Abstract
The majority of hepatitis C virus (HCV)-infected individuals progress from acute to chronic disease, despite the presence of a strong humoral immune response to the envelope glycoproteins E1 and E2. When expressed in mammalian cells, E1 and E2 form both noncovalently linked E1E2 heterodimers, believed to be properly folded, and disulfide-linked, high-molecular-weight aggregates that are misfolded. Previously, we identified 10 human monoclonal antibodies (HMAbs) that bind E2 glycoproteins from different genotypes. Here we demonstrate that one of these HMAbs, CBH-2, is unique in its ability to distinguish between properly folded and misfolded envelope proteins. This HMAb recognizes HCV-E2 only when complexed with E1. The E1E2 complexes recognized by CBH-2 are noncovalently linked heterodimers and not misfolded disulfide-linked, high-molecular-weight aggregates. The E1E2 heterodimers seen by CBH-2 no longer associate with the endoplasmic reticulum chaperone calnexin and are likely to represent the prebudding form of the HCV virion.
Hepatitis C virus (HCV) is the causal agent of hepatitis C, which is a major health problem worldwide (28). HCV is a positive-strand RNA virus (2) that belongs to the Flaviviridae family. Its genome encodes two membrane-associated envelope glycoproteins, E1 and E2, which are N glycosylated in their large N-terminal ectodomains and are anchored into membranes by their C-terminal transmembrane domains (31). These latter domains have been shown to be endoplasmic reticulum (ER) retention signals (5, 7, 17, 20). When expressed in cell culture, the E1 and E2 glycoproteins assemble into noncovalently linked E1E2 heterodimers. These noncovalent E1E2 complexes have been proposed as functional subunits of the HCV particle. In addition, a significant amount of E1 and E2 is also present in high-molecular-weight, disulfide-linked aggregates, thought to result from a nonproductive folding pathway leading to misfolded protein complexes (for review see reference 31).
Because of the lack of a suitable cell culture system for in vitro propagation of HCV and the unavailability of virions in sufficient quantities, truncated, secreted versions of E2 have been used as soluble surrogates for native virus particles. Indeed, the identification of CD81 as the putative cellular receptor for HCV is based on its binding to a truncated form of E2 (36). Intriguingly, intracellular forms of truncated E2, enriched for the presence of monomeric, nonaggregated E2, were found to bind CD81 with greater affinity than did the secreted forms (18, 26), suggesting that antigenic or structural differences exist between intracellular and secreted forms of the E2 glycoprotein. Several murine monoclonal antibodies (MAbs) have been shown to recognize conformation-dependent epitopes within E2. Studies using these antibodies (Abs) (including MAb H53) have provided additional insight into the conformational state of the envelope glycoproteins during intracellular processing and folding and have helped to define a native, prebudding form of the HCV glycoprotein complex (7, 12, 34).
CBH-2 human MAb (HMAb) specifically recognizes E2 complexed with E1.
Abs that arise in HCV-infected individuals in response to viral infection are anticipated to react with the truly native conformation of the viral envelope structure. Recently, several HMAbs have been identified that react with conformational epitopes within E2 (1, 11, 23, 24). Moreover, some of these HMAbs have been shown to have neutralization-of-binding (NOB) activity (1, 23, 24) defined by their ability to neutralize binding of recombinant, truncated HCV-E2 to human cells (37). Previously, we identified 10 HMAbs that bind to full-length HCV-E2 glycoproteins from genotypes 1a, 1b, 2a, and 2b. Nine of these Abs reacted with conformational epitopes, six of which were NOB positive based on their ability to block E2 binding to cells or to CD81-coated plates (24). Additionally, two of the NOB-positive HMAbs inhibited binding of infectious HCV virus particles (genotype 1a) to CD81 immobilized on polystyrene beads (24), suggesting that these two HMAbs recognize important conformational epitopes within E2.
Preliminary experiments using this panel of Abs indicated that CBH-2, one of the two NOB-positive HMAbs that inhibited binding of infectious virus particles, reacted selectively with E2 glycoprotein only when coexpressed with E1. To follow up on this observation, HEK 293 cells cultured in Dulbecco’s modified Eagle medium-10% fetal calf serum were transiently transfected (using the GenePORTER 2 transfection reagent; Gene Therapy System, San Diego, Calif.) with 10 μg of plasmid encoding different forms of HCV glycoproteins from genotype 1a, H strain: full-length E1 (E1; amino acids 171 to 383), truncated E2 (E2 661; amino acids 364 to 661), full-length E2 (E2; amino acids 364 to 746), E1 and E2 (E1E2; amino acids 171 to 746), and E1E2p7NS2 (amino acids 171 to 1026). Transfected cells were lysed in lysis buffer, 4% Triton X-100 (Sigma, St. Louis, Mo.), 100 mM Tris-HCl (pH 8.0), 1 mM EDTA, and Complete Mini protease inhibitor cocktail tablets (Roche Diagnostics, Mannheim, Germany), for 30 min on ice and were clarified by centrifugation at 20,000 × g for 30 min at 4°C. Protein A-immobilized CBH-2 was incubated 2 h at 4°C with clarified cell lysates. Ab-antigen complexes were washed four times with phosphate-buffered saline containing 0.2% Triton X-100. CBH-2 immunoprecipitations and aliquots of each lysate were boiled for 3 min in reducing sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) sample buffer and were electrophoresed on 10% polyacrylamide gels (Invitrogen, Carlsbad, Calif.). Proteins were transferred to polyvinyl difluoride (Immobilon-P; Millipore, Bedford, Mass.) and were immunoblotted with an anti-E2 MAb (3/11 [19]) and detected with a mouse horseradish peroxidase-conjugated anti-rat (human-adsorbed) Ab (BioSource, Camarrillo, Calif.). Detection was performed using enhanced chemiluminescence detection reagents (Amersham Pharmacia Biotech, Little Chalfont, United Kingdom).
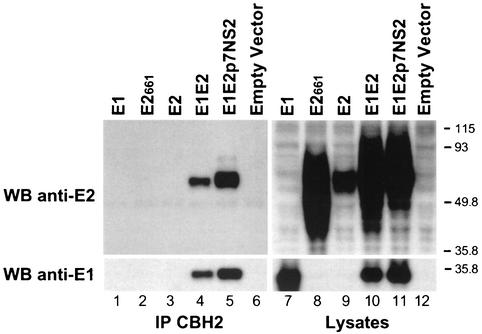
Interestingly, CBH-2 did not precipitate (Fig. 1) truncated E2661 (upper panel, lane 2) or full-length E2 (upper panel, lane 3) glycoprotein, although cell lysates contained E2 proteins as determined by Western blotting (WB) with the anti-E2 MAb (Fig. 1, upper panel, lanes 8 and 9). However, CBH-2 was able to precipitate E2 when coexpressed with E1 (Fig. 1, upper panel, lanes 4 and 5). The additional faint higher-molecular-weight band observed in lane 5 is probably the unprocessed E2-NS2 precursor. Negative controls included empty vector-transfected cell lysates (Fig. 1, upper panel, lanes 6 and 12) or lysates prepared from cells expressing E1 alone (Fig. 1, upper panel, lanes 1 and 7). The blots were subsequently stripped by incubating in stripping buffer (62.5 mM Tris-HCl [pH 6.7], 100 mM β-mercaptoethanol, and 2% SDS) for 30 min at 50°C and were reprobed with an anti-E1 MAb (A4 [15]), which was used to confirm the presence of E1 in the immunoprecipitates and lysates. As previously observed with the conformation-dependent anti-E2 MAb, H53 (7), CBH-2 was able to coprecipitate E1 with E2 when the HCV envelope glycoproteins were coexpressed (lower panel, lanes 4 and 5). The requirement of CBH-2 to bind E2 only when expressed in the presence of E1 suggests that this HMAb may be recognizing a conformation of E2 that closely resembles native E2 glycoproteins as they exist on virus particles.
FIG. 1.
CBH-2 HMAb immunoprecipitates (IP) E2 only when coexpressed intracellularly with E1. HEK 293 cells transfected with plasmids encoding E1 (lane 1), truncated E2661 (lane 2), E2 (lane 3), E1E2 (lane 4), E1E2p7NS2 (lane 5), or an empty vector (lane 6) were lysed 72 h posttransfection and were immunoprecipitated with CBH-2 as described in the text. Immunoprecipitations (lanes 1 to 6) and lysates (lanes 7 to 12) were separated by SDS-10% PAGE under reducing conditions, and immunoblots were analyzed with the anti-E2 MAb 3/11 (upper panels). The blot was stripped and reprobed with the anti-E1 MAb A4 (lower panels). Sizes (in kilodaltons) of protein molecular mass markers are indicated on the right. Results are representative of data obtained in three independent experiments.
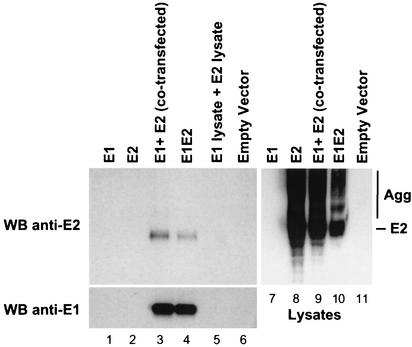
Properly folded E1 and E2 glycoproteins interact to form noncovalently linked heterodimers (12), which are believed to be the native, prebudding form of the virus (31). Previous studies have shown that E1E2 heterodimers can be formed when HCV envelope proteins are expressed both in cis and in trans (6, 9). In order to know whether CBH-2 recognizes these heterodimers, 293 cells were transfected with plasmids encoding E1 alone, E2 alone, or E1E2 (cis) or were cotransfected with a plasmid expressing E1 and a plasmid expressing E2 (trans). Following transfection, cells were lysed and intracellular HCV glycoproteins were immunoprecipitated with CBH-2, separated, and detected as detailed above (Fig. 2). To control for postlysis heterodimer formation, lysates from cells transfected separately with E1 or E2 were mixed prior to immunoprecipitation with CBH-2 (Fig. 2, lane 5). Cell lysates contained similar amounts of E2 glycoprotein as determined by WB with 3/11 (Fig. 2, lanes 8 to 10). The cell lysates contained additional diffuse bands in the upper part of the gel that likely represent aggregates (Fig. 2, lanes 8 to 10). Again, CBH-2 failed to precipitate E2 molecules when expressed in the absence of E1 (Fig. 2, upper panel, lane 2). In contrast, coexpression of E1 and E2, either in cis (E1E2) or in trans (E1 plus E2), resulted in efficient binding of E2 by the HMAb (upper panel, lanes 3 and 4). Proper folding and formation of E1E2 complexes did not appear to occur postlysis, as the Ab did not precipitate an appropriately sized band when incubated with a mixture of E1 and E2 lysates (Fig. 2, upper panel, lane 5). The blot was subsequently stripped and reprobed with A4, which was used to confirm the presence of E1 in the immunoprecipitates. As previously observed with the conformation-dependent anti-E2 MAb H53 (6, 9), CBH-2 was able to coprecipitate E1 with E2 when the HCV envelope glycoproteins were cotranslationally expressed (lower panel, lanes 3 and 4) but not when they were mixed postlysis (Fig. 2, lower panel, lane 5). Taken together, these results demonstrate that CBH-2 binds only to E2 glycoproteins that are complexed with E1, suggesting that this HMAb recognizes a prebudding, possibly native form of E2 envelope protein.
FIG. 2.
Cotranslational expression of E1 and E2 is required for immunoprecipitation of E2 by CBH-2. HEK 293 cells were transfected with plasmids expressing E1 (lane 1), E2 (lane 2), E1E2 (lane 4), and empty vector (lane 6) or were cotransfected with a plasmid expressing E1 and a plasmid expressing E2 (lane 3). After 72 h, cells were lysed and immunoprecipitated with CBH-2. In addition, lysates from cells transfected separately with E1 or E2 were mixed prior to immunoprecipitation (lane 5). Immunoprecipitations and lysates were analyzed by SDS-10% PAGE under nonreducing conditions and were immunoblotted with the anti-E2 MAb 3/11 (upper panels). The blot was stripped and reprobed with the anti-E1 MAb A4 (lower panel). Results are representative of data obtained in three independent experiments. High-molecular-weight aggregates (Agg) are indicated.
CBH-2 requires cotranslational expression of E1 to efficiently bind to E2 glycoproteins. This result was surprising, since this HMAb was originally identified by assays that required its binding to full-length E2 expressed in the absence of E1 (24). The assays used previously detected reactivity with immobilized E2, whereas we used immobilized Abs to precipitate the envelope proteins from a soluble cell lysate. Under such conditions, it is likely that only high-affinity interactions (>108/mol) could be detected (25). It is also possible that the binding of E2 on solid phase may mimic the interaction with E1 to create the CBH-2 epitope.
The effect of HCV envelope proteins on folding of each other has been addressed by several studies. E1 has been shown to fold improperly, remaining in the reduced state, in the absence of E2 (30, 35). Insertion of alanine substitutions within the transmembrane domains of E1 and E2 disrupted E1E2 heterodimer formation and decreased the amount of properly folded E1 (32). Similarly, glycosylation of E1 was dramatically improved when it was coexpressed with E2, indicating that glycosylation is also dependent on the presence of polypeptide sequences downstream of E1 (14). These results indicate that E2 possesses a chaperone-like function to facilitate proper folding of E1 (for review, see reference 31). In contrast to E1, E2 expressed in the absence of E1 was shown to fold properly (30). Yet, coexpression of E1 either in cis or in trans was required for stable association of E2 with the ER membrane, suggesting that interaction between the hydrophobic transmembrane domains of these proteins is required for efficient ER membrane insertion and complex formation (6). The requirement of CBH-2 to bind E2 only when coexpressed with E1 suggests once more that the presence of E1 may also influence the folding of E2, such that the epitope recognized by CBH-2 is formed only when E1 and E2 are in a complex.
The unique ability of CBH-2 to react with E1E2 heterodimers is seen only in the context of genotype 1a. Recently, Triyatni et al. have shown that the ability of CBH-2 to bind HCV-like particles (HCV-LPs) is restricted to genotype 1a (H77 strain); CBH-2 did not react with HCV-LPs from genotype 1b (J strain) (38). In that study other conformation-dependent mouse MAbs, H2 and H53, also showed preferential binding to the HCV-LP 1a. The interaction of HCV-LP 1b with Abs was broader, exhibiting reactivity with Abs to both conformational and to linear epitopes (38).
CBH-2 HMAb selectively binds noncovalently associated E1E2 heterodimers and not disulfide-linked aggregates.
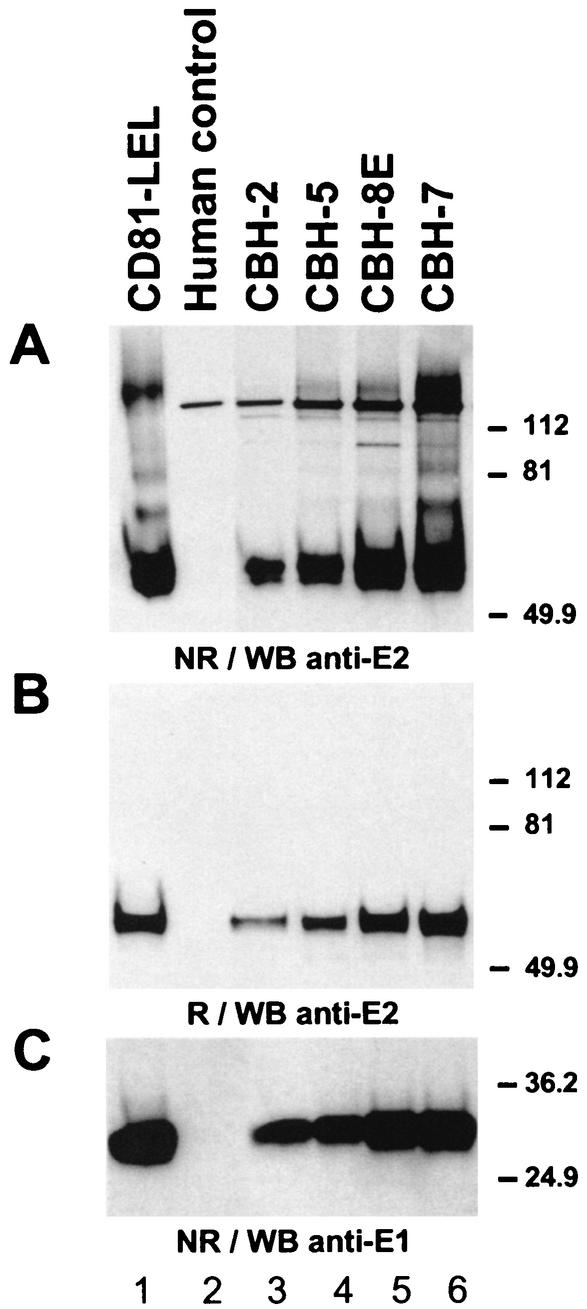
Studies using transient-expression systems have shown that E2 interacts with E1 to form oligomers. In the presence of nonionic detergents, two forms of E1E2 complexes are detected: a heterodimer of E1 and E2 stabilized by noncovalent interactions and heterogeneous disulfide-linked aggregates (15). Aggregates were first observed when the proteins were expressed by virus vectors that can induce a high level of protein synthesis (15, 22). However, aggregates have also been reported when HCV proteins are expressed in a nonvirus vector that drives a lower level of protein synthesis (3). In our study, we analyzed the assembly state of E1E2 complexes precipitated by CBH-2. For this purpose, we immunoprecipitated HCV glycoproteins with CBH-2 and compared its reactivity to three other NOB-positive HMAbs (CBH-5, CBH-8E, and CBH-7 [24]). Cells expressing E1E2 were lysed in the presence of 20 mM iodoacetamide (Sigma). The cell lysate was subsequently divided into equal fractions, immunoprecipitated with each of the HMAbs, and analyzed on an SDS-PAGE gel under nonreducing and reducing conditions followed by WB with 3/11 (Fig. 3A and B, respectively). Interestingly, CBH-2 preferentially bound the nonaggregated form of E2 (lane 3), whereas the other NOB-positive HMAbs bound additional aggregated E2 (lanes 4 to 6). It is worth noting that these other HMAbs immunoprecipitated a more intense band corresponding to E2 that likely contains both properly and improperly folded monomeric forms of E2. These results suggest that CBH-2 probably recognizes only properly folded E2 glycoproteins. When the same samples were analyzed under reducing conditions (Fig. 3B), the precipitated E2 molecules bound by each of the HMAbs collapsed to a single band, supporting the hypothesis that all HMAbs, with the exception of CBH-2, bind both nonaggregated forms of E2 and high- molecular-weight, disulfide-linked aggregates. The presence of E1 in each of the immunoprecipitates was confirmed by stripping the blot and reprobing with A4 (Fig. 3C). The HMAbs that bound more total E2 molecules also coprecipitated higher levels of E1. This experiment confirms that, although all HMAbs bind to E2 molecules, only CBH-2 specifically binds to noncovalently linked E1E2 complexes, which are believed to be the native, prebudding form of the virus.
FIG. 3.
Comparison of the ability of HMAbs and soluble CD81 to precipitate noncovalent E1E2 heterodimers. HEK 293 cells transfected with a plasmid encoding E1E2 were lysed in buffer containing iodoacetamide (20 mM). The cell lysate was divided into equal fractions and immunoprecipitated with each of the NOB-positive (lanes 3 to 6) or with a human immunoglobulin G1 isotype control MAb (lane 2). Precipitation with an immobilized CD81-LEL fusion protein (lane 1) was performed as detailed in the text. Samples were run under nonreducing (NR) (A) and reducing (R) (B) conditions and were analyzed by immunoblotting with the anti-E2 MAb, 3/11 (A and B). Blot A was stripped and reprobed with the anti-E1 MAb A4 (C). Results are representative of data obtained in two independent experiments.
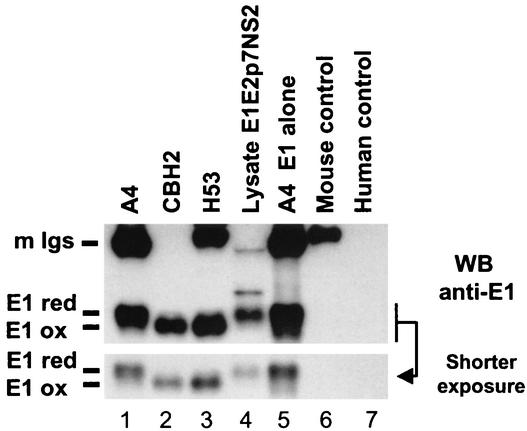
It has been previously shown that the folding of E1 is slow and that coexpression of E2 is necessary for the proper folding of E1 (30). In the absence of E2, E1 remains in a reduced state and forms aggregates containing improperly folded oligomeric structures of E1 (30, 35). Since acquisition of oxidized form correlates with proper folding and assembly of native heterodimers (12, 16, 30), we analyzed the folding of E1 to further characterize E1E2 complexes recognized by CBH-2. Since no MAb that recognizes properly folded E1 glycoprotein is presently available, we monitored disulfide bond formation in E1 by SDS-PAGE under nonreducing conditions, as described previously (16). This method takes advantage of an increase in mobility as a protein acquires a compact conformation, stabilized by the formation of intramolecular disulfide bonds. The increase in migration of proteins with a low molecular weight, such as E1, is more apparent than when higher-molecular-weight proteins, such as E2, are analyzed. We therefore focused on the conformation of E1 in the E1E2 heterodimeric complex. HEK 293 cells were transfected with plasmids expressing E1E2p7NS2 or E1 alone and were lysed in the presence of iodoacetamide. Proteins from E1E2p7NS2 lysate were immunoprecipitated with A4, CBH-2, or the well-characterized mouse conformation-dependent anti-E2 MAb, H53 (7, 12). Proteins from E1 lysate were also immunoprecipitated with A4. Bound proteins were analyzed under nonreducing conditions in WB with A4. As shown in Fig. 4 (lane 5 and shorter exposure), a reduced form of E1 was clearly immunoprecipitated by A4 in the context of E1 expressed alone, as previously observed (30, 35). In our experiments, this Ab that recognizes a linear epitope seems to preferentially recognize the reduced form of E1 proteins from E1E2p7NS2 lysate, either in immunoprecipitation (Fig. 4, lane 1) or WB (Fig. 4, lane 4). Similar to previous findings (7, 12), the mouse conformation-dependent anti-E2 MAb, H53, recognized only the oxidized form of E1 (Fig. 4, lane 3). Similar to the case for H53, only the oxidized form of E1 was immunoprecipitated by CBH-2 (Fig. 4, lane 2), indicating that E1 proteins coimmunoprecipitated by CBH-2 have acquired a compact configuration and are properly folded. Since precipitation of the oxidized form of E1 correlates with proper folding and assembly of native E1E2 heterodimers (12, 16, 30), these results show that CBH-2 recognizes only properly folded, native E1E2 heterodimers.
FIG. 4.
CBH-2 HMAb precipitates only properly folded E1E2 complexes. HEK 293 cells were transfected with plasmid expressing E1E2p7NS2 or E1 alone and were lysed in the presence of iodoacetamide. Proteins from E1E2p7NS2 lysate were immunoprecipitated with CBH-2, the mouse anti-E1 (A4), or the mouse conformation-dependent anti-E2 (H53). Irrelevant mouse and human immunoglobulins were used as negative controls. Proteins from E1 lysate were immunoprecipitated with A4. Bound proteins and lysate E1E2p7NS2 (3 × 105 cell equivalents, 10 times less than in immunoprecipitations) were analyzed by SDS-12% PAGE under nonreducing conditions followed by immunoblotting with A4. Results are representative of data obtained in two independent experiments. Mouse immunoglobulins (m Igs) are indicated. The reduced form (E1 red) and oxidized form (E1 ox) of E1 are indicated.
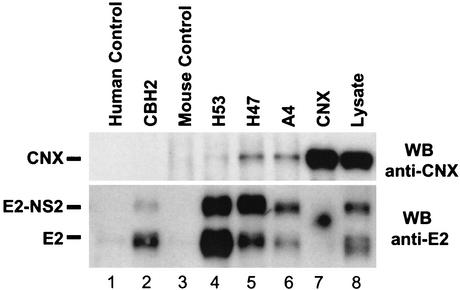
The involvement of ER chaperones in HCV glycoprotein assembly has been studied. It showed that calreticulin and BiP interact preferentially with aggregates of E1 and E2. In contrast, calnexin (CNX) has been shown to have a higher affinity for noncovalently linked complexes (4). Immediately after their synthesis, monomeric forms of E1 and E2 interact with CNX. Intramolecular disulfide bonds form rapidly in E2, leading to the folding of at least one subdomain. This partially folded form of E2 interacts with the nonoxidized form of E1, forming an intermediate E1E2 complex that is associated with CNX. E1 and E2 acquire their final state of folding and finally dissociate from CNX as the prebudding form of HCV glycoprotein complex (13, 31). In our study, we examined whether E1E2 complexes recognized by CBH-2 still interact or not with CNX. The state of folding and assembly of E1E2 immunoprecipitated by CBH-2 was analyzed by monitoring the presence or the absence of coimmunoprecipitation of CNX, as described previously for H2, a mouse conformation-dependent anti-E2 MAb (12). For this purpose, COS-7 cells were infected with a recombinant adenovirus expressing E1E2p7NS2 (amino acids 171 to 1026) at a multiplicity of infection of 25 PFU per cell. This recombinant adenovirus was produced by the Adeno-X expression system (BD Biosciences Clontech, Palo Alto, Calif.). Forty-eight hours postinfection, cells were lysed and intracellular HCV glycoproteins were immunoprecipitated with CBH-2, H53, A4, and a linear anti-E2 MAb, H47 (8). A rabbit polyclonal anti-CNX Ab (Transduction Laboratories, Lexington, Calif.) was used as positive control. Proteins were analyzed under reducing conditions and were revealed by WB with the rabbit anti-CNX Ab (Fig. 5, upper panel). As expected, cell lysate contained large amounts of CNX that can be detected by immunoprecipitation followed by a WB with the anti-CNX Ab (Fig. 5, upper panel, lane 7) or directly by WB with the same Ab (Fig. 5, upper panel, lane 8). CNX was coimmunoprecipitated with the mouse linear anti-E1 (Fig. 5, upper panel, lane 6) and anti-E2 (Fig. 5, upper panel, lane 5) MAbs, A4 and H47, respectively, indicating that these MAbs recognize E1E2 complexes associated with CNX. MAb H53 coprecipitated minuscule amounts of CNX (Fig. 5, upper panel, lane 4), as described earlier (J. Dubuisson, personal communication). Interestingly, as observed with the conformation-dependent anti-E2 MAb, H2 (12), CBH-2 did not coprecipitate CNX (Fig. 5, upper panel, lane 2), indicating that E1E2 heterodimers recognized by CBH-2 have dissociated from CNX and are properly folded. The blot was subsequently stripped and reprobed with 3/11, which was used to confirm the immunoprecipitation of E2 (Fig. 5, lower panel). E2 glycoprotein and E2-NS2 precursor were efficiently detected in immunoprecipitation with the anti-E1 and anti-E2 MAbs (Fig. 5, lower panel, lanes 2, 4, 5, and 6) and in lysate (Fig. 5, lower panel, lane 8). The anti-CNX Ab seems to preferentially recognize cellular CNX that does not interact with HCV glycoproteins (Fig. 5, lower panel, lane 7). High proportions of E2-NS2 precursor are probably due to a less efficient cleavage in the adenovirus expression system. Both E2 and E2-NS2 precursor were recognized by MAb CBH-2 (Fig. 5, lower panel, lane 2, and Fig. 1). It is not known whether one or both of these E2 forms are present in mature HCV particles. Taken together, these results show that CBH-2 binds E1E2 heterodimers in their final state of folding, dissociated from CNX, indicating that this HMAb recognizes the prebudding form of the HCV glycoprotein complex. CBH-2 preferentially binds noncovalent E1E2 heterodimers and is able to inhibit the binding of HCV infectious particles. These data suggest to us that CBH-2 may be a useful tool for identification of native HCV envelope complexes.
FIG. 5.
CBH-2 binds E1E2 heterodimers dissociated from CNX. COS-7 cells were infected with a recombinant adenovirus expressing E1E2p7NS2; intracellular HCV glycoproteins were immunoprecipitated with CBH-2, H53, A4, a linear anti-E2 MAb (H47), or a polyclonal anti-CNX Ab. Irrelevant mouse and human immunoglobulins were used as negative controls. Bound proteins (lanes 1 to 7) and lysate (lane 8) were analyzed by SDS-10% PAGE under reducing conditions followed by immunoblotting with the anti-CNX Ab (upper panel). The blot was stripped and reprobed with the anti-E2 MAb 3/11 (lower panel). Results are representative of data obtained in two independent experiments.
Interaction of E1E2 heterodimers with CD81.
The ability of recombinant versions of HCV-E2 to bind human CD81 has been confirmed in several studies using recombinant fusion proteins containing the large extracellular loop (LEL) of CD81 (CD81-LEL) (10, 19-21, 24, 27, 33, 34, 36, 38-41). It was also shown to react with E1 and E2 glycoproteins reconstituted into liposomes (29) and with HCV-LP 1a (38). Most importantly, the CD81-LEL fusion protein was shown to interact with HCV genome-containing particles (36), demonstrating that CD81 does indeed bind native E2 glycoproteins as they exist on HCV virions. We therefore wanted to investigate the reactivity of the CD81-LEL fusion protein with intracellular forms of the HCV glycoproteins. For this purpose, a recombinant carboxy-terminal fusion protein containing the LEL of human CD81 fused to glutathione-S-transferase (19) was preadsorbed onto glutathione-Sepharose 4B beads (Pharmacia Biotech, Uppsala, Sweden) and incubated with cell lysates expressing E1E2 containing or not containing 20 mM iodoacetamide as described above. Bound proteins were analyzed under nonreducing and reducing conditions and were immunoblotted with an anti-E2 MAb (Fig. 3A and B, lane 1, respectively). CD81-LEL preferentially bound a high proportion of nonaggregated E2, as shown by the intense band that comigrated with the CBH-2-precipitated E2 molecules (Fig. 3A, compare lanes 1 and 3). The soluble CD81 also precipitated high-molecular-weight aggregates of E2, which collapsed to a single band when the samples were reduced (Fig. 3B, lane 1). Similar to what was shown with the HMAbs, the interaction between CD81-LEL and E1E2 complexes was analyzed by stripping blot A and reprobing with A4 (Fig. 3C). CD81-LEL coprecipitated high levels of E1 glycoprotein. These data demonstrate that soluble CD81 recognizes noncovalently linked E1E2 complexes, similar to those recognized by CBH-2. It is presently unknown whether the reactivity with misfolded and aggregated E2 is due to misfolded, soluble CD81 or whether aggregated material could bind to cell surface-expressed native CD81.
In conclusion, CBH-2 is a conformation-sensitive HMAb that recognizes E2 only when coexpressed with E1. This exclusive specificity indicates that coexpression of E1 probably affects the conformation of E2. CBH-2 specifically binds E1E2 heterodimers from genotype 1a that are noncovalently linked and properly folded and no longer associate with the ER chaperone CNX (Fig. 5) and are believed to be the prebudding form of the HCV virion. Besides providing a novel tool for the analysis of HCV virion assembly and entry, CBH-2 is useful for optimizing production and isolation of native HCV envelope complexes for vaccine applications.
Acknowledgments
This work was supported by National Institutes of Health grants CA34233 (S.L.) and DA06596 and AI47355 (S.K.H.F.) and by grant 2110046 from the California Cancer Research Program (S.L.) under Interagency Agreement 97-12013 (University of California Davis contract no. 98-00924V) with the Department of Health Sciences, Cancer Research Section.
We thank J. A. McKeating for the E2-encoding plasmids and for anti-E2 MAb 3/11. We thank J. Dubuisson for anti-E1 MAb A4 and anti-E2 MAbs H53 and H47.
REFERENCES
- 1.Burioni, R., P. Plaisant, A. Manzin, D. Rosa, V. Delli Carri, F. Bugli, L. Solforosi, S. Abrignani, P. E. Varaldo, G. Fadda, and M. Clementi. 1998. Dissection of human humoral immune response against hepatitis C virus E2 glycoprotein by repertoire cloning and generation of recombinant Fab fragments. Hepatology 28:810-814. [DOI] [PubMed] [Google Scholar]
- 2.Choo, Q.-L., G. Kuo, A. J. Weiner, L. R. Overby, D. W. Bradley, and M. Houghton. 1989. Isolation of a cDNA clone derived from a blood-borne non-A, non-B viral hepatitis genome. Science 244:359-362. [DOI] [PubMed] [Google Scholar]
- 3.Choukhi, A., A. Pillez, H. Drobecq, C. Sergheraert, C. Wychowski, and J. Dubuisson. 1999. Characterization of aggregates of hepatitis C virus glycoproteins. J. Gen. Virol. 80:3099-3107. [DOI] [PubMed] [Google Scholar]
- 4.Choukhi, A., S. Ung, C. Wychowski, and J. Dubuisson. 1998. Involvement of endoplasmic reticulum chaperones in the folding of hepatitis C virus glycoproteins. J. Virol. 72:3851-3858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cocquerel, L., S. Duvet, J. C. Meunier, A. Pillez, R. Cacan, C. Wychowski, and J. Dubuisson. 1999. The transmembrane domain of hepatitis C virus glycoprotein E1 is a signal for static retention in the endoplasmic reticulum. J. Virol. 73:2641-2649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cocquerel, L., J. C. Meunier, A. Op de Beeck, D. Bonte, C. Wychowski, and J. Dubuisson. 2001. Coexpression of hepatitis C virus envelope proteins E1 and E2 in cis improves the stability of membrane insertion of E2. J. Gen. Virol. 82:1629-1635. [DOI] [PubMed] [Google Scholar]
- 7.Cocquerel, L., J. C. Meunier, A. Pillez, C. Wychowski, and J. Dubuisson. 1998. A retention signal necessary and sufficient for endoplasmic reticulum localization maps to the transmembrane domain of hepatitis C virus glycoprotein E2. J. Virol. 72:2183-2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cocquerel, L., A. Op De Beeck, M. Lambot, J. Roussel, D. Delgrange, A. Pillez, C. Wychowski, F. Penin, and J. Dubuisson. 2002. Topological changes in the transmembrane domains of hepatitis C virus envelope glycoproteins. EMBO J. 21:2893-2902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cocquerel, L., C. Wychowski, F. Minner, F. Penin, and J. Dubuisson. 2000. Charged residues in the transmembrane domains of hepatitis C virus glycoproteins play a major role in the processing, subcellular localization, and assembly of these envelope proteins. J. Virol. 74:3623-3633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Crotta, S., A. Stilla, A. Wack, A. D'Andrea, S. Nuti, U. D'Oro, M. Mosca, F. Filliponi, R. M. Brunetto, F. Bonino, S. Abrignani, and N. M. Valiante. 2002. Inhibition of natural killer cells through engagement of CD81 by the major hepatitis C virus envelope protein. J. Exp. Med. 195:35-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.da Silva Cardoso, M., K. Siemoneit, D. Sturm, C. Krone, D. Moradpour, and B. Kubanek. 1998. Isolation and characterization of human monoclonal antibodies against hepatitis C virus envelope glycoproteins. J. Med. Virol. 55:28-34. [PubMed] [Google Scholar]
- 12.Deleersnyder, V., A. Pillez, C. Wychowski, K. Blight, J. Xu, Y. S. Hahn, C. M. Rice, and J. Dubuisson. 1997. Formation of native hepatitis C virus glycoprotein complexes. J. Virol. 71:697-704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dubuisson, J., L. Cocquerel, C. Wychowski, A. Choukhi, J.-C. Meunier, and S. Duvet. 1999. Assembly of hepatitis C virus glycoproteins. Recent Res. Dev. Virol. 1:29-39. [Google Scholar]
- 14.Dubuisson, J., S. Duvet, J. C. Meunier, A. Op De Beeck, R. Cacan, C. Wychowski, and L. Cocquerel. 2000. Glycosylation of the hepatitis C virus envelope protein E1 is dependent on the presence of a downstream sequence on the viral polyprotein. J. Biol. Chem. 275:30605-30609. [DOI] [PubMed] [Google Scholar]
- 15.Dubuisson, J., H. H. Hsu, R. C. Cheung, H. B. Greenberg, D. G. Russell, and C. M. Rice. 1994. Formation and intracellular localization of hepatitis C virus envelope glycoprotein complexes expressed by recombinant vaccinia and Sindbis viruses. J. Virol. 68:6147-6160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dubuisson, J., and C. M. Rice. 1996. Hepatitis C virus glycoprotein folding: disulfide bond formation and association with calnexin. J. Virol. 70:778-786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Duvet, S., L. Cocquerel, A. Pillez, R. Cacan, A. Verbert, D. Moradpour, C. Wychowski, and J. Dubuisson. 1998. Hepatitis C virus glycoprotein complex localization in the endoplasmic reticulum involves a determinant for retention and not retrieval. J. Biol. Chem. 273:32088-32095. [DOI] [PubMed] [Google Scholar]
- 18.Flint, M., J. Dubuisson, C. Maidens, R. Harrop, G. R. Guile, P. Borrow, and J. A. McKeating. 2000. Functional characterization of intracellular and secreted forms of a truncated hepatitis C virus E2 glycoprotein. J. Virol. 74:702-709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Flint, M., C. Maidens, L. D. Loomis-Price, C. Shotton, J. Dubuisson, P. Monk, A. Higginbottom, S. Levy, and J. A. McKeating. 1999. Characterization of hepatitis C virus E2 glycoprotein interaction with a putative cellular receptor, CD81. J. Virol. 73:6235-6244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Flint, M., and J. A. McKeating. 1999. The C-terminal region of the hepatitis C virus E1 glycoprotein confers localization within the endoplasmic reticulum. J. Gen. Virol. 80:1943-1947. [DOI] [PubMed] [Google Scholar]
- 21.Forns, X., T. Allander, P. Rohwer-Nutter, and J. Bukh. 2000. Characterization of modified hepatitis C virus E2 proteins expressed on the cell surface. Virology 274:75-85. [DOI] [PubMed] [Google Scholar]
- 22.Grakoui, A., C. Wychowski, C. Lin, S. M. Feinstone, and C. M. Rice. 1993. Expression and identification of hepatitis C virus polyprotein cleavage products. J. Virol. 67:1385-1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Habersetzer, F., A. Fournillier, J. Dubuisson, D. Rosa, S. Abrigniani, C. Wychowski, I. Nakano, C. Trépo, C. Desgranges, and G. Inchauspé. 1998. Characterization of human monoclonal antibodies specific of the hepatitis C virus glycoprotein E2 with in vitro binding neutralization properties. Virology 249:32-41. [DOI] [PubMed] [Google Scholar]
- 24.Hadlock, K. G., R. E. Lanford, S. Perkins, J. Rowe, Q. Yang, S. Levy, P. Pileri, S. Abrignani, and S. K. H. Foung. 2000. Human monoclonal antibodies that inhibit binding of hepatitis C virus E2 protein to CD81 and recognize conserved conformational epitopes. J. Virol. 74:10407-10416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Harlow, E., and D. P. Lane. 1988. Antibodies: a laboratory manual. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 26.Heile, J. M., Y.-L. Fong, D. Rosa, K. Berger, G. Saletti, S. Campagnoli, G. Bensi, S. Capo, S. Coates, K. Crawford, C. Dong, M. Wininger, G. Baker, L. Cousens, D. Chien, P. Ng, P. Archangel, G. Grandi, M. Houghton, and S. Abrignani. 2000. Evaluation of hepatitis C virus glycoprotein E2 for vaccine design: an endoplasmic reticulum-retained recombinant protein is superior to secreted recombinant protein and DNA-based vaccine candidates. J. Virol. 74:6885-6892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Higginbottom, A., E. R. Quinn, C.-C. Kuo, M. Flint, L. H. Wilson, E. Bianchi, A. Nicosia, P. N. Monk, J. A. McKeating, and S. Levy. 2000. Identification of amino acid residues in CD81 critical for interaction with hepatitis C virus envelope glycoprotein E2. J. Virol. 74:3642-3649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Houghton, M. 1996. Hepatitis C viruses, p. 1035-1058. In B. N. Fields, D. M. Knipe, and P. M. Howley (ed.), Fields virology, 3rd ed. Lippincott-Raven Publishers, Philadelphia, Pa.
- 29.Lambot, M., S. Fretier, A. Op De Beeck, B. Quatannens, S. Lestavel, V. Clavey, and J. Dubuisson. 2002. Reconstitution of hepatitis C virus envelope glycoproteins into liposomes as a surrogate model to study virus attachment. J. Biol. Chem. 277:20625-20630. [DOI] [PubMed] [Google Scholar]
- 30.Michalak, J. P., C. Wychowski, A. Choukhi, J. C. Meunier, S. Ung, C. M. Rice, and J. Dubuisson. 1997. Characterization of truncated forms of hepatitis C virus glycoproteins. J. Gen. Virol. 78:2299-2306. [DOI] [PubMed] [Google Scholar]
- 31.Op De Beeck, A., L. Cocquerel, and J. Dubuisson. 2001. Biogenesis of hepatitis C virus envelope glycoproteins. J. Gen. Virol. 82:2589-2595. [DOI] [PubMed] [Google Scholar]
- 32.Op De Beeck, A., R. Montserret, S. Duvet, L. Cocquerel, R. Cacan, B. Barberot, M. Le Maire, F. Penin, and J. Dubuisson. 2000. The transmembrane domains of hepatitis C virus envelope glycoproteins E1 and E2 play a major role in heterodimerization. J. Biol. Chem. 275:31428-31437. [DOI] [PubMed] [Google Scholar]
- 33.Owsianka, A., R. F. Clayton, L. D. Loomis-Price, J. A. McKeating, and A. H. Patel. 2001. Functional analysis of hepatitis C virus E2 glycoproteins and virus-like particles reveals structural dissimilarities between different forms of E2. J. Gen. Virol. 82:1877-1883. [DOI] [PubMed] [Google Scholar]
- 34.Patel, A. H., J. Wood, F. Penin, J. Dubuisson, and J. A. McKeating. 2000. Construction and characterization of chimeric hepatitis C virus E2 glycoproteins: analysis of regions critical for glycoprotein aggregation and CD81 binding. J. Gen. Virol. 81:2873-2883. [DOI] [PubMed] [Google Scholar]
- 35.Patel, J., A. H. Patel, and J. McLauchlan. 2001. The transmembrane domain of the hepatitis C virus E2. glycoprotein is required for correct folding of the E1 glycoprotein and native complex formation. Virology 279:58-68. [DOI] [PubMed] [Google Scholar]
- 36.Pileri, P., Y. Uematsu, S. Campagnoli, G. Galli, F. Falugi, R. Petracca, A. J. Weiner, M. Houghton, D. Rosa, G. Grandi, and S. Abrignani. 1998. Binding of hepatitis C virus to CD81. Science 282:938-941. [DOI] [PubMed] [Google Scholar]
- 37.Rosa, D., S. Campagnoli, C. Moretto, E. Guenzi, L. Cousens, M. Chin, C. Dong, A. J. Weiner, J. Y. Lau, Q. L. Choo, D. Chien, P. Pileri, M. Houghton, and S. Abrignani. 1996. A quantitative test to estimate neutralizing antibodies to the hepatitis C virus: cytofluorimetric assessment of envelope glycoprotein 2 binding to target cells. Proc. Natl. Acad. Sci. USA 93:1759-1763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Triyatni, M., J. Vergalla, A. R. Davis, K. G. Hadlock, S. K. Foung, and T. J. Liang. 2002. Structural features of envelope proteins on hepatitis C virus-like particles as determined by anti-envelope monoclonal antibodies and CD81 binding. Virology 298:124-132. [DOI] [PubMed] [Google Scholar]
- 39.Tseng, C. T., and G. R. Klimpel. 2002. Binding of the hepatitis C virus envelope protein E2 to CD81 inhibits natural killer cell functions. J. Exp. Med. 195:43-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wack, A., E. Soldaini, C. Tseng, S. Nuti, G. Klimpel, and S. Abrignani. 2001. Binding of the hepatitis C virus envelope protein E2 to CD81 provides a co-stimulatory signal for human T cells. Eur. J. Immunol. 31:166-175. [DOI] [PubMed] [Google Scholar]
- 41.Wunschmann, S., J. D. Medh, D. Klinzmann, W. N. Schmidt, and J. T. Stapleton. 2000. Characterization of hepatitis C virus (HCV) and HCV E2 interactions with CD81 and the low-density lipoprotein receptor. J. Virol. 74:10055-10062. [DOI] [PMC free article] [PubMed] [Google Scholar]