Abstract
The detection of small amounts of viral pathogens in infected cells by classical PCR is hampered by a partial loss of virus nucleic acid due to extraction and by difficulties in discrimination between truly intracellular virus genome material and that possibly adhered to the cell surface. These impediments limit reliable identification of virus traces within infected cells, which are typically encountered in latent and persistent occult infections. In this study, hepadnavirus-specific in situ PCR combined with the enzymatic elimination of extracellular virus and flow cytometry permitted detection of viral genomes in lymphoid cells without nucleic acid isolation and allowed quantification of infected cells during the course of persistent infection with woodchuck hepatitis virus (WHV). The validity of the procedure was confirmed by hybridization analysis of the in situ-amplified viral sequences. The results showed that hepadnavirus can be directly detected within lymphoid cells not only in serologically accountable infection, but also years after recovery from viral hepatitis and in the course of primary occult virus carriage. Percentages of infected peripheral lymphoid cells in symptomatic WHV hepatitis fluctuate between 3.4 and 20.4% (mean ± standard error of the mean, 9.6% ± 1.7%), whereas those in persistent, serologically mute WHV infection range from 1.1 to 14.6% (mean ± standard error of the mean, 4.8% ± 0.8%) (P = 0.005). The data obtained provide further evidence that WHV infection continues indefinitely in the lymphatic system independently of whether it is symptomatic or concealed. They document that hepadnavirus can be detected in a significant proportion of circulating lymphoid cells in both immunovirologically apparent as well as occult persistent infection.
The PCR has proven to be one of the greatest advances in molecular diagnostics that is now widely applied to detect pathogens and to identify clinical conditions. In its classical form, this technique requires the extraction of DNA or, eventually, RNA that has to be reverse transcribed to cDNA prior to amplification. The isolation step is inevitably associated with a partial loss of nucleic acid, and therefore, it may pose a serious impediment when identification of traces of genomic material of pathogens occurring in minute quantities is required. This is particularly important in diagnosis of latent or persistent occult viral infections, where very low virus loads are normally encountered within infected cells. In these infections, even a relatively negligible decrease during virus nucleic acid recovery or its minor degradation may predetermine whether a pathogen is detected or not.
Hepatitis B virus (HBV) inflicts lifelong, serologically detectable (i.e., HBV surface antigen reactive) infection in an estimated 400 million people worldwide (11, 24). However, small quantities of this noncytopathic DNA virus, which are not identifiable by currently used and otherwise sensitive serological immunoassays, also commonly occur in the absence of clinical symptoms and biochemical evidence of liver injury. It was documented that patients who resolved acute hepatitis B continue to carry small amounts of HBV genomes in serum, circulating lymphoid cells, and, most likely, the liver for decades after recovery despite production of potentially protective antiviral antibodies and a virus-specific vigorous cytotoxic T-cell response (2, 4, 15, 20, 22, 25).
Investigations in the woodchuck model of hepatitis B, which is the closest natural system for study of HBV pathobiology and the efficacy of anti-HBV agents (reviewed in references 6, 12, and 23), confirmed these observations and revealed that scant amounts of infectious woodchuck hepatitis virus (WHV) consistently persist for life after resolution of acute hepatitis or acquisition of a primary serologically occult infection (5, 7, 14; reviewed in reference 13). They also showed that the silently carried WHV is transmissible from mothers recovered from viral hepatitis to offspring, in which it induces a long-term, serologically mute infection (5). These and other studies proved that WHV invariably infects the host's lymphatic system even when the liver is seemingly not infected (5, 10), that the lymphoid cells support replication of pathogenic virus (9, 10), and that they are a reliable source of genomic material for detection of persisting virus (10, 14, 16).
The protocols currently applied for identification of small numbers of hepadnavirus genomes require isolation of DNA and a multistep and contamination-prone procedure that involves nested PCR followed by hybridization analysis of the amplified virus sequences. This approach, although of superior sensitivity, is time-consuming, does not permit easy determination of the number of infected cells, and does not allow reliable discrimination between the intracellular virus sequences, which might be indicative of virus replication and those nonspecifically attached to the cell surface. To circumvent these drawbacks and to establish a simplified method facilitating further studies on the role of the lymphatic system in hepadnavirus infection, we developed a direct in situ PCR procedure employing intact lymphoid cells which, in conjunction with enzymatic elimination of extracellular virus, detects viral amplicons within the cells by flow cytometry.
Although in situ PCR coupled with flow cytometry has been shown to be applicable for identification of human immunodeficiency virus and hepatitis C virus in peripheral blood mononuclear cells (PBMC) (17, 21), our approach overcomes an ambiguity posed by possible identification of contaminating extracellular viral sequences and permits specific detection of small quantities of viral genomes in both freshly isolated and archival, cryopreserved PBMC samples.
In the present study, the woodchuck model of HBV infection was employed to develop the procedure and to assess its applicability for identification of lymphotropic virus during chronic infection accompanied by either abundant or minuscule loads of circulating virus.
MATERIALS AND METHODS
Animals and lymphoid cell separation.
Samples of peripheral lymphoid cells examined in this study were derived from four groups of animals. Group 1 included PBMC isolated from four randomly selected WHV carriers with chronic hepatitis confirmed by histological examination of liver biopsy. All the animals had naturally acquired chronic infection and were positive WHV surface antigen in serum and reactive for antibodies to WHV core antigen for at least 6 months prior to PBMC collection. Multiple PBMC samples were available for analysis from one of the animals (1/F) (Table 1). In all woodchucks, serum WHV loads were in the range of 1011 virus genome equivalents/ml, as determined by a dot blot hybridization assay (sensitivity, ∼106 virus genome equivalents/ml) (Table 1).
TABLE 1.
