Abstract
Membrane fusion plays a key role in many biological processes including vesicle trafficking, synaptic transmission, fertilization or cell entry of enveloped viruses. As a common feature the fusion process is mediated by distinct membrane proteins. We describe here ‘Fusoselect’, a universal procedure allowing the identification and engineering of molecular determinants for cell–cell fusion-activity by directed evolution. The system couples cell–cell fusion with the release of retroviral particles, but can principally be applied to membrane proteins of non-viral origin as well. As a model system, we chose a γ-retroviral envelope protein, which naturally becomes fusion-active through proteolytic processing by the viral protease. The selection process evolved variants that, in contrast to the parental protein, mediated cell–cell fusion in absence of the viral protease. Detailed analysis of the variants revealed molecular determinants for fusion competence in the cytoplasmic tail (CT) of retroviral Env proteins and demonstrated the power of Fusoselect.
INTRODUCTION
Nature has evolved a variety of cellular events that are based on membrane fusion. These include vesicle–cell fusion, virus–cell fusion and cell-to-cell fusion such as in synaptic transmission, viral cell entry and fertilization, respectively. All these processes differ not only in their intra or extracellular environment, but also in the involved molecules and the time span required for the fusion of the two separate membranes (1). However, the molecular events underlying membrane fusion occur in a consensus manner including two membrane-inserted moieties on opposed membranes, brought into close proximity through the formation of a stable helical bundle (2,3). According to the nomenclature originally used to specify compartments in vesicle trafficking we will refer to these moieties as fusion donor proteins (proteins or protein complexes harboring a hydrophobic fusion peptide) and fusion acceptor proteins (proteins or protein complexes that mediate comprehensive conformational changes of the fusion donor protein upon binding) (1,4). Once the membranes are brought into close proximity by the fusion donor and acceptor proteins, mixing of the proximal membrane leaflets (hemifusion) occurs. Subsequently, the membrane distal leaflets interact resulting in a rapidly expanding fusion pore.
While membrane fusion during vesicular trafficking involves supra-molecular complexes (SNARE protein family) instead of single fusion donor and acceptor proteins, some viruses have evolved envelope (Env) proteins facilitating this process without any accessory factors (5,6). In case of orthomyxoviruses and retroviruses the fusion machinery has been particularly well characterized and consists of the viral Env protein (fusion donor) on the virus membrane and a single virus receptor molecule (fusion acceptor) on the cellular membrane (7). In both cases, the envelope proteins are synthesized as meta-stable fusion-incompetent precursor molecules. Subsequently, fusion-activity is triggered either by low pH (endocytosis of virus particles) or, in case of retroviruses, by receptor contact (8).
Most retroviral Env proteins consist of two non-covalently linked protein domains, the surface unit (SU) and the transmembrane protein (TM), which are expressed as a single precursor polyprotein that is cleaved by a cellular protease in the Golgi. Env proteins of γ-retroviruses, like the murine leukemia virus (MLV) or the gibbon ape leukemia virus (GaLV), contain a further regulatory element. This so-called R peptide, the membrane distal domain of the cytoplasmic tail (CT) of the TM protein, inhibits the membrane fusion-activity, thus preventing cytotoxic effects within the producer cell (9). Accordingly, it is cleaved off by the viral protease during particle budding thus giving rise to the fusion-active form of the Env protein. Genetic truncation of the R peptide hence results in efficient syncytia formation of cells expressing such Env variants.
Given the broad impact of membrane fusion on different biological processes there is great interest in the engineering of fusion proteins in order to improve or alter their functions and to understand the structure/function relationships of these molecules. Here we used the GaLV Env protein to set up a system that allows the identification of molecular determinants mediating cell–cell fusion-activity by directed evolution. During selection highly fusogenic Env variants accumulated revealing different sequence motifs that achieve fusion competence.
MATERIALS AND METHODS
Cell lines
HEK-293T and NIH-3T3 cells were obtained from the American Type Culture Collection. Phoenix-Eco cells (www.stanford.edu/group/nolan) were kindly provided by Stefan Stein (Georg-Speyer-Haus, Frankfurt, Germany). All cells were maintained in high glucose (4.5 g/l) DMEM (Gibco, Eggenstein, Germany) supplemented with 10% fetal calf serum (FCS; Biochrom, Berlin, Germany), benzylpenicillin (60 µg/ml) and streptomycin (100 µg/ml) at 37°C in an atmosphere of 5% CO2.
