Abstract
The mechanisms by which AP endonucleases recognize AP sites have not yet been determined. Based on our previous study with Escherichia coli exonuclease III (ExoIII), the ExoIII family AP endonucleases probably recognize the DNA-pocket formed at an AP site. The indole ring of a conserved tryptophan residue in the vicinity of the catalytic site presumably intercalates into this pocket. To test this hypothesis, we constructed a series of mutants of ExoIII and human APE1. Trp-212 of ExoIII and Trp-280 of APE1 were critical to the AP endonuclease activity and binding to DNA containing an AP site. To confirm the ability of the tryptophan residue to intercalate with the AP site, we examined the interaction between an oligopeptide containing a tryptophan residue and an oligonucleotide containing AP sites, using spectrofluorimetry and surface plasmon resonance (SPR) technology. The tryptophan residue of the oligopeptide specifically intercalated into an AP site of DNA. The tryptophan residue in the vicinity of the catalytic site of the ExoIII family AP endonucleases plays a key role in the recognition of AP sites.
INTRODUCTION
Cellular DNA is spontaneously and continuously damaged by environmental and internal factors such as X-rays, ultraviolet (UV) light, alkylating agents and oxygen radicals (1,2). DNA damage contributes to mutagenesis, carcinogenesis and aging (1,2). Apurinic/apyrimidinic (AP) sites form with the repair of damaged bases during base excision repair (BER) (3,4). Damage-specific DNA N-glycosidases cleave the N-glycosidic linkage between the damaged base and deoxyribose ring, producing an AP site (4,5). Furthermore, AP sites are generated by the spontaneous loss of normal bases (1,2). In human lung fibroblasts, the number of AP sites in the steady state is about 0.67/106 nt (∼2000 AP sites/human cell) (6). Unless the AP site is repaired, transcription and replication are disrupted (1,7). In BER, the AP endonuclease is an important player in the repair of AP sites. This enzyme recognizes a AP site in DNA and cleaves on the 5′ side of the site through AP endonuclease activity. In mammals, after cleavage of the AP site by APE1 AP endonuclease, the nucleotide gap is filled, the 5′-abasic residue is removed, and the nick is sealed in the short-patch or long-patch pathway. In these pathways, DNA polymeraseβ and DNA ligase III act in the short-patch pathway and DNA polymeraseβ/ɛ, flap endonuclease (FEN1) and DNA ligase I act in the long-patch pathway (3,4).
Based on the homology of the amino acid sequences and structures, AP endonucleases are divided into the Escherichia coli exonuclease III (ExoIII) family and E.coli endonuclease IV family. From prokaryotes to eukaryotes, a number of genes which encode ExoIII family AP endonucleases have been cloned (8–21). The major AP endonuclease of Homo sapiens, which is termed APE1 (also APEX, HAP1 or Ref-1), belongs to the ExoIII family. The E.coli xthA mutant lacking ExoIII was shown to be hypersensitive to hydrogen peroxide and UV (22,23). Homozygous mutant mice lacking the APE1 AP endonuclease died early in embryonic development (24). Experiments with these deletion mutants have suggested that the AP endonucleases play an important role in the life of organisms from prokaryotes to eukaryotes.
Despite many studies of the ExoIII family AP endonucleases, how they recognize AP sites has not yet been elucidated. Four mechanisms of recognition have been proposed. Hypothesis I: based on the structure of E.coli ExoIII, it has been suggested that the AP endonuclease recognizes an extrahelical non-pairing base opposite an AP site (25). Hypothesis II: based on the crystal structure of APE1 with or without substrate DNA, it has been suggested that the AP endonuclease recognizes an extrahelical abasic deoxyribose ring (26,27). Hypothesis III: a specific local distortion produced by the AP site might be recognized by the AP endonuclease (28,29). Hypothesis IV: we have proposed that the space produced by deletion of the nucleic acid base is recognized by the AP endonuclease through intercalation of the aromatic side chain (indole ring) of the tryptophan residue, which is located in the vicinity of the active site (30). In the case of ExoIII, this tryptophan residue is Trp-212. The tryptophan residue protrudes from the open face of the active site (25). This hypothesis was based on our previous experimental results.
In our previous experiments, ExoIII cleaved single-stranded DNA containing an AP site, and DNA containing an AP site analog such as propanediol, which had no ring structure, was a good substrate for the endonuclease (30). From these results, it has been suggested that an orphan base opposite an AP site and abasic ribose ring are not critical for recognition of the AP site by the ExoIII family AP endonuclease. Moreover, it was clarified that ExoIII and APE1 do not cleave bulged DNA, which is similar in structure to AP-DNA (30,31). This result has indicated that a site-specific structure, namely a specific local distortion, is not recognized by the AP endonuclease. Therefore, hypotheses I, II and III seem not to explain how the AP site is recognized by the ExoIII family AP endonucleases. According to hypothesis IV, the tryptophan residue in the vicinity of the catalytic site is conserved from bacteria to mammals (Figure 1). Furthermore, the tryptophan side chain of the tripeptide (Lys-Trp-Lys) intercalates into the space of the AP site in double-stranded DNA supports this hypothesis (32,33).
Figure 1.
Sequence alignment of active sites of AP endonucleases. The amino acid sequences of AP endonuclease were aligned using EX3_ECOLI (E.coli ExoIII), EX3_SALTY (Salmonella typhimurium ExoIII), EX3_HAEIN (Haemophilus influenzae ExoIII), EXOA_BACSU (B.subtilis ExoA), EXOA_STRPN (Streptococcus pneumoniae ExoA), RRP1_DROME (Drosophila melanogaster Rrp1), APEA_DICDI (Dictyostelium discoideum APEA), ARP_ARATH (Arabidopsis thaliana ARP), APEX1_RAT (Rattus norvegicus APE1), APEX1_MOUSE (Mus musculus APE1), APEX1_BOVIN (Bos taurus APE1) and APEX1_HUMAN (H.sapiens APE1). The reversed and the shaded characters indicate conserved aromatic amino acid residues and active amino acid residues, respectively. The amino acid residue numbers are given at the left and right of each line. The corresponding numbers of the amino acid residues in EX3_ECOLI and APEX1_HUMAN are shown above and below the sequences, respectively. Asterisks indicate the AP endonucleases used in this study.
The present study was undertaken to elucidate the mechanism by which the ExoIII family AP endonucleases recognize AP sites. First, based on hypothesis IV, we constructed a series of mutants of ExoIII and APE1 with substitutions of the aromatic amino acid residues, which exist in the vicinity of the active site. The AP endonuclease activity and binding ability of the mutants were examined using oligonucleotides containing AP sites. Second, to confirm that tryptophan has the ability to interact specifically with the AP site, we examined the specific binding of an oligopeptide containing a tryptophan residue to the oligonucleotides containing AP sites using spectrofluorimetry and the surface plasmon resonance (SPR) technique.
MATERIALS AND METHODS
Plasmid construction and site-directed mutagenesis
The overexpression plasmid for E.coli ExoIII was produced by cloning a PCR-generated xthA fragment into the EcoRI and HindIII restriction sites of pKP1500 (34). The resulting plasmid was termed pKPE3. The overexpression plasmid for the N-terminal-truncated human APE1 (Δ40APE1: 40–318 residues) was constructed by cloning a PCR-amplified DNA fragment into the NdeI and XhoI sites of pET-15b (Novagen). The DNA fragment coding the N-terminal-truncated human APE1 was amplified by PCR using pUAEH1 (21) as a template. The sense and antisense primers for PCR were d-CTGCTGCAT|TAATGAGGGCCCAGCCCTGTATG (PshBI recognition sequence, underlined; PshBI cleavage site, |) and d-GGATCC|TCGAGTCACAGTGCTAGGTATAGGGTG (XhoI recognition site, underlined; XhoI cleavage site, |), respectively. The PCR product was digested with PshBI and XhoI restriction endonucleases and then cloned into the NdeI and XhoI sites of pET-15b. The resulting plasmid was termed pETΔ40APE1.