Immunovirological characteristics of WHV infection at the time of analysis of WHV DNA in circulating lymphoid cells by in situ PCR combined with flow cytometry
| Group and animal no. | Time of PBMC acquisition (mo)a | Serology
|
WHV DNA in serum (vge/ml)b | WHV DNA in PBMC (% positive)c | ||
|---|---|---|---|---|---|---|
| Surface antigen | Anti-surface antigen | Anti-core antigen | ||||
| Group 1 (serologically evident chronic infection) | ||||||
| 1/F | 1 | + | − | + | 4.7 × 1011 | 20.4 |
| 2 | + | − | + | 4.7 × 1011 | 9.3 | |
| 4 | + | − | + | 4.1 × 1011 | 6.6 | |
| 6 | + | − | + | 3.4 × 1011 | 10.6 | |
| 8 | + | − | + | 1.7 × 1011 | 6.3 | |
| 11 | + | − | + | 4.8 × 1011 | 6.6 | |
| 15 | + | − | + | 1.8 × 1011 | 19.1 | |
| 19 | + | − | + | 2.7 × 1011 | 16.5 | |
| 2/M | 9 | + | − | + | 5.5 × 1011 | 3.4 |
| 3/F | 13 | + | − | + | 3.5 × 1011 | 7.1 |
| 4/F | 33 | + | − | + | 5.2 × 1011 | 5.4 |
| 35 | + | − | + | 4.8 × 1011 | 4.1 | |
| Mean ± SEM | 9.6 ± 1.7 | |||||
| Group 2 (serologically silent residual infection)d | ||||||
| 5/F | 5.5 | − | + | + | 103 | 3.8 |
| 17.5 | − | + | + | ∼10 | 3.2 | |
| 21 | − | + | + | ∼10 | 2.1 | |
| 6/F | 8e | − | − | + | 103 | 8.9 |
| 7/F | 8e | − | − | + | ∼10 | 3.6 |
| 20 | − | − | + | ∼10 | 4.7 | |
| 8/F | 16 | − | − | + | ∼10 | 5.6 |
| 30 | − | − | + | 103 | 2.1 | |
| 9/M | 20e | − | − | + | ∼10 | 11.2 |
| 31f | − | − | + | 103 | 12.9 | |
| 10/F | 26e | − | + | + | ∼10 | 8.0 |
| 66f | − | − | + | ∼10 | 2.6 | |
| 11/M | 31 | − | + | + | ∼10 | 1.9 |
| 43 | − | + | + | 103 | 8.0 | |
| 44 | − | + | + | ∼10 | 3.7 | |
| 51f | − | − | + | ∼10 | 1.2 | |
| 12/M | 58 | − | − | + | ∼10 | 1.2 |
| 13/M | 42 | − | − | + | 103 | 0.9 |
| 60f | − | − | + | ∼10 | 1.2 | |
| Mean ± SEM | 4.5 ± 0.8 | |||||
| Group 3 (serologically silent primary occult infection)g | ||||||
| 14/F | 6 | − | − | − | ∼10 | 1.3 |
| 18 | − | − | − | ∼10 | 1.2 | |
| 26 | − | − | − | ∼10 | 2.6 | |
| 30 | − | − | − | ∼10 | 4.8 | |
| 15/F | 12 | − | − | − | ∼10 | 2.4 |
| 16/F | 17 | − | − | − | ∼10 | 14.6 |
| 17/F | 45 | − | − | − | 103 | 5.7 |
| 52h | − | − | − | ∼10 | 8.0 | |
| Mean ± SEM | 5.1 ± 1.6 | |||||
From inoculation with WHV, from birth to a mother convalescent from acute hepatitis, or from time of arrival at colony for animals with serologically evident, chronic WHV infection.
WHV DNA detected by dot blot hybridization (sensitivity, ∼106 viral genome equivalents [vge]/ml) and, when negative, by nested PCR with WHV core gene-specific primers and Southern blot hybridization of the amplified virus sequences (sensitivity, ∼10 vge/ml).
Positive by in situ PCR combined with flow cytometry.
Adult animals who recovered after an acute episode of WHV hepatitis and carried trace amounts of WHV for life.
PBMC samples tested for WHV cccDNA by nested PCR followed by Southern blot hybridization and found reactive.
PBMC samples collected at the end of the natural life span of the animals examined.
Animals born to mothers who resolved acute WHV hepatitis and carried trace amounts of WHV for life.
PBMC samples tested for WHV cccDNA by nested PCR followed by Southern blot hybridization and found negative.
Group 2 included PBMC obtained from nine animals with a history of a self-limiting episode of acute WHV hepatitis (Table 1). These animals were initially WHV naive, and they developed a transient acute hepatitis when infected as adults with WHV (7, 13). The woodchucks in this study group were serum WHV surface antigen negative and anti-WHV core antigen reactive, and some of them demonstrated antibodies to WHV surface antigen at the time of or prior to PBMC collection (Table 1). After recovery from acute hepatitis, all animals carried trace amounts of WHV DNA in serum, lymphoid cells, and the liver for life. The levels of WHV DNA in serum did not exceed 103 virus genome equivalents/ml, although they were usually around 10 virus genome equivalents/ml, as determined by a WHV-specific nested PCR/Southern blot hybridization assay (sensitivity, <10 virus genome equivalents/ml) (10). PBMC were collected for up to 5.5 years after clearance of serum WHV surface antigen, at the time when liver histology was normal, or occasionally demonstrated features of minimal, intermittent inflammation. The molecular and pathological features of this residual infection were described in detail in our previous works (7, 14).
Group 3 consisted of PBMC obtained from four woodchucks born to mothers who resolved WHV hepatitis (Table 1). These offspring were consistently WHV surface antigen, anti-WHV surface antigen, and anti-WHV core antigen negative, but they carried low levels of WHV in serum, circulating lymphoid cells, lymphatic organs, and frequently, but not always, the liver, as described previously (5). In the present study, circulating lymphoid cells acquired from three offspring (14/F, 16/F, and 17/F) with WHV DNA expression in the liver and one offspring (15/F) without hepatic expression were examined. Multiple PBMC samples were available for analysis from woodchuck 14/F (Table 1). Only PBMC found reactive for WHV DNA by classical nested PCR/Southern blot hybridization analysis were examined in the current study.
Group 4 comprised PBMC derived from healthy, WHV-naive animals. These cells were used to optimize the assay conditions and as negative controls.
Tests applied for identification of serological markers of WHV infection and dot blot hybridization and PCR/Southern hybridization assays used for determination of WHV DNA loads were described in detail in our previous works (5, 10, 14).
PBMC were isolated from blood treated with sodium EDTA drawn with Vacutainers (lavender top; Becton Dickinson, Franklin Lanes, N.J.). They were separated by standard density gradient centrifugation on Ficoll-Paque Plus (Amersham Pharmacia Biotech AB, Uppsala, Sweden) (15) and washed in phosphate-buffered saline (PBS, pH 7.4) containing 1 mM EDTA (PBS-EDTA). Red blood cells were lysed by incubation with 0.83% NH4Cl for 15 min. PBMC were washed extensively in PBS-EDTA, and then their number and viability were determined by trypan blue exclusion in a hemacytometer. Finally, cells were suspended at 107 cells in 1-ml aliquots of heat-inactivated fetal calf serum (Gibco-BRL, Grand Island, N.Y.) with 10% dimethyl sulfoxide (Fisher Chemicals, Fair Lawn, N.J.) and stored in liquid nitrogen for up to 5.5 years prior to use.
Optimization of assay conditions.
In order to detect solely intracellular virus sequences and, at the same time, to preserve the integrity of cells through extended PCR cycling, various enzyme treatments and cell fixation, permeabilization, and washing protocols were examined. This included evaluations of different schemes of limited cell digestion with DNase and trypsin to remove WHV virions and virus DNA fragments potentially attached to the cell surface (see below), the use of different fixatives to stabilize the structural integrity of the cells, and testing of various washing protocols. In addition, since detection of virus DNA was intended in the cells stored frozen for a prolonged period of time, procedures for handling these cells, which are more fragile than those freshly isolated, were examined. The steps outlined below are the results of these thorough investigations.
In a series of preliminary experiments, the conditions for effective removal of extracellular WHV particles and protein-free WHV DNA sequences by DNase/trypsin/DNase digestion were tested. In these experiments, PBMC from healthy animals were exposed to WHV virions which were semipurified from the serum of a woodchuck chronic WHV carrier by ultracentrifugation over a 60% sucrose cushion, as previously described (5), or spiked with recombinant, full-length WHV DNA (10, 19). For this purpose, 106 cells per sample were incubated for 30 min at room temperature with approximately 300 DNase-protected WHV particles or with 300 virus genome equivalents of recombinant WHV DNA per cell. In both situations, the cells were washed with 0.25% Tween 20 in PBS and then treated by applying different DNase/trypsin/DNase protocols.
In another trial, it was established that fixation of PBMC in cold acetone for 3 min was as efficient as that with 1% or 4% paraformaldehyde for 2 h or 30 min, respectively. However, acetone fixation gave evidently lower background signals than paraformaldehyde when PBMC were examined by in situ PCR/flow cytometry.
Furthermore, examination by light microscopy of PBMC before and after permeabilization with proteinase K and the enzyme inactivation revealed that the morphology of cells remained intact, although their overall number decreased by up to 20%. This was without any effect on the final results since only 104 cells after in situ PCR were required for flow cytometric analysis. It was also found that extensive washing of fixed or unfixed PBMC with 0.25% Tween 20 in PBS did not perturb cell morphological integrity and that omission of this washing noticeably increased background binding of the fluorescein isothiocyanate (FITC)-labeled primer.
Cell surface DNase/trypsin/DNase treatment.
Under standard conditions, cells were removed from liquid nitrogen, thawed on ice, and washed twice with 0.25% Tween 20 in PBS by centrifugation at 660 × g for 5 min to remove fetal calf serum and dimethyl sulfoxide. The resulting cell pellet was resuspended in 900 μl of sterile PBS. The cells were supplemented with 100 μl of 10× DNase buffer (500 mM MgCl2 in 100 mM Tris-HCl buffer, pH 8.0) and 10 μl of 1-mg/ml DNase I (type IV from bovine pancreas; activity, 2 U/μg; Sigma Chemical Co., St. Louis, Mo.) and incubated for 30 min at 37°C.
After DNase digestion, 10 μl of 0.1 M CaCl2 and 10 μl of 10-mg/ml trypsin (type IX from bovine pancreas; activity, 7.3 U/μg; Sigma Chemical Co.) were added, and cells were incubated on ice for 30 min. Subsequently, 20 μl of 10-mg/ml trypsin inhibitor (type II-O from chicken egg white; Sigma Chemical Co.) was added, and the cells were briefly mixed at room temperature. In the next step, DNase digestion was repeated by adding 10 μl of 1-mg/ml DNase I under the conditions described above. The treated cells were washed twice with 0.25% Tween 20 in PBS and counted after trypan blue staining. They were aliquoted at 106 viable cells per 0.6-ml, thick-walled PCR tube (Fisher Chemicals), in which they were kept throughout all subsequent steps.
Cell fixation and permeabilization.
Samples containing 106 DNase/trypsin/DNase-treated cells were fixed on ice in acetone precooled at −20°C (molecular grade; Sigma Chemical Co.) (1, 18) for 3 min and then washed twice with 0.25% Tween 20 in PBS by centrifugation at 660 × g for 5 min at room temperature. In the following step, the cells were permeabilized by incubation with 200 μl of 10-μg/ml proteinase K (activity, >20 U/mg; Gibco-BRL) in PBS at 37°C for 5 min. The enzyme was inactivated by boiling for 2 min to prevent excessive digestion, as in in situ hybridization protocols described by others (1, 18). Then, the cells were washed in two changes of 0.25% Tween 20 in PBS.
PCR primers and amplification conditions.
Cells obtained after this procedure were suspended in a 100 μl volume containing 300 ng of each primer, 200 μmol of each deoxynucleotide triphosphate per liter, 1 unit of Taq DNA polymerase, 1.5 mM MgCl2, and 10 μl of 10× reaction buffer (all reaction components from Gibco-BRL). The PCR mixture was covered with 80 μl of molecular-grade mineral oil to prevent evaporation.
In this study, oligonucleotide primers homologous to the WHV core antigen gene were employed for the detection of virus-specific DNA. Thus, primer PPCC conjugated at the 5′ end with FITC with the sequence 5′-TAGGAGGCTGTAGGCAT (2033 to 2049) was used as the sense primer (FITC-PPCC), whereas primer CCOV with the sequence 5′-TCTCAATCGCCGCGTCGCAGA (2439 to 2460) was used as the antisense primer. Numbers specified above denote the position of the primer sequence in the WHV genome (GenBank accession no. M11082) (8). In some instances, unlabeled PPCC and CCOV primers were employed in control reactions. All primers were synthesized by Gibco-BRL.
To amplify viral DNA, cells were initially treated for 5 min at 95°C, cooled to 52°C for 2 min, and subjected to 72°C for 3 min. Then, PCR amplification was carried out for 50 cycles at 95°C for 30 s, 52°C for 30 s, and 72°C for 30 s. In the final step, cells were heated for 10 min at 72°C. In all PCR runs, at least three samples of the same cell preparation, each containing 106 cells, were processed. One underwent PCR amplification as a test sample, and two others served as controls. In one control reaction, both Taq DNA polymerase and the FITC-labeled primer were omitted to assess background cell autofluorescence. In the second sample, Taq DNA polymerase was omitted to determine whether the FITC-labeled primer may directly hybridize to intracellular WHV DNA under the thermocycling conditions used.
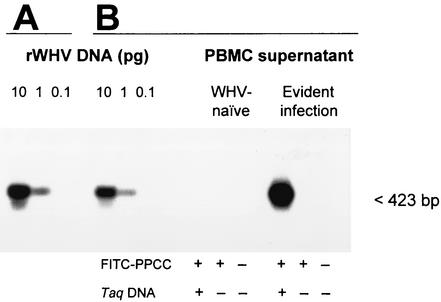
In preliminary experiments, it was established that the 5′-end FITC label on PPCC did not affect the efficiency of WHV DNA sequence amplification, as proven in parallel reactions with the primer pair comprised of unlabeled PPCC and CCOV and recombinant WHV DNA (see Fig. 1). All amplifications were performed in a TwinBlock thermal cycler (Ericomp Inc., San Diego, Calif.).
FIG. 1.
Sensitivity and specificity of WHV DNA detection by PCR with the oligonucleotide primer pair homologous to the virus core gene sequence containing a fluorescein-conjugated sense primer. Serial 10-fold dilutions of full-length, recombinant WHV DNA (rWHV DNA) were amplified with either PPCC-CCOV (A) or FITC-PPCC-CCOV (B) primer pairs under the same PCR conditions. In parallel, supernatants from WHV-naive PBMC or PBMC derived from a chronic WHV carrier obtained after in situ PCR with FITC-PPCC-CCOV or, as controls, FITC-PPCC in the absence of Taq DNA polymerase or in the absence of both FITC-PPCC and Taq DNA polymerase were analyzed (B). The amplified WHV DNA sequences were identified by Southern blot hybridization to 32P-labeled recombinant WHV DNA as a probe. The results show that either standard PCR procedure employing labeled or unlabeled PPCC primer or in situ PCR method with the FITC-PPCC primer generate DNA fragments of identical molecular size (423 bp) and specificity and that the use of the FITC-PPCC primer does not modify the sensitivity of WHV core gene detection.
In selected PBMC samples examined in this study, the presence of WHV covalently closed circular DNA (cccDNA), indicative of replicating virus, was assessed. This was accomplished following a PCR-based procedure previously established in this laboratory (10). Briefly, DNA from PBMC was isolated with the proteinase K-phenol-chloroform method and digested with a single-strand-specific mung bean nuclease to eliminate WHV DNA interrupted molecules (10). PCR primers spanning the nick region of the WHV genome were used to specifically amplify WHV cccDNA, as reported previously (10).
Flow cytometry analysis.
Following PCR, cells were spun down at 660 × g for 5 min, and the supernatant was collected and saved for molecular hybridization analysis. The resulting cell pellet was gently suspended in 0.5 ml of 0.25% Tween 20 in PBS and washed by centrifugation under the same conditions as above. The final pellet was suspended in 200 μl of PBS, transferred to a sterile 5-ml polypropylene tube, covered with parafilm, and kept on ice until analyzed in a FACS Star-Plus flow cytometer (Becton Dickinson, Palo Alto, Calif.). This examination was done within 12 h after completion of PCR.
Cells were gated by forward and side scatter to exclude cellular debris. The data generated from a flow of 104 events were analyzed with the help of Cell Quest software (Becton Dickinson). Trace signals occasionally given by control cell samples subjected to thermocycling in the absence of a FITC-labeled primer and Taq DNA polymerase were subtracted from the signals given by test cells. These signals never exceeded the mean value of 0.7% (standard error of the mean, 0.08%) (see below) of the total counted events. In some experiments, supernatants from cells after in situ PCR and cells not utilized for cytometric analysis were examined by dot blot hybridization, as described below.
Hybridization analysis of amplified virus sequences in cells and in their PCR supernatants.
To confirm the validity of the flow cytometry results, a dot blot hybridization assay was adopted to examine the presence of virus amplicons in both PBMC and their PCR supernatants. For this purpose, the cells after in situ PCR were spun down, counted, and adjusted to the same cell concentration (3 to 5 × 105 cells/dot). Then, they were disrupted by three cycles of freezing and thawing with vortexing, which eliminated almost all (∼99%) cells. The resulting suspension was blotted onto a nylon membrane (Hybond-N; Amersham) with a dot blot microfiltration apparatus (Bio-Rad Laboratories, Hercules, Calif.) and then washed twice with 200 μl of sterile PBS.
When cell PCR supernatants were examined, 90 μl of each supernatant recovered after PCR was blotted onto a nylon membrane following the same procedure as above. Subsequently, the nucleic acid was denatured and neutralized, and the membrane was baked and then hybridized overnight at 65°C with 32P-labeled recombinant WHV DNA as a probe (14). After washing in 0.2× SSC (standard saline citrate) containing 0.1% sodium dodecyl sulfate, the blots were exposed for autoradiography to Kodak X-Omat film (Eastman Kodak Co., Rochester, N.Y.). In all instances, twofold serial dilutions of recombinant WHV DNA were blotted as quantitative standards. In addition, in preliminary experiments, PBMC supernatants obtained after in situ PCR were subjected to electrophoresis on 1% agarose gels and analysis by Southern blot hybridization (14). These experiments showed that the amplified PCR products were of the expected molecular sizes and had WHV DNA specificity (Fig. 1).
Statistical analysis.
Based on the mean and standard error of the mean of background signals detected in WHV-naive cells (mean ± standard error of the mean, 0.7% ± 0.08%; n = 16), readings equal to or less than 1.0% were accepted as negative. Statistical comparison of background signals from WHV-negative cell samples to the low positive signals (1.1 to 2.1%) in samples obtained from serologically silent residual or serologically silent primary occult WHV infection gave a highly significant difference (P = 0.0001), indicating that values above 1.0% were in fact positive. The significance of differences between percentages of WHV DNA-negative and weakly WHV DNA-positive PBMC and between WHV DNA-positive PBMC detected in the groups studied (Table 1) was determined by the Mann-Whitney nonparametric, unpaired test. Two-tailed P values of ≤0.05 were considered significant.
RESULTS
Validation of enzymatic elimination of cell surface-adhered viral DNA.
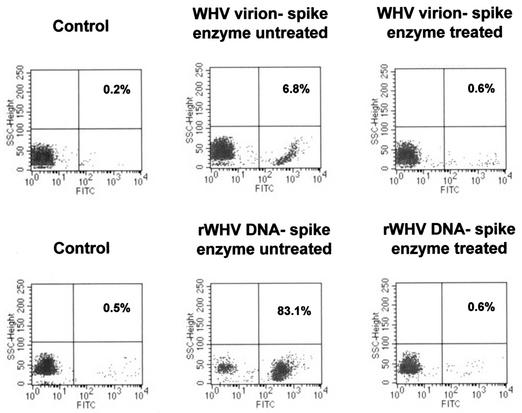
To validate the effectiveness of DNase/trypsin/DNase treatment on the removal of extracellular attached WHV virions and/or WHV DNA and at the same time to optimize conditions for sole detection of intracellular virus sequences, PBMC from WHV-naive animals were incubated with an excess of either WHV virions or recombinant WHV DNA and then subjected to the stepwise enzyme treatment described. As illustrated in Fig. 2, when cells were exposed to approximately the same amounts of virus genomes contained in either virions or protein-free recombinant WHV DNA preparation, they showed WHV DNA signals, as detected by in situ PCR/flow cytometry. However, the percentage of WHV DNA-positive PBMC was much greater after incubation with free virus DNA (83.1%) than with virions (6.8%). These signals were removed following DNase/trypsin/DNase treatment, indicating that the treatment was effective and ensured that only intracellular viral DNA was subsequently detected. Comparable results were obtained when either freshly isolated or previously frozen WHV-naive PBMC were examined and when the cells were incubated with recombinant WHV DNA suspended in PBS or normal woodchuck serum (data not shown).
FIG. 2.
Flow cytometry analysis of WHV-naive PBMC exposed to either WHV virions or cloned recombinant WHV (rWHV) DNA and subjected or not to stepwise DNase/trypsin/DNase digestion and WHV DNA-specific PCR amplification. PBMC isolated from a healthy woodchuck were incubated with either 3 × 108 WHV virions (approximately 300 virions/cell) (upper panel) or 1 ng of recombinant WHV DNA (approximately 300 virus genome equivalents/cell) (lower panel), washed, and treated or not with DNase/trypsin/DNase before PCR and flow cytometric examination. Adsorption of virions or protein-free WHV DNA to PBMC is evident in the absence of enzyme treatment, whereas following treatment, no positive cell signal was observed, indicating successful elimination of the attached WHV particles or nucleic acid sequences.
In general, the data demonstrated that WHV virions or viral DNA fragments may in fact attach to the lymphoid cell surface and, therefore, compromise the specificity of PCR aimed at exclusive detection of intracellular virus sequences. They also showed that washing cells alone is not sufficient to dissociate the surface bound viral material and that the enzymatic digestion applied in this study was effective at its elimination.
In situ PCR coupled with flow cytometry detects WHV in PBMC of animals with either high or minuscule levels of circulating virus.
Peripheral lymphoid cells isolated from serum WHV surface antigen-reactive, chronically infected woodchucks with serum loads of WHV of around 1011 virus genome equivalents/ml (group 1; Table 1) showed readily detectable intracellular WHV DNA when examined by in situ PCR-flow cytometry. In this disease situation, percentages of WHV genome containing cells ranged between 3.4 and 20.4% (mean 9.6% ± SEM 1.7; n = 12) (Table 1), or, in other words, there were 3.4 × 104 to 20.4 × 104 infected cells per 106 PBMC analyzed.
Considering individual samples obtained during follow-up of chronically infected animals, the number of WHV DNA-positive PBMC did not fittingly correlate with the load of circulating virus. This fact is well illustrated for woodchuck 1/F, from which eight PBMC and parallel serum samples obtained during a 19-month observation period were available for analysis (Table 1). Thus, although the level of circulating WHV did not fluctuate meaningfully (1.7 × 1011 to 4.8 × 1011 virus genome equivalents/ml) in this animal, the percentage of infected PBMC ranged between 6.3 and 20.4% during the same time period.
The proportion of WHV DNA-reactive PBMC in woodchucks with a serologically concealed infection that continued after recovery from acute hepatitis (group 2) or with a primary occult WHV infection (group 3), where the serum levels of WHV were usually around 10 virus genome equivalents/ml, ranged between 1.1 and 14.6% (mean 4.8% ± SEM 0.8; n = 27). When the percentage values of WHV-positive PBMC determined for woodchucks with residual or occult WHV infection (groups 2 and 3) were compared to those detected in animals with WHV surface antigen-positive chronic hepatitis (group 1), the difference was highly significant (P = 0.005). This indicates that although some animals with very low levels of circulating virus and serologically silent infection may carry numbers of WHV-positive PBMC comparable to those seen in woodchucks with high WHV loads and serologically demonstrable infection, the population of WHV DNA-positive PBMC is overall significantly greater in serologically evident than serologically silent infection. PBMC samples derived from WHV-naive animals showed minimal background signals, of which the mean was 0.7% ± SEM 0.08.
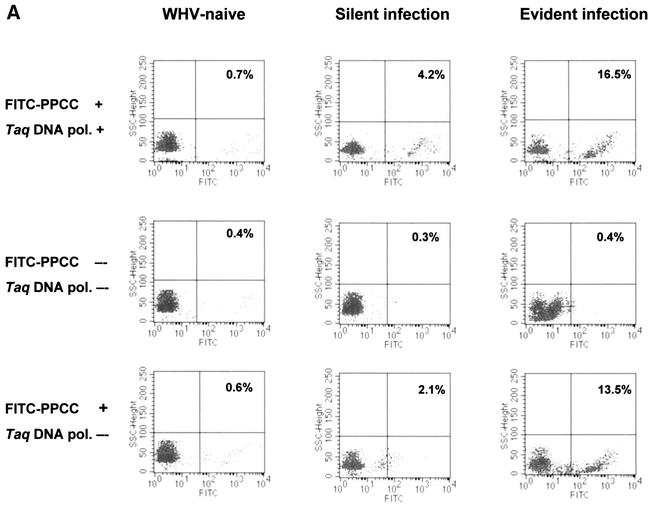
Figure 3A shows representative plots from flow cytometry analyses of PBMC isolated from animals with actually progressing, WHV surface antigen-positive chronic hepatitis or with a past episode of self-limited acute infection. In addition, this figure illustrates the results obtained from control reactions. As expected, PBMC derived from a healthy animal were negative for WHV DNA. In contrast, populations of WHV DNA-positive cells were readily detectable in PBMC taken from woodchucks with either active or residual WHV infection. As it is also shown in Fig. 3A, middle row, PBMC treated under the thermocycling conditions in the absence of both FITC-labeled, virus-specific primer and Taq DNA polymerase were WHV DNA nonreactive. Occasionally, these control reactions gave a trace background autofluorescence which did not exceed 1% of the total events counted. These signals, if occurring, were subtracted from the values given by relevant test samples.
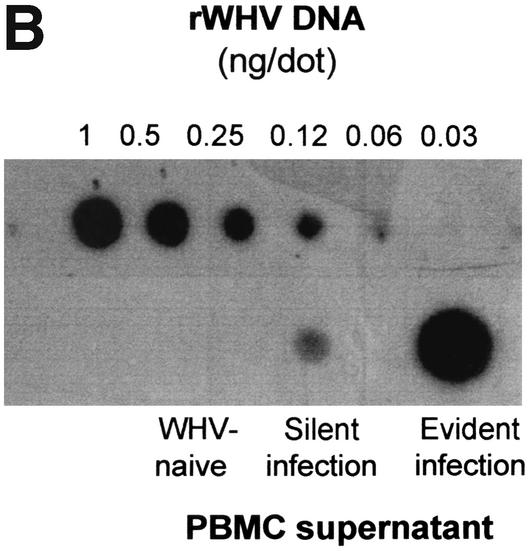
FIG. 3.
Detection of WHV DNA in lymphoid cells from WHV-infected woodchucks and a healthy control by in situ PCR coupled with flow cytometry or with hybridization analysis of cell supernatants obtained after PCR amplification. (A) Representative flow cytometry results showing WHV DNA-positive populations of PBMC from an animal with progressive WHV surface antigen-positive chronic hepatitis (Evident infection) and from a woodchuck with silent WHV infection continuing for 12 months after resolution of acute hepatitis (Silent infection) and from a control, WHV-naive woodchuck (top row). The same cell samples treated under identical cycling conditions but in the absence of FITC-PPCC primer and Taq DNA polymerase (pol.) showing almost nonexisting background autofluorescence (middle row) or after omitting Taq DNA polymerase illustrating signals obtained due to hybridization of the FITC-labeled PCCC primer with WHV DNA in PBMC from chronic and residual WHV infections but not from a healthy animal (bottom row). (B) Dot blot hybridization analysis of supernatants collected after PBMC underwent in situ PCR, confirming the validity of positive flow cytometry signals and negative controls shown in panel A.
Interestingly, when infected PBMC were subjected to the standard thermocycling conditions in the presence of FITC-labeled, WHV-specific primer but without Taq DNA polymerase, WHV genome positive cells could be detected occasionally, as illustrated in Fig. 3A, bottom row. Overall, the percentages of WHV DNA reactive cells detected by this method were on average 5% lower than those identified by the standard in situ PCR/flow cytometry protocol. Therefore, this approach did not allow detection of infected cells where low numbers (<5%) of WHV-positive PBMC were identified by in situ PCR/flow cytometry. It became evident that the signals generated in the absence of Taq DNA polymerase were due to a specific hybridization of the FITC-PPCC primer with intracellular WHV DNA. This conclusion was based on the facts that neither PBMC from WHV-naive animals treated under the same thermocycling conditions in the presence of FITC-PPCC nor WHV-infected PBMC treated with FITC-labeled HCV-specific primer produced positive signals (data not shown). It is conceivable that in situ hybridization, utilizing a fluorochrome-labeled oligonucleotide as a probe, might constitute a base for future development of even more simplified flow cytometry technique applicable for specific detection of pathogens when they are present at moderate to high levels within infected cells, as observed in this study.
In order to validate the results obtained by the in situ PCR-flow cytometry and, at the same time, to test whether hybridization of the amplified sequences with a radiolabeled probe may increase sensitivity of WHV DNA detection, PBMC supernatants collected after in situ PCR were probed by a dot blot hybridization with 32P-labeled, cloned WHV DNA. This approach was based on our observation that enzymatic permeabilization of cells leads to the escape of some amplicons during the PCR procedure and, therefore, may allow for assessment of the cell positivity by analyzing solely reaction mixtures obtained after PCR and removal of PBMC. As illustrated in Fig. 3B, the intracellularly amplified WHV DNA was detectable by this dot blot hybridization method, validating the positive and negative signals obtained for PBMC presented in Fig. 3A. However, although this approach gave results closely comparable to those generated by in situ PCR-flow cytometry in regard to positivity or negativity of individual PBMC samples tested, it overall did not improve the sensitivity of WHV DNA detection. Similarly, in another set of experiments, when either infected PBMC alone or together with their supernatants were analyzed by a dot blot hybridization after standard in situ PCR, there was no meaningful enhancement in the sensitivity of WHV genome detection over the standard in situ PCR-flow cytometry procedure developed in the current study (data not shown). Nevertheless, the data revealed that the in situ amplified sequences can be alternatively detected by a DNA hybridization method in situations where a flow cytometer might not be available.
Frequency of virus genome-containing PBMC in residual and primary occult WHV infections.
Two groups of animals known from our previous studies to persistently carry low levels of WHV in the lymphatic system, as determined by classical PCR amplification techniques (5, 7, 14), were examined by in situ PCR-flow cytometry. Group 2, composed of PBMC from animals who spontaneously overcame an acute episode of hepatitis and continued to have lifelong residual WHV infection, showed percentages of infected PBMC ranging from 1.1 to 12.9% (mean 4.5% ± SEM 0.8; n = 19). Five of the PBMC samples tested were collected at the end of the natural life span of the recovered animals (i.e., approximately 3 to 5.5 years after clearance of WHV surface antigen from serum) (Table 1). The percentages of positive cells in these particular samples ranged between 1.1 and 12.9%, indicating that the population of infected PBMC may significantly vary even in the very late period after resolving acute infection.
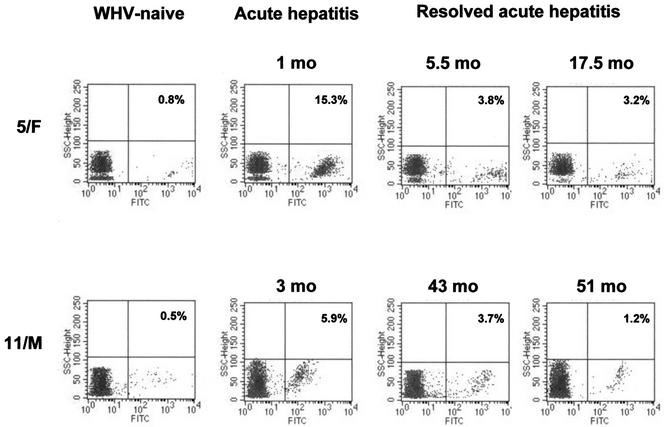
Figure 4 illustrates representative plots of WHV DNA-positive lymphoid cells from two woodchucks (5/F and 11/M; Table 1) with apparent complete resolution of experimentally induced acute hepatitis. Animal 5/F had finally developed hepatocellular carcinoma at 36.5 months after clearance of WHV surface antigen and seroconversion to anti-WHV surface antigen, whereas 11/M has been euthanized due to crippling senility at 51 months after seroconversion. As shown, approximately 15% of PBMC collected during acute hepatitis from 5/F were WHV DNA reactive, whereas 3.8% and 3.2% of cells were infected at 5.5 and 17.5 months after WHV surface antigen clearance, respectively. For 11/M, 5.9% of PBMC demonstrated detectable levels of virus DNA during the late phase of acute hepatitis, and 3.7% and 1.2% cells were infected at approximately 3.5 and 4 years after disappearance of serum WHV surface antigen, respectively. As expected, PBMC obtained prior to inoculation with WHV were WHV DNA nonreactive.
FIG. 4.
Representative plots from flow cytometry analysis of in situ-amplified WHV DNA in PBMC samples obtained from two woodchucks with residual serologically silent infection persisting after termination of acute hepatitis. PBMC obtained during acute phase of WHV hepatitis and at the indicated time points after clearance of serum WHV surface antigen and seroconversion to anti-WHV surface antigen were subjected to DNase/trypsin/DNase treatment, PCR amplification and flow cytometry analysis. The percentage of WHV DNA reactive PBMC is indicated in each panel. PBMC collected prior to inoculation with WHV from the same animals are shown as negative controls.
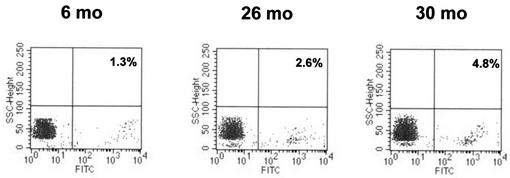
In group 3, PBMC collected from animals with a primary occult infection acquired by vertical transmission of WHV from mothers who resolved hepatitis were included (Table 1). The percentages of WHV infected cells detected in these animals by in situ PCR-flow cytometry ranged between 1.2 and 14.6% (mean 5.1% ± SEM 1.6; n = 8). Three representative plots of PBMC obtained at approximately 6 months and 2 and 3 years after birth from the same offspring (14/F) are shown in Fig. 5.
FIG. 5.
In situ PCR-flow cytometry analysis of PBMC from a woodchuck with primary occult infection who was born to a mother convalescent from acute WHV hepatitis. Peripheral lymphoid cells obtained at the indicated time points after birth were examined following the standard protocol. The percentages of WHV DNA-reactive cells detected in individual samples are indicated in each panel.
No significant difference in the percentages of WHV-infected PBMC was observed between animals with residual infection continuing after resolution of acute hepatitis and those with a primary occult infection. Statistically significant differences were found between animals with serum WHV surface antigen-positive chronic infection and those with silent residual infection (P = 0.008) and primary occult infection (P = 0.04).
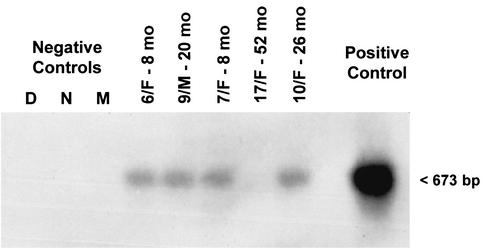
As an indicator of active hepadnavirus replication, the presence of WHV cccDNA was assessed by the nick region-specific PCR in selected PBMC samples which were found WHV DNA reactive by in situ PCR/flow cytometry. WHV cccDNA was detected in four of five PBMC samples tested, as confirmed by Southern blot hybridization analysis and noted in Table 1 and illustrated in Fig. 6.
FIG. 6.
Detection of cccDNA in representative PBMC samples which were obtained from woodchucks with serologically silent, persistent infection found WHV DNA reactive by in situ PCR/flow cytometry. DNA isolated from PBMC was subjected to mung bean nuclease digestion and PCR amplification. PCR products were analyzed by Southern blot hybridization to confirm molecular size (673 bp) and specificity. For immunovirological characteristics of animals from which the PBMC samples shown originated, see Table 1. Contamination controls included water added instead of DNA and amplified by a direct (D) or a nested (N) reaction and mock (M) treated as test DNA. The positive control consisted of DNA derived from PBMC of a woodchuck with WHV surface antigen-positive, chronic WHV infection.
DISCUSSION
This study shows that hepadnavirus genome can be readily detected in lymphoid cells by a direct PCR method combined with flow cytometry. This one-step PCR procedure detects hepadnavirus DNA with a sensitivity (∼102 virus genome equivalents/ml) comparable to that previously achieved only by nested PCR and allows reliable unveiling of the virus genomes in the lymphatic system during persistent, serologically concealed infection. The PCR-flow cytometry technique established in this study offers several important advantages over the classical PCR protocols utilized for identification of hepadnaviruses in lymphoid cells. It abrogates the need for extraction of nucleic acid and, by minimizing manipulations, maximizes preservation of virus genomes within essentially intact cells decreasing, at the same time, the risk of an accidental contamination.
By including limited enzyme treatment of cells with DNase/trypsin/DNase prior to PCR, detection of intracellular viral DNA but not that potentially nonspecifically attached to the cell surface from plasma or other body fluids is secured. Furthermore, for the first time, quantification of infected lymphoid cells in the course of hepadnaviral infection becomes feasible. This method might be further adopted to multiparametric analysis of infected lymphoid cells, their isolation, and identification of cell subsets in which hepadnavirus preferentially replicates, as well as to detection of replicative forms of hepadnavirus genomes.
The results obtained in this study validate previous findings demonstrating that silent WHV persistence continuing after recovery from hepatitis or as a primary occult infection is associated with the sustained infection of the host's lymphatic system (reviewed in reference 13). Comparing, on a case-by-case basis, the levels of serum WHV DNA determined by the classical PCR protocol with the percentages of infected PBMC identified by the in situ PCR-flow cytometry (Table 1), it is evident that a barely detectable level of virus in serum can be accompanied by a relatively large population of infected PBMC.
This situation has been encountered both in the residual infection continuing after resolution of acute WHV hepatitis and in primary occult WHV carriage. In these animals, the percentages of WHV-positive PBMC were occasionally as high as those detected in WHV surface antigen-reactive, chronic hepatitis. This indicates that serologically mute infection does not necessarily imply a small number of infected lymphoid cells, although it is highly likely that the amount of virus per individual infected cell is considerably greater in serologically evident than in serologically silent infection. Furthermore, as the present study suggests, the proportion of infected PBMC may fluctuate significantly, even in a short period of time, during the course of both silent and serologically detectable infections. This is consistent with another observation from our study, indicating that the high proportion of WHV-infected PBMC may appear even years after apparently complete resolution of hepatitis and in the late stage of the lifelong, primary occult infection. Nevertheless, taken together, the percentages of infected PBMC were found to be significantly greater (P = 0.005) in the serologically evident than in the silent virus infection.
In addition, the presence of WHV cccDNA in PBMC samples found positive by in situ PCR/flow cytometry supports the conclusion that virus replication occurs in these cells. This is consistent with our previous studies, in which expression of WHV DNA and cccDNA in the lymphatic system and infectivity of lymphoid cell-derived WHV were documented (5, 10, 14). The current study describes a new approach by which intracellular WHV DNA in lymphoid cells can be detected.
The percentages of WHV-positive lymphoid cells identified in our study appear to be similar to those delineated for other infections with lymphotropic viruses, when the in situ PCR-flow cytometry method was applied for detection of infected PBMC. In one reported study, HCV-specific sequences were detected in 0.2 to 8.1% of PBMC, often in the absence of overt clinical symptoms (17). In another work, the percentages of virus-positive PBMC identified in HIV-infected patients did not exceed 20%, with a range of positivity from 0.6 to 20% (21). This study also showed that, when the data were viewed on a case-by-case basis, highly variable populations of HIV-positive cells occurred regardless of the CD4+ T-cell count and viral load estimated by measuring HIV-1 p24 antigenemia. This observation may corroborate our findings in WHV infection. Considering the data from the in situ PCR-flow cytometry analyses mentioned above, it can be perceived that the WHV lymphotropic potential, seen as virus ability to establish infection in lymphoid cells, is comparable to that displayed by other known lymphotropic viruses.
The present study reinforces the notion that serologically silent hepadnavirus carriage should be monitored not only by evaluation of virus genome in serum, but also by examining its expression in peripheral lymphoid cells. It is evident that testing of circulating lymphoid cells may offer an important diagnostic advantage and identify virus in apparently negative cases. We have previously shown that the systematic analysis of serial sera, PBMC and liver tissue samples collected during the natural life span of woodchucks with residual WHV infection progressing after termination of hepatitis allows for detection of WHV DNA with a much higher frequency in PBMC (78%) than in sera (68%), although the liver remains the most reliable source for the identification of silently persisting virus (14; reviewed in reference 13).
The close virological and pathobiological similarities between WHV and HBV suggest that the same could be true for serologically silent HBV infection. Cumulative data from a growing number of studies documenting the persistence of HBV traces following spontaneous or therapeutically induced resolution of hepatitis B and the appearance of HBV infection in recipients of organs from apparently HBV negative donors (reviewed in references 3 and 13) strengthen the epidemiological and pathogenic significance of the cryptic form of HBV infection. In the course of this study, a relatively simple approach to the specific detection of intracellular hepadnavirus has been established. This procedure should provide a valuable tool for further investigations on hepadnaviral lymphotropism and the role of the lymphatic system in the natural course of hepadnaviral infection. This new method could also be applied to monitor the progression of symptomatic or concealed infection and to assess the effectiveness of antiviral agents against hepadnavirus propagating within lymphoid cells.
Acknowledgments
This research was supported by grant MOP-14818 provided to T.I.M. from the Canadian Institutes of Health Research. P.M.M. is a recipient of a Doctoral Fellowship Award from the Canadian Blood Services. T.I.M. is the Canada Research Chair (Tier 1) in Viral Hepatitis/Immunology sponsored by the Canada Research Chair Program and funds from the Canadian Institutes of Health Research and the Canada Foundation for Innovation.
