Abstract
Retroviral insertional mutagenesis in mouse hematopoietic tumors provides a powerful cancer gene discovery tool. Here, we describe a high-throughput, single nucleotide polymorphism (SNP)-based method, for mapping retroviral integration sites cloned from mouse tumors, and a bacterial artificial chromosome (BAC) hybridization method, for localizing these retroviral integration sites to common sites of retroviral integration (CISs). Several new CISs were identified, including one CIS that mapped near Notch1, a gene that has been causally associated with human T-cell tumors. This mapping method is applicable to many different species, including ones where few genetic markers and little genomic sequence information are available. It can also be used to map endogenous proviruses.
Retroviruses induce leukemia or lymphoma by randomly integrating into the genome and activating cellular proto-oncogenes or inactivating tumor suppressor genes. The retroviral integration sites in these tumors thus provide powerful genetic tags for cancer gene discovery (2, 8). Several disease genes identified by retroviral tagging in mouse tumors have also been causally associated with human disease (5, 12, 14, 16), validating the usefulness of this method for human cancer gene discovery.
Recently, an inverse PCR (IPCR) method was developed for cloning retroviral integration sites from mouse leukemias and lymphomas that dramatically increases the throughput of retroviral tagging for cancer gene discovery (12). This IPCR method was used to clone 419 retroviral integration sites from BXH2 myeloid leukemias and a few AKXD B- and T-cell tumors. Approximately 1.2 to 1.4 kb of mouse genomic DNA was then sequenced from each IPCR product, and the sequences, termed proviral tagged sequences (PTSs), were compared with one another to look for sequence overlaps. Sequence overlaps for PTSs cloned from different tumors indicate a common viral integration site (CIS). The probability that two or more PTSs among the 419 PTSs scored would be located in the same small region of the genome by chance alone is very low and suggests that this region contains a disease gene that is mutated by viral integration. The PTSs were also BLAST searched against the nonredundant and expressed sequence tag databases to look for coding regions of known genes or for homology with already published CISs. In this way, 13% of the PTSs could be localized to known CISs, while 11% could be localized to novel CISs. Another 10% of sequences identified known genes, but these genes were hit only once in the tumor panel, while 8% hit expressed sequence tags of unknown function. Fifty-eight percent of the sequences were noninformative.
Since retroviral integrations can cause disease by affecting gene expression over long distances (tens or even hundreds of kilobases) (11), it is possible that some of the noninformative PTSs in these experiments were also located at CISs but were missed because only a limited amount of mouse genomic sequence information was available for each IPCR product. To determine whether any of the noninformative PTSs are also located at CISs and to better estimate the percentages of PTSs that are located at CISs, we determined the chromosomal location of 131 of these noninformative PTSs with the Frederick Interspecific Backcross Mapping Panel (IBMP) (4). Cosegregation of two or more noninformative PTSs in the Frederick IBMP provides suggestive evidence of a CIS. Physical mapping with mouse bacterial artificial chromosomes (BACs) was then used to further localize these sequences in the mouse genome.
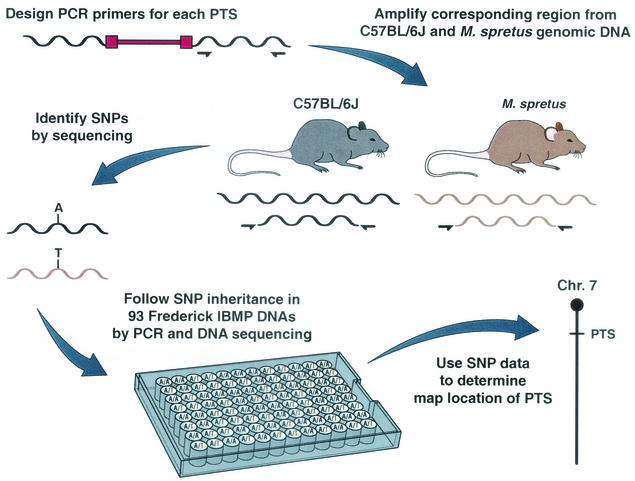
Due to the large number of noninformative PTSs that needed to be mapped, we employed a high-throughput, single nucleotide polymorphism (SNP)-based mapping method. PCR primer pairs were generated for 240 of the noninformative PTSs. The primer pairs, designed from nonrepetitive sequences, were then used to amplify 250- to 300-bp fragments of genomic DNA from C57BL/6J and Mus spretus mice, the two parents of the Frederick IBMP (Fig. 1). Of the 240 primer pairs used in these experiments, 185 (77%) amplified products from DNAs of both species (Table 1). For the other 55 primer pairs, either no PCR products were generated or they were obtained from only one DNA (Table 1). Sequence analysis of the amplification products identified sequence polymorphisms for 148 of the 185 informative primer pairs (80%). SNPs were the most common polymorphism, but insertion-deletion polymorphisms were also identified at lower frequencies (Table 1). In total, about 1 bp was polymorphic for every 101 bp sequenced. This is consistent with the long evolutionary distance between these two species (3). Each polymorphic primer pair was then used to amply 93 DNAs from the Frederick IBMP along with C57BL/6J and M. spretus control DNAs. The amplification products were then sequenced with an ABI 3700 capillary electrophoresis sequence machine. By monitoring the inheritance of the SNPs, or insertion-deletion polymorphisms, in the backcross DNAs, it was then possible to accurately position these PTSs on the Frederick IBMP map.
FIG. 1.
High-throughput, SNP-based mapping of PTSs cloned from mouse hematopoietic tumors. PCR primer pairs were generated for 240 noninformative PTSs (12) and used to amplify 250- to 350-bp fragments from C57BL/6J and M. spretus genomic DNA (10). Proviral DNA is shown in red, and the locations of amplification primers in the flanking cellular DNA are shown by half-arrows. Amplification reaction mixtures contained 4 ng of mouse DNA, 1 μg of forward and reverse primers, 2 U of Platinum Taq polymerase (Gibco BRL), and the buffer supplied by the manufacturer. To improve the specificity of the PCR, we used a touchdown PCR program for the first 12 cycles. The annealing temperature was decreased by 1°C from 65 to 60°C after every two cycles. This was followed by 25 cycles of 94°C for 20 s, 56°C for 20 s, and 68°C for 1 min and a final extension of 68°C for 5 min on a GeneAmp PCR System 9700 (PE Applied Biosystems). The amplification products were then sequenced to identify SNPs or insertion-deletion polymorphisms. PCR products were pretreated to remove excess primers and deoxynucleoside triphosphates before they were sequenced. Each sample was incubated with 1 U of shrimp alkaline phosphatase (Amersham)/μl and 10 U of exonuclease (Amersham)/μl at 37°C for 5 min, followed by 72°C for 15 min to denature the enzyme. The amplification products were sequenced with the ABI PRISM BigDye Terminator Cycle Sequencing kit (PE Applied Biosystems) with the forward primer. Sequence files were analyzed with PolyPhred (Washington University) for simple SNPs or with MultiALN for polymorphisms involving insertions and deletions. Polymorphic primer pairs were then used to amplify 93 DNAs from the Frederick IBMP along with C57BL/6J and M. spretus control DNA. The 205 Frederick IBMP progeny were generated by mating (C57BL/6J × M. spretus)F1 females and C57BL/6J males as previously described (4). By monitoring the inheritance of these polymorphisms in the backcross DNAs, it was possible to accurately position each PTS on the Frederick IBMP.
TABLE 1.
SNP frequency in C57BL/6J (BL/6) and M. spretus DNAs
| Primer pair analysis | |
| Total PTS primer pairs | 240 |
| PCR products | |
| Producing both types | 185 |
| BL/6 only | 19 |
| M. spretus only | 5 |
| None | 31 |
| Sequence analysis | |
| Total bases sequenced | 35,960 |
| Point mutations | |
| No. | 311 |
| Frequency | 1/116 |
| Insertions-deletions | |
| No. | 46 |
| Frequency | 1/782 |
| Frequency of all polymorphisms | 1/101 |
| Polymorphic primer pairs | 148 |
| Nonpolymorphic primer pairs | 37 |
Among the 145 polymorphic amplification products, 131 could be mapped on the Frederick IBMP (Table 2). The other 14 primer pairs preferentially amplified the C57BL/6J allele, apparently due to primer competition (data not shown). C57BL/6J and the leukemic strains BXH2 and AKXD are highly related to one another but only distantly related to M. spretus (1). Therefore, primer pairs generated from BXH2 and AKXD genomic sequence will often have mismatches with respect to M. spretus, but not C57BL/6J, genomic DNA (Table 1). These mismatches are not a problem when C57BL/6J DNA is amplified but become a problem when backcross DNA carrying both C57BL/6J and M. spretus alleles is amplified. Interestingly, 22 of the 131 PTSs mapped to known common sites that had already been localized on the panel (Table 2). Another 18 PTSs appeared to be located at novel CISs, defined as two or more noninformative PTSs that mapped together on the panel but which did not cosegregate with a known CIS or, more frequently, a noninformative PTS that cosegregated with a PTS that had already been mapped on the panel in previous experiments by Southern analysis but which had not yet been localized to a CIS (Table 2).
TABLE 2.
CISs defined by genetic and physical mapping
| Total PTSs mapped | 131 |
| Total mapped to CISs | 40 |
| Mapped to known CISs | 22 |
| Mapped to new CISs | 18 |
| Total confirmed by BAC analysis | 22 |
| Mapped to known CISs | 9 (5)a |
| Mapped to new CISs | 13 (8)a |
Numbers in parentheses are the numbers of CISs to which the PTSs confirmed by BAC analysis localized.
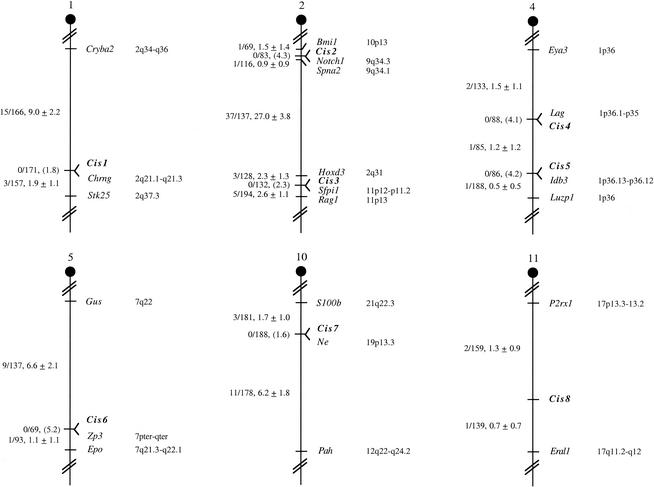
Two PTSs that cosegregate in the Frederick IBMP could still be located far apart in physical terms. Genetic cosegregation in 93 backcross animals implies that the two PTSs are located within 3.9 centimorgans of each other at the 95% confidence limit. This corresponds to ∼7 Mb of mouse genome DNA, well beyond what would normally be considered to represent a CIS. To further localize these PTSs in the mouse genome, one of the PTSs that defined each CIS was used as a probe to isolate mouse BAC(s). Mouse BAC clones were isolated from two BAC libraries: the RPCI-23 C57BL/6J BAC library (Roswell Park Cancer Institute) and the CITB 129/Sv BAC library (Research Genetics). The PCR products used for the SNP analysis were also used as probes for BAC screening. We then asked whether the other PTSs that defined the CIS were also located on the same BAC. The average insert size of these BAC libraries is ∼190 kb. Therefore, on average, this type of analysis localizes the PTSs to an ∼190-kb genomic region. Since proviral integrations can affect gene expression over distances approaching 300 kb (11), this is well within the range of what would normally be considered to constitute a CIS. By use of BAC hybridization, 22 of the 40 PTSs could be confirmed to be located at CISs (Table 2). When extrapolated across the 58% of PTSs that were noninformative in previous experiments and combined with the 24% of PTSs that were located at CISs in previous experiments, these results suggest that at least 35% of the PTSs in this tumor panel are located at CISs. Among these 22 PTSs, 9 were located at five previously published CISs (Evi2, Evi8, Pim1, Evi29, and Evi31) (12), while 13 mapped to eight novel CISs (designated Cis1 through Cis8) (Fig. 2).
FIG. 2.
Chromosomal location of eight novel CISs identified by SNP-based mapping. CIS loci were mapped by monitoring the inheritance of SNPs, or insertion-deletion polymorphisms, in the Frederick IBMP mice (Fig. 1). The number of recombinant N2 animals over the total number of N2 animals typed for each polymorphism is shown to the left of the chromosome maps between each pair of loci. The recombination frequencies, expressed as genetic distance in centimorgans (±1 standard deviation), are also shown. The upper 95% confidence limit of the recombination distance is given in parentheses when no recombinants were found between loci. When recombination between genes was observed, gene order was determined by minimizing the number of recombination events required to explain the allele distribution pattern. The positions of loci on human chromosomes, where known, are shown to the right of the chromosome maps. References for the map positions of human loci can be obtained from LocusLink (http://www.ncbi.nlm.gov/LocusLink).
The eight novel Cis loci mapped to six different mouse chromosomes. Two Cis loci mapped to chromosome 2 (Cis2 and Cis 3), and two Cis loci mapped to chromosome 4 (Cis4 and Cis5). Cis1, Cis6, Cis7, and Cis8 mapped to chromosomes 1, 5, 10, and 11, respectively. The Cis3 locus on chromosome 2 cosegregated with Sfpi1. Sfpi1 is a gene that was originally identified at a common site of retroviral integration in murine erythroleukemias induced by the acute leukemogenic retrovirus spleen focus-forming virus (13, 15). Sfpi1, also known as PU.1, is an ETS-domain transcription factor essential for the development of myeloid and B-lymphoid cells (6). BAC hybridization, however, failed to show that Sfpi1 and Cis3 are located on the same BAC-sized fragment of genomic DNA. It is therefore unlikely that viral integrations at Cis3 affect Sfpi1 expression. Likewise, the Cis2 locus on chromosome 2 cosegregated with Notch1. Notch1 has not yet been associated with a CIS in hematopoietic tumors from mice or other species; however, human NOTCH1 transcripts are truncated by fusion to the beta T-cell receptor gene in T-lymphoblastic leukemia patients carrying a t(7;9)(q34;q34.3) chromosomal translocation (7). Notch1 is thus an excellent candidate for a mouse leukemia disease gene. Consistent with this hypothesis, Notch1 and Cis2 are located on the same BAC, and database searches showed that the two PTSs that defined Cis2 are located 12 and 47 kb upstream of Notch1.
In the studies described here we were able to map 55% of the noninformative PTSs tested by using one set of PCR primers for each noninformative PTS. However, some primer pairs failed to amplify C57BL/6J or M. spretus DNA or, alternatively, preferentially amplified the C57BL/6J allele in interspecific backcross DNA. Both of these problems can largely be eliminated by designing a second set of PCR primers from regions of the genome that are identical in C57BL/6J and M. spretus mice. In addition, 20% of the amplification products were not polymorphic in C57BL/6J and M. spretus DNA. Again, amplifying other regions that flank these proviral integration sites can eliminate this problem.
Nearly 98% of the mouse genome sequence has recently been deposited in GenBank. This sequence information has been assembled into scaffolds averaging 16.5 Mbp in size, and the scaffolds have been mapped to chromosomes by using genetic and radiation hybrid markers. Many of the retroviral integration sites cloned from the mouse can therefore now be mapped on the mouse genome by using this sequence information. For other species, however, where retroviral insertional mutagenesis has been used to identify disease genes, it will be some time before CISs can be identified by sequence alone. The mapping method described here, combined with BAC hybridization, provides an alternative method besides Southern analysis or radiation hybrids for identifying CISs in these species. The primary requirement is that SNPs can be identified in the DNA flanking the proviral integration sites and that individuals segregating for these SNPs are available. SNP-based mapping, combined with BAC hybridization, can detect CISs that would be difficult to detect by conventional Southern analysis, because, for example, the proviral integrations at the CIS are spread out over a large genomic region. Radiation hybrid analysis also has the potential to generate false positives as well as false negatives due to PCR artifacts. These abnormal typings could obscure the identification of novel CISs. SNP-based mapping is especially applicable to species like chickens and cats, where interspecific crosses have already been produced (9, 17). Finally, SNP-based mapping can be used to determine the chromosomal location of endogenous proviruses, provided again that SNPs can be identified in the DNA flanking the endogenous provirus.
Acknowledgments
This research was supported by the National Cancer Institute, Department of Health and Human Services (N.A.J. and N.G.C.), and SAIC-Frederick under National Cancer Institute contract N01-CO-12400 (D.J.M.).
REFERENCES
- 1.Beck, J. A., S. Lloyd, M. Hafezparast, M. Lennon-Pierce, J. T. Eppig, M. F. Festing, and E. M. Fisher. 2000. Genealogies of mouse inbred strains. Nat. Genet. 24:23-25. [DOI] [PubMed] [Google Scholar]
- 2.Bedigian, H. G., D. A. Johnson, N. A. Jenkins, N. G. Copeland, and R. Evans. 1984. Spontaneous and induced leukemias of myeloid origin in recombinant inbred BXH mice. J. Virol. 51:586-594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bonhomme, F. 1986. Evolutionary relationships in the genus Mus. Curr. Top. Microbiol. Immunol. 127:19-34. [DOI] [PubMed] [Google Scholar]
- 4.Copeland, N. G., and N. A. Jenkins. 1991. Development and applications of a molecular genetic linkage map of the mouse genome. Trends Genet. 7:113-118. [DOI] [PubMed] [Google Scholar]
- 5.Copeland, N. G., and N. A. Jenkins. 1999. Myeloid leukemia: disease genes and mouse models. Prog. Exp. Tumor Res. 35:53-63. [DOI] [PubMed] [Google Scholar]
- 6.DeKoter, R. P., and H. Singh. 2000. Regulation of B lymphocyte and macrophage development by graded expression of PU.1. Science 288:1439-1441. [DOI] [PubMed] [Google Scholar]
- 7.Ellisen, L. W., J. Bird, D. C. West, A. L. Soreng, T. C. Reynolds, S. D. Smith, and J. Sklar. 1991. TAN-1, the human homolog of the Drosophila notch gene, is broken by chromosomal translocations in T lymphoblastic neoplasms. Cell 66:649-661. [DOI] [PubMed] [Google Scholar]
- 8.Gilbert, D. J., P. E. Neumann, B. A. Taylor, N. A. Jenkins, and N. G. Copeland. 1993. Susceptibility of AKXD recombinant inbred mouse strains to lymphomas. J. Virol. 67:2083-2090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Groenen, M. A., H. H. Cheng, N. Bumstead, B. F. Benkel, W. E. Briles, T. Burke, D. W. Burt, L. B. Crittenden, J. Dodgson, J. Hillel, S. Lamont, A. P. de Leon, M. Soller, H. Takahashi, and A. Vignal. 2000. A consensus linkage map of the chicken genome. Genome Res. 10:137-147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jenkins, N. A., N. G. Copeland, B. A. Taylor, and B. K. Lee. 1982. Organization, distribution, and stability of endogenous ecotropic murine leukemia virus DNA sequences in chromosomes of Mus musculus. J. Virol. 43:26-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lazo, P. A., J. S. Lee, and P. N. Tsichlis. 1990. Long-distance activation of the Myc protooncogene by provirus insertion in Mlvi-1 or Mlvi-4 in rat T-cell lymphomas. Proc. Natl. Acad. Sci. USA 87:170-173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li, J., H. Shen, K. L. Himmel, A. J. Dupuy, D. A. Largaespada, T. Nakamura, J. D. Shaughnessy, Jr., N. A. Jenkins, and N. G. Copeland. 1999. Leukaemia disease genes: large-scale cloning and pathway predictions. Nat. Genet. 23:348-353. [DOI] [PubMed] [Google Scholar]
- 13.Moreau-Gachelin, F., A. Tavitian, and P. Tambourin. 1988. Spi-1 is a putative oncogene in virally induced murine erythroleukaemias. Nature 331:277-280. [DOI] [PubMed] [Google Scholar]
- 14.Ogawa, S., M. Kurokawa, T. Tanaka, K. Mitani, J. Inazawa, A. Hangaishi, K. Tanaka, Y. Matsuo, J. Minowada, T. Tsubota, Y. Yazaki, and H. Hirai. 1996. Structurally altered Evi-1 protein generated in the 3q21-q26 syndrome. Oncogene 13:183-191. [PubMed] [Google Scholar]
- 15.Paul, R., S. Schuetze, S. L. Kozak, C. A. Kozak, and D. Kabat. 1991. The Sfpi-1 proviral integration site of Friend erythroleukemia encodes the ets-related transcription factor Pu.1. J. Virol. 65:464-467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Roberts, T., O. Chernova, and J. K. Cowell. 1998. NB4S, a member of the TBC1 domain family of genes, is truncated as a result of a constitutional t(1;10)(p22;q21) chromosome translocation in a patient with stage 4S neuroblastoma. Hum. Mol. Genet. 7:1169-1178. [DOI] [PubMed] [Google Scholar]
- 17.Sun, S., W. J. Murphy, M. Menotti-Raymond, and S. J. O'Brien. 2001. Integration of the feline radiation hybrid and linkage maps. Mamm. Genome 12:436-441. [DOI] [PubMed] [Google Scholar]