Abstract
The Bartha strain of pseudorabies virus has several recognized mutations, including a deletion in the unique short region encompassing the glycoprotein I (gI), gE, Us9, and Us2 genes and point mutations in the gC, gM, and UL21 genes. We have determined that Bartha has mutations in the serine/threonine kinase encoded by the Us3 gene relative to the wild-type Becker strain. Our analysis revealed that Becker virions contain the Us3 protein, whereas Bartha virions do not. To test whether the mutations in the Bartha Us3 protein were responsible for this observation, we constructed a recombinant Bartha strain, PRV632, which expresses the Becker Us3 protein. PRV632 failed to package Us3 into the tegument, indicating that mutations other than those in the Us3 primary amino acid sequence were responsible for the failure of Bartha to package its Us3 protein. A recombinant Becker strain, PRV634, which expresses the Bartha Us3 protein, was constructed to test whether it was capable of being packaged into virions. The Bartha Us3 protein was not incorporated into PRV634 virions efficiently, suggesting that the primary sequence of the Bartha Us3 protein affects packaging into the tegument. To determine whether the packaging of other tegument proteins was affected in the Bartha strain, we examined VP22. Whereas Becker packaged VP22 into virions, Bartha had a severe deficiency in VP22 incorporation. Analysis of VP22 expression in Bartha-infected cells revealed that Bartha VP22 had a slower mobility on sodium dodecyl sulfate-polyacrylamide gels, indicating either primary sequence differences and/or different posttranslational modifications relative to Becker VP22. Taken together, these data indicate that, while the primary sequence of the Us3 protein does affect its incorporation into the tegument, other factors are involved. Furthermore, our data suggest that one or more of the gI, gE, Us9, or Us2 genes influences the localization of the Us3 protein in infected cells, and this effect may be important for the proper incorporation of Us3 into virions.
The Alphaherpesvirinae subfamily of the Herpesviridae family contains neurotropic viruses, such as herpes simplex virus type 1 (HSV-1), HSV-2, varicella-zoster virus, and the swine pathogen pseudorabies virus (PRV) (39). All herpesvirus virions consist of four distinct structures: an inner core that contains the linear double-stranded DNA genome, an icosahedral capsid, a proteinaceous layer termed the tegument, and a host-derived lipid envelope decorated with viral glycoproteins (38, 39). The tegument is a complex network of over 15 proteins that may act as a “bridge” between the viral capsid and viral glycoproteins in the lipid envelope (reviewed in reference 30). In support of this notion, deleting the UL36, UL37, and UL48 genes (coding for the tegument proteins VP1/2, UL37, and VP16, respectively) resulted in the accumulation of nonenveloped capsids in the cytoplasm of infected cells (6, 20, 22, 30). From these studies, it has become clear that certain tegument proteins are required for the subsequent addition of other tegument proteins as well as secondary envelopment into vesicles of the trans-Golgi network. However, several tegument proteins have been shown to be nonessential for virion formation. These proteins are VP18.8, virion host shutoff factor, VP11/12, VP13/14, VP22, and Us3, products of the UL13, UL41, UL46, UL47, UL49, and Us3 genes, respectively (30, 33, 34, 38). Despite the requirement of some key proteins, the tegument displays flexibility with regard to what proteins are packaged and in what stoichiometry.
The Us3 gene encodes a serine/threonine kinase (11, 28) that has many roles during viral infection. Several studies have reported that the Us3 gene product is involved in virion morphogenesis, specifically aiding in the deenvelopment of perinuclear virions (21, 44). Other reports have shown that the Us3 protein plays a role in the inhibition of both HSV-1- and HSV-2-induced apoptosis (14, 15, 19, 24). Furthermore, it has been shown that the Us3 protein is involved in the cell-to-cell spread of virus infection in a cell-type-dependent manner (5).
In the process of characterizing the Us3 protein and its involvement in cell-to-cell spread, Demmin et al. noted that the attenuated PRV Bartha strain was unable to spread between cells efficiently when the levels of Us3 expression were reduced; in contrast, the wild-type PRV Becker strain was able to tolerate reduced levels of Us3 expression and spread between cells efficiently (5). These observations prompted us to examine the Us3 gene of Bartha. Here we report that, unlike the wild-type Becker strain, the attenuated Bartha strain is unable to incorporate the Us3 protein into virions. Evidence is provided that while the primary sequence of the Us3 protein is important for incorporation into the tegument, other factors present in virus-infected cells are required for this process. Upon further analysis, it became evident that the tegument protein VP22 is not packaged into PRV Bartha virions either. The results of this study demonstrate that PRV virions can tolerate the loss of multiple tegument proteins without a substantial effect on virus infectivity and further support the notion that the incorporation of “nonessential” tegument proteins is likely mediated by complex protein-protein interactions.
MATERIALS AND METHODS
Viruses and cells.
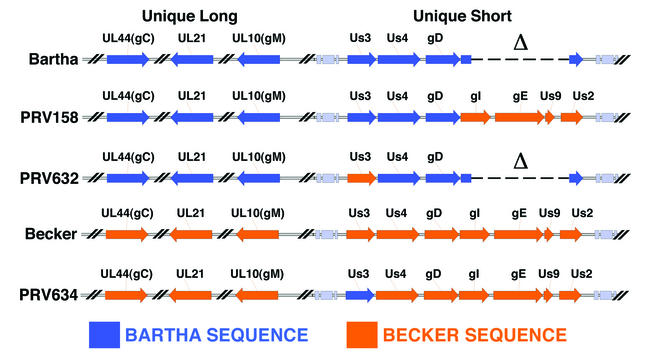
The virus strains used in this study are shown in Fig. 1. The attenuated live-vaccine strain Bartha (1) has mutations in the glycoprotein C (gC), gM, and UL21 genes and a deletion in the unique short region spanning the gI, gE, Us9, and Us2 genes (7, 23, 26, 37). The wild-type Becker strain was kindly provided by L. W. Enquist, Princeton University. PRV158 was constructed by homologous recombination between plasmid pALM94 (3) containing the BamHI 7 fragment from Becker and the Bartha genome. Virus produced after cotransfection was plated on PK15 cells, and recombinant virus was screened for gE expression by a “black-plaque assay” with a pool of gE monoclonal antibodies (17, 41). Briefly, virus from cotransfection experiments was plated on cells growing on a 100-mm dish (200 to 300 plaques/dish) and incubated for 2 days. The medium was then removed from the dish, and the cells were washed three times with phosphate-buffered saline (PBS). Next, 1 ml of a cocktail of monoclonal antibodies specific to gE was added to the dish and incubated for 1 h at room temperature. The primary antibodies then were removed from the dish, and the cells were washed three times with PBS. Next, horseradish peroxidase-conjugated goat anti-mouse secondary antibodies (1 ml) were added to the dish and incubated at room temperature for 1 h. The secondary antibodies then were removed, and the cells were washed three times with PBS. The substrate was prepared by adding 10 mg of 4-chloro-1-naphthol to 1 ml of 100% ethanol. This mixture was added slowly to 99 ml of PBS, and 0.1 ml of 3% H2O2 was added to the solution. Five milliliters of the substrate was added to the cells and allowed to incubate until color developed (15 min). After color development, the substrate was removed and the monolayer was washed once with PBS. Black plaques were then picked and subjected to three rounds of purification by using the above-mentioned assay. Southern blot analysis was performed to verify that recombination of the BamHI fragment into the PRV Bartha genome had occurred as expected.
FIG. 1.
PRV strains used in this study. Sequences of PRV vaccine strain Bartha and PRV wild-type strain Becker are shown. The PRV genome is divided into two regions: the unique long and the unique short. Bartha contains mutations in the gC, UL21, and gM genes in the unique long region and a large deletion spanning the gI, gE, Us9, and Us2 genes in the unique short region.
