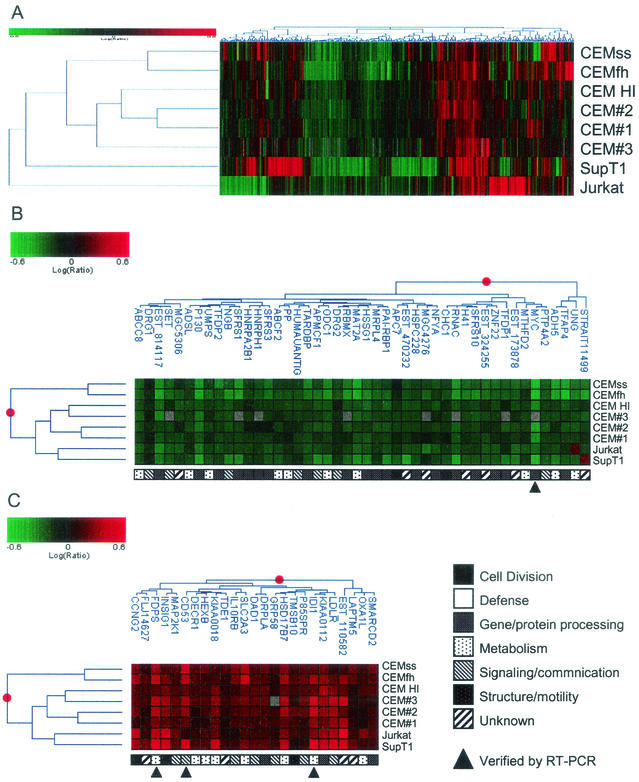
To control for cell-specific effects, we examined four additional CD4+-T-cell lines—CEMss, CEMfh, Jurkat, and SupT1—24 h after infection with HIV-1BRU. Overall, 1,484 genes were regulated (P ≤ 0.005) in at least one cell line 24 h postinfection (see supplemental data S3 for common and anti-correlated responses between genes regulated in each cell line). Figure 4A shows the 319 genes that were twofold differentially expressed (P ≤ 0.005) in at least one cell line, illustrating both the common and cell-specific regulation observed. Responses in CEM, CEMss, and CEMfh cells were more similar to each other than to those in Jurkat or SupT1 cells, corresponding to the different origins of the various cell lines (see supplemental data S3 for correlation coefficients between the 24 h infections of each cell line). Of the 1,484 regulated genes, 71 genes were significantly differentially expressed in all eight experiments (46 down- and 25 up-regulated), shown in Fig. 4B and C (P ≤ 0.10).
FIG. 4.
Gene clusters impacted by HIV-1BRU infection of five CD4+-T-cell lines. Log10 ratios are depicted in green (down-regulated) or red (up-regulated). The magnitude of the regulation is illustrated by the intensity of the color. HI, heat-inactivated BRU versus BRU. (A) Cell-type-specific responses to HIV-1 infection. Three hundred nineteen genes exhibited >2-fold change in expression in at least one of the eight HIV-1-infected cell line experiments (P ≤ 0.005). (B and C) Homogeneous responses to HIV-1 infection. Seventy-one genes were regulated in all cell lines with greater than 90% confidence (P ≤ 0.10); 46 genes were down-regulated (B), and 25 genes were up-regulated (C).
Verification of differentially expressed genes.
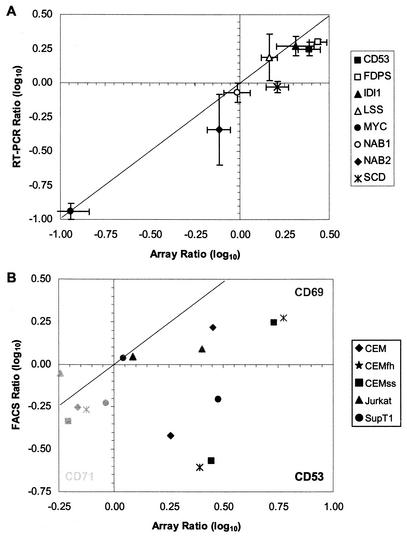
We next sought to verify the array results by assessing the expression of a sample of eight genes by semiquantitative RT-PCR. Figure 5A compares the results obtained by the two techniques, with the RT-PCR data corresponding to a single experiment read at two different PCR cycles, whereas the array data corresponded to duplicate slide experiments with fluorescent labels reversed. Overall, the RT-PCR results were consistent with the array results, providing confidence for the interpretation of the array data set.
