Abstract
The genome of Bombyx mori nucleopolyhedrovirus (BmNPV) is predicted to contain six RING finger proteins: IAP1, ORF35, IAP2, CG30, IE2, and PE38. Several other members of the RING finger family have recently been shown to have the ubiquitin-ligase (E3) activity. We thus examined whether BmNPV RING finger proteins have the E3 activity. In vitro ubiquitination assay with the rabbit reticulocyte lysates and BmNPV RING finger proteins fused with maltose-binding protein (MBP) showed that four of them (IAP2, IE2, PE38, and CG30) were polyubiquitinated in the presence of zinc ion. Furthermore, MBP-IAP2, MBP-IE2, and MBP-PE38 were able to reconstitute ubiquitination activity in cooperation with the Ubc4/5 subfamily of ubiquitin-conjugating enzymes. Mutational analysis also showed that ubiquitination activity of MBP-IAP2, MBP-IE2, and MBP-PE38 were dependent on their RING finger motif. Therefore, these results suggest that IAP2, IE2, and PE38 may function as E3 enzymes during BmNPV infection.
The modification of proteins by ubiquitin is essential for numerous cellular processes. For example, ubiquitin-dependent protein degradation is involved in the regulation of cell cycle progression, signal transduction, transcription, DNA repair, and protein quality control (35). Modification of proteins with ubiquitin is also important in nondegradative processes, such as endocytosis (23).
Ubiquitination also plays key roles during viral infection. For instance, human adenovirus and papillomavirus direct degradation of host p53 by altering the activity of host ubiquitin ligase (E3) (16, 54, 58). Moreover, ubiquitination is required for the budding of retroviruses from infected cells (46, 50, 59, 60). Herpesvirus-encoded E3 ligase promotes ubiquitination and subsequent degradation of immune recognition molecules to evade the host immune system (2, 6, 22, 37). Baculoviruses encode a ubiquitin gene, although it is not known whether they use similar mechanisms to control their own budding or the activity of host proteins (10, 12, 19). Baculoviral ubiquitin is 75% homologous to eukaryotic ubiquitin. Guarino et al. reported that baculoviral ubiquitin is coexpressed with viral coat protein (13). In addition, baculoviral ubiquitin may be involved in the formation of viral particles because deletion of viral ubiquitin gene reduced yield of infectious virus (55).
The ubiquitin pathway begins with the activation of the C terminus of ubiquitin by the ubiquitin-activating enzyme (E1) in an ATP-dependent manner. This activation results in a high-energy thioester bond between ubiquitin and E1. Ubiquitin is subsequently transferred to a ubiquitin-conjugating enzyme (E2) through the thioester linkage. Finally, ubiquitin ligase (E3) attaches ubiquitin to its substrate by interacting with E2 and the target protein (5, 20). Ubiquitinated proteins are then subject to different fates, such as protein degradation, transport, or functional alteration, depending on the structure of the ubiquitin chain added (24, 35, 51).
The E3 ligases fall into two different families: the HECT and RING finger types (30). The RING finger motif of RING finger protein is Cys-X2-Cys-Xn-Cys-X1-3-His-X2-3-Cys-X2-Cys-Xn-Cys-X2-Cys (C3HC4) and binds two zinc atoms per RING finger motif (57). The E3 activity of RING finger proteins is dependent on the RING finger domain, which binds an E2 ubiquitin-conjugating enzyme (38, 67).
The baculovirus Bombyx mori nucleopolyhedrovirus (BmNPV) potentially encodes 136 proteins (10). Among these 136 genes, six (iap1, orf35, iap2, cg30, ie2, and pe38) are predicted to encode proteins containing a RING finger motif. The functions of ORF35 and CG30 are unknown. IE2 (11, 63, 66) and PE38 (33, 49) are transregulators of viral transcription and viral DNA replication. IAP1 and IAP2 are believed to function as inhibitors of apoptosis because they are homologues of known antiapoptotic factors (10). However, the exact mechanisms of action of these proteins remain unclear. The goal of the present study is to determine whether BmNPV RING finger proteins could function as ubiquitin ligases.
MATERIALS AND METHODS
Construction of plasmids.
To express fusion proteins with the maltose-binding protein (MBP), plasmid pMAL-p2 (New England Biolabs) was utilized. The six BmNPV genes encoding RING finger proteins (IAP1, ORF35, IAP2, CG30, IE2 and PE38) were amplified by PCR with BmNPV genome DNA and the primers listed in Table 1. The resulting PCR products, which encoded the entire protein minus start codon, were then digested with BamHI and EcoRI and inserted into the BamHI-EcoRI sites of pMAL-p2. The constructed plasmids were designated pMAL-IAP1, pMAL-ORF35, pMAL-IAP2, pMAL-CG30, pMAL-IE2, and pMAL-PE38.