Envelope expression constructs
All plasmids were amplified in Escherichia coli Top F10 and GM2163 strains. The MLV-packagable vector pMSCV-NEO has been described previously (10), and a similar construct with modified restriction sites was purchased from Clontech (Palo Alto, USA) and is referred to as pMSCV-Clontech-Neo. The BstEII site in pMSCV-Neo was deleted by BstEII restriction, Klenow fill in and religation. The resulting plasmid was then used to insert an EcoRI-digested PCR fragment encoding the HA-tagged GaLV Env protein (11) which was amplified using the following primers: GaLV-HA-EcoRI(+), 5′-CTTAGAATTCATGGTATTGCTGCCTGGGTC-3′, and GaLV-HA-EcoRI(−), 5′-TCATGAATTCTTACAGAATTTTAACCGCGGATATCC-3′. The resulting plasmid pMSCV-GaLV-HA was used as a parental plasmid to generate the GaLV envelope library encoding construct pMSCV-GaLV-X3 and the control vector pMSCV-GaLV-ΔR by introduction of double stranded oligonucleotides via SacII/BstEII. These were generated in a Klenow fill in reaction using forward primers GaLV-X3(+), 5′-GATATCCGCGGTT(G/C/A)NN(G/C/A)NNCTGTGATAA(G/C/A)NNCAGAAATATCAGGCCCTAGAGAACGAAGGTAACCTTT-3′ or GaLV-ΔR(+), 5′-GATATCCGCGGTTAAAATTCTGTGATAAAGACAGAAATATCAGGCCCTAGAGAACGAAGGTAACCTTT-3′ and the reverse primer GaLV-X(−), 5′-GTTACCTTCGTTCTCTAGAGC-3′. After digestion of pMSCV-GaLV-X3 with EcoRI, a diversified GaLV-HA envelope encoding fragment was obtained which was subsequently inserted into pMSCV-Clontech-NEO resulting in the bicistronic vector pMSCV-GaLV-X3-NEO. Ligation and cloning conditions for the libraries were described previously with the exception that ElectroTen-Blue bacterial cells were used in 0.1 cm cuvettes at 1.7 kV, 200 W and 25 µF (12). For both libraries, ∼1 × 106 independent bacterial clones were obtained. In theory, each library encoded 4.1 × 103 different variants, which were thus well covered by the number of clones obtained. Sequence analysis of single clones confirmed diverse sequences with no obvious bias (data not shown). After plating, cells were scraped off, resuspended and subsequently expanded in LB medium for purification of the plasmid DNA.
Generation of retroviral vectors and target cell transduction
Twenty four hours prior to transfection 3 × 107 Phoenix-Eco cells were seeded into T175 culture flasks. Transfection was performed using 45 µg of the respective MLV-packagable transfer vector and Lipofectamine Plus (Gibco, Eggenstein, Germany) according to the manufacturer's instructions. Two days post transfection the supernatant was collected and filtrated through a 0.45 µm filter. Subsequently, the supernatant was incubated with NIH-3T3 target cells for 2 h in presence of 8 µg/µl polybrene.
Library selection
Prior to selection, retroviral vectors having the library packaged were used to transduce 5 × 106 NIH-3T3 cells. One day post transduction 4 × 106 of these cells were mixed with the same amount of Phoenix-Eco cells and seeded on to the bottom of a transwell chamber (Corning, Schiphol-Rijkand, The Netherlands). Simultaneously, a permeable membrane coated with 3 × 106 NIH-3T3 cells was inserted into the chamber and incubated for 3 days. To initiate the next selection cycle, the transduced cells were detached from the membrane by trypsinization and expanded for two days followed by cultivation in DMEM supplemented with 1 mg/ml G418 (Gibco, Eggenstein, Germany) and 10% FCS for at least 5 days. With each selection cycle, the co-cultivation periods within the transwell system were decreased from 72 h (first round of selection) to 24 h (second round) and 12 h for the third selection cycle.
For sequence analysis, vector particle RNA, or in most cases, genomic DNA was isolated from transduced cells. PCR was performed using Primers Seq2(+), 5′-CCCCTATTACTCCTCCTTCTGTTGCTCATCCTC-3′ and GaLV-R?(−), 5′-CGATAAGCTTGATATCGAATTCTT-3′, or Neo(−), 5′-GGCGAACAGTTCGGCTGG-3′. The resulting PCR products were sub-cloned using the pGEM-TEasy kit (Promega, Mannheim, Germany) and plasmid DNA from single bacterial clones was sequenced using a standard T7 primer (MWG, Ebersberg, Germany).
Fluorescence-activated cell sorting (FACS)
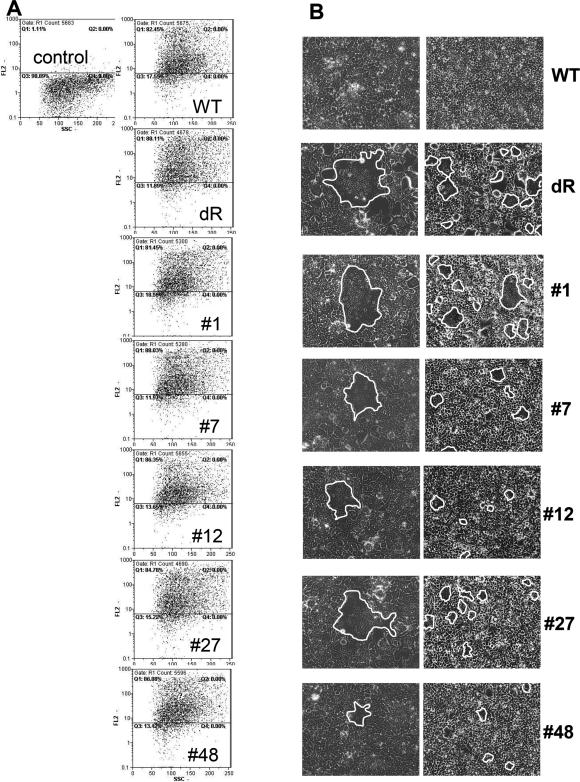
For FACS analysis 5 × 105 transfected cells were washed with PBA [phosphate-buffered saline (PBS) with 2% FCS and 0.1% sodium azide] and incubated with α-HA antibodies (Sigma, Deisenhofen, Germany) in a 1:200 dilution for 45 min at 4°C. After washing with PBA, cells were incubated with 1:50 diluted phycoerythrin (PE)-labeled secondary antibodies directed against mouse IgG (Sigma, Deisenhofen, Germany) for 30 min. After washing, cells were fixed in PBS/1% paraformaldehyde and subjected to FACS analysis (FACScan, Becton Dickinson, San Jose, USA).