FIG. 5.
Verification of RNA expression and comparison to changes in protein levels. Array results were compared to semiquantitative RT-PCR for eight genes (A) and for three membrane antigens by FACS (B). (A) Total RNA samples from a 24-h infection of CEMfh cells with HIV-1BRU or mock infection were tested with primers for 18S internal control and gene-specific primers for eight different genes at two different cycles. The graph depicts results obtained using microarrays on the x axis and those obtained by RT-PCR on the y axis. (B) Five CD4+-T-cell lines were infected with HIV-1BRU or mock infected with culture supernatant from uninfected cells. After 24 h, cells were washed and stained with specific purified monoclonal antibodies or isotype controls. The level of regulation corresponded with the level of p24 expression in each cell line (data not shown). The graph depicts results obtained using microarrays on the x axis and those obtained by FACS on the y axis.
Regulation of gene transcription does not ensure a corresponding change in gene product levels. Because HIV-1 infection is known to down-regulate surface expression of several membrane markers (including CD4 and major histocompatibility complex [31]), FACS analysis was performed (Fig. 5B) to examine the impact of HIV infection on the expression of cell surface markers observed to be differentially expressed by array analysis (CD53, CD69, and CD71). In all five CD4+-T-cell lines tested 24 h after infection with HIV-1BRU, CD69 was observed to be up-regulated and CD71 was observed to be down-regulated, in concordance with the array results. However, CD53 gene expression was shown to be up-regulated on the arrays and by RT-PCR (Fig. 5A), but protein expression was down-regulated as measured by FACS (Fig. 5B).
Specificity of gene expression changes induced by HIV-1BRU infection.
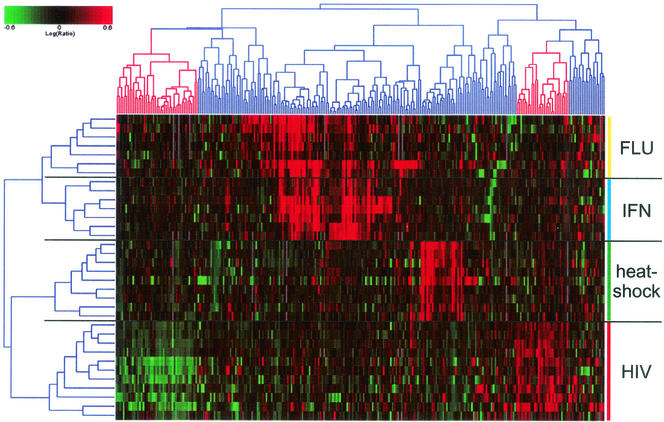
To assess the specificity of the changes in gene expression induced by HIV-1 infection, we performed a compendium analysis, comparing genes impacted by HIV-1 and influenza A virus infection, as well as by interferon and heat shock treatment of cells. Figure 6 shows a two-dimensional cluster analysis of genes versus experiments depicting 271 genes that were twofold regulated (P ≤ 0.005) in at least 2 of the 34 experiments analyzed. In general, genes impacted by each of the four infections or treatments clustered separately, indicating that they were differentially regulated in response to these agents. However, a set of genes was coordinately regulated in the interferon and influenza virus experiments, reflecting the interferon response in influenza virus-infected cells. Comparing the heat shock, interferon, and influenza virus- and HIV-1-induced expression profiles, it is clear that genes regulated in one group were either not regulated or regulated in the opposite direction than in most of the other experiment groups. This indicates that HIV-1 infection largely induces changes specific to HIV-1 infection, rather than simply activation of stress-response or cytokine response pathways.
FIG. 6.
Impact of HIV infection compared to influenza A virus infection, heat shock, and interferon treatment. Two-dimensional cluster analysis of 271 genes in 34 array experiments comparing treatment of A549 cells with influenza A virus (16); HeLa, HFH, or HH2 cells with interferon (Geiss et al., unpublished observations); heat shock in CD4+-T-cell lines (van 't Wout et al., unpublished observations); and HIV-1 infection in CD4+-T-cell lines. Ratios were expressed as described in Fig. 4. Highlighted branches depict genes specifically regulated in HIV-1 infection experiments.