Site-directed mutagenesis of the overexpression plasmids pKPE3 (ExoIII) and pETΔ40APE1 (APE1) was performed using the QuickChange site-directed mutagenesis kit (Stratagene). Five ExoIII (Y109S, W212S, F213W, F213S and W212S/L226W) and four APE1 (F266S, W280S, F266W/W280S, F266S/W280S) mutant proteins were designed to investigate the role of the aromatic amino acid residues. Two complementary mutagenic DNA oligomer sets as the sense and antisense primers were designed for the substitution of each amino acid residue of the AP endonuclease. The mutagenic sense primers were d-GATCAACGGTTCCTTCCCGCAGG, d-GATCGTTTCTCATCGTTTGATTACCGCTC, d-GATCGTTTCTCATGGTGGGATTACCGCTC, d-GATCGTTTCTCATGGTCTGATTACCGCTC, d-GATAACCGTGGTTGGCGCATCGACCTG, d-CTATGCCTACACCTCTTGGACTTATATG, d-CTATGCCTACACCTGGTGGACTTATATG and d-CAAGAATGTTGGTTCGCGCCTTGATTAC for Y109S, W212S, F213W, F213S, L226W, F266S, F266W and W280S, respectively (substituted amino acid codon, underlined; substituted nucleotide, boldfaced).
Purification of ExoIII and its mutant proteins
In the cases of the wild-type ExoIII and its mutant proteins (W212S, F213W and Y109S), E.coli XL-1 Blue carrying the plasmid corresponding to individual proteins was grown in Luria–Bertani (LB) medium (500 ml) containing ampicillin (50 µg/ml) at 28°C. When the cell density reached 0.5 at 600 nm, 500 µl of 1 M isopropyl-β-d-thiogalactopyranoside (IPTG) was added to the culture and the incubation temperature was raised to 42°C. After 4 h or when the inclusion bodies began to appear in the cells, the cells were harvested by centrifugation. The harvested cells were suspended in 10 ml of the sonication buffer [50 mM Tris–HCl (pH 8.0), 50 mM KCl, 1 mM EDTA and 0.002% phenylmethylsulfonyl fluoride (PMSF)] and then sonicated with 3 g of glass beads on ice. The supernatant was collected by centrifugation. The target proteins in the supernatant were fractionated by selective ammonium sulfate precipitation. The target proteins were soluble at 50% saturation and precipitated at 70% saturation. The precipitated proteins were redissolved in 40 ml of 20 mM Tris–HCl (pH 8.0), 0.1 mM DTT and 0.002% PMSF and then dialyzed three times with 2 l of 20 mM Tris–HCl (pH 8.0) and 0.1 mM DTT. The resulting solution was injected on to a DEAE cellulose column (Whatman DE52, 1.5 × 10 cm) and the proteins were eluted with a linear gradient of 0–1 M NaCl in 20 mM Tris–HCl (pH 8.0) and 0.2 mM DTT. The fractions (∼30 ml) containing the target proteins were dialyzed three times against 2 l of 20 mM potassium phosphate (pH 6.5) and 0.1 mM DTT. The dialyzed solution was loaded on to a phosphocellulose column (Whatman P11, 1.5 × 5 cm), and the proteins were eluted with a linear gradient of 20–500 mM potassium phosphate (pH 6.5) and 0.1 mM DTT. The fractions (∼45 ml) containing the target protein were dialyzed three times against 2 l of 20 mM Tris–HCl (pH 8.0) and 0.1 mM DTT. The dialyzed solution was loaded into a Resource Q column (Amersham Biosciences, 0.64 × 3 cm) and the proteins were eluted with a linear gradient of 0–500 mM NaCl in 20 mM Tris–HCl (pH 8.0) and 0.1 mM DTT. The fractions (∼2 ml) containing the target proteins were dialyzed three times against 1 l of 25 mM Tris–HCl (pH 8.0), 0.5 mM DTT, 50 mM KCl and 50% glycerol. The dialyzed solution was then stored at −20°C.
In the cases of the F213S and W212S/L226W mutant proteins, cultivations were carried out as described above, except that they were continued until complete production of the inclusion bodies. The wild-type ExoIII could form inclusion bodies in cells on cultivation for a long time. The cells harboring the inclusion bodies were collected by centrifugation. The harvested cells were suspended in 4 ml of 30 mM Tris–HCl (pH 7.2) and sonicated on ice without glass beads. The precipitate containing the inclusion bodies was collected by centrifugation (4000 r.p.m., 4°C, 5 min) using MX-300 micro centrifuge (Tomy). To wash the inclusion bodies, the precipitate was suspended in 8 ml of 10 mM Tris–HCl (pH 7.5) and 1 mM EDTA and then centrifuged (4000 r.p.m., 4°C, 5 min) using MX-300. This washing step was repeated four times. The purified inclusion bodies were dissolved in 250 µl of 12.3 mM Tris–HCl (pH 6.8), 4.6% SDS, 20% glycerol and 20% 2-mercaptethanol by heating at 95°C for 5 min. An equal volume of 10 mM Tris–HCl (pH 7.4) and 25% 2-propanol was added to the solution of the inclusion bodies at room temperature, and then the resulting solution was dialyzed three times against 1 l of 10 mM Tris–HCl (pH 7.4) and 25% 2-propanol. Finally, the solution was dialyzed three times against 20 mM Tris–HCl (pH 8.0) and 0.1 mM DTT.
Purification of APE1 and its mutant proteins
The hexahistidine-tagged APE1 (Δ40APE1) and its mutant proteins were overproduced in E.coli BL21 (DE3) (Novagen). The competent E.coli cells were transformed by the plasmid corresponding to individual proteins. Transformants were grown in LB medium (500 ml) containing ampicillin (50 µg/ml) at 37°C. When the cell density reached 0.5 at 600 nm, 250 µl of 1 M IPTG was added to the culture and further cultivation was carried out at 25°C. After 2 h, the cells were harvested by centrifugation. The harvested cells were suspended in 10 ml of 20 mM sodium phosphate (pH 7.4), 1 M NaCl and 20 mM imidazole and then sonicated with 3 g of glass beads on ice. The supernatant was collected by centrifugation and loaded on to a HisTrap HP column (Amersham Biosciences, 0.7 × 2.5 cm). The target proteins were eluted with a stepwise gradient of 40–500 mM imidazole in 20 mM phosphate (pH 7.4) and 1 M NaCl. The eluates containing the target protein were collected and dialyzed three times against 1 l of 20 mM Tris–HCl (pH 8.0), 300 mM NaCl, 1 mM DTT and 50% glycerol. The resulting solution was stored at −20°C.
AP endonuclease assays and gel shift assays
The AP endonuclease assays and gel shift assays were carried out as described previously (30). The oligonucleotide sequences used in this study were d-GCGATGACTAACGHTACTAGGCTTCCGAGCC (AP-ssDNA, H: 1′, 2′-dideoxyribofranose residue) and d-CGGAAGCCTAGTATCGTTAGTCATCGCCATG (complimentary strand). The 5′ end of the AP-ssDNA was labeled with [γ-32P]ATP (Amersham Biosciences) by T4 polynucleotide kinase (Nippon Gene). The markers were 5′ 32P-labeled d-GCGATGACTAACG and d-GCGATGACTAACGATACTAGGCTTCCGAGCC. The AP endonuclease and gel shift assays were quantified based on the detection of radioactivity using STORM860 (Amersham Biosciences) and ImageQuant software (Amersham Biosciences).
SPR analysis
SPR-based analyses were performed using a Biacore X (Biacore). All analyses were performed at a flow rate of 20 µl/min and 25°C. The biotinylated DNA oligomers were immobilized on the sensor chip SA. The oligonucleotide containing three AP sites (3AP-ssDNA) was d-GGGCGGCCTGTCGHTCTAGGCGGCTACGHTCAAGGCCTCGACGHTCTACGCGCGCGCCTTT-biotin (H: 1′, 2′-dideoxyribofranose). The oligonucleotide not containing the AP site (3Native-ssDNA) was d-GGGCGGCCTGTCGATCTAGGCGGCTACGATCAAGGCCTCGACGATCTACGCGCGCGCCTTT-biotin. For formation of the double-stranded oligonucleotides, the oligonucleotides 3AP-ssDNA and 3Native-ssDNA were annealed with the complementary strand, d-GGCGCGCGCGTAGATCGTCGAGGCCTTGATCGTAGCCGCCTAGATCGACAGGCCGCCC before immobilization on the SA chip. APE1 and its mutant proteins used as analytes were dialyzed three times against 1 l of 10 mM HEPES–NaOH (pH 7.4), 150 mM NaCl and 10 mM EDTA buffer immediately before use. Each concentration of protein was then adjusted to 297 nM. The dialysis buffer after dialysis was used as the running buffer of Biacore X. The association and dissociation phases each lasted for 120 s. The sequences of the peptides used in the SPR analysis were KSRGKWKGRSK (11KWK) and KSRGKAKGRSK (11KAK) (difference between 11KWK and 11KAK: boldfaced). The concentrations of these peptides were adjusted to 100 µM in 10 mM HEPES–NaOH (pH 7.4), 150 mM NaCl and 10 mM EDTA.
Spectrofluorimetric analyses
The fluorescence analyses were performed using a RF-1500 (Shimazu) spectrofluorometer. The excitation wavelength was 275 nm. The emission spectra were obtained by scanning from 320 to 500 nm. The double-stranded DNA and the peptide were dissolved in 1 mM sodium cacodylate (pH 6.0), 1 mM NaCl and 0.2 mM EDTA. The concentrations of the double-stranded DNA and the peptides were 600 and 60 nM, respectively. The double-stranded DNA containing an AP site was the same as that used in the AP endonuclease assays. The double-stranded DNA without an AP site (Native-dsDNA) was prepared by annealing d-GCGATGACTAACGATACTAGGCTTCCGAGCC and a complementary strand. The peptides used in the fluorescence analyses were the same as those used in the SPR analyses.
Measurements of melting temperature (Tm) of oligonucleotide
Using a V-560 (JASCO) spectrophotometer, the Tm was measured at 260 nm from 20 to 70°C at a heating rate of 1°C/min. The oligonucleotides (2 nmol) were heated at 90°C for 1 min in 2 mM NaCl and 20 mM sodium cacodylate (pH 7.0), before addition of the peptide. After the solution had cooled, the solution with the peptides (1.5 nmol) was added to the DNA solution, and then the volume of the sample was adjusted to 2 ml by the addition of ultrapure water. The peptides used in this analysis were the same as those used in the spectrofluorimetric measurements. The double-stranded oligonucleotide containing an AP site (AP-17dsDNA) was prepared by annealing d-GCCAGACGHTACGACCG and d-CGGTCGTATCGTCTGGC. The native-dsDNA (Native-17dsDNA) was prepared by annealing d-GCCAGACGATACGACCG and d-CGGTCGTATCGTCTGGC. The DNA sequences of AP-17dsDNA and Native-17dsDNA were the same excluding the presence of an AP site, namely the 1′, 2′-dideoxyribofranose residue used in this study.
RESULTS
Comparison of the amino acid sequence in the vicinity of the active sites of AP endonucleases
To select the key residue for the recognition of AP sites by the ExoIII family AP endonucleases, a multiple sequence alignment of the AP endonucleases was performed (Figure 1). The molecular sizes and sequences of the AP endonuclease domains are highly conserved from bacteria to mammals.
The crystal structures of E.coli ExoIII and human APE1 have already been reported (25,26). Figure 1 indicates the amino acid sequences in the vicinity of the catalytic centers. Three residues, Glu-34, Asp-229 and His-259 of E.coli ExoIII and Glu-96, Asp-283 and His-309 of human APE1, are known to be essential for AP endonuclease activity as catalytic residues, but not for the recognition of DNA containing an AP site (AP-DNA). The crystallographic studies showed that the three residues are located in close proximity to each other at the bottom of a putative DNA-binding cleft. The sequence alignment of twelve homologs shows that some aromatic amino acid residues are conserved (Figure 1, the reverse displayed amino acids). These residues reside in the vicinity of the catalytic centers of these AP endonucleases. We have proposed that an aromatic amino acid residue near the catalytic center of the AP endonuclease plays a key role in the recognition of AP sites (30). Hence, three conserved aromatic amino acid residues in the vicinity of the catalytic center of ExoIII, Tyr-109, Trp-212 and Phe-213, were selected for site-directed mutagenesis. Mutational analyses of the aromatic amino acid residues of APE1 were performed (32). The mutation of Phe-266, which corresponds to Trp-212 of ExoIII, had only minor effects on the cleavage and binding abilities of APE1. The mutation of Trp-267, which corresponds to Phe-213 of ExoIII, also only slightly affected function. From these results, it was considered that the specific intercalation of the aromatic amino acid into the AP site was unlikely. However, based on the crystal structure of human APE1, we speculate that Phe-266 and Trp-267 do not serve as the intercalator, but Trp-280 intercalates into the AP site, as well as Trp-212 of ExoIII. Trp-212 of ExoIII and Trp-280 of APE1 protrude from the surface in the vicinity of the catalytic center (25,26). Therefore, Trp-280 of APE1 was selected as the target of mutagenesis.
Preparation of mutant ExoIII and APE1 proteins
The ExoIII mutant proteins, Y109S, W212S and F213W, were purified from the supernatants of cell extracts. Since the mutant protein, F213S, was expressed as an inclusion body, soluble F213S was obtained from the purified inclusion body by a renaturation procedure. It was confirmed that the method used in this study was suitable for ExoIII by renaturation of the inclusion body of the wild-type ExoIII. The wild-type ExoIII proteins purified from the inclusion body and from the supernatant had almost the same AP endonuclease activity (data not shown). The wild-type and mutant proteins of APE1 were purified as soluble proteins. All these proteins were detected as a clear single band by SDS–PAGE followed by Coomassie brilliant blue staining (data not shown).
Cleavage activities and complex formation abilities of the ExoIII mutant proteins
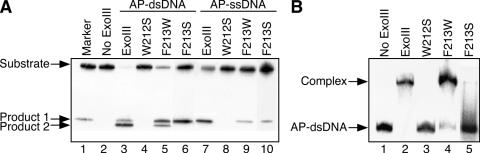
All the mutant proteins were assayed for the ability to cleave DNA containing an AP site, which was a 1′, 2′-dideoxyribose residue (tetrahydrofuran residue). As shown in Figure 2A, the double-stranded AP-DNA substrate was almost completely cleaved by the wild-type ExoIII (lane 3). Under the same conditions, no cleaved product was detected from the W212S mutant protein (lane 4). The mutant proteins, F213W (lane 5), F213S (lane 6) and Y109S (data not shown in Figure 2A), cleaved the double-stranded AP-DNA by 90, 72 and 14%, respectively. Products smaller than product 1 were generated by exonucleolytic cleavage of the AP endonucleolytic product from the 3′ end to the 5′ side (product 2). In our previous study, it has been elucidated that ExoIII cleaves single-stranded DNA containing an AP site (30). The single-stranded AP-DNA was cleaved by the wild-type ExoIII (lane 7) and the mutant proteins F213W (lane 9), F213S (lane 10) and Y109S (data not shown in Figure 2A) by 65, 19, 17 and 4%, respectively. However, the W212S mutant did not cleave the single-stranded AP-DNA at all (lane 8).
Figure 2.
Influence of substitutions at the conserved aromatic amino acid residues around the active site of ExoIII. (A) Detection of products of cleavage by ExoIII and its mutant proteins. Proteins used in each reaction are shown above each lane. The oligonucleotide containing an AP site (AP-ssDNA) was d-GCGATGACTAACGHTACTAGGCTTCCGAGCC. AP-dsDNA was constructed by annealing AP-ssDNA and d-CGGAAGCCTAGTATCGTTAGTCATCGCCATG. The substrate DNA (4.4 pmol), in which the oligonucleotide containing an AP site was 5′ 32P-labeled, was incubated with the wild-type ExoIII or its mutant (0.04 pmol) in buffer containing 75 mM NaCl, 5 mM MgCl2, 10 mM DTT and 65 mM Tris–HCl (pH 8.0) at 23°C. The DNA products were analyzed using a 20% denaturing (7 M urea) polyacrylamide gel. The product 1 was produced by AP endonucleolytic cleavage at the 5′ side of the AP site. The product 2 was produced by exonucleolytic cleavage from 3′ end of the product 1. The markers were 5′ 32P-labeled d-GCGATGACTAACG. (B) Abilities of ExoIII and its mutant proteins to bind the dsDNA containing an AP site. The proteins used in each reaction are shown above each lane. AP-dsDNA (0.45 pmol), in which the oligonucleotide containing an AP site was 5′ 32P-labeled, was incubated with the wild-type ExoIII or its mutant (4.5 pmol) in buffer containing 77 mM NaCl, 10 mM EDTA and 66 mM Tris–HCl (pH 8.3) at 4°C. The protein–DNA complex was analyzed by 15% native polyacrylamide gel electrophoresis.
To find the cause of the decrease in the cleavage activity of the mutant ExoIII proteins, substrate-binding ability was examined using a gel retardation assay with a 32P-labeled AP-DNA. As shown in Figure 2B, unambiguous retarded bands, which correspond to the AP-DNA•ExoIII complex, were observed in the case of the wild-type ExoIII and the F213W mutant. The binding ability of the F213W mutant (lane 4) relative to that of the wild-type ExoIII (lane 2) was 90%. In the case of the F213S mutant protein, a subtle retarded band of the AP-DNA•F213S complex (lane 5) was detected (6%) and a smeared band was observed. This phenomenon indicates that the F213S mutant has a weak, but obvious ability to bind AP-DNA. A slight decrease in the intensity of the complex's band was observed for the Y109S protein (data not shown in Figure 2B, 40%). In the case of W212S, the complex was not detected at all (lane 3). The tendency for ExoIII and its mutant proteins to form a complex was nearly identical to the tendency of their AP endonuclease activities. Based on the results described above, it has been elucidated that the Trp-212 residue of E.coli ExoIII is critical for the AP endonuclease activity and also the ability to bind AP-DNA. It has already been clarified that Glu-34, Asp-229 and His-259 are the catalytic residues (25). Though Trp-212 is not among the catalytic residues, it is obvious that it too plays an important role.
Cleavage activities and complex formation abilities of the APE1 mutant proteins
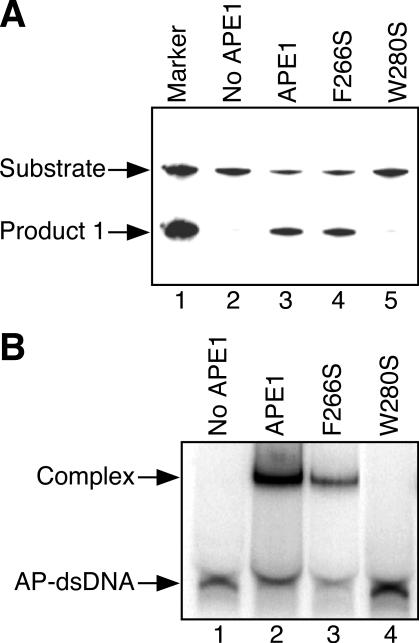
To ascertain the importance of the tryptophan residue for the functions of the ExoIII family AP endonucleases, mutational experiments of APE1 were conducted similar to the ExoIII mutants. There are four aromatic amino acid residues, Tyr-171, Phe-266, Trp-267 and Trp-280, in the vicinity of the catalytic center of human APE1 (Figure 1). Because Tyr-171 of APE1 corresponds to Tyr-109 of ExoIII which is not important for AP endonuclease activity (Figure 2), a Tyr-171 mutant of APE1 was not prepared. In addition, it has already been demonstrated that Trp-267 is not related to the AP endonuclease activity with the use of the W267A mutant (32). Thus, we prepared F266S and W280S mutants in this study. The position of Phe-266 of APE1 corresponds to that of Trp-212 of ExoIII, which is a critical amino acid residue for the activity of ExoIII (Figure 2). ExoIII does not have an aromatic amino acid residue at the position corresponding to Trp-280 of APE1. However, it has been elucidated from an X-ray crystallographic analysis that Trp-280 of APE1 extrudes from the surface of the active site as does Trp-212 of ExoIII. As shown in Figure 3A, the double-stranded AP-DNA substrate was almost completely cleaved by the wild-type APE1 (lane 3). Under the same conditions, the F266S mutant cleaved the AP-DNA as much as the wild-type APE1 (lane 4, 82% of the wild-type cleavage). On the other hand, the W280S mutant protein (lane 5) did not cleave the double-stranded AP-DNA at all. A similar tendency was observed for the binding of the mutant proteins to the double-stranded AP-DNA (Figure 4B). The F266S mutant bound to AP-DNA like the wild-type APE1 (93% of the wild-type binding). However, W280S did not form a complex (lane 4). These results have suggested that the tryptophan residue protruding from the surface of the catalytic center of the AP endonuclease, such as Trp-212 of ExoIII and Trp-280 of APE1, plays an important role in the function of the AP endonuclease.
Figure 3.
Influence of substitutions of the aromatic amino acid residues around the active site of human APE1. (A) Detection of products of cleavage by wild-type APE1 and its mutant proteins. Substrate DNA (AP-dsDNA: 4.4 pmol), in which the oligonucleotide containing an AP site was 5′ 32P-labeled, was incubated with the wild-type APE1 or its mutant (0.04 pmol). The DNA products were analyzed by 20% denaturing polyacrylamide gel electrophoresis. (B) Binding of APE1 and its mutant proteins to the dsDNA containing an AP site (AP-dsDNA). AP-dsDNA (0.45 pmol) was incubated with the wild-type APE1 or its mutants (4.5 pmol). Protein–DNA complex was analyzed by 15% native polyacrylamide gel electrophoresis.
Figure 4.
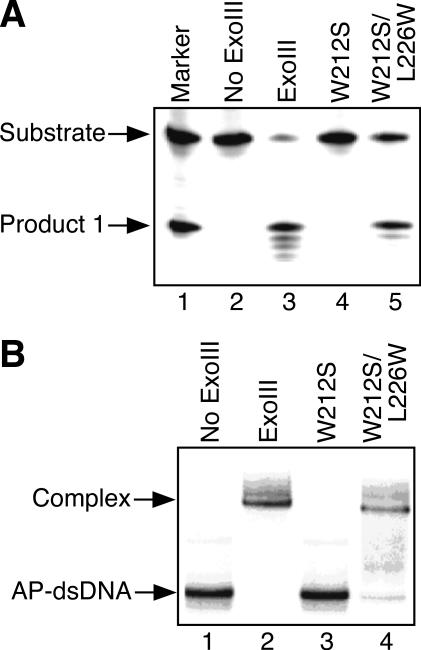
Effects of translocating the tryptophan residue protruding from the surface of the ExoIII active site. (A) Detection of AP endonucleolytic activity of ExoIII mutants. DNA substrate (AP-dsDNA: 4.4 pmol), in which the oligonucleotide containing an AP site was 5′ 32P-labeled, was incubated with the wild-type ExoIII or its mutant (0.04 pmol). The DNA products were analyzed using a 20% denaturing polyacrylamide gel. (B) Binding of ExoIII mutants to the dsDNA containing an AP site (AP-dsDNA). AP-dsDNA (0.45 pmol) was incubated with the wild-type ExoIII or its mutants (4.5 pmol). The protein–DNA complex was analyzed using a 15% polyacrylamide gel.
Effect of translocating tryptophan in the vicinity of the active site on the functions of the AP endonuclease
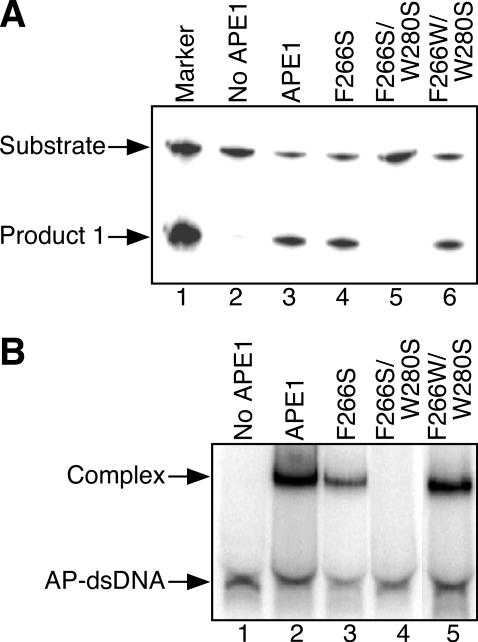
The tryptophan residues in the vicinity of the catalytic sites of the ExoIII family AP endonucleases are conserved at the position corresponding to Trp-212 of ExoIII and/or Trp-280 of APE1 (Figure 1). The W212S mutant of ExoIII had neither AP endonuclease activity nor any ability to bind AP-DNA as described above (lanes 4 and 3 in Figure 4A and B, respectively). Then we prepared the W212S/L226W mutant of ExoIII, in which the tryptophan residue was translocated from position 212 to 226. Namely, the Leu-226 of ExoIII was changed to Trp-226 because Leu-226 of ExoIII corresponds to the position of the critical Trp-280 of APE1 (Figure 1). Surprisingly, the W212S/L226W mutant had almost the same level of AP endonuclease activity (Figure 4A, lane 5, 66%) and the same binding ability (Figure 4B, lane 4, 82%) as the wild-type ExoIII. In a similar manner, we tried to recover the AP endonuclease activity of the inactivated W280S APE1 mutant, which lost the critical Trp-280 residue. The F266S mutant of APE1 has AP endonuclease activity and can bind AP-DNA as described previously [lane 4 (82%) and lane 3 (93%) in Figures 5A and 4B, respectively]. These results indicate that Phe-266 does not play an important role in the enzyme activity of APE1. The F266S/W280S mutant which lacked the critical residue Trp-280 also lost all AP endonuclease activity and ability to bind AP-DNA (lanes 5 and 4 in Figure 5A and B, respectively). We then prepared a F266W/W280S mutant, in which Phe-266 of the W280S protein was replaced with Trp-266. In this mutant, it can be considered that Trp-280 and Ser-266 of the F266S protein are exchanged. The F266W/W280S mutant had almost the same level of AP endonuclease activity and binding ability as the wild-type APE1 or F266S mutant [lane 6 (82%) and lane 5 (82%) in Figure 5A and B, respectively]. Consequently, if there is a tryptophan residue in the position that corresponds to Trp-212 of ExoIII or Trp-280 of APE1, the ExoIII family AP endonuclease functions as an AP endonuclease.
Figure 5.
Effects of translocating the tryptophan residue protruding from the surface of the APE1 active site. (A) Detection of AP endonucleolytic activity of APE1 mutants. DNA substrate (AP-dsDNA: 4.4 pmol), in which the oligonucleotide containing an AP site was 5′ 32P-labeled, was incubated with the wild-type APE1 or its mutant (0.04 pmol). The DNA products were analyzed using a 20% denaturing polyacrylamide gel. (B) Ability of APE1 mutants to form a complex with the dsDNA containing an AP site. AP-dsDNA (0.45 pmol) was incubated with the wild-type APE1 or its mutant (4.5 pmol). The protein–DNA complexes were analyzed using a 15% native polyacrylamide gel.
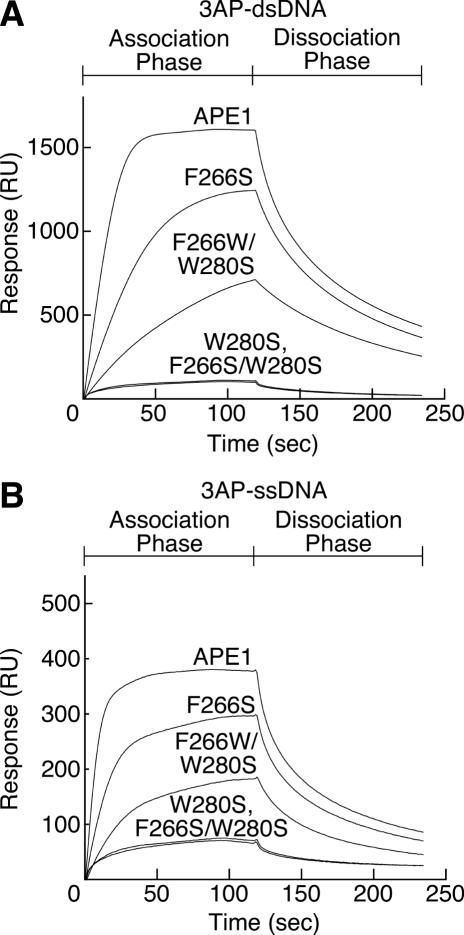
In order to detect the interactions between APE1 mutants and AP-DNA on a condition, that was nearer in vivo than the condition of the gel retardation assays, direct binding experiments using SPR-based Biacore technology were performed. The response was directly proportional to the bound mass. APE1 and its mutants as analytes were dialyzed against 150 mM NaCl, 10 mM EDTA and 10 mM HEPES–NaOH (pH 7.4) immediately before use. Consistent with the results obtained from the gel retardation assay, the direct binding analyses using the SPR-based Biacore technology clearly demonstrated that the tryptophan residue in the vicinity of the catalytic center played an important role in the formation of a complex with the double-stranded AP-DNA. As shown in Figure 6, the W280S and F266S/W280S mutants lacking the tryptophan residue near the catalytic center showed only week interaction with the oligonucleotide containing three AP sites. These weak responses were also observed in the SPR analyses of the wild-type APE1 and its mutants with the DNA not containing an AP site (data not shown). It seems that the subtle binding is due to non-specific interactions, so-called electrostatic interactions. There was no response at all when BSA was used as an analyte (data not shown). Therefore, it has been speculated that these weak responses due to non-specific interaction correspond to the palpation of the DNA to search for an AP site by the AP endonuclease. The wild-type APE1 with Trp-280, the F266S mutant with Trp-280 and the F266W/W280S mutant with Trp-266 were bound to the double-stranded AP-DNA (Figure 6A). Using Biacore technology, significant binding responses of the wild-type APE1, F266S and F266W/W280S to the single-stranded AP-DNA (3AP-ssDNA) were observed (Figure 6B), although a complex of APE1 and the single-stranded AP-DNA was not detected by the gel retardation assay (data not shown). The response of the wild-type APE1, F266S and F266W/W280S to the double-stranded AP-DNA decreased in strength in that order. It has been elucidated that the function of the AP endonuclease is maintained even if the critical tryptophan residue migrates to a different position within the catalytic center. We could not obtain kinetic parameters for the native APE1 and its mutants because the observed sensorgrams did not fit any model. These unfitted sensorgrams could be due to the AP-DNA containing three AP sites.
Figure 6.
Sensorgrams for the binding of APE1 and its mutants to single- or double-stranded DNA containing AP sites. The Overlay of sensorgrams represents interactions of the wild-type APE1 and mutant proteins with immobilized dsDNA (1620 RU) containing AP sites (A), and immobilized ssDNA (989 RU) containing AP sites (B). There were three AP sites in the immobilized single-stranded oligonucleotide (3AP-ssDNA). The immobilized double-stranded oligonucleotide containing three AP sites (3AP-dsDNA) was prepared by annealing 3AP-ssDNA and its complementary strand. The flow rate of the enzyme solution was 20 µl/min. Association and dissociation phases were both taken as 120 s each. For all the sensorgrams, the injection point was set as the zero time and the baseline prior to the injection was set to zero RU.
Interaction between peptides containing a tryptophan residue and AP-DNA
It has been clarified that the conserved tryptophan residues near the catalytic sites of ExoIII and APE1 play an important role in the formation of the complex and AP endonuclease activity. This finding implies that the tryptophan residue interacts with the AP site. Thus, we have assumed that the tryptophan residue in the vicinity of the catalytic center of the AP endonuclease intercalates into the space of the AP site. To investigate the possibility that the tryptophan residue is inserted into the AP site, the interaction between the peptide containing a tryptophan residue and the double-stranded DNA containing AP sites was examined (Figure 7 and Table 1).
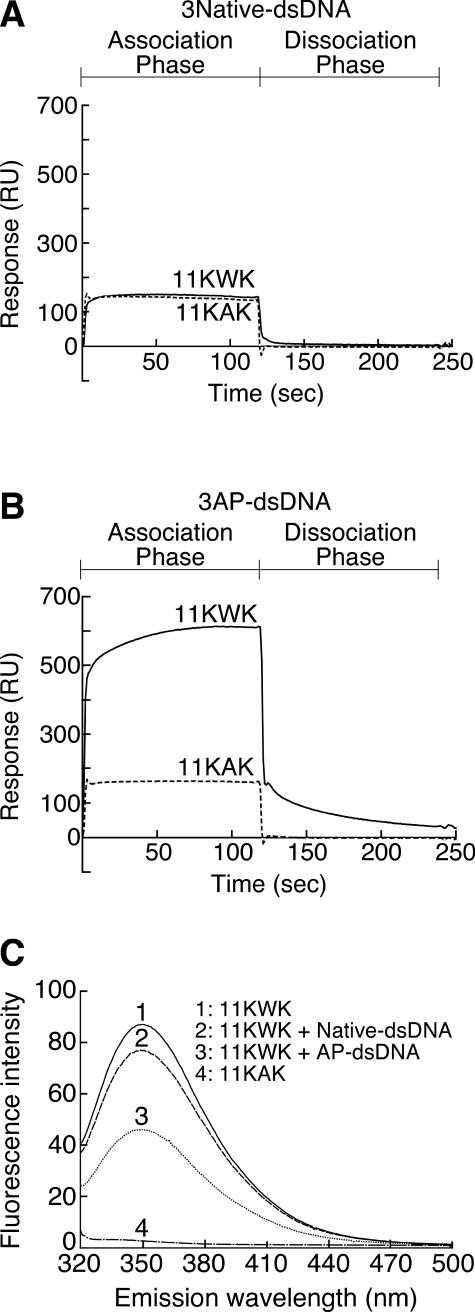
Figure 7.
Effect of tryptophan residues on interaction between peptides and double-stranded oligonucleotides containing AP sites or not. (A) SPR analyses of the interaction between a peptide containing a tryptophan residue (11KWK, solid line) or not (11KAK, dashed line) and dsDNA not containing an AP site. The amount of immobilized oligonucleotides not containing AP sites (3Native-dsDNA) was 1515 RU. For all the sensorgrams, the injection point was set as the zero time and the baseline prior to the injection was set to zero RU. The 11KWK and 11KAK oligopeptides were KSRGKWKGRSA and KSRGKAKGRSK, respectively. (B) SPR analyses of the interaction between a peptide containing a tryptophan residue (11KWK, solid line) or not (11KAK, dashed line) and dsDNA containing three AP sites. The amount of immobilized oligonucleotide containing three AP sites (3AP-dsDNA) was 1500 RU. This analysis was carried out under the same experimental conditions as in (A), except for the immobilized oligonucleotide. (C) Fluorescence spectra of the peptide containing a tryptophan residue in the presence or absence of dsDNA containing an AP site. AP-dsDNA was the substrate used in the AP endonuclease assay (Figure 2). The DNA sequences of AP-dsDNA and Native-dsDNA were the same excluding the presence of an AP site. Peptide and dsDNA were mixed at a molar ratio of 1:10. The excitation wavelength was 275 nm.
Table 1.
Influence of peptide containing a tryptophan residue on Tm of dsDNA containing an AP site
| DNAa | Peptideb | Tm (°C)c | ΔTm (°C)d |
|---|---|---|---|
| AP-17dsDNA | — | 33.8 | |
| 11KWK | 38.6 | 4.8 | |
| 11KAK | 35.7 | 1.9 | |
| Native-17dsDNA | — | 45.3 | |
| 11KWK | 46.1 | 0.8 | |
| 11KAK | 46.8 | 1.5 |
aDouble-stranded oligodeoxyribonucleotides (AP-17dsDNA and Native-17dsDNA) containing an AP site or not were d-GCCAGACGHTACGACCG/d-CGGTCGTATCGTCTGGC and d-GCCAGACGATACGACCG/d-CGGTCGTATCGTCTGGC, respectively.
bPeptides (11KWK and 11KAK) were KSRGKWKGRSA and KSRGKAKGRSK, respectively. M-dash indicate the absence of the peptide in the sample.
cThe double-stranded oligonucleotides (17mer) were mixed with/without the peptide (11KWK or 11KAK). The mixture of the oligonucleotide (1 µM) and the peptide (0.75 µM) was dissolved in 1 mM NaCl, 10 mM sodium cacodylate (pH 7.0). Absorbance at 260 nm was measured from 20 to 70°C at the heating rate of 1°C/min.
dΔTm indicates the difference in which Tm (without peptide) is subtracted from Tm (with peptide).
To examine the interaction between the peptide and AP-DNA, experiments using SPR-based Biacore technology were performed (Figure 7A and B). Because of the insolubility of the oligopeptide that referred to the amino acid sequence surrounding Trp-212 of ExoIII, it was not used. Two peptides which are rich in basic amino acids in order to enhance the electrostatic interaction between the peptide and AP-DNA were prepared. The peptide sequences used as the analytes were KSRGKWKGRSK (11KWK) and KSRGKAKGRSK (11KAK). The amino acid sequence of 11KWK is the same as that of 11KAK, except that a tryptophan residue is substituted for the alanine residue at the center of the peptide. Both peptides bound to the native double-stranded DNA (3Native-dsDNA) to the same degree (Figure 7A). Thus, this binding seems to be attributable to a non-specific interaction such as an electrostatic interaction. To the double-stranded DNA containing three AP sites (3AP-dsDNA), 11KWK showed an obviously large response (Figure 7B). On the other hand, the strength of the interaction of 11KAK with 3AP-dsDNA was almost the same as that of a non-specific interaction with the native DNA (Figure 7B). It has been understood that a process obviously different from an electrostatic interaction occurs during the interaction between 11KWK and 3AP-dsDNA.
To clarify how 11KWK interacts with AP-DNA, the interaction between 11KWK and AP-DNA was examined using a fluorescent spectrophotometer (Figure 7C). The environment around the tryptophan residue influences the emission of the residue. The emission spectrum of the tryptophan residue in a mixture of 11KWK and 3Native-dsDNA was slightly decreased when compared with 11KWK alone (∼88%). In the case of a mixture of 11KWK and AP-dsDNA, an unambiguous quenching of ∼53% was observed in the emission spectrum of the tryptophan residue of 11KWK (Figure 7C). This phenomenon indicates that the tryptophan residue of 11KWK is embedded in a hydrophobic pocket, that is, the AP site.
Thermal stabilization of AP-DNA by a peptide containing a tryptophan residue
The stabilization of the nucleic acid structures by proteins or peptides can be readily monitored as an elevation in the nucleic acid Tm. As shown in Table 1, 11KWK significantly elevated the Tm of AP-17dsDNA containing an AP site from 33.8 to 38.6°C (ΔTm; 4.8°C). On the other hand, 11KAK only partially elevated the Tm of AP-17dsDNA (ΔTm; 1.9°C). Similarly, the Tm's of the native 17mer duplex (Native-17dsDNA) was slightly increased by the addition of 11KWKor 11KAK. The ΔTm's of the native-17dsDNA in both cases was 0.8 and 1.5°C, respectively. Based on these Tm's, it has been elucidated that a peptide containing a tryptophan residue significantly stabilizes the double-stranded DNA containing an AP site. Thus, these results have suggested that the tryptophan residue intercalates into the pocket of the AP site instead of the deleted nucleic acid base.
DISCUSSION
AP sites are generated spontaneously under physiological conditions and produced enzymatically during the process of BER. How to repair these AP sites is an essential problem for all organisms. AP endonucleases seem to be evolutionarily stable, and the homology of these enzymes from bacteria to mammals is surprisingly high. Many studies have been performed to characterize AP endonucleases. However, the mechanism by which the ExoIII family AP endonucleases recognize AP sites has not yet been clarified. Many complex structures formed between DNA-repair enzymes and substrate DNAs have already been reported. Such structures are stable Michaelis complexes. Because the initial step in the recognition of a DNA-damaged site by a DNA-repair enzyme occurs before the formation of the ultimate Michaelis complex, the early-stage recognition mechanisms cannot be determined from these complex structures. With respect to how the AP sites are identified by the ExoIII family AP endonuclease, four hypotheses have been proposed (25–30). In this study, we focused on a number of aromatic amino acid residues in the vicinity of the catalytic site that may play a definitive role in the recognition of AP sites.
Previously, we speculated that an indole ring of the tryptophan residue or a side chain of another aromatic amino acid residue in the vicinity of the catalytic site intercalates into the DNA-pocket formed at an abasic site (30). In the present study, site-directed mutagenesis of the aromatic amino acid residues of E.coli ExoIII (Figures 2 and 4) and human APE1 (Figures 3, 5 and 6) demonstrated that Trp-212 of ExoIII and Trp-280 of APE1 are critical for the AP endonuclease activity and binding to AP-DNA. Each of these site-directed mutants, W212S of ExoIII and W280S of APE1, lost all AP endonuclease activity and the ability to bind AP-DNA, although these essential tryptophan residues are not catalytic amino acid residues. It was shown that these tryptophan residues protrude from the hollow catalytic sites by the crystal structures of the AP endonucleases (25,26). It was reported that the mutation of Trp-267 of APE1, which is at almost the same position as Trp-212 of ExoIII, did not influence its activity (32). This Trp-267 is buried at the surface in the vicinity of the catalytic site. Therefore, we speculate that the tryptophan residue protruding from the surface near the catalytic center of the AP endonuclease is a key residue at the initial stage of in the recognition of a substrate. The ExoIII and APE1 AP endonucleases belong to the DNase I superfamily and retain the tryptophan residue near the catalytic site. Members of this superfamily such as DNase I (35), the cytolethal distending toxin subunit B (36), the endonuclease domain of LINE-1 reverse transcriptase homolog (37) and the endonuclease domain of TRAS1 retrotransposon (38), lack AP endonuclease activity, and do not have a tryptophan residue in the vicinity of the catalytic sites. These findings support that the tryptophan residue near the catalytic site is indispensable for AP endonuclease activity.
To investigate the function of the tryptophan residue, experiments involving interaction between an oligopeptide containing a tryptophan residue and an oligonucleotide containing an AP site were performed (Figure 7). Basic peptides containing a tryptophan residue, KSRGKWKGRSK (11KWK) and lacking a tryptophan residue, KSRGKAKGRSK (11KAK), were used. Using SPR-based Biacore technology, the interaction between the peptide and AP-DNA was investigated. Both the peptides lacking and containing a tryptophan residue weakly interacted with the native DNA duplex to the same degree (Figure 7A). The interaction seems to be an electrostatic interaction between the basic peptide and the oligonucleotide. As for the oligopeptide 11KWK containing a tryptophan residue, a characteristic interaction obviously different from an electrostatic interaction was observed with the oligonucleotide duplex containing an AP site (Figure 7B). Furthermore, specific interaction between 11KWK and the oligonucleotide duplex (AP-DNA) was observed in the experiment with the mixture of 11KWK and AP-DNA (Figure 7C). These observations provided further support for our hypothesis that the indole ring of the tryptophan residue in the vicinity of the catalytic site of the AP endonuclease intercalates into the DNA-pocket formed at an abasic site.
In the sequence of DNA-repair processes, DNA-repair enzymes recognize, hold and excise the damaged parts in the huge DNA duplex. Recent studies have discussed a long-standing question with regard to DNA recognition, namely, how do site-specific DNA-binding proteins find their target sites in DNA? Many studies have been carried out to answer this question, and three proposals (sliding, hopping and intersegmental transfer) have been made (39). In all these processes, the protein initially binds at a random non-specific site in the DNA, then translocates from the initial binding site to the final specific target site. It seems that the weak responses between the APE1 mutants (W280S, F266S/W280S) and the AP-DNA, as shown in Figure 6, are attributed to a weak non-specific binding between the mutants and DNA during the translocational search. This non-specific binding was also observed in the SPR analyses of the wild-type APE1 and its mutants with the native DNA (data not shown).
The many complex structures between DNA-repair enzymes and damaged DNA have been solved. These structures seem to be stable and to be the ultimate Michaelis complexes immediately before or after the reaction. Solving the complex structures has provided the structural basis for understanding the catalytic mechanism and the manner in which the substrate is hold. However, the initial recognition of the target site in DNA, which occurs before the formation of the Michaelis complex, is a dynamic process with an induced-fit (conformational change) of the enzyme and the target DNA. Therefore, the initial damage-encountering complex is not equivalent to the stable complex structures analyzed by NMR or X-ray. In the X-ray structure of APE1•AP-DNA complex, the essential tryptophan residue does not intercalate into the AP site pocket (27). However, it is almost certainly correct that the intercalation of the tryptophan residue occurs in the first step of the complex's formation prior to the development of the Michaelis complex.
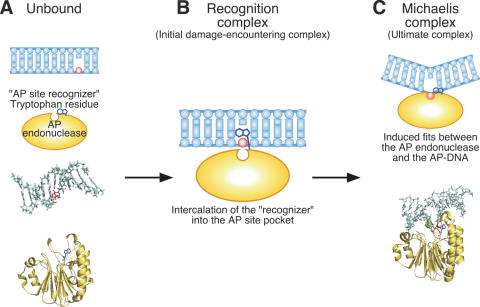
We speculate about the process by which the AP site is recognized by a tryptophan residue as follows. First of all, the AP endonuclease searches for an AP site while translocating on the DNA strand. It recognizes such a site through the insertion of a tryptophan residue into the AP site pocket and then becomes fixed there (Figure 8B). We term such a tryptophan residue a ‘recognizer’. In addition, the induced-fit from this state to the Michaelis complex seems to happen (Figure 8C). Finally, the AP endonuclease cleaves the AP site, and then migrates from the cleaved site.
Figure 8.
The interaction of double-stranded DNA and ExoIII family AP endonuclease. A model for recognition of an AP site by the ExoIII family AP endonuclease. (A) Unbound forms of dsDNA containing an AP site (PDB ID: 1A9I) and human APE1 (PDB ID: 1BIX). The negative charge of the substrate DNA attracts the positively charged region of the enzyme. (B) An AP site recognition complex. When AP endonuclease encounters an AP site, the tryptophan residue in the vicinity of the catalytic site intercalates into an AP site pocket as an AP site ‘recognizer’. This recognition complex would immediately change into a reaction complex. (C) A cleavage reaction complex (PDB ID: 1DEW). To cleave on the 5′ side of the AP site, an abasic ribose ring is flipped out from the duplex interior and accommodated into a catalytic pocket with DNA kinking at the AP site.
There are many ExoIII homologue proteins, which have two tryptophan residues at the positions corresponding to Trp-212 of ExoIII and Trp-280 of APE1. Such ExoIII family AP endonucleases widely exist in bacteria. ExoA of Bacillus subtilis is an AP endonuclease of this type (namely ExoA type AP endonuclease). We have already clarified the properties of this ExoA (40). It has been elucidated that ExoA is a multifunctional DNA-repair enzyme in B.subtilis that is very similar to E.coli ExoIII except with less 3′–5′ exonuclease activity. Further evidence in support of the role of the tryptophan residue in the vicinity of the catalytic site of the ExoA type AP endonucleases from Thermoplasma volcanium and Lactobacillus plantarum will be reported elsewhere.
Acknowledgments
We thank Prof. S. Ikeda of the Okayama University of Science and Prof. S. Seki of Chugokugakuen University for providing the pUAEH1 plasmid. For the spectrofluorimetric analyses, we are indebted to the Food Technology Department of Nagano Prefecture General Industrial Technology Center for providing the facilities. For the radioisotope experiments, we are indebted to the Gene Research Center, Shinshu University, for providing the facilities. This work was supported by the Ministry of Education, Culture, Sports, Science and Technology and by the Shinshu University 21st Century COE Program. Funding to pay the Open Access publication charges for this article was provided by the Ministry of Education, Culture, Sports, Science and Technology of Japan.
Conflict of interest statement. None declared.
REFERENCES
- 1.Lindahl T. Instability and decay of the primary structure of DNA. Nature. 1993;362:709–715. doi: 10.1038/362709a0. [DOI] [PubMed] [Google Scholar]
- 2.Hoeijmakers J.H. Genome maintenance mechanisms for preventing cancer. Nature. 2001;411:366–374. doi: 10.1038/35077232. [DOI] [PubMed] [Google Scholar]
- 3.Dianov G.L., Sleeth K.M., Dianova I.I., Allinson S.L. Repair of abasic sites in DNA. Mutat. Res. 2003;531:157–163. doi: 10.1016/j.mrfmmm.2003.09.003. [DOI] [PubMed] [Google Scholar]
- 4.Fortini P., Pascucci B., Parlanti E., D'Errico M., Simonelli V., Dogliotti E. The base excision repair: mechanisms and its relevance for cancer susceptibility. Biochimie. 2003;85:1053–1071. doi: 10.1016/j.biochi.2003.11.003. [DOI] [PubMed] [Google Scholar]
- 5.Schärer O.D., Jiricny J. Recent progress in the biology, chemistry and structural biology of DNA glycosylases. Bioessays. 2001;23:270–281. doi: 10.1002/1521-1878(200103)23:3<270::AID-BIES1037>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- 6.Atamna H., Cheung I., Ames B.N. A method for detecting abasic sites in living cells: age-dependent changes in base excision repair. Proc. Natl Acad. Sci. USA. 2000;97:686–691. doi: 10.1073/pnas.97.2.686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shibutani S., Takeshita M., Grollman A.P. Translesional synthesis on DNA templates containing a single abasic site. A mechanistic study of the ‘A rule’. J.Biol.Chem. 1997;272:13916–13922. doi: 10.1074/jbc.272.21.13916. [DOI] [PubMed] [Google Scholar]
- 8.Saporito S.M., Smith-White B.J., Cunningham R.P. Nucleotide sequence of the xth gene of Escherichia coli K-12. J.Bacteriol. 1988;170:4542–4547. doi: 10.1128/jb.170.10.4542-4547.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Parkhill J., Dougan G., James K.D., Thomson N.R., Pickard D., Wain J., Churcher C., Mungall K.L., Bentley S.D., Holden M.T., et al. Complete genome sequence of a multiple drug resistant Salmonella enterica serovar Typhi CT18. Nature. 2001;413:848–852. doi: 10.1038/35101607. [DOI] [PubMed] [Google Scholar]
- 10.McClelland M., Sanderson K.E., Spieth J., Clifton S.W., Latreille P., Courtney L., Porwollik S., Ali J., Dante M., Du F., et al. Complete genome sequence of Salmonella enterica serovar Typhimurium LT2. Nature. 2001;413:852–856. doi: 10.1038/35101614. [DOI] [PubMed] [Google Scholar]
- 11.Fleischmann R.D., Adams M.D., White O., Clayton R.A., Kirkness E.F., Kerlavage A.R., Bult C.J., Tomb J.F., Dougherty B.A., Merrick J.M. Whole-genome random sequencing and assembly of Haemophilus influenzae Rd. Science. 1995;269:496–512. doi: 10.1126/science.7542800. [DOI] [PubMed] [Google Scholar]
- 12.Ogasawara N., Nakai S., Yoshikawa H. Systematic sequencing of the 180 kilobase region of the Bacillus subtilis chromosome containing the replication origin. DNA Res. 1994;1:1–14. doi: 10.1093/dnares/1.1.1. [DOI] [PubMed] [Google Scholar]
- 13.Puyet A., Greenberg B., Lacks S.A. The exoA gene of Streptococcus pneumoniae and its product, a DNA exonuclease with apurinic endonuclease activity. J. Bacteriol. 1989;171:2278–2286. doi: 10.1128/jb.171.5.2278-2286.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sander M., Lowenhaupt K., Lane W.S., Rich A. Cloning and characterization of Rrp1, the gene encoding Drosophila strand transferase: carboxy-terminal homology to DNA repair endo/exonucleases. Nucleic Acids Res. 1991;19:4523–4529. doi: 10.1093/nar/19.16.4523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Freeland T.M., Guyer R.B., Ling A.Z., Deering R.A. Apurinic/apyrimidinic (AP) endonuclease from Dictyostelium discoideum: cloning, nucleotide sequence and induction by sublethal levels of DNA damaging agents. Nucleic Acids Res. 1996;24:1950–1953. doi: 10.1093/nar/24.10.1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Babiychuk E., Kushnir S., Van Montagu M., Inz D. The Arabidopsis thaliana apurinic endonuclease Arp reduces human transcription factors Fos and Jun. Proc. Natl Acad. Sci. USA. 1994;91:3299–3303. doi: 10.1073/pnas.91.8.3299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wilson T.M., Carney J.P., Kelley M.R. Cloning of the multifunctional rat apurinic/apyrimidinic endonuclease (rAPEN)/redox factor from an immature T cell line. Nucleic Acids Res. 1994;22:530–531. doi: 10.1093/nar/22.3.530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Seki S., Akiyama K., Watanabe S., Hatsushika M., Ikeda S., Tsutsui K. cDNA and deduced amino acid sequence of a mouse DNA repair enzyme (APEX nuclease) with significant homology to Escherichia coli exonuclease III. J. Biol. Chem. 1991;266:20797–20802. [PubMed] [Google Scholar]
- 19.Robson C.N., Milne A.M., Pappin D.J., Hickson I.D. Isolation of cDNA clones encoding an enzyme from bovine cells that repairs oxidative DNA damage in vitro: homology with bacterial repair enzymes. Nucleic Acids Res. 1991;19:1087–1092. doi: 10.1093/nar/19.5.1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Demple B., Herman T., Chen D.S. Cloning and expression of APE, the cDNA encoding the major human apurinic endonuclease: definition of a family of DNA repair enzymes. Proc. Natl Acad. Sci. USA. 1991;88:11450–11454. doi: 10.1073/pnas.88.24.11450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Seki S., Hatsushika M., Watanabe S., Akiyama K., Nagao K., Tsutsui K. cDNA cloning, sequencing, expression and possible domain structure of human APEX nuclease homologous to Escherichia coli exonuclease III. Biochim. Biophys. Acta. 1992;1131:287–299. doi: 10.1016/0167-4781(92)90027-w. [DOI] [PubMed] [Google Scholar]
- 22.Demple B., Halbrook J., Linn S. Escherichia coli xth mutants are hypersensitive to hydrogen peroxide. J. Bacteriol. 1983;153:1079–1082. doi: 10.1128/jb.153.2.1079-1082.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sammartano L.J., Tuveson R.W. Escherichia coli xthA mutants are sensitive to inactivation by broad-spectrum near-UV (300 to 400 nm) radiation. J. Bacteriol. 1983;156:904–906. doi: 10.1128/jb.156.2.904-906.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xanthoudakis S., Smeyne R.J., Wallace J.D., Curran T. The redox/DNA repair protein, Ref-1, is essential for early embryonic development in mice. Proc. Natl Acad. Sci. USA. 1996;93:8919–8923. doi: 10.1073/pnas.93.17.8919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mol C.D., Kuo C.F., Thayer M.M., Cunningham R.P., Tainer J.A. Structure and function of the multifunctional DNA-repair enzyme exonuclease III. Nature. 1995;374:381–386. doi: 10.1038/374381a0. [DOI] [PubMed] [Google Scholar]
- 26.Gorman M.A., Morera S., Rothwell D.G., de La Fortelle E., Mol C.D., Tainer J.A., Hickson I.D., Freemont P.S. The crystal structure of the human DNA repair endonuclease HAP1 suggests the recognition of extra-helical deoxyribose at DNA abasic sites. EMBO J. 1997;16:6548–6558. doi: 10.1093/emboj/16.21.6548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mol C.D., Izumi T., Mitra S., Tainer J.A. DNA-bound structures and mutants reveal abasic DNA binding by APE1 and DNA repair coordination. Nature. 2000;403:451–456. doi: 10.1038/35000249. [DOI] [PubMed] [Google Scholar]
- 28.Weiss B. Endonuclease II of Escherichia coli is exonuclease III. J. Biol. Chem. 1976;251:1896–1901. [PubMed] [Google Scholar]
- 29.Singer B., Hang B. What structural features determine repair enzyme specificity and mechanism in chemically modified DNA? Chem. Res. Toxicol. 1997;10:713–732. doi: 10.1021/tx970011e. [DOI] [PubMed] [Google Scholar]
- 30.Shida T., Noda M., Sekiguchi J. Cleavage of single- and double-stranded DNAs containing an abasic residue by Escherichia coli exonuclease III (AP endonuclease VI) Nucleic Acids Res. 1996;24:4572–4576. doi: 10.1093/nar/24.22.4572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Behmoaras T., Toulme J.J., Helene C. A tryptophan- containing peptide recognizes and cleaves DNA at apurinic sites. Nature. 1981;292:858–859. doi: 10.1038/292858a0. [DOI] [PubMed] [Google Scholar]
- 32.Erzberger J.P., Barsky D., Schärer O.D., Colvin M.E., Wilson D.M., III Elements in abasic site recognition by the major human and Escherichia coli apurinic/apyrimidinic endonucleases. Nucleic Acids Res. 1998;26:2771–2778. doi: 10.1093/nar/26.11.2771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Behmoaras T., Toulme J.J., Helene C. Specific recognition of apurinic sites in DNA by a tryptophan-containing peptide. Proc. Natl Acad. Sci. USA. 1981;78:926–930. doi: 10.1073/pnas.78.2.926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Miki T., Yasukochi T., Nagatani H., Furuno M., Orita T., Yamada H., Imoto T., Horiuchi T. Construction of a plasmid vector for the regulatable high level expression of eukaryotic genes in Escherichia coli: an application to overproduction of chicken lysozyme. Protein Eng. 1987;1:327–332. doi: 10.1093/protein/1.4.327. [DOI] [PubMed] [Google Scholar]
- 35.Weston S.A., Lahm A., Suck D. X-ray structure of the DNase I-d(GGTATACC)2 complex at 2.3 Å resolution. J. Mol. Biol. 1992;226:1237–1256. doi: 10.1016/0022-2836(92)91064-v. [DOI] [PubMed] [Google Scholar]
- 36.Nešić D., Hsu Y., Stebbins C.E. Assembly and function of a bacterial genotoxin. Nature. 2004;429:429–433. doi: 10.1038/nature02532. [DOI] [PubMed] [Google Scholar]
- 37.Weichenrieder O., Repanas K., Perrakis A. Crystal structure of the targeting endonuclease of the human LINE-1 retrotransposon. Structure. 2004;12:975–986. doi: 10.1016/j.str.2004.04.011. [DOI] [PubMed] [Google Scholar]
- 38.Maita N., Anzai T., Aoyagi H., Mizuno H., Fujiwara H. Crystal structure of the endonuclease domain encoded by the telomere-specific long interspersed nuclear element, TRAS1. J. Biol. Chem. 2004;279:41067–41076. doi: 10.1074/jbc.M406556200. [DOI] [PubMed] [Google Scholar]
- 39.Halford S.E., Marko J.F. How do site-specific DNA-binding proteins find their targets? Nucleic Acids Res. 2004;32:3040–3052. doi: 10.1093/nar/gkh624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shida T., Ogawa T., Ogasawara N., Sekiguchi J. Characterization of Bacillus subtilis ExoA protein: a multifunctional DNA-repair enzyme similar to the Escherichia coli exonuclease III. Biosci. Biotechnol. Biochem. 1999;63:1528–1534. doi: 10.1271/bbb.63.1528. [DOI] [PubMed] [Google Scholar]