TABLE 1.
Oligonucleotides used for PCR
| Purpose and oligonucleotide | Sequencea |
|---|---|
| MBP fusion | |
| pIAP1-1 | 5′-CAGAATTCTTGAACGAGGACACTCCTCC-3′ |
| pIAP1-2 | 5′-CAGGATCCAACACCATTTTATTACACCAC-3′ |
| pORF35-1 | 5′-CAGAATTCCACGACGGTCGCGTTAAG-3′ |
| pORF35-2 | 5′-CAGGATCCATGTTACAAAGTTTTGTATTTC-3′ |
| pIAP2-1 | 5′-TAGAATTCAATTTGAGGCAATTTAATTTTTTG-3′ |
| pIAP2-2 | 5′-CAGGATCCTTACTGAGGTAATGTTTCG-3′ |
| pCG30-1 | 5′-CAGAATTCGAGTTTGTCAAATTGCAATGC-3′ |
| pCG30-2 | 5′-CAGGAATCCTTAATCTACATTTATTGTAAC-3′ |
| pIE2-1 | 5′-CAGAATTCAGTCGCCAAATCAACGCCGTC-3′ |
| pIE2-2 | 5′-CAGGATCCTTAAGGTTTAGACATCTC-3′ |
| pPE38-1 | 5′-CAGAATTCCCAAGGGACACCAACAATCG-3′ |
| pPE38-2 | 5′-CAGGATCCATCACATTAATTTTCAAACC-3′ |
| Mutation in RING finger motif | |
| pIAP2cs-f | 5′-GTGTTTCATGCCTTCCCGTCACCTGGC-3′ |
| pIAP2cs-r | 5′-GCCAGGTGACGGGAAGGCATGAAACAC-3′ |
| pIE2cs-f | 5′-GACTTCGATTGATTCTAACCATGCTG-3′ |
| pIE2cs-r | 5′-CAGCATGGTTAGAATCAATCGAAGTC-3′ |
| pPE38cs-f | 5′-GATTCCGACTACGTCCGACCACGGTTTTTG-3′ |
| pPE38cs-r | 5′-CAAAAACCGTGGTCGGACGTAGTCGGAATC-3′ |
Solid and broken underlines indicate the recognition sites of EcoRI and BamHI, respectively. Double underlines show the substituted base.
To demonstrate that the observed activities were due to the ubiquitin ligase activity, a series of plasmids encoding mutant versions of the RING finger proteins was also constructed. In these proteins, one of the zinc-chelating cysteine in the RING finger (57) was mutated to serine (CS). To obtain MBP-RING-CS mutant proteins, site-specific substitutions were introduced by overlapping PCR (25) with one of the pMAL fusion plasmids and the corresponding mutagenic primers shown in Table 1 and flanking pMAL-p2 specific primers. The resulting PCR products were digested with BamHI and EcoRI and then inserted into the BamHI and EcoRI sites of pMAL-p2 to express the fusion protein.
Purification of recombinant MBP fusion proteins.
All recombinant proteins were expressed in Escherichia coli and purified according to the manufacturer's recommended protocol (New England Biolabs) with some minor modifications. ZnSO4 was added as the source of zinc to the culture to a final concentration of 10 or 100 μM at the time of induction with isopropylthiogalactoside. In addition, all purification buffers lacked EDTA and contained ZnSO4 at a final concentration of 10 or 100 μM, as indicated.
In vitro ubiquitination assay with rabbit reticulocyte lysate.
The ubiquitination assay with rabbit reticulocyte lysate was performed as described previously (40) with minor modifications. The reaction mixture contained 40 mM Tris-HCl (pH 7.5), 5 mM MgCl2, 2 mM ATP, 2 mM dithiothreitol, 300 μg of ubiquitin (Sigma)/ml, 25 μM MG132 (Sigma), 10% (vol/vol) rabbit reticulocyte lysate (Promega), and 2.5 μg of MBP-RING finger protein/ml. The reaction mixtures were incubated at 30°C for 3 h and subjected to immunoblotting with anti-MBP antisera (New England Biolabs).
Reconstitution of ubiquitination activity.