Complete data sets are available through EAM at http://expression.microslu.washington.edu. Supplemental data are available at http://ubik.microbiol.washington.edu/HIV/array1/.
DISCUSSION
In this study, the response of the human CD4+-T-cell line CCRF-CEM to HIV-1 infection was explored using DNA microarrays. A total of 409 of 4,608 genes were significantly regulated at one or more time points postinfection: 165 had increased and 244 had decreased levels of stable RNA expression (Fig. 3). We found the overall level of signaling and activation to be increased, as many genes associated with cell activation, cell signaling, and communication were up-regulated, including several within 1 to 8 h postinfection, before significant levels of virus production were detectable (Fig. 1). This increased level of activation was also found in cells infected with HIV-1 at a lower MOI (8) or exposed to HIV-1 Nef (41).
Transcription of several membrane-bound receptors was regulated by HIV-1 infection, notably CD53 and CD69 (both up-regulated) and CD71 (down-regulated). Down-regulation of the transferrin receptor (CD71) has been shown to involve Nef (33, 37) and may be related to observations of iron accumulation in patients (4, 38). Iron metabolism in HIV-1-infected cells could also be affected by the strong down-regulation of the transcription factor MYC we observed (49).
Interestingly, FACS analysis of membrane-bound receptor proteins revealed down-regulation of one of the three proteins tested (CD53) despite increases in the levels of RNA (Fig. 5). CD53 has been implicated in mediating CD2 signaling pathways and thus is involved in immune responses (1). Moreover, patients with genetic CD53 deficiency show increased susceptibility to viral infections (29), suggesting that HIV-1 might interfere with this potentially antiviral pathway. The viral protein Nef has been shown to down-regulate several membrane bound receptors, including CD4 and major histocompatibility complex molecules (31). CD53 may be another target for Nef-mediated down-regulation of surface expression because of its close association with CD4 (10). Thus, the increased mRNA expression levels of CD53 measured on arrays (and confirmed by RT-PCR) may reflect a cellular feedback loop sensing low surface expression.
Infected cells had decreased levels of proliferation or cycling, as cell cycle repressors were up-regulated and inducers were down-regulated. This could reflect the impact of the viral protein Vpr, which has been shown to induce G2 arrest (19, 21, 34, 36). Genes involved in gene and protein expression, including transcription factors, DNA helicases, and splice factors, were mostly down-regulated. A small group of transcription factors was up-regulated, suggesting a specific role in increasing HIV-1 production. Indeed, some of these factors have been previously implicated in HIV-1 infection (6, 11, 12, 25, 51).
Although many genes seem to be specific for the particular system used, a small group of genes has been consistently found to be regulated in most studies of the impact of HIV-1 infection on host cell gene expression. This group includes thymosin beta 10, prothymosin alpha, and T-cell receptor alpha chain (8, 15, 45). The majority of metabolic enzymes regulated in this study were decreased in HIV-1-infected cells. However, a group of enzymes known to be involved in cholesterol biosynthesis and regulated by the sterol responsive element binding protein 2 (SREBP2) were up-regulated (van 't Wout et al., submitted for publication) in all cell lines under study.
The changes induced by HIV-1 infection were most pronounced at 24 h postinfection. By this time, clear differences were observed between different CD4+-T-cell lines infected with HIV-1. Target cells have long been known to play an important role in HIV infection and pathogenesis, although the specific interactions and the importance of these different interactions to the disease process are unknown. Species barriers to infection are partly explained by the absence or mutation of host proteins essential for the viral life cycle in nonpermissive cells (44, 47). Similarly, certain permissive cells lack antiviral pathways that inhibit HIV-1 infection and can be infected with mutant strains lacking the viral protein Vif, thought to inhibit an antiviral pathway (27, 42). CD4+ T cells have been classified as permissive (e.g., CEMss and SupT1) and nonpermissive (e.g., CEM and PBMC) to infection with viruses lacking Vif (27, 42). Indeed, our gene expression studies found several groups of genes regulated by HIV-1 infection in only specific cell lines. Studying the differences between host-cell responses in different target cells provides an opportunity to identify host proteins essential for HIV-1 infection.