To construct PRV631, a region of approximately 500 bp upstream and downstream of the PRV Us3 gene was amplified by PCR with the Bartha genome as a template. The upstream fragment was amplified by using the upstream forward primer with the sequence 5′-CGGAATTCCGAGAGCGTTTATTGTTAAAGTTGTTG-3′ (see below for explanation of lettering) and the downstream reverse primer with the sequence 5′-CAAAGGTGTGTGTGTCCTACCGCTCGATGCATTGACCCATCTCCACATCGCCAACAT-3′. These primers introduced an EcoRI site into the 5′ end and an NsiI site into the 3′ end of the PCR product. The downstream fragment was amplified by using the upstream forward primer with the sequence 5′-ATGTTGGCGATGTGGAGATGGGTCAATGCATCGAGCGGTAGGACACACACACCTTTG-3′ and the downstream reverse primer with the sequence 5′-GGGGTACCCCAAAGACGAGCACGACGATGTACAGG-3′. Roman type represents sequences homologous to the virus, and italic type represents sequences introduced for cloning purposes. Both blunt-ended PCR products were cloned separately into the pCR-Blunt II-TOPO vector (TOPO cloning kit; Invitrogen, Carlsbad, Calif.), creating plasmids pMLTO1 (upstream PCR product) and pMLTO2 (downstream PCR product). Next, pMLTO1 was digested with EcoRI and NsiI and pMLTO2 was digested with NsiI and KpnI. Both PCR products were gel purified and cloned into pBluescript KS(+) via a three-piece ligation to generate plasmid pML6. Next, a 2.3-kbp NsiI fragment obtained from pEGFP-C1 (Clontech, Palo Alto, Calif.) and containing the cytomegalovirus immediate-early promoter, enhanced green fluorescent protein (EGFP) sequences, and a simian virus 40 poly(A) signal was cloned into a unique NsiI site to create plasmid pML7. The latter plasmid was digested with SpeI (to linearize the plasmid) and cotransfected with purified Bartha genomic DNA into PK15 cells. Virus produced after cotransfection was plated on PK15 cells, and plaques expressing EGFP were identified by using a Nikon TE200 inverted epifluorescence microscope. Virus was isolated from EGFP-expressing plaques and subjected to three rounds of purification. This recombinant virus was designated PRV631.
To construct PRV632, purified PRV631 DNA was cotransfected with pGD7. Briefly, pGD7 was constructed by digesting Becker genomic DNA with NotI and cloning the NotI fragment that contains 200 bp upstream of Us3, the entire Us3 open reading frame (ORF), and approximately 1 kbp of the 5′ end of the gG gene into pBluescript KS(+). Following cotransfection of purified PRV631 genomic DNA and linearized pGD7 into PK15 cells, virus was plated on PK15 cells; plaques not expressing EGFP (non-green) were identified by using a Nikon TE200 inverted epifluorescence microscope and subjected to three rounds of plaque purification. Southern blot analysis was performed with both PRV631 and PRV632 genomic DNAs to verify that appropriate recombination had occurred. Us3 protein expression in PRV631- or PRV632-infected cell lysates was examined by Western blotting to confirm the absence or presence of Us3 expression, respectively.
To construct PRV633, pML7 was cotransfected with purified PRV Becker DNA into PK15 cells. Virus produced after transfection was plated on PK15 cells; EGFP-expressing plaques were isolated and subjected to three rounds of purification. This recombinant virus was designated PRV633. To construct PRV634, purified PRV633 DNA was cotransfected with the NotI/AspEI fragment from pGD2 which contains a NotI fragment from the Bartha strain cloned into pBluescript. This fragment contains 200 bp upstream of Us3, the Bartha Us3 ORF, and 235 bp of the 5′ end of the gG gene. Following cotransfection of PRV633 genomic DNA and linearized pGD2, virus was plated on PK15 cells; plaques not expressing EGFP (non-green) were identified and subjected to three rounds of plaque purification. Southern blot analysis was performed with PRV633 and PRV634 genomic DNAs to verify that the correct recombination events had occurred. Us3 protein expression in PRV633- or PRV634-infected cell lysates was examined by Western blotting to confirm the absence or presence of Us3 expression, respectively.
All virus strains were propagated and titers were determined on PK15 cells. PK15 cells were maintained at 37°C in Dulbecco's modified Eagle's medium supplemented with 10% fetal calf serum (Gibco/BRL, Grand Island, N.Y.) in a 5% CO2 environment.
Antisera.
Antisera against a Us3 peptide were raised in rabbits and affinity purified by Bethyl Laboratories (Montgomery, Tex.) as described previously (5). Antisera to gB, gE, and VP22 were kindly provided by L. W. Enquist. The VP22 antisera were recently described by del Rio et al. (4).
Virus purification.
Virus was purified as described previously (2). Briefly, three confluent 150-mm-diameter dishes of PK15 cells were infected with virus (multiplicity of infection [MOI], 10). At 16 h postinfection, the medium was collected and centrifuged at 3,000 rpm in a Sorvall ST-H750 rotor (Sorvall Super T21 centrifuge) to remove cellular debris. The clarified supernatant was layered on a 30% sucrose cushion and centrifuged in an SW28 rotor at 23,000 rpm for 3 h. The sucrose cushion was removed, and the virion pellet was resuspended in 0.5 ml of PBS by 10 1-s pulses in a bath sonicator. Virions were then centrifuged through a 1-ml 30% sucrose cushion at 28,000 rpm for 90 min in an SW55ti rotor. The pelleted virions were resuspended in PBS and stored in aliquots at −80°C.
Protease treatment of virions.
Isolated virions were treated with 10 μg of proteinase K (PK) (Fisher, Fair Lawn, N.J.) per ml in either the presence or the absence of 1% IGEPAL (NP-40). After incubation for 60 min at room temperature, phenylmethylsulfonyl fluoride was added to each sample to a final concentration of 2 mM to inhibit further proteolysis (2). Samples were immediately loaded onto sodium dodecyl sulfate (SDS)- 12% polyacrylamide gels and transferred to Immobilon-P membranes (Millipore, Bedford, Mass.). Us3 or VP22 was detected with specific antiserum.
Western blot analysis.
Purified virus and cellular extracts were electrophoresed on SDS-12% polyacrylamide gels and transferred to Immobilon-P membranes by using a Bio-Rad (Hercules, Calif.) semidry transfer apparatus in accordance with the manufacturer's instructions. Membranes were blocked by using 2% bovine serum albumin (BSA). Proteins were visualized by using rabbit (for Us3 and VP22) or goat (for gB) polyclonal antibodies, horseradish peroxidase-conjugated secondary antibodies, and enhanced chemiluminescence detection as recommended by the manufacturer (Amersham Biosciences, Little Chalfont, United Kingdom).
Cloning and sequencing of Bartha Us3 and Becker Us3.
Bartha and Becker genomic DNAs were digested with NotI, and 2.4-kbp NotI fragments containing the entire Us3 ORFs were cloned into pBluescript KS(+). Overlapping primers were then used to sequence Becker and Bartha 1.1-kbp Us3 ORFs in both directions. Sequencing was performed by the UCHSC Cancer Center DNA Sequencing and Analysis Core.
Indirect immunofluorescence microscopy.
PK15 cells were grown on glass coverslips to 30 to 40% confluence. Cells were then infected with Becker, Bartha, PRV158, and PRV632 and incubated for 2, 3, 5, or 6 h. At the appropriate time after infection, cells were rinsed three times with PBS and fixed in 4% paraformaldehyde in PBS for 10 min. Cells were rinsed gently with PBS and permeabilized in PBS containing 1% BSA (PBS/BSA) and 0.1% Triton X-100 at room temperature for 3 min. Cells were washed and incubated for 45 min with anti-Us3 antibodies diluted 1:2,000 in PBS/BSA. Anti-Us3 antibodies were removed, and the cells were rinsed three times with PBS/BSA and incubated for 30 min with Alexa-488 goat anti-rabbit antibodies (Molecular Probes, Eugene, Oreg.) diluted 1:500 in PBS/BSA. The secondary antibodies were removed, and the cells were rinsed three times with PBS/BSA and then three times with PBS. Coverslips were mounted on glass slides, and images were captured by using a Ziess Axiophot epifluorescence microscope equipped with a mechanical stage and a cooled charge-coupled device camera. Series of z images were deconvolved by using Slidebook 3.0.7.3 software (Intelligent Imaging Innovations Inc., Denver, Colo.), and images of 0.5-μm optical sections through the middle of cells were exported into Adobe Photoshop 6.0 (Adobe Systems Inc., San Jose, Calif.) for the construction of image composites.
RESULTS
Packaging of Us3 into PRV virions.
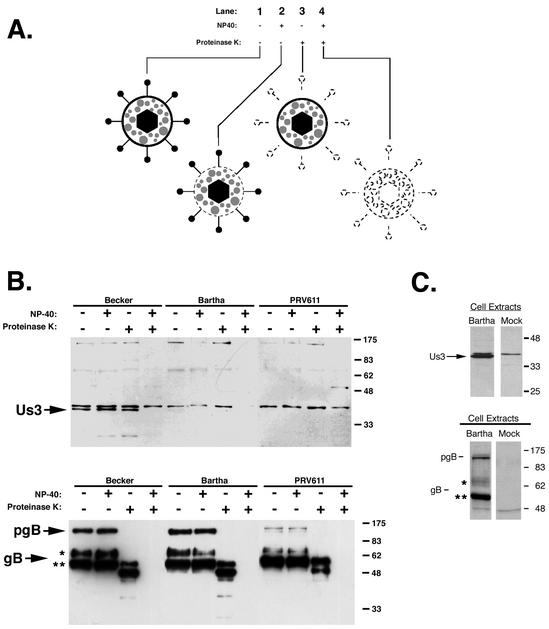
The alphaherpesvirus Us3 gene encodes a serine/threonine kinase that is packaged into purified virions (47). Us3 has been reported to be a tegument protein, although, to our knowledge, no data confirming this report have been published (30). To confirm that the Us3 protein was packaged into the tegument of PRV strains Becker and Bartha, we subjected purified virions to a protease protection assay, which is summarized in Fig. 2A, lane 2). Purified virions were treated with PBS, NP-40 (a mild detergent), PK, or both NP-40 and PK. Treated virions were then subjected to SDS-polyacrylamide gel electrophoresis (PAGE), and the Us3 protein was detected by Western blotting. In Fig. 2A, lane 1, purified virions were treated with PBS alone to serve as a control for virion integrity. In lane 2, virions were treated with NP-40 alone to solubilize the lipid envelope. In lane 3, virions were treated with PK alone to degrade proteins exterior to the lipid envelope, namely, the ectodomains of viral glycoproteins. As shown in Fig. 2A, lanes 1 to 3, treatment with PBS, NP-40 alone, or PK alone did not affect the tegument. As shown in Fig. 2A, lane 4, where virions were treated with both NP-40 and PK, NP-40 treatment made tegument proteins susceptible to PK digestion. Therefore, Us3 (and all tegument proteins) should be detected by Western blotting under the first three conditions but not under the fourth condition.
FIG. 2.
The Us3 protein is found in the tegument of purified virions. (A) Diagram of the protease protection assay used in this study. Purified virions were treated with PBS (lane 1), NP-40 (lane 2), PK (lane 3), or both NP-40 and PK (lane 4). Effects of treatment on individual virus particles are shown. Note that tegument proteins remain unaffected by the first three treatments and are degraded by the fourth treatment. (B) Purified Becker, Bartha, and PRV611 (Us3 null) virions were treated with NP-40 and PK, separated on 12% polyacrylamide gels, and analyzed by Western blotting with rabbit polyclonal antiserum specific for Us3 or goat polyclonal antiserum specific for gB. The preprocessed form of gB (pgB) is labeled, as are the 69-kDa (*) and the 58-kDa (**) subunits. (C) PK15 cells were infected with PRV Bartha at an MOI of 10. At 6 h postinfection, extracts from infected and mock-infected cells were prepared and subjected to Western blot analysis with antiserum against Us3 or gB.
An analysis of Us3 protein packaging into Becker and Bartha virions revealed that wild-type Becker virions packaged Us3, whereas the attenuated vaccine strain Bartha did not (Fig. 2B). PRV611 is a Us3-null virus and was used as a negative control in this experiment (J. Randall and B. Banfield, unpublished data). It is noteworthy that the Us3 protein was synthesized in abundance in Bartha-infected cells, despite not being packaged into mature virions (Fig. 2C). A Western blot with gB served as a virus loading control and a control for PK activity in the absence of NP-40.
PRV gB is a type I membrane protein (36) that is cleaved in the Golgi apparatus (45) from the preprocessed monomer into two smaller subunits that are linked by disulfide bonds. These subunits have molecular masses of 69 and 58 kDa and represent the amino-terminal and carboxy-terminal “halves” of the molecule, respectively (27, 31, 46). Both the preprocessed monomer and the two smaller subunits are packaged into mature virions, albeit mature virions are enriched for the processed gB subunits. When purified virions were treated with either PBS or NP-40, the preprocessed and processed forms of gB remained unaffected (Fig. 2B, lower panel). In the presence of PK alone, the preprocessed gB form was cleaved, as was the 69-kDa amino-terminal subunit, which constitutes most of the gB ectodomain. The 58-kDa subunit, part of which contains the transmembrane domain and the cytoplasmic tail, remained intact but was degraded in the presence of both NP-40 and PK (Fig. 2B, lower panel, +NP-40/+PK). It is noteworthy that the gB antiserum reacted strongly with the 58-kDa gB subunit. However, the 69-kDa gB subunit did not react as strongly with our antiserum and also ran as a smear on Western blots (Fig. 2C, lower panel). We believe this result was due to the fact that the 69-kDa subunit is heavily glycosylated.
Our Us3 antiserum also cross-reacted with a host cell protein that was slightly larger than the Us3 protein on SDS-polyacrylamide gels. This band was detected in our mock-infected cell lysates (Fig. 2C). The “mock” band was not consistently found in all of our samples, but when it was present, it was resistant to PK treatment.
These data confirmed that the Us3 protein was found in the tegument of mature PRV virions. They also indicated that the Us3 protein was packaged into wild-type Becker virions but was not packaged into Bartha virions.
Us3 localization in Becker- and Bartha-infected cells.
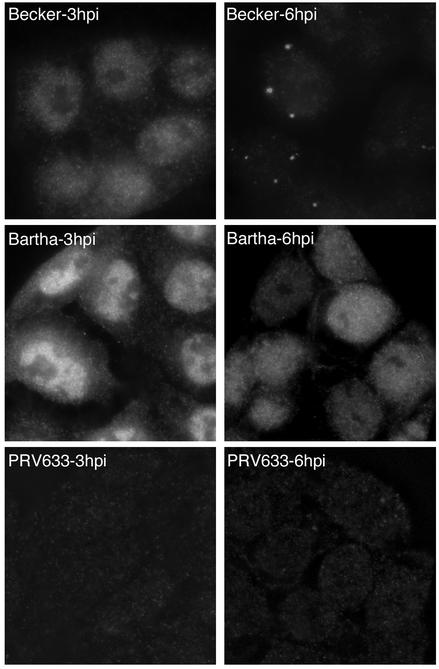
To further characterize the Us3 protein, we examined the localization of Us3 in infected PK15 cells by indirect immunofluorescence microscopy. At 3 h postinfection, Us3 localized primarily to the nuclei of both Becker- and Bartha-infected cells (Fig. 3). At 6 h postinfection, Becker Us3 localized to small, punctate structures in the cytoplasm. However, Bartha Us3 was found diffusely dispersed throughout the nuclei and the cytoplasm. Cells infected with a Becker mutant, PRV633, which has a deletion of Us3, failed to react with our Us3 antiserum, indicating the specificity of this reagent. We found these punctate structures intriguing, and we hypothesized that these structures represent areas of tegument assembly or, more specifically, areas where the Us3 protein was incorporated into the tegument of maturing virus. This hypothesis is consistent with Becker being able to package Us3 into the tegument and Bartha not being able to package Us3. Becker Us3 may be targeted to these areas for incorporation, whereas Bartha Us3 remains dispersed throughout the cytoplasm.
FIG. 3.
Localization of the Us3 protein in Becker-, Bartha-, and PRV633-infected cells by indirect immunofluorescence microscopy. PK15 cells grown on glass coverslips were infected with PRV Becker or PRV Bartha at an MOI of 10 for 3 or 6 h. Cells were fixed and stained with rabbit antiserum to Us3 followed by an Alexa-488-conjugated secondary antibody. PRV633 is a Becker derivative that has a deletion of Us3.
Roles of gI, gE, Us9, and Us2 in the packaging of Us3 into the tegument.
The largest genomic difference between the Becker and Bartha PRV strains is a substantial deletion in the unique short region of Bartha spanning the gI, gE, Us9, and Us2 genes. A virus isogenic relative to Bartha, PRV158, was constructed previously (5) by repairing this large deletion with a Becker sequence (Fig. 1). In this study, we used PRV158 to determine the roles of gI, gE, Us9, and Us2 in Us3 localization and packaging into virions.
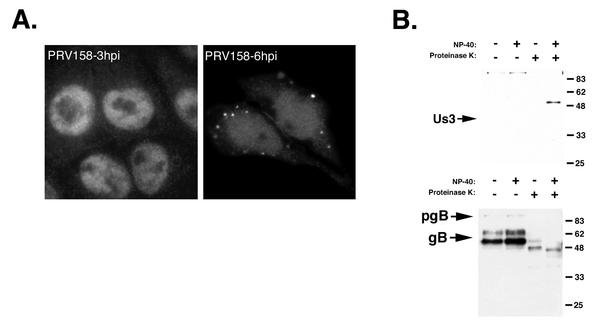
Indirect immunofluorescence microscopy was performed on PRV158-infected cells to determine Us3 localization at 3 and 6 h postinfection. Interestingly, repairing gI, gE, Us9, and Us2 expression restored Us3 localization to punctate cytoplasmic structures at 6 h postinfection, similar to what was observed in Becker-infected cells (Fig. 4A). In contrast, repairing the Bartha deletion did not restore Us3 protein packaging into virions (Fig. 4B). These data indicated that one or more of gI, gE, Us9, and Us2 may be responsible for targeting the Us3 protein to the site of tegument assembly and suggested that the Bartha Us3 protein may be incapable of incorporation into virions.
FIG. 4.
Localization of the Us3 protein in PRV158-infected cells and Us3 incorporation into PRV158 virions. (A) PK15 cells growing on glass coverslips were infected with PRV158 at an MOI of 10 for 3 or 6 h. Cells were fixed and stained with rabbit antiserum to Us3 followed by an Alexa-488-conjugated secondary antibody. (B) Purified PRV158 virions were treated with NP-40 and/or PK, separated on a 12% polyacrylamide gel, and analyzed by Western blotting with rabbit antiserum specific for Us3 or goat antiserum specific for gB. The band that migrates at approximately 55 kDa in the lane in which NP-40 and PK were used on the Us3 blot is a degradation product of a nonspecific band with which our antiserum cross-reacts and that migrates at approximately 100 kDa in the other three lanes. This larger protein is cleaved in the presence of NP-40 and PK, therefore allowing the detection of this smaller degradation product. pgB, preprocessed form of gB.
Bartha has mutations in Us3 relative to Becker.
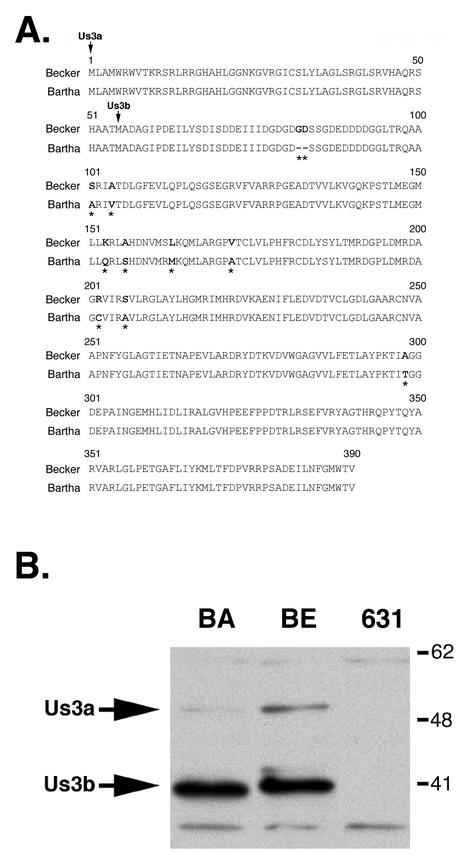
We considered that there could be amino acid sequence differences between the Becker and Bartha Us3 proteins. To further explore this idea, the Us3 ORFs were cloned from both Becker and Bartha and sequenced. Sequence analysis revealed nine amino acid changes and a two-amino-acid deletion in Bartha Us3 compared to Becker Us3 (Fig. 5A). The Us3 ORF codes for two proteins, Us3a and Us3b. Us3a has a molecular mass of approximately 53 kDa and is less abundant in infected cells than the smaller Us3b, which has a molecular mass of 41 kDa (Fig. 5B) (43). The smaller form, Us3b, has been detected in mature virions (47). Furthermore, Bartha Us3b migrated faster than Becker Us3b on SDS-polyacrylamide gels (Fig. 5B). Primary amino acid sequence differences between the Us3 proteins from the two strains may account for the difference in electrophoretic mobility. Upon discovering the primary amino acid differences between Becker and Bartha, we wondered whether the primary amino acid sequence determines whether Us3 is packaged into the tegument. In order to answer this question, we constructed a recombinant Bartha virus that expressed the wild-type Us3 protein.
FIG. 5.
Amino acid sequence comparison between Becker and Bartha Us3 proteins. (A) Amino acid differences between Becker and Bartha Us3 proteins are shown in bold type and highlighted by an asterisk under the Bartha Us3 sequence. The start sites of the Us3a and Us3b sequences are indicated by vertical arrows. (B) Extracts from Bartha (BA)-, Becker (BE)-, and PRV631 (Us3 null) (631)-infected cells were prepared at 6 h postinfection. Extracts were subjected to SDS-PAGE with a 12% gel and subsequently analyzed by Western blotting with Us3-specific antiserum. Note the difference in mobility between Bartha and Becker Us3 proteins. Numbers at right are in kilodaltons.
PRV632 and Us3 packaging into the tegument.
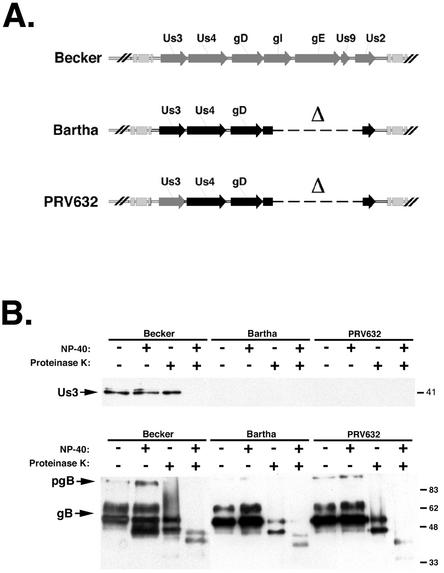
We wished to determine whether the primary amino acid sequence differences between the Becker and Bartha Us3 proteins were responsible for the Us3 packaging difference. To test this hypothesis, we constructed PRV632. PRV632 is isogenic relative to Bartha, with the only difference being a replacement of the Bartha Us3 gene with the Becker Us3 gene (Fig. 6A). Western blot analysis of PRV632-infected cell lysates confirmed that PRV632 expressed the more slowly migrating Becker Us3 (data not shown). However, PRV632 did not package Us3 into virions (Fig. 6B). In this experiment, Becker was used as a positive control and Bartha was used as a negative control. We concluded from this experiment that the primary amino acid sequence alone does not determine whether the Us3 protein is packaged into the tegument of mature virus. These data also suggested that in order to be packaged into mature virions, Us3 may need to interact with other proteins.
FIG. 6.
Us3 protein incorporation into PRV632 virions. (A) Diagram of the PRV unique short regions of Becker, Bartha, and PRV632. PRV632 is isogenic relative to Bartha and expresses Becker Us3. Bartha sequences are shown in black, and Becker sequences are shown in gray. (B) Purified Becker, Bartha, and PRV632 virions were treated with NP-40 and PK and subjected to SDS-PAGE, and Us3 and gB were analyzed by Western blotting. pgB, preprocessed form of gB. Numbers at right are in kilodaltons.
Us3 localization in PRV632-infected cells.
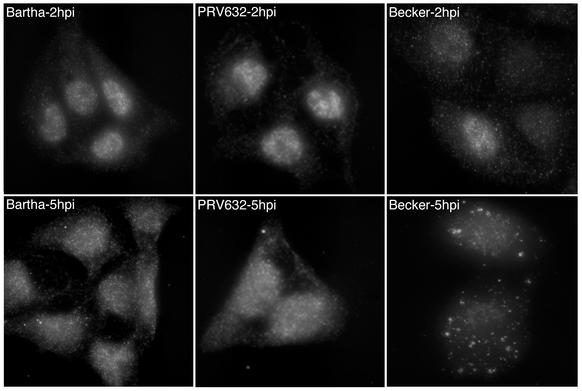
We examined Us3 localization in PRV632-infected cells to determine whether it was consistent with our hypothesis that the punctate cytoplasmic structures observed in infected cells are important for tegument assembly (Fig. 7). The Us3 protein was found in the nuclei of Bartha-, PRV632-, and Becker-infected cells at 2 h postinfection. At 5 h postinfection, PRV632 Us3 localization was spread diffusely throughout the nucleus and the cytoplasm and was indistinguishable from the localization of Bartha Us3 at this time postinfection. In contrast, Us3 in Becker-infected cells localized to punctate cytoplasmic structures at 5 h postinfection. We concluded from these data that PRV632, despite expressing a “packageable” Us3 protein, is unable to localize Us3 to sites of tegument assembly in the cytoplasm.
FIG. 7.
Localization of the Us3 protein in Bartha-, PRV632-, and Becker-infected cells by indirect immunofluorescence microscopy. PK15 cells were grown on glass coverslips and infected with Bartha, PRV632, and Becker at an MOI of 10 for 2 or 5 h. Cells were fixed and stained with rabbit antiserum to Us3 followed by an Alexa-488-conjugated secondary antibody.
Us3 packaging in PRV634.
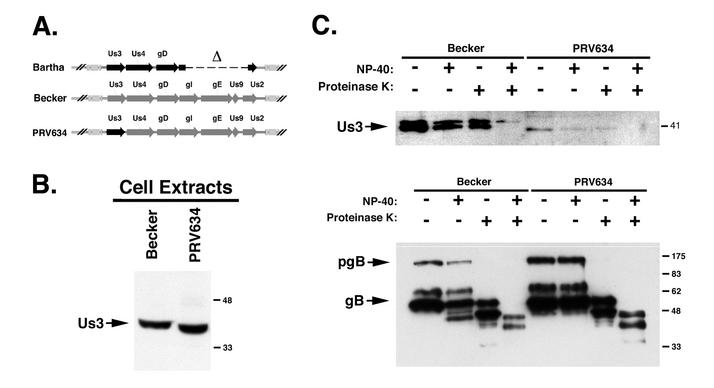
Repairing the deletion in the unique short region of Bartha (PRV158) restored the localization of the Us3 protein to the punctate cytoplasmic structures, where we believe Us3 is incorporated into virus particles. Because PRV158 did not package Us3 into mature virions and because of the differences between Bartha Us3 and Becker Us3, we tested whether Bartha Us3 was capable of being packaged. To do so, we constructed a new virus, PRV634, which is isogenic relative to Becker but expresses the Bartha Us3 gene at the Us3 locus (Fig. 8A). As mentioned above, the Bartha Us3 protein migrated faster in SDS-PAGE than the Becker Us3 protein. This difference in electrophoretic mobility provided a convenient assay to confirm that the correct Us3 protein was expressed in our recombinant viruses. PRV634 was shown to express Bartha Us3 in infected cell lysates (Fig. 8B). As shown in Fig. 8C, the Bartha Us3 protein was not packaged into mature PRV634 virions. From these data, we concluded that Bartha is unable to package Us3 into virions for two reasons: (i) the Bartha Us3 protein has mutations in the primary amino acid sequence that affect packaging, and (ii) other proteins that interact with Us3 and assist in tegument incorporation are absent or mutated in Bartha.
FIG. 8.
Us3 protein incorporation into PRV634 virions. (A) Diagram of the unique short regions of Bartha, Becker, and PRV634. PRV634 is isogenic relative to Becker and expresses Bartha Us3. Bartha sequences are shown in black, and Becker sequences are shown in gray. (B) Extracts from Becker- and PRV634-infected cells were prepared at 6 h postinfection. Extracts were subjected to Western blot analysis with Us3-specific antiserum. Note the shift in Us3 mobility between Becker and PRV634, a recombinant Becker virus expressing Bartha Us3. Numbers at right are in kilodaltons. (C) Purified Becker and PRV634 virions were treated with NP-40 and PK, separated on a 12% polyacrylamide gel, and analyzed by Western blotting with Us3-specific antiserum. The mock band is the top band (resistant to both NP-40 and PK), while Us3 is the lower band. The Western blot for gB served as a loading control and as a control for protease activity. pgB, preprocessed form of gB.
Packaging of the major tegument protein VP22.
To determine whether the packaging of other tegument proteins was affected in the Bartha strain, we examined the incorporation of VP22 into Becker and Bartha virions. VP22 is a major tegument protein that has received much interest because of the remarkable ability of the HSV-1 homolog to spread between cells (8, 32, 40). PRV has a VP22 homolog that was characterized recently by del Rio et al. (4).
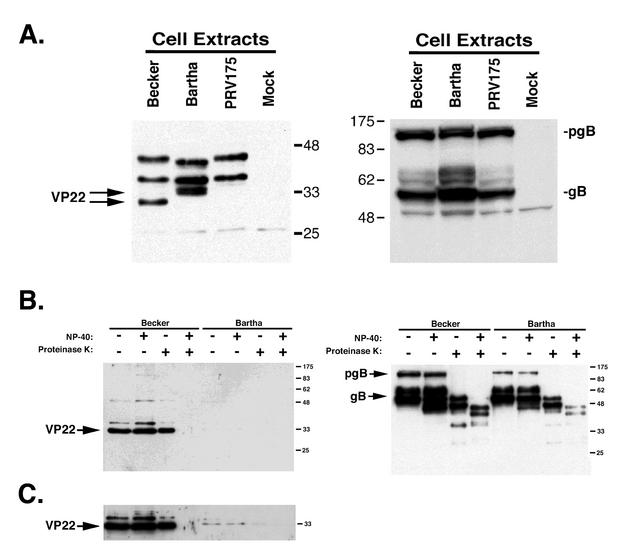
VP22 was detected in Becker- and Bartha-infected cell lysates at 6 h postinfection (Fig. 9A). Interestingly, Becker VP22 migrated faster in SDS-PAGE than Bartha VP22. Bartha VP22 also migrated as two distinct forms, which likely represent different phosphorylated forms of the protein (4). Becker VP22 has been shown to exist in three isoforms, two of which are phosphorylated and one of which, a smaller form, is not phosphorylated (4). However, in Becker-infected cell lysates, the two phosphorylated forms of VP22 were not detected at 6 h postinfection but were present by 16 h postinfection (4). PRV175 is a Becker VP22-null virus (4) and was used as a negative control in this experiment. Western blots with gB served as both loading and infection controls.
FIG. 9.
VP22 incorporation into Becker and Bartha virions. (A) Extracts from Becker-, Bartha-, PRV175 (VP22 null)-, and mock-infected cells were prepared at 6 h postinfection. Bartha VP22 runs more slowly in SDS-PAGE and migrates as a tight doublet, whereas Becker VP22 runs faster and migrates as a single band. The Western blot for gB served as a loading control. pgB, preprocessed form of gB. Numbers beside gels are in kilodaltons. (B) Purified Becker and Bartha virions were treated with NP-40 and PK and subjected to SDS-PAGE and Western blot analysis with VP22- or gB-specific antiserum. This film was exposed for 30 s. (C) Same blot as in panel B but with film exposed for 3 min.
We tested whether Becker and Bartha incorporated similar amounts of VP22 into mature, cell-free virions. Becker and Bartha were grown in PK15 cells and purified as described in Materials and Methods. Purified virus was subjected to a protease protection assay (Fig. 9B). Becker packaged VP22 efficiently; however, Bartha had a major deficiency in packaging VP22 into virions. It is noteworthy that we were able to detect minute amounts of VP22 in Bartha virions after a longer exposure of the Western blot (Fig. 9C). From these data, we concluded that Bartha is unable to package VP22 efficiently.
Roles of gE and VP22 packaging in Bartha.
It has been reported that VP22 interacts with the cytoplasmic tails of gE and gM in a yeast two-hybrid analysis (13, 30). It has also been reported that gE or gM must be present for VP22 to be incorporated into mature virions (12, 13). Bartha, as mentioned above, has a deletion of gE and also has mutations within the N glycosylation motif of gM that result in the expression of nonglycosylated gM (7). We reasoned that Bartha might not package VP22 because of the absence or mutation of two key glycoproteins that have been shown to interact with VP22. We hypothesized that by repairing gE in Bartha, we might restore VP22 incorporation into the tegument.
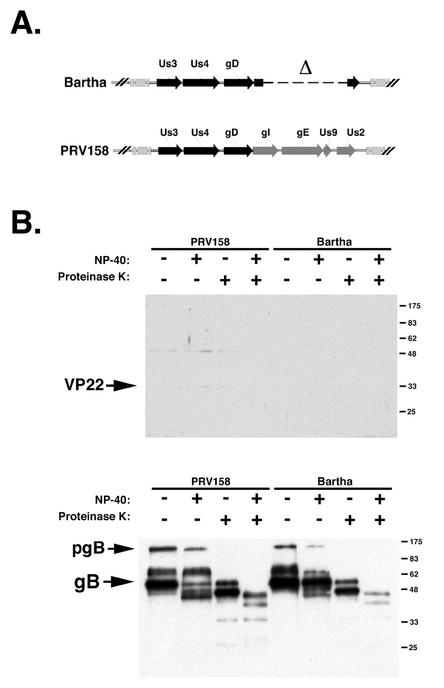
To test this hypothesis, we investigated the incorporation of VP22 into PRV158. Recall that PRV158 is a Bartha derivative with a repair of the large deletion in the unique short region spanning the gI, gE, Us9, and Us2 genes (Fig. 10A). The results showed that repairing the gE deletion in Bartha did not restore VP22 packaging (Fig. 10B). Becker was also examined in this experiment and served as a positive control for VP22 packaging (Fig. 9B). The gB Western blot confirmed that enough virus was loaded into all lanes to detect VP22, if present. We concluded from these data that the inability of Bartha to package VP22 was not related to the absence of gE expression.
FIG. 10.
VP22 incorporation into PRV158 virions. (A) Diagram of the unique short regions of Bartha and PRV158. Bartha sequences are shown in black, and Becker sequences are shown in gray. (B) Purified PRV158 and Bartha virions were treated with NP-40 and PK, separated by SDS-PAGE, and subjected to Western blot analysis with VP22- or gB-specific antiserum. pgB, preprocessed form of gB. Numbers at right are in kilodaltons.
DISCUSSION
The alphaherpesvirus Us3 protein is believed to be a tegument protein and has been detected in purified virions (47). Mettenleiter and colleagues have performed an immunoelectron microscopy study of mutant viruses with defects in envelopment and have detected the Us3 protein associated with the tegument material in large cytoplasmic aggregates (T. Mettenleiter, personal communication). The results of this study support previous observations suggesting that Us3 is found in the tegument of purified PRV virions (Fig. 2). Furthermore, we determined that vaccine strain Bartha did not package Us3, whereas wild-type strain Becker did package Us3 (Fig. 2). We also found that Bartha had a major deficiency in packaging of a second tegument protein, VP22 (Fig. 9B). The importance of these findings is discussed below.
The PRV Bartha strain has several known mutations in its genome. Mutations exist in the gM (7), gC (37), and UL21 (23) genes, and there is a large deletion spanning the gI, gE, Us9, and Us2 genes (26). We sequenced the Us3 gene from the Bartha strain and discovered several mutations that result in nine amino acid differences and a two-amino acid deletion relative to the wild-type Becker strain (Fig. 5A). Thus, we add the Us3 gene to the growing list of Bartha genes that contain mutations. We further hypothesized that Bartha VP22 (encoded by the UL49 gene) contains mutations as well, on the basis of the different mobilities of the Bartha and Becker VP22 proteins observed in SDS-PAGE (Fig. 9A). However, we are unsure whether this difference in mobilities is due to the primary amino acid sequence or posttranslational modifications. We are in the process of cloning and sequencing the Becker and Bartha UL49 genes to address these issues.
Because of the predicted amino acid sequence differences between the Becker and Bartha Us3 proteins, we hypothesized that the wild-type Us3 primary amino acid sequence was important for Us3 incorporation into virions. We tested this idea by expressing the Becker Us3 gene in a Bartha genetic background (Fig. 6). To our surprise, the resulting virus, PRV632, failed to package the Us3 protein. When we expressed the Bartha Us3 gene in a Becker genetic background, the resulting virus, PRV634, also failed to package the Us3 protein. Thus, we conclude that the wild-type Us3 amino acid sequence is necessary but not sufficient for incorporation into the tegument of assembling virions.
How and where the tegument is assembled upon alphaherpesvirus egress remain complex issues. Several studies have indicated that tegument proteins are assembled into mature virions in the cytoplasm via an interaction with the viral capsid, other tegument proteins, or the cytoplasmic tails of viral glycoproteins in the trans-Golgi network (reviewed in reference 30; 12, 20, 29). In Becker-infected cells, the Us3 protein localized to punctate structures in the cytoplasm (Fig. 3 and 8). We believe these structures to be sites where the Us3 protein is incorporated into maturing virions. A preliminary characterization of these structures showed that viral membrane proteins gB and Us9 are present (Randall and Banfield, unpublished). Furthermore, these punctate structures are sensitive to treatment with brefeldin A, a fungal metabolite that causes the Golgi apparatus to redistribute to the endoplasmic reticulum (data not shown). Thus, these structures likely represent vesicles of the secretory pathway. Interestingly, Reynolds and colleagues recently reported that the HSV-1 Us3 homolog localizes to membranous organelles in the cytoplasm of infected cells (35). It is possible that the structures that we observed in PRV-infected cells are similar in nature to those observed during HSV-1 infection. The Us3 protein does not localize to these structures in Bartha-infected cells (Fig. 3). When we repaired the large deletion in the unique short region of Bartha (PRV158), we restored Us3 localization to these punctate structures, but we did not restore Us3 packaging into the tegument of mature virus (Fig. 4). These results may be explained by the fact that Bartha Us3 is not packaged efficiently, even when it is present in a wild-type background (Fig. 8C). One interpretation of these data is that PRV158 targets the Us3 protein to the correct site for packaging, but mutations in the Bartha Us3 protein exclude it from being packaged. We predict that both the wild-type Us3 protein and targeting of Us3 to the correct site are important for the incorporation of Us3 into the tegument. To test these predictions, we are constructing a recombinant Bartha virus that expresses the Becker Us3 gene in conjunction with the Becker gI, gE, Us9, and Us2 genes to determine whether it will package the Us3 protein into mature virions.
To test whether Bartha had deficiencies in the packaging of other tegument proteins, we examined VP22 incorporation into virions. Studies with HSV-1 have shown that VP22 is one of four major structural proteins that contribute the majority of the mass to mature virions (16, 18, 42). To our surprise, there was a striking difference in the amounts of VP22 packaged into Bartha virions and Becker virions (Fig. 9B). At present, the reason for this difference is unclear. We believe that it may be a direct result of mutations in the Bartha VP22 gene rather than the absence of or mutations in another protein that interacts with VP22. As mentioned above, it has been shown that VP22 interacts with the cytoplasmic tails of gE and gM and that the presence of at least one of these glycoproteins is required for VP22 incorporation into virions (12, 13). However, when we restored a functional gE to Bartha (PRV158), there was no increase in the amount of VP22 that was packaged (Fig. 10). We are in the process of cloning and sequencing the Bartha and Becker UL49 genes (VP22) to identify any differences between them. Another possible explanation for the difference in VP22 packaging between Becker and Bartha is that Bartha VP22 may be hyperphosphorylated in infected cells and therefore not incorporated into assembling virions. Studies have shown for both HSV-1 and PRV that VP22 is phosphorylated in infected cells; however, only the nonphosphorylated form is packaged into the tegument (4, 9, 10). The phosphorylated forms of VP22 in Becker-infected cells are not detected at 6 h postinfection (4). However, two forms of Bartha VP22 are detected in infected cell lysates at 6 h postinfection (Fig. 9). This finding is significant because at 6 h postinfection, mature virus is already being released from infected cells. The possibility that the majority of Bartha VP22 is phosphorylated and excluded from maturing virions may account for the inability of Bartha to package VP22. Phosphatase experiments to determine whether the two forms observed in Bartha-infected cell lysates are due to phosphorylation will be required to test this hypothesis.
We report that the PRV Bartha strain does not package two nonessential tegument proteins, Us3 and VP22, despite efficient synthesis in infected cells. Despite the loss of Us3 and VP22, Bartha replicates as well as wild-type Becker and shows no obvious defects in virus maturation or infectivity. These findings raise several interesting questions. Is Bartha unable to package other tegument proteins? How does Bartha compensate for the loss of these tegument proteins? It has been reported that the loss of VP22 from extracellular virions in HSV-1 is compensated for by the increased incorporation of several minor virion proteins (33). Conversely, other studies have shown that the overexpression of VP22 results in a decrease in VP13/14 incorporation into mature virions (25). These studies, in conjunction with our data, highlight the flexibility of alphaherpesviruses in compensating for the loss of tegument proteins.
Acknowledgments
We thank L.W. Enquist and T. del Rio, Princeton University, for helpful discussions and for providing antisera and strain PRV175. We thank Amanda Clase and Renée Finnen for helpful comments and Jessica Randall for expert technical assistance.
This work was supported by grant 1RO1-AI48626 from the NIAID, NIH, to B.W.B. M.G.L. was supported in part by NIH/NIAID training grant AI52066-01.
REFERENCES
- 1.Bartha, A., S. Belak, and J. Benyeda. 1969. Trypsin- and heat-resistance of some strains of the herpesvirus group. Acta Vet. Acad. Sci. Hung. 19:97-99. [PubMed] [Google Scholar]
- 2.Brideau, A. D., B. W. Banfield, and L. W. Enquist. 1998. The Us9 gene product of pseudorabies virus, an alphaherpesvirus, is a phosphorylated, tail-anchored type II membrane protein. J. Virol. 72:4560-4570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Card, J. P., M. E. Whealy, A. K. Robbins, and L. W. Enquist. 1992. Pseudorabies virus envelope glycoprotein gI influences both neurotropism and virulence during infection of the rat visual system. J. Virol. 66:3032-3041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.del Rio, T., H. C. Werner, and L. W. Enquist. 2002. The pseudorabies virus VP22 homologue (UL49) is dispensable for virus growth in vitro and has no effect on virulence and neuronal spread in rodents. J. Virol. 76:774-782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Demmin, G. L., A. C. Clase, J. A. Randall, L. W. Enquist, and B. W. Banfield. 2001. Insertions in the gG gene of pseudorabies virus reduce expression of the upstream Us3 protein and inhibit cell-to-cell spread of virus infection. J. Virol. 75:10856-10869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Desai, P. J. 2000. A null mutation in the UL36 gene of herpes simplex virus type 1 results in accumulation of unenveloped DNA-filled capsids in the cytoplasm of infected cells. J. Virol. 74:11608-11618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dijkstra, J. M., A. Brack, A. Jons, B. G. Klupp, and T. C. Mettenleiter. 1998. Different point mutations within the conserved N-glycosylation motif of pseudorabies virus glycoprotein M result in expression of a nonglycosylated form of the protein. J. Gen. Virol. 79:851-854. [DOI] [PubMed] [Google Scholar]
- 8.Elliott, G., and P. O'Hare. 1997. Intercellular trafficking and protein delivery by a herpesvirus structural protein. Cell 88:223-233. [DOI] [PubMed] [Google Scholar]
- 9.Elliott, G., D. O'Reilly, and P. O'Hare. 1996. Phosphorylation of the herpes simplex virus type 1 tegument protein VP22. Virology 226:140-145. [DOI] [PubMed] [Google Scholar]
- 10.Elliott, G. D., and D. M. Meredith. 1992. The herpes simplex virus type 1 tegument protein VP22 is encoded by gene UL49. J. Gen. Virol. 73:723-726. [DOI] [PubMed] [Google Scholar]
- 11.Frame, M. C., F. C. Purves, D. J. McGeoch, H. S. Marsden, and D. P. Leader. 1987. Identification of the herpes simplex virus protein kinase as the product of viral gene US3. J. Gen. Virol. 68:2699-2704. [DOI] [PubMed] [Google Scholar]
- 12.Fuchs, W., H. Granzow, B. G. Klupp, M. Kopp, and T. C. Mettenleiter. 2002. The UL48 tegument protein of pseudorabies virus is critical for intracytoplasmic assembly of infectious virions. J. Virol. 76:6729-6742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fuchs, W., B. G. Klupp, H. Granzow, C. Hengartner, A. Brack, A. Mundt, L. W. Enquist, and T. C. Mettenleiter. 2002. Physical interaction between envelope glycoproteins E and M of pseudorabies virus and the major tegument protein UL49. J. Virol. 76:8208-8217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Galvan, V., and B. Roizman. 1998. Herpes simplex virus 1 induces and blocks apoptosis at multiple steps during infection and protects cells from exogenous inducers in a cell-type-dependent manner. Proc. Natl. Acad. Sci. USA 95:3931-3936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hata, S., A. H. Koyama, H. Shiota, A. Adachi, F. Goshima, and Y. Nishiyama. 1999. Antiapoptotic activity of herpes simplex virus type 2: the role of US3 protein kinase gene. Microbes Infect. 1:601-607. [DOI] [PubMed] [Google Scholar]
- 16.Heine, J. W., R. W. Honess, E. Cassai, and B. Roizman. 1974. Proteins specified by herpes simplex virus. XII. The virion polypeptides of type 1 strains. J. Virol. 14:640-651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Holland, T. C., R. M. Sandri-Goldin, L. E. Holland, S. D. Marlin, M. Levine, and J. C. Glorioso. 1983. Physical mapping of the mutation in an antigenic variant of herpes simplex virus type 1 by use of an immunoreactive plaque assay. J. Virol. 46:649-652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Honess, R. W., and B. Roizman. 1973. Proteins specified by herpes simplex virus. XI. Identification and relative molar rates of synthesis of structural and nonstructural herpes virus polypeptides in the infected cell. J. Virol. 12:1347-1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jerome, K. R., R. Fox, Z. Chen, A. E. Sears, H. Lee, and L. Corey. 1999. Herpes simplex virus inhibits apoptosis through the action of two genes, Us5 and Us3. J. Virol. 73:8950-8957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Klupp, B. G., W. Fuchs, H. Granzow, R. Nixdorf, and T. C. Mettenleiter. 2002. Pseudorabies virus UL36 tegument protein physically interacts with the UL37 protein. J. Virol. 76:3065-3071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Klupp, B. G., H. Granzow, and T. C. Mettenleiter. 2001. Effect of the pseudorabies virus US3 protein on nuclear membrane localization of the UL34 protein and virus egress from the nucleus. J. Gen. Virol. 82:2363-2371. [DOI] [PubMed] [Google Scholar]
- 22.Klupp, B. G., H. Granzow, E. Mundt, and T. C. Mettenleiter. 2001. Pseudorabies virus UL37 gene product is involved in secondary envelopment. J. Virol. 75:8927-8936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Klupp, B. G., B. Lomniczi, N. Visser, W. Fuchs, and T. C. Mettenleiter. 1995. Mutations affecting the UL21 gene contribute to avirulence of pseudorabies virus vaccine strain Bartha. Virology 212:466-473. [DOI] [PubMed] [Google Scholar]
- 24.Leopardi, R., C. Van Sant, and B. Roizman. 1997. The herpes simplex virus 1 protein kinase US3 is required for protection from apoptosis induced by the virus. Proc. Natl. Acad. Sci. USA 94:7891-7896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Leslie, J., F. J. Rixon, and J. McLauchlan. 1996. Overexpression of the herpes simplex virus type 1 tegument protein VP22 increases its incorporation into virus particles. Virology 220:60-68. [DOI] [PubMed] [Google Scholar]
- 26.Lomniczi, B., M. L. Blankenship, and T. Ben-Porat. 1984. Deletions in the genomes of pseudorabies virus vaccine strains and existence of four isomers of the genomes. J. Virol. 49:970-979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lukacs, N., H. J. Thiel, T. C. Mettenleiter, and H. J. Rziha. 1985. Demonstration of three major species of pseudorabies virus glycoproteins and identification of a disulfide-linked glycoprotein complex. J. Virol. 53:166-173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McGeoch, D. J., and A. J. Davison. 1986. Alphaherpesviruses possess a gene homologous to the protein kinase gene family of eukaryotes and retroviruses. Nucleic Acids Res. 14:1765-1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McNabb, D. S., and R. J. Courtney. 1992. Characterization of the large tegument protein (ICP1/2) of herpes simplex virus type 1. Virology 190:221-232. [DOI] [PubMed] [Google Scholar]
- 30.Mettenleiter, T. C. 2002. Herpesvirus assembly and egress. J. Virol. 76:1537-1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nixdorf, R., B. G. Klupp, A. Karger, and T. C. Mettenleiter. 2000. Effects of truncation of the carboxy terminus of pseudorabies virus glycoprotein B on infectivity. J. Virol. 74:7137-7145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Phelan, A., G. Elliott, and P. O'Hare. 1998. Intercellular delivery of functional p53 by the herpesvirus protein VP22. Nat. Biotechnol. 16:440-443. [DOI] [PubMed] [Google Scholar]
- 33.Pomeranz, L. E., and J. A. Blaho. 2000. Assembly of infectious Herpes simplex virus type 1 virions in the absence of full-length VP22. J. Virol. 74:10041-10054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Purves, F. C., R. M. Longnecker, D. P. Leader, and B. Roizman. 1987. Herpes simplex virus 1 protein kinase is encoded by open reading frame US3 which is not essential for virus growth in cell culture. J. Virol. 61:2896-2901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Reynolds, A. E., E. G. Wills, R. J. Roller, B. J. Ryckman, and J. D. Baines. 2002. Ultrastructural localization of the herpes simplex virus type 1 UL31, UL34, and Us3 proteins suggests specific roles in primary envelopment and egress of nucleocapsids. J. Virol. 76:8939-8952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Robbins, A. K., D. J. Dorney, M. W. Wathen, M. E. Whealy, C. Gold, R. J. Watson, L. E. Holland, S. D. Weed, M. Levine, J. C. Glorioso, et al. 1987. The pseudorabies virus gII gene is closely related to the gB glycoprotein gene of herpes simplex virus. J. Virol. 61:2691-2701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Robbins, A. K., J. P. Ryan, M. E. Whealy, and L. W. Enquist. 1989. The gene encoding the gIII envelope protein of pseudorabies virus vaccine strain Bartha contains a mutation affecting protein localization. J. Virol. 63:250-258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Roizman, B., and D. M. Knipe. 2001. Herpes simplex viruses and their replication, p. 2399-2459. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology, 4th ed. Lippincott Williams & Wilkins, Philadelphia, Pa.
- 39.Roizman, B., and P. E. Pellet. 2001. The family Herpesviridae: a brief introduction, p. 2381-2397. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology, 4th ed. Lippincott Williams & Wilkins, Philadelphia, Pa.
- 40.Sciortino, M. T., B. Taddeo, A. P. Poon, A. Mastino, and B. Roizman. 2002. Of the three tegument proteins that package mRNA in herpes simplex virions, one (VP22) transports the mRNA to uninfected cells for expression prior to viral infection. Proc. Natl. Acad. Sci. USA 99:8318-8323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Smith, K. O., W. L. Kennell, and D. L. Lamm. 1981. Visualization of minute centers of viral infection in unfixed cell cultures by an enzyme-linked antibody assay. J. Immunol. Methods 40:297-305. [DOI] [PubMed] [Google Scholar]
- 42.Spear, P. G., and B. Roizman. 1972. Proteins specified by herpes simplex virus. V. Purification and structural proteins of the herpesvirion. J. Virol. 9:143-159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.van Zijl, M., H. van der Gulden, N. de Wind, A. Gielkens, and A. Berns. 1990. Identification of two genes in the unique short region of pseudorabies virus; comparison with herpes simplex virus and varicella-zoster virus. J. Gen. Virol. 71:1747-1755. [DOI] [PubMed] [Google Scholar]
- 44.Wagenaar, F., J. M. Pol, B. Peeters, A. L. Gielkens, N. de Wind, and T. G. Kimman. 1995. The US3-encoded protein kinase from pseudorabies virus affects egress of virions from the nucleus. J. Gen. Virol. 76:1851-1859. [DOI] [PubMed] [Google Scholar]
- 45.Whealy, M. E., A. K. Robbins, and L. W. Enquist. 1990. The export pathway of the pseudorabies virus gB homolog gII involves oligomer formation in the endoplasmic reticulum and protease processing in the Golgi apparatus. J. Virol. 64:1946-1955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wolfer, U., V. Kruft, D. Sawitzky, H. Hampl, B. Wittmann-Liebold, and K. O. Habermehl. 1990. Processing of pseudorabies virus glycoprotein gII. J. Virol. 64:3122-3125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang, G., R. Stevens, and D. P. Leader. 1990. The protein kinase encoded in the short unique region of pseudorabies virus: description of the gene and identification of its product in virions and in infected cells. J. Gen. Virol. 71:1757-1765. [DOI] [PubMed] [Google Scholar]