His-tagged recombinant mouse E1 was purified from Sf9 insect cells. E. coli lysates expressing recombinant human E2 (E2-20K, E2-25K, Ubc3, Ubc4, UbcH5a, Ubc8, Ubc7, and UbcH7) were prepared as described previously (40). Ubc7 (31) was a kind gift from T. Mizushima of Tokyo Metropolitan Institute of Medical Science, and UbcH7 was purchased from Affiniti (Exeter, Devon, United Kingdom). The reaction mixture contained 40 mM Tris-HCl (pH 7.5), 5 mM MgCl2, 2 mM ATP, 2 mM dithiothreitol, 300 μg of ubiquitin (Sigma)/ml, 2.5 μg of E1/ml, 0.2 mg of E2-expressing E. coli lysate/ml, and 2.5 mg of MBP-RING finger protein/ml. Reaction mixtures were incubated at 30°C for 3 h and subjected to immunoblotting with anti-MBP antisera (New England Biolabs).
RESULTS
MBP-fused BmNPV RING finger proteins show ubiquitination activities, which depend on zinc ion.
Six genes of BmNPV—orf18, orf35, orf58, orf71, orf127, and orf128—are predicted to encode RING finger proteins. The goal of the present study was to determine whether BmNPV RING finger proteins could function as ubiquitin ligases (E3). These enyzmes are substrate specific; therefore, it is often necessary to know which proteins are authentic substrates (20). Recently, however, it was shown that Rma1 and EL5 ubiquitinate an in-frame fused MBP domain (40, 62). This result suggests that the fused MBP could be a test substrate for any E3 enzyme, and this information is useful when the authentic substrate is not known. Thus, we constructed MBP fusions of all six BmNPV RING finger proteins to check for the E3 activity.
To examine whether the viral RING finger proteins have the E3 activity, we first performed an in vitro ubiquitination assay with rabbit reticulocyte lysate as a source of ubiquitin-activating enzyme E1 and ubiquitin-conjugating enzyme E2, since reticulocyte lysates have previously been shown to contain high ubiquitination activity (8, 21, 65). Each MBP-fused RING finger protein (MBP-IAP1, MBP-ORF35, MBP-IAP2, MBP-CG30, MBP-IE2, or MBP-PE38) was incubated with ATP, ubiquitin, and rabbit reticulocyte lysate at 30°C. After 3 h, the modification of MBP fusion protein was analyzed by immunoblotting with anti-MBP antibody.
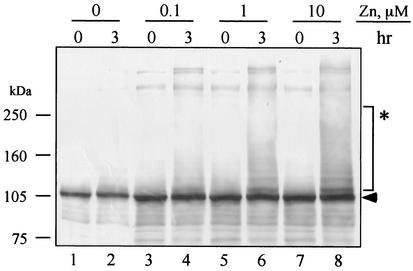
We first expressed the RING finger proteins in the absence of zinc as previously reported (40, 62), but the proteins did not show the E3 activity (data not shown). We suspected that this was due to misfolding of the proteins, possibly because the amount of zinc in the culture was insufficient. The unique structure of RING finger domain is dependent upon chelation of zinc atoms (57). Therefore, the dependence on zinc was examined with the MBP-IE2 protein. MBP-IE2 was expressed and purified in the presence of 0, 1, 10, or 100 μM zinc and then tested for ubiquitination activity. Additional zinc ion was not added to the ubiquitination assay, and the final concentrations of zinc were 0, 0.1, 1, and 10 μM, respectively (Fig. 1). In the absence of zinc ion, the migration of MBP-IE2 was unchanged after a 3-h incubation. This result suggests that the protein lacked ubiquitin ligase activity, since the covalent attachment of ubiquitin should result in a ladder of bands in multiples of ∼8 kDa. When MBP-IE2 was expressed in the presence of zinc higher-molecular-mass bands appeared, and the intensity of the ubiquitin ladders was dependent on the zinc concentration (lanes 4, 6, and 8). This finding strongly suggested that zinc ion was required for correct folding of the RING finger domain and the E3 ligase activity. Therefore, we expressed and purified other MBP-RING finger proteins in the presence of zinc ion. We tested all six proteins with two different concentrations of zinc: 10 and 100 μM (data not shown).
FIG. 1.
Analysis of zinc requirement for ubiquitination of MBP-IE2. MBP-IE2 proteins were expressed and purified with a zinc concentration of 0, 1, 10, or 100 μM and then incubated with ATP, ubiquitin, and rabbit reticulocyte lysate for 3 h. Since the presence of zinc ions was originated only from the expression and purification steps, the final concentrations of zinc were 0 (lanes 1 and 2), 0.1 (lanes 3 and 4), 1 (lanes 5 and 6), and 10 μM (lanes 7 and 8), respectively. After incubation, MBP-IE2 was subjected to immunoblotting with anti-MBP antibodies. The arrowhead indicates the predicted molecular mass of MBP-IE2. The asterisk indicates the modified MBP-IE2.