Gene expression profiling using microarrays is increasingly being used to study host-pathogen interactions. By determining the pattern of gene expression at different times, it is possible to elucidate which host genes are impacted, and how, over the course of infection. Identification of genes that are differentially regulated and the characterization of their functions provide a promising window on the understanding of pathogenicity and design of novel therapeutic agents.
Acknowledgments
We thank Angela McKay, David Upchurch, and Ananta Ghosh for assisting in the generation of array data; Hongxia He for primer design; Nick Llewellyn for data entry; and all members of the Microbiology Array Group for comments and suggestions. We are indebted to Erick Hammersmark for the Spot-On spot-finding software, Jeff Furlong for design and implementation of the EAM, Ted Holzman for maintaining Resolver software, and Mark Jensen for careful reading of the manuscript.
This work was supported by grants from the American Foundation for AIDS Research (A.B.V.W.) and from the U.S. Public Health Service (J.I.M.).
REFERENCES
- 1.Bell, G. M., W. E. Seaman, E. C. Niemi, and J. B. Imboden. 1992. The OX-44 molecule couples to signaling pathways and is associated with CD2 on rat T lymphocytes and a natural killer cell line. J. Exp. Med. 175:527-536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Blader, I. J., I. D. Manger, and J. C. Boothroyd. 2001. Microarray analysis reveals previously unknown changes in Toxoplasma gondii-infected human cells. J. Biol. Chem. 276:24223-24231. [DOI] [PubMed] [Google Scholar]
- 3.Blazquez, M. V., A. Macho, C. Ortiz, C. Lucena, M. Lopez-Cabrera, F. Sanchez-Madrid, and E. Munoz. 1999. Extracellular HIV type 1 Tat protein induces CD69 expression through NF-κB activation: possible correlation with cell surface Tat-binding proteins. AIDS Res. Hum. Retrovir. 15:1209-1218. [DOI] [PubMed] [Google Scholar]
- 4.Boelaert, J. R., G. A. Weinberg, and E. D. Weinberg. 1996. Altered iron metabolism in HIV infection: mechanisms, possible consequences, and proposals for management. Infect. Agents Dis. 5:36-46. [PubMed] [Google Scholar]
- 5.Brown, M. S., and J. L. Goldstein. 1997. The SREBP pathway: regulation of cholesterol metabolism by proteolysis of a membrane-bound transcription factor. Cell 89:331-340. [DOI] [PubMed] [Google Scholar]
- 6.Chirmule, N., H. Goonewardena, S. Pahwa, R. Pasieka, and V. S. Kalyanaraman. 1995. HIV-1 envelope glycoproteins induce activation of activated protein-1 in CD4+ T cells. J. Biol. Chem. 270:19364-19369. [DOI] [PubMed] [Google Scholar]
- 7.Chomczynski, P., and N. Sacchi. 1987. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal. Biochem. 162:156-159. [DOI] [PubMed] [Google Scholar]
- 8.Corbeil, J., D. Sheeter, D. Genini, S. Rought, L. Leoni, P. Du, M. Ferguson, D. R. Masys, J. B. Welsh, J. L. Fink, R. Sasik, D. Huang, J. Drenkow, D. D. Richman, and T. Gingeras. 2001. Temporal gene regulation during HIV-1 infection of human CD4+ T cells. Genome Res. 11:1198-1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Demirov, D. G., A. Ono, J. M. Orenstein, and E. O. Freed. 2002. Overexpression of the N-terminal domain of TSG101 inhibits HIV-1 budding by blocking late domain function. Proc. Natl. Acad. Sci. USA 99:955-960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dianzani, U., M. Bragardo, D. Buonfiglio, V. Redoglia, A. Funaro, P. Portoles, J. Rojo, F. Malavasi, and A. Pileri. 1995. Modulation of CD4 lateral interaction with lymphocyte surface molecules induced by HIV-1 gp120. Eur. J. Immunol. 25:1306-1311. [DOI] [PubMed] [Google Scholar]