Western blot analysis
Cells were harvested 48 h after transfection and lysed in RIPA lysis buffer [25 mM Tris (pH 8), 137 mM NaCl, 10% glycerol, 0.1% SDS, 0.5% NaDOC, 1% NP40, 2 mM EDTA]. Upon immunoprecipitation using the monoclonal α-HA antibody (Sigma, Deisenhofen, Germany), TM protein was detected by western blot analysis using hybridoma supernatants containing the rat monoclonal antibody 42/114 10 at a 1:12.5 dilution (11). Anti-rat IgG (rabbit; DAKO, Glostrup, Denmark) horseradish peroxidase (HRP) conjugate was used as secondary antibody in a 1:2000 dilution. Detected proteins were visualized using the SuperSignal chemoluminescence kit (Pierce, Rockford, USA).
Quantitative cell-to-cell fusion assays
Measurement of Env protein-mediated syncytia formation was performed as described elsewhere (13). In brief, 5 × 105 murine NIH-3T3 cells were transfected with pCMV-α, encoding the α-subunit of β-galactosidase, and the corresponding pMSCV-GaLV expression plasmid (1.5 mg, each). In parallel, the same number of HEK-293T target cells was transfected with 3 µg of pCMV-ω, expressing the ω-subunit of β-galactosidase. Two days post transfection, the cells were harvested and ∼2.5 × 105 cells of each type were mixed and incubated in thin-walled PCR tubes at 37°C for the desired time period. Subsequently, cells were lysed in 100 ml lysis buffer [100 mM potassium phosphate (pH 7.8), 0.2% Triton X-100] and 10 ml of this solution were used for luminescent measurement of β-galactosidase activity using GalactoStar reagents (Applied Biosystems) and a microplate luminometer.
RESULTS
Establishing Fusoselect
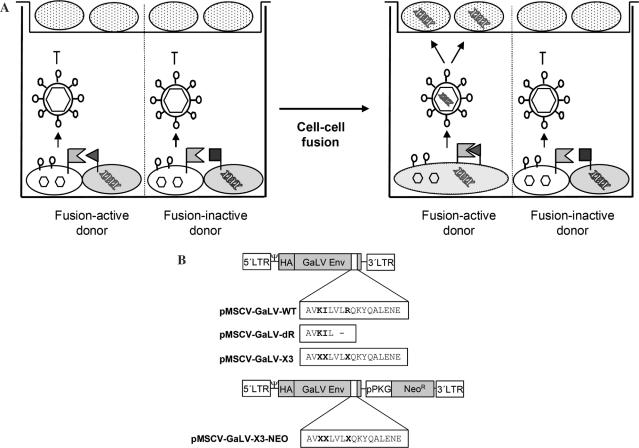
The key element of Fusoselect (Figure 1A) is the coupling of cell–cell fusion with the release of retroviral vector particles that specifically package and transfer genes encoding fusion-competent membrane proteins. For selection a retroviral packagable library encoding fusion donor protein variants is expressed in a suitable cell type (donor cell) and co-cultivated with a retroviral packaging cell line expressing the respective fusion acceptor protein (acceptor packaging cell). The expression of a fusion-competent library member in a given donor cell facilitates fusion with a neighboring acceptor packaging cell which produces retroviral particles able to package and transfer genetic information linked to a retroviral packaging signal. Upon cell-to-cell fusion, the resulting syncytium releases retroviral vector particles that have packaged the coding sequence of the fusion-competent donor protein variant. Cells transduced with these particles can be used to amplify the selected genes and to initiate the next selection cycle by repeating co-cultivation with acceptor packaging cells.
Figure 1.
Experimental design. (A) Principle of Fusoselect. Fusion donor cells (grey) expressing active (filled triangle) or inactive (filled square) fusion donor protein variants from a retroviral (MLV) packagable transfer vector (shown as double helices) are co-cultivated with an acceptor packaging cell (white) expressing the fusion acceptor, the retroviral (MLV) Gag/Pol proteins (hexagons) and the Env protein of the ecotropic MoMLV (lollipops) in the bottom chamber of a transwell system. A fusion-competent donor variant will mediate cell–cell fusion. Thus, upon syncytia formation, the vector encoding the fusion-competent donor variant (bright helix) is packaged into vector particles and will be transferred into fusion acceptor-negative target cells (dotted) seeded on to the membrane of the top transwell chamber. The coding sequence for fusion-active donor variants can be rescued directly from the target cells or upon amplification following G418 selection (in case the transfer vector confers resistance). Note that fusion protein variants that might become incorporated into the released particles (data not shown) do not affect the transduction efficiency of the target cells since the corresponding fusion acceptor protein is not expressed. (B) Schematic representation of the plasmids used. The MLV-packagable pMSCV vector encodes the HA-tagged GaLV Env protein under control of the 5′-LTR. The plasmid pMSCV-Clontech features an additional Neomycin resistance cassette (NeoR) driven by a murine phosphoglycerate kinase promoter (pPKG). The diversified amino acid residues of the X3-libraries are depicted in bold.
