Abstract
The genome of the prototypic arenavirus lymphocytic choriomeningitis virus (LCMV) consists of two negative-sense, single-strand RNA segments designated L and S. Arenavirus genomes exhibit high sequence conservation at their 3′ ends. All arenavirus genomes examined to date have a conserved terminal sequence element (3′-terminal 20 nucleotides [nt]) thought to be a highly conserved viral promoter. Terminal complementarity between the 5′ and 3′ ends of the L and S RNAs predicts the formation of a thermodynamically stable panhandle structure that could contribute to the control of RNA synthesis. We investigated these issues by using a transcription- and replication-competent minireplicon system. A series of overlapping deletions spanning the 3′-terminal 20-nt region of an LCMV minigenome (MG) was generated, and the mutant MGs were analyzed for their activity as templates for RNA synthesis by the LCMV polymerase. The minimal LCMV genomic promoter was found to be contained within the 3′-terminal 19 nt. Substitution of C for G at the last 3′-end nucleotide position in the MG resulted in nondetection of RNA transcription or replication, whereas the addition of a C at the 3′ end did not have any significant affect on RNA synthesis mediated by the LCMV polymerase. All other mutations introduced within the 3′-terminal 19 nt of the MG resulted in undetectable levels of promoter activity. Deletions and nucleotide substitutions within the MG 5′ end that disrupted terminal complementarity abolished chloramphenicol acetyltransferase expression and RNA synthesis mediated by the LCMV polymerase. These findings indicate that both sequence specificity within the 3′-terminal 19 nt and the integrity of the predicted panhandle structure appear to be required for efficient RNA synthesis mediated by the LCMV polymerase.
The prototypic arenavirus lymphocytic choriomeningitis virus (LCMV) is one of the most widely used model systems to study virus-host interactions, such as viral persistence and associated disease (8, 39). The LCMV genome is composed of two negative-sense single-stranded RNA segments, called S (3.2 kb) and L (7.2 kb) (46, 49). Both segments use an ambisense coding strategy to direct synthesis of two proteins from two open reading frames with opposite orientation and separated by an intergenic region (IGR) (2, 3, 60). The S RNA encodes the nucleoprotein NP (ca. 63 kDa) and the glycoprotein precursor GP-C (75 kDa). GP-C is posttranslationally cleaved to yield the mature glycoproteins GP-1 (40 to 46 kDa) and GP-2 (35 kDa) (45, 54, 62). Tetramers of GP-1 and GP-2 form the spikes on the virion envelope and mediate virus interaction with the cellular receptor (9, 11). The L RNA encodes the virus RNA-dependent RNA polymerase (RdRp) (L, ca. 200 kDa) (21, 30, 51) and a small (11-kDa) RING finger protein (Z) (49). NP and L are associated with the viral RNA to form ribonucleoprotein (RNP) complexes, which are active in transcription and replication (14, 22). As with other negative-strand RNA viruses, this RNP is the minimal infectious unit.
All cis-acting sequences needed for initiation of RNA synthesis by the polymerases of negative-strand RNA viruses seem to be located within the 3′-terminal untranslated regions (UTR) of genome and antigenome RNA species (13). Negative-strand RNA viruses also exhibit complementarity between the 5′ and 3′ ends of their genomes and antigenomes. This terminal complementarity may reflect the presence at the 5′ ends of cis-acting signal sequences that provide a nucleation site for RNA encapsidation, required to generate the nucleocapsid templates recognized by the virus polymerase. Terminal complementarity may also be a consequence of strong similarities between the genome and antigenome promoters used by the virus polymerases.
Arenaviruses exhibit a high degree of sequence conservation at the 3′ end of the L and S RNA segments (17 out of 19 nucleotides [nt] are identical). It has been proposed that this conserved terminal sequence element constitutes the virus promoter for polymerase entry. The almost exact inverted complement of the 3′-end 19 nt is found at the 5′ termini of genomes and antigenomes of arenaviruses. Thus, the 5′ and 3′ ends of both L and S genome segments are predicted to form panhandle structures (49). This prediction is supported by electron microscopy data showing the existence of circular RNPs within arenavirus virion particles (65). An additional nontemplated G residue has been detected on the 5′ end of the arenavirus genomic S RNAs (10, 26, 42). As with other RdRp, the LCMV polymerase carries on two different processes, transcription and replication. In its transcriptional mode the LCMV polymerase produces subgenomic mRNAs terminating at the IGR (53). In contrast, during replication the polymerase reads through the transcription termination signal provided by the IGR and generates uncapped full-length antigenomic and genomic RNA species. These RNAs are encapsidated by the NP and serve as templates for further mRNA transcription and for production of virus progeny. Both processes are thought to be initiated by binding of the virus polymerase to the promoter at the 3′ end of the genomic and antigenomic templates.
We have developed a reverse genetic system that recreates intracellularly the biosynthetic steps of LCMV replication and transcription (30). Here we describe studies aimed at the molecular characterization of the LCMV genomic virus promoter. We carried out a mutational analysis of the 3′- and 5′-terminal sequences of a minigenome (MG) of the S segment of strain ARM of LCMV (ARM/S-MG). We present evidence that both transcription and replication by the LCMV polymerase requires a promoter region located within the first 19 nt of the 3′ end of the virus genome. In addition, our results indicate that the first 19 nt of the genome 5′ end contributing to the predicted panhandle are also strictly required for promoter activity. These findings suggest that both sequence specificity and integrity of the predicted panhandle structure formed between the 3′ and the 5′ termini are required for the activity of the LCMV genomic promoter.
MATERIALS AND METHODS
Cells and viruses.
Baby hamster kidney BHK-21 cells (ATCC CCL 10) were maintained in high-glucose Dulbecco's modified Eagle medium (DMEM) supplemented with 10% heat-inactivated fetal calf serum (Life Technologies), 2 mM glutamine, 1× tryptose phosphate broth (Life Technologies), 1 mM sodium pyruvate (Life Technologies), and 0.5% glucose. VTF7-3 was kindly provided by B. Moss (National Institutes of Health, Bethesda, Md.) (20).
Plasmids.
Plasmid pLCMVSCAT2 (renamed MG#3 in this work) has been described previously (30). To construct MG#6, the LCMV-specific 3′ hairpin ribozyme (HR) was replaced by the hepatitis delta ribozyme (HDR). The HDR sequence was amplified by PCR from plasmid pSA1-2 (41) by using primers OHDRRBss (TTGGCGCGCCTCCCTTAGCCATCCGAGTGGACG) and OLCMVHDRF (CTAGGATCCACTGTGCCGGGTCGGCATGGCATCTCCACCTCC). Restriction sites are underlined, and mutated positions appears in bold. The PCR product was digested with BamHI and BssHII and was cloned in pLCMVSCAT2 digested with BamHI and BssHII. Digestion with these enzymes removed the HR sequence from the plasmid. The mutation of the 3′ G to C was introduced by using the Site-Directed Mutagenesis kit (Stratagene) following the recommendations of the manufacturer. The same system was used to generate all the mutants reported in this article. Sequences of the primers used for each individual mutagenesis are available upon request. Mutations were verified by DNA sequencing with an AbiPrism automatic sequencer.
DNA transfections.
BHK-21 cells (106) growing in 35-mm-diameter wells were infected with vTF7-3 at a multiplicity of infection (MOI) of 3 PFU/cell. Cells were transfected by using Lipofectamine (Life Technologies) with plasmids encoding NP (pCITE-NP, 1.5 μg), L (pGEM-L, 0.1 μg) (30), and the different model genomes (0.5 μg). Control transfections lacking L or all support plasmids received empty pUC19 plasmid so that the total amount of DNA transfected in each well was kept constant. 1-β-d arabinofuranosylcytosine (AraC) was added to a final concentration of 50 μg/ml to inhibit the late phase of vaccinia virus expression. All plasmid DNA was prepared by using Qiagen technology.
CAT assays.