- 11.Dron, M., L. Hameau, L. Benboudjema, J. Guymarho, C. Cajean-Feroldi, P. Rizza, C. Godard, C. Jasmin, M. G. Tovey, and M. C. Lang. 1999. Cloning of a long HIV-1 readthrough transcript and detection of an increased level of early growth response protein-1 (Egr-1) mRNA in chronically infected U937 cells. Arch. Virol. 144:19-28. [DOI] [PubMed] [Google Scholar]
- 12.Estable, M. C., B. Bell, A. Merzouki, J. S. Montaner, M. V. O'Shaughnessy, and I. J. Sadowski. 1996. Human immunodeficiency virus type 1 long terminal repeat variants from 42 patients representing all stages of infection display a wide range of sequence polymorphism and transcription activity. J. Virol. 70:4053-4062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Garrus, J. E., U. K. von Schwedler, O. W. Pornillos, S. G. Morham, K. H. Zavitz, H. E. Wang, D. A. Wettstein, K. M. Stray, M. Cote, R. L. Rich, D. G. Myszka, and W. I. Sundquist. 2001. Tsg101 and the vacuolar protein sorting pathway are essential for HIV-1 budding. Cell 107:55-65. [DOI] [PubMed] [Google Scholar]
- 14.Geiss, G. K., M. C. An, R. E. Bumgarner, E. Hammersmark, D. Cunningham, and M. G. Katze. 2001. Global impact of influenza virus on cellular pathways is mediated by both replication-dependent and -independent events. J. Virol. 75:4321-4331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Geiss, G. K., R. E. Bumgarner, M. C. An, M. B. Agy, A. B. van 't Wout, E. Hammersmark, V. S. Carter, D. Upchurch, J. I. Mullins, and M. G. Katze. 2000. Large-scale monitoring of host cell gene expression during HIV-1 infection using cDNA microarrays. Virology 266:8-16. [DOI] [PubMed] [Google Scholar]
- 16.Geiss, G. K., M. Salvatore, T. M. Tumpey, V. S. Carter, X. Wang, C. F. Basler, J. K. Taubenberger, R. E. Bumgarner, P. Palese, M. G. Katze, and A. Garcia-Sastre. 2002. Cellular transcriptional profiling in influenza A virus-infected lung epithelial cells: the role of the nonstructural NS1 protein in the evasion of the host innate defense and its potential contribution to pandemic influenza. Proc. Natl. Acad. Sci. USA 99:10736-10741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guatelli, J. C., T. R. Gingeras, and D. D. Richman. 1990. Alternative splice acceptor utilization during human immunodeficiency virus type 1 infection of cultured cells. J. Virol. 64:4093-4098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hartigan, J. A. 1975. Clustering algorithms. John Wiley and Sons, New York, N.Y.
- 19.He, J., S. Choe, R. Walker, P. Di Marzio, D. O. Morgan, and N. R. Landau. 1995. Human immunodeficiency virus type 1 viral protein R (Vpr) arrests cells in the G2 phase of the cell cycle by inhibiting p34cdc2 activity. J. Virol. 69:6705-6711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hrimech, M., X. J. Yao, P. E. Branton, and E. A. Cohen. 2000. Human immunodeficiency virus type 1 Vpr-mediated G(2) cell cycle arrest: Vpr interferes with cell cycle signaling cascades by interacting with the B subunit of serine/threonine protein phosphatase 2A. EMBO J. 19:3956-3967. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 21.Jowett, J. B., V. Planelles, B. Poon, N. P. Shah, M. L. Chen, and I. S. Chen. 1995. The human immunodeficiency virus type 1 vpr gene arrests infected T cells in the G2 + M phase of the cell cycle. J. Virol. 69:6304-6313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kalpana, G. V., S. Marmon, W. Wang, G. R. Crabtree, and S. P. Goff. 1994. Binding and stimulation of HIV-1 integrase by a human homolog of yeast transcription factor SNF5. Science 266:2002-2006. [DOI] [PubMed] [Google Scholar]
- 23.Kato-Maeda, M., Q. Gao, and P. M. Small. 2001. Microarray analysis of pathogens and their interaction with hosts. Cell Microbiol. 3:713-719. [DOI] [PubMed] [Google Scholar]