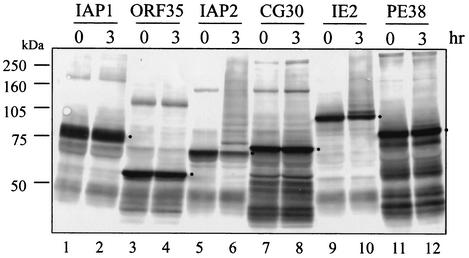
Each MBP-RING finger protein was tested for ubiquitination activity (Fig. 2). MBP-IAP2 exhibited the highest degree of ubiquitin ligase activity when induced in the presence of 10 μM zinc. After 3 h of incubation, most of the fusion protein was shifted to slower-migrating forms, and little unmodified protein remained (lane 6). Significant amounts of MBP-IE2 were also shifted into molecular-mass ladders (lane 10). However, it was difficult to determine whether the other proteins contain ubiquitination activity or not because of high background levels. Therefore, detection of monoubiquitinated protein was used as a standard for determination. MBP-CG30 and MBP-PE38 also showed higher-molecular-mass bands, which is indicative of ubiquitination although to a lesser extent (lanes 8 and 12). The preparations of MBP-CG30, MBP-IE2, and MBP-PE38 shown in Fig. 2 were expressed in the presence of 100 μM zinc. This concentration resulted in higher levels of activity than did the 10 μM concentration (data not shown and Fig. 1). Ladders of ubiquitinated protein were not detected with MBP-IAP1 and MBP-ORF35, whether the enzymes were expressed in the presence of 10 μM zinc (lanes 2 and 4) or 100 μM zinc (data not shown).
FIG. 2.
In vitro ubiquitination assay of BmNPV RING finger proteins with rabbit reticulocyte lysate. For expression and purification, 10 μM zinc was used for MBP-IAP1 (lanes 1 and 2), MBP-ORF35 (lanes 3 and 4), and MBP-IAP2 (lanes 5 and 6), whereas 100 μM was used for MBP-CG30 (lanes 7 and 8), MBP-IE2 (lanes 9 and 10), and MBP-PE38 (lanes 11 and 12). MBP-fused proteins were analyzed as described in Fig. 2. Dots on the panel indicate the predicted molecular mass of each MBP-RING finger protein.
IAP2, IE2, and PE38 are E3 ligases.
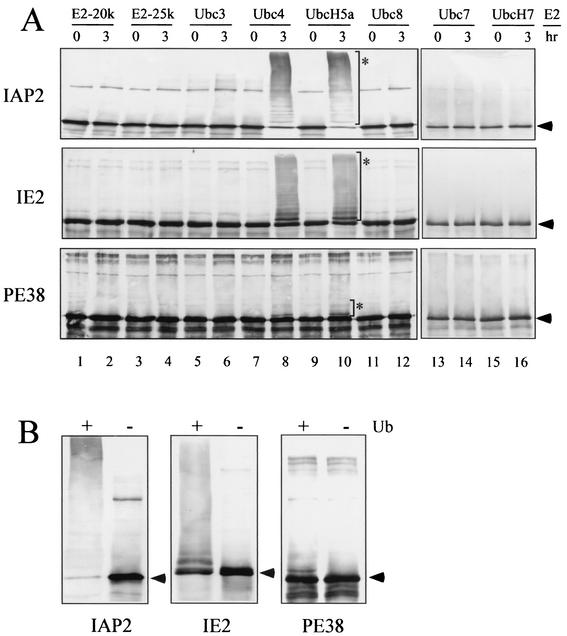
To further characterize the activity of the viral RING finger proteins, we performed a ubiquitination reconstitution assay with only the essential components in purified forms. In addition, we tested eight different E2 enzymes (E2-20K, E2-25K, Ubc3, Ubc4, UbcH5a, Ubc7, UbcH7, and Ubc8) because E3 enzymes require specific E2s to catalyze ubiquitination (51). Therefore, each MBP-RING finger protein was incubated with ATP, ubiquitin, recombinant E1, and one of the recombinant E2 enzymes described above. After 3 h of incubation, reaction products were separated on sodium dodecyl sulfate-polyacrylamide gel electrophoresis and probed with the MBP antibody. MBP-IAP2, MBP-IE2, and MBP-PE38 clearly exhibited slower-migrating ladders in some lanes (Fig. 3A), indicating that these MBP-RING finger proteins possess intrinsic ubiquitination activity. Ubiquitin ladders were obtained only from the reaction with Ubc4 or UbcH5a (lanes 8 and 10), whereas the other E2 enzymes did not support this modification. Although ubiquitinated MBP-CG30 was observed in the assay with reticulocyte lysate (Fig. 2), the reconstitution assay did not produce high-molecular-mass ladders (data not shown). This result suggests that MBP-CG30 was ubiquitinated by an E3 enzyme in the reticulocyte lysate or that CG30 requires a different E2 for activity. Neither MBP-IAP1 nor MBP-ORF35 showed any ubiquitinated ladders (data not shown). This finding was consistent with the previous result from the assay with reticulocyte lysate (Fig. 2).
