Abstract
Cellular changes have been monitored during the suppression, mediated by the overproduction of tRNALys, of thermosensitivity in Escherichia coli strain AA7852 carrying a mutation in peptidyl-tRNA hydrolase (Pth) encoded by the pth(Ts) gene. The presence in AA7852 cells of a plasmid bearing lysV gene helped to maintain low levels of the unstable Pth(Ts) protein and to preserve the viability of the mutant line at 41°C whereas plasmids bearing other tRNA genes were ineffective. At 32°C the excess of tRNALys did not alter the percentages of the free-, charged- or peptidyl-tRNALys species compared with those found in strains that did not overproduce tRNALys. At 41°C, however, despite increases in the level of peptidyl-tRNALys, the excess tRNALys helped to maintain the concentration of charged-tRNALys at a level comparable with that found in non-overproducer cells grown at a permissive temperature. In addition, the excess tRNALys at 41°C provoked a reduction in the concentrations of various peptidyl-tRNAs, which normally accumulate in pth(Ts) cells, and a proportional increase in the concentrations of the corresponding aminoacyl-tRNAs. The possible mechanism of rescue due to the overexpression of tRNALys and the causes of tRNALys starvation in pth(Ts) strains grown at non-permissive temperatures are considered.
INTRODUCTION
Peptidyl-tRNA hydrolase (Pth) hydrolyses the peptidyl-tRNAs (p-tRNAs) that are prematurely dissociated from ribosomes, thus regenerating free aminoacylable tRNAs necessary for new rounds of protein synthesis. In Escherichia coli this enzyme, which is essential for the synthesis of proteins and for the viability of the bacterium, is encoded by the gene pth. The mutant gene pth(Ts) encodes a mature protein with a single amino acid substitution at residue 100 (Gly to Asp) giving rise to a temperature sensitive phenotype (1,2). Whilst Pth(Ts) protein is quite unstable in vivo even at permissive temperatures, an increase in temperature to 43°C causes the protein concentration to fall rapidly to undetectable levels within 10 min (3). Indeed, soon after a shift to temperatures higher than 39°C, p-tRNAs accumulate, protein synthesis is arrested, and the mutant pth(Ts) cells die (4,5).
A number of different mechanisms are known by which the thermosensitivity of a pth(Ts) strain may be suppressed, mediated either through a variation in the level of Pth(Ts) protein or an alteration in the metabolism of p-tRNA in the cell. The first group includes the over expression of pth(Ts) per se (3). Thus, chromosomal duplication of pth(Ts) has been shown to increase the level of Pth(Ts) protein and to maintain the viability of the mutant (6). Moreover, the over expression of the chaperonins GroEL and GroES, and a defect in the dnaJ gene, can stabilize the Pth(Ts) protein and therefore enhance its level (7,3). The second group of suppressors comprises factors involved in the dissociation of p-tRNA from ribosomes. Thus, mutations in the factors RRF and RF3 restore the growth of pth(Ts) cells through a reduction in drop-off (8), whilst the overproduction of tmRNA (10Sa RNA) suppresses thermosensitivity by reducing stalled p-tRNA–ribosome complexes (9).
Of particular note is the finding of Heurgué-Hamard et al. (7) that the overproduction of tRNALys, but not of certain other tRNAs, is also able to suppress the Pth(Ts) phenotype. It has been shown that pth(Ts) mutant cells, when incubated at 40°C, accumulate different families of isoacceptors as p-tRNAs at various rates, but that p-tRNALys accumulates most rapidly (4). Overproduction of tRNALys seems to compensate for the defective levels of Pth protein thus reducing the thermosensitivity of the mutant cells, although the mechanism of this suppression is presently unknown. In the present study, we have quantified the cellular levels of Pth(Ts) protein and of a number of tRNA species in transformed pth(Ts) cell lines in order to gain an insight into the suppression of thermosensitivity of pth(Ts) mutants through tRNALys overproduction.
MATERIALS AND METHODS
Strains and plasmids
E.coli strain AA7852, a pth(Ts) derivative of strain CP-78 [F−, arg−, leu−, thr−, his−, thiamine−, T1s] (1) was employed in the study. The plasmid constructs harbouring tRNA genes were (i) pVH119, bearing valUα, valUβ, valUγ and lysV genes; (ii) pVH125, a derivative of pVH119 carrying valUα, valUβ, valUγ and partly deleted lysV genes; (iii) pVH124, a derivative in which all of these tRNA genes had been deleted (7); (iv) ptRNAHisCCA, carrying argX, hisR, leuT and proM genes; and (v) pTH2, containing the thrW gene (7).
Growth assays
Isolated colonies of E.coli strain AA7852, that had been independently transformed with different plasmid constructs, were streaked onto plates of Luria–Bertani (LB) medium containing 100 µg/ml ampicillin (LB-Ap) and incubated overnight at 32, 41 or 43°C (Figure 1A). For the determination of bacterial growth in liquid culture (Figure 2A), an aliquot of a 32°C overnight culture in LB-Ap was diluted 1:100 with fresh medium and further incubated at 32°C to an OD600 of 0.2. Equal portions of the resulting culture were incubated at 32 or 41°C, and cellular growth followed by determination of OD600 with respect to time.
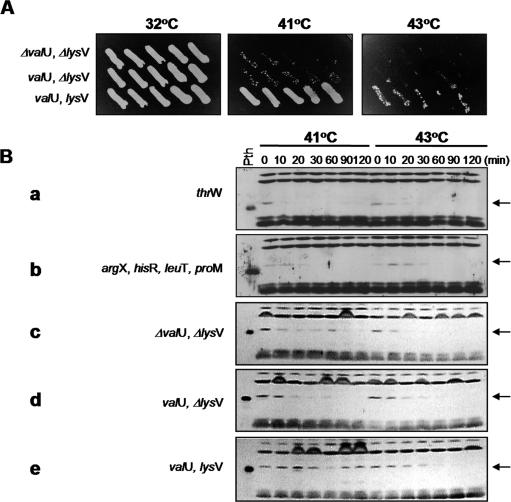
Figure 1.
Suppression of the Pth(Ts) phenotype mediated by the overproduction of tRNALys maintains moderate levels of the Pth(Ts) protein. (A) Depicts the cellular growth of the pth(Ts) mutant strain AA7852 separately transformed with pVH124 (ΔvalU, ΔlysV), pVH125 (valU, ΔlysV) or pVH119 (valU, lysV) incubated at different temperatures. Isolated colonies of the independent transformants were streaked onto LB-Ap plates and incubated overnight at the indicated temperatures. (B) Presents the immunodetection of Pth(Ts) in the pth(Ts) mutant strain AA7852 separately transformed with pVH124, pVH125, pVH119, ptRNAHisCCA (argX, hisR, leuT, proM) or pTH2 (thrW) and grown at 32°C prior to transfer at time = 0 min at 41 or 43°C. The concentration of Pth(Ts) protein was estimated by immunoblot analysis. The left lane shows purified wild-type Pth protein, which migrates slightly faster in SDS–PAGE than the Pth(Ts) variant (arrowed) (3).
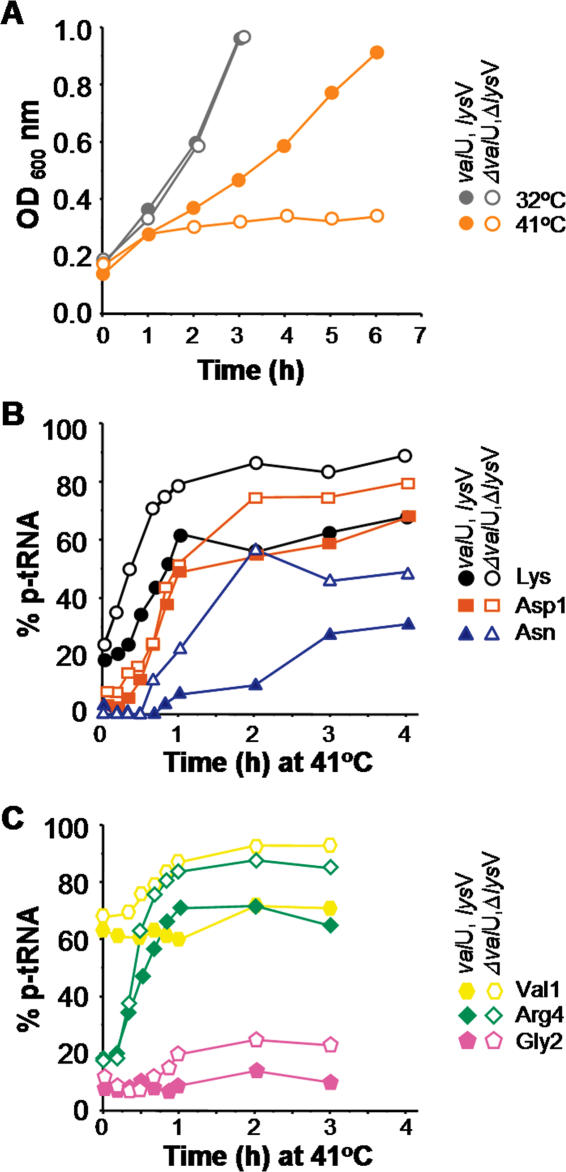
Figure 2.
Suppression of the Pth(Ts) phenotype mediated by the overproduction of tRNALys promotes growth and reduces only moderately the concentration of p-tRNAs at 41°C. (A) Presents the growth in liquid medium of the pth(Ts) mutant strain AA7852 separately transformed with pVH124 (ΔvalU, ΔlysV; open symbols) or pVH119 (valU, lysV; closed symbols) at 32 and 41°C. Cells were grown at 32°C to an OD600 of 0.2 and further incubated at 32°C (grey symbols) or transferred to 41°C at time = 0 min (orange symbols). (B and C) show the percentages of p-tRNAs accumulated during growth at 41°C as described in (A). The relative fractions of the various p-tRNAs [p-tRNALys (closed circles), p-tRNAAsp1 (dark orange squares), p-tRNAAsn (blue triangles), p-tRNAArg4 (green diamonds), p-tRNAGly2 (pink pentagons) and p-tRNAVal1 (yellow hexagons)] were quantified by northern blotting of acid urea gels. The curves show representative experiments. Note: half of the material extracted from the cells that banded as aminoacyl- or p-tRNAVal1 (yellow hexagons) in the acid urea gels was refractory to treatments with purified Pth protein or with CuSO4 (data not shown) and may correspond to a partially modified variant of tRNAVal1 (10). Therefore, the actual concentration of p-tRNAVal1 represented 30% of total tRNAVal1 at the non-permissive temperature.
Concentration of Pth(Ts) protein
Non-transformed and transformed strains of E.coli AA7852 were incubated at 32°C to an OD600 of 0.4 in liquid LB medium containing antibiotic as appropriate. Cultures were then incubated at 41 or 43°C and samples taken at the times indicated in Figure 1B. For each sample, cells were harvested by centrifugation, resuspended in lysis buffer [62.5 mM Tris–HCl (pH 6.8), 2% SDS, 10% glycerol and 5% 2-mercaptoethanol] and boiled for 5 min. Total protein was resolved through 15% SDS–PAGE, and the gel electroblotted onto a nitrocellulose membrane. Pth(Ts) was detected by western blot using rabbit polyclonal anti-Pth serum (diluted 1:8000) (11), the signal being amplified by treatment with rabbit anti-IgA coupled to horseradish peroxidase (diluted 1:10,000) and revealed using Western blotting fluorescent detection reagents (ECL, Amersham Pharmacia Biotech, Amersham, UK).
Levels of aminoacyl- and peptidyl-tRNAs
The levels of uncharged, aminoacylated and peptidylated tRNA were determined by northern blot assays as described previously (12). Liquid cultures (200 ml) of non-transformed and transformed strains of E.coli AA7852 were grown at 32°C to an OD600 of 0.2 in LB-Ap medium and then transferred to an incubator at 41°C. Samples (10 ml) were taken at the times indicated in Figures 2–4, the cells were pelleted at 4°C and total RNA isolated under acidic conditions (13). A portion of RNA was resuspended in 166 µl of diethyl pyrocarbonate-water and divided into two fractions, one of which, the control sample, was mixed with 6.7 µl of 3 M NaOAc (pH 5.0) and used to resolve the aminoacylated and peptidylated tRNA forms from the uncharged forms. The second sample was submitted to cupric ion-mediated hydrolysis in which aminoacylated tRNAs, but not the p-tRNA derivatives, were cleaved (14): this sample was mixed with 6.7 µl of 3 M NaOAc (pH 5.0) and 10 µl of 100 mM CuSO4. Each sample was incubated for 30 min at 37°C and adjusted to 5 mM with respect to Na2EDTA prior to precipitation with 2 vol of absolute ethanol at −20°C.
Two additional control samples were prepared, in the presence or absence of 100 ng of a purified preparation of Pth(Ts) protein (Figure 3), each containing 3 µg of total tRNA (obtained 60 min after the temperature shift of the cells) dissolved in 25 µl of reaction buffer (Tris–HCl 100 mM and NH4Cl 100 mM) and 50 µl of a buffer containing 10 mM Tris–HCl (pH 7.6), 10 mM MgOAc, 6 mM β-mercaptoethanol and 20 mM NH4Cl. These samples were incubated for 20 min at 37°C and precipitated with 2 vol of absolute ethanol and 0.1 vol of 3 M NaOAc.
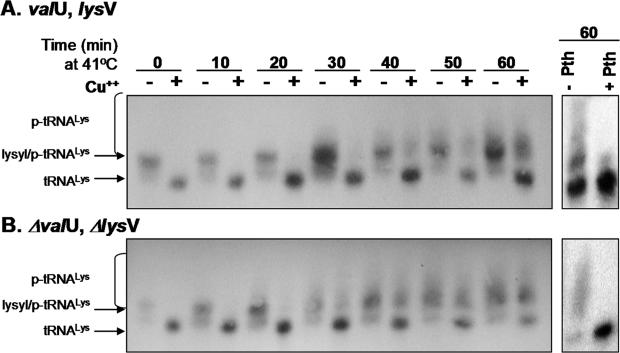
Figure 3.
Suppression of the Pth(Ts) phenotype mediated by the overproduction of tRNALys allows the accumulation of heterogeneous p-tRNALys at 41°C. Changes in the concentrations of the tRNALys forms with time in the presence (A) [showing strain AA7852 transformed with pVH119 (valU, lysV)] or in the absence (B) [showing strain AA7852 transformed with pVH124 (ΔvalU, ΔlysV)] of excess tRNALys are presented. Transformed cells were grown at 32°C to an OD600 of 0.2 prior to transfer at time = 0 min to 41°C, and samples were taken at the times indicated. p-tRNALys was revealed by northern blotting using a specific 32P-labelled oligo-probe in samples of total RNA. The sample in the right lane was treated with a preparation of Pth protein prior to electrophoresis. The relative locations of the various tRNALys derivatives are arrowed, whilst each brace indicates the extent of the smear corresponding to heterogeneous p-tRNALys. The absolute amount of material applied to each lane varied according to the efficiency of recovery of the tRNA in each manipulation. Band intensities of the different tRNA forms should, strictly, only be compared within lanes, thus only approximate indications of amounts of tRNA forms can be derived between independent lanes (compare 30 min lanes treated and untreated with CuSO4).
The RNAs obtained from the untreated, the CuSO4 treated and the Pth treated samples were each dissolved in 5 µl of 10 mM NaOAc (pH 5.0) and 1 mM Na2EDTA, and an aliquot (1 µl) of each sample used to determine RNA concentration at 260 nm. Samples containing 4 µg of extracted RNA were resolved by overnight electrophoresis in a 6% polyacrylamide gel containing 8M urea in 0.07 M NaOAc buffer (pH 5.0) at 16°C. The RNA was electroblotted onto Hybond-N+ nylon membranes (Amersham Biosciences, Amersham, UK), in a BioRad (Hercules, CA) Trans-Blot Semi-Dry Electrophoretic Transfer Cell for 18 min at 380 mA using 1× TAE as transfer buffer. tRNA-specific oligodeoxyribonucleotide probes were synthesized according to a published method (15). The blotted membranes were pre-hybridized with denatured salmon sperm DNA for 3 h at 42°C, and hybridized to 5′-32P end-labelled oligonucleotides for 16 h at 42°C. Finally, the membranes were washed twice for 1 h at room temperature (13) and the radioactive signals quantified using a Typhoon Scan (Amersham Biosciences). The membranes were routinely probed at least twice with other oligonucleotides. The respective percentages of uncharged, aminoacylated or peptidylated tRNAs were estimated relative to the total tRNA for each electrophoretic lane using the following formula:
RESULTS
Overproduction of tRNALys suppresses the effects of pth(Ts) mutation at 41°C but not at 43°C
E.coli strain AA7852 was transformed with the construct pVH119, harbouring the gene lysV encoding tRNALys, and control constructs pVH124 and pVH125 in which lysV was either partly or fully deleted (7). Growth assays, performed by streaking independent colonies on LB-Ap medium (Figure 1A), demonstrated that cells containing pVH119 exhibited significant suppression of the Pth(Ts) phenotype at 41°C, but much lower growth at 43°C. In contrast, there was hardly any detectable growth of cells containing the control constructs at either of these temperatures.
Overproduction of tRNALys maintains the levels of Pth(Ts) protein at 41°C
E.coli strain AA7852 was transformed with constructs harbouring different tRNA-encoding genes and incubated at 41 or 43°C: the concentrations of Pth(Ts) protein were determined in extracts of these cells by western blot analysis (Figure 1B). The control construct pVH124, and those bearing the tRNA-encoding genes valU, argX, hisR, leuT, proM and thrW, were ineffective in suppressing the thermosensitivity of the pth(Ts) strain at 41°C (data not shown) as has been demonstrated previously (7). Accordingly, these constructs were unable to maintain detectable levels of Pth(Ts) antigen at 41°C (Figure 1B, a–d). However, overproduction of tRNALys gave rise to levels of Pth(Ts) antigen that were clearly detectable for at least 2 h following transfer of the cells to 41°C, but only for the first 30 min or so after cells were transferred to 43°C (Figure 1B, e). It may be inferred from the restored capacity of the cells to grow at 41°C that the presence of Pth(Ts) protein at levels compatible with viability should continue indefinitely. In AA7852 cells incubated at 32°C, neither the level (Figure 1B) nor the half-life of Pth(Ts) changed with tRNALys overproduction (data not shown). Thus, the overproduction of tRNALys may rise the rate of Pth(Ts) protein synthesis at a non-permissive temperature to levels that are just high enough to maintain the amount and activity of Pth(Ts) required for protein synthesis and cell viability.
Overproduction of tRNALys slightly reduces the relative concentration of accumulated peptidyl-tRNAs in pth(Ts) cells at 41°C
It was of interest to determine whether the steady-state level of Pth(Ts) mediated by the overproduction of tRNALys at a non-permissive temperature could relieve the accumulation of p-tRNA in the mutant cells. For this purpose the relative concentrations of various p-tRNAs in pVH119-transformed AA7852 cells, incubated at 41°C, were compared with those found in AA7852 cells transformed with the pVH124 control construct. The results (Figure 2B and C) show that, after 60 min at 41°C, the relative concentrations of p-tRNAs of tRNALys, tRNAAsp1, tRNAAsn, tRNAVal1, tRNAArg4 and tRNAGly2 were consistently lower in pth(Ts) cells transformed with pVH119 compared with control transformants. In nearly all of the assays, the concentration of p-tRNA remained low for the first few minutes after the cells had been transferred to 41°C, then increased for another 30 min, and began to level off after 60 min. This finding was confirmed for all of the tRNAs analysed. It would thus appear that the limited levels of Pth(Ts) protein maintained during suppression of the effects of pth(Ts) mutation mediated by the overproduction of tRNALys can only partially handle the increased rate of accumulation of p-tRNA at 41°C. However, whilst the levels of p-tRNAs built up significantly at 41°C, the cells were still able to grow although at a reduced rate in comparison with that observed at 32°C (Figure 2A).
The complexity of the p-tRNALys increases with the time of incubation at 41°C
The complexity of the p-tRNALys species that accumulated in the presence of limited levels of Pth(Ts) protein was evaluated by northern blots of samples extracted from pVH119- and pVH124-transformed AA7852 cells at different times after transference from 32 to 41°C. As shown in Figure 3A and B, during the first 30 min at 41°C, the tRNALys species observed were mainly in the aminoacylated form. However, the level and the complexity of the accumulated p-tRNALys increased in a time-dependent manner as revealed by the increasing intensities and range of electrophoretic mobilities of the species above the free-tRNALys bands in the northern blots (Figure 3). This increase in accumulated p-tRNALys, irrespective of tRNALys overproduction, may result from an increased rate of translation at 41°C (3), given that the levels of Pth(Ts) protein in the overproducing cells were comparable at both 32 and 41°C (Figure 1B). Larger amounts of free-tRNA were generated by the cupric ion-mediated hydrolysis of RNA extracts obtained from pVH119-transformed pth(Ts) cells incubated at 41°C than from the pVH124-transformed strain (Figure 3A and B, compare the pairs of 60 min). RNA samples, obtained 60 min after the temperature shift of the pth(Ts) transformed cells, were treated with a purified preparation of Pth protein (Figure 3, right panels). It was observed that the tRNALys forms that had been concentrated in the smear localized above the aminoacylated form disappeared, whilst the levels of free-tRNALys increased proportionally (Figure 3A and B, right panels) indicating the p-tRNA nature of the material in the smear. The faint band of aminoacylated-tRNALys (Figure 3A, right panel, +Pth lane) did not correspond with the relatively intensity material produced by cupric ion-mediated hydrolysis of the 60 min sample (Figure 3A, left panel), although both bands should represent the same proportion of aminoacylated-tRNALys. Since this was not the case, it is likely that part of the aminoacylated-tRNA was hydrolysed during the preparation of samples for treatment with Pth.
Overproduction of tRNALys increases the levels of aminoacyl-tRNALys
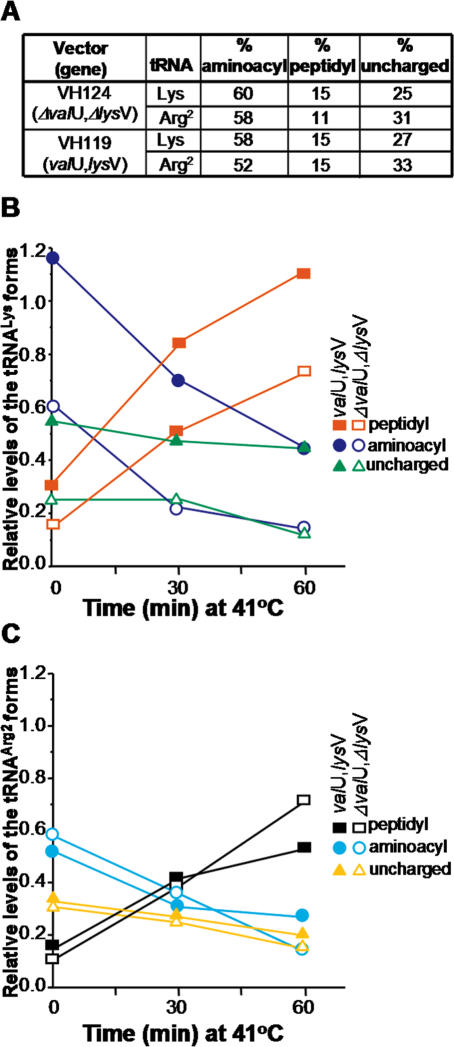
Since the levels of aminoacylation determine the availability of precursors for protein synthesis in the cell, it was important to assess the extent by which transformation of pth(Ts) cells with pVH119 changed the concentrations of the tRNALys species present. For this purpose, the levels and relative proportions of tRNALys found as uncharged-, aminoacyl- and p-tRNA were determined in pth(Ts) cells harbouring pVH119 and pVH124 constructs (Figure 4). Whilst the level of total tRNALys detected in pVH119-transformed cells was 2-fold greater than that found in the control transformants at 32°C (data not shown) (7), the relative proportions of the different tRNALys species and of the control tRNAArg2 were comparable in both cell lines (Figure 4A). Following the temperature rise to 41°C, the levels of aminoacylated tRNALys and tRNAArg2 in both pVH119- and pVH124-transformed cells decreased with time, whilst the amounts of the respective p-tRNAs increased proportionally (Figure 4B and C). Taking into consideration that the levels of both lysyl-tRNALys and p-tRNALys increased 2-fold in the pVH119-transformed cells, the suppression of the temperature-sensitivity of the pth(Ts) strain may be a consequence of an increase in the absolute concentration of lysyl-tRNALys rather than of a change in the concentration of an inhibitory p-tRNA (5).
Figure 4.
The concentration of lysyl-tRNALys in transformed pth(Ts) cells decreases during incubation at 41°C. (A) Presents the relative amounts of aminoacyl-, peptidyl- and uncharged-tRNA in strain AA7852 transformed with pVH124 (ΔvalU, ΔlysV) or pVH119 (valU, lysV) and incubated at 32°C. The percentage of each of the tRNA forms relative to total tRNA (uncharged plus aminocylated plus peptidylated tRNA) was estimated by comparing the signal intensities before and after treatment with CuSO4 (Figure 3). In (B and C), changes in the relative fractions of the Lys and Arg2 tRNA forms, respectively, during suppression of the Pth(Ts) phenotype mediated by the overproduction of tRNALys are shown. Experiments conducted with AA7852 cells transformed with pVH119 (valU, lysV) are indicated by closed symbols, whilst those that involved pVH124 (ΔvalU, ΔlysV) transformants are shown by open symbols: in each case p-tRNALys is represented by red squares, aminoacyl-tRNALys by blue circles, uncharged-tRNALys by green triangles, p-tRNAArg2 by black squares, aminoacyl-tRNAArg2 by cyan circles, and uncharged-tRNAArg2 by yellow triangles. The additional amount of tRNALys produced by the pVH119 transformant was estimated with respect to the concentration of tRNAArg2, which was considered to be unaffected by the overproduction of tRNALys. Two successive hybridisations were conducted on the same membrane, first using a tRNALys probe and then with a tRNAArg2 probe, in order to normalize for the RNA load in different lanes. These data indicate that the total concentration of tRNALys in cells transformed with pVH119 was 2-fold higher than that in cells transformed with pVH124. The relative levels of the tRNALys forms in (B) have been multiplied by 2 to take this account. The data correspond to a representative experiment.
DISCUSSION
In this work we showed that the overproduction of tRNALys was able to suppress the thermosensitivity of E.coli strain AA7852, the original pth(Ts) isolate (1), at 41°C but not at 43°C, a result which differs from that obtained previously for C600 pth(Ts) in which suppression was observed even at 43°C (7). Suppression of the effects of pth(Ts) mutation was accompanied by a rise in the rate of Pth(Ts) protein synthesis that was clearly just sufficient to maintain cell viability at 41°C. Furthermore, the levels of Pth(Ts) protein maintained by tRNALys overproduction permitted linear growth of the mutant cells at 41°C (Figure 2A), but not exponential growth as would be expected if the mutation were fully suppressed. The levels of Pth(Ts) protein at 41°C were comparable with those present in pth(Ts) cells at 32°C (compare t = 0 and 120 min lanes in Figure 1B, e) namely, some 5-fold lower than the wild-type concentration (3). The maintenance of the concentration of Pth(Ts) protein in the presence of tRNALys overproduction was not due to an extended stability of the protein (data not shown), although an effect on the levels of pth(Ts) mRNA cannot be ruled out owing to the difficulty in assaying the low concentrations of mRNA in untransformed cells.
It is argued that the higher availability of charged-tRNALys, rather than the reduced level of p-tRNALys, provokes a general enhancement in protein synthesis that maintains low but effective concentrations of Pth(Ts) protein at 41°C. The results showed that the restoration of growth of the pth(Ts) mutant at a restrictive temperature mediated by the overproduction of tRNALys was correlated with significant increases in the levels of both charged-tRNALys and p-tRNALys (Figure 4B). If p-tRNALys were poisoning translation one would not expect a higher level to be correlated with the observed restoration of growth. The concentration of lysyl-tRNALys in pVH119-transformed cells decreased with the time of incubation at 41°C (Figure 4B); however, the proportion of this species with respect to the total tRNALys always remained higher than that present in the pVH124-transformants. The fractions of the aminoacylated-tRNAArg2 species decreased in both pVH119- and pVH124-transformants after transfer of the cells to 41°C (Figure 4C). After 60 min incubation, however, the relative concentration of aminoacylated-tRNAArg2 in pVH119-transformed cells was somewhat higher than in the pVH124-transformed counterparts, apparently at the expense of the p-tRNAArg2 fraction. Also, the pth(Ts) transformant that overproduced tRNALys contained 20–50% less p-tRNA of other species than the non-overproducing control cells after 2 h incubation at 41°C (Figure 2B and C). This behaviour may be explained by assuming that the overproduction of tRNALys maintains the levels of Pth(Ts) protein by enhancing its synthesis. As a result Pth(Ts) would hydrolyse the p-tRNAs thus raising the amounts of other aminoacylable tRNAs. The limited increase in the relative concentrations of the charged tRNAs may help in maintaining the cell viability indefinitely.
The proportion of charged-tRNALys to free-forms remained unchanged following transformation of pth(Ts) cells with pVH119 and further growth at 32°C, in spite of a 2-fold increase in the levels of tRNALys (see legend to Figure 4) (7). However, upon incubation at 41°C, the concentration of charged-tRNALys fell gradually over 60 min to levels comparable with those observed in the control cells incubated at 32°C, (Figure 4B, compare overproduced tRNALys at 60 min with control at time 0 min). Apparently, once this critical phase of 60 min is overcome, the high levels of Lys-tRNALys mediated by tRNALys overproduction are not essential for cell viability. This result suggests that the surplus charged-tRNALys overcomes the transient effect of the heat shock response on protein synthesis that is occasioned by a temperature shift to 41°C.
The increasing intensity and extent of the smear located above the free-tRNA bands in the northern blots (Figure 3) suggested that the concentration of the accumulated material and the apparent complexity of the p-tRNALys species increased after the first 30 min following transfer of the cells to 41°C. Analogous patterns of change with respect to time were observed for the p-tRNA derivatives of tRNAArg2, tRNAArg4, tRNAAsn, tRNAAsp1, tRNAHis, tRNALeu3, tRNAPhe, tRNAThr4 and tRNAVal1 (data not shown). Similar smears located above tRNA bands have been taken as evidence of p-tRNA heterogeneity in previous studies (16). However, since the p-tRNALys species considered in the present study were only those recovered in the aqueous phase of the phenol extract and those that migrated close to the free and aminoacylated tRNA forms in acid gel electrophoresis, it may be assumed that they carried peptide chains containing only a few amino acid residues (17). Presumably, those p-tRNAs in which the protein nature of the peptide predominates over the RNA character of the molecule are not present in the aqueous phase but precipitate with protein during phenol extraction (17).
Why is p-tRNALys most rapidly accumulated upon transfer of the pth(Ts) mutant to 40°C (4)? The propensity of p-tRNALys to drop-off readily either upon expression of a minigene (12) or of a gene in which a lysine codon precedes the termination triplet (18), may be ascribed mainly to the long pause at the termination codon before the ribosome terminates (18). However, the soluble heterogeneous p-tRNALys that accumulated in the presence of limited levels of Pth(Ts) protein could result from drop-off at the lysine codons frequently located in the initial open reading frame positions of E.coli genes (19). Based on the relative high frequency of the AAA codon at positions 2, 3, 4 and 5 in the E.coli reading frames (19), we favour that the heterogeneous and short p-tRNALys species originate by drop-off early after translation initiation of bacterial mRNAs. In contrast, the infrequent location of glycine and cysteine codons at early positions in the reading frames, correlates with the slow accumulation of p-tRNAGly and p-tRNACys (19,4). This proposal is consistent with observations, namely, that the propensity of the p-tRNAs to drop-off correlates inversely with the length of the coding sequence as the oligopeptidyl moiety increases in size from 2 to 6 amino acid residues (20), that drop-off occurs irrespective of whether the codon in the ribosomal A-site is a sense or stop codon (21). It is also possible that a variety of minigenes containing lysine codons exist in the bacterial chromosome the expression of which would result in the accumulation of heterogeneous p-tRNALys. Further experiments designed to decide which, if either, of these hypotheses is correct are required in order to establish the origin of the accumulated p-tRNALys in the pth(Ts) cells incubated at non-permissive temperatures.
Acknowledgments
The authors wish to thank Richard Buckingham for providing plasmid constructs, Marco A. Magos-Castro, José G. Bueno-Martínez and Gloria León Avila for technical support and an anonymous referee for critical review and comments on the manuscript. This work was supported by grants from the Consejo Nacional de Ciencia y Tecnología (CONACyT, Mexico) to G.G. One of us (S.V.D) was supported by grants from CONACyT and Consejo del Sistema Nacional de Educación Technológica (COSNET). Funding to pay the Open Access publication charges for this article was provided by Centro de Investigación y de Estudios Avanzados.
Conflict of interest statement. None declared.
REFERENCES
- 1.Atherly A.G., Menninger J.R. Mutant E.coli strain with temperature sensitive peptidyl-transfer RNA hydrolase. Nature New Biol. 1972;240:245–246. doi: 10.1038/newbio240245a0. [DOI] [PubMed] [Google Scholar]
- 2.García-Villegas M.R., De la Vega F.M., Galindo J.M., Segura M., Buckingham R.H., Guarneros G. Peptidyl-tRNA hydrolase is involved in inhibition of host protein λ synthesis. EMBO J. 1991;10:3549–3555. doi: 10.1002/j.1460-2075.1991.tb04919.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cruz-Vera L.R., Toledo I., Hernández-Sánchez J., Guarneros G. Molecular basis for the temperature sensitivity of Escherichia coli pth(Ts) J. Bacteriol. 2000;182:1523–1528. doi: 10.1128/jb.182.6.1523-1528.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Menninger J.R. The accumulation as peptidyl-transfer RNA of isoaccepting transfer RNA families in Escherichia coli with temperature-sensitive peptidyl-transfer RNA hydrolase. J. Biol. Chem. 1978;253:6808–6813. [PubMed] [Google Scholar]
- 5.Menninger J.R. Accumulation of peptidyl tRNA is lethal to Escherichia coli. J. Bacteriol. 1979;137:694–696. doi: 10.1128/jb.137.1.694-696.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Menez J., Remy E., Buckingham R.H. Suppression of thermosensitive peptidyl-tRNA hydrolase mutation in Escherichia coli by gene duplication. Microbiology. 2001;147:1581–1589. doi: 10.1099/00221287-147-6-1581. [DOI] [PubMed] [Google Scholar]
- 7.Heurgué-Hamard V., Mora L., Guarneros G., Buckingham R.H. The growth defect in Escherichia coli deficient in peptidyl-tRNA hydrolase is due to starvation for Lys-tRNALys. EMBO J. 1996;15:2826–2833. [PMC free article] [PubMed] [Google Scholar]
- 8.Heurgué-Hamard V., Karimi R., Mora L., MacDougall J., Leboeuf C., Grentzmann G., Ehrenberg M., Buckingham R.H. Ribosome release factor RF4 and termination factor RF3 are involved in dissociation of peptidyl-tRNA from the ribosome. EMBO J. 1998;17:808–816. doi: 10.1093/emboj/17.3.808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Singh N.S., Varshney U. A physiological connection between tmRNA and peptidyl-tRNA hydrolase functions in Escherichia coli. Nucleic Acids Res. 2004;32:6028–6037. doi: 10.1093/nar/gkh924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Derrick W.B., Horowitz J. Probing structural differences between native and in vitro transcribed Escherichia coli valine transfer RNA: evidence for stable modification-dependent conformers. Nucleic Acids Res. 1993;21:4948–4953. doi: 10.1093/nar/21.21.4948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hernández-Sánchez J., Valadez J.G., Vega-Herrera J., Ontiveros C., Guarneros G. λ bar minigene-mediated inhibition of protein synthesis involves accumulation of peptidyl-tRNA and starvation for tRNA. EMBO J. 1998;17:3758–3765. doi: 10.1093/emboj/17.13.3758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cruz-Vera L.R., Hernández-Ramón E., Pérez-Zamorano B., Guarneros G. The rate of peptidyl-tRNA dissociation from the ribosome during minigene expression depends on the nature of the last decoding interaction. J. Biol. Chem. 2003;278:26065–26070. doi: 10.1074/jbc.M301129200. [DOI] [PubMed] [Google Scholar]
- 13.Varshney U., Lee C.P., RajBhandary U.L. Direct analysis of aminoacylation levels of tRNAs in vivo. J. Biol. Chem. 1991;266:24712–24718. [PubMed] [Google Scholar]
- 14.Schofield P., Zamecnik P.C. Cupric ion catalysis in hydrolysis of aminoacyl-tRNA. Biochim. Biophys. Acta. 1968;155:410–416. doi: 10.1016/0005-2787(68)90185-8. [DOI] [PubMed] [Google Scholar]
- 15.Dong H., Nilsson L., Kurland C.G. Co-variation of tRNA abundance and codon usage in Escherichia coli at different growth rates. J. Mol. Biol. 1996;260:649–663. doi: 10.1006/jmbi.1996.0428. [DOI] [PubMed] [Google Scholar]
- 16.Rao A.R., Varshney U. Specific interaction between the ribosome recycling factor and the elongation factor G from Mycobacterium tuberculosis mediates peptidyl-tRNA release and ribosome recycling in Escherichia coli. EMBO J. 2001;20:2977–2986. doi: 10.1093/emboj/20.11.2977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Slabaugh R.C., Morris A.J. Purification of peptidyl transfer ribonucleic acid from rabbit reticulocyte ribosomes. J. Biol. Chem. 1970;245:6182–6189. [PubMed] [Google Scholar]
- 18.Menez J., Heurgué-Hamard V., Buckingham R.H. Sequestration of specific tRNA species cognate to the last sense codon of an overproduced gratuitous protein. Nucleic Acids Res. 2000;28:4725–4732. doi: 10.1093/nar/28.23.4725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sato T., Terabe M., Watanabe H., Gojobori T., Hori-Takemoto C., Miura K. Codon and base biases after the initiation codon of the open reading frames in the Escherichia coli genome and their influence on the translation efficiency. J. Biochem. 2001;129:851–860. doi: 10.1093/oxfordjournals.jbchem.a002929. [DOI] [PubMed] [Google Scholar]
- 20.Heurgué-Hamard V., Dinçbas V., Buckingham R.H., Ehrenberg M. Origins of minigene-dependent growth inhibition in bacterial cells. EMBO J. 2000;19:2701–2709. doi: 10.1093/emboj/19.11.2701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Karimi R., Pavlov M.Y., Heurgué-Hamard V., Buckingham R.H., Ehrenberg M. Initiation factors IF1 and IF2 synergistically remove peptidyl-tRNAs with short polypeptides from the P-site of translating Escherichia coli ribosomes. J. Mol. Biol. 1998;281:241–252. doi: 10.1006/jmbi.1998.1953. [DOI] [PubMed] [Google Scholar]