- 24.Kim, S., R. Byrn, J. Groopman, and D. Baltimore. 1989. Temporal aspects of DNA and RNA synthesis during human immunodeficiency virus infection: evidence for differential gene expression. J. Virol. 63:3708-3713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kumar, A., S. K. Manna, S. Dhawan, and B. B. Aggarwal. 1998. HIV-Tat protein activates c-Jun N-terminal kinase and activator protein-1. J. Immunol. 161:776-781. [PubMed] [Google Scholar]
- 26.Luban, J., K. L. Bossolt, E. K. Franke, G. V. Kalpana, and S. P. Goff. 1993. Human immunodeficiency virus type 1 Gag protein binds to cyclophilins A and B. Cell 73:1067-1078. [DOI] [PubMed] [Google Scholar]
- 27.Madani, N., and D. Kabat. 1998. An endogenous inhibitor of human immunodeficiency virus in human lymphocytes is overcome by the viral Vif protein. J. Virol. 72:10251-10255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Martin-Serrano, J., T. Zang, and P. D. Bieniasz. 2001. HIV-1 and Ebola virus encode small peptide motifs that recruit Tsg101 to sites of particle assembly to facilitate egress. Nat. Med. 7:1313-1319. [DOI] [PubMed] [Google Scholar]
- 29.Mollinedo, F., G. Fontan, I. Barasoain, and P. A. Lazo. 1997. Recurrent infectious diseases in human CD53 deficiency. Clin. Diagn. Lab. Immunol. 4:229-231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nekhai, S., R. R. Shukla, A. Fernandez, A. Kumar, and N. J. Lamb. 2000. Cell cycle-dependent stimulation of the HIV-1 promoter by Tat-associated CAK activator. Virology 266:246-256. [DOI] [PubMed] [Google Scholar]
- 31.Piguet, V., O. Schwartz, S. Le Gall, and D. Trono. 1999. The downregulation of CD4 and MHC-I by primate lentiviruses: a paradigm for the modulation of cell surface receptors. Immunol. Rev. 168:51-63. [DOI] [PubMed] [Google Scholar]
- 32.Preiser, P. R., S. Khan, F. T. Costa, W. Jarra, E. Belnoue, S. Ogun, A. A. Holder, T. Voza, I. Landau, G. Snounou, and L. Renia. 2002. Stage-specific transcription of distinct repertoires of a multigene family during Plasmodium life cycle. Science 295:342-345. [DOI] [PubMed] [Google Scholar]
- 33.Pugliese, A., C. Cantamessa, A. Saini, A. Piragino, L. Gennero, C. Martini, and D. Torre. 1999. Effects of the exogenous Nef protein on HIV-1 target cells. Cell Biochem. Funct. 17:183-192. [DOI] [PubMed] [Google Scholar]
- 34.Re, F., D. Braaten, E. K. Franke, and J. Luban. 1995. Human immunodeficiency virus type 1 Vpr arrests the cell cycle in G2 by inhibiting the activation of p34cdc2-cyclin B. J. Virol. 69:6859-6864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Reddy, B., and J. Yin. 1999. Quantitative intracellular kinetics of HIV type 1. AIDS Res. Hum. Retrovir. 15:273-283. [DOI] [PubMed] [Google Scholar]
- 36.Rogel, M. E., L. I. Wu, and M. Emerman. 1995. The human immunodeficiency virus type 1 vpr gene prevents cell proliferation during chronic infection. J. Virol. 69:882-888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Savarino, A., L. Calosso, A. Piragino, C. Martini, L. Gennero, G. P. Pescarmona, and A. Pugliese. 1999. Modulation of surface transferrin receptors in lymphoid cells de novo infected with human immunodeficiency virus type-1. Cell Biochem. Funct. 17:47-55. [DOI] [PubMed] [Google Scholar]
- 38.Savarino, A., G. P. Pescarmona, and J. R. Boelaert. 1999. Iron metabolism and HIV infection: reciprocal interactions with potentially harmful consequences? Cell Biochem. Funct. 17:279-287. [DOI] [PubMed] [Google Scholar]
- 39.Scheuring, U. J., J. Corbeil, D. E. Mosier, and A. N. Theofilopoulos. 1998. Early modification of host cell gene expression induced by HIV-1. AIDS 12:563-570. [DOI] [PubMed] [Google Scholar]
- 40.Selig, L., S. Benichou, M. E. Rogel, L. I. Wu, M. A. Vodicka, J. Sire, R. Benarous, and M. Emerman. 1997. Uracil DNA glycosylase specifically interacts with Vpr of both human immunodeficiency virus type 1 and simian immunodeficiency virus of sooty mangabeys, but binding does not correlate with cell cycle arrest. J. Virol. 71:4842-4846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Simmons, A., V. Aluvihare, and A. McMichael. 2001. Nef triggers a transcriptional program in T cells imitating single-signal T cell activation and inducing HIV virulence mediators. Immunity 14:763-777. [DOI] [PubMed] [Google Scholar]
- 42.Simon, J. H., N. C. Gaddis, R. A. Fouchier, and M. H. Malim. 1998. Evidence for a newly discovered cellular anti-HIV-1 phenotype. Nat. Med. 4:1397-1400. [DOI] [PubMed] [Google Scholar]
- 43.Speth, C., I. Pichler, G. Stockl, M. Mair, and M. P. Dierich. 1998. Urokinase plasminogen activator receptor (uPAR; CD87) expression on monocytic cells and T cells is modulated by HIV-1 infection. Immunobiology 199:152-162. [DOI] [PubMed] [Google Scholar]
- 44.Stivahtis, G. L., M. A. Soares, M. A. Vodicka, B. H. Hahn, and M. Emerman. 1997. Conservation and host specificity of Vpr-mediated cell cycle arrest suggest a fundamental role in primate lentivirus evolution and biology. J. Virol. 71:4331-4338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vahey, M. T., M. E. Nau, L. L. Jagodzinski, J. Yalley-Ogunro, M. Taubman, N. L. Michael, and M. G. Lewis. 2002. Impact of viral infection on the gene expression profiles of proliferating normal human peripheral blood mononuclear cells infected with HIV type 1 RF. AIDS Res. Hum. Retrovir. 18:179-192. [DOI] [PubMed] [Google Scholar]
- 46.VerPlank, L., F. Bouamr, T. J. LaGrassa, B. Agresta, A. Kikonyogo, J. Leis, and C. A. Carter. 2001. Tsg101, a homologue of ubiquitin-conjugating (E2) enzymes, binds the L domain in HIV type 1 Pr55(Gag). Proc. Natl. Acad. Sci. USA 98:7724-7729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wei, P., M. E. Garber, S. M. Fang, W. H. Fischer, and K. A. Jones. 1998. A novel CDK9-associated C-type cyclin interacts directly with HIV-1 Tat and mediates its high-affinity, loop-specific binding to TAR RNA. Cell 92:451-462. [DOI] [PubMed] [Google Scholar]
- 48.Withers-Ward, E. S., J. B. Jowett, S. A. Stewart, Y. M. Xie, A. Garfinkel, Y. Shibagaki, S. A. Chow, N. Shah, F. Hanaoka, D. G. Sawitz, R. W. Armstrong, L. M. Souza, and I. S. Chen. 1997. Human immunodeficiency virus type 1 Vpr interacts with HHR23A, a cellular protein implicated in nucleotide excision DNA repair. J. Virol. 71:9732-9742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wu, K. J., A. Polack, and R. Dalla-Favera. 1999. Coordinated regulation of iron-controlling genes, H-ferritin and IRP2, by c-MYC. Science 283:676-679. [DOI] [PubMed] [Google Scholar]
- 50.Yang, X., M. O. Gold, D. N. Tang, D. E. Lewis, E. Aguilar-Cordova, A. P. Rice, and C. H. Herrmann. 1997. TAK, an HIV Tat-associated kinase, is a member of the cyclin-dependent family of protein kinases and is induced by activation of peripheral blood lymphocytes and differentiation of promonocytic cell lines. Proc. Natl. Acad. Sci. USA 94:12331-12336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yang, Z., and J. D. Engel. 1993. Human T cell transcription factor GATA-3 stimulates HIV-1 expression. Nucleic Acids Res. 21:2831-2836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zimmerman, C., K. C. Klein, P. K. Kiser, A. R. Singh, B. L. Firestein, S. C. Riba, and J. R. Lingappa. 2002. Identification of a host protein essential for assembly of immature HIV-1 capsids. Nature 415:88-92. [DOI] [PubMed] [Google Scholar]