Twenty four hours after transfection, cells were washed once with ice-cold phosphate-buffered saline and lysed in 250 mM Tris-HCl (pH 7.5) by three cycles of freezing and thawing. Cell lysates were clarified by centrifugation, and protein concentration was estimated by using the Protein Assay kit (Bio-Rad). For the chloramphenicol acetyltransferase (CAT) assay, 2 μg of protein was diluted in 0.5 M Tris-HCl (pH 7.5) containing 1 mM acetyl coenzyme A (Boehringer Mannheim Biochemicals) and mixed with 0.25 μCi of [14C]chloramphenicol (Amersham). Samples were incubated for 30 min at 37°C, and the products of the reaction were analyzed on a thin-layer chromatography (TLC) developed with chloroform-methanol (19:1).
Analysis of RNA by Northern blot hybridization.
RNA was isolated from transfected cells by using TriReagent (Molecular Research Center) according to the manufacturer's instructions and was analyzed by Northern blot by using either CAT sense or antisense strand-specific riboprobes. CAT RNA riboprobes were prepared by runoff transcription of plasmids pCRII-CAT with T7 RNA polymerase (T7RP) in the presence of [32P]UTP. RNA was separated on 1.6% agarose gels containing 2.2 M formaldehyde and was transferred to nylon membranes (Schleicher & Schuell). Blots were hybridized with 32P-labeled riboprobes in Zip hybridization solution (Ambion), yeast tRNA (100 μg/ml), and single-stranded DNA (100 μg/ml) at 65°C overnight. Filters were washed at 68°C two times for 15 min each in 2× SSC (1 × SSC is 0.15 M NaCl plus 0.015 M sodium citrate)-0.2% sodium dodecyl sulfate and twice again in 0.2× SSC-0.2% sodium dodecyl sulfate. Bands were visualized by autoradiography. Quantitation was done on a Molecular Dynamics PhosphorImager.
Isolation and analysis of encapsidated RNA.
Cytosolic extracts from transfected cells were prepared by incubating them on ice for 15 min in hypotonic buffer (10 mM Tris-HCl, pH 7.5; 1.5 mM MgCl2; 10 mM KCl; 5 mM dithiothreitol; 0.5% NP-40; 500 U of RNAsin/ml). Lysates were then clarified and adjusted to a final concentration of 0.5 M NaCl. To disrupt polysomes, EDTA was added to a concentration of 10 mM and extracts were incubated for a further 30 min at 0°C. Extracts were loaded onto a 5.7 and 0.5 M discontinuous CsCl2 gradient. After 3 h of centrifugation, the RNP containing interphase was collected and recentrifuged to collect the RNP (14). RNA was extracted from RNP by using TriReagent.
RESULTS
Effect of replacing the 3′-terminal nucleotide (G) with a C on promoter activity.
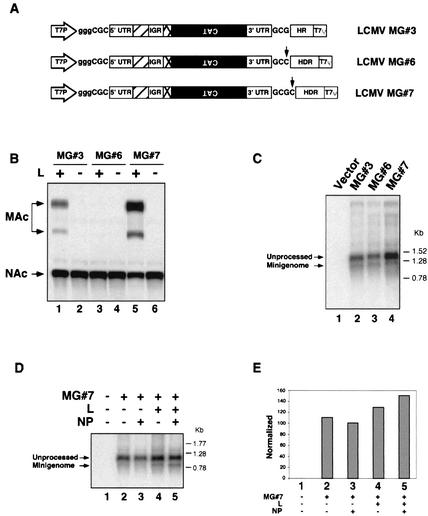
We sought to define the cis-acting sequences that constitute the minimal LCMV genome promoter located at the 3′ end of the LCMV S genomic RNA (48). For this purpose we used the LCMV MG system previously described (30). In the original LCMV MG, called LCMV MG#3 in this paper, generation of the authentic viral 3′ end was mediated by an LCMV-specific HR inserted downstream to the 3′-end UTR of the S MG RNA and was followed by T7 terminator sequences (T7T) (Fig. 1A). Transfection with plasmid pLCMVSCAT2 carrying LCMV MG#3 allows for T7RP-mediated intracellular synthesis of an RNA that will be subsequently processed by the HR. This processed RNA (LCMV MG#3) will serve as a template for transcription and replication mediated by the LCMV polymerase. The use of plasmid pLCMVSCAT2 to generate deletion mutants within the 3′ end of LCMV MG#3 would have required adapting the HR sequence to the 3′-end sequence of each new construct. To overcome this problem, we replaced the HR by the sequence of the HDR. The autolytic activity of the HDR is usually not influenced by sequences upstream of the cleavage site. However, it has been reported that its efficiency follows the pattern C > U > A > G, with respect to the nucleotide located immediately 5′ to the site of cleavage (41). The last nucleotide of the 3′ end of LCMV S RNA is a G (position 3376 in the S RNA) (1, 48), and we observed that less than 1% of the intracellularly synthesized MG RNA was autolytically processed by the activity of the HDR (30). We therefore decided to mutate this last nucleotide residue from G to C to generate plasmid LCMV MG#6 (Fig. 1A). We expected this change to dramatically enhance the processing by the autolytic activity of the HDR of the intracellularly produced MG#6 RNA. We tested whether plasmid-supplied MG#6 RNA was efficiently used as template by the virus polymerase. For this, BHK-21 cells were infected with vFT7-3 at an MOI of 3 PFU/cell and were cotransfected with plasmids carrying MG#6 and the trans-acting factors NP and L. Twenty-four hours after transfection, cell extracts were prepared and analyzed for CAT activity (Fig. 1B). Unexpectedly, we did not detect CAT activity in cells transfected with LCMV MG#6 together with NP and L (Fig. 1B, lane 3). This result indicated that the G residue present at the very 3′ end of the S RNA is required for efficient RNA synthesis mediated by the LCMV polymerase. We then generated a new MG (LCMV MG#7) identical to LCMV MG#6, except that it contained an additional C residue after the G residue at position 3376; therefore, MG#7 is 1 nt longer than LCMV MG#3 and MG#6. Cotransfection of MG#7 together with NP and L resulted in levels of CAT expression comparable to those obtained with LCMV MG#3 (compare lanes 1 and 5 in Fig. 1B). Impaired T7RP-mediated intracellular synthesis of MG#6 RNA, or an inefficient processing of this MG#6 RNA by the HDR, or both, could account for the observed lack of CAT activity. To address this question, we analyzed by Northern blot hybridization RNA isolated from BHK-21 cells infected with vFT7-3 and transfected with the indicated model genome (Fig. 1C). Hybridization with a CAT sense riboprobe detected two main RNA species with approximate sizes of 1,237 nt (unprocessed RNA) and 1,021 nt (self cleaved). We observed similar levels of T7RP-mediated intracellular primary transcription for the three LCMV MG tested (Fig. 1C). Moreover, the efficiency of RNA processing mediated by HDR was similar to that seen with the LCMV-specific HR. Thus, the amounts of MG#3 and MG#6 processed RNA species were very similar (compare lanes 2 and 3 in Fig. 1C).
FIG. 1.
Schematic diagram and characterization of LCMV MGs. (A) Plasmid LCMV MG#3 corresponds to previously described ARM/S-MG (30). LCMV MG contains the following elements: the minimal T7 RNA polymerase promoter (T7P) followed by three extra G residues; the 5′ UTR of the LCMV S RNA (nt 1 to 78); the IGR of the LCMV S RNA (nt 1484 to 1694); a DNA encoding the full-length CAT open reading frame in antisense orientation with respect to the T7P; the 3′ UTR of the LCMV S RNA (nt 1484 to 3376); an LCMV-specific HR; and a T7 terminator (T7ψ). Hatched and crossed boxes represent the 3′ end of the glycoprotein and NP genes, respectively. In plasmid LCMV MG#6 the HR was replaced by the HDR, and the G at position 3376 in LCMV S RNA was replaced by a C (arrow) to maximize the cleavage by the HDR (41). In LCMV MG#7 an extra C was added after the last 3′-terminal nucleotide. (B) Analysis of CAT activity produced by LCMV MGs. BHK-21 cells were infected with vFT7-3 at an MOI of 3 PFU/cell and were transfected with 0.5 μg of the indicated LCMV MG, 1.5 μg of pCITE-NP, and 0.1 μg of pGEM-L. Cells were harvested at 24 h postinfection and were analyzed for CAT activity by TLC as described in Materials and Methods. Pluses and minuses indicate the presence or absence of plasmid, respectively. NAc and MAc indicate the position of nonacetylated or monoacetylated forms of chloramphenicol in the TLC, respectively. (C) T7RP-mediated intracellular production of LCMV MG RNAs. BHK-21 cells were infected with vFT7-3 (MOI, 3 PFU/cell) and were transfected with 2 μg of the MG plasmid indicated at the top of the panel. Twenty-four hours postinfection total cellular RNA was prepared by using TriReagent, and 7 μg of RNA was analyzed by Northern blotting with a CAT sense riboprobe. Unprocessed and HDR-processed RNA species are indicated by arrows at the left. Molecular markers are indicated on the right. (D) Amplification of MG RNA by the LCMV polymerase. BHK-21 cells were infected with vFT7-3 (MOI, 3 PFU/cell) and transfected with 0.5 μg of MG#7. Plasmids pCITE-NP (1.5 μg) and pGEM-L (0.1 μg) were included in (+) or excluded from (−) the transfection mix. Total cell RNA was isolated, and 5 μg of RNA was analyzed by Northern blot hybridization by using a CAT sense RNA probe. Long exposure time of the film (data not shown) allowed for clear visualization of MG RNA species in lanes 2 and 3. (E) Quantification of MG RNA amplification by the LCMV polymerase. RNA species shown in panel D were quantified by PhosphorImager as described in Materials and Methods. The fraction of correctly processed MG RNA was determined for each sample (MG/MG + unprocessed) and was normalized to the amount of MG RNA synthesized by MG#7 in the presence of NP but in the absence of L (lane 3).
We next examined to what extent MG#7 RNA could be amplified by the LCMV polymerase. For this, BHK-21 cells infected with vTF7-3 were transfected with plasmid LCMV MG#7 alone or in combination with each of the supporting plasmids NP or L or both. Total RNA was prepared 24 h after transfection, and equal amounts were analyzed by Northern blot by using a positive-sense CAT riboprobe. In the absence of trans-acting factors, levels of MG#7 RNA detected corresponded to those resulting from the cleavage by the HDR of the T7RP-derived transcript (Fig. 1D, lane 2). In the absence of polymerase, no amplification was detected. Unexpectedly, some amplification was observed when L, but not NP, was present (Fig. 1D, lane 4). This result probably reflects a low level of unspecific encapsidation of the MG RNA by proteins produced as a result of the infection with vTF7-3. This encapsidated RNA would then be a suitable substrate for transcription and replication driven by the L polymerase. In the presence of both supporting plasmids, the amount of MG increased only by 50% (Fig. 1D, lane 5, and E). These data indicated that under the experimental conditions used in this study LCMV MG#7 could most likely undergo one round of replication.
Effects of 3′-terminal deletions on genome promoter activity.
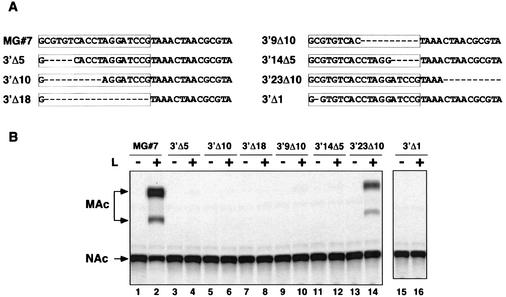
All arenaviruses examined to date show a high degree of sequence conservation at the 3′ termini of the L and S RNAs (17 of the first 19 nt are identical between the L and S RNAs), and the inverted complement of this sequence is located at the 5′ termini of the genomic RNAs (1). Twenty-one (L segment) or 20 (S segment) out of the first 23 nt of the 3′ and 5′ ends are complementary. Therefore, we hypothesized that these conserved 3′-terminal sequences contain the cis-acting signals that constitute the LCMV genome promoter. To test this hypothesis, we generated a collection of deletion mutants lacking increasing sequences of the 3′ end of MG#7 RNA (Fig. 2A). The effects of these mutations on transcription and replication mediated by the LCMV polymerase were determined by using the MG system. For all the deletion mutants we kept the G residue present at the end of the 3′ terminus of the S RNA. This was done on the basis of our finding that a change of G to C resulted in complete abrogation of CAT activity mediated by LCMV MG#6 (Fig. 1B).
FIG. 2.
Activity of mutant MGs with deletions within the 3′-terminal sequences of LCMV S RNA. (A) Sequences of the 3′ termini of the MG deletion mutants. Deleted nucleotides are indicated by a dash. The box enclosed the 19 conserved nucleotides at the 3′ end of the S RNA. (B) CAT expression by MGs with mutated 3′ termini. BHK-21 cells (106) were infected with vTF7-3 (MOI, 3 PFU/cell) and subsequently were transfected with 1.5 μg of pCITE-NP and 0.5 μg of the indicated MGs. Plasmid pGEM-L (0.1 μg) was included or omitted as indicated by the plus and minus signs. Total cell extracts were prepared 24 h postinfection, and 2 μg of protein was assayed with our standard CAT assay (see Materials and Methods). NAc and MAc indicate the position of nonacetylated or monoacetylated forms of chloramphenicol in the TLC, respectively.
BHK-21 cells infected with vTF7-3 were transfected with the indicated MG, L, and NP. L was omitted in duplicated samples as a negative control for RNA synthesis. Cell extracts were prepared 24 h after transfection, and the amount of protein was quantified. To obtain levels of CAT activity within the linear range (<30% conversion), 2 μg of total protein was used in a CAT reaction mixture that was incubated for 30 min a 37°C. MGs lacking the first 5, 10, or 18 3′-terminal nt were not competent in transcription on the basis of the lack of detectable levels of CAT activity (Fig. 2B). Likewise, mutants with deletions within the 3′ terminus lacking nt 10 to 19 (3′9Δ10) or nt 15 to 19 (3′14Δ5) were not competent in transcription (Fig. 2B, lanes 9 to 12).
The deletion of only 1 nt (3′Δ1) in the 3′ end of LCMV S RNA was sufficient to completely abolish CAT expression by the LCMV MG (Fig. 2B, lane 16). When the last 23 nt of the 3′ end were kept and the next 10 nt were deleted (3′23Δ10), the levels of CAT activity obtained were comparable to those of LCMV MG#7 (Fig. 2B, lane 14). These results indicated that in the S RNA the first 23 3′-terminal nucleotides contain the minimal LCMV genome promoter.
Contribution of the predicted panhandle structure to promoter activity.
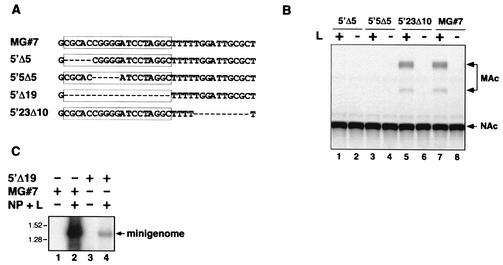
Studies on the influenza A virus promoter have provided strong evidence about the contribution of both 3′ and 5′ termini on initiation of transcription and replication. This process involves regions of single- and double-stranded RNA, and a RNA-fork model has been proposed for the initiation of transcription in influenza A (4, 16-19, 27, 38, 63). In contrast, a hook-like model has been proposed for Thogoto virus (29, 57). In LCMV, both the S and L RNA segments exhibit terminal complementarity resulting in a predicted panhandle structure between the respective 3′ and 5′ ends. However, there is no information about the contribution of the panhandle structure to the LCMV genome promoter activity. For this reason we examined the effects of 5′-terminal deletions on RNA synthesis. Mutants were generated (Fig. 3A) that lacked the first 5 nt of the 5′-terminal sequence (5′Δ5), that lacked nt 6 to 10 (5′5Δ5), or that contained the first 23 nt but lacked the following 10 nt (5′23Δ10). When these mutant MGs were tested for promoter activity, CAT activity was detected only when the first 23 nt of the 5′ end were retained (Fig. 3B).
FIG. 3.
Activity of mutant MGs with deletions within the 5′-terminal sequences of LCMV S RNA. (A) Sequence of the 5′ end of the MG deletion mutants. Deleted nucleotides are represented by a dash. (B) CAT expression by MGs with mutated 5′ ends. Transfection with the indicated MGs and analysis of CAT activity were done as described in the legend to Fig. 2. (C) Encapsidation of 5′Δ19 MG RNA. BHK-21 cells were transfected with either 5′Δ19 mutant or wild-type MG, alone or in combination with plasmids expressing NP and L. Cytoplasmic extracts were prepared and RNPs were purified as described in Materials and Methods. RNA was extracted from purified RNPs and was analyzed by Northern blot by using a CAT sense riboprobe. NAc and MAc indicate the position of nonacetylated or monoacetylated forms of chloramphenicol in the TLC, respectively.
For the negative-stranded RNA viruses, the encapsidation signal(s) is presumed to reside at the 5′ terminus of the RNA (28). Therefore, the lack of CAT activity observed with mutant MGs 5′Δ5 and 5′5Δ5 could reflect a disruption in a putative encapsidation signal located within or in the proximity of the deleted nucleotides. To address this issue we generated a mutant MG that lacked the first 19 nt of the 5′ end (MG 5′Δ19). As expected, this construct did not produce detectable levels of CAT activity when tested in the minigenome assay in the presence of NP and L (data not shown). We then assessed the ability of MG 5′Δ19 to be encapsidated by the viral nucleoprotein. For this, BHK-21 cells were cotransfected with MG 5′Δ19 and a plasmid that expresses the T7RP under the control of a Pol II promoter (pCAGGs-T7pol). Transcription-supporting plasmids pCITE-NP and pGEM-L were included in or excluded from the transfection mix as indicated (Fig. 3C). Cytoplasmic extracts were prepared 24 h after transfection. Encapsidated RNA was isolated as described in Materials and Methods. Encapsidated RNA (Fig. 3C) was analyzed by Northern blot by using a CAT sense riboprobe. The introduction of an extra C residue at the 3′ end of LCMV MG#7 did not affect the encapsidation of the plasmid-encoded minigenome (Fig. 3C, lane 2). Transcription of plasmid MG 5′Δ19 by T7RP consistently resulted in levels of MG 5′Δ19 RNA that were significantly lower than those of MG#7, and therefore levels of encapsidated 5′Δ19 MG appeared to be significantly lower than those of MG#7. However, when normalized for their corresponding levels of intracellular synthesis (compare lanes 2 and 9 of Fig. 6A), both MG RNA species were encapsidated with similar efficiency in the presence of NP and L (Fig. 3C, lanes 2 and 4).
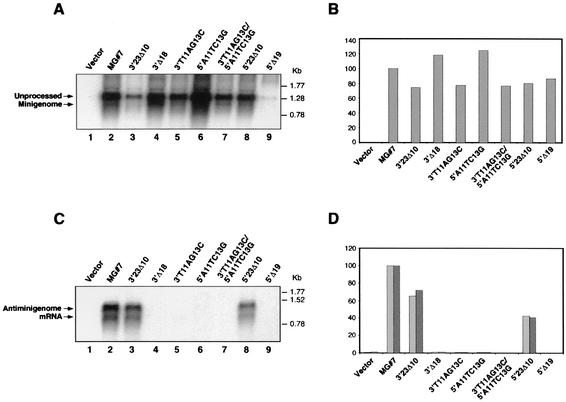
FIG. 6.
Northern blot analysis of the genomic and antigenomic RNA species and mRNA generated from different mutant MGs. (A) Indicated wild-type and mutant MGs were transfected in the absence of trans-acting factors. Total RNA (5 μg) was analyzed by Northern blot by using a CAT sense riboprobe. (B) Quantification of the levels of T7RP-mediated primary transcription and of the amount of processed MG RNA generated intracellularly. The amount of MG RNA was normalized with respect to the total amount of RNA synthesized as described for Fig. 1E. (C) Each MG plasmid was transfected together with plasmids expressing NP and L. Total RNA was analyzed by Northern blot by using a CAT antisense riboprobe that hybridized to the CAT mRNA and antigenomic RNA species. (D) Quantification of the amounts of antigenome (light bars) and mRNA (dark bars) synthesized. The abundance of each RNA is expressed as a percentage of that of the wild-type MG.
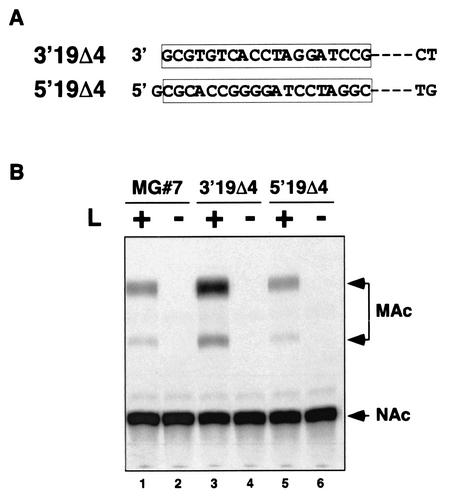
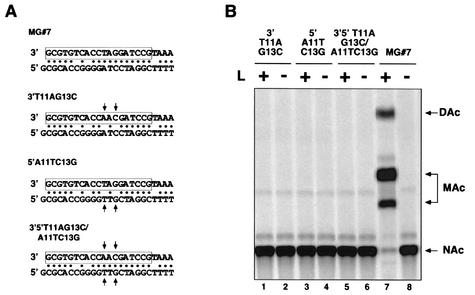
To characterize further the cis-acting sequences involved in RNA synthesis, we generated two new mutant MGs. These MG RNAs retained the first 19 nt of the 3′ or 5′ termini and lacked the following 4 nt that are largely conserved between LCMV L and S RNA segments. These mutants were named 3′19Δ4 and 5′19Δ4, respectively (Fig. 4A). Both mutant MGs promoted levels of CAT activity similar to those seen with wild-type MG#7 (Fig. 4B). Likewise, to further examine the role of complementarity in LCMV promoter activity we generated additional MG mutants. We mutated the 3′-terminal sequence at positions 11 and 13. The change of the T residue at position 11 to an A residue and of the G residue at position 13 to a C (3′T11AG13C) modified the linear sequence of the virus promoter and at the same time prevented the formation of two of the base pairings (Fig. 5A) between the 3′ and the 5′ termini of the S RNA. In mutant 5′A11TC13G two positions were mutated in the 5′-terminal sequence, abrogating the formation of two base pairs without affecting the putative 3′ promoter sequence. Both mutants failed to express CAT, as measured by the CAT assay shown in Fig. 5B (lanes 1 and 3). In this particular experiment, the amount of protein used for the CAT reaction mixture was increased from 2 to 100 μg in an attempt to detect any residual activity the promoter might have retained.
FIG. 4.
The first and last 19 nt of the 5′ and 3′ termini, respectively, are sufficient for promoter activity. (A) Sequence of the MG mutants. The 19 conserved nucleotides at either the 3′ or 5′ end were kept, and the next four nucleotides were deleted. (B) Analysis of CAT activity. BHK-21 cells were infected with vTF7-3 and were transfected as indicated in the legend of Fig. 2B in the presence (lanes 1, 3, and 5) or absence (lanes 2, 4, and 6) of L. NAc and MAc indicate the position of nonacetylated or monoacetylated forms of chloramphenicol in the TLC, respectively.
FIG. 5.
Role of the complementary between the 3′ and the 5′ termini of the S RNA on promoter activity. (A) Sequence complementarity between the 3′ and 5′ termini. An arrow indicates positions mutated. (B) Detection of CAT activity. Equal amounts of extracts prepared from BHK-21 cells infected with vTF7-3 and transfected with the indicated MG together with NP and in the presence (+) or absence (−) of L were analyzed for CAT activity. NAc and MAc indicate the position of nonacetylated or monoacetylated forms of chloramphenicol in the TLC, respectively. DAc, diacetylated chloramphenicol.
We considered that if initiation of transcription was mainly related to panhandle formation, then 3′T11AG13C mutant promoter activity could be rescued by introducing the appropriate compensatory mutations within the 5′ end to restore the structure of the panhandle. With this aim we constructed mutant 3′5′T11AG13C/A11TC13G in which all four positions were mutated and therefore base pairing was reestablished, yet the sequences of the 3′ genomic and 5′ antigenomic promoters were altered. In contrast with the findings reported for influenza A virus (18), restoring the structure of the panhandle did not recover the activity of the LCMV promoter (Fig. 5B). The estimated free energy of the maximally stable double-stranded structures formed between the 3′- and 5′-terminal sequences were very similar for the wild-type and mutated 3′5′T11AG13C/A11TC13G MG RNAs and would be very unlikely to account for the lack of promoter activity.
These results prove that the 5′ end of the viral RNA plays a substantial role as part of the viral promoter and that both sequence and structure of the 3′- and 5′-terminal regions are important for transcription initiation.
Analysis of RNA expression of LCMV MGs with mutated promoters.
Low levels of T7RP-mediated intracellular transcription of MG RNA or inefficient processing of the MG RNA by the HDR could have contributed to undetectable levels of CAT activity derived from some of the mutant MGs. This, in turn, would have important consequences for the interpretation of the experiments aimed at defining the minimal LCMV genome promoter. To address this issue we isolated RNA from BHK-21 cells infected with vFT7-3 and transfected with the indicated MG mutants. Equal amounts of each RNA were analyzed by Northern blot hybridization by using a CAT sense riboprobe (Fig. 6A). Levels of T7RP-mediated transcription were similar for most mutant MGs compared to that of MG#7. The reduced amount of both unprocessed and processed RNA, shown in lane 3 of Fig. 6 for MG 3′23Δ10, corresponded to a difference in the amount of RNA sample loaded and not to a real decrease in RNA synthesis and was not observed in repeats of the same experiment. The only mutant MG that consistently expressed very low levels of RNA was 5′Δ19. It is possible that the new sequence downstream of the T7 promoter affected efficient synthesis by T7RP, as has been described for Rous sarcoma virus (RSV) genome analogs (40). In all the cases, a similar percentage of the unprocessed RNA was cleaved by the HDR to render the processed MG RNA (Fig. 6B).
We also examined levels of RNA species corresponding to replication and transcription by the virus polymerase of mutant MGs. For this, vTF7-3-infected BHK-21 cells were transfected with the indicated mutant MG constructs together with plasmids encoding NP and L. RNA was isolated 24 h after transfection and was analyzed by Northern blot hybridization by using a CAT antisense riboprobe. Two species of RNA were detected, the bigger one corresponding to the product of MG replication and the smaller one corresponding to the CAT mRNA (Fig. 6C). Only those MGs that were competent in replication were able to serve as template for the transcription of CAT mRNA. Only mutant MGs that displayed detectable levels of CAT activity supported levels of replication and transcription mediated by the viral polymerases that were detectable by Northern blot hybridization.
DISCUSSION
The observation of a high degree of sequence conservation among the 3′ termini of arenavirus genomes and terminal complementarity between the 3′ and 5′ ends of genome and antigenome RNA species led to the proposal that these sequences likely play key roles in the regulation of RNA synthesis by the virus polymerase (6). However, the inability to genetically manipulate the arenavirus genomes has prevented testing this hypothesis experimentally. The development of a reverse genetic system for the prototypic arenavirus LCMV (30) has provided us with a new powerful tool to examine this question. By using an MG rescue assay based on the S segment of LCMV (ARM strain) we have obtained evidence that the conserved 3′-terminal 19 nt of the S RNA contain the core of the LCMV genome promoter. Minimal disruption of the terminal complementarity between the 3′ and 5′ ends of the LCMV MG abolished RNA synthesis by the virus polymerase. This finding suggests that the predicted panhandle structure formed between the 3′ and 5′ ends of the viral genome is required for efficient RNA synthesis mediated by the LCMV polymerase. Furthermore, the LCMV promoter appears to be exquisitely sensitive to minor sequence alterations. Thus, all nucleotide substitutions we have so far introduced within the MG 3′-terminal 19 nt resulted in abrogation of promoter activity, as determined by the lack of detectable CAT activity and synthesis of both CAT mRNA and anti-genome RNA species. Deletion of a single nucleotide (3′Δ1) within the 3′-terminal 19 nt of LCMV MG S RNA resulted in undetectable levels of CAT expression (Fig. 2B, lane 16). In contrast, for several other negative-strand RNA viruses it has been possible to introduce changes within the minimal viral genome promoter without abrogating its activity. For example, deletion of the 3′-terminal first 3 nt of the genome promoter of RSV had no effect on promoter activity (12), whereas the influenza A genome promoter lacking the first 3′-end nucleotide retained about 60% of wild-type promoter activity (32). Moreover, the 3′-terminal first two nucleotides of the S RNA of the phlebovirus Rift Valley Fever (RVF) were not required for the transcriptional activity of the RVF S genome promoter (43). Our data do not allow us to rule out that deletions or substitutions of more than one nucleotide at a time are not permitted in the case of the LCMV promoter. In our study, most of the deletions removed a minimum of 5 nt, whereas mutants generated by site-directed mutagenesis involved two different nucleotide residues at the same time. To address this question, a more detailed analysis of the promoter sequence would be required, including saturation mutagenesis of each nucleotide position that conforms to the core LCMV genome promoter.
All mutant MGs we examined, with the exception of 5′Δ19, were efficiently transcribed by the T7RP, and the efficiency of HDR-mediated processing was similar for all T7RP-derived primary transcripts. Therefore, the lack of CAT activity observed for some of the mutants cannot be explained by a reduction in the amount of RNA template available to the LCMV polymerase. The low levels of T7RP-driven transcription observed with mutant 5′Δ19 could be explained if the sequence at the 5′ end of 5′Δ19 MG, located immediately after the T7RP promoter, is unfavorable for T7RP transcription. Peeples and Collins have reported that the magnitude of T7RP transcription in transfected cells is sensitive to substitutions in the region adjoining the T7RP promoter located beyond the three G residues that are required for maximal promoter activity (40).
Signals involved in encapsidation of the genomic and antigenomic RNAs of nonsegmented negative-stranded RNA virus are thought to be located near the 5′ end of each RNA (28). There is evidence supporting this hypothesis for rabies virus (64) and vesicular stomatitis virus (VSV) (7, 37, 52) but not for RSV (40). Moreover, mutant mini-antigenomes of VSV containing 6-nt deletions spanning the 5′-terminal 47-nt region were encapsidated with efficiency similar to that of the wild-type mini-antigenome (31). Recently, a 5′ encapsidation signal has also been described for the S segment of the tripartite, negative-stranded Hantaan viruses (50). In our case, deletion of the 5′-terminal 19 nt did not appear to significantly affect the encapsidation of the LCMV MG RNA synthesized intracellularly by the T7RP. Nevertheless, a more detailed analysis of the 5′-terminal sequence of LCMV genome RNAs will be required before ruling out the presence of an encapsidation signal within the 5′ end. The differences observed in the amounts of encapsidated 5′Δ19 and MG#7 RNAs (Fig. 3C) correlated with the substantial reduction in the amount of 5′Δ19 primary transcript synthesized intracellularly by the T7RP (Fig. 6A).
As with other minireplicon systems described for negative-strand RNA viruses, only a very small percentage of the LCMV MG RNA synthesized by the T7RP was encapsidated, becoming a suitable template for L-driven replication and transcription. We observed only a minor increase (less than twofold) in the amount of correctly sized MG#7 RNA produced in the presence versus absence of the trans-acting factors NP and L (Fig. 1D, compare lanes 2 and 5). This result likely reflects the low amount of MG#7 RNA synthesized de novo by the LCMV polymerase. Encapsidation of only a very small fraction of the MG#7 RNA synthesized intracellularly by T7RP, together with high stability of unencapsidated MG RNA, could also contribute to mask the amplification of MG#7 RNA by the LCMV polymerase. However, a plasmid encoding an MG identical to MG#7 but lacking two of the three G residues at the end of the core T7RP promoter sequence produced very low intracellular levels of T7RP-derived MG RNA, but it was amplified very efficiently (about 50-fold) by the LCMV polymerase (data not shown). Therefore, we favor the explanation that under the experimental conditions used in this study LCMV MG#7 appears to be restricted mostly to the synthesis of positive-sense RNAs (anti-MG RNA and CAT mRNA) and produces very little progeny MG RNA. The 5′ end of arenavirus genomic S RNA segments appears to have one nontemplated G residue (24, 44). The presence of one nontemplated G nucleotide at the 5′ end of the genomic S RNA segment has been reported for LCMV (34), Tacaribe virus (23, 44), Pichinde virus (2, 42), and Lassa fever virus (3) but not for LCMV strain WE (46, 47). It is possible that the presence of two additional nonvirally coded G residues at the 5′ end of the MG#7 would result in mini-antigenome RNA species with a 3′ end that is not recognized by the virus polymerase as efficiently as the original wild-type 3′-end sequence. It is also worth mentioning that although CAT assays have a high sensitivity, levels of MG replication below the sensitivity of our Northern blot assay might occur but go undetected.
The MG used in this study contains the 3′- and 5′-terminal sequences of the S segment of strain ARM of LCMV. Nevertheless, the high degree of sequence conservation observed between the 3′ end of the S and the L segments likely reflects a conservation of the promoter sequences, and thereby the conclusions of this study are likely extrapolative to the transcription and replication of the L segment. The 3′-terminal nucleotide sequence of the LCMV ARM L RNA differs from the S RNA nucleotide sequence by four base substitutions (positions 6, 8, 9, and 17) (1). It is plausible that these positions, together with the mismatches in the base pairing of the inverted terminal repeats, represent regulatory elements that modulate the activity of each S and L genomic promoters.
A prime-and-realign mechanism has been proposed for the initiation of arenavirus RNA replication (23, 24). This model accounts for the presence of a nontemplated G at the 5′ ends of the arenavirus genomic and antigenomic RNAs. This model assumes that arenavirus polymerases, like many other viral RdRps, can initiate RNA synthesis de novo only with ATP or GTP (5). Therefore, arenavirus RNA initiation is thought to take place from an internal (+2) templated cytidilate. Once the first phosphodiester bond has been formed, the pppGpC will slip backwards on the template and realign, creating a nascent chain whose 5′ end is at position −1 with respect to the template, before the polymerase can continue the downstream synthesis. The length of the RNA is kept constant despite a nontemplated nucleotide being added at the 5′ end, because the polymerase is supposed to terminate RNA synthesis by removing the last base (24). A similar mechanism has been proposed for RNA initiation of Hantaan viruses (Bunyaviridae) (25). According to this model, the replacement of the 3′-terminal G by C made in MG#6 should allow the LCMV polymerase to initiate at the very 3′ end of the template without requirement for realignment. The lack of the 5′-end nontemplated G should result in the synthesis of an anti-MG#6 one nucleotide shorter than its template, and the shortening of the genome ends with continued replication. In contrast, the addition of a C residue to the 3′ end of MG#7 would not be expected to have an impact in MG and anti-MG RNA sizes during replication mediated by the LCMV polymerase. These differences between the 3′ ends of MG#6 and MG#7 might have contributed to the lack of CAT activity by MG#6. MG#7 is unlikely to be engaged in multiple rounds of replication (Fig. 1D), but it exhibited wild-type levels of promoter activity (Fig. 1B). These findings are more consistent with the view that the sequence change G to C in MG#6 altered the activity of the promoter and thereby its ability to drive synthesis of CAT mRNA and subsequent production of CAT activity.
Genomic and antigenomic viral RNA species with terminal deletions or nontemplated nucleotide extensions have been documented during persistent LCMV infections. These truncated RNAs have been proposed to be a new type of defective interfering genome that contributes to the establishment and maintenance of LCMV persistence (35). It has been hypothesized that truncated RNAs are replication competent but are not used as templates for transcription (35, 36). An increased ratio of truncated to full-length RNAs would be expected to cause a decrease in viral gene expression. On the other hand, addition of nontemplated nucleotides could repair some truncated RNAs to full-length sequence, thus leading to a transient increase in virus gene expression. This hypothesis is compatible with the observation that LCMV persistence is characterized by cyclical changes in numbers of antigen-positive cells and production of infectious virus. A corollary of this proposed hypothesis would be that the arenavirus highly conserved 3′-terminal 19-nt sequence is not strictly required for initiation of replication. Nevertheless, by using our MG system we found that deletions affecting the 3′ end of the MG resulted in a complete lack of promoter activity. One possible explanation for this apparent discrepancy between our data and the observations by Meyer and Southern (35) would be that truncated RNAs detected in LCMV persistence are continuously generated de novo from full-length templates by an unknown mechanism. On the other hand, we cannot rule out the possibility that cellular factors induced or modified by LCMV persistence could influence the activity of the virus polymerase in ways that are not recreated by our MG rescue assay.
The structure of a predicted panhandle formed between the 3′- and 5′-terminal genomic sequences has been proved to be strictly required for promoter function of several viruses. This is the case for orthomyxoviruses, like influenza (16, 19, 38), and Thogoto virus (57), where both 3′- and 5′-terminal sequences have been involved in the initiation of transcription. Terminal complementarity has also been shown to affect transcription and replication by VSV (58, 59). In contrast to these examples, formation of a panhandle does not appear to play any role in the replication of RSV (15) or Sendai virus (56). Our results indicate that in the case of LCMV, preservation of terminal complementarity is required for promoter activity. Lack of complementarity at two nucleotide positions within the predicted panhandle of MG#7 was sufficient to abolish promoter activity. Estimated free energy (ΔG) for predicted panhandles of MG#7, 3′T11AG13C, and 5′A11TC13G MG RNAs were very similar, suggesting that panhandle instability was unlikely to be the reason for the lack of promoter activity associated with 3′T11AG13C and 5′A11TC13G MGs. Moreover, the introduction of compensatory mutations in MG 3′5′T11AG13C/A11TC13G did not restore promoter activity (Fig. 5). Mutations within the 5′ end of MG#7 could affect the ability of the antigenomic promoter and thereby the complete cycle of RNA replication (i.e., MG to anti-MG to MG). Ensuing decreased levels of MG production by the LCMV polymerase would result in lower levels of template for production of anti-MG RNA species. This, in turn, could introduce a confounding factor in our assessment of the influence of mutations within the MG 5′ end on LCMV genomic promoter activity. However, as we discussed earlier, in our assay LCMV MG RNA replication is mostly restricted to one step of the full replication cycle. Therefore, both sequence specificity at the 3′ and 5′ ends together with structural constraints associated with the predicted panhandle formed between the 3′- and 5′-terminal sequences are critical for the activity of the LCMV genomic promoter. The presence of the IGR of the LCMV S RNA in MG#7 raises the possibility that the IGR could play an essential role in the activity of the arenavirus genome promoter. However, the activity of the Tacaribe virus genome promoter was not affected in a Tacaribe virus MG devoid of IGR sequences (33).
There is an increasing interest in the potential of RNA as a new drug target (55, 61). The apparent lack of sequence flexibility within the arenavirus core promoter, if validated, raises the possibility of novel antiviral strategies against arenaviruses based on the use of small molecules to target the predicted panhandle structure required for arenavirus promoter activity.
Acknowledgments
We thank P. Southern for very valuable criticisms and comments.
This work was supported by NIH grant AI47140 (J.C.D.L.T.).
Footnotes
This is publication 15213-NP from The Scripps Research Institute.
REFERENCES
- 1.Auperin, D. D., R. W. Compans, and D. H. Bishop. 1982. Nucleotide sequence conservation at the 3′ termini of the virion RNA species of New World and Old World arenaviruses. Virology 121:200-203. [DOI] [PubMed] [Google Scholar]
- 2.Auperin, D. D., V. Romanowski, M. Galinski, and D. H. Bishop. 1984. Sequencing studies of pichinde arenavirus S RNA indicate a novel coding strategy, an ambisense viral S RNA. J. Virol. 52:897-904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Auperin, D. D., D. R. Sasso, and J. B. McCormick. 1986. Nucleotide sequence of the glycoprotein gene and intergenic region of the Lassa virus S genome RNA. Virology 154:155-167. [DOI] [PubMed] [Google Scholar]
- 4.Bae, S. H., H. K. Cheong, J. H. Lee, C. Cheong, M. Kainosho, and B. S. Choi. 2001. Structural features of an influenza virus promoter and their implications for viral RNA synthesis. Proc. Natl. Acad. Sci. USA 98:10602-10607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Banerjee, A. K. 1980. 5′-Terminal cap structure in eucaryotic messenger ribonucleic acids. Microbiol. Rev. 44:175-205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bishop, D. H., and D. D. Auperin. 1987. Arenavirus gene structure and organization. Curr. Top. Microbiol. Immunol. 133:5-17. [DOI] [PubMed] [Google Scholar]
- 7.Blumberg, B. M., C. Giorgi, and D. Kolakofsky. 1983. N protein of vesicular stomatitis virus selectively encapsidates leader RNA in vitro. Cell 32:559-567. [DOI] [PubMed] [Google Scholar]
- 8.Borrow, P., and M. B. Oldstone. 1997. Lymphocytic choriomeningitis virus, p. 593-627. In N. Nathaanson (ed.), Viral pathogenesis, vol. 1. Lippincott-Raven, Philadelphia, Pa.
- 9.Borrow, P., and M. B. Oldstone. 1994. Mechanism of lymphocytic choriomeningitis virus entry into cells. Virology 198:1-9. [DOI] [PubMed] [Google Scholar]
- 10.Bowen, M. D., C. J. Peters, and S. T. Nichol. 1996. The phylogeny of New World (Tacaribe complex) arenaviruses. Virology 219:285-290. [DOI] [PubMed] [Google Scholar]
- 11.Cao, W., M. D. Henry, P. Borrow, H. Yamada, J. H. Elder, E. V. Ravkov, S. T. Nichol, R. W. Compans, K. P. Campbell, and M. B. Oldstone. 1998. Identification of alpha-dystroglycan as a receptor for lymphocytic choriomeningitis virus and Lassa fever virus. Science 282:2079-2081. [DOI] [PubMed] [Google Scholar]
- 12.Collins, P. L., M. A. Mink, and D. S. Stec. 1991. Rescue of synthetic analogs of respiratory syncytial virus genomic RNA and effect of truncations and mutations on the expression of a foreign reporter gene. Proc. Natl. Acad. Sci. USA 88:9663-9667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Conzelmann, K. K. 1996. Genetic manipulation of non-segmented negative-strand RNA viruses. J. Gen. Virol. 77:381-389. [DOI] [PubMed] [Google Scholar]
- 14.Cornu, T. I., and J. C. de la Torre. 2001. RING finger Z protein of lymphocytic choriomeningitis virus (LCMV) inhibits transcription and RNA replication of an LCMV S-segment minigenome. J. Virol. 75:9415-9426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fearns, R., P. L. Collins, and M. E. Peeples. 2000. Functional analysis of the genomic and antigenomic promoters of human respiratory syncytial virus. J. Virol. 74:6006-6014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Flick, R., G. Neumann, E. Hoffmann, E. Neumeier, and G. Hobom. 1996. Promoter elements in the influenza vRNA terminal structure. RNA 2:1046-1057. [PMC free article] [PubMed] [Google Scholar]
- 17.Fodor, E., P. Palese, G. G. Brownlee, and A. Garcia-Sastre. 1998. Attenuation of influenza A virus mRNA levels by promoter mutations. J. Virol. 72:6283-6290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fodor, E., D. C. Pritlove, and G. G. Brownlee. 1995. Characterization of the RNA-fork model of virion RNA in the initiation of transcription in influenza A virus. J. Virol. 69:4012-4019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fodor, E., D. C. Pritlove, and G. G. Brownlee. 1994. The influenza virus panhandle is involved in the initiation of transcription. J. Virol. 68:4092-4096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fuerst, T. R., E. G. Niles, F. W. Studier, and B. Moss. 1986. Eukaryotic transient-expression system based on recombinant vaccinia virus that synthesizes bacteriophage T7 RNA polymerase. Proc. Natl. Acad. Sci. USA 83:8122-8126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fuller-Pace, F. V., and P. J. Southern. 1989. Detection of virus-specific RNA-dependent RNA polymerase activity in extracts from cells infected with lymphocytic choriomeningitis virus: in vitro synthesis of full-length viral RNA species. J. Virol. 63:1938-1944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fuller-Pace, F. V., and P. J. Southern. 1988. Temporal analysis of transcription and replication during acute infection with lymphocytic choriomeningitis virus. Virology 162:260-263. [DOI] [PubMed] [Google Scholar]
- 23.Garcin, D., and D. Kolakofsky. 1990. A novel mechanism for the initiation of Tacaribe arenavirus genome replication. J. Virol. 64:6196-6203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Garcin, D., and D. Kolakofsky. 1992. Tacaribe arenavirus RNA synthesis in vitro is primer dependent and suggests an unusual model for the initiation of genome replication. J. Virol. 66:1370-1376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Garcin, D., M. Lezzi, M. Dobbs, R. M. Elliott, C. Schmaljohn, C. Y. Kang, and D. Kolakofsky. 1995. The 5′ ends of Hantaan virus (Bunyaviridae) RNAs suggest a prime-and-realign mechanism for the initiation of RNA synthesis. J. Virol. 69:5754-5762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gonzalez, J. P., M. D. Bowen, S. T. Nichol, and R. Rico-Hesse. 1996. Genetic characterization and phylogeny of Sabia virus, an emergent pathogen in Brazil. Virology 221:318-324. [DOI] [PubMed] [Google Scholar]
- 27.Kim, H. J., E. Fodor, G. G. Brownlee, and B. L. Seong. 1997. Mutational analysis of the RNA-fork model of the influenza A virus vRNA promoter in vivo. J. Gen. Virol. 78:353-357. [DOI] [PubMed] [Google Scholar]
- 28.Lamb, R. A., and D. Kolkofsky. 2001. Paramyxoviridae: the viruses and their replication, p. 1305-1340. In D. M. Knipe and P. M. Howley (ed.), Fields virology, vol. 1, 4th ed. Lippincott Williams & Wilkins, Philadelphia, Pa.
- 29.Leahy, M. B., J. T. Dessens, and P. A. Nuttall. 1997. Striking conformational similarities between the transcription promoters of Thogoto and influenza A viruses: evidence for intrastrand base pairing in the 5′ promoter arm. J. Virol. 71:8352-8356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lee, K. J., I. S. Novella, M. N. Teng, M. B. Oldstone, and J. C. de La Torre. 2000. NP and L proteins of lymphocytic choriomeningitis virus (LCMV) are sufficient for efficient transcription and replication of LCMV genomic RNA analogs. J. Virol. 74:3470-3477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li, T., and A. K. Pattnaik. 1999. Overlapping signals for transcription and replication at the 3′ terminus of the vesicular stomatitis virus genome. J. Virol. 73:444-452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li, X., and P. Palese. 1992. Mutational analysis of the promoter required for influenza virus virion RNA synthesis. J. Virol. 66:4331-4338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lopez, N., R. Jacamo, and M. T. Franze-Fernandez. 2001. Transcription and RNA replication of Tacaribe virus genome and antigenome analogs require N and L proteins: Z protein is an inhibitor of these processes. J. Virol. 75:12241-12251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Meyer, B. J., and P. J. Southern. 1993. Concurrent sequence analysis of 5′ and 3′ RNA termini by intramolecular circularization reveals 5′ nontemplated bases and 3′ terminal heterogeneity for lymphocytic choriomeningitis virus mRNAs. J. Virol. 67:2621-2627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Meyer, B. J., and P. J. Southern. 1997. A novel type of defective viral genome suggests a unique strategy to establish and maintain persistent lymphocytic choriomeningitis virus infections. J. Virol. 71:6757-6764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Meyer, B. J., and P. J. Southern. 1994. Sequence heterogeneity in the termini of lymphocytic choriomeningitis virus genomic and antigenomic RNAs. J. Virol. 68:7659-7664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Moyer, S. A., S. C. Baker, and J. L. Lessard. 1986. Tubulin: a factor necessary for the synthesis of both Sendai virus and vesicular stomatitis virus RNAs. Proc. Natl. Acad. Sci. USA 83:5405-5409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Neumann, G., and G. Hobom. 1995. Mutational analysis of influenza virus promoter elements in vivo. J. Gen. Virol. 76:1709-1717. [DOI] [PubMed] [Google Scholar]
- 39.Oldstone, M. B., R. Ahmed, J. Byrne, M. J. Buchmeier, Y. Riviere, and P. Southern. 1985. Virus and immune responses: lymphocytic choriomeningitis virus as a prototype model of viral pathogenesis. Br. Med. Bull. 41:70-74. [DOI] [PubMed] [Google Scholar]
- 40.Peeples, M. E., and P. L. Collins. 2000. Mutations in the 5′ trailer region of a respiratory syncytial virus minigenome which limit RNA replication to one step. J. Virol. 74:146-155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Perrotta, A. T., and M. D. Been. 1991. A pseudoknot-like structure required for efficient self-cleavage of hepatitis delta virus RNA. Nature 350:434-436. [DOI] [PubMed] [Google Scholar]
- 42.Polyak, S. J., S. Zheng, and D. G. Harnish. 1995. 5′ Termini of Pichinde arenavirus S RNAs and mRNAs contain nontemplated nucleotides. J. Virol. 69:3211-3215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Prehaud, C., N. Lopez, M. J. Blok, V. Obry, and M. Bouloy. 1997. Analysis of the 3′ terminal sequence recognized by the Rift Valley fever virus transcription complex in its ambisense S segment. Virology 227:189-197. [DOI] [PubMed] [Google Scholar]
- 44.Raju, R., L. Raju, D. Hacker, D. Garcin, R. Compans, and D. Kolakofsky. 1990. Nontemplated bases at the 5′ ends of Tacaribe virus mRNAs. Virology 174:53-59. [DOI] [PubMed] [Google Scholar]
- 45.Riviere, Y., R. Ahmed, P. J. Southern, M. J. Buchmeier, F. J. Dutko, and M. B. Oldstone. 1985. The S RNA segment of lymphocytic choriomeningitis virus codes for the nucleoprotein and glycoproteins 1 and 2. J. Virol. 53:966-968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Romanowski, V., and D. H. Bishop. 1985. Conserved sequences and coding of two strains of lymphocytic choriomeningitis virus (WE and ARM) and Pichinde arenavirus. Virus Res. 2:35-51. [DOI] [PubMed] [Google Scholar]
- 47.Romanowski, V., Y. Matsuura, and D. H. Bishop. 1985. Complete sequence of the S RNA of lymphocytic choriomeningitis virus (WE strain) compared to that of Pichinde arenavirus. Virus Res. 3:101-114. [DOI] [PubMed] [Google Scholar]
- 48.Salvato, M., E. Shimomaye, P. Southern, and M. B. Oldstone. 1988. Virus-lymphocyte interactions. IV. Molecular characterization of LCMV Armstrong (CTL+) small genomic segment and that of its variant, clone 13 (CTL-). Virology 164:517-522. [DOI] [PubMed] [Google Scholar]
- 49.Salvato, M. S., and E. M. Shimomaye. 1989. The completed sequence of lymphocytic choriomeningitis virus reveals a unique RNA structure and a gene for a zinc finger protein. Virology 173:1-10. [DOI] [PubMed] [Google Scholar]
- 50.Severson, W. E., X. Xu, and C. B. Jonsson. 2001. cis-acting signals in encapsidation of Hantaan virus S-segment viral genomic RNA by its N protein. J. Virol. 75:2646-2652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Singh, M. K., F. V. Fuller-Pace, M. J. Buchmeier, and P. J. Southern. 1987. Analysis of the genomic L RNA segment from lymphocytic choriomeningitis virus. Virology 161:448-456. [DOI] [PubMed] [Google Scholar]
- 52.Smallwood, S., and S. A. Moyer. 1993. Promoter analysis of the vesicular stomatitis virus RNA polymerase. Virology 192:254-263. [DOI] [PubMed] [Google Scholar]
- 53.Southern, P. J. 1996. Arenaviridae: the virus and their replication, p. 1505-1519. In B. N. Fields, D. M. Knipe, and P. M. Howley (ed.), Fields virology, vol. 2, 3rd ed. Lippincott-Raven, Philadelphia, Pa.
- 54.Southern, P. J., M. K. Singh, Y. Riviere, D. R. Jacoby, M. J. Buchmeier, and M. B. Oldstone. 1987. Molecular characterization of the genomic S RNA segment from lymphocytic choriomeningitis virus. Virology 157:145-155. [DOI] [PubMed] [Google Scholar]
- 55.Sucheck, S. J., and C. H. Wong. 2000. RNA as a target for small molecules. Curr. Opin. Chem. Biol. 4:678-686. [DOI] [PubMed] [Google Scholar]
- 56.Tapparel, C., and L. Roux. 1996. The efficiency of Sendai virus genome replication: the importance of the RNA primary sequence independent of terminal complementarity. Virology 225:163-171. [DOI] [PubMed] [Google Scholar]
- 57.Weber, F., O. Haller, and G. Kochs. 1997. Conserved vRNA end sequences of Thogoto-orthomyxovirus suggest a new panhandle structure. Arch. Virol. 142:1029-1033. [DOI] [PubMed] [Google Scholar]
- 58.Wertz, G. W., S. Whelan, A. LeGrone, and L. A. Ball. 1994. Extent of terminal complementarity modulates the balance between transcription and replication of vesicular stomatitis virus RNA. Proc. Natl. Acad. Sci. USA 91:8587-8591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Whelan, S. P., and G. W. Wertz. 1999. Regulation of RNA synthesis by the genomic termini of vesicular stomatitis virus: identification of distinct sequences essential for transcription but not replication. J. Virol. 73:297-306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wilson, S. M., and J. C. Clegg. 1991. Sequence analysis of the S RNA of the African arenavirus Mopeia: an unusual secondary structure feature in the intergenic region. Virology 180:543-552. [DOI] [PubMed] [Google Scholar]
- 61.Wilson, W. D., and K. Li. 2000. Targeting RNA with small molecules. Curr. Med. Chem. 7:73-98. [DOI] [PubMed] [Google Scholar]
- 62.Wright, K. E., R. C. Spiro, J. W. Burns, and M. J. Buchmeier. 1990. Post-translational processing of the glycoproteins of lymphocytic choriomeningitis virus. Virology 177:175-183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yamanaka, K., N. Ogasawara, H. Yoshikawa, A. Ishihama, and K. Nagata. 1991. In vivo analysis of the promoter structure of the influenza virus RNA genome using a transfection system with an engineered RNA. Proc. Natl. Acad. Sci. USA 88:5369-5373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yang, J., H. Koprowski, B. Dietzschold, and Z. F. Fu. 1999. Phosphorylation of rabies virus nucleoprotein regulates viral RNA transcription and replication by modulating leader RNA encapsidation. J. Virol. 73:1661-1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Young, P. R., and C. R. Howard. 1983. Fine structure analysis of Pichinde virus nucleocapsids. J. Gen. Virol. 64:833-842. [DOI] [PubMed] [Google Scholar]