Abstract
The recently developed hepatitis C virus (HCV) subgenomic replicon system was utilized to evaluate the efficacy of several known antiviral agents. Cell lines that persistently maintained a genotype 1b replicon were selected. The replicon resident in each cell line had acquired adaptive mutations in the NS5A region that increased colony-forming efficiency, and some replicons had acquired NS3 mutations that alone did not enhance colony-forming efficiency but were synergistic with NS5A mutations. A replicon constructed from the infectious clone of the HCV-1 strain (genotype 1a) was not capable of inducing colony formation even after the introduction of adaptive mutations identified in the genotype 1b replicon. Alpha interferon (IFN-α), IFN-γ, and ribavirin exhibited antiviral activity, while double-stranded RNA (dsRNA) and tumor necrosis factor alpha did not. Analysis of transcript levels for a series of genes stimulated by IFN (ISGs) or dsRNA following treatment with IFN-α, IFN-γ, and dsRNA revealed that both IFNs increased ISG transcript levels, but that some aspect of the dsRNA response pathway was defective in Huh7 cells and replicon cell lines in comparison to primary chimpanzee and tamarin hepatocytes. The colony-forming efficiency of the replicon was reduced or eliminated following replication in the presence of ribavirin, implicating the induction of error-prone replication. The potential role of error-prone replication in the synergy observed between IFN-α and ribavirin in attaining sustained viral clearance is discussed. These studies reveal characteristics of Huh7 cells that may contribute to their unique capacity to support HCV RNA synthesis and demonstrate the utility of the replicon system for mechanistic studies on antiviral agents.
Chronic hepatitis C virus (HCV) infections are one of the leading causes of liver disease worldwide (2). The prevalence of HCV infections is 1 to 2%, although certain geographical regions, age groups, and ethnic groups have much higher rates of infection (3). Although symptoms may be mild for decades, 20% of persistently infected individuals may eventually develop serious liver disease including cirrhosis and liver cancer (2). HCV infection is the leading cause for liver transplantation in the United States (12). Although the initial use of interferon (IFN) for treatment of chronic infections yielded marginal results, the current therapeutic regimen of pegylated alpha 2b IFN (IFN-α2b) and ribavirin provides substantially improved rates of sustained viral clearance of 42 and 82% for genotype 1 and genotype 2 and 3, respectively (45). Treatment of acute infections with standard IFN therapy without ribavirin is highly efficacious and approaches 100% sustained viral clearance (33). Nonetheless, a great need exists for improved antiviral agents, since many patients still do not benefit from IFN therapy, and IFN therapy is associated with undesirable side effects. The lack of a suitable tissue culture system has previously hampered the development of antiviral agents, but the recent development of a replicon system for HCV (43) has partially fulfilled this need.
HCV is a member of the Flaviviridae family. The genome is single-stranded, positive-sense RNA (Fig. 1). Since the cloning of the viral genome (1, 11), rapid advances have been attained in defining viral functions (reviewed in references 5 and 56). The 5′ noncoding region (NCR) contains an internal ribosome entry site (IRES). The amino terminus of the viral polyprotein contains the structural proteins, the capsid and two envelope proteins, E1 and E2. The function of p7 is not known. NS2/NS3 is a metalloprotease that cleaves NS2 from NS3. NS3 is a serine protease and the viral helicase. NS4A is a cofactor for the serine protease. The function of NS4B is unknown. NS5B is the viral RNA polymerase. NS5A is a phosphoprotein that contains a sequence known as the IFN sensitivity-determining region (ISDR). Enomoto et al. (17) first demonstrated a relationship to sequence variation in this region and resistance to IFN therapy. Gale and colleagues have shown that this region interacts with PKR, providing a plausible mechanism for the modulation of the host response to IFN (22, 23). However, the precise function of NS5A is still unknown, and whether PKR binding accounts for viral resistance to IFN is controversial (54, 61). NS5A induces interleukin 8 synthesis that may contribute to IFN resistance (25, 55), and NS5A has been shown to interact with grb2 (62) and a SNARE-like protein (66). E2 has been shown to interact with PKR and may be involved in IFN resistance as well (51, 65). Following the polyprotein open reading frame is a 3′ NCR that has a variable region, a long poly(U)-polypyrimidine stretch, and a highly conserved 98-nucleotide terminus.
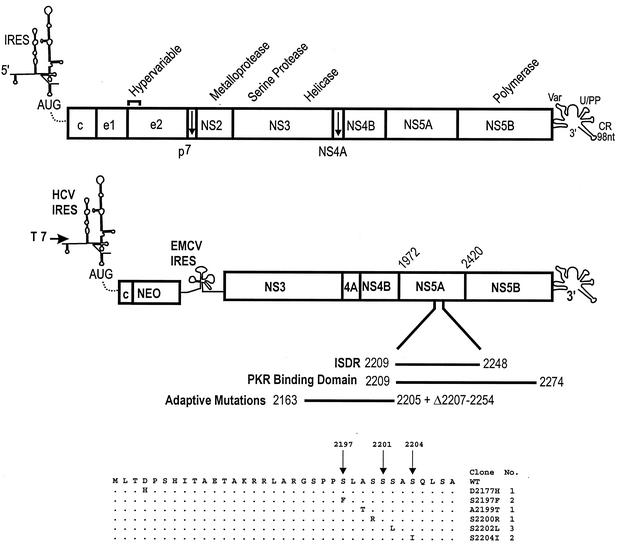
FIG. 1.
Schematic of HCV genome and replicon design. The HCV genome is depicted with the 5′ NCR containing an IRES and the 3′ NCR including a variable region (Var), a polyuridine or polypyrimidine stretch (U/PP), and a 98-nucleotide (nt) conserved region (CR). The depiction of the HCV IRES was adapted from the structural study of Honda et al. (28). The open reading frame of the polyprotein is depicted as a rectangle with demarcation of the individual viral protein domains, and the positions of some of the viral functions are depicted above. c, capsid protein; e1 and e2, envelope proteins E1 and E2. The structure of the bicistronic replicon is illustrated below the HCV genome, with the HCV IRES and EMCV IRES regulating translation of the neomycin phosphotransferase (NEO) gene and the nonstructural proteins of HCV, respectively. A T7 promoter is fused to the HCV 5′ terminus for the production of synthetic RNA. A domain of the NS5A protein is expanded to illustrate the ISDR, PKR binding domain, and the region of the most common adaptive replicon mutations, including the deletion from amino acids 2207 to 2254 observed by Blight et al. (8). The amino acid changes for the adaptive mutations detected in the resident replicons isolated from 10 independent G418-resistant colonies are indicated at the bottom of the figure, and the sites of NS5A hyperphosphorylation (amino acids 2197, 2201, and 2204) are indicated by arrows. WT, wild type.
Although HCV does not replicate in conventional tissue culture systems, a surrogate system has been created on the basis of a bicistronic replicon constructed by Lohmann and coworkers (43). The 5′ end of the HCV genome, including the IRES and 12 codons of the core protein, is fused in frame with the neomycin phosphotransferase gene, the encephalomyocarditis virus (EMCV) IRES drives translation of the HCV nonstructural proteins, and the construct terminates with the 3′ NCR of HCV. When synthetic RNA from this construct is transfected into the human liver cell line Huh7, G418-resistant colonies which persistently maintain the replicon RNA can be isolated. The utility of this system was markedly enhanced when Blight and coworkers (8) demonstrated that adaptive mutations arise that permit highly efficient colony formation. This has been reproduced in a number of studies including this one (8, 27, 35, 42). A replicon has been developed from a second genotype 1b strain (27, 32) that did not require adaptive mutations for high colony-forming efficiency (32), and recently replicons containing the full-length genome have been developed (32, 53).
In this study, we have utilized the replicon system for analysis of several compounds for potential antiviral effect, including IFN-α, IFN-γ, tumor necrosis factor alpha (TNF-α), poly(I)-poly(C), and ribavirin. Although no antiviral effect was observed with TNF-α or poly(I)-poly(C), a synthetic double-stranded RNA (dsRNA) and known inducer of IFN and IFN-stimulated genes (ISGs), both IFN-α and IFN-γ exhibited antiviral effects. The antiviral effect of ribavirin in this system could be ascribed to the induction of error-prone replication similar to recent findings with GB virus B (GBV-B) (38), a surrogate model for HCV. The expression levels for a number of ISGs were monitored before and after antiviral treatments to begin a characterization of the virus-host interactions involved in this system.
MATERIALS AND METHODS
Replicon constructs.
Rep1bNeo was constructed from synthetic oligonucleotides to be an exact copy of the replicon described by Lohmann et al. (43) (GenBank accession no. AJ242652). The Rep1aNeo construct is identical to the Rep1bNeo construct except that all HCV components were derived from the HCV-1 infectious clone (40) (GenBank accession no. AF271632) including the 5′ NCR, the region encoding NS3-NS5B, and the 3′ NCR. The Rep1b/aNeo construct was derived from Rep1aNeo by substituting 228 nucleotides (positions 1927 to 2155) from Rep1bNeo into Rep1aNeo, which altered 12 of the first 73 amino acids of NS3. Adaptive mutations were detected by direct sequencing of PCR products from NS3, NS5A, and NS5B which spanned nucleotides 3414 to 5312, 6828 to 7118, and 8855 to 9029, respectively. The specific adaptive mutations NS5A-S2204I and NS3-D1431Y were introduced into replicons using PCR-directed mutagenesis. Synthetic replicon RNA was prepared from DNA linearized with ScaI (Rep1bNeo) and XbaI (Rep1aNeo and Rep1b/aNeo) using the T7 Megascript kit (Ambion, Austin, Tex.) and was purified by DNase treatment, RNazol (Leedo, Houston, Tex.) extraction, and ethanol precipitation. RNA was quantified by optical density, and the concentration and quality were confirmed by agarose gel electrophoresis.
Cells and transfections.
Huh7 cells were cultivated in a 1:1 mixture of Dulbecco's modified Eagle's medium and Ham's F12 medium supplemented with 10% fetal bovine serum (FBS) and 50 μg of gentamicin sulfate per ml. RNA transfections were performed using DMRIE-C transfection reagent (GIBCO/BRL, Rockville, Md.) at a ratio of 5 μg of lipid to 1 μg of RNA. Replicons with low or no colony-forming efficiency were transfected using 10 μg of replicon RNA per 100-mm-diameter culture dish; however, replicons with adaptive mutations were transfected at 1 to 0.01 μg of replicon RNA diluted into Huh7 RNA. The RNA and lipid were diluted individually into 4.5 ml of medium without FBS, combined, incubated for 15 min at room temperature, and added to the cultures. Cultures were washed three times with medium without FBS prior to transfection and two times with medium containing FBS after transfection. Cultures were exposed to the RNA-lipid mixture for 6 h at 37°C. Culture medium was supplemented with 250 μg of G418 per ml beginning 1 day after transfection. This transfection protocol routinely yielded approximately 45% transfection efficiency for RNA, as evaluated by transfection of a Sindbis virus replicon expressing β-galactosidase (8). Measurement of replicon copy number and antiviral studies were conducted with cultures 48 h after plating (cultures were approximately 60% confluent). None of the antiviral treatments employed induced noticeable toxicity by microscopic inspection of the cultures or by measurement of glyceraldehyde-3-phosphate dehydrogenase (GAPDH) transcript levels. This was true even when treatment was extended for 7 days. Primary chimpanzee and tamarin hepatocytes were cultivated in a hormonally defined, serum-free medium as previously described (39).
TaqMan quantitative RT-PCR for replicon RNA and host transcripts.
Total cell RNA was isolated from cell cultures using RNazol (Leedo). Replicon RNA was quantified by a real-time, 5′ exonuclease reverse transcription-PCR (RT-PCR) (TaqMan) assay (40) using the ABI 7700 sequence detector (Perkin-Elmer Biosystems, Foster City, Calif.). The primers and probe were derived from the 5′ NCR and were selected using the Primer Express software designed for this purpose (Perkin-Elmer Biosystems). The forward primer contains nucleotides 149 to 167 (5′-TGCGGAACCGGTGAGTACA-3′), the reverse primer contains nucleotides 210 to 191 (5′-CGGGTTTATCCAAGAAAGGA-3′), and the probe contains nucleotides 189 to 169 (5′-CCGGTCGTCCTGGCAATTCCG-3′). The fluorogenic probe was labeled with FAM (6-carboxyfluorescein) and TAMRA (6-carboxytetramethylrhodamine) and was obtained from Synthegen (Houston, Tex.). The primers and probe were used at 10 pmol per 50-μl reaction mixture. The reactions were performed using the Brilliant Plus Single Step RT-PCR kit (Stratagene, La Jolla, Calif.) and included a 30-min 48°C RT step, followed by 10 min at 95°C, and then 40 cycles of amplification using the universal TaqMan RT-PCR standardized conditions: 15 s at 95°C for denaturation and 1 min at 60°C for annealing and extension. The standards used to establish genome equivalents (ge) were synthetic RNAs transcribed from a clone of the 5′ NCR of the HCV-1 strain (40). Synthetic RNA was prepared using the T7 Megascript kit and was purified by DNase treatment, RNazol extraction, and ethanol precipitation. RNA was quantified by optical density, and 10-fold serial dilutions were prepared from 1 million to 10 copies using tRNA as a carrier. These standards were run in all TaqMan RT-PCR assays in order to calculate the number of ge in the experimental samples. The conditions for quantification of transcripts from ISGs were identical to those described above for replicon RNA. Most assays were multiplexed using GAPDH. The primer and probe sets used in these assays will be presented elsewhere (C. Bigger, B. Guerra, K. M. Brasky, G. B. Hubbard, M. Beard, and R. E. Lanford, submitted for publication).
Antiviral treatments.
Poly(I)-poly(C) was obtained from ICN (Costa Mesa, Calif.) and Sigma (St. Louis, Mo.). A highly purified preparation of ribavirin (1-β-d-ribofuranosyl-1H-1,2,4-triazole-3-carboximide) was a gift from Schering Plough Research Institute (Kenilworth, N.J.). Some commercially available preparations of ribavirin contain trace contaminants that are often toxic in tissue culture studies. This preparation was specifically purified and selected for low toxicity in vitro. Human IFN-α-2b (intron A) was obtained from Schering Plough Research Institute. IFN-γ was obtained from R&D Systems (Minneapolis, Minn.). Antiviral treatments with IFN-α, IFN-γ, and poly(I)-poly(C) were initiated using subconfluent cultures, but after 24 h, the cultures reached confluency. Treatment with ribavirin used confluent cultures to reduce the adverse effects of ribavirin seen on rapidly dividing cells.
In vitro translation.
In vitro translations were preformed with 1 μg of replicon RNA prepared as described above, heated to 70°C for 10 min, and cooled on ice. Translation reactions were performed in a total volume of 25 μl using the Promega (Madison, Wis.) nuclease-treated rabbit reticulocyte lysate systems supplemented to contain 90 mM KCl to enhance for IRES-driven translation. Reaction mixtures contained 20 μM amino acids lacking methionine, 10 μCi of [35S]methionine (Express 35S; Perkin-Elmer, Boston, Mass.), and RNasin (Promega). Reactions were performed for 1 h at 30°C and the products were examined directly by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE).
PKR assays.
For PKR assays, Huh7 cells were pretreated with 100 μg of poly(I)-poly(C) per ml for 2 h or with 1,000 U of IFN-α2b per ml for 16 h or left untreated. Cultures were washed twice in phosphate-buffered saline (PBS), and cell lysates were prepared using PEB (PBS containing 1% Nonidet P-40 [NP-40], 10% glycerol, 1 mM dithiothreitol, and protease inhibitors) and were clarified in a microcentrifuge for 20 min at 4°C. PKR was immunoprecipitated for 2 h at 4°C using protein G beads containing 5 μg of monoclonal antibody clone 13 to PKR (BD Biosciences, San Diego, Calif.). The beads were washed five times with PEB and once with PKR buffer (20 mM Tris [pH 7.5], 0.1 mM EDTA, 80 mM KCl, 5% glycerol, 2 mM MgCl2, 2 mM MnCl2, and 0.2 mg of bovine serum albumin per ml). Kinase reactions were conducted for 15 min at 30°C with PKR bound to the beads in 50 μl of PKR buffer supplemented with 5 μCi of [γ-32P]ATP (Perkin-Elmer). PKR was eluted from the beads with SDS gel sample buffer and analyzed by SDS-PAGE and phosphorimage analysis.
Northern blot hybridization.
For analysis of replicon RNA by Northern blot hybridization, total cellular RNA was isolated from replicon lines using RNazol. RNA was analyzed by electrophoresis on a 1% agarose-formaldehyde gel at 30 V for 16 h at 16°C and was transferred to Gene Screen Plus hybridization transfer membranes (Perkin-Elmer) using downward capillary transfer. Membranes were prehybridized in SDS hybridization buffer {6× SSPE [1× SSPE is 0.18 M NaCl, 10 mM NaH2PO4, and 1 mM EDTA (pH 7.7)], 10% SDS, 200 μg of salmon sperm DNA per ml, and 50% formamide} for 5 h at 59°C and were hybridized in the same buffer containing 106 counts of 32P-labeled riboprobe per ml for 16 h at 59°C. Riboprobes were prepared from a linearized vector containing the neomycin phosphotransferase gene downstream of a T7 RNA polymerase promoter using the Promega Riboprobe System as described by the manufacturer. Membranes were washed twice with 1× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) with 0.1% SDS at 23°C for 30 min and twice with 0.1× SSC with 0.1% SDS at 65°C for 30 min. Membranes were analyzed by phosphorimage analysis and autoradiography.
RESULTS
Isolation of cell lines with genotype 1b replicons with adaptive mutations.
Our studies were initiated in a manner previously described using an exact copy of the original genotype 1b replicon described by Lohmann et al. (43) (designated Rep1bNeo in these studies). Huh7 cells were transfected with synthetic RNA produced from the construct using DMRIE-C, a RNA transfection reagent. The colony-forming efficiency of this replicon in the presence of G418 selection was frequently zero and never exceeded five colonies per μg of transfected RNA. Ten colonies from a single experiment were expanded for further analysis. Replicon RNA was quantified using a TaqMan, real-time RT-PCR assay with a primer and probe set directed at the HCV 5′ NCR. The level of replicon RNA varied from 1.7 × 106 to 1.3 × 107 ge/μg of cellular RNA (Table 1) or approximately 17 to 130 ge/cell, assuming that copy number is uniform in cells of the same line. Clone 45 cells were plated in identical plates at 60% confluency and analyzed at multiple times over a 10-day period. Although a 50% decline in replicon RNA (number of ge per microgram of cell RNA) occurred between days 1 and 2, no further decline was observed over 10 days, suggesting that the differences in replicon number in different cell lines was not due to minor variations in culture confluency (data not shown). Three regions (a portion of NS3, NS5A, and NS5B) of the replicon present in each cell line were PCR amplified and sequenced to determine the consensus sequence of the resident replicon. Previously, most adaptive mutations have been detected in a region adjacent to the ISDR of NS5A (Fig. 1) (8, 27, 35, 42); however, adaptive mutations have also been detected in NS3, NS4B, and NS5B (27, 35, 42). In this study, the replicon present in each cell line contained a single mutation within NS5A, and 2 of the 10 replicons contained mutations in NS3, while no mutations were noted within NS5B. The NS5A mutations spanned nucleotides 6861 to 6951 and amino acids 2177 to 2204 (nucleotide and amino acid numbers are based on HCV genomic RNA sequence) (Fig. 1 and Table 1). A total of six unique mutations were observed; two were present in two different replicons each (S2197F and S2204I), and one was present in three replicons (S2202L). One aspect that was noteworthy was the mutation of two of the serine residues associated with hyperphosphorylation of NS5A (amino acids 2197 and 2204) (64); 4 of the 10 mutations detected involved these two serine residues (Fig. 1 and Table 1). Clones 35 (NS5A S2204I) and 38 (NS5A A2199T) also carried mutations in NS3 at I1278L and Q1335E, respectively. In addition, a mutation at NS3 D1431Y was observed in combination with NS5A S2204I in a cell line not shown in Table 1. Transfection of total cellular RNA derived from each cell line resulted in a much higher colony-forming efficiency than the parental replicon, suggesting that the changes were adaptive mutations. Extrapolation of the TaqMan RT-PCR results for replicon copy number (number of ge per microgram of cellular RNA) to the number of colonies formed per microgram of transfected cell RNA revealed that colony-forming efficiencies ranged from 52,000 to 500,000 colonies/μg of replicon RNA (Table 1). Since limited regions of each replicon were sequenced, the presence of additional adaptive mutations cannot be excluded.
TABLE 1.
NS5A adaptive mutations of Rep1bNeoa
| Clone | Replicon RNA (106 ge/μg of cell RNA)b | Amino acidc
|
Colony-forming efficiency (103 colonies/μg of replicon RNA)d | |
|---|---|---|---|---|
| Position | Change | |||
| 2 | 1.7 | 2177 | D→H | 500 |
| 42 | 10.1 | 2197 | S→F | 79 |
| 45 | 11.9 | 2197 | S→F | 87 |
| 38 | 9.5 | 2199 | A→T | 167 |
| 40 | 9.2 | 2200 | S→R | 53 |
| 13 | 7.4 | 2202 | S→L | 94 |
| 39 | 1.8 | 2202 | S→L | 154 |
| 43 | 5.3 | 2202 | S→L | 52 |
| 35 | 13.1 | 2204 | S→I | 72 |
| 37 | 2.6 | 2204 | S→I | 147 |
Huh7 cells were transfected with Rep1bNeo, and G418-resistant colonies were established as cell lines.
The replicon copy number was determined by TaqMan RT-PCR and expressed as ge per microgram of cellular RNA. To determine the number of ge per microgram of RNA, cells were plated at a low density and harvested 48 h later, when the cultures were still subconfluent.
Regions of NS3, NS5A, and NS5B were amplified and sequenced to determine adaptive mutations. The amino acids are given in the single-letter code.
To determine the colony-forming efficiency of the adapted replicons, Huh7 cells were transfected with 10 μg of total cell RNA isolated from each cell line, and the colony-forming efficiency in the presence of G418 was determined. The colony-forming efficiency is presented as the number of colonies per microgram of transfected replication RNA that was extrapolated from the estimated number of replicon ge per microgram of cell RNA.
Synergy of NS3 mutations with NS5A mutations and failure of adaptive mutations to enhance the colony-forming efficiency of genotype 1a replicons.
To construct a Rep1bNeo replicon with high colony-forming efficiency, one of the common NS5A mutations at amino acid position 2204 (serine to isoleucine; S2204I) was introduced into Rep1bNeo by PCR-directed mutagenesis. While the parental replicon often yielded no colonies in individual assays, Rep1bNeo-2204 produced up to 800 colonies/μg of replicon RNA. Although a significant improvement over the parental replicon, the transfection of total cell RNA (Table 1) from selected colonies suggested that additional mutations must account for the very high efficiency observed in the adapted replicons. Next, we tested the NS3 mutation at amino acid D1431Y by itself and in concert with the S2204I mutation. Rep1bNeo-1431 exhibited no significant improvement in colony-forming efficiency over the parental replicon, while the NS3 mutation was highly synergistic with the NS5A mutation (Rep1bNeo-1431/2204 [Table 2 ]), producing up to 80,000 colonies/μg of replicon RNA. This colony-forming efficiency is comparable to that determined for the replicons present in selected colonies and suggests that NS3 mutations complement NS5A mutations in a nonadditive mechanism.
TABLE 2.
Colony-forming efficiency of replicon constructsa
| Replicon RNA | Colony-forming efficiency (colonies/μg of replicon RNA) |
|---|---|
| Rep1bNeo | ≤5 |
| Rep1bNeo-2204 | 800 |
| Rep1bNeo-1431 | 0 |
| Rep1bNeo-1431/2204 | 80,000 |
| Rep1aNeo | 0 |
| Rep1b/aNeo | 0 |
| Rep1b/aNeo-2204 | 0 |
| Rep1b/aNeo-1431/2204 | 0 |
Huh7 cells were transfected with various replicons, and the colony-forming efficiency in the presence of G418 was determined. Replicons were transfected at 10 μg/100-mm-diameter dish, except for Rep1bNeo-1431/2204, which was transfected at 10 ng of replicon RNA mixed with 10 μg of Huh7 RNA as carrier. Each replicon was tested in multiple experiments. Rep1bNeo frequently failed to yield colonies, but in a single experiment yielded 50 colonies/10 μg of transfected RNA, and these colonies were the source of the cell lines in Table 1.
Previous studies have suggested that genotype 1a replicons are not capable of forming colonies in Huh7 cells at detectable levels (8, 27). Since these studies were conducted with a single strain of genotype 1a (H77), it is not clear whether this is a general property of genotype1a isolates or unique to this strain. We constructed genotype 1a replicons using our infectious clone of the HCV-1 prototype sequence (40), the only other genotype 1a infectious clone. The construct design was identical to that used for Rep1bNeo, except that all HCV sequences were derived from the HCV-1 clone. The resulting replicon RNA, Rep1aNeo, was not capable of inducing colony formation following transfection into Huh7 cells in multiple attempts (Table 2). Analysis of the in vitro translation pattern from this RNA in rabbit reticulolysates revealed that expression of the HCV nonstructural proteins from the EMCV IRES was very low in comparison to Rep1bNeo, despite the fact that translation of the neo gene from the HCV IRES was equivalent for the two constructs (Fig. 2A). A series of chimeric constructs revealed that a short sequence at the amino terminus of NS3 adjacent to the EMCV IRES was responsible for the differences in translation efficiencies of the two constructs. To provide more-efficient translation of the HCV nonstructural proteins from the Rep1aNeo replicon, a portion (228 nucleotides) of the genotype 1a sequence was replaced with the genotype 1b sequence to create Rep1b/aNeo, which differed from Rep1aNeo by 12 amino acids (see Materials and Methods for details). Presumably, a RNA structure within the genotype 1a sequence inhibited translation from the EMCV IRES. Although this chimeric construct exhibited in vitro translation properties equivalent to Rep1bNeo (Fig. 2A), it was not capable of inducing colony formation in Huh7 cells (Table 2). Since the Rep1bNeo replicon itself exhibited marginal colony-forming ability prior to acquisition of adaptive mutations, the NS5A-S2204I mutation was introduced alone or in combination with D1431Y into Rep1b/aNeo. These mutations did not increase the colony-forming efficiency of Rep1b/aNeo to detectable levels following transfection of Huh7 cells. These results suggest that replicons based on the HCV-1 genotype 1a sequence do not possess the capacity to induce detectable levels of colony formation in Huh7 cells even with mutations known to enhance genotype 1b strains. Part of our analysis of these constructs included in vitro translation, as shown in Fig. 2A. When the replicons containing the NS3 mutations were compared to the parental replicons and those containing the NS5A mutation alone, NS3 exhibited a significantly increased mobility by SDS-PAGE. This was true whether the D1431Y mutation was present in the Rep1bNeo or Rep1b/aNeo background and whether the S2204I mutation was present or not (Fig. 2B). At this time, the reason for the increased mobility of NS3 is not understood, but it may be indicative of an altered posttranslational modification, which in turn may play a role in the synergy of this mutation with the NS5A mutation.
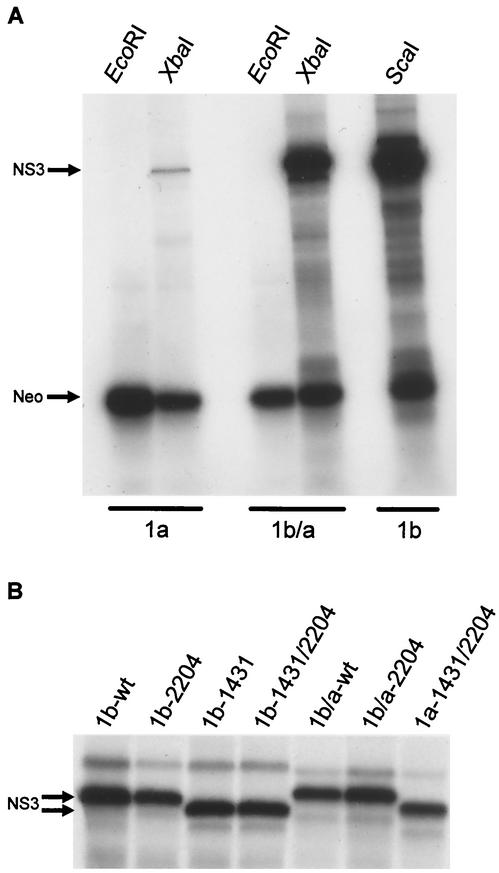
FIG. 2.
In vitro translation of bicistronic HCV replicons. (A) Rep1aNeo (1a), Rep1b/a (1b/a), and Rep1b (1b) synthetic RNAs were produced by in vitro transcription reactions utilizing vectors linearized at EcoRI, a site downstream of the neomycin phosphotransferase gene (Fig. 1), or at XbaI or ScaI to produce full-length replicon RNA. RNAs were in vitro translated using rabbit reticulocyte lysates in the presence of [35S]methionine, and the products were analyzed by SDS-PAGE. Rep1a produced reduced levels of NS3 compared to both Rep1b and Rep1b/a, while translation of the Neo protein was similar for all constructs. (B) The impact of the NS3 D1431Y mutation on the migration of NS3 by SDS-PAGE was examined following in vitro translation. All replicon RNAs containing the D1431Y mutation exhibited an increased mobility for NS3 regardless of whether the mutation was in the presence of the NS5A S2204I mutation or in the Rep1b (1b) or Rep1a (1a) background. wt, wild type.
Antiviral effects of IFN-α, IFN-γ, poly(I)-poly(C), and TNF-α.
The potential antiviral activities of IFN-α, IFN-γ, poly(I)-poly(C), and TNF-α were examined using the replicon system. In these studies, a replicon cell line was treated with various concentrations of the compound, and the level of replicon RNA in cell lysates was quantified using TaqMan RT-PCR assays. All assays were multiplexed for GAPDH mRNA to ensure that the treatment had no overt adverse effect on cellular mRNA levels. Initially, a time course study was performed with each antiviral agent. Clone 45 cultures (Table 1) were treated for 24, 48, and 72 h with 100 to 1,000 U of IFN-α2b or IFN-γ per ml, 100 to 1,000 μg of poly(I)-poly(C) per ml, or 200 to 1,000 U of TNF-α per ml. Dramatic reductions in replicon RNA levels were observed at both concentrations of IFN-α2b at 72 h, i.e., 40- and 172-fold reduction at 100 and 1,000 U/ml, respectively (Fig. 3). Although substantial antiviral activity was also observed with IFN-γ, the reduction in replicon RNA was approximately three- to fourfold less (14.4- and 44.9-fold reduction at 100 and 1,000 U/ml, respectively, at 72 h). No significant reduction was observed with poly(I)-poly(C), even at 1,000 μg/ml (Fig. 3), or with TNF-α at 1,000 U/ml (data not shown).
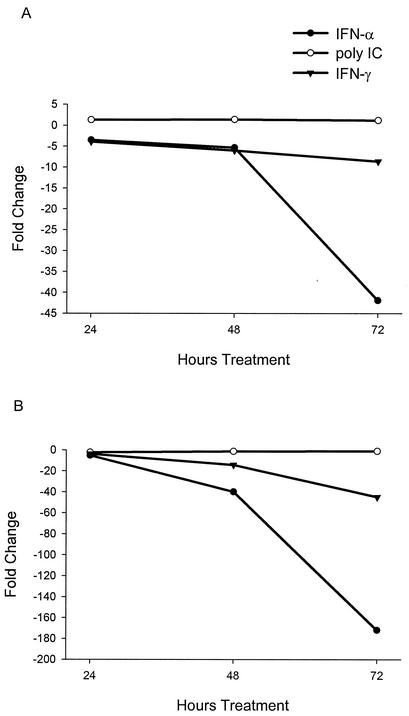
FIG. 3.
Kinetics of HCV replicon RNA decline following treatment with IFN-α, IFN-γ, and poly(I)-poly(C). Clone 45 cells were treated with IFN-α or IFN-γ at 100 or 1,000 U/ml and with poly(I)-poly(C) at 100 or 1,000 μg/ml and harvested at 24, 48, and 72 h. Results from 100 (A) and 1,000 (B) U/ml or μg/ml. Replicon RNA levels were quantified by TaqMan RT-PCR and expressed as the fold change from the levels of untreated cultures harvested at the same time points. All values are averages from duplicate cultures.
In part to confirm these findings and to determine the maximum reduction observed with each antiviral following a more prolonged treatment, cultures were treated for 7 to 9 days with each antiviral. Replicon levels were reduced by 2,300- and 23,000-fold at 100 and 1,000 U/ml of IFN-α2b, respectively (Table 3). This was not an unanticipated finding, since other studies have observed a reduction in replicon RNA following IFN-α treatment (8, 20, 27). However, extended treatment with poly(I)-poly(C), a known inducer of dsRNA response genes including type 1 IFN genes and ISGs, still had no antiviral effect. This was an unanticipated finding, since poly(I)-poly(C) has a high level of antiviral activity for many viruses, including GBV-B replication in primary tamarin hepatocytes (38), which is a closely related surrogate model for HCV. Poly(I)-poly(C) treatment was performed by direct addition of the compound to the medium at concentrations as high at 1,000 μg/ml, as well as transfection with lipid reagents without detectable antiviral effect. Although IFN-γ again exhibited antiviral activity, the prolonged treatment did not result in significantly greater reduction of replicon RNA than treatment for 72 h (100 or 1,000 U/ml of IFN-γ for 7 days resulted in a 2.7- and 31.9-fold reduction in replicon RNA, respectively).
TABLE 3.
Antiviral effects of IFN-α, poly(I)-poly(C), and IFN-γa
| Treatment | Fold reductionb | GAPDH Ctc |
|---|---|---|
| IFN-α (U/ml) | ||
| 0 | 0 | 18.0 |
| 100 | 2,300 | 17.6 |
| 1,000 | 23,000 | 17.6 |
| Poly(I)-poly(C) (μg/ml) | ||
| 0 | 0 | 16.8 |
| 50 | 1.4 | 16.5 |
| IFN-γ (U/ml) | ||
| 0 | 0 | 15.9 |
| 100 | 2.7 | 15.6 |
| 1,000 | 31.9 | 15.2 |
Replicon lines were treated with IFN-α, poly(I)-poly(C), or IFN-γ for 7 to 9 days. In this experiment, clone 8 cells were treated with IFN-α, while clone 45 cells were treated with IFN-γ and poly(I)-poly(C). Treatments were initiated on confluent cultures, and the medium was changed every 2 days. No overt toxicity was observed.
Replicon RNA was quantified by TaqMan RT-PCR, and values are expressed as fold reduction compared to the values for untreated cells.
The TaqMan RT-PCR assay for replicon RNA was multiplexed for the GAPDH mRNA to demonstrate a lack of effect of treatments on cellular mRNA, and all values were normalized for GAPDH. GAPDH values are expressed as Ct, the amplification cycle at which the values exceeded the background threshold.
Host cell response to antiviral treatments.
To further explore the differences in response to IFN-α and poly(I)-poly(C), induction of dsRNA-IFN-stimulated gene was examined using TaqMan RT-PCR assays for the transcripts of STAT1α, STAT1β, PKR, ISG12, IRF-1, and IP10. Huh7 cells were compared to replicon cells following 24 to 48 h of treatment with IFN-α, IFN-γ, or poly(I)-poly(C) (Table 4). In this experiment, IFN-α resulted in 11.8- and 104-fold reduction in replicon RNA at 24 and 48 h, respectively, while IFN-γ resulted in 6.1- and 31.2-fold reduction in replicon RNA at 24 and 48 h. Poly(I)-poly(C) again failed to induce a significant decline in replicon RNA. All of the ISGs were elevated in IFN-α-treated Huh7 and replicon cells; however, IRF-1 was increased by only 2.3- to 3.3-fold, and it is not clear that these values represent significant increases over the baseline levels of the untreated cultures.
TABLE 4.
Induction of ISG transcripts by IFN-α, IFN-γ, and poly(I)-poly(C)a
| Cellb | Treatment | Fold changec
|
|||||
|---|---|---|---|---|---|---|---|
| STAT1α | STAT1β | PKR | ISG12 | IRF-1 | IP10 | ||
| Huh7 | IFN-α | 41.1 | 38.8 | 20.3 | 312 | 3.1 | 10.0 |
| Rep | IFN-α | 18.6 | 24.1 | 18.7 | 2,374 | 3.3 | 111.5 |
| Huh7 | IFN-γ | 80.8 | 70.6 | 3.4 | 80.0 | 47.9 | 198.8 |
| Rep | IFN-γ | 31.3 | 33.5 | 4.5 | 74.3 | 45.5 | 123.1 |
| Huh7 | Poly(I)-poly(C) | 1.3 | −1.3 | 1.0 | 1.6 | −1.2 | −1.6 |
| Rep | Poly(I)-poly(C) | −1.1 | 1.1 | −1.5 | 2.2 | −1.1 | 1.3 |
| CH | IFNα | 2.0 | 3.6 | 3.8 | 9.3 | 1.4 | 12.6 |
| CH | Poly(I)-poly(C) | 3.7 | 8.9 | 5.3 | 40.4 | 2.2 | 549.0 |
Huh7 and clone 45 cell lines were treated with IFN-α (1,000 U/ml), polyIC (100 μg/ml), or IFN-γ (1,000 U/ml). Primary chimpanzee hepatocytes were treated with IFN-α (100 U/ml) or polyIC (50 μg/ml).
Rep, replicon clone 45 cell line; CH, chimpanzee hepatocytes.
Transcripts for various ISGs were quantified using TaqMan RT-PCR assays. The data are averages from duplicate cultures and are expressed as the fold change compared to the values for untreated cultures. The data for IFN-α and IFN-γ treatment are from the 24-h harvest, since this time point had the highest values. For polyIC, similar values were obtained for the 24- and 48-h time points. The 48-h values are shown to demonstrate that a delayed response was not missed. The chimpanzee hepatocytes were harvested at 72 h for both treatments. Additional cells were not available to repeat the chimpanzee hepatocyte study at multiple time points. The level of the replicon RNA was decreased by 104- and 31.2-fold in IFN-α- and IFN-γ-treated cells at the 48-h time point. No decrease in replicon RNA was observed following polyIC treatment.
Although the basal levels of the ISGs were similar in both Huh7 cells and clone 45 cells, some variation was noted in the degree of induction of ISGs in the two cell types following treatment with IFN-α and IFN-γ. While PKR was induced to similar levels in the presence and absence of the replicon and STAT1α and STAT1β were increased approximately twofold more in the absence of the replicon, ISG12 and IP10 induction by IFN-α was increased by approximately 8- to 10-fold in the presence of the replicon (Table 4). The greater induction of ISG12 and IP10 by IFN-α in the presence of the replicon was not observed following treatment with IFN-γ. As expected, some of the IFN response genes were induced differentially by IFN-α and IFN-γ. While IRF-1 was minimally induced by IFN-α in comparison to IFN-γ, the opposite was true for PKR.
The increased induction of ISG12 and IP10 by IFN-α in clone 45 cells in comparison to Huh7 cells could be due to clonal variation of individual cells selected from the Huh7 population; alternatively, it could represent an important difference induced by either viral dsRNA or an HCV protein. To determine the degree of variation in the IFN induction of ISGs in Huh7 cells and different replicon lines, clones 2, 40, and 45 were compared to Huh7 cells following 24 h of treatment with IFN-α and IFN-γ. Stat1α, ISG12, and IP10 transcripts were examined by TaqMan RT-PCR assays. Although in all cases the transcripts were induced in both Huh7 and replicon cell lines in the presence of IFN-α or IFN-γ, the differential induction of IP10 and ISG12 in cells containing the replicon in comparison to Huh7 cells was less apparent. The induction of IP10 transcripts in IFN-α-treated cells was greater in all replicon lines than in Huh7 cells. In two lines (clones 40 and 45), it was again increased by >10-fold more than in Huh7 cells, but in a third line (clone 2), IP10 transcripts were induced only about 2.5-fold more than in Huh7 cells (Table 5). No pattern was observed for either Stat1α or ISG12, but in general, replicon lines had higher levels of ISG induction in the presence of IFN-α. Interestingly, the pattern for IP10 in the two experiments remained constant with regard to Huh7 cells and clone 45 cells treated with IFN-α and IFN-γ. IP10 transcripts were induced to 10-fold-greater levels by IFN-α in clone 45 cells in comparison to Huh7 cells, while IFN-γ induced IP10 to a greater extent in Huh7 cells than in clone 45 cells (Table 5). These data indicate that (i) ISG induction occurs in the presence of both IFN-α and IFN-γ, (ii) some differential regulation of ISGs is observed between the two IFNs, and (iii) although some differences were observed in the presence and absence of the replicon, some of the variation may be due to the clonal selection of cell lines.
TABLE 5.
Induction of ISGs by IFN-α and IFN-γ in multiple replicon lines
| Treatment and cella | Fold changeb
|
|||
|---|---|---|---|---|
| Stat-1α | ISG12 | IP10 | HCV | |
| IFN-α | ||||
| Huh7 | 4.5 | 104.5 | 3.0 | |
| Clone 2 | 6.9 | 403.6 | 7.6 | 9.5 |
| Clone 40 | 7.0 | 275.0 | 52.9 | 8.1 |
| Clone 45 | 4.8 | 130.6 | 43.0 | 4.8 |
| IFN-γ | ||||
| Huh7 | 8.4 | 25.0 | 227.3 | |
| Clone 2 | 8.0 | 47.1 | 33.1 | 7.5 |
| Clone 40 | 8.2 | 21.9 | 109.7 | 8.7 |
| Clone 45 | 6.5 | 15.3 | 75.2 | 4.7 |
Huh7 and clone 2, 40, and 45 cells were treated for 24 h with IFN-α or IFN-γ at 1,000 U/ml.
Levels of HCV replicon RNA and transcripts for STAT-1α, IP10, and ISG12 were quantified by TaqMan RT-PCR assays. The data are averages from duplicate cultures and are expressed as the fold change compared to the values for untreated cultures.
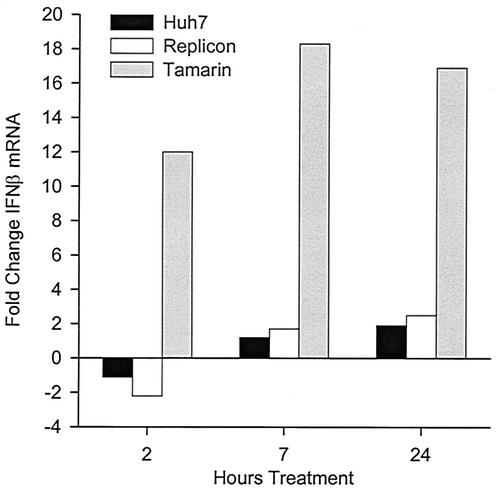
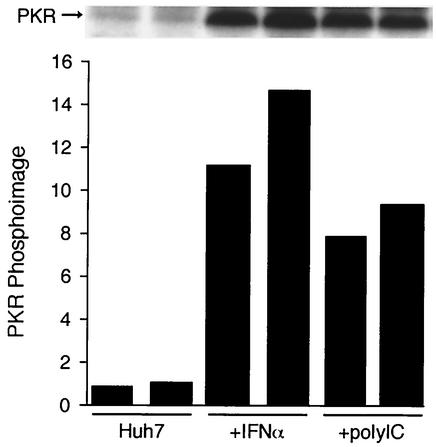
In contrast to IFN-α and IFN-γ, poly(I)-poly(C) treatment did not induce the expression of ISGs in either Huh7 cells or clone 45 cells (Table 4). These data suggest that the parental Huh7 cells are altered in regulation of some component of the dsRNA response pathway, which may explain the lack of antiviral effect by poly(I)-poly(C) on the replicon in these cells. This contrasts sharply with primary chimpanzee hepatocytes, which were highly responsive to poly(I)-poly(C). Primary chimpanzee hepatocytes were treated with either IFN-α or poly(I)-poly(C) and analyzed for the induction of ISGs. In chimpanzee hepatocytes, 50 μg of poly(I)-poly(C) per ml was superior to 100 U of IFN-α per ml in the induction of ISGs (Table 4). We had previously demonstrated the potent antiviral properties of poly(I)-poly(C) in the GBV-B/tamarin hepatocyte model. Since this system permitted the direct comparison of two cell types in which poly(I)-poly(C) had either no antiviral activity against HCV replicons or potent antiviral activity against an HCV surrogate, we examined poly(I)-poly(C) treatment in Huh7 cells and primary tamarin hepatocytes. TaqMan RT-PCR assays were used to quantify the level of induction of IFN-β transcripts at 2, 7, and 24 h after poly(I)-poly(C) addition. We chose IFN-β, since it is known to be responsive to dsRNA and has potent antiviral activity. No induction of IFN-β transcripts was noted in Huh7 cells, while 12- to 18-fold induction occurred in tamarin hepatocytes at all three time points (Fig. 4). Since numerous different approaches to induction of ISG transcription with poly(I)-poly(C) were attempted in Huh7 cells without success, we questioned whether poly(I)-poly(C) was entering the cells and inducing latent dsRNA binding proteins. To test this, Huh7 cells were treated with poly(I)-poly(C) for 2 h or with IFN-α for 16 h, and PKR was immunoprecipitated and used in kinase assays. In the presence of dsRNA, PKR dimerizes and undergoes autophosphorylation, and PKR kinase activity can also be induced by IFN-α. In comparison to untreated Huh7 cells, both poly(I)-poly(C) and IFN-α dramatically induced PKR kinase activity (Fig. 5). These data suggest that Huh7 cells are responsive to poly(I)-poly(C) but that they do not respond in a typical fashion to dsRNA, since no antiviral activity or induction of ISG transcription was apparent using this compound.
FIG. 4.
IFN-β transcription in response to poly(I)-poly(C). Huh7, clone 45, and primary tamarin hepatocyte cultures were harvested after 2, 7, and 24 h of treatment with 100 μg of poly(I)-poly(C) per ml. The levels of IFN-β transcripts were quantified in total cell RNA by TaqMan RT-PCR and were expressed as fold change in comparison to the levels of untreated cells. All values are averages of duplicate cultures.
FIG. 5.
PKR activation by poly(I)-poly(C) in Huh7 cells. Huh7 cells were treated with 100 μg of poly(I)-poly(C) per ml for 2 h or with 1,000 U of IFN-α per ml for 16 h prior to harvest. PKR was immunoprecipitated from cell lysates, and kinase reactions were conducted with [γ-32P]ATP with PKR still bound to the antibody beads. Phosphorylated PKR was analyzed by SDS-PAGE and autoradiography (top) or by phosphorimage analysis (bottom). Phosphorimage values are expressed as arbitrary units and represent the volume in individual PKR bands shown in the blot.
Ribavirin induces error-prone replication of HCV replicon RNA.
Ribavirin has an antiviral effect for a number of viruses in tissue culture (14, 47, 58). The monophosphate form of ribavirin is an IMP dehydrogenase (IMPDH) inhibitor, and at least some of the in vitro antiviral effect has been ascribed to the reduction of GTP pools by inhibition of IMPDH. Although this has deleterious effects on cellular metabolism, these effects are tolerated in short-term studies in nondividing cell cultures. Ribavirin is used in combination with IFN-α for the treatment of HCV infections and provides a significant improvement in the rate of sustained viral clearance in comparison to the rate for IFN-α alone. However, ribavirin monotherapy does not result in a significant reduction in the level of viremia despite a marked improvement in liver disease (9, 16). One mechanism suggested to explain the improvement with ribavirin therapy involves an immunomodulatory activity possessed by ribavirin that promotes a Th1-biased immune response (19, 30, 50, 60). However, the mechanism of the synergistic effect that ribavirin has with IFN-α for increasing the percentage of patients with sustained viral clearance remains uncertain. We recently demonstrated a direct antiviral effect of ribavirin on GBV-B virus that involves induction of error-prone replication due to ribavirin triphosphate incorporation (38). The following experiments were conducted to determine whether ribavirin has an antiviral effect on the HCV replicon and whether this effect could be ascribed to induction of error-prone replication.
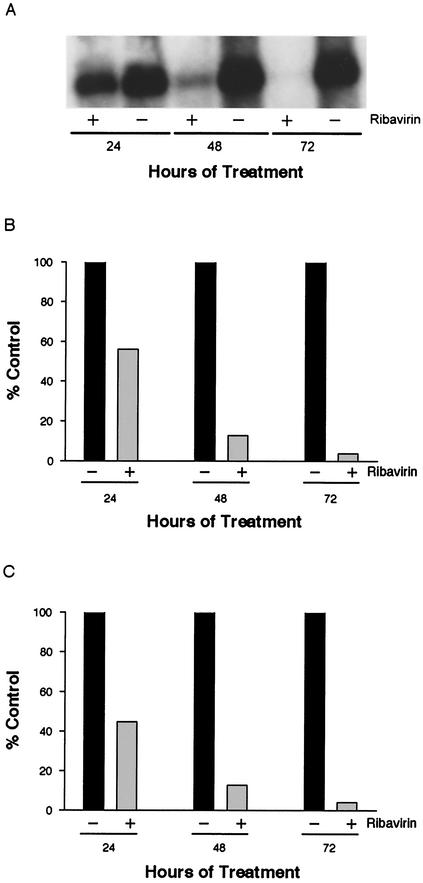
Experiments with ribavirin were conducted with confluent nondividing cultures to minimize the adverse effects from GTP pool reduction. In addition, a highly purified preparation of ribavirin was used; the preparation was devoid of trace contaminants that often cause toxicity in tissue culture studies unrelated to the IMPDH inhibition by ribavirin (see Materials and Methods). Initially, the level of ribavirin was titrated to determine the optimal dose for antiviral effect with minimal cellular toxicity. A replicon cell line was treated with 50 to 400 μM ribavirin for 9 days, and total cell RNA was examined by TaqMan RT-PCR assay to determine the effects on replicon RNA and GAPDH mRNA levels. Little effect was observed at 50 or 100 μM, while treatment with 200 and 400 μM ribavirin resulted in 19.3- and 2,900-fold reduction in replicon RNA, respectively (Table 6). No overt toxicity or decrease in the level of GAPDH mRNA was observed at any treatment level, but some decline in the secretion of apolipoprotein B was observed at the higher ribavirin concentrations (data not shown). A time course study revealed that treatment of cultures with 400 μM ribavirin reduced replicon RNA from 2.2-fold at 24 h to 24.3-fold at 72 h when analyzed by TaqMan RT-PCR (Fig. 6C). Analysis of the same cellular RNAs for replicon RNA levels by Northern blot hybridization and quantification by phosphorimager analysis of the blot provided essentially identical results for the percent decrease in replicon RNA in ribavirin-treated cultures (Fig. 6B). Since only the area of the gel representing intact replicon RNA is used in phosphorimage analysis, these results suggest that the replicon RNA was not degraded in ribavirin-treated cultures. The autoradiogram of the Northern also demonstrated that the replicon RNA from treated cultures was not detectably degraded (Fig. 6A); however, due to the 24.3-fold decrease in replicon RNA at 72 h, overexposure of the autoradiogram was required to confirm that the RNA at this time point was not degraded (data not shown).
TABLE 6.
Titration of antiviral effect of ribavirina
| Ribavirin concn (μM) | Replicon (106 ge/μg of cell RNA)b | Fold reductionc | GAPDH Ctd |
|---|---|---|---|
| 0 | 5.8 | 0 | 17.4 |
| 50 | 3.4 | 1.7 | 17.6 |
| 100 | 2.5 | 2.3 | 17.6 |
| 200 | 0.3 | 19.3 | 17.6 |
| 400 | 0.002 | 2,900 | 17.6 |
Clone 8 cells were plated and allowed to reach confluency for 3 days and then were treated with ribavirin for 9 days.
Replicon RNA was quantified by TaqMan RT-PCR and expressed as copy number (ge) per microgram of cell RNA.
The data are expressed as the fold change compared to the values for untreated cultures.
The TaqMan RT-PCR assay for replicon RNA was multiplexed for the GAPDH mRNA to demonstrate a lack of overt toxicity or decrease in cellular mRNA in ribavirin-treated cells. GAPDH values are expressed as Ct, the amplification cycle at which the values exceeded the background threshold.
FIG. 6.
Antiviral effect of ribavirin on HCV replicon RNA. Clone 24 cells were cultivated with (+) or without (−) 400 μM ribavirin for 24, 48, or 72 h. Replicon RNA was analyzed by agarose gel electrophoresis, Northern blot hybridization, and autoradiography (A). RNA in the same blot was quantified by phosphorimage analysis (B), or the replicon RNA was quantified by TaqMan RT-PCR (C). Values in panels B and C are expressed as percentages of untreated, control cultures at each time point.
The cellular RNAs from the same time course study were used to evaluate whether the antiviral effect was due to error-prone replication. To test this hypothesis, Huh7 cells were transfected with total cellular RNA derived from treated and untreated cultures to determine whether a decrease occurred in the colony-forming capacity of the replicon in treated cultures. This test for error-prone replication avoids the issue of ribavirin toxicity and is similar to the approach used for GBV-B (38) where we demonstrated that virus from treated cultures had a reduced specific infectivity when used to infect new cultures. In the current studies, cells were transfected with equivalent numbers of ge of replicon RNA derived from treated and untreated cultures. For each time point, cells were transfected with 10 μg of total cell RNA from the treated cultures (the maximum amount of cellular RNA that can be efficiently transfected), and the RNA from untreated cultures was adjusted with Huh7 cell RNA such that cells were transfected with equal amounts of cell RNA containing equal replicon copy numbers from treated and untreated cultures (Table 7).
TABLE 7.
Effect of ribavirin on replicon colony-forming efficiencya
| Harvest time (h) | Ribavirinb | Transfected replicon (ge) | Colony-forming efficiencyc (colonies/μg of replicon) |
|---|---|---|---|
| 24 | − | 2.6 × 108 | 140,000 |
| + | 2.6 × 108 | 45,333 | |
| 48 | − | 8.3 × 107 | 93,750 |
| + | 8.3 × 107 | 14,583 | |
| 72 | − | 3.1 × 107 | 138,888 |
| + | 3.1 × 107 | 0 |
Clone 24 cells were treated with or without 400 μM ribavirin for 24 to 72 h. Cell RNA was harvested at each time point, and the replicon copy number was determined (Fig. 6). RNA from untreated cultures was adjusted with Huh7 RNA such that transfections of treated and untreated RNAs were performed with identical replicon copy number (ge) and such that each transfection was performed with 10 μg of total cell RNA per 100-mm-diameter dish.
Symbols: −, no ribavirin; +, 400 μM ribavirin.
Colony-forming efficiency was expressed as the number of colonies per microgram of transfected replicon RNA which was extrapolated from the ge of replicon transfected at each time point as shown in the table.
Colony-forming activity for the RNA from treated cultures was reduced by 3.1- and 6.4-fold at 24 and 48 h of treatment, respectively, while RNA from cultures treated for 72 h was devoid of colony-forming activity (Table 7). These data demonstrate a progressive loss in colony-forming activity for the replicon RNA with increased exposure to ribavirin and support the conclusion that ribavirin has the capacity to affect HCV RNA replication through a mechanism of error-prone replication.
DISCUSSION
The development of a replicon system for HCV has provided a means to approach a number of biological questions previously not possible. One of the primary uses of this system will be the evaluation of antiviral agents and virus-host interactions. Persistence of the replicon is very sensitive to IFN-α, confirming that IFN-α has a direct antiviral effect on the virus (8, 20, 27). In this study, we have extended these observations to include IFN-γ and ribavirin, and we have demonstrated the absence of antiviral effect for poly(I)-poly(C) and TNF-α.
The successful application of the replicon system to HCV research is dependent in part upon the realization that specific adaptive mutations dramatically increase the ability of the replicon to persist and induce colony formation (8). Adaptive mutations have been described by several laboratories (8, 27, 35, 42), and the scope of these mutations was increased in this study. As in previous studies, the primary adaptive mutations were localized to a small domain adjacent to the ISDR and PKR binding sites of NS5A (Fig. 1), although adaptive mutations have been observed in NS3, NS4B, and NS5B (27, 35, 42). We found that mutations in NS3 frequently arise in conjunction with the NS5A mutations. Although the NS3 mutation at D1431Y did not increase colony-forming efficiency of the parental replicon, it was highly synergistic with the S2204I mutation in NS5A. The synergy between NS3 and NS5A mutations has been observed previously (35), but this specific combination of mutations has not been previously observed. Previously described mutations in NS3 span a region of 407 amino acids (positions 1202 to 1609) (27, 42) within the helicase domain of NS3. In our studies, a single NS5A S2204I mutation was associated with NS3 mutations separated by 153 amino acids (positions 1278 to 1431). The mechanisms of adaptive mutations and the synergy between NS3 and NS5A mutations are not currently understood but may involve changes in the interactions among viral proteins and in the interactions of viral proteins with specific host factors. The NS3 D1431Y mutation increased the mobility of NS3 by SDS-PAGE, whether it was in the genotype 1b or 1a background, which is suggestive of an altered posttranslational modification. The adaptive mutations for genotype 1b did not confer an increased colony-forming ability to genotype 1a replicons in Huh7 cells, and they did not extend the host range of the genotype 1b replicon to cell lines other than Huh7 (data not shown). Previous studies that examined genotype 1a replicons were restricted to the use of the H77 strain (8, 27). In this study, we extended the apparent inactivity of genotype 1a replicons to include the infectious clone derived from the HCV-1 sequence (40), suggesting that this may be a general attribute of genotype 1a strains in Huh7 cells, rather than an isolated observation with a single clone. Currently, the replicon model is limited to the original genotype 1b strain employed by Lohmann et al. (43) and one other genotype 1b isolate (27, 32), the HCV-N infectious clone (6). Replicons based on the HCV-N sequence do not require adaptive mutations for high colony-forming efficiency. A four-amino-acid insertion in the ISDR appears to be responsible for the inherent colony-forming ability of this clone (32). In addition to being restricted to two genotype 1b isolates, colony formation with HCV replicons is limited to a single human liver cell line, Huh7. Undoubtedly, these limitations will not persist for long. Recently, full-length bicistronic and monocistronic constructs that replicate in Huh7 cells have been developed (32, 53), but the production of infectious particles has not been demonstrated.
In this study, we have extended our observations on the antiviral activity of ribavirin to include the subgenomic replicons of HCV. We previously demonstrated a reduction in specific infectivity for GBV-B produced in the presence of ribavirin that was attributed to the incorporation of ribavirin triphosphate by the GBV-B polymerase and an accompanying increase in error-prone replication (38). This effect has also been observed with poliovirus (13). HCV replicon RNA obtained from cultures exposed to ribavirin exhibited a decrease in colony-forming efficiency when transfected into untreated cells, and the decrease was proportional to the duration of ribavirin exposure. Recent in vitro observations with HCV NS5B protein suggest that the HCV RNA polymerase can incorporate ribavirin triphosphate utilizing a synthetic template (44).
Because ribavirin monotherapy does not result in a significant reduction in viral load, the improvement in liver disease realized during ribavirin monotherapy cannot be attributed to the antiviral properties of ribavirin demonstrated in these studies. The immunomodulatory activity of ribavirin may be involved in both the improvement of liver disease during monotherapy and the increased rate of sustained viral clearance during combination therapy (for a recent review on ribavirin, see reference 41). However, the possibility that the synergism with IFN-α is due in part to ribavirin-induced error-prone replication cannot be dismissed. The small increase in error rate that may occur at the levels of ribavirin used in the clinic may result in too few lethal mutations to impact a large pool of replicating viral RNA. In contrast, this error rate may have a significant impact on HCV survival once IFN has reduced the viral load to the extent where the viral RNA is undetectable by RT-PCR and thus facilitate sustained viral clearance (37). This would suggest that three phases are involved in sustained viral clearance in IFN-α-ribavirin therapy. The initial rapid decline of virus (phase I) is most likely due to the direct antiviral effect of IFN-α. A second more gradual and variable decline in viral load (phase II) may be due to the death of infected hepatocytes. Ribavirin does not appear to play a significant role in either of the first two phases. A third phase in which ribavirin induces biologically significant mutagenesis may occur after replicating viral RNA has been reduced to very low levels. If this hypothesis is correct, then treatment with higher levels of ribavirin may lead to an increase in sustained viral clearance; however, at higher levels, the toxic side effects of ribavirin necessitate short-term therapy. Indeed, induction of error-prone replication following short-term, high-dose, intravenous ribavirin therapy may be the mechanism responsible for the successful treatment of hemorrhagic fevers induced by arenaviruses (Lassa, Junin, and Machupo viruses) and bunyaviruses (Hantaan virus) (18, 29, 34, 48). For HCV, improved ribavirin therapy may be possible by specific targeting of ribavirin to the liver to decrease extrahepatic toxicity and potentially increase efficacy if error-prone replication is indeed a mechanism for synergy with IFN-α. The results of future clinical studies with drugs that separate the antiviral and immunomodulatory effects of ribavirin likely will resolve this mechanistic uncertainty.
In this study, we also observed antiviral activity with IFN-α and IFN-γ but not with poly(I)-poly(C) and TNF-α. A number of laboratories have observed the direct antiviral effect of IFN-α in the replicon system (8, 20, 27), and while this paper was under review, a publication by Frese et al. (21) demonstrated the antiviral effect with IFN-γ, as well. This antiviral effect was independent of the production of nitric oxide or the depletion of tryptophan, pathways known to be involved in the antiviral activity of IFN-γ in some systems. We examined IFN-γ because of its known antiviral effect in many systems and our observations that IFN-γ transcripts are increased in the livers of chimpanzees undergoing HCV viral clearance (R. E. Lanford, unpublished data). Following binding to distinct receptors, both types of IFNs utilize the JAK-STAT signal transduction pathway to activate ISG transcription (for reviews, see references 10, 57, and 59). The primary IFN-α/β response is mediated through the binding of ISRE promoter elements by the transcription factor ISGF3, a complex of STAT-1, STAT-2, and IRF-9, while the primary type II IFN response is accomplished by the binding of GAS promoter elements by GAF, a homodimer of STAT-1. Many similarities exist in the signal transduction pathways induced by type I and II IFNs, including the induction of transcription of large, partially overlapping sets of ISGs (15), most of which have poorly understood or no known function. Thus, although the best-characterized antiviral mechanisms of type I and II IFNs differ, in some systems they may actually involve similar ISG responses. IFN-γ can activate ISGF3, and this response is dependent upon activation of the type I IFN pathway (63). We initiated studies into the common induction of genes in Huh7 cells by IFN-α and IFN-γ. The basal level of ISG transcripts did not differ significantly between Huh7 cells and replicon lines, suggesting a lack of significant response to the presence of viral dsRNA. Most ISGs were induced by both IFN-α and IFN-γ, although some differences were observed. For example, IRF-1 transcription is known to be induced to a greater extent by IFN-γ than by IFN-α (15, 49), and this difference was observed in this study in the presence or absence of the replicon. IFN-α induced IP10 to a much greater extent in replicon lines than in Huh7 cells; however, some of the differences in the level of induction of specific genes may be due to the clonal nature of the replicon lines. At this time, the exact mechanisms of antiviral activity of IFN-α and IFN-γ in the replicon system remain unresolved.
We had previously demonstrated a potent antiviral activity for poly(I)-poly(C) in GBV-B-infected primary tamarin hepatocytes (38). The finding that the replicon system was sensitive to IFN-α, but not to poly(I)-poly(C), prompted an exploration of the dsRNA response in these cells. ISG transcript levels did not increase in Huh7 cells or replicon lines following poly(I)-poly(C) treatment. In contrast, the induction of ISGs in primary chimpanzee hepatocytes was greater for poly(I)-poly(C) than for IFN-α. Since poly(I)-poly(C) has a high level of antiviral activity against GBV-B in primary tamarin hepatocytes, we compared this model system directly with Huh7 cells for the induction of IFN-β transcripts following poly(I)-poly(C) treatment. Although tamarin hepatocytes exhibited 12- to 18-fold increases in IFN-β transcripts 2 to 24 h after poly(I)-poly(C) treatment, no significant increase in IFN-β transcripts was noted in Huh7 cells. Some response to dsRNA was detected in Huh7 cells and replicon lines, since PKR became phosphorylated following poly(I)-poly(C) treatment, so the defect in dsRNA-induced transcription of ISGs in Huh7 cells is independent of, or downstream of, PKR activation. After the initial review of this manuscript, Pflugheber et al. (52) published similar observations with regard to poly(I)-poly(C)-induced phosphorylation of PKR in Huh7 cells, as well as activation of IRF-1. Although some induction of ISG transcription was observed in their study, the antiviral impact of poly(I)-poly(C) on replicon RNA was not examined. It is of interest that the poly(I)-poly(C)-induced degradation of HBV transcripts in transgenic mice replicating HBV was not altered in PKR and IRF-1 knockout mice, indicating that each of these antiviral pathways is dispensable in this system (26).
Our findings indicate that Huh7 cells are defective in some portion of the signaling pathway for dsRNA but that this defect does not prevent response to IFN-α or IFN-γ. Whether this defect is related to the permissiveness of Huh7 cells for HCV replicons is not known, but the defect in dsRNA response suggests that information gained from the replicon system with regard to the antiviral mechanism of IFN-α may not be entirely applicable to HCV-infected individuals. Microarray analysis of serial liver samples from an acute resolving HCV infection in the chimpanzee model demonstrated high levels of ISG transcript induction within 2 days of infection (7). We interpreted this response to be indicative of a robust dsRNA response in infected cells that resulted in the secretion of type I IFNs, which in turn would create zones of cells with ISG induction adjacent to each infected cell. Since cells within each zone would be resistant to HCV infection, a reduction in available replication space would occur as the infection spread in the liver (for greater discussion of these observations, see references 7 and 37). In this manner, the innate antiviral response may control HCV infections until a T-cell response can eliminate infected cells.
The cellular response to dsRNA has been extensively characterized but is still only partially understood. Microarray analysis of the response to dsRNA revealed that 175 genes exhibited increased expression and 95 genes exhibited decreased expression (24). This study was conducted in a cell type defective for the production of type 1 IFNs; therefore, an autocrine loop from secreted IFN did not complicate interpretation of the results. Several latent proteins are activated by dsRNA. The antiviral pathway of 2′,5′ oligoadenylate synthetase is activated by dsRNA, and it in turn activates RNase L, which possesses potent antiviral activity. MxA possesses antiviral activity and is activated by dsRNA, but MxA does not appear to be involved in the antiviral activity of IFN-α in the replicon system (20). PKR is activated by dsRNA, which leads to the phosphorylation and inactivation of IκB, the inhibitor of NF-κB. Activation of NF-κB is required for the dsRNA induction of IFN-β and some ISGs (36, 68). Under some conditions, the response to dsRNA requires both IRF-1 and STAT1α (4) but is not dependent on PKR (68) or ISGF3 (4). At least some of the activity of dsRNA is conveyed by the latent dsRNA-activated factor transcription factor, or DRAF1, which is comprised, in part, of IRF-3 and CBP/p300 (67). Recent studies have also demonstrated a requirement for p53 in the dsRNA response (31). At this time, the defect in the dsRNA response pathway in Huh7 cells is not known, and it is not known whether this influences the permissiveness for HCV replicons. In some replicon lines, IP10 and ISG12 were induced to a greater extent by IFN-α in comparison to Huh7 cells. These data imply that IFN-α and the presence of the replicon may act together to increase the level of expression of these genes. The role of the replicon in this induction could be the presence of dsRNA, since IFN-α and dsRNA are known to behave synergistically (46). As with the PKR activation by poly(I)-poly(C), this would imply that at least portions of the dsRNA response pathway are intact in Huh7 cells, at least under the conditions of IFN-α stimulation.
Our studies did not detect suppression of ISG induction by IFN-α in replicon cells in comparison to Huh7 cells. Although in some studies sequence variation in NS5A has been implicated in the degree of IFN-α responsiveness in patients, the exact mechanism by which NS5A impacts IFN-α responsiveness has not been determined. In vitro studies have suggested that NS5A suppresses the IFN response by virtue of its interaction with PKR (23) or by induction of interleukin 8 expression (25, 55). The lack of detectable suppression of the IFN-α response in the replicon cells may have been due to a number of factors, including the high level of IFN exposure, deficiencies in the dsRNA response in Huh7 cells, or the effects of adaptive mutations present in the replicons. However, the results of these studies do suggest that further investigation of the interaction of subgenomic replicons with the cellular factors involved in viral resistance may improve our understanding of the mechanism(s) of IFN resistance among different genotypes of HCV. Examination of the newly developed full-length replicons (32, 53) will be important as well, since multiple HCV proteins may modulate the host response to dsRNA and IFNs.
Acknowledgments
This work was supported in part by NIH grants U19 AI40035, RO1 AI49574, and P51 RR13986.
We thank Stuart Ray and David Thomas for insightful discussions on the potential antiviral role of ribavirin once the replicating viral population has been reduced by IFN treatment.
REFERENCES
- 1.Alter, H. J., and M. Houghton. 2000. Hepatitis C virus and eliminating post-transfusion hepatitis. Nat. Med. 6:1082-1086. [DOI] [PubMed] [Google Scholar]
- 2.Alter, H. J., and L. B. Seeff. 2000. Recovery, persistence, and sequelae in hepatitis C virus infection: a perspective on long-term outcome. Semin. Liver Dis. 20:17-35. [DOI] [PubMed] [Google Scholar]
- 3.Alter, M. J., D. Kruszon-Moran, O. V. Nainan, G. M. McQuillan, F. X. Gao, L. A. Moyer, R. A. Kaslow, and H. S. Margolis. 1999. The prevalence of hepatitis C virus infection in the United States, 1988 through 1994. N. Engl. J. Med. 341:556-562. [DOI] [PubMed] [Google Scholar]
- 4.Bandyopadhyay, S. K., G. T. Leonard, Jr., T. Bandyopadhyay, G. R. Stark, and G. C. Sen. 1995. Transcriptional induction by double-stranded RNA is mediated by interferon-stimulated response elements without activation of interferon-stimulated gene factor 3. J. Biol. Chem. 270:19624-19629. [DOI] [PubMed] [Google Scholar]
- 5.Bartenschlager, R., and V. Lohmann. 2000. Replication of hepatitis C virus. J. Gen. Virol. 81:1631-1648. [DOI] [PubMed] [Google Scholar]
- 6.Beard, M. R., G. Abell, M. Honda, A. Carroll, M. Gartland, B. Clarke, K. Suzuki, R. Lanford, D. V. Sangar, and S. M. Lemon. 1999. An infectious molecular clone of a Japanese genotype 1b hepatitis C virus. Hepatology 30:316-324. [DOI] [PubMed] [Google Scholar]
- 7.Bigger, C. B., K. M. Brasky, and R. E. Lanford. 2001. DNA microarray analysis of chimpanzee liver during acute resolving hepatitis C virus infection. J. Virol. 75:7059-7066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Blight, K. J., A. A. Kolykhalov, and C. M. Rice. 2000. Efficient initiation of HCV RNA replication in cell culture. Science 290:1972-1974. [DOI] [PubMed] [Google Scholar]
- 9.Bodenheimer, H. C., Jr., K. L. Lindsay, G. L. Davis, J. H. Lewis, S. N. Thung, and L. B. Seeff. 1997. Tolerance and efficacy of oral ribavirin treatment of chronic hepatitis C: a multicenter trial. Hepatology 26:473-477. [DOI] [PubMed] [Google Scholar]
- 10.Boehm, U., T. Klamp, M. Groot, and J. Howard. 1997. Cellular responses to interferon-γ. Annu. Rev. Immunol. 15:749-795. [DOI] [PubMed] [Google Scholar]
- 11.Choo, Q.-L., G. Kuo, A. J. Weiner, L. J. Overby, D. W. Bradley, and M. Houghton. 1989. Isolation of a cDNA clone derived from blood-borne non-A, non-B viral hepatitis genome. Science 244:359-362. [DOI] [PubMed] [Google Scholar]
- 12.Conry-Cantilena, C., M. VanRaden, J. Gibble, J. Melpolder, A. O. Shakil, L. Viladomiu, L. Cheung, A. DiBisceglie, J. Hoofnagle, J. W. Shih, R. Kaslow, P. Ness, and H. J. Alter. 1996. Routes of infection, viremia, and liver disease in blood donors found to have hepatitis C virus infection. N. Engl. J. Med. 334:1691-1696. [DOI] [PubMed] [Google Scholar]
- 13.Crotty, S., C. E. Cameron, and R. Andino. 2001. RNA virus error catastrophe: direct molecular test by using ribavirin. Proc. Natl. Acad. Sci. USA 98:6895-6900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.De Clercq, E. 1993. Antiviral agents: characteristic activity spectrum depending on the molecular target with which they interact. Adv. Virus Res. 42:1-55. [DOI] [PubMed] [Google Scholar]
- 15.Der, S. D., A. Zhou, B. R. Williams, and R. H. Silverman. 1998. Identification of genes differentially regulated by interferon alpha, beta, or gamma using oligonucleotide arrays. Proc. Natl. Acad. Sci. USA 95:15623-15628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dusheiko, G., J. Main, H. Thomas, O. Reichard, C. Lee, A. Dhillon, S. Rassam, A. Fryden, H. Reesink, M. Bassendine, G. Norkrans, T. Cuypers, N. Lelie, P. Telfer, J. Watson, C. Weegink, P. Sillikens, and O. Weiland. 1996. Ribavirin treatment for patients with chronic hepatitis C: results of a placebo-controlled study. J. Hepatol. 25:591-598. [DOI] [PubMed] [Google Scholar]
- 17.Enomoto, N., I. Sakuma, Y. Asahina, M. Kurosaki, T. Murakami, C. Yamamoto, and N. Izumi. 1996. Interferon sensitivity determining sequence of the hepatitis C virus genome. Hepatology 24:460-461. [DOI] [PubMed] [Google Scholar]
- 18.Enria, D. A., and J. I. Maiztegui. 1994. Antiviral treatment of argentine hemorrhagic fever. Antiviral Res. 23:23-31. [DOI] [PubMed] [Google Scholar]
- 19.Fang, S. H., L. H. Hwang, D. S. Chen, and B. L. Chiang. 2000. Ribavirin enhancement of hepatitis C virus core antigen-specific type 1 T helper cell response correlates with the increased IL-12 level. J. Hepatol. 33:791-798. [DOI] [PubMed] [Google Scholar]
- 20.Frese, M., T. Pietschmann, D. Moradpour, O. Haller, and R. Bartenschlager. 2001. Interferon-α inhibits hepatitis C virus subgenomic RNA replication by an MxA-independent pathway. J. Gen. Virol. 82:723-733. [DOI] [PubMed] [Google Scholar]
- 21.Frese, M., V. Schwarzle, K. Barth, N. Krieger, V. Lohmann, S. Mihm, O. Haller, and R. Bartenschlager. 2002. Interferon-gamma inhibits replication of subgenomic and genomic hepatitis C virus RNAs. Hepatology 35:694-703. [DOI] [PubMed] [Google Scholar]
- 22.Gale, M., C. M. Blakely, B. Kwieciszewski, S. L. Tan, M. Dossett, N. M. Tang, M. J. Korth, S. J. Polyak, D. R. Gretch, and M. G. Katze. 1998. Control of PKR protein kinase by hepatitis C virus nonstructural 5A protein: molecular mechanisms of kinase regulation. Mol. Cell. Biol. 18:5208-5218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gale, M. J., M. J. Korth, N. M. Tang, S. L. Tan, D. A. Hopkins, T. E. Dever, S. J. Polyak, D. R. Gretch, and M. G. Katze. 1997. Evidence that hepatitis C virus resistance to interferon is mediated through repression of the PKR protein kinase by the nonstructural 5A protein. Virology 230:217-227. [DOI] [PubMed] [Google Scholar]
- 24.Geiss, G., G. Jin, J. Guo, R. Bumgarner, M. G. Katze, and G. Sen. 2001. A comprehensive view of regulation of gene expression by double-stranded RNA-mediated cell signaling. J. Biol. Chem. 276:30178-30182. [DOI] [PubMed] [Google Scholar]
- 25.Girard, S., P. Shalhoub, P. Lescure, A. Sabile, D. E. Misek, S. Hanash, C. Brechot, and L. Beretta. 2002. An altered cellular response to interferon and up-regulation of interleukin-8 induced by the hepatitis C viral protein NS5A uncovered by microarray analysis. Virology 295:272-283. [DOI] [PubMed] [Google Scholar]
- 26.Guidotti, L. G., A. Morris, H. Mendez, R. Koch, R. H. Silverman, B. R. G. Williams, and F. V. Chisari. 2002. Interferon-regulated pathways that control hepatitis B virus replication in transgenic mice. J. Virol. 76:2617-2621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Guo, J. T., V. Bichko, and C. Seeger. 2001. Effect of alpha interferon on the hepatitis C virus replicon. J. Virol. 75:8516-8523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Honda, M., M. R. Beard, L. H. Ping, and S. M. Lemon. 1999. A phylogenetically conserved stem-loop structure at the 5′ border of the internal ribosome entry site of hepatitis C virus is required for cap-independent viral translation. J. Virol. 73:1165-1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Huggins, J. W., C. M. Hsiang, T. M. Cosgriff, J. Y. Guang, J. I. Smith, Z. O. Wu, J. W. DeDuc, Z. M. Zheng, J. M. Meegan, Q. N. Wang, D. D. Oland, X. E. Gui, P. H. Gibbs, G. H. Yuan, and T. M. Zhang. 1991. Prospective, double-blind, concurrent, placebo-controlled clinical trial of intravenous ribavirin therapy of hemorrhagic fever with renal syndrome. J. Infect. Dis. 164:1119-1127. [DOI] [PubMed] [Google Scholar]
- 30.Hultgren, C., D. R. Milich, O. Weiland, and M. Sällberg. 1998. The antiviral compound ribavirin modulates the T helper (Th)1/Th2 subset balance in hepatitis B and C virus-specific immune responses. J. Gen. Virol. 79:2381-2391. [DOI] [PubMed] [Google Scholar]
- 31.Hummer, B. T., X. L. Li, and B. A. Hassel. 2001. Role for p53 in gene induction by double-stranded RNA. J. Virol. 75:7774-7777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ikeda, M., M. K. Yi, K. Li, and S. A. Lemon. 2002. Selectable subgenomic and genome-length dicistronic RNAs derived from an infectious molecular clone of the HCV-N strain of hepatitis C virus replicate efficiently in cultured Huh7 cells. J. Virol. 76:2997-3006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jaeckel, E., M. Cornberg, H. Wedemeyer, T. Santantonio, J. Mayer, M. Zankel, G. Pastore, M. Dietrich, C. Trautwein, and M. Manns. 2001. Treatment of acute hepatitis C with interferon alpha-2b. N. Engl. J. Med. 345:1-6. [DOI] [PubMed] [Google Scholar]
- 34.Kilgore, P. E., T. G. Ksiazek, P. E. Rollin, J. N. Mills, M. R. Villagra, M. J. Montenegro, M. A. Costales, L. C. Paredes, and C. J. Peters. 1997. Treatment of bolivian hemorrhagic fever with intravenous ribavirin. Clin. Infect. Dis. 24:718-722. [DOI] [PubMed] [Google Scholar]
- 35.Krieger, N., V. Lohmann, and R. Bartenschlager. 2001. Enhancement of hepatitis C virus RNA replication by cell culture-adaptive mutations. J. Virol. 75:4614-4624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kumar, A., Y. L. Yang, V. Flati, S. Der, S. Kadereit, A. Deb, J. Haque, L. Reis, C. Weissmann, and B. R. Williams. 1997. Deficient cytokine signaling in mouse embryo fibroblasts with a targeted deletion in the PKR gene: role of IRF-1 and NF-κB. EMBO J. 16:406-416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lanford, R. E., and C. Bigger. 2002. Advances in model systems for hepatitis C virus research. Virology 293:1-9. [DOI] [PubMed] [Google Scholar]
- 38.Lanford, R. E., D. Chavez, B. Guerra, J. Y. N. Lau, Z. Hong, K. M. Brasky, and B. Beames. 2001. Ribavirin induces error-prone replication of GB virus B in primary tamarin hepatocytes. J. Virol. 75:8074-8081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lanford, R. E., and L. E. Estlack. 1998. A cultivation method for highly differentiated primary chimpanzee hepatocytes permissive for hepatitis C virus replication. Methods Mol. Med. 19:501-516. [DOI] [PubMed] [Google Scholar]
- 40.Lanford, R. E., H. Lee, D. Chavez, B. Guerra, and K. M. Brasky. 2001. Infectious cDNA clone of the hepatitis C virus genotype 1 prototype sequence. J. Gen. Virol. 82:1291-1297. [DOI] [PubMed] [Google Scholar]
- 41.Lau, J. Y., R. C. Tam, T. J. Liang, and Z. Hong. 2002. Mechanism of action of ribavirin in the combination treatment of chronic HCV infection. Hepatology 35:1002-1009. [DOI] [PubMed] [Google Scholar]
- 42.Lohmann, V., F. Korner, A. Dobierzewska, and R. Bartenschlager. 2001. Mutations in hepatitis C virus RNAs conferring cell culture adaptation. J. Virol. 75:1437-1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lohmann, V., F. Körner, J. O. Koch, U. Herian, L. Theilmann, and R. Bartenschlager. 1999. Replication of subgenomic hepatitis C virus RNAs in a hepatoma cell line. Science 285:110-113. [DOI] [PubMed] [Google Scholar]
- 44.Maag, D., C. Castro, Z. Hong, and C. E. Cameron. 2001. Hepatitis C virus RNA-dependent RNA polymerase (NS5B) as a mediator of the antiviral activity of ribavirin. J. Biol. Chem. 276:46094-46098. [DOI] [PubMed] [Google Scholar]
- 45.Manns, M. P., J. G. McHutchison, S. C. Gordon, V. K. Rustgi, M. Shiffman, R. Reindollar, Z. D. Goodman, M. H. Ling, and J. K. Albrecht. 2001. Peginterferon alfa-2b plus ribavirin compared with interferon alfa-2b plus ribavirin for initial treatment of chronic hepatitis C: a randomised trial. Lancet 358:958-965. [DOI] [PubMed] [Google Scholar]
- 46.Marcus, P. I., and M. J. Sekellick. 2001. Combined sequential treatment with interferon and dsRNA abrogates virus resistance to interferon action. J. Interferon Cytokine Res. 21:423-429. [DOI] [PubMed] [Google Scholar]
- 47.Markland, W., T. J. McQuaid, J. Jain, and A. D. Kwong. 2000. Broad-spectrum antiviral activity of the IMP dehydrogenase inhibitor VX-497: a comparison with ribavirin and demonstration of antiviral additivity with alpha interferon. Antimicrob. Agents Chemother. 44:859-866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.McCormick, J. B., I. J. King, P. A. Webb, C. L. Scribner, R. B. Craven, K. M. Johnson, L. H. Elliott, and R. Belmont-Williams. 1986. Lassa fever/Effective therapy with ribavirin. N. Engl. J. Med. 314:20-26. [DOI] [PubMed] [Google Scholar]
- 49.Melen, K., P. Keskinen, A. Lehtonen, and I. Julkunen. 2000. Interferon-induced gene expression and signaling in human hepatoma cell lines. J. Hepatol. 33:764-772. [DOI] [PubMed] [Google Scholar]
- 50.Ning, Q., D. Brown, J. Parodo, M. Cattral, R. Gorczynski, E. Cole, L. Fung, J. W. Ding, M. F. Liu, O. Rotstein, M. J. Phillips, and G. Levy. 1998. Ribavirin inhibits viral-induced macrophage production of TNF, IL-1, the procoagulant fgl2 prothrombinase and preserves Th1 cytokine production but inhibits Th2 cytokine response. J. Immunol. 160:3487-3493. [PubMed] [Google Scholar]
- 51.Pavio, N., D. R. Taylor, and M. M. C. Lai. 2002. Detection of a novel unglycosylated form of hepatitis C virus E2 envelope protein that is located in the cytosol and interacts with PKR. J. Virol. 76:1265-1272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pflugheber, J., B. Fredericksen, R. Sumpter, Jr., C. Wang, F. Ware, D. L. Sodora, and M. Gale, Jr. 2002. Regulation of PKR and IRF-1 during hepatitis C virus RNA replication. Proc. Natl. Acad. Sci. USA 99:4650-4655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pietschmann, T., V. Lohmann, A. Kaul, N. Krieger, G. Rinck, G. Rutter, D. Strand, and R. Bartenschlager. 2002. Persistent and transient replication of full-length hepatitis C virus genomes in cell culture. J. Virol. 76:4008-4021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Podevin, P., A. Sabile, R. Gajardo, N. Delhem, A. Abadie, P. Y. Lozach, L. Beretta, and C. Brechot. 2001. Expression of hepatitis C virus NS5A natural mutants in a hepatocytic cell line inhibits the antiviral effect of interferon in a PKR-independent manner. Hepatology 33:1503-1511. [DOI] [PubMed] [Google Scholar]
- 55.Polyak, S. J., K. S. A. Khabar, D. M. Paschal, H. J. Ezelle, G. Duverlie, G. N. Barber, D. E. Levy, N. Mukaida, and D. R. Gretch. 2001. Hepatitis C virus nonstructural 5A protein induces interleukin-8, leading to partial inhibition of the interferon-induced antiviral response. J. Virol. 75:6095-6106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Reed, K. E., and C. M. Rice. 1998. Molecular characterization of hepatitis C virus. Curr. Stud. Hematol. Blood Transfus. 62:1-37. [DOI] [PubMed] [Google Scholar]
- 57.Samuel, C. E. 2001. Antiviral actions of interferons. Clin. Microbiol. Rev. 14:778-809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sidwell, R. W., J. H. Huffman, G. P. Khare, L. B. Allen, J. T. Witkowski, and R. K. Robins. 1972. Broad-spectrum antiviral activity of virazole: 1-β-d-ribofuranosyl-1,2,4-triazole-3-carboxamide. Science 177:705-706. [DOI] [PubMed] [Google Scholar]
- 59.Stark, G. R., I. M. Kerr, B. R. Williams, R. H. Silverman, and R. D. Schreiber. 1998. How cells respond to interferons. Annu. Rev. Biochem. 67:227-264. [DOI] [PubMed] [Google Scholar]
- 60.Tam, R. C., B. Pai, J. Bard, C. Lim, D. R. Averett, U. T. Phan, and T. Milovanovic. 1999. Ribavirin polarizes human T cell responses towards a type 1 cytokine profile. J. Hepatol. 30:376-382. [DOI] [PubMed] [Google Scholar]
- 61.Tan, S. L., and M. G. Katze. 2001. How hepatitis C virus counteracts the interferon response: the jury is still out on NS5A. Virology 284:1-12. [DOI] [PubMed] [Google Scholar]
- 62.Tan, S. L., H. Nakao, Y. He, S. Vijaysri, P. Neddermann, B. L. Jacobs, B. J. Mayer, and M. G. Katze. 1999. NS5A, a nonstructural protein of hepatitis C virus, binds growth factor receptor-bound protein 2 adaptor protein in a Src homology 3 domain/ligand-dependent manner and perturbs mitogenic signaling. Proc. Natl. Acad. Sci. USA 96:5533-5538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Taniguchi, T., and A. Takaoka. 2001. A weak signal for strong responses: interferon-alpha/beta revisited. Nat. Rev. Mol. Cell Biol. 2:378-386. [DOI] [PubMed] [Google Scholar]
- 64.Tanji, Y., T. Kaneko, S. Satoh, and K. Shimotohno. 1995. Phosphorylation of hepatitis C virus-encoded nonstructural protein NS5A. J. Virol. 69:3980-3986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Taylor, D. R., S. T. Shi, P. R. Romano, G. N. Barber, and M. M. Lai. 1999. Inhibition of the interferon-inducible protein kinase PKR by HCV E2 protein. Science 285:107-110. [DOI] [PubMed] [Google Scholar]
- 66.Tu, H., L. Gao, S. T. Shi, D. R. Taylor, T. Yang, A. K. Mircheff, Y. M. Wen, A. E. Gorbalenya, S. B. Hwang, and M. M. Lai. 1999. Hepatitis C virus RNA polymerase and NS5A complex with a SNARE-like protein. Virology 263:30-41. [DOI] [PubMed] [Google Scholar]
- 67.Weaver, B. K., K. P. Kumar, and N. C. Reich. 1998. Interferon regulatory factor 3 and CREB-binding protein/p300 are subunits of double-stranded RNA-activated transcription factor DRAF1. Mol. Cell. Biol. 18:1359-1368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yang, Y. L., L. F. Reis, J. Pavlovic, A. Aguzzi, R. Schafer, A. Kumar, B. R. Williams, M. Aguet, and C. Weissmann. 1995. Deficient signaling in mice devoid of double-stranded RNA-dependent protein kinase. EMBO J. 14:6095-6106. [DOI] [PMC free article] [PubMed] [Google Scholar]