REFERENCES
- 1.Bagasra, O., S. Thikkavarapu, R. Pomerantz, and J. Hansen. 1995. In situ PCR and hybridization to detect low-abundance nucleic acid targets, p. 14.8.18-14.8.24. In F. A. Ausubel, R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl (ed.), Current protocols in molecular biology. John Wiley & Sons, Inc., New York, N.Y.
- 2.Bläckberg, J., and K. Kidd-Ljunggren. 2001. Occult hepatitis B virus after acute self-limited infection persisting for 30 years without sequence variation. J. Hepatol. 33:992-997. [DOI] [PubMed] [Google Scholar]
- 3.Bréchot, C., V. Thiers, D. Kremsdorf, B. Naplas, S. Pol, and P. Paterlini-Bréchot. 2001. Persistent hepatitis B virus infection in subjects without hepatitis B surface antigen: clinically significant or purely “occult”? Hepatology 34:194-203. [DOI] [PubMed] [Google Scholar]
- 4.Cabrerizo, M., J. Bartolomé, C. Caramelo, G. Barril, and V. Carreño. 2000. Molecular analysis of hepatitis B virus DNA in serum and peripheral blood mononuclear cells from hepatitis B surface antigen-negative cases. Hepatology 32:116-123. [DOI] [PubMed] [Google Scholar]
- 5.Coffin, C. S., and T. I. Michalak. 1999. Persistence of infectious hepadnavirus in the offspring of woodchuck mothers recovered from viral hepatitis. J. Clin. Investig. 104:203-212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Colacino, J. M., and K. A. Staschke. 1998. The identification and development of antiviral agents for the treatment of chronic hepatitis B virus infection, p. 259-321. In E. Jucker (ed.), Progress in drug research. Birkhauser Verlag, Basel, Switzerland. [DOI] [PubMed]
- 7.Hodgson, P. D., and T. I. Michalak. 2001. Augmented hepatic interferon gamma expression and T-cell influx characterize acute hepatitis progressing to recovery and residual lifelong virus persistence in experimental adult woodchuck hepatitis virus infection. Hepatology 34:1049-1059. [DOI] [PubMed] [Google Scholar]
- 8.Kodoma, K., N. Ogasawara, H. Yoshikawa, and S. Murakami. 1985. Nucleotide sequence of a cloned woodchuck hepatitis virus genome: evolutional relationship between hepadnaviruses. J. Virol. 56:978-986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Korba, B. E., F. Wells, B. C. Tennant, P. J. Cote, and J. L. Gerin. 1987. Lymphoid cells in the spleens of woodchuck hepatitis virus-infected woodchucks are a site of active viral replication. J. Virol. 61:1318-1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lew, Y-Y., and T. I. Michalak. 2001. In vitro and in vivo infectivity and pathogenicity of the lymphoid cell-derived woodchuck hepatitis virus. J. Virol. 75:1770-1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Margolis, H. S., M. J. Alter, and S. C. Hadler. 1991. Hepatitis B: evolving epidemiology and implications for control. Semin. Liver Dis. 11:84-92. [DOI] [PubMed] [Google Scholar]
- 12.Michalak, T. I. 1998. The woodchuck animal model of hepatitis B. Viral Hepatitis Rev. 4:139-165. [Google Scholar]
- 13.Michalak, T. I. 2000. Occult persistence and lymphotropism of hepadnaviral infection: insights from the woodchuck viral hepatitis model. Immunol. Rev. 174:98-111. [DOI] [PubMed] [Google Scholar]
- 14.Michalak, T. I., I. U. Pardoe, C. S. Coffin, N. D. Churchill, D. S. Freake, P. Smith, and C. L. Trelegan. 1999. Occult life-long persistence of infectious hepadnavirus and residual liver inflammation in woodchucks convalescent from acute viral hepatitis. Hepatology 29:928-938. [DOI] [PubMed] [Google Scholar]
- 15.Michalak, T. I., C. Pasquinelli, S. Guilhot, and F. V. Chisari. 1994. Hepatitis B virus persistence after recovery from acute viral hepatitis. J. Clin. Investig. 93:230-239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Michalak, T. I., P. D. Hodgson, and N. D. Churchill. 2000. Posttranscriptional inhibition of class I major histocompatibility complex presentation on hepatocytes and lymphoid cells in chronic woodchuck hepatitis virus infection. J. Virol. 74:4483-4494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Muratori, L., D. Gibellini, M. Lenzi, M. Cataleta, P. Muratori, M. C. Morelli, and F. B. Bianchi. 1996. Quantification of hepatitis C virus-infected peripheral blood mononuclear cells by in situ reverse transcriptase polymerase chain reaction. Blood 88:2768-2774. [PubMed] [Google Scholar]
- 18.Nuovo, G. J. 1995. In situ PCR, p. 235-248. In C. W. Dieffenbach and G. S. Dveksler (ed.), PCR primer: a laboratory manual. Cold Spring Harbor Laboratory Press, New York, N.Y.
- 19.Pardoe, I. U., and T. I. Michalak. 1995. Detection of hepatitis B and woodchuck hepatitis viral DNA in plasma and mononuclear cells from heparinized blood by the polymerase chain reaction. J. Virol. Methods 51:277-288. [DOI] [PubMed] [Google Scholar]
- 20.Penna, A., M. Artini, A. Cavalli, M. Levero, A. Bertoletti, M. Pilli, F. V. Chisari, B. Rehermann, G. Del Prete, F. Fiaccadori, and C. Ferrari. 1996. Long-lasting memory T-cell responses following self-limited acute hepatitis B. J. Clin. Investig. 98:1185-1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Re, M. C., G. Furlini, D. Gibellini, M. Vignoli, E. Ramazzotti, S. Lolli, S. Ranieri, and M. La Placa. 1994. Quantification of human immunodeficiency virus type-1-infected mononuclear cells in peripheral blood of seropositive subjects by newly developed flow cytometry analysis of the product of an in situ PCR assay. J. Clin. Microbiol. 32:2152-2157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rehermann, B., C. Ferrari, C. Pasquinelli, and F. V. Chisari. 1996. The hepatitis B virus persists for decades after patients' recovery from acute viral hepatitis despite active maintenance of a cytotoxic T-lymphocyte response. Nat. Med. 2:1104-1108. [DOI] [PubMed] [Google Scholar]
- 23.Tennant, B. C., and J. L. Gerin. 1994. The woodchuck model of hepatitis B virus infection, p. 1455-1466. In I. M. Arias, J. L. Boyer, N. Fausto, W. B. Jakoby, D. A. Schachter, and D. A. Shafritz (ed.), The liver: biology and pathobiology. Raven Press, New York, N.Y.
- 24.World Health Organization. 2000. Hepatitis B fact sheet, October 2000 revision date. W.H.O./204. World Health Organization. Geneva, Switzerland. (Online, http://www.who.int/inf-fs/en/fact204.html.)
- 25.Yotsuyanagi, H., K. Yasuda, S. Iino, K. Moriya, Y. Shintani, H. Fujie, T. Tsutsumi, S. Kimura, and K. Koike. 1998. Persistent viremia after recovery from self-limited acute hepatitis B. Hepatology 27:1377-1382. [DOI] [PubMed] [Google Scholar]