FIG. 3.
Analysis of ubiquitination activity. (A) Reconstitution assay of ubiquitination activity. MBP-fused IAP2, IE2, or PE38 was incubated with ATP, ubiquitin, E1, and one of the E2 enzymes as indicated and then subjected to immunoblotting with anti-MBP antibody. For the E2 enzymes, E2-20K (lanes 1 and 2), E2-25K (lanes 3 and 4), Ubc3 (lanes 5 and 6), Ubc4 (lanes 7 and 8), UbcH5a (lanes 9 and 10), Ubc8 (lanes 11 and 12), Ubc7 (lanes 13 and 14), and UbcH7 (lanes 15 and 16) were tested. Arrowheads indicate the predicted molecular mass of each MBP-RING finger protein. Asterisks indicate the reconstituted ubiquitination bands. (B) Conjugation assay with or without ubiquitin. MBP-IAP2, MBP-IE2, or MBP-PE38 was incubated with ATP, E1, and Ubc4 in the presence (+) or absence (−) of ubiquitin.
In order to demonstrate that the modification of MBP-IAP2, MBP-IE2, and MBP-PE38 was really due to the addition of ubiquitin, we repeated the conjugation assay with ATP, E1, and Ubc4 in the presence or absence of ubiquitin. The slower-migrating ladders were not detected in the absence of ubiquitin (Fig. 3B). Taken together, we concluded that IAP2, IE2, and PE38 catalyze ubiquitination as ubiquitin ligases in cooperation with Ubc4 and UbcH5a.
The RING finger motif is required for the E3 activity of IAP2, IE2, and PE38.
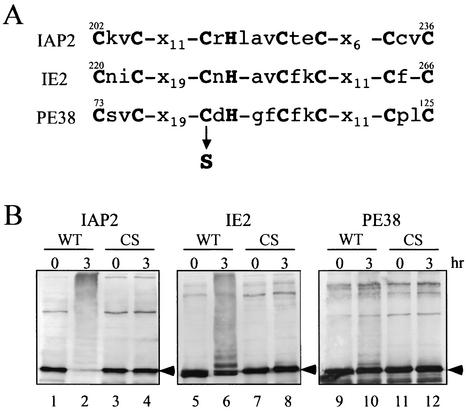
The RING finger motif of RING finger type E3 ligases is required for their ubiquitin ligase activity (29). In order to examine whether IAP2, IE2, and PE38 fall under this rule, we introduced a point mutation in the RING finger motif of the three viral proteins that had been shown to have E3 ligase activity in the previous assays. The third cysteine residue in the RING finger motif was replaced with a serine residue (Fig. 4A). Then, each CS mutant protein (MBP-IAP2CS, MBP-IE2CS, or MBP-PE38CS) was incubated with ATP, ubiquitin, E1, and Ubc4 and used for Western analysis. None of these CS mutants exhibited slower-migrating ladders (Fig. 4B, lanes 4, 8, and 12), whereas wild-type proteins produced high-molecular-mass bands (Fig. 4B, lanes 2, 6, and 10). Thus, these results confirmed that the RING finger motif is essential for the E3 activities of IAP2, IE2, and PE38.
FIG. 4.
Analysis of RING finger motif for ubiquitination activity. (A) The amino acid sequences of RING finger motif in IAP2, IE2, and PE38. The boldface letters show the zinc-coordinating residues comprising a RING finger motif. The third cysteine (C) of each motif was mutated to serine (S). (B) Assay of ubiquitination. The wild type (WT) or mutated version (CS) of MBP-IAP2, MBP-IE2, or MBP-PE38 was analyzed as described in Fig. 4. Ubc4 was used as an E2 enzyme. Arrowheads indicate the predicted molecular mass of each MBP-RING finger protein.
DISCUSSION
In the present study, we examined whether the BmNPV RING finger proteins could function as ubiquitin ligases. There are six RING finger proteins in BmNPV, and we showed that three of them, i.e., IAP2, IE2, and PE38, were able to catalyze ubiquitination in cooperation with Ubc4/5 subfamily of E2 enzymes. In addition, mutational analysis confirmed that E3 activities of these proteins were dependent on their RING finger motif. This is consistent with previous studies since the RING finger of RING-type E3 is a binding site for E2 enzymes (29, 38). Many RING finger-type E3s have been reported to function with Ubc4/5; however, preference for specific E2 by RING finger E3s does not appear to be present at this point. For instance, UbcH1, UbcH5, UbcH7, and Ubc13 have been shown to function with RING finger E3s (51). Several E2 enzymes except for Ubc4/5 did not support the ubiquitination in our study. Among these, Ubc7 and UbcH7 were shown to be functional with other RING finger E3 (unpublished data), suggesting that these are not in cooperation with BmNPV RING finger proteins. However, same preparation of the other E2 enzymes was not tested with other E3 to confirm their activity; thus, the possibility that they might function with BmNPV RING finger proteins still remains. Two proteins, IAP1 and ORF35 did not show ubiquitination activity in either assay. CG30 was active in the reticulocyte lysate but not in the reconstituted extract, so these results are inconclusive. The negative results obtained with these three proteins could just be due to problems with the expression or assay conditions, or they could truly reflect the lack of E3 ligase function in these proteins. Recent reports indicate that not all RING finger proteins have the E3 activity (17). Crystallization analyses showed that the RING finger structure of V(D)J recombination-activating protein RAG1 lacks the groove for binding to E2 compared to the structure of c-Cbl (E3) (1, 67), and thus V(D)J is probably not an E3 ligase. Such differences in the structure of the RING finger domain may explain why IAP1, ORF35, and CG30 did not show ubiquitin ligase activity. It is also possible that these proteins require additional coactivators for activity. For example, BRCA1 is a RING finger ubiquitin ligase that needs BARD1 as a coactivator for its activity (18).
In other studies, MBP-fused RING finger proteins were able to exhibit ubiquitination activity without additional zinc ions (40, 62). However, our study showed that zinc ions are required for ubiquitination of MBP-fused BmNPV RING finger proteins, although we used same system but different proteins were analyzed. The requirement for additional zinc during overexpression may be caused by exhaustion of zinc because bacterial growth media contain only trace amounts of zinc. Therefore, it is possible that the binding affinity to zinc is unstable in our MBP-fused RING finger proteins and that additional zinc ions are required for their E3 activities since the RING finger domain requires zinc to form its unique structure (57).
It is tempting to speculate when and why these BmNPV RING finger proteins need to function as E3 enzymes during BmNPV infection cycle. Inhibitor of apoptosis proteins (IAP) was originally identified in the baculovirus system. They are highly conserved in many species and play an important role in regulating apoptosis. Some of them also contain a RING finger motif (43, 56). It was reported that mammalian X-linked IAP and c-IAP2 target for caspase-3 as E3 enzymes via the RING finger domain (26, 61). In addition, c-IAP1 mediates ubiquitination of TRAF2, which is also dependent on its RING finger domain (36). Therefore, it may be possible that BmNPV IAP2 catalyzes ubiquitination of apoptotic proteins during viral infection (10).
Prikhod'ko et al. reported that IE2 of Autographa californica NPV (AcNPV) blocks host cell cycle progression (53). Moreover, a single mutation in the RING finger motif abolished the ability to block cell division (53). Therefore, it may be possible that IE2 ubiquitinates one or more host factors involved in cell cycle regulation. Prikhod'ko et al. also reported that AcNPV PE38 augments apoptosis of host cells induced by IE1 (52). However, it was shown that mutation on the RING finger motif of PE38 had little or no effect on this ability (52). Therefore, ubiquitination activity of PE38 may be involved in a function other than augmentation of apoptosis.
Another possible role of IE2 and PE38 is related to their function as transregulators. ND10, alternately referred as the PML body, is a nuclear site for transcription and replication of DNA viruses (41). Human cytomegalovirus IE72 is responsible for the dispersion of ND10 in the early stage of infection (28, 32, 34). E4ORF3 of adenovirus 5, which is essential for viral DNA replication, induces a redistribution of protein in ND10 (4, 7, 27). A RING finger protein of herpesvirus 1, ICP0, also exhibits ND10 dispersing activity (42). These observations imply that host protein(s) are transported to and from specific ND10 sites in the nucleus and that this pathway might be altered by viral factors during infection. In support of this model, it has been reported that ICP0 possesses ubiquitin ligase activity (3, 14, 15, 64). In addition, it was found that ICP0 induces the proteasome-dependent degradation of many proteins, including PML and Sp100, which are major components of ND10 in a RING finger-dependent manner (9, 47, 48). ND10-like structures have also been shown as sites for DNA replication and transcription of BmNPV (45). AcNPV IE2 colocalizes with PML at the time of DNA replication (39), whereas PE38 was partially present in nuclear dots at a very early infection stage (44). Therefore, it could be possible that IE2 and PE38 might ubiquitinate some factor(s) of the specific nuclear sites to set up the viral replication factories in the nucleus.
We demonstrated that at least three RING finger proteins of BmNPV (IAP2, IE2, and PE38) possess ubiquitin ligase E3 activities. This is the first report showing that baculovirus RING finger proteins have ubiquitin ligase activity. To more completely understand the molecular function of ubiquitination by these proteins during infection, it is now important to identify their authentic substrates.
Acknowledgments
We are grateful to Tsunehiro Mizushima for kind gift of Ubc7.
This research was supported by grants from Bioarchitect program of the Science and Technology Agency of Japan, a Grant-in Aid for Encouragement of Young Scientists from Japan Society for the promotion of science (N.I.), and special postdoctoral fellowship of RIKEN (N.I. and N.M.).
REFERENCES
- 1.Bellon, S. F., K. K. Rodgers, D. G. Schatz, J. E. Coleman, and T. A. Steitz. 1997. Crystal structure of the RAG1 dimerization domain reveals multiple zinc-binding motifs including a novel zinc binuclear cluster. Nat. Struct. Biol. 4:586-591. [DOI] [PubMed] [Google Scholar]
- 2.Boname, J. M., and P. G. Stevenson. 2001. MHC class I ubiquitination by a viral PHD/LAP finger protein. Immunity 15:627-636. [DOI] [PubMed] [Google Scholar]
- 3.Boutell, C., S. Sadis, and R. D. Everett. 2002. Herpes simplex virus type 1 immediate-early protein ICP0 and its isolated RING finger domain act as ubiquitin E3 ligases in vitro. J. Virol. 76:841-850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Carvalho, T., J. S. Seeler, K. Ohman, P. Jordan, U. Pettersson, G. Akusjarvi, M. Carmo-Fonseca, and A. Dejean. 1995. Targeting of adenovirus E1A and E4-ORF3 proteins to nuclear matrix-associated PML bodies. J. Cell Biol. 131:45-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ciechanover, A., A. Orian, and A. L. Schwartz. 2000. Ubiquitin-mediated proteolysis: biological regulation via destruction. Bioessays 22:442-451. [DOI] [PubMed] [Google Scholar]
- 6.Coscoy, L., D. J. Sanchez, and D. Ganem. 2001. A novel class of herpesvirus-encoded membrane-bound E3 ubiquitin ligases regulates endocytosis of proteins involved in immune recognition. J. Cell Biol. 155:1265-1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Doucas, V., A. M. Ishov, A. Romo, H. Juguilon, M. D. Weitzman, R. M. Evans, and G. G. Maul. 1996. Adenovirus replication is coupled with the dynamic properties of the PML nuclear structure. Genes Dev. 10:196-207. [DOI] [PubMed] [Google Scholar]
- 8.Etlinger, J. D., and A. L. Goldberg. 1977. A soluble ATP-dependent proteolytic system responsible for the degradation of abnormal proteins in reticulocytes. Proc. Natl. Acad. Sci. USA 74:54-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Everett, R. D., W. C. Earnshaw, J. Findlay, and P. Lomonte. 1999. Specific destruction of kinetochore protein CENP-C and disruption of cell division by herpes simplex virus immediate-early protein Vmw110. EMBO J. 18:1526-1538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gomi, S., K. Majima, and S. Maeda. 1999. Sequence analysis of the genome of Bombyx mori nucleopolyhedrovirus. J. Gen. Virol. 80:1323-1337. [DOI] [PubMed] [Google Scholar]
- 11.Gomi, S., C. E. Zhou, W. Yih, K. Majima, and S. Maeda. 1997. Deletion analysis of four of eighteen late gene expression factor gene homologues of the baculovirus, BmNPV. Virology 230:35-47. [DOI] [PubMed] [Google Scholar]
- 12.Guarino, L. A. 1990. Identification of a viral gene encoding a ubiquitin-like protein. Proc. Natl. Acad. Sci. USA 87:409-413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guarino, L. A., G. Smith, and W. Dong. 1995. Ubiquitin is attached to membranes of baculovirus particles by a novel type of phospholipid anchor. Cell 80:301-309. [DOI] [PubMed] [Google Scholar]
- 14.Hagglund, R., and B. Roizman. 2002. Characterization of the novel E3 ubiquitin ligase encoded in exon 3 of herpes simplex virus-1-infected cell protein 0. Proc. Natl. Acad. Sci. USA 99:7889-7894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hagglund, R., C. Van Sant, P. Lopez, and B. Roizman. 2002. Herpes simplex virus 1-infected cell protein 0 contains two E3 ubiquitin ligase sites specific for different E2 ubiquitin-conjugating enzymes. Proc. Natl. Acad. Sci. USA 99:631-636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Harada, J. N., A. Shevchenko, D. C. Pallas, and A. J. Berk. 2002. Analysis of the adenovirus E1B-55K-anchored proteome reveals its link to ubiquitination machinery. J. Virol. 76:9194-9206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hardtke, C. S., H. Okamoto, C. Stoop-Myer, and X. W. Deng. 2002. Biochemical evidence for ubiquitin ligase activity of the Arabidopsis COP1 interacting protein 8 (CIP8). Plant J. 30:385-394. [DOI] [PubMed] [Google Scholar]
- 18.Hashizume, R., M. Fukuda, I. Maeda, H. Nishikawa, D. Oyake, Y. Yabuki, H. Ogata, and T. Ohta. 2001. The RING heterodimer BRCA1-BARD1 is a ubiquitin ligase inactivated by a breast cancer-derived mutation. J. Biol. Chem. 276:14537-14540. [DOI] [PubMed] [Google Scholar]
- 19.Herniou, E. A., T. Luque, X. Chen, J. M. Vlak, D. Winstanley, J. S. Cory, and D. R. O'Reilly. 2001. Use of whole genome sequence data to infer baculovirus phylogeny. J. Virol. 75:8117-8126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hershko, A., and A. Ciechanover. 1998. The ubiquitin system. Annu. Rev. Biochem. 67:425-479. [DOI] [PubMed] [Google Scholar]
- 21.Hershko, A., A. Ciechanover, H. Heller, A. L. Haas, and I. A. Rose. 1980. Proposed role of ATP in protein breakdown: conjugation of protein with multiple chains of the polypeptide of ATP-dependent proteolysis. Proc. Natl. Acad. Sci. USA 77:1783-1786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hewitt, E. W., L. Duncan, D. Mufti, J. Baker, P. G. Stevenson, and P. J. Lehner. 2002. Ubiquitylation of MHC class I by the K3 viral protein signals internalization and TSG101-dependent degradation. EMBO J. 21:2418-2429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hicke, L. 2001. A new ticket for entry into budding vesicles-ubiquitin. Cell 106:527-530. [DOI] [PubMed] [Google Scholar]
- 24.Hicke, L. 2001. Protein regulation by monoubiquitin. Nat. Rev. Mol. Cell. Biol. 2:195-201. [DOI] [PubMed] [Google Scholar]
- 25.Higuchi, R. 1990. Recombinant PCR, p. 177-183. In M. A. Gelfand, J. J. Sninsky, and T. J. White (ed.), PCR protocol: a guide to methods and application. Academic Press, Inc., San Diego, Calif.
- 26.Huang, H., C. A. Joazeiro, E. Bonfoco, S. Kamada, J. D. Leverson, and T. Hunter. 2000. The inhibitor of apoptosis, cIAP2, functions as a ubiquitin-protein ligase and promotes in vitro monoubiquitination of caspases 3 and 7. J. Biol. Chem. 275:26661-26664. [DOI] [PubMed] [Google Scholar]
- 27.Ishov, A. M., and G. G. Maul. 1996. The periphery of nuclear domain 10 (ND10) as site of DNA virus deposition. J. Cell Biol. 134:815-826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ishov, A. M., R. M. Stenberg, and G. G. Maul. 1997. Human cytomegalovirus immediate early interaction with host nuclear structures: definition of an immediate transcript environment. J. Cell Biol. 138:5-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jackson, P. K., A. G. Eldridge, E. Freed, L. Furstenthal, J. Y. Hsu, B. K. Kaiser, and J. D. Reimann. 2000. The lore of the RINGs: substrate recognition and catalysis by ubiquitin ligases. Trends Cell Biol. 10:429-439. [DOI] [PubMed] [Google Scholar]
- 30.Joazeiro, C. A., and A. M. Weissman. 2000. RING finger proteins: mediators of ubiquitin ligase activity. Cell 102:549-552. [DOI] [PubMed] [Google Scholar]
- 31.Katsanis, N., and E. M. Fisher. 1998. Identification, expression, and chromosomal localization of ubiquitin conjugating enzyme 7 (UBE2G2), a human homologue of the Saccharomyces cerevisiae ubc7 gene. Genomics 51:128-131. [DOI] [PubMed] [Google Scholar]
- 32.Kelly, C., R. Van Driel, and G. W. Wilkinson. 1995. Disruption of PML-associated nuclear bodies during human cytomegalovirus infection. J. Gen. Virol. 76:2887-2893. [DOI] [PubMed] [Google Scholar]
- 33.Kool, M., C. H. Ahrens, R. W. Goldbach, G. F. Rohrmