Abstract
We have investigated whether nonneutralizing monoclonal antibodies (MAbs) to the gp120 subunit of the envelope glycoprotein (Env) complex of human immunodeficiency virus type 1 (HIV-1) can interfere with HIV-1 neutralization by another anti-gp120 MAb. We used neutralizing (b12) and nonneutralizing (205-42-15, 204-43-1, 205-46-9) MAbs to the epitope cluster overlapping the CD4-binding site (CD4BS) on gp120. All the MAbs, neutralizing or otherwise, cross-competed for binding to monomeric gp120, indicating the close topological proximity of their epitopes. However, the nonneutralizing CD4BS MAbs did not interfere with the neutralization activity of MAb b12. In contrast, in a binding assay using oligomeric Env expressed on the surface of Env-transfected cells, the nonneutralizing MAbs did partially compete with b12 for Env binding. The surface of Env-transfected cells contains two categories of binding site for CD4BS MAbs. One type of site is recognized by both b12 and nonneutralizing CD4BS MAbs; the other is recognized by only b12. Binding assays for Env-gp120 interactions based on the use of monomeric gp120 or Env-transfected cells do not predict the outcome of HIV-1 neutralization assays, and they should therefore be used only with caution when gauging the properties of anti-Env MAbs.
Neutralization of human immunodeficiency virus type 1 (HIV-1) involves the binding of antibodies to the native, fusion-competent envelope glycoprotein (Env) complex on the surface of infectious virions (27, 45, 48, 56). Most of the known neutralizing antibodies whose epitopes have been characterized and whose mechanisms of action have been explored work by inhibiting the interactions of the virus with its receptors, CD4 and the CCR5 or CXCR4 coreceptor (45, 66, 70, 71). In the case of the 2G12 monoclonal antibody (MAb) to a glycan epitope on gp120, inhibition of coreceptor binding probably occurs indirectly (52, 57, 70, 72). The inhibitory actions of MAbs can occur prior to attachment of the virus to the cell surface or subsequent to a semispecific absorption of the virus to ancillary receptors such as heparin sulfate proteoglycans (62, 70). An exception is the 2F5 MAb to a conserved epitope in the C-terminal region of the gp41 ectodomain, which probably neutralizes HIV-1 infectivity by interfering with receptor-mediated conformational changes in the envelope glycoproteins subsequent to the virus-receptor interactions (70). By analogy, the 4E10 and Z13 antibodies to epitopes proximal to the 2F5 site are likely to have a similar mechanism of action (63, 74, 75).
Neutralization assays performed with primary cells in vitro have now been shown to predict, with reasonable confidence, whether or not an animal can be protected from a viral challenge by antibodies in plasma (1, 18, 19, 29, 30, 31, 41, 45, 47, 59). In general, nonneutralizing antibodies are not protective in these studies whereas neutralizing antibodies can be protective if they are present at a sufficiently high titer in the plasma of a test animal at the time of challenge. The protective concentration of a test MAb can also usually be approximated from an in vitro neutralization assay performed against the challenge isolate: typically, sterile protection is achieved in vivo if the plasma MAb concentration is around 100-fold greater than that required to cause a 10-fold reduction in viral infectivity (i.e., 90% neutralization) in vitro. Lower antibody concentrations can sometimes provide partial protection (31, 47). This may be useful in the context of a vaccine intended to induce both cellular and humoral immune responses (8, 45).
Most antibodies raised against the HIV-1 envelope glycoproteins during natural infection or after vaccination with gp120 subunits are, however, nonneutralizing (4, 10, 30, 35, 36, 38, 45, 46, 49). Although these antibodies are ineffective, might they somehow interfere with virus neutralization? Antagonism has recently been reported between a nonneutralizing antibody to the CD4-binding site (MAb F105) and the neutralizing MAbs 2F5 and 2G12 (22). Furthermore, why nonneutralizing antibodies do not neutralize HIV-1 has become controversial because of the suggestion that such antibodies can successfully bind to virions but do not then hinder the function of the envelope glycoprotein complexes that mediate fusion and infection (73). The basis for this argument is that nonneutralizing antibodies can bind to envelope glycoprotein epitopes on the surface of Env-transfected cells or on virus particles (32, 42, 43, 73). The theoretical dangers of overinterpreting Env-binding assays have been noted elsewhere (39); here, we address the issue experimentally by studying the interactions between neutralizing (MAb b12) and nonneutralizing MAbs directed at the epitope cluster that overlaps the CD4-binding site (CD4BS) on gp120.
We chose to study the CD4BS epitope cluster for several reasons. The CD4BS is important for receptor attachment, and antibodies to this region prevent virus binding to the surface of susceptible target cells (25, 50, 70). The site is highly conserved across the genetic subtypes of HIV-1 group M, exemplified by the ability of MAb b12 to neutralize multiple isolates (9, 68). However, the binding sites for neutralizing and nonneutralizing MAbs within the CD4BS are only subtly different and there is much overlap between them (53, 65). The influence of distal amino acid polymorphisms on the CD4BS is another indicator of the structural complexity of this region of gp120 (24, 44). Finally, gp120 vaccines tested to date have not elicited CD4BS-directed antibodies with significant neutralizing activity against primary isolates (4, 10, 30, 38, 39). A better understanding of the CD4BS epitope cluster could therefore be useful for the rational design of HIV-1 vaccines.
MATERIALS AND METHODS
Reagents and plasmids.
MAbs 205-43-1, 205-42-15, and 205-46-9 (Tanox) have all been described previously (16, 17), as have MAb b12 (9, 53, 54) and MAb 2G12 (52, 57, 69). Note that MAbs 205-43-1, 205-42-15, and 205-46-9 were designated HT5, HT6, and HT7 in reference 17.
The pDsRed2-N1 plasmid, expressing DsRed2, a variant of Discosoma sp. red fluorescent protein, was obtained from Clontech (Palo Alto, Calif.). Paraformaldehyde, bovine serum albumin (BSA), and glycine were from Sigma Chemical Co. (St. Louis, Mo.).
Purified, monomeric gp120 from HIV-1 JR-FL was expressed in CHO cells and was a gift from Paul Maddon and William Olson of Progenics Pharmaceuticals, Inc. (Tarrytown, N.Y.) (66), as was the CD4-immunoglobulin G2 (IgG2) molecule (68). The JR-FL SOS gp140 protein and the pPPI4-based plasmid used to express the JR-FL Env protein gp140Twt have both been described elsewhere (6, 7, 58). Briefly, the gp140Twt Env insert, encoding an envelope glycoprotein lacking most of the cytoplasmic domain of gp41, was generated from a JR-FL gp160 template by PCR (sense primer, 5′-GTCTATTATGGGGTACCTGTGTGGAAAGAAGC, antisense primer, 5′-CGCAGACGCAGATTCGAATTAAACTCTATTCACTATAGAAAGTAC) and cloned into pPPI4 with the KpnI and BstBI restriction enzymes. The Env glycoprotein produced was truncated after position 708 due to the addition of a stop codon at position 709 (Env amino acid numbering is based on the HXB2 sequence). This cytoplasmic tail truncation is similar, but not identical, to the one described by Si et al. (60). Full-length Env sequencing revealed two conservative changes, T415I and E647D, compared to the database version of JR-FL (accession number U63632), but these are not expected to have a significant impact on Env structure or function.
Cells and culture conditions.
All cell cultures were maintained at 37°C in an atmosphere containing 5% CO2. Human epithelial kidney (HEK) 293T cells were grown in Dulbecco's minimal essential medium (GIBCO, Grand Island, N.Y.) containing 10% fetal calf serum (FCS), 2 mM l-glutamine, antibiotics (100 U of penicillin/ml, 100 μg of streptomycin/ml), and 0.5 mg of the neomycin analog G-418/ml. Peripheral blood mononuclear cells (PBMC) were isolated from healthy HIV-seronegative donors by Ficoll-Hypaque centrifugation, mitogen stimulated as previously described (67), and maintained in RPMI 1640 medium containing 10% FCS, 2 mM l-glutamine, antibiotics (100 U of penicillin/ml, 100 μg of streptomycin/ml), and 100 U of interleukin 2/ml. HeLa-CD4-CCR5 cells were cultured in Dulbecco's minimal essential medium supplemented with 10% FCS, l-glutamine, and antibiotics.
The source of the virus JR-FL and the construction of pseudotyped luciferase reporter viruses bearing JR-FL envelope have been described elsewhere (20, 68).
Binding of MAbs to monomeric gp120.
The binding of biotin-labeled MAbs to gp120 and its inhibition by unlabeled MAbs was measured by enzyme-linked immunosorbent assay essentially as described elsewhere (16, 40). Briefly, gp120 was immobilized on plastic via adsorbed sheep antibody D7324 (Cliniqa Inc., Fallbrook, Calif.) to a conserved C-terminal epitope. Unlabeled MAb was added at variable concentrations, and its binding was detected by using a goat anti-human alkaline phosphatase conjugate and the AMPAK colorimetric detection system. For cross-competition analysis, an unlabeled MAb was added to the immobilized gp120 for 30 min, followed by a fixed amount of the same MAb or a different biotin-labeled MAb. The binding of the labeled MAb was detected by the use of a streptavidin alkaline phosphatase conjugate and the AMPAK system (16, 40).
Conjugation of MAbs to fluorochromes.
The protein content of purified MAb preparations was quantified by the bicinchoninic acid method (Pierce, Rockford, Ill.) with BSA as standard (61). MAbs were dialyzed against labeling buffer (50 mM H3BO3, 200 mM NaCl, pH 9.2) at 4°C, and then a 20-μl aliquot of fluorescein isothiocyanate (FITC) at 5 mg/ml in dimethyl sulfoxide was added for each milligram of antibody present. After incubation for 2 h at room temperature, unbound fluorochrome was removed from the labeled MAb by gel filtration with a Sephadex G-25 column (Amersham Biosciences, Uppsala, Sweden).
Envelope glycoprotein expression and immunostaining assays.
HEK 293T cells were transiently cotransfected with plasmids by the calcium phosphate precipitation method (Profection mammalian transfection system-calcium phosphate; Promega, Madison, Wis.). Briefly, 10 μg of the pPPI4-Env and 1 μg of pDsRed2-N1 plasmids were cotransfected into HEK 293T cells in a six-well plate containing 3 ml of culture medium without G-418. The cells were washed 15 h later and then refed with 3 ml of the same culture medium. The immunostaining procedure was performed 36 h after transfection. The transfected HEK 293T cells (2 × 106 per sample) were harvested, washed with phosphate-buffered saline (PBS), and incubated for 1 h at room temperature with 50 μl of a solution containing a MAb. The cells were then washed with cold PBS and fixed in 2% paraformaldehyde-60 mM sucrose in PBS, pH 7.4, for 15 min at room temperature. After two washes with PBS containing 20 mM glycine (buffer A), the cells were incubated for 15 min at room temperature with buffer A containing 1% BSA and 0.05% NaN3 (buffer B). For indirect immunostaining, samples were stained with 10 μg of goat F(ab′)2 anti-human IgG-FITC (Biosource International, Camarillo, Calif.)/ml in buffer B (45 min at room temperature), washed three times, and resuspended in the same buffer for flow cytometry analysis. For competition experiments, the unlabeled MAb (20 μg/ml) was added to the cells for 30 min before the addition of FITC-labeled MAb (20 μg/ml) for 1 h at room temperature. Stained cell suspensions were analyzed by using an Epics Elite flow cytometer (Coulter Corporation, Hialeah, Fla.). FITC and DsRed2 excitation was achieved by the use of a 488-nm Argon laser lamp, and the fluorescence emissions were collected by using 525- and 575-nm band-pass filters, respectively. The parameters used to select cell populations for analysis were forward- and side-light scatter, followed by double-positive fluorescence. A total of 8,000 events within the double-positive population were collected for analysis. The Env- and DsRed2-expressing plasmids were transfected at a ratio (10:1) previously determined to cause negligible interference between the two fluorescence emission spectra while ensuring that ≥90% of the DsRed2-positive cells were also Env positive (data not shown).
The cell surface Env-binding assays were performed at room temperature in all cases, similar to the procedure used by Si et al. (60). While binding reactions performed at 37°C might be preferable from a biological perspective, internalization or other mechanisms of down-regulation reduced the levels of surface-expressed Env and generally precluded the generation of meaningful data at 37°C. When small-scale studies were performed at 4°C, room temperature, and 37°C, the absolute levels of 2G12 and b12 binding were temperature dependent. However, only minor influences of temperature on the b12/2G12 binding ratio were observed.
HIV-1 neutralization assays.
Neutralization of the HIV-1 JR-FL primary isolate by MAbs was determined by measuring the inhibition of virus replication in mitogen-stimulated PBMC from normal donors, essentially as described elsewhere (16, 67). Briefly, 50 μl of a nonneutralizing antibody was mixed with 50 μl of b12 at appropriate concentrations and then incubated for 1 h at 37°C with 100 50% tissue culture infective doses of HIV-1 JR-FL (50 μl) before addition of the virus-MAb mixture to 2 × 105 mitogen-stimulated PBMC (50 μl). The extent of virus replication in the cultures was determined by measuring the extracellular p24 antigen concentration after 6 to 7 days.
The luciferase-expressing Env-pseudotype virus HIV-1JR-FL was also used in neutralization assays involving HeLa-CD4-CCR5 cells. Luciferase activity was measured 72 h postinfection and used to determine the neutralization end point, as described elsewhere (20).
RESULTS
MAbs 205-43-1, 205-42-15, 205-46-9, and b12 react with related epitopes that overlap the CD4-binding site on monomeric gp120.
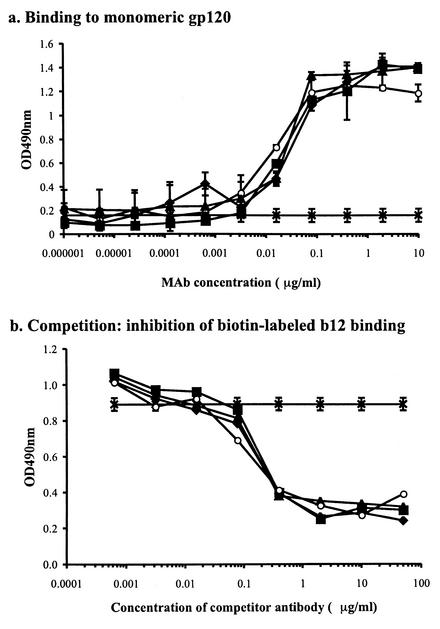
The four MAbs 205-43-1, 205-42-15, 205-46-9, and b12 all bind to epitopes that overlap the CD4BS on monomeric gp120, each efficiently blocks the binding of soluble CD4 to gp120, and each of them reciprocally cross-block the binding of the other three (16) (data not shown). An example of this cross-inhibition is that 205-43-1, 205-42-15, and 205-46-9 each bound equivalently to monomeric gp120 from the HIV-1 JR-FL strain and by doing so inhibited the binding of b12 to the same protein (Fig. 1). Similar results were obtained for the SOS gp140 protein, captured onto plastic via lectin (data not shown). Thus, all four MAbs bind to topologically related epitopes that are in close proximity to the CD4-binding site on monomeric gp120. Moreover, the pattern of amino acid substitutions in HxBc2 gp120 that disrupts the binding of each MAb is characteristic of that for antibodies designated CD4BS MAbs (16).
FIG. 1.
Inhibition of MAb b12 binding to monomeric gp120 by MAbs 205-42-15, 205-43-1, and 205-46-9. (a) The binding of MAbs b12 (○), 205-42-15 (⧫), 205-43-1 (▪), and 205-46-9 (▴) to gp120 was measured. An optical density at 490 nm (OD490nm) of 0.20 in this assay represents an assay background (∗). (b) The inhibition of biotin-labeled b12 binding to gp120 by MAbs 205-43-1, 205-42-15, and 205-46-9 was measured. The effect of the absence of competitor in this assay is also noted (×).
Nonneutralizing MAbs to CD4BS epitopes do not interfere with HIV-1 neutralization by the b12 MAb to an overlapping epitope.
It was reported previously that MAbs 205-43-1, 205-42-15, and 205-46-9 do not efficiently neutralize primary HIV-1 isolates, although they strongly neutralize T-cell-line-adapted (TCLA) viruses (16, 17). In contrast, MAb b12 can neutralize most primary isolates (9, 68). Since all four MAbs recognize related epitopes, we determined whether the nonneutralizing antibodies could interfere with the action of the neutralizing antibody.
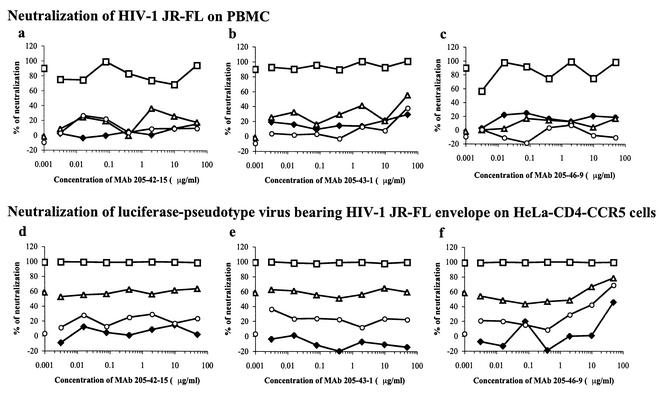
Preliminary experiments established that MAbs 205-43-1, 205-42-15, and 205-46-9, at concentrations up to 100 μg/ml, had little or no neutralizing activity against the primary HIV-1 JR-FL isolate in PBMC, whereas b12 caused potent neutralization with 50 and 90% infectious concentrations of approximately 0.25 and 1 μg/ml, respectively (Fig. 2 and data not shown). We next mixed a fixed concentration of b12 (0.01, 0.1, or 1 μg/ml) with HIV-1 JR-FL, together with a variable concentration of either 205-43-1, 205-42-15, or 205-46-9, and added the MAb-virus mixture to the PBMC. In no case did the presence of a nonneutralizing MAb either increase or decrease the neutralization activity of b12 (Fig. 2a through c). This was the case whether the nonneutralizing MAb was mixed with the virus simultaneously with b12 (Fig. 2a through c) or added to the virus 1 h prior to the addition of b12 to give additional time for a binding reaction to occur (data not shown).
FIG. 2.
MAbs 205-42-15, 205-43-1, and 205-46-9 do not interfere with neutralization of HIV-1 JR-FL by MAb b12. MAbs 205-42-15, 205-43-1, and 205-46-9 at the concentration indicated, with or without (⧫) MAb b12 at 0.01 (○), 0.1 (▵), or 1 (□) μg/ml, were mixed with HIV-1 JR-FL for 1 h prior to addition to PBMC (a, b, and c) or mixed with the HIV-1JR-FL Env-pseudotype virus for 1 h prior to addition to HeLa-CD4-CCR5 cells (d, e, and f). The outcome of infection was determined, and the percentage neutralization by the input antibodies was determined. The data points overlapping the y axis were derived in the absence of MAb (i.e., at 0 μg/ml).
The above experiment was repeated on a clonal population of luciferase-expressing pseudoviruses (HIV-1JR-FL) and HeLa-CD4-CCR5 target cells. The results were similar to those obtained in the PBMC assay with the HIV-1 JR-FL primary isolate (Fig. 2d through f), except that a small amount of additional neutralization occurred when the highest concentration of MAb 205-46-9 was mixed with a low concentration of b12 (Fig. 2f). This is probably a reflection of the limited amount of neutralizing activity possessed by MAb 205-46-9 (16). However, we could find no evidence whatsoever that any of the three nonneutralizing MAbs interfered with the activity of the neutralizing MAb.
Binding of CD4BS MAbs to oligomeric Env on the surface of Env-transfected cells.
HIV-1 neutralization was achieved by antibody binding to native oligomeric Env complexes on the surface of infectious virions. The inability of MAbs 205-43-1, 205-42-15, and 205-46-9 to interfere with the neutralizing activity of b12 would be most simply explained by their inability to compete with b12 for binding to their related epitopes on the virion surface. This would be consistent with their own lack of neutralizing activity.
As an initial approach to investigating why the different CD4BS MAbs behave differently in neutralization assays, we have studied their reactivity with Env-expressing cells by using flow cytometry to quantitate the extent of antibody binding (44, 50, 60, 73, 75). Preliminary experiments showed that there was a significant overlap between the fluorescence peak corresponding to the specific labeling of Env by MAb and a peak of nonspecific fluorescence, as noted previously (73). A procedure similar to that developed by York et al. was therefore adopted (73); we gated on the double-positive population of cells that expressed both Env proteins and the coexpressed DsRed2 fluorophore.
The b12 and 2G12 MAbs were titrated, and their binding to gp140Twt Env on the surface of transfected HEK 293 cells was determined (Fig. 3). Similar binding curves were generated for the 205-42-15, 205-43-1, and 205-46-9 MAbs (Fig. 3). The binding of each of the CD4BS MAbs to cell surface Env was saturable. The three nonneutralizing CD4BS MAbs had very similar half-maximal binding concentrations of 0.27 to 0.57 μg/ml, whereas the neutralizing b12 MAb had a somewhat lower affinity for cell surface Env, with half-maximal binding occurring at 2.83 μg/ml (Fig. 3).
FIG. 3.
Titration curves for CD4BS MAb binding to cell surface Env. HEK 293T cells were cotransfected with pPPI4-gp140Twt and pDsRed2-N1. The immunostaining procedure was performed with different concentrations of b12 (•) or 2G12 (○), 205-46-9 (▪), 205-42-15 (⧫), or 205-43-1 (▴) followed by 10 μg of FITC-labeled goat F(ab′)2 anti-human IgG/ml. Nonspecific fluorescence due to the secondary antibody was subtracted from the mean of the fluorescence intensity values. The data shown were obtained from two independent experiments of similar design. The half-maximal binding concentrations for each MAb were 2.83 μg/ml for b12, 0.57 μg/ml for 205-46-9, 0.27 μg/ml for 205-42-15, 0.46 μg/ml for 205-43-1, and 2.43 μg/ml for 2G12.
The gp140Twt Env protein contains gp120, the gp41 ectodomain, the membrane-spanning domain, and a truncated form of the gp41 cytoplasmic tail. It possesses an unmodified cleavage site between gp120 and gp41, although the extent to which Env cleavage occurs in Env-transfected cells is quite limited and rarely exceeds 25% (6, 7, 73; S. Beddows and J. P. Moore, unpublished data).
Competition between neutralizing and nonneutralizing CD4BS MAbs for binding to oligomeric Env on the surface of Env-transfected cells.
Preliminary experiments in which the various labeled and unlabeled MAbs were titrated showed that inhibition of the binding of the labeled MAb was dose dependent and saturable (data not shown). Maximal inhibition of the FITC-labeled MAb was achieved when the unlabeled MAb (20 μg/ml) was added to the cells 30 min before the labeled MAb. When the labeled and unlabeled MAbs were added to the cells simultaneously, the maximal level of competition was achieved only when the unlabeled MAb was present at a fivefold excess over that of the labeled MAb. The binding experiments enabled us to determine that 20 μg/ml was an appropriate saturating concentration to use for each MAb in subsequent competition experiments.
Env-transfected cells were then incubated with 20 μg of each competitor MAb/ml for 30 min before the addition of 20 μg of FITC-labeled b12/ml and the measurement of b12 binding (Table 1). Unlabeled b12 was an efficient inhibitor of the binding of FITC-b12, the mean value for b12 inhibition in each experiment being defined as 100% for normalization purposes (Table 1). As expected, MAb 2G12 did not interfere with FITC-b12 binding, the extent of inhibition (0 to 25%) being negligible in an assay of this type (Table 1). In contrast, the CD4-IgG2 molecule, which interacts with the CD4BS on gp120, was nearly as efficient as b12 at inhibiting FITC-b12 binding (Table 1).
TABLE 1.
Competition of FITC-conjugated CD4BS MAbs
| Epitope cluster and unlabeled MAb | Inhibition of indicated FITC-conjugated MAb binding to surface-expressed JR-FL Env by unlabeled MAbsa:
|
||
|---|---|---|---|
| b12 | 205-46-9 | 205-43-1 | |
| Neutralizing CD4BS | |||
| b12 | 100 | 106 (30) | 89 (47) |
| CD4-IgG2 | 80 (7) | 101 (18) | 105 (16) |
| Nonneutralizing CD4BS | |||
| 205-46-9 | 51 (18) | 100 | 119 (16) |
| 205-42-15 | 44 (18) | 126 (2) | 113 (12) |
| 205-43-1 | 53 (8) | 91 (4) | 100 |
| Neutralizing-complex epitope | |||
| 2G12 | 9 (14) | 20 (19) | 0 (0) |
Competition between CD4BS MAbs and other agents for binding to cell surface Env. The binding signals were normalized so that the binding of each FITC-labeled MAb by the same unlabeled MAb represented 100% competition. The extent of inhibition achieved by the other test MAbs was expressed relative to this value. The percentage inhibition values shown are the means and standard deviations (in parentheses) derived from two independent experiments. The actual levels of competition for FITC-b12 by b12, of FITC-205-46-9 by 205-46-9, and of FITC-205-43-1 by 205-43-1 were 60, 30, and 30%, respectively.
The three nonneutralizing CD4BS MAbs, 205-42-15, 205-43-1, and 205-46-9, only partially inhibited the binding of FITC-b12 to cell surface Env. The extent of inhibition varied from 44 to 53% depending upon the particular MAb used (Table 1) and was markedly less than the inhibition achieved by b12 and CD4-IgG2 in the same experiments (Table 1). Thus, despite their having an affinity higher than that for b12 for cell surface Env (a lower half-maximal binding concentration [Fig. 3]), the nonneutralizing CD4BS MAbs are less effective than b12 at blocking FITC-b12 binding.
We next determined how the binding of the nonneutralizing CD4BS MAbs was affected by b12, by CD4-IgG2, and by each other. To do this, we performed competition experiments similar to those described above but with FITC-labeled 205-46-9 or 205-43-1. The b12 MAb and the CD4-IgG2 molecule each efficiently inhibited the binding of FITC-205-43-1 and FITC-205-46-9, whereas 2G12 had no significant effect (Table 1). However, in contrast with their limited ability to compete with FITC-b12 for cell surface Env binding, the three nonneutralizing CD4BS MAbs each completely inhibited the Env binding of FITC-205-43-1 and FITC-205-46-9 or nearly so (Table 1).
We treated the three nonneutralizing CD4BS MAbs as a single competition group to compare the extent to which they and b12 could inhibit binding of the FITC-labeled MAbs. Thus, the average inhibition of FITC-b12 binding by the three nonneutralizing MAbs was 49% ± 15%, that of FITC-205-46-9 binding was 97% ± 18%, and that of FITC-205-43-1 binding was 121% ± 32%. The inhibition of FITC-b12 binding by the nonneutralizing MAbs was significantly different from their inhibition of FITC-205-46-9 or FITC-205-43-1 binding (P < 0.01; Mann-Whitney U test). In contrast, the inhibition of FITC-b12 binding by b12 was 100% (by the normalization definition), that of FITC-205-46-9 binding was 106%, and that of FITC-205-43-1 binding was 89%. These values are not significantly different from each other.
Thus, binding of the neutralizing MAb b12 and nonneutralizing CD4BS MAbs to cell surface Env can be discriminated by their sensitivity to inhibition by nonneutralizing CD4BS MAbs. The binding of b12 is only partially sensitive to the nonneutralizing CD4BS MAbs; that of the nonneutralizing is fully sensitive.
These various observations are best explained by there being two categories of binding sites for CD4BS MAbs present on the surface of Env-transfected cells: one category is unique to b12 (and CD4-IgG2); the second is shared with MAbs to nonneutralizing CD4BS MAbs. A likely explanation for the presence of the two categories of binding site involves the incomplete cleavage of Env.
DISCUSSION
In this study, we show that three nonneutralizing MAbs to CD4BS-related epitopes do not interfere with the neutralization of a HIV-1 primary isolate by a MAb, b12, to an overlapping, closely related epitope. The epitopes for all four of the MAbs are proximal to the CD4-binding site on gp120, all four antibodies efficiently inhibit the binding of soluble CD4 to monomeric gp120, and all cross-block each other's binding to gp120. Hence, all four MAbs recognize very similar overlapping epitopes on monomeric gp120 yet only MAb b12 efficiently neutralizes primary HIV-1 isolates. All four MAbs, however, do neutralize TCLA viruses (36), so the potential for HIV-1 primary isolate neutralization is clearly present in all four. Why, then, do primary isolates, exemplified here by HIV-1 JR-FL, resist inhibition by MAbs 205-43-1, 205-42-15, and 205-46-9 while remaining susceptible to MAb b12?
It has long been argued that the resistance of primary isolates to neutralization by anti-Env MAbs lies at the level of the native oligomeric envelope glycoprotein complex (15, 27, 36, 38, 45, 46, 48, 50, 51, 55, 60, 64). Specifically, we have proposed that the structure of this complex has evolved to resist the binding of MAbs to most of its surface, including epitopes that are proximal to the CD4BS (37, 38, 48). Moreover, we have argued that resistance to neutralizing antibodies is essential for the persistence of a lentivirus such as HIV-1 in vivo (45, 48). This argument is supported by observations that primary isolates of several other lentiviruses are neutralization resistant (2, 11, 33, 34) and that neutralization-sensitive lentiviruses revert to a resistance phenotype in vivo (3, 5, 14, 23, 60). HIV-1 variants with a global neutralization-resistance phenotype have also arisen after the passage of neutralization-sensitive viruses in the presence of neutralizing serum in vitro (24, 44). In both cases, the phenotypic determinants were located in the envelope glycoprotein complex, affecting the overall topology of the native Env oligomer rather than any one specific neutralization epitope (24, 44).
There is good evidence that antibodies to the CD4BS function by preventing attachment of the virion to CD4 on the target cell (26, 50, 70). We believe that the reason why the nonneutralizing MAbs to CD4BS-related epitopes neither neutralize HIV-1 JR-FL nor interfere with the neutralizing activity of b12 is simply because the nonneutralizing CD4BS MAbs fail to bind to the native envelope complexes on the virion that mediates fusion. On this argument, the nonneutralizing CD4BS MAbs are irrelevant to the neutralization process because they remain in solution, unable to interfere with the virus-cell attachment and fusion process. Whether the same is true of nonneutralizing MAbs to other epitopes on gp120 and gp41 will require additional studies in this or different experimental systems, but we believe that the CD4BS will not turn out to be a special case among the known neutralization epitopes. It will also be necessary to further study why TCLA strains are sensitive to neutralizing antibodies that fail to neutralize closely related primary viruses; how Env proteins from TCLA viruses compare with those from primary isolates in Env-binding and competition assays such as those we have used here is presently under investigation. We expect that it will be difficult to discriminate between the binding of b12 and the nonneutralizing CD4BS MAbs to TCLA Env on the surface of Env-transfected cells (assuming all the test MAbs neutralize the particular TCLA virus under study). However, this supposition will require experimental confirmation and clearly other outcomes are possible.
An alternative perspective has, however, been provided: it is argued that nonneutralizing MAbs bind to their epitopes on fusion-competent envelope glycoprotein complexes but that this binding does not interfere with the function of these complexes (73). According to this hypothesis, attachment and fusion either take place irrespective of the presence of the bound MAb or else the envelope glycoprotein complex in some way shakes off the MAb as it attaches to its cellular receptors and then undergoes the conformational changes which drive the fusion process (73). The data that support such an argument are derived from binding assays that employ virions or Env-transfected cells. In such assays, the binding of nonneutralizing MAbs to envelope glycoproteins from neutralization-resistant primary isolates can clearly be demonstrated (73). Indeed, we show here that nonneutralizing CD4BS MAbs not only bind Env-transfected cells but can also partially reduce the binding of the neutralizing MAb b12 to the same cells (Fig. 3). The reactivity of MAbs with Env on the surface of Env-transfected cells clearly does not predict how they perform in neutralization assays. For TCLA HIV-1 strains, a good correlation between cell surface MAb binding and virus neutralization can, however, be obtained with virus-infected cells (55).
An observation that helps explain the limitations of cell surface Env-binding assays is that nonneutralizing CD4BS MAbs only partially inhibit the Env binding of FITC-b12, whereas b12 and CD4-IgG2 are more effective competitors (Table 1). This is despite the fact that the nonneutralizing MAbs actually have a higher affinity than b12 for cell surface Env (Fig. 3). Moreover, in the reciprocal competition, b12 and the nonneutralizing CD4BS MAbs behave equivalently as inhibitors of the binding of the FITC-labeled, nonneutralizing MAbs. One explanation for these related observations is that there are two categories of b12-binding site on cell surface Env: one category is accessible to all CD4BS MAbs, including b12, and to CD4-IgG2, whereas the other site is available only for the binding of b12 and CD4-IgG2. If this is the case, a logical extension is that only the latter, b12-unique binding sites are relevant to Env function whereas the sites accessible to the other CD4BS MAbs (and b12) are present on defective forms of Env that are irrelevant to Env function (39).
The most obvious, but not the only, Env defect that could affect MAb binding is the lack of cleavage of gp120 from gp41. This process occurs inefficiently in Env-transfected cells (6, 7, 60, 73), and only approximately 25% of the Env species present on the cell surface are cleaved under the conditions of our experiments. Indeed, there is a correlation between the binding of a MAb to cleaved Env on the cell surface and its neutralization activity, but this is not observed when cleavage-defective Env is expressed instead (60; Z. Si, C. Phan, E. Kiprilov, and J. Sodroski, unpublished results). Moreover, nonneutralizing CD4BS MAbs bind preferentially to recombinant, oligomeric uncleaved gp140 proteins whereas b12 binds preferentially to cleaved Env on the surface of virus-infected cells (46). Further studies will be required to resolve exactly how Env cleavage affects antibody binding.
An additional influence on how antibodies react with Env may be the interaction between Env and Gag on the cell or virion surface. The presence of Gag is known to affect how Env is stained by antibodies on the surface of transfected cells (21), and the cytoplasmic domain of gp41, which contains the Gag interaction site, can modify the reactivity of MAbs with both the gp120 and gp41 components of the Env ectodomain (13). Studies in other experimental systems have also shown that assays which rely on Env expression on the cell surface can yield results different from those derived by using assays of viral infectivity (12; R. W. Doms, personal communication). Overall, conclusions derived from Env-binding assays cannot always be extrapolated to cover what occurs when an infectious virus encounters its cellular receptors.
Concerns can also be expressed about the use of virion-binding assays as surrogates for neutralization assays (39, 48), although we have not addressed these issues experimentally here. If virions contain binding sites for antibodies that are present either on defective forms of Env or on Env complexes that have already transited from the native to postfusion form, such binding sites would have no relevance to antibody-mediated neutralization. They would have no function for an antibody to impair. However, alternative arguments can also be made about why virion-binding and neutralizing assays could give different results; for example, variation in the rates at which different antibodies bind to virion-associated Env would probably influence binding and neutralization assays to different extents.
HIV-1 neutralization requires that an antibody binds to the surface of a fusion-competent envelope glycoprotein complex on the virion surface; when sufficient such complexes have been occupied, the infectivity of the virion is compromised (25, 26, 28, 45). This is particularly so for antibodies directed to the CD4BS (26, 45, 50, 70). We believe the preponderance of accumulated evidence suggests that nonneutralizing antibodies have no involvement in this process simply because they are unable to bind to functional, untriggered Env complexes on virions. Whether additional or alternative mechanisms also apply will require further studies.
Acknowledgments
C.H. and C.S. contributed equally to this work.
We are grateful to Linnea Schiffner, Shivani Kake, and Aditi Master for skilled technical assistance. We also appreciate the technical advice received from the Weill-Cornell flow cytometry core facility. We thank Paul Maddon and William Olson for the gift of several reagents and Bob Doms and Joe Sodroski for the communication of unpublished results.
This work was funded by NIH grants AI39420 and AI36082 to J.P.M. and AI33292 to D.R.B. and by the International AIDS Vaccine Initiative. J.P.M. is a Stavros S. Niarchos Scholar. The Department of Microbiology and Immunology at the Weill Medical College gratefully acknowledges the support of the William Randolph Hearst Foundation.
REFERENCES
- 1.Baba, T. W., V. Liska, R. Hofmann-Lehmann, J. Vlasak, W. Xu, S. Ayehunie, L. A. Cavacini, M. R. Posner, H. Katinger, G. Stiegler, B. J. Bernacky, T. A. Rizvi, R. Schmidt, L. R. Hill, M. E. Keeling, Y. Lu, J. E. Wright, T. C. Chou, and R. M. Ruprecht. 2000. Human neutralizing monoclonal antibodies of the IgG1 subtype protect against mucosal simian-human immunodeficiency virus infection. Nat. Med. 6:200-206. [DOI] [PubMed] [Google Scholar]
- 2.Baldinotti, F., D. Matteucci, P. Mazzetti, C. Gianelli, P. Bandecchi, F. Tozzini, and M. Bendinelli. 1994. Serum neutralization of feline immunodeficiency virus is markedly dependent on passage history of the virus and host system. J. Virol. 68:4572-4579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beaumont, T., A. van Neusen, S. Broersen, W. A. Blattner, V. V. Lukashov, and H. Schuitemaker. 2001. Reversal of human immunodeficiency virus type 1 IIIB to a neutralization-resistant phenotype in an accidentally infected laboratory worker with a progressive clinical course. J. Virol. 75:2246-2252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Belshe, R. B., G. J. Gorse, M. J. Mulligan, T. G. Evans, M. C. Keefer, J.-L. Excler, A. M. Duliege, J. Tartaglia, W. I. Cox, J. McNamara, K.-L. Hwang, A. Bradney, D. C. Montefiori, and K. J. Weinhold. 1998. Induction of immune responses to HIV-1 by canarypox virus (ALVAC) HIV-1 and gp120 SF-2 recombinant vaccines in uninfected volunteers. AIDS 12:2407-2415. [DOI] [PubMed] [Google Scholar]
- 5.Bendinelli, M., M. Pistello, D. Del Mauro, G. Cammarota, F. Maggi, A. Leonildi, S. Giannecchini, C. Bergamini, and D. Matteucci. 2001. During readaptation in vivo, a tissue culture-adapted strain of feline immunodeficiency virus reverts to broad neutralization resistance at different times in individual hosts but through changes at the same position of the surface glycoprotein. J. Virol. 75:4584-4593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Binley, J. M., R. W. Sanders, B. Clas, N. Schuelke, A. Master, Y. Guo, F. Kajumo, D. J. Anselma, P. J. Maddon, W. C. Olson, and J. P. Moore. 2000. A recombinant human immunodeficiency virus type 1 envelope glycoprotein complex stabilized by an intermolecular disulfide bond between the gp120 and gp41 subunits is an antigenic mimic of the trimeric virion-associated structure. J. Virol. 74:627-643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Binley, J. M., R. W. Sanders, A. Master, C. S. Cayanan, C. L. Wiley, L. Schiffner, B. Travis, S. Kuhmann, D. R. Burton, S.-L. Hu, W. C. Olson, and J. P. Moore. 2002. Enhancing the proteolytic maturation of human immunodeficiency virus type 1 envelope glycoproteins. J. Virol. 76:2606-2616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Burton, D. R., and J. P. Moore. 1998. Why do we not have an HIV vaccine and how can we make one? Nat. Med. 4:495-498. [DOI] [PubMed] [Google Scholar]
- 9.Burton, D. R., J. Pyati, R. Koduri, G. B. Thornton, L. S. W. Sawyer, R. M. Hendry, N. Dunlop, P. L. Nara, M. Lamacchia, E. Garratty, E. R. Stiehm, Y. J. Bryson, J. P. Moore, D. D. Ho, and C. F. Barbas III. 1994. Efficient neutralization of primary isolates of HIV-1 by a recombinant human monoclonal antibody. Science 266:1024-1027. [DOI] [PubMed] [Google Scholar]
- 10.Connor, R. I., B. T. M. Korber, B. S. Graham, B. H. Hahn, D. D. Ho, B. D. Walker, A. U. Neumann, S. H. Vermund, J. Mestecky, S. Jackson, E. Fenamore, Y. Cao, F. Gao, S. Kalams, K. Kuntsman, D. McDonald, N. McWilliams, A. Trkola, J. P. Moore, and S. M. Wolinsky. 1998. Immunological and virological analyses of persons infected by human immunodeficiency virus type 1 while participating in trials of recombinant gp120 subunit vaccines. J. Virol. 72:1552-1576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cook, R. F., S. L. Berger, K. E. Rushlow, J. M. McManus, S. J. Cook, S. Harrold, M. L. Raabe, R. C. Montelaro, and C. J. Issel. 1995. Enhanced sensitivity to neutralizing antibodies in a variant of equine infectious anemia virus is linked to amino acid substitutions in the surface unit envelope glycoprotein. J. Virol. 69:1493-1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dragic, T., A. Trkola, S. W. Lin, K. A. Nagashima, F. Kajumo, L. Zhao, W. C. Olson, L. Wu, C. R. Mackay, G. P. Allaway, T. P. Sakmar, J. P. Moore, and P. J. Maddon. 1998. Amino-terminal substitutions in the CCR5 coreceptor impair gp120 binding and human immunodeficiency virus type 1 entry. J. Virol. 72:279-285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Edwards, T. G., S. Wyss, J. D. Reeves, S. Zolla-Pazner, J. A. Hoxie, R. W. Doms, and F. Baribaud. 2002. Truncation of the cytoplasmatic domain induces exposure of conserved regions in the ectodomain of human immunodeficiency virus type 1 envelope protein. J. Virol. 76:2683-2691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Etemad-Moghadam, B., G. B. Karlsson, M. Halloran, Y. Sun, D. Schenten, M. Fernandes, N. L. Letvin, and J. Sodroski. 1998. Characterization of simian-human immunodeficiency virus envelope glycoprotein epitopes recognized by neutralizing antibodies from infected monkeys. J. Virol. 72:8437-8445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Etemad-Moghadam, B., Y. Sun, E. K. Nicholson, G. B. Karlsson, D. Schenten, and J. Sodroski. 1999. Determinants of neutralization resistance in the envelope glycoproteins of a simian-human immunodeficiency virus passaged in vivo. J. Virol. 73:8873-8879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fouts, T. R., J. M. Binley, A. Trkola, J. E. Robinson, and J. P. Moore. 1997. Neutralization of the human immunodeficiency virus type 1 primary isolate JR-FL by human monoclonal antibodies correlates with antibody binding to the oligomeric form of the envelope glycoprotein complex. J. Virol. 71:2779-2785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fouts, T. R., A. Trkola, M. S. Fung, and J. P. Moore. 1998. Interactions of polyclonal and monoclonal anti-glycoprotein 120 antibodies with oligomeric glycoprotein 120-glycoprotein 41 complexes of a primary HIV type 1 isolate: relationship to neutralization. AIDS Res. Hum. Retrovir. 14:591-598. [DOI] [PubMed] [Google Scholar]
- 18.Gauduin, M. C., P. W. H. I. Parren, R. Weir, C. F. I. Barbas, D. R. Burton, and R. A. Koup. 1997. Passive immunization with a human monoclonal antibody protects hu-PBL-SCID mice against challenge by primary isolates of HIV-1. Nat. Med. 3:1389-1393. [DOI] [PubMed] [Google Scholar]
- 19.Gauduin, M. C., J. T. Safrit, R. Weir, M. S. Fung, and R. A. Koup. 1995. Pre- and post-exposure protection against human immunodeficiency virus type 1 infection mediated by a monoclonal antibody. J. Infect. Dis. 171:1203-1209. [DOI] [PubMed] [Google Scholar]
- 20.Gordon, C. J., M. A. Muesing, A. E. Proudfoot, C. A. Power, J. P. Moore, and A. Trkola. 1999. Enhancement of human immunodeficiency virus type 1 infection by the CC-chemokine RANTES is independent of the mechanism of virus-cell fusion. J. Virol. 73:684-694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Herrida-Matsumoto, L., and M. D. Resh. 2000. Localization of human immunodeficiency virus type 1 Gag and Env at the plasma membrane by confocal imaging. J. Virol. 74:8670-8679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hofmann-Lehmann, R., J. Vlasak, R. A. Rasmussen, B. A. Smith, T. W. Baba, V. Liska, F. Ferrantelli, D. C. Montefiori, H. M. McClure, D. C. Anderson, B. J. Bernacky, T. A. Rizvi, R. Schmidt, L. R. Hill, M. E. Keeling, H. Katinger, G. Stiegler, L. A. Cavacini, M. R. Posner, T. C. Chou, J. Andersen, and R. M. Ruprecht. 2001. Postnatal passive immunization of neonatal macaques with a triple combination of human monoclonal antibodies against oral simian-human immunodeficiency virus challenge. J. Virol. 75:7470-7480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Johnson, W. E., and R. C. Desrosiers. 2002. Viral persistence: HIV's strategies of immune system evasion. Annu. Rev. Med. 53:499-518. [DOI] [PubMed] [Google Scholar]
- 24.Klasse, P. J., J. A. McKeating, M. Schutten, M. S. Reitz, Jr., and M. Robert-Guroff. 1993. An immune-selected point mutation in the transmembrane protein of human immunodeficiency virus type 1 [HXB2-Env: Ala582(Thr)] decreases viral neutralization by monoclonal antibodies to the CD4-binding site. Virology 196:332-337. [DOI] [PubMed] [Google Scholar]
- 25.Klasse, P. J., and J. P. Moore. 1996. Quantitative model of antibody- and soluble CD4-mediated neutralization of primary isolates and T-cell line-adapted strains of human immunodeficiency virus type 1. J. Virol. 70:3668-3677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Klasse, P. J., and Q. J. Sattentau. 2002. Occupancy and mechanism in antibody-mediated neutralization of animal viruses. J. Gen. Virol. 83:2091-2108. [DOI] [PubMed] [Google Scholar]
- 27.Kwong, P., R. Wyatt, Q. J. Sattentau, J. Sodroski, and W. A. Hendrickson. 2000. Oligomeric modeling and electrostatic analysis of the gp120 envelope glycoprotein of human immunodeficiency virus. J. Virol. 74:1961-1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Layne, S. P., M. J. Merges, M. Dembo, J. L. Spouge, and P. L. Nara. 1990. HIV requires multiple gp120 molecules for CD4-mediated infection. 346:277-279. [DOI] [PubMed]
- 29.Mascola, J. R., M. G. Lewis, G. Stiegler, D. Harris, T. C. VanCott, D. Hayes, M. K. Louder, C. Brown, C. V. Sapan, S. S. Frankel, Y. Lu, M. L. Robb, H. Katinger, and D. L. Birx. 1999. Protection of macaques against pathogenic SHIV-89.6PD by passive transfer of neutralizing antibodies. J. Virol. 73:4009-4018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mascola, J. R., S. W. Snyder, O. S. Weislow, S. M. Belay, R. B. Belshe, D. H. Schwartz, M. L. Clements, R. Dolin, B. S. Graham, G. J. Gorse, M. C. Keefer, M. J. McElrath, M. C. Walker, K. F. Wagner, J. G. McNeil, F. E. McCutchan, D. S. Burke and the National Institute of Allergy Infectious Diseases AIDS Vaccine Evaluation Group. 1996. Immunization with envelope subunit vaccine products elicits neutralizing antibodies against laboratory adapted but not primary isolates of human immunodeficiency virus type 1. J. Infect. Dis. 173:340-348. [DOI] [PubMed] [Google Scholar]
- 31.Mascola, J. R., G. Stiegler, T. C. VanCott, H. Katinger, C. B. Carpenter, C. E. Hanson, H. Beary, D. Hayes, S. S. Frankel, D. L. Birx, and M. G. Lewis. 2000. Protection of macaques against vaginal transmission of a pathogenic HIV-1/SIV chimeric virus by passive infusion of neutralizing antibodies. Nat. Med. 6:207-210. [DOI] [PubMed] [Google Scholar]
- 32.Mbah, H. A., S. Burda, M. K. Gorny, C. Williams, K. Revesz, S. Zolla-Pazner, and P. N. Nyambi. 2001. Effect of soluble CD4 on exposure of epitopes on primary, intact, native human immunodeficiency virus type 1 virions of different genetic clades. J. Virol. 75:7785-7788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Means, R. E., T. Greenough, and R. C. Desrosiers. 1997. Neutralization sensitivity of cell culture-passaged simian immunodeficiency virus. J. Virol. 71:7895-7902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Montefiori, D. C., K. A. Reimann, M. S. Wyand, K. Manson, M. G. Lewis, R. G. Collman, J. G. Sodroski, D. P. Bolognesi, and N. L. Letvin. 1998. Neutralizing antibodies in sera from macaques infected with chimeric simian-human immunodeficiency virus containing the envelope glycoproteins of either a laboratory-adapted variant or a primary isolate of human immunodeficiency virus type 1. J. Virol. 72:3427-3431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Moore, J. P., and D. R. Burton. 1999. HIV-1 neutralizing antibodies: how full is the bottle? Nat. Med. 5:142-144. [DOI] [PubMed] [Google Scholar]
- 36.Moore, J. P., Y. Cao, L. Qing, Q. J. Sattentau, J. Pyati, R. Koduri, J. Robinson, C. F. Barbas III, D. R. Burton, and D. D. Ho. 1995. Primary isolates of human immunodeficiency virus type 1 are relatively resistant to neutralization by monoclonal antibodies to gp120 and their neutralization is not predicted by studies with monomeric gp120. J. Virol. 69:101-109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Moore, J. P., and D. D. Ho. 1993. HIV tropism. Nature 361:309-310. [DOI] [PubMed] [Google Scholar]
- 38.Moore, J. P., and D. D. Ho. 1995. HIV-1 neutralization: the consequences of viral adaptation to growth on transformed T cells. AIDS 9(Suppl. A):S117-S136. [PubMed] [Google Scholar]
- 39.Moore, J. P., P. W. H. I. Parren, and D. R. Burton. 2001. Genetic subtypes, humoral immunity, and human immunodeficiency virus type 1 vaccine development. J. Virol. 75:5721-5729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Moore, J. P., and J. Sodroski. 1996. Antibody cross-competition analysis of the human immunodeficiency virus type 1 exterior envelope glycoprotein. J. Virol. 70:1863-1872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nishimura, Y., T. Igarashi, N. Haigwood, R. Sadjadpour, R. J. Plishka, A. Buckler-White, R. Shibata, and M. A. Martin. 2002. Determination of a statistically valid neutralization titer in plasma that confers protection against simian-human immunodeficiency virus challenge following passive transfer of high-titered neutralizing antibodies. J. Virol. 76:2123-2130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nyambi, P. N., H. A. Mbah, S. Burda, C. Williams, M. K. Gorny, A. Nadas, and S. Zolla-Pazner. 2000. Conserved and exposed epitopes on intact, native, primary human immunodeficiency virus type 1 virions of group M. J. Virol. 74:7096-7107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nyambi, P. N., A. Nadas, H. A. Mbak, S. Burda, C. Williams, M. K. Gorny, and S. Zolla-Pazner. 2000. Immunoreactivity of intact virions of human immunodeficiency virus type 1 (HIV-1) reveals the existence of fewer HIV-1 immunotypes than genotypes. J. Virol. 74:10670-10680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Park, E. J., M. K. Gorny, S. Zolla-Pazner, and G. V. Quinnan, Jr. 2000. A global neutralization resistance phenotype of human immunodeficiency virus type 1 is determined by distinct mechanisms mediating enhanced infectivity and conformational change of the envelope complex. J. Virol. 74:4183-4191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Parren, P. W. H. I., and D. R. Burton. 2001. The antiviral activity of antibodies in vitro and in vivo. Adv. Immunol. 77:195-262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Parren, P. W. H. I., P. Fiscaro, A. F. Labrijn, J. M. Binley, W.-P. Yang, H. J. Ditzel, C. F. Barbas I. II, and D. R. Burton. 1996. In vitro antigen challenge of human antibody libraries for vaccine evaluation: the human immunodeficiency virus type 1 envelope. J. Virol. 70:9046-9050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Parren, P. W. H. I., P. A. Marx, A. J. Hessell, A. Luckay, J. Harouse, C. Cheng-Mayer, J. P. Moore, and D. R. Burton. 2001. Antibody protects macaques against vaginal challenge with a pathogenic R5 simian/human immunodeficiency virus at serum levels giving complete neutralization in vitro. J. Virol. 75:8340-8347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Parren, P. W. H. I., J. P. Moore, D. R. Burton, and Q. J. Sattentau. 1999. The neutralizing antibody response to HIV-1: viral evasion and escape from humoral immunity. AIDS 13(Suppl. A):S137-S162. [PubMed] [Google Scholar]
- 49.Parren, P. W. H. I., Q. J. Sattentau, and D. R. Burton. 1997. HIV-1 antibody—debris or virion? Nat. Med. 3:366-367. [DOI] [PubMed] [Google Scholar]
- 50.Parren, P. W. H. I., I. Mondor, D. Naniche, P. J. Klasse, D. Burton, and Q. J. Sattentau. 1998. Neutralization of HIV-1 by antibody to gp120 is determined primarily by occupancy of sites on the virion irrespective of epitope specificity. J. Virol. 72:3512-3519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Roben, P., J. P. Moore, M. Thali, J. Sodroski, C. F. Barbas III, and D. R. Burton. 1994. Recognition properties of a panel of human recombinant Fab fragments to the CD4 binding site of gp120 showing differing ability to neutralize human immunodeficiency virus type 1. J. Virol. 68:4821-4828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sanders, R. W., M. Venturi, L. Schiffner, R. Kalyanaraman, H. Katinger, K. O. Lloyd, P. D. Kwong, and J. P. Moore. 2002. The mannose-dependent epitope for neutralizing antibody 2G12 on human immunodeficiency virus type 1 glycoprotein gp120. J. Virol. 76:7293-7305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Saphire, E. O., P. W. Parren, R. Pantophlet, M. B. Zwick, G. M. Morris, P. M. Rudd, R. A. Dwek, R. L. Stanfield, D. R. Burton, and I. A. Wilson. 2001. Crystal structure of a neutralizing human IgG against HIV-1: a template for vaccine design. Science 293:1155-1159. [DOI] [PubMed] [Google Scholar]
- 54.Saphire, E. O., R. L. Stanfield, M. D. Max Crispin, P. W. Parren, P. M. Rudd, R. A. Dwek, D. R. Burton, and I. A. Wilson. 2002. Contrasting IgG structures reveal extreme asymmetry and flexibility. J. Mol. Biol. 319:9-18. [DOI] [PubMed] [Google Scholar]
- 55.Sattentau, Q. J., and J. P. Moore. 1995. Human immunodeficiency virus type 1 neutralization is determined by epitope exposure on the glycoprotein gp120 oligomer. J. Exp. Med. 182:185-196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sattentau, Q. J., M. Moulard, B. Brivet, F. Botto, J. C. Guillemot, I. Mondor, P. Poignard, and S. Ugolini. 1999. Antibody neutralization of HIV-1 and the potential for vaccine design. Immunol. Lett. 66:143-149. [DOI] [PubMed] [Google Scholar]
- 57.Scanlan, C. N., R. Pantophlet, M. R. Wormald, E. Ollmann Saphire, R. Stanfield, I. A. Wilson, H. Katinger, R. A. Dwek, P. M. Rudd, and D. R. Burton. 2002. The broadly neutralizing anti-human immunodeficiency virus type 1 antibody 2G12 recognizes a cluster of α1→2 mannose residues on the outer face of gp120. J. Virol. 76:7306-7321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Schülke, N., M. S. Vesanen, R. W. Sanders, P. Zhu, M. Lu, D. J. Anselma, A. R. Villa, P. W. H. I. Parren, J. M. Binley, K. H. Roux, P. J. Maddon, J. P. Moore, and W. C. Olson. 2002. Structural and antigenic properties of a proteolytically mature, disulfide-stabilized human immunodeficiency virus type 1 gp140 envelope glycoprotein. J. Virol. 76:7760-7776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Shibata, R., T. Igarashi, N. Haigwood, A. Buckler-White, R. Ogert, W. Ross, R. Willey, M. W. Cho, and M. A. Martin. 1999. Neutralizing antibody directed against the HIV-1 envelope glycoprotein can completely block HIV-1/SIV chimeric virus infections of macaque monkeys. Nat. Med. 5:204-210. [DOI] [PubMed] [Google Scholar]
- 60.Si, Z., M. Cayabyab, and J. Sodroski. 2001. Envelope glycoprotein determinants of neutralization resistance in a simian-human immunodeficiency virus (SHIV HXBc2p 3.2) derived by passage in monkeys. J. Virol. 75:4208-4218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sorensen, K., and U. Brodbeck. 1986. A sensitive protein assay using microtiter plates. Experientia 42:161-162. [Google Scholar]
- 62.Spenlehauer, C., A. Kirn, A.-M. Aubertin, and C. Moog. 2001. Antibody-mediated neutralization of primary human immunodeficiency virus type 1 isolates: investigation of the mechanism of inhibition. J. Virol. 75:2235-2245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Stiegler, G., R. Kunert, M. Purtscher, S. Wolbank, R. Voglauer, F. Steindl, and H. Katinger. 2001. A potent cross-clade neutralizing human monoclonal antibody against a novel epitope on gp41 of human immunodeficiency virus type 1. AIDS Res. Hum. Retrovir. 17:1757-1765. [DOI] [PubMed] [Google Scholar]
- 64.Sullivan, N., Y. Sun, J. Binley, J. Lee, C. F. Barbas III, P. W. Parren, D. R. Burton, and J. Sodroski. 1998. Determinants of human immunodeficiency virus type 1 envelope glycoprotein activation by soluble CD4 and monoclonal antibodies. J. Virol. 72:6332-6338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Thali, M., C. Furman, D. D. Ho, J. Robinson, S. Tilley, A. Pinter, and J. Sodroski. 1992. Discontinuous, conserved neutralization epitopes overlapping the CD4-binding region of human immunodeficiency type I gp120 envelope glycoprotein. J. Virol. 66:5635-5641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Trkola, A., T. Dragic, J. Arthos, J. M. Binley, W. C. Olson, G. P. Allaway, C. Cheng-Mayer, J. Robinson, P. J. Maddon, and J. P. Moore. 1996. CD4 dependent, antibody-sensitive interactions between HIV-1 and its co-receptor CCR-5. Nature 384:184-187. [DOI] [PubMed] [Google Scholar]
- 67.Trkola, A., T. Ketas, V. N. KewalRamani, F. Endorf, J. M. Binley, H. Katinger, J. Robinson, D. R. Littman, and J. P. Moore. 1998. Neutralization sensitivity of human immunodeficiency virus type 1 primary isolates to antibodies and CD4-based reagents is independent of co-receptor usage. J. Virol. 72:1876-1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Trkola, A., A. B. Pomales, H. Yuan, B. Korber, P. J. Maddon, G. P. Allaway, H. Katinger, C. F. Barbas III, D. R. Burton, D. D. Ho, and J. P. Moore. 1995. Cross-clade neutralization of primary isolates of human immunodeficiency virus type 1 by human monoclonal antibodies and tetrameric CD4 IgG. J. Virol. 69:6609-6617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Trkola, A., M. Purtscher, T. Muster, C. Ballaun, A. Buchacher, N. Sullivan, K. Srinivasan, J. Sodroski, J. P. Moore, and H. Katinger. 1996. Human monoclonal antibody 2G12 defines a distinctive neutralization epitope on the gp120 glycoprotein of human immunodeficiency virus type 1. J. Virol. 70:1100-1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ugolini, S., I. Mondor, P. W. H. I. Parren, D. Burton, S. Tilley, P. J. Klasse, and Q. J. Sattentau. 1997. Inhibition of virus attachment to CD4+ target cells is a major mechanism of T cell line-adapted HIV-1 neutralization. J. Exp. Med. 186:1287-1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wu, L., N. P. Gerard, R. Wyatt, H. Choe, C. Parolin, N. Ruffing, A. Borsetti, A. A. Cardoso, E. Desjardin, W. Newman, C. Gerard, and J. Sodroski. 1996. CD4-induced interaction of primary HIV-1 gp120 glycoproteins with the chemokine receptor CCR-5. Nature 384:179-183. [DOI] [PubMed] [Google Scholar]
- 72.Wyatt, R., P. D. Kwong, E. Desjardins, R. W. Sweet, J. Robinson, W. A. Hendrickson, and J. G. Sodroski. 1998. The antigenic structure of the HIV gp120 envelope glycoprotein. Nature 393:705-711. [DOI] [PubMed] [Google Scholar]
- 73.York, J., K. E. Follis, M. Trahey, P. N. Nyambi, S. Zolla-Pazner, and J. Nunberg. 2001. Antibody binding and neutralization of primary and T-cell line-adapted isolates of human immunodeficiency virus type 1. J. Virol. 75:2741-2752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zwick, M. B., A. F. Labrijn, M. Wang, C. Spenlehauer, E. Ollmann Saphire, J. M. Binley, J. P. Moore, G. Stiegler, H. Katinger, D. R. Burton, and P. W. H. I. Parren. 2001. Broadly neutralizing antibodies targeted at the membrane-proximal external region of the human immunodeficiency virus type 1 glycoprotein gp41. J. Virol. 75:10892-10905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zwick, M. B., M. Wang, P. Poignard, G. Stiegler, H. Katinger, D. R. Burton, and P. W. H. I. Parren. 2001. Neutralization synergy of human immunodeficiency virus type 1 primary isolates by cocktails of broadly neutralizing antibodies. J. Virol. 75:12198-12208. [DOI] [PMC free article] [PubMed] [Google Scholar]