Abstract
The adenovirus (Ad) DNA-binding protein (DBP) is essential for the elongation phase of Ad DNA replication by unwinding the template in an ATP-independent fashion, employing its capacity to form multimers. DBP also enhances the rate of initiation, with the highest levels obtained at low concentrations of Ad DNA polymerase (Pol). Here, we show that stimulation of initiation depends on the template conformation. Maximal stimulation, up to 15-fold, is observed on double-stranded or viral TP-containing origins. The stimulation is reduced on partially single-stranded origins and DBP does not enhance initiation any more once the origin is completely unwound. This suggests a role for DBP in origin unwinding that is comparable to its unwinding capacity during elongation. However, mutant DBP proteins defective in unwinding and elongation can still enhance initiation on ds templates. DBP also stimulates the binding of nuclear factor I (NFI) to the origin and lowers the Km for coupling of the first nucleotide to the precursor terminal protein by Pol. Mobility shift experiments reveal that DBP stimulates the binding of Pol on double-stranded origin and nonorigin DNA but not on single-stranded DNA. This effect is specific for DBP and is also seen with other DNA Pols. Our results suggest that, rather than by origin unwinding, DBP enhances initiation by modulating the origin conformation such that DNA Pol can bind more efficiently.
DNA-binding protein (DBP) plays an important role in the adenovirus (Ad) life cycle, in which it is involved in DNA replication, transcriptional control, and mRNA stability (8, 36), host range specificity (1, 13, 20), transformation (12), and virus assembly (26).
Ad type 5 (Ad5) DBP is a 529-amino-acid (aa) protein with a molecular mass of 59,049 Da, which binds nucleic acids with high affinity. DBP binds cooperatively to single-stranded DNA (ssDNA) with a binding site size of ca. 12 nucleotides in a sequence-independent manner (21). Mild chymotrypsin treatment cleaves DBP into two domains: an N-terminal part (aa 1 to 173) and a C-terminal part (aa 174 to 529) that is well conserved among different serotypes and harbors most of the biological functions ascribed to DBP, including its nucleic acid-binding and replication functions (32). The outermost C-terminal part of DBP (aa 510 to 529) forms a protruding C-terminal arm and contains a hook (33). This C-terminal arm is involved in the formation of a DBP protein chain, where the C-terminal arm hooks into another DBP molecule to form a multiprotein complex. DBP binds to ssDNA in a cooperative manner, whereas binding to double-stranded DNA (dsDNA) is noncooperative (11). Two crystal structures of the C-terminal part of DBP have been resolved and show a remarkable difference in the orientation of this C-terminal arm, suggesting flexibility around the so-called hinge region (aa 512 to 515) (15). Deletion of the C-terminal arm results in a loss of cooperative DBP binding to ssDNA and also abolishes the DNA unwinding activity of DBP (11). Recently, we determined that the flexibility of the DBP molecule is important for the DNA unwinding activity of DBP as it functions to adapt the ssDNA and dsDNA structures present at the replication fork during initiation (34).
The role of DBP during adenovirus DNA replication is well established. Efficient DNA replication can be reconstituted with five proteins only. Three of these proteins are of viral origin: precursor terminal protein (pTP), polymerase (Pol), and DBP. pTP and Pol form a tight heterodimer in solution and bind to the origin located at the molecular ends of adenovirus DNA (23, 28, 31). Two other proteins, nuclear factor I (NFI) and Oct-1, originate from the host cell. NFI and Oct-1 are not absolutely required for in vitro DNA replication, but addition of these two transcription factors results in a 200-fold stimulation of DNA replication (for reviews, see references 10, 14, and 35).
Two phases can be distinguished in Ad DNA replication. During initiation the first nucleotide, a dCTP residue is coupled opposite the fourth base of the template strand to the serine(580)-OH of pTP. Subsequently, the pTP-C product is elongated to form the trinucleotide intermediate pTP-CAT (19). The pTP-CAT then shifts in position and jumps back to residues 1 to 3 of the template strand. During or right after the jumping back step, pTP and Pol dissociate, after which further elongation can take place (18).
DBP is essential during the elongation phase of DNA replication. Here, DBP is responsible for the unwinding of the dsDNA template and enhancing the processivity of Pol via the removal of secondary structures (11, 22).
DBP enhances the initiation of DNA replication in at least two ways. It enhances the binding of NFI to its recognition site within the auxiliary origin (7, 30), which in turn results in the stimulation of initiation by NFI (3). Also, DBP lowers the Km for the coupling of the first nucleotide to pTP (24). This suggests a specific DBP-Pol interaction, as was also suggested from protection of Pol by DBP against thermal inactivation (22). However, no direct interaction could be established by immunoprecipitation experiments (data not shown). We examined here other alternatives that can explain the specific function of DBP during the initiation of adenovirus replication. DBP may facilitate origin unwinding, thereby allowing more efficient binding of the pTP-Pol complex. Alternatively, DBP may increase binding of pTP-Pol in another way, either via a direct interaction of DBP with Pol or indirectly via a change in dsDNA structure. Our results show that DBP is involved in the stimulation of pol binding to the origin, employing the latter mechanism.
MATERIALS AND METHODS
DNA templates and oligonucleotides.
All oligonucleotides were purchased from Amersham Pharmacia Biotech. T50 (5′-CTCATTATCATATTGGCTTCAATCCAAAATAAGGTATATTATTGATGATG-3′) represented the first 50 nucleotides of the template strand of the Ad5 genome, and D50 (5′-CATCATCAATAATATACCTTATTTTGGATTGAAGCCAATAT-GATAATGAG-3′) represented the complementary (displaced) strand of T50. T50 and D50 were hybridized to form TD50. TDΔ5, TDΔ10, TDΔ15, and TDΔ20 are all derivatives from TD50 in which the number following the “Δ” represents the number of nucleotides deleted from the 5′ end of the D50 complementary strand. TD50fork was created after hybridization of T50 and D50fork (5′-GTAGTAGTTATTATATGGAAATTTTGGATTGAAGCCAATATGATAATGAG-3′) to form a partially double-stranded DNA template in which the first 20 nucleotides are noncomplementary to create a forked structure. TD50random was formed after hybridization of T50random (5′-TGGCTTGCTTGGTGGTCGTCTTCTATGTTGTCTCCACTCCGCTAGTCATA-3′) and D50random (5′-TATGACTAGCGGAGTGGAGACAACATAGAAGACGACCACCAAGCAAGCCA-3′) to create a non-Ad dsDNA template.
Labeling of the oligonucleotides was performed with T4 polynucleotide kinase (Amersham Pharmacia Biotech) and [γ-32P]ATP. All oligonucleotides (both single and double stranded) were purified by 12.5% polyacrylamide-Tris-borate-EDTA gel electrophoresis.
Proteins.
The adenovirus proteins ΔN-DBP, PPP-DBP, Pol, Pol exo mutant (D422A), and pTP were expressed in baculovirus-infected Sf9 cells and purified to near homogeneity, as verified by silver staining as described previously (5, 6, 9, 34). Phage T4 gp32 and T4 Pol were purchased from Amersham Pharmacia Biotech. Rabbit polyclonal antibodies were raised against Ad5 DBP (αDBP; a kind gift of J. Dekker) and Ad5 Pol (αPol) (6) and used in electrophoretic mobility shift assay (EMSA) studies.
Initiation assay.
Initiation of replication assays on origin-based templates were performed in a final reaction volume of 25 μl containing 25 mM HEPES-KOH (pH 7.5), 50 mM NaCl, 1.5 mM MgCl2, 1 mM dithiothreitol, 50 nM [α-32P]dCTP, 0.3 pmol of template, 0.35 pmol of Pol, and 0.35 pmol of pTP. The amounts of ΔN-DBP are indicated in the figure legends. Reactions were performed for 45 min at 37°C and were stopped by the addition of 80 mM EDTA. The samples were precipitated with 20% trichloroacetic acid for 30 min on ice. Precipitates were washed with 5% trichloroacetic acid, redissolved in sample buffer, analyzed on a sodium dodecyl sulfate-7.5% polyacrylamide gel, and autoradiographed. Data were quantified by densitometric analysis with a PhosphorImager.
EMSA.
For the EMSAs, ssDNA or dsDNA [γ-32P]ATP-labeled probes (∼0.05 ng) were incubated with purified proteins in 25 mM HEPES-KOH (pH 7.5), 4 mM MgCl2, 0.4 mM dithiothreitol, 4% Ficoll, and 50 mM NaCl in a total volume of 25 μl for 30 min at 4°C. Bound and unbound DNA were separated on a 9% polyacrylamide gel at 4°C in Tris-borate-EDTA and 0.01% Nonidet P-40 and then analyzed by autoradiography or densitometric scanning by using a phosphorimager.
RESULTS
Stimulation of initiation by DBP depends on template conformation.
Previously, it was shown that the C-terminal part of DBP (aa 174 to 529) containing the DNA binding domain is sufficient to function in Ad DNA replication in vitro (2). The DBP deletion mutant ΔN-DBP lacking the first 173 aa will be referred to as DBP in the present study.
Because stimulation of in vitro initiation was shown to be dependent on the Pol concentration (25), we first determined the optimal Pol concentration to analyze stimulatory effects of additional components. For 0.3 pmol of the dsDNA or ssDNA templates, the optimal concentration of Pol for stimulation of initiation was determined to be 0.35 pmol. pTP was added to the initiation reactions in a 1:1 molar ratio with Pol, to produce pTP-Pol complexes necessary for initiation. Addition of higher pTP concentrations by up to a fivefold molar excess over Pol did not influence the initiation reaction. Under the given conditions, the basal level of initiation produces a detectable signal.
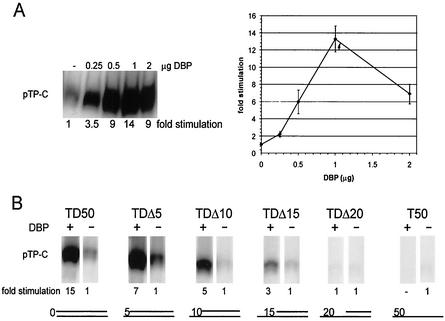
Optimal conditions for stimulation of initiation by DBP were determined by using a dsDNA Ad5 origin consisting of the first 50 bp (TD50) as a template. A DBP concentration range from 0 to 33.8 pmol (0 to 2 μg) was tested in an initiation assay (Fig. 1A). A linear range of stimulation of initiation is observed up to 1 μg of DBP, whereas higher amounts inhibit the initiation. At 1 μg of DBP the maximal level of stimulation was 14-fold (±1.5). Similar titrations were performed for all templates used in the initiation assays, all showing optimal stimulation with 1 μg of DBP. Furthermore, the levels of stimulation of initiation by DBP were comparable for both TP-DNA and TP-less DNA templates (11).
FIG. 1.
(A) Initiation assay on 0.3 pmol of the TD50 template with various concentrations of DBP. A representative initiation assay is shown. The average level of initiation based on three independent experiments is graphically represented. (B) Initiation assay on origin-containing templates with increasing 5′ gaps. Equimolar amounts of DNA templates (0.3 pmol) were used. The fold stimulation of initiation by DBP of a representative experiment is shown. The pTP-C signal without the addition of DBP was set to 1 for each template. The basal level of initiation (no DBP) on TD50 was set to 1, and the basal levels of initiation of the other templates were determined as follows: TDΔ5, 2.5; TDΔ10, 0.6; TDΔ15, 0.7; TDΔ20, 0.3; and T50, 0.3.
Since DBP is a helix-destabilizing protein, it may be capable of unwinding the dsDNA terminus of the origin, thereby allowing efficient binding of the pTP-Pol complex, leading to stimulation of initiation. The partial unwinding of the terminus can be mimicked by creating templates with progressive deletions of the displaced strand. Assuming that the major role of DBP is unwinding of the DNA terminus, one would expect that the stimulation of initiation by DBP is lost when the opening is large enough to facilitate efficient binding of the pTP-Pol complex to the origin.
To test this possibility partially, ssDNA templates were designed with progressive 5′ deletions. The stimulation of initiation in the presence or absence of DBP was determined for templates with an single-stranded moiety ranging from 5 to 20 nucleotides, as well as for the completely single-stranded template strand (Fig. 1B). The stimulation of initiation by DBP was optimal when the DNA template is in a complete double-stranded configuration. Upon increasing deletions of the displaced strand, the stimulation of initiation by DBP gradually decreases, and beyond 15 nucleotides of ssDNA the stimulation by DBP is lost completely (Fig. 1B). This indicates that the role of DBP in stimulating the initiation requires the presence of dsDNA. An obvious possibility is that this is related to the unwinding capability of DBP.
It should be noted that the basal level of initiation in the absence of DBP, is higher for the TDΔ5 template than for the TD50. Similar observation of enhanced basal activity with partially ssDNA templates were reported by Kenny et al. (16). The most likely explanation for this observation is that the pTP-Pol complex can bind more efficiently to the already partially unwound template.
Helix destabilization by DBP does not play a role in the stimulation of initiation.
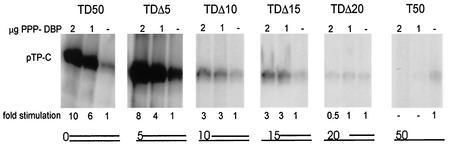
To investigate whether DNA unwinding by DBP is the determining factor that stimulates initiation, a similar experiment with partially ssDNA was performed with an unwinding-negative DBP mutant called PPP-DBP (34). In PPP-DBP three amino acids (512N, 513V, and 514S) in the hinge region of the flexible C-terminal arm are substituted by proline, thereby destroying the flexibility, which in turn results in loss of unwinding activity on TD50 and smaller dsDNA templates, whereas binding of PPP-DBP to ssDNA and dsDNA was only slightly affected (34). This mutant was still capable of stimulating initiation on the various templates (Fig. 2). Although the stimulation on TD50 by PPP-DBP is slightly lower than that by DBP in this experiment, this result is due to the variation of individual experiments. An average of three independent experiments showed that PPP-DBP stimulated initiation on TD50 is (12.3 ± 1.7)-fold, and the average of DBP stimulated initiation is 13.0 ± 1.5 (n = 3), demonstrating that the stimulation of initiation is not affected by the PPP-DBP mutant. It should be noted that due to the slightly lower DNA binding affinity of PPP-DBP, a twofold-higher concentration is required to obtain optimal stimulation of initiation (34). Therefore, we conclude that the stimulation of initiation is independent of the unwinding activity. Apparently, optimal stimulation of initiation requires the first 15 nucleotides of the origin to be in a double-stranded form.
FIG. 2.
Initiation assay with the unwinding defective PPP-DBP mutant on 0.3 pmol of origin-containing templates. Indicated are the 5′ gaps ranging from 0 to 50 nucleotides. The amount of stimulation of initiation is shown. The pTP-C signal without the addition of PPP-DBP was set to 1 for each template. The basal level of initiation (no PPP-DBP) on TD50 was set to 1, and the basal levels of initiation of the other templates were determined as follows: TDΔ5, 3.3; TDΔ10, 0.4; TDΔ15, 0.3; TDΔ20, 0.3; and T50, 0.4.
Binding of DBP to the displaced strand does not stimulate initiation.
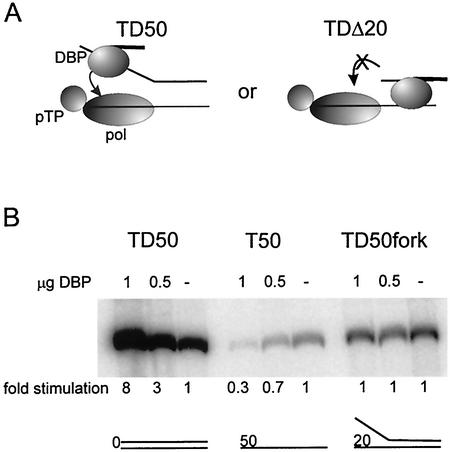
Since the unwinding of dsDNA by DBP does not seem to play a major role in stimulation of initiation, we considered an alternative mechanism. The stimulatory effect of DBP on initiation could be caused by binding of DBP to the displaced strand, which might lead to a transient interaction of DBP with the pTP-Pol complex, thus stimulating initiation. At partially single-stranded templates such as TDΔ20, the distance between the pTP-Pol complex on the template strand and DBP on the displaced strand might become too long, preventing this putative interaction and leading to loss of the stimulation (Fig. 3A). To test this hypothesis, a forked template was constructed similar to TD50, but with the first 20 nucleotides of the displaced strand noncomplementary to the template strand (TD50fork). Stimulation of initiation by DBP of the forked template was compared with the stimulation of TD50 and with a completely single-stranded template strand, T50 (Fig. 3B). A clear stimulation of initiation by DBP is observed on the TD50 template, whereas DBP had a slightly inhibiting effect on the T50 single-stranded template strand. When using the TD50fork, initiation of replication could still take place, but DBP did not stimulate the reaction. This suggests that mere binding of DBP to the displaced strand does not determine the stimulation of initiation.
FIG. 3.
(A) Model for transient DBP-Pol interaction via the displaced strand. On the left, DBP is able to contact Pol via the displaced strand and thereby stimulates the initiation. On the right, DBP is unable to contact Pol when the ssDNA gap becomes too large, such as in TDΔ20; hence, no stimulation of initiation can be observed. (B) Initiation assay on 0.3 pmol of origin-containing templates. Assay conditions are as described in the legends to Fig. 1 and 2. The fold stimulation of initiation by DBP is shown.
DBP stimulates binding of Pol to the dsDNA origin.
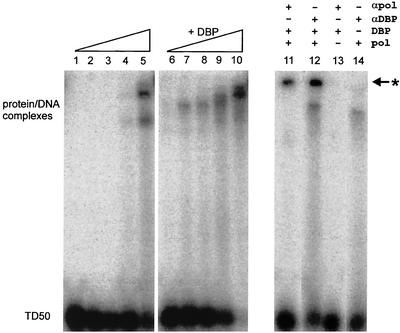
Since stimulation of initiation was neither dependent on the binding of DBP to the displaced strand nor dependent on unwinding of the displaced strand, we examined another possibility. Does DBP bind to the dsDNA template in such a way that it facilitates the binding of the pTP-Pol complex to the dsDNA origin? One indication could be that a similar mechanism was described for the effect of DBP on the binding of NFI to its cognate site in the auxiliary origin. This is accompanied by DBP-induced conformational changes in the dsDNA, as observed by hydroxyl radical footprinting and circular dichroism experiments (29). In the case of NFI, DBP alters the conformation of dsDNA, leading to more efficient NFI binding. To examine whether DBP is able to stimulate Pol binding, an EMSA was performed with TD50 DNA and increasing amounts of Pol in the presence or absence of DBP (Fig. 4, lanes 1 to 10). Complex formation between TD50 and Pol was visible from 100 ng of polymerase onward (Fig. 4, lane 4). Increasing amounts of Pol showed additional complexes containing multiple Pol molecules bound to one TD50 template (Fig. 4, lane 5), as was previously described (6). In the presence of DBP, a protein-DNA complex was already visible when only 25 ng of Pol was used in the reaction (Fig. 4, lane 7). Interestingly, the complex seems to run at a slightly higher mobility when DBP is present. This suggests that DBP is included in the DNA-protein complex. A similar experiment was performed to determine whether this stimulation also occurs when pTP-Pol is used instead of Pol alone. Indeed, binding of DNA by pTP-Pol is also stimulated in the presence of DBP (data not shown).
FIG. 4.
EMSA with TD50 and increasing Pol concentrations (0, 25, 50, 100, and 200 ng). Lanes 1 to 5 contain no DBP and show DNA-Pol complexes only. The reactions in lanes 6 to 10 contain an additional 20 ng of DBP. Supershifted complexes with antibodies to Pol (lane 11) or DBP (lane 12) are indicated with an asterisk. The “+” symbol indicates the addition of DBP (20 ng), Pol (50 ng), or antibodies.
To determine the composition of the multiprotein complex, supershift experiments were performed under conditions comparable to those of lane 8, with antibodies specifically raised against Ad5 Pol or DBP. When using the anti-Pol antibody (αPol) (lane 11), a supershift was observed. Likewise, when the anti-DBP antibody (αDBP) was used (lane 12), a supershift with a similar mobility was observed. In addition, complexes with a higher mobility are present in lanes 12 and 14 that are most likely caused by contaminants in the αDBP sample. Two conclusions can be drawn from these experiments. First, binding of Pol to the dsDNA origin is significantly stimulated by DBP. Second, DBP and Pol form a multiprotein complex at the dsDNA origin. Although we show that both proteins are bound, we do not know whether this is due to a direct Pol-DBP interaction or if both proteins bind to separate positions on dsDNA. Since we were not able to show under replication assay conditions a direct interaction between DBP and Pol in immunoprecipitation assays (data not shown), we favor the model in which Pol and DBP bind to separate dsDNA regions.
Enhanced binding of Pol correlates with enhanced stimulation of initiation.
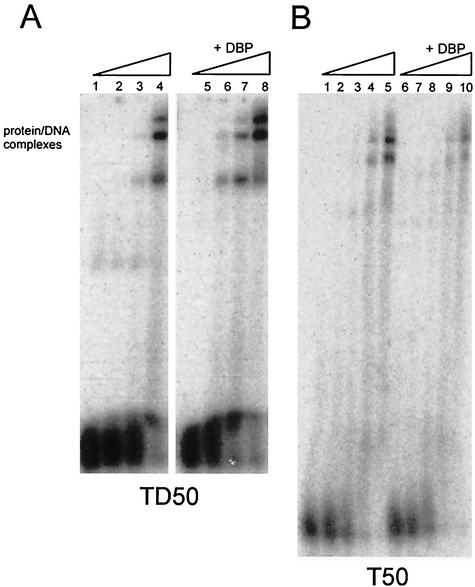
Is the enhanced stimulation of initiation by DBP, as observed in Fig. 1, reflected in enhanced Pol binding? To address this issue we compared templates TD50 and T50, showing the two outermost situations in stimulation, in an EMSA (Fig. 5). To examine the specificity of the stimulation of Pol binding by DBP on the ssDNA template T50, we used an exonuclease-deficient Pol mutant (Pol exo mutant, D422A) because wild-type Pol is known to degrade ssDNA in the absence of deoxynucleoside triphosphates (5). Stimulation of Pol binding to TD50 with the Pol exo mutant was similar to wild-type Pol binding (compare Fig. 5A, lanes 1 to 8, with Fig. 6, lanes 1 to 10). However, incubation of T50 with the Pol exo mutant in the absence or presence of DBP revealed no stimulation of Pol binding (Fig. 5B, lanes 1 to 10). In fact, a decreased Pol binding is observed in the presence of DBP, possibly due to competition between Pol and DBP for ssDNA binding.
FIG. 5.
(A) EMSA with TD50 and increasing Pol (exo mutant) concentrations (0, 25, 50, and 100 ng). Lanes 1 to 4 contain no DBP and show DNA-Pol complexes only. The reactions in lanes 5 to 8 contain an additional 15 ng of DBP. (B) EMSA with increasing Pol (exo mutant) concentrations (0, 25, 50, 100, or 200 ng) preincubated with T50 ssDNA either without DBP (lanes 1 to 5) or with 0.6 ng of DBP (lanes 6 to 10). Note that the DBP concentration is lower in EMSAs on ssDNA because the affinity of DBP for ssDNA is much higher than that for dsDNA.
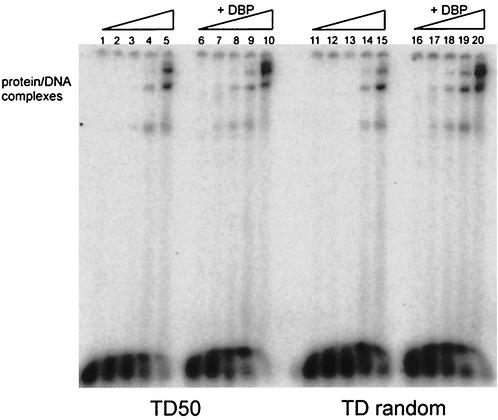
FIG. 6.
EMSA with increasing Pol concentrations (0, 12.5, 25, 50, or 100 ng) preincubated either with TD50 or a 50-bp random dsDNA probe (TD random), without DBP (lanes 1 to 5 and 11 to 15) or with 15 ng of DBP (lanes 6 to 10 and 16 to 20).
Specificity of DBP-stimulated Pol binding.
How specific are the interactions between DNA, Pol, and DBP? To address this issue, we compared the binding of Ad5 Pol and DBP to another dsDNA template. Incubation of the origin containing dsDNA template TD50 and Pol in the absence or presence of DBP resulted in stimulation of Pol binding when DBP was present (Fig. 6, lanes 1 to 10). Because it is known that Pol can bind nonspecifically to any dsDNA fragment (31), we examined whether the DBP stimulation was based on the nucleotide sequence of the Ad origin. Using a 50-bp random dsDNA probe (TD random), the experiment was repeated. In the absence of DBP, Pol bound to the dsDNA nonspecifically, as expected (Fig. 6, lanes 11 to 15). The addition of DBP still stimulated Pol binding to random dsDNA (Fig. 6, lanes 16 to 20). The experiment was also performed with other 50-bp dsDNA templates that were either GC-rich or TA-rich, and again Pol binding was stimulated by DBP (data not shown). Our results suggest that the enhanced binding of Pol in the presence of DBP is not dependent on the specific sequence of the template.
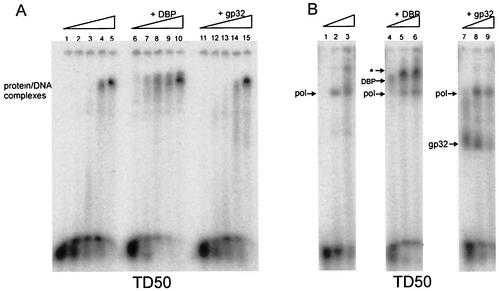
We further examined the specificity of DBP for another Pol. TD50 template was incubated with Ad5 Pol or with phage T4 Pol in the absence or presence of DBP (Fig. 7). In the absence of DBP T4 Pol can bind to TD50 (Fig. 7A, lanes 1 to 5), and in the presence of DBP the binding of T4 Pol is stimulated (Fig. 7A, lanes 6 to 10). The results indicate that the effect of DBP is not specific for Ad5 Pol.
FIG. 7.
(A) EMSA of the template TD50, preincubated with increasing phage T4 Pol concentrations (0, 3.5, 7, 14, or 28 ng) without SSB (lanes 1 to 5), in the presence of 15 ng of DBP (lanes 6 to 10), or in the presence of 40 ng of phage T4 gp32 (lanes 11 to 15). (B) EMSA of the template TD50 preincubated with increasing Ad5 Pol concentrations (0, 50, and 100 ng) without SSB (lanes 1 to 3) or in the presence of 15 ng of DBP (lanes 4 to 6) or 40 ng of phage T4 gp32. Arrows indicate the single-protein DNA complexes. Only in lanes 5 and 6 is a complex consisting of DNA, DBP and Pol present (✽).
Finally, we examined the specificity of DBP and compared its stimulatory properties with that of phage T4 single-stranded DNA-binding protein (SSB), gp32. TD50 was incubated with Ad5 Pol in the absence or presence of gp32 (Fig. 7B, lanes 1 to 3 and lanes 7 to 9) or with DBP (lanes 4 to 6). Furthermore, we tested whether enhanced Pol binding could occur with phage T4 DNA Pol and its native SSB gp32 (Fig. 7A, lanes 11 to 15). No stimulation of phage T4 Pol binding to TD50 was detected when gp32 was added. In addition, no stimulation of Ad5 Pol was detected, since no gp32-Pol-DNA complexes with lower mobility than the Pol-DNA complex were observed (Fig. 7B, lanes 7 to 9). This suggests that the enhanced Pol binding is DBP specific and that functionally related SSB proteins such as gp32 are not capable of stimulating Pol binding to dsDNA.
DISCUSSION
The major role of DBP in Ad5 DNA replication is helix destabilization of the dsDNA template (11, 37). Previously, an indirect role of DBP during initiation was described, showing that DBP stimulates NFI binding to the origin of replication (7, 30). Here we describe an additional role for DBP during initiation of DNA replication based on the fact that initiation can also be stimulated by DBP without NFI. Two possible models for the stimulation of initiation by DBP were investigated in detail.
The first model proposes that DBP partially unwinds the termini, thereby creating a partially unwound duplex origin required for optimal pTP-Pol binding. This model is based on an observation by Kenny et al. (16, 17), who showed that optimal initiation activity of Ad5 replication was dependent on the function of a 5′→3′ exonuclease, factor pL. Factor pL, purified from noninfected HeLa cells, was shown to degrade the 5′ end of the nontemplate (displaced) strand of the Ad5 origin. This partially duplex DNA is an efficient template for initiation of DNA replication (16).
We hypothesized that the helix destabilization activity of DBP could create a partially unwound origin that similarly would stimulate initiation of DNA replication. In this model DBP would destabilize the dsDNA origin in such a way that it allows optimal access of the replication complex, consisting of pTP-pol, NFI, and Oct-1.
Initiation assays on partially ssDNA templates were performed with DBP or with a DBP mutant defective in its dsDNA destabilization activity (Fig. 1 and 2). No helix-destabilizing activity of DBP was required for the stimulation of initiation. In fact, the binding of pTP to dsDNA inhibits unwinding (R. N. de Jong, unpublished results), strengthening our hypothesis that during initiation unwinding of the dsDNA template by DBP does not play a role. In addition, it was shown with a forked template that mere binding of DBP to the displaced strand is also not sufficient for stimulation of initiation (Fig. 3). From these results we can conclude that initiation of replication is not dependent on either the binding of DBP to the displaced strand or the destabilization of the origin.
It might be argued that the conformational differences among the various partially dsDNA templates influence the kinetics of the initiation assays and thus the stimulation by DBP. Although we cannot exclude this completely, we have no indications that the template conformation changes the kinetics. The incubation time of 45 min is well beyond the reaction time. Incubation at shorter time points up to 30 min gave a similar linear response for all templates tested.
In a second model we hypothesized that DBP can bind to dsDNA in such a way that it alters the dsDNA structure, thereby facilitating binding of the pTP-Pol complex to the origin. This assumption was based on experiments of Stuiver et al. (29) showing that DBP can change the structure of dsDNA upon binding. Due to DBP binding the dsDNA becomes more rigid and adapts a more regular conformation devoid of tertiary structures, as was shown by hydroxyl radical footprinting and circular dichroism. The effect of DBP binding on dsDNA results in stimulation of NFI binding to the dsDNA origin, which in turn leads to stimulation of initiation.
The specific role of DBP in initiation could thus be the stimulation of Pol binding to the origin in a fashion similar to that described for NFI (29). To examine DBP stimulated Pol binding, EMSAs were performed with Pol and DBP with the Ad5 origin as a dsDNA probe. Upon addition of DBP, Pol binding to dsDNA was clearly enhanced, strongly suggesting that DBP can stimulate Pol binding to the origin (Fig. 4).
We wondered whether the enhanced stimulation of initiation by DBP is reflected in stabilization of Pol binding to dsDNA or if it is due to enhanced Pol binding. After Pol binding in the presence or absence of DBP in competitor experiments to study the stability of the preformed complex, no changes in stability of the complex were observed (data not shown). In addition, when comparing templates TD50 and T50 in an EMSA, stimulation of Pol binding was only observed with the dsDNA template (Fig. 5). This strengthens the assumption that the origin needs to be in a dsDNA conformation and that DBP changes this conformation in such a way that Pol binding is enhanced. An obvious next step would be to also test the partial duplex templates for their ability to function in the DBP-stimulated Pol binding to dsDNA. However, due to the nonspecific nature of Pol and DBP binding to dsDNA and the fact that all partially single-stranded templates also contain a dsDNA region, these templates are able to sustain enhanced Pol binding by DBP.
Additional experiments were performed to examine the specificity of DBP-stimulated Pol binding. Enhanced Pol binding was not dependent on the specific sequence of the template (Fig. 6). In addition, the binding of phage T4 polymerase to dsDNA is also enhanced by DBP (Fig. 7A).
The specificity and affinity of Pol for the Ad dsDNA origin is predominantly determined by two cellular transcription factors, NFI and Oct-1, that are part of the preinitiation complex. Both NFI and Oct-1 target the pTP-Pol complex to the origin and then dissociate from the pTP-Pol complex early during initiation. In addition, increased specificity of the pTP-Pol complex for the origin was reported (31). We report an additional function of DBP, which is the stimulation of the Pol binding affinity to the origin.
Stimulation of Pol binding to dsDNA was only observed with DBP. When a related SSB, phage T4 gp32 was used no stimulated Pol binding was detected (Fig. 7A, lanes 12 to 15, and Fig. 7B). This suggests that the conformational changes induced by DBP on dsDNA are specific for DBP. These specific changes in the dsDNA structure might increase the binding affinity of proteins such as Pol and NFI to dsDNA.
One could argue that the increased Pol binding is caused by a direct interaction between DBP and Pol (22). Interactions between SSBs and their cognate Pol have been described in other systems such as phage T4 Pol and gp32 (27). However, we do not favor this alternative because we have never been able to demonstrate a direct interaction between DBP and Ad5 Pol. In addition, since stimulation of Pol binding by DBP was also shown for other Pols, it is unlikely that such a specific interaction exists between Pol and DBP. Functionally, such a direct interaction could keep the pTP-Pol complex trapped to the origin since direct binding of DBP to Pol might interfere with elongation. Similarly, no direct interaction could be demonstrated between DBP and NFI, also suggesting that direct interaction would interfere with elongation (4, 29).
Based on the presented data, we propose the following model for the role of DBP in the initiation of DNA replication. In vivo DBP is present in large quantities and, consequently, it can cover the entire Ad genome. DBP binding changes the dsDNA structure. First, DBP stimulates binding of NFI to its dsDNA recognition site in the Ad origin. Since NFI is part of a multiprotein complex consisting of pTP-Pol, Oct-1, and NFI, stimulated NFI binding enhances the recruitment of pTP-Pol to the origin. Second, due to the DBP-induced altered dsDNA structure of the origin also, the pTP-Pol complex binds with higher affinity to its binding site. Once the entire complex is formed on the DNA, initiation can take place. During elongation, DBP employs its helix-destabilizing activity and unwinds the dsDNA template ahead of Pol.
Acknowledgments
We thank Richard Heideman for isolation and purification of Ad DNA Pol and the Pol exo mutant and Monika Mysiak for the purification of DBP.
This work was supported in part by The Netherlands Foundation for the Chemical Research (SON).
REFERENCES
- 1.Anderson, C. W., M. M. Hardy, J. J. Dunn, and D. F. Klessig. 1983. Independent, spontaneous mutants of adenovirus type 2-simian virus 40 hybrid Ad2+ND3 that grow efficiently in monkey cells possess identical mutations in the adenovirus type 2 DNA-binding protein gene. J. Virol. 48:31-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ariga, H., H. Klein, A. J. Levine, and M. S. Horwitz. 1980. A cleavage product of the adenovirus DNA binding protein is active in DNA replication in vitro. Virology 101:307-310. [DOI] [PubMed] [Google Scholar]
- 3.Armentero, M. T., M. Horwitz, and N. Mermod. 1994. Targeting of DNA polymerase to the adenovirus origin of DNA replication by interaction with nuclear factor I. Proc. Natl. Acad. Sci. USA 91:11537-11541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bosher, J., I. R. Leith, S. M. Temperley, M. Wells, and R. T. Hay. 1991. The DNA-binding domain of nuclear factor I is sufficient to cooperate with the adenovirus type 2 DNA-binding protein in viral DNA replication. J. Gen. Virol. 72:2975-2980. [DOI] [PubMed] [Google Scholar]
- 5.Brenkman, A. B., E. C. Breure, and P. C. van der Vliet. 2002. Molecular architecture of adenovirus DNA polymerase and location of the protein primer. J. Virol. 76:8200-8207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brenkman, A. B., M. R. Heideman, V. Truniger, M. Salas, and P. C. van der Vliet. 2001. The (I/Y)XGG motif of adenovirus DNA polymerase affects template DNA binding and the transition from initiation to elongation. J. Biol. Chem. 276:29846-29853. [DOI] [PubMed] [Google Scholar]
- 7.Cleat, P. H., and R. T. Hay. 1989. Co-operative interactions between NFI and the adenovirus DNA binding protein at the adenovirus origin of replication. EMBO J. 8:1841-1848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cleghon, V., K. Voelkerding, N. Morin, C. Delsert, and D. F. Klessig. 1989. Isolation and characterization of a viable adenovirus mutant defective in nuclear transport of the DNA-binding protein. J. Virol. 63:2289-2299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.de Jong, R. N., M. E. Mysiak, L. A. Meijer, M. van der Linden, and P. C. van der Vliet. 2002. Recruitment of the priming protein pTP and DNA binding occur by overlapping Oct-1 POU homeodomain surfaces. EMBO J. 21:725-735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.de Jong, R. N., and P. C. van der Vliet. 1999. Mechanism of DNA replication in eukaryotic cells: cellular host factors stimulating adenovirus DNA replication. Gene 236:1-12. [DOI] [PubMed] [Google Scholar]
- 11.Dekker, J., P. N. Kanellopoulos, A. K. Loonstra, J. A. van Oosterhout, K. Leonard, P. A. Tucker, and P. C. van der Vliet. 1997. Multimerization of the adenovirus DNA-binding protein is the driving force for ATP-independent DNA unwinding during strand displacement synthesis. EMBO J. 16:1455-1463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ginsberg, H. S., M. J. Ensinger, R. S. Kauffman, A. J. Mayer, and U. Lundholm. 1975. Cell transformation: a study of regulation with types 5 and 12 adenovirus temperature-sensitive mutants. Cold Spring Harbor Symp. Quant. Biol. 39(Pt. 1):419-426. [DOI] [PubMed] [Google Scholar]
- 13.Harfst, E., and K. N. Leppard. 1999. A comparative analysis of the phosphorylation and biochemical properties of wild type and host range variant DNA binding proteins of human adenovirus 5. Virus Genes 18:97-106. [DOI] [PubMed] [Google Scholar]
- 14.Hay, R. T., A. Freeman, I. Leith, A. Monaghan, and A. Webster. 1995. Molecular interactions during adenovirus DNA replication. Curr. Top. Microbiol. Immunol. 199:31-48. [DOI] [PubMed] [Google Scholar]
- 15.Kanellopoulos, P. N., D. Tsernoglou, P. C. van der Vliet, and P. A. Tucker. 1996. Alternative arrangements of the protein chain are possible for the adenovirus single-stranded DNA binding protein. J. Mol. Biol. 257:1-8. [DOI] [PubMed] [Google Scholar]
- 16.Kenny, M. K., L. A. Balogh, and J. Hurwitz. 1988. Initiation of adenovirus DNA replication. I. Mechanism of action of a host protein required for replication of adenovirus DNA templates devoid of the terminal protein. J. Biol. Chem. 263:9801-9808. [PubMed] [Google Scholar]
- 17.Kenny, M. K., and J. Hurwitz. 1988. Initiation of adenovirus DNA replication. II. Structural requirements using synthetic oligonucleotide adenovirus templates. J. Biol. Chem. 263:9809-9817. [PubMed] [Google Scholar]
- 18.King, A. J., W. R. Teertstra, and P. C. van der Vliet. 1997. Dissociation of the protein primer and DNA polymerase after initiation of adenovirus DNA replication. J. Biol. Chem. 272:24617-24623. [DOI] [PubMed] [Google Scholar]
- 19.King, A. J., and P. C. van der Vliet. 1994. A precursor terminal protein-trinucleotide intermediate during initiation of adenovirus DNA replication: regeneration of molecular ends in vitro by a jumping back mechanism. EMBO J. 13:5786-5792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Klessig, D. F., and T. Grodzicker. 1979. Mutations that allow human Ad2 and Ad5 to express late genes in monkey cells map in the viral gene encoding the 72K DNA binding protein. Cell 17:957-966. [DOI] [PubMed] [Google Scholar]
- 21.Kuil, M. E., H. van Amerongen, P. C. van der Vliet, and R. van Grondelle. 1989. Complex formation between the adenovirus DNA-binding protein and single-stranded poly(rA). Cooperativity and salt dependence. Biochemistry 28:9795-9800. [DOI] [PubMed] [Google Scholar]
- 22.Lindenbaum, J. O., J. Field, and J. Hurwitz. 1986. The adenovirus DNA binding protein and adenovirus DNA polymerase interact to catalyze elongation of primed DNA templates. J. Biol. Chem. 261:10218-10227. [PubMed] [Google Scholar]
- 23.Mul, Y. M., and P. C. van der Vliet. 1992. Nuclear factor I enhances adenovirus DNA replication by increasing the stability of a preinitiation complex. EMBO J. 11:751-760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mul, Y. M., and P. C. van der Vliet. 1993. The adenovirus DNA binding protein effects the kinetics of DNA replication by a mechanism distinct from NFI or Oct-1. Nucleic Acids Res. 21:641-647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mul, Y. M., C. P. Verrijzer, and P. C. van der Vliet. 1990. Transcription factors, NFI, and NFIII/Oct-1 function independently, employing different mechanisms to enhance adenovirus DNA replication. J. Virol. 64:5510-5518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nicolas, J. C., P. Sarnow, M. Girard, and A. J. Levine. 1983. Host range temperature-conditional mutants in the adenovirus DNA binding protein are defective in the assembly of infectious virus. Virology 126:228-239. [DOI] [PubMed] [Google Scholar]
- 27.Nossal, N. G. 1992. Protein-protein interactions at a DNA replication fork: bacteriophage T4 as a model. FASEB J. 6:871-878. [DOI] [PubMed] [Google Scholar]
- 28.Schilling, R., B. Weingartner, and E. L. Winnacker. 1975. Adenovirus type 2 DNA replication. II. Termini of DNA replication. J. Virol. 16:767-774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stuiver, M. H., W. G. Bergsma, A. C. Arnberg, H. van Amerongen, R. van Grondelle, and P. C. van der Vliet. 1992. Structural alterations of double-stranded DNA in complex with the adenovirus DNA-binding protein. Implications for its function in DNA replication. J. Mol. Biol. 225:999-1011. [DOI] [PubMed] [Google Scholar]
- 30.Stuiver, M. H., and P. C. van der Vliet. 1990. Adenovirus DNA-binding protein forms a multimeric protein complex with double-stranded DNA and enhances binding of nuclear factor I. J. Virol. 64:379-386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Temperley, S. M., and R. T. Hay. 1992. Recognition of the adenovirus type 2 origin of DNA replication by the virally encoded DNA polymerase and preterminal proteins. EMBO J. 11:761-768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tsernoglou, D., A. Tsugita, A. D. Tucker, and P. C. van der Vliet. 1985. Characterization of the chymotryptic core of the adenovirus DNA-binding protein. FEBS Lett. 188:248-252. [DOI] [PubMed] [Google Scholar]
- 33.Tucker, P. A., D. Tsernoglou, A. D. Tucker, F. E. Coenjaerts, H. Leenders, and P. C. van der Vliet. 1994. Crystal structure of the adenovirus DNA binding protein reveals a hook-on model for cooperative DNA binding. EMBO J. 13:2994-3002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.van Breukelen, B., P. N. Kanellopoulos, P. A. Tucker, and P. C. van der Vliet. 2000. The formation of a flexible DNA-binding protein chain is required for efficient DNA unwinding and adenovirus DNA chain elongation. J. Biol. Chem. 275:40897-40903. [DOI] [PubMed] [Google Scholar]
- 35.van der Vliet, P. C. 1995. Adenovirus DNA replication. Curr. Top. Microbiol. Immunol. 199:1-30. [DOI] [PubMed] [Google Scholar]
- 36.Zijderveld, D. C., F. d'Adda di Fagagna, M. Giacca, H. T. Timmers, and P. C. van der Vliet. 1994. Stimulation of the adenovirus major late promoter in vitro by transcription factor USF is enhanced by the adenovirus DNA binding protein. J. Virol. 68:8288-8295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zijderveld, D. C., and P. C. van der Vliet. 1994. Helix-destabilizing properties of the adenovirus DNA-binding protein. J. Virol. 68:1158-1164. [DOI] [PMC free article] [PubMed] [Google Scholar]