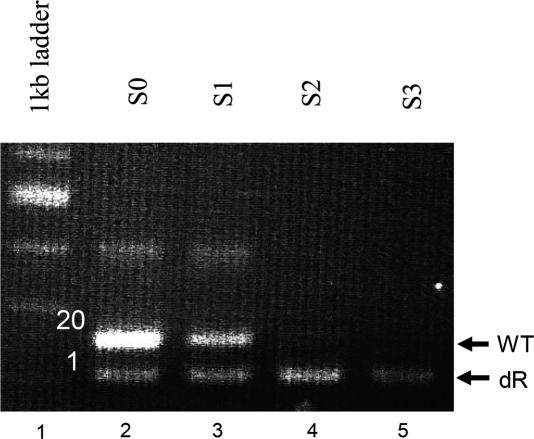
To establish this system we chose the GaLV Env protein and its receptor (GLVR1) as fusion donor/acceptor pair (14) and performed a model selection (Figure 2). In particular, we assayed the selective pressure by applying the procedure to a mixture of plasmids encoding the fusion-competent mutant EnvΔR and the fusion-incompetent wild-type (wt) Env (15). To avoid fusion among the donor cells expressing the ΔR variant, the GLVR1-negative (GLVR1−) murine cell line NIH-3T3 was used for expression. As acceptor cells we used the GLVR1-positive (GLVR+) Phoenix-Eco cell line which releases ‘empty’ (no transfer vector packaged) MLV particles displaying the ecotropic MLV Env protein (www.stanford.edu/group/nolan/retroviral_systems/retsys.html). The coding sequences for both GaLV Env variants were provided by the retroviral transfer vector plasmid pMSCV-GaLV (10) (Figure 1B). To start the selection, retroviral vector particles were generated having packaged the corresponding genes at a ratio of 20:1 (wt/ΔR) and were used to transduce murine NIH-3T3 cells at a multiplicity of infection (MOI) of <1. Transduced cells expressing a single copy of the genes were subsequently co-cultivated with Phoenix-Eco cells according to the Fusoselect procedure. Cell-free supernatants harvested after each selection cycle were used to purify viral vector RNA subsequently serving as a template for RT–PCR to determine the ratio of the packaged genes. Prior to selection, a strong signal corresponding to the non-fusogenic wt Env was obtained whereas the signal corresponding to the fusogenic variant ΔR appeared much weaker (Figure 2, lane 2), thus being in good agreement with the initial 20:1 ratio. With ongoing selection cycles, the wt GaLV Env-derived signal decreased and disappeared completely during the second selection cycle (Figure 2, lanes 2–5). In contrast, the EnvΔR-derived signal persisted over all three selection cycles performed. Thus, proof of principle for Fusoselect was obtained.
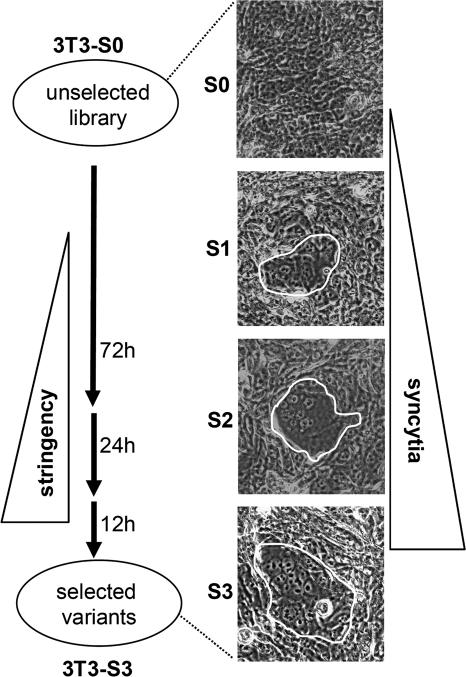
Figure 2.
Selection of a model library. A mixture of NIH-3T3 cells expressing the non-fusogenic wt Env or the fusogenic EnvΔR in a ratio of 20:1 was applied to the selection procedure. Particles from each selection cycle (S0–S3) were used for RT–PCR of the packaged genes. Primers were designed to result in two different PCR fragments for the wt and the ΔR Env (indicated with arrows).
Selection of fusion-active variants of retroviral Env proteins
To demonstrate the utility of Fusoselect, we addressed the question if Env variants can be selected that are fusion-competent in absence of the viral protease. Amino acid residues K618, I619 and R623, all located within the R peptide cleavage site, were diversified by insertion of randomized oligonucleotides into the C-tail of GaLV env (11). Two types of plasmid libraries, pMSCV-GaLV-X3 and pMSCV-GaLV-X3-NEO, differing in the presence or absence of a neomycin resistance gene were generated (Figure 1B). Prior to selection, the plasmid libraries were transfected into viral packaging cells thereby generating VSV-G pseudotyped MLV vector particles. These were then used to transduce the packaged libraries into murine NIH-3T3 cells, thus becoming potential fusion donor cells. Subsequently, the selection procedure was initiated. Vector particles released from the co-cultures with Phoenix-Eco cells were incubated with NIH-3T3 cells. These transduced cells were either used to prepare genomic DNA for sequence analysis of any selected variants or, in case of the pMSCV-GaLV-X3-NEO library, selectively amplified by expansion in G418-supplemented media. The G418-resistant cells were then used as fusion donor cells to initiate another selection cycle. In this setting, the selective pressure on fusion competence was increased with each selection cycle by decreasing the time of co-cultivation and thus the time period allowed to complete cell–cell fusion.
From the first cycle of selection of the pMSCV-GaLV-X3 library nine independent clones were obtained showing a high rate of amino acid alterations in addition to the diversified residues (data not shown; Table 1). Yet, about 30% of these clones were found to be fusion-competent (data not shown). However, the selection procedure including G418 selection of the transduced cells was even more efficient. After three rounds of selection almost 100% fusion-competent variants were obtained (see below). To monitor the progress of the selection G418-resistant bulk populations of transduced NIH-3T3 cells from each selection cycle were co-cultivated with GLVR+ HEK-293T cells. Subsequently, syncytia formation was determined using light-microscopy (Figure 3). NIH-3T3 cells expressing the unselected library did not form any visible syncytia. In contrast, NIH-3T3 cells recovered from all other selection cycles induced the formation of multinucleated cells. Furthermore, the number of nuclei within each syncytium increased from approximately seven after the first selection round to about 15 after the third selection round. This also correlated with an increased number of syncytia within the culture flask from only a few to several thousands (data not shown). Sequence analysis of the selected clones revealed the obvious accumulation of distinct motifs. Clone #1 appeared most prevalently represented by 17 out of 34 clones analyzed. Surprisingly, it contained further mutations in addition to the diversified positions. Consequently, amino acid residues 617 to 626 differed from the parental Env protein (Table 1). Noteworthy, these mutations could not have resulted from simple frameshifts but most likely originated from recombination events during reverse transcription of the vector particle packaged RNA. Clone #48 was obtained through the selection of both libraries (once from pMSCV-GaLV-X3 and twice from pMSCV-GaLV-X3-NEO). Strikingly, this clone contains an insertion of an additional nucleotide resulting in the deletion of the R peptide. Nevertheless, sequences that differed from the parental Env exclusively at the diversified residues were also selected. (Table 1, clones #7 and #12).
Table 1.
Selected variants from the GaLV-X3 librariesa
| Clone | Selected sequenceb | Libraryc | Frequency |
|---|---|---|---|
| ↓ | |||
| WT | RISAV K I LVL R QKYQALENEGNL | control | control |
| #1 | RISAGC STGPCAEIQALENEGNL- | n | 17/34 |
| #7 | RISAV R K LVL D QKYQALENEGNL- | n | 1/34 |
| #48 | RISAV P G LGPSPEISGSRERR- | m and n | 1/9 and 2/34 |
| #12 | RISAV R R LVL P QKYQALENEGNL- | m | 1/9 |
| #27 | RISAV R L LGP- | m | 1/9 |
aOnly those variants that were used for further analysis are listed.
bDiversified positions are depicted in bold, additional mutations are underlined, the R peptide cleavage site is indicated by an arrow.
cClones obtained from selection of the pMSCV-GaLV-X3-NEO library (n) or pMSCV-GaLV-X3 library (m).
Figure 3.
Increasing fusogenicity of selected variants during selection of the pMSCV-Clontech-X3 library. Transduced NIH-3T3 cells from each selection cycle were co-cultivated with GLVR1+ HEK-293T cells. The stringency of the selection was increased with ongoing cycles by decreasing the time available for cell-to-cell fusion. Syncytia formation was analyzed under the light-microscope at 200-fold magnification. Multinucleated cells are labeled by white lines.
Characterization of the selected Env variants
Selected variants from both libraries were re-inserted into the pMSCV-GaLV-WT backbone to exclude an influence of any possible mutations outside the sequenced part of the selected env genes on the phenotype. The resulting constructs and the wt- and ΔR-encoding plasmids were then transfected into HEK-293T cells to allow comprehensive characterization of the selected variants. First, the expression rates were determined by staining the transfected cells with antibodies directed against the HA epitope and subsequent FACS analysis (Figure 4A). All constructs, including the wt Env, showed similar signal intensities revealing highly efficient surface expression. In parallel, the transfected cells were analyzed by microscopy for cell-to-cell fusion. While the wt Env did not mediate any detectable syncytia formation, expression of the ΔR-mutant and the selected variants resulted in the formation of syncytia with up to 25 nuclei (Figure 4B). Next, the cell–cell fusion-activity of the selected variants was quantified using a fusion assay in which syncytia formation results in β-galactosidase activity (13). The wt Env protein was negative in this assay resulting in comparable values as Env-negative control cells. Allowing only 5 h for cell-to-cell fusion of NIH-3T3 cells expressing the selected variants and the GLVR+ HEK-293T cells, all selected variants showed fusion-activities which were at least two orders of magnitude higher than the background level (Figure 5A). After co-cultivation for 22 h the signals of all variants increased further by more than one order of magnitude without significant changes in the relative signal intensities (Figure 5A). Thus, the selected variants mediated cell-to-cell fusion as efficient as the ΔR positive control while the parental wt Env showed significantly less activity.
Figure 4.
Characterization of selected variants. (A) Surface expression of selected variants on transfected HEK-293T cells determined by FACS analysis using anti-HA antibodies. Untransfected cells served as control. (B) Syncytia formation of transfected HEK-293T cells mediated by selected variants or controls. Multinucleated cells were observed under the light-microscope at different magnifications (left column = 200-fold, right column = 100-fold). Syncytia are framed in white lines.
Figure 5.
(A) Quantitative cell-to-cell fusion assay. NIH-3T3 cells transfected with pCMV-α and the corresponding pMSCV-GaLV construct were mixed with an equal number of HEK-293T target cells expressing pCMV-ω and incubated at 37°C for either 5 h (dark bars) or 22 h (bright bars). Subsequently, β-galactosidase activity in cell lysates was determined using a luminometric assay. (B) R peptide cleavage of selected variants and controls. Cell lysates of transfected HEK-293T cells were subjected to immunoprecipitation with an anti-HA antibody followed by western blot analysis using anti-TM antibodies. Untransfected cells served as a negative control (lane 9). As positive control for R peptide cleavage, purified MLV particles having the wild-type GaLV Env incorporated were loaded in lane 1. The positions of the unprocessed (TM) and processed (TM-ΔR) form of the TM protein are indicated. The light chains of the anti-HA antibody used for immunoprecipitation are labeled by an asterisk. Arrows point to the positions of the selected TM protein variants.
Since cell entry of retroviruses is based on membrane fusion, variants surviving this selection procedure should mediate virus–cell fusion. To prove this, we generated MLV and HIV vector particles displaying the selected Env protein variants, respectively. While the MLV protease is able to cleave off the GaLV-R peptide, the HIV protease is not (11,15). Therefore, infectious HIV vector particles cannot be formed with the parental Env protein. However, as all the selected variants developed R peptide inactivation we anticipated also the formation of infectious HIV vector particles. In context of MLV particles, the fusion-active variants #1, #12 and #27 mediated on average 5-fold higher titers than the parental Env protein (data not shown). However, HIV particles pseudotyped with the five variants tested exerted at least low infectivity (>102 U/ml). Remarkably, employing Env variants #7 and #27 for HIV vector generation resulted in much higher titers (>105 U/ml) suggesting their applicability for HIV vector production.
Since no viral protease capable of cleaving the R peptide was expressed in the fusion donor cells during selection, we tested if the newly evolved variants were processed by a cellular protease. For this purpose, plasmids encoding the selected variants or controls were transfected into HEK-293T cells. Two days post transfection cell lysates were prepared and subsequently subjected to a combined immunoprecipitation and western blot analysis using TM protein-specific antibodies. While only the background signal derived from the light chain of the antibody used for the immunoprecipitation step was detectable in the negative control sample, distinct bands were obtained for the different variants (Figure 5B). As expected, the TM protein of the parental Env migrated as a single band at ∼15 kDa, whereas a band corresponding to a lower molecular mass was obtained for the EnvΔR variant (lanes 2 and 3). Clones #1, #7 and #12 migrated as single bands at the same position as the parental TM protein. Thus, there was no evidence for further processing of these variants, while the parental Env was cleaved when coexpressed with the viral protease (compare lanes 4–6 with lane 1). Clone #27 showed a band at a lower molecular weight resulting from the deletion of the R peptide. This variant encodes only two more amino acids than the TM protein of the EnvΔR variant (lane 7). Clone #48 revealed an electrophoretic mobility between that of the parental TM and that of the EnvΔR variant probably due to its unique length and three additional positively charged arginine residues (lane 8). Overall these results demonstrated that the selected variants did not require any further processing to become fusion-active. The latter is especially remarkable for clones #7 and #12 since these variants encompassed the complete R peptide.
DISCUSSION
We report here a novel directed evolution strategy, termed ‘Fusoselect’ allowing for comprehensive analysis and engineering of cell–cell fusion-activity within TMs. The system is based on coupling cell-to-cell fusion, mediated by a fusion-active protein variant, with the release of retroviral particles that have packaged the coding region of the respective protein. This strategy was instrumental to set up an in vivo system that selects for the formation of lethal syncytia, i.e. a cytotoxic property. This has so far only been possible via the generation of replica-libraries and the screening of small library pool fractions, as described for the identification of the HIV co-receptor (16,17). However, replica-libraries only allow low diversities and require time consuming sub-fractionation steps. Due to its integrated signal amplification Fusoselect circumvents these limitations. The selected genes can easily be amplified, expressed and analyzed upon transduction of fusion acceptor-negative indicator cells with the released vector particles.
Since the determinants for membrane fusion activation are well characterized for Env proteins of retroviruses, we used these to demonstrate the power of Fusoselect by showing that fusion-inactive Env proteins can be converted to fusion-active ones. Even without amplification of the packaged genes, fusion-active variants were obtained from libraries of Env proteins, diversified at three amino acid residues located in their CTs, after just one round of selection. By employing retroviral transfer vectors that encoded an additional antibiotic resistance gene several selection rounds could be performed. Thereby, an additional level of diversification mediated by the reverse transcription process was introduced. In fact, most of the selected variants carried such mutations (see below). After three selection rounds all of the characterized clones were fusion-competent revealing the high stringency of the selection procedure.
The selected variants not only gained cell–cell fusion competence but were also active in mediating cell entry of retroviral and lentiviral vector particles. This clearly shows the potential of Fusoselect in optimizing gene transfer systems e.g. for gene therapy purposes.
A more detailed look at the selected Env protein variants revealed how fusogenicity has been evolved. The most frequently selected clone #1 had accumulated several glycine and serine residues. Previous investigations of the MLV Env protein revealed that fusion-activity can be achieved in the absence of R peptide cleavage by introduction of glycine and serine residues upstream of the R peptide, most likely rendering the CT more flexible and releasing the inhibitory constraint (18). Our results indicate that this is a general mechanism of γ-retroviral Env proteins. Another way to gain fusion-activity is either by truncation or by complete exchange of the R peptide as seen for selected clones #27 and #48. Interestingly, variants that differed from the parental fusion-inactive protein within the diversified sites only were also selected. These variants (clone #7 and #12) had positively charged amino acids at residues 618 and 619 in common. As R peptide cleavage and improved cell surface trafficking could be excluded for these variants, altered folding of the C-tail due to the additional charges resulting in a non-functional R peptide appears to be the most likely explanation. Taken together, the selected variants gained fusion-activity either by truncation, deletion or altered folding of the R peptide but not by proteolytical cleavage through cellular proteases. This might be due to the fact that only three residues of the viral cleavage site were diversified initially, making the generation of motifs that are recognized by cellular proteases rather unlikely.
An immediate application of Fusoselect will be the evolution of highly specific suicide genes for cancer gene therapy. It has been shown previously that fusogenic envelope proteins exhibit a strong bystander effect facilitating cell death of tumor cells at high efficiency (19). These proteins can be engineered, e.g. by fusion to scFvs or receptor ligands, to mediate cell–cell fusion by binding to specific cell surface markers acting as fusion acceptors (20,21). Fusoselect will allow the screening of highly diverse libraries of such modified envelope proteins. Alternatively, targeted cell–cell fusion-activity might be achieved by evolving specific cleavage sites within the TM cytoplasmic domain for calpain, a protease known to be overexpressed in a variety of cancer cells (22,23).
Beyond that, we expect Fusoselect to become applicable for proteins involved in cellular membrane fusion events as well, since there is no requirement for the protein of interest to be of viral origin or to become incorporated into the retroviral particles. The selection of variants not covered by the library design as shown here for variants #1 and #48 shows that very rare single events, occurring through reverse transcription during the screening procedure, within the library can be identified by Fusoselect. The scale up of Fusoselect required for this purpose should be easily achievable. Plenty examples exist demonstrating that retroviral cDNA libraries covering complexities greater than 1 × 106 can be successfully generated and screened on a single cell level (24,25). Then, yet unknown proteins mediating cell–cell fusion directly, such as fusion-active envelope proteins originating from endogenous retroviruses and their cognate receptors (26,27), or indirectly, such as proteins involved in vesicle fusion (28,29) could be identified by subjecting retroviral cDNA libraries to the Fusoselect screening procedure.
Acknowledgments
C.A.M. was supported by a Ph.D. scholarship of the Fonds der Chemischen Industrie and the Bundesministerium für Bildung und Forschung, J.S. was partly supported by the European Molecular Biology Organization. Funding to pay the Open Access publication charges for this article was provided by Paul-Ehrlich-Institut.
Conflict of interest statement. None declared.
REFERENCES
- 1.Jahn R, Lang T., Sudhof T.C. Membrane fusion. Cell. 2003;112:519–533. doi: 10.1016/s0092-8674(03)00112-0. [DOI] [PubMed] [Google Scholar]
- 2.Waters M.G., Hughson F.M. Membrane tethering and fusion in the secretory and endocytic pathways. Traffic. 2000;1:588–597. doi: 10.1034/j.1600-0854.2000.010802.x. [DOI] [PubMed] [Google Scholar]
- 3.Noguchi H., Takasu M. Self-assembly of amphiphiles into vesicles: a Brownian dynamics simulation. Phys. Rev. E Stat. Nonlin Soft Matter Phys. 2001;64:041913. doi: 10.1103/PhysRevE.64.041913. [DOI] [PubMed] [Google Scholar]
- 4.Bonifacino J.S., Glick B.S. The mechanisms of vesicle budding and fusion. Cell. 2004;116:153–166. doi: 10.1016/s0092-8674(03)01079-1. [DOI] [PubMed] [Google Scholar]
- 5.Zhang F., Chen Y., Su Z., Shin Y.K. SNARE assembly and membrane fusion, a kinetic analysis. J. Biol. Chem. 2004;279:38668–38672. doi: 10.1074/jbc.M404904200. [DOI] [PubMed] [Google Scholar]
- 6.Ungar D., Hughson F.M. SNARE protein structure and function. Annu. Rev. Cell Dev. Biol. 2003;19:493–517. doi: 10.1146/annurev.cellbio.19.110701.155609. [DOI] [PubMed] [Google Scholar]
- 7.Fass D., Harrison S.C., Kim P.S. Retrovirus envelope domain at 1.7 angstrom resolution. Nature Struct. Biol. 1996;3:465–469. doi: 10.1038/nsb0596-465. [DOI] [PubMed] [Google Scholar]
- 8.Smith A.E., Helenius A. How viruses enter animal cells. Science. 2004;304:237–242. doi: 10.1126/science.1094823. [DOI] [PubMed] [Google Scholar]
- 9.Ragheb J.A., Anderson W.F. pH-independent murine leukemia virus ecotropic envelope-mediated cell fusion: implications for the role of the R peptide and p12E TM in viral entry. J. Virol. 1994;68:3220–3231. doi: 10.1128/jvi.68.5.3220-3231.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Grez M., Akgun E., Hilberg F., Ostertag W. Embryonic stem cell virus, a recombinant murine retrovirus with expression in embryonic stem cells. Proc. Natl Acad. Sci. USA. 1990;87:9202–9206. doi: 10.1073/pnas.87.23.9202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Merten C.A., Stitz J., Braun G., Poeschla E.M., Cichutek K., Buchholz C.J. Directed evolution of retrovirus envelope protein cytoplasmic tails guided by functional incorporation into lentivirus particles. J. Virol. 2005;79:834–840. doi: 10.1128/JVI.79.2.834-840.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Buchholz C.J., Peng K.W., Morling F.J., Zhang J., Cosset F.L., Russell S.J. In vivo selection of protease cleavage sites from retrovirus display libraries. Nat. Biotechnol. 1998;16:951–954. doi: 10.1038/nbt1098-951. [DOI] [PubMed] [Google Scholar]
- 13.Holland A.U., Munk C., Lucero G.R., Nguyen L.D., Landau N.R. Alpha-complementation assay for HIV envelope glycoprotein-mediated fusion. Virology. 2004;319:343–352. doi: 10.1016/j.virol.2003.11.012. [DOI] [PubMed] [Google Scholar]
- 14.O'Hara B., Johann S.V., Klinger H.P., Blair D.G., Rubinson H., Dunn K.J., Sass P., Vitek S.M., Robins T. Characterization of a human gene conferring sensitivity to infection by gibbon ape leukemia virus. Cell Growth Differ. 1990;1:119–127. [PubMed] [Google Scholar]
- 15.Christodoulopoulos I., Cannon P.M. Sequences in the cytoplasmic tail of the gibbon ape leukemia virus envelope protein that prevent its incorporation into lentivirus vectors. J. Virol. 2001;75:4129–38. doi: 10.1128/JVI.75.9.4129-4138.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Singhi A.D., Kondratov R.V., Neznanov N., Chernov M.V., Gudkov A.V. Selection-subtraction approach (SSA): a universal genetic screening technique that enables negative selection. Proc. Natl Acad. Sci. USA. 2004;101:9327–9332. doi: 10.1073/pnas.0403080101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Feng Y., Broder C.C., Kennedy P.E., Berger E.A. HIV-1 entry cofactor: functional cDNA cloning of a seven-transmembrane, G protein-coupled receptor. Science. 1996;272:872–877. doi: 10.1126/science.272.5263.872. [DOI] [PubMed] [Google Scholar]
- 18.Li M., Yang C., Compans R.W. Mutations in the cytoplasmic tail of murine leukemia virus envelope protein suppress fusion inhibition by R peptide. J. Virol. 2001;75:2337–2344. doi: 10.1128/JVI.75.5.2337-2344.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bateman A., Bullough F., Murphy S., Emiliusen L., Lavillette D., Cosset F.L., Cattaneo R., Russell S.J., Vile R.G. Fusogenic membrane glycoproteins as a novel class of genes for the local and immune-mediated control of tumor growth. Cancer Res. 2000;60:1492–1497. [PubMed] [Google Scholar]
- 20.Fielding A.K., Chapel-Fernandes S., Chadwick M.P., Bullough F.J., Cosset F.L., Russell S.J. A hyperfusogenic gibbon ape leukemia envelope glycoprotein: targeting of a cytotoxic gene by ligand display. Hum. Gene Ther. 2000;11:817–826. doi: 10.1089/10430340050015437. [DOI] [PubMed] [Google Scholar]
- 21.Nakamura T., Peng K.W., Vongpunsawad S., Harvey M., Mizuguchi H., Hayakawa T., Cattaneo R., Russell S.J. Antibody-targeted cell fusion. Nat. Biotechnol. 2004;22:331–336. doi: 10.1038/nbt942. [DOI] [PubMed] [Google Scholar]
- 22.Carragher N.O., Frame M.C. Calpain: a role in cell transformation and migration. Int. J. Biochem. Cell Biol. 2002;34:1539–1543. doi: 10.1016/s1357-2725(02)00069-9. [DOI] [PubMed] [Google Scholar]
- 23.Lakshmikuttyamma A., Selvakumar P., Kanthan R., Kanthan S.C., Sharma R.K. Overexpression of m-calpain in human colorectal adenocarcinomas. Cancer Epidemiol. Biomarkers Prev. 2004;13:1604–1609. [PubMed] [Google Scholar]
- 24.Kitamura T., Onishi M., Kinoshita S., Shibuya A., Miyajima A., Nolan G.P. Efficient screening of retroviral cDNA expression libraries. Proc. Natl Acad. Sci. USA. 1995;92:9146–9150. doi: 10.1073/pnas.92.20.9146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stitz J., Krutzik P.O., Nolan G.P. Screening of retroviral cDNA libraries for factors involved in protein phosphorylation in signaling cascades. Nucleic Acids Res. 2005;33:e39. doi: 10.1093/nar/gni039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Blond J.-L., Lavillette D., Cheynet V., Bouton O., Oriol G., Chapel-Fernandes S., Mandrand B., Mallet F., Cosset F.-L. An envelope glycoprotein of the human endogenous retrovirus HERV-W is expressed in the human placenta and fuses cells expressing the type d mammalian retrovirus receptor. J. Virol. 2000;74:3321–3329. doi: 10.1128/jvi.74.7.3321-3329.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lavillette D., Marin M., Ruggieri A., Mallet F., Cosset F.-L., Kabat D. The envelope glycoprotein of the human endogenous retrovirus type W uses a divergent family of amino acid transporters/cell surface receptors. J. Virol. 2002;76:6442–6452. doi: 10.1128/JVI.76.13.6442-6452.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hu C., Mahiuddin A., Melia T.J., Söllner T.H., Mayer T., Rothman J.E. Fusion of cells by flipped SNAREs. Science. 2003;300:1745–1749. doi: 10.1126/science.1084909. [DOI] [PubMed] [Google Scholar]
- 29.Witze E., Rothman J.E. Cell fusion: an EFFicient sculptor. Curr. Biol. 2002;12:R467–R479. doi: 10.1016/s0960-9822(02)00948-x. [DOI] [PubMed] [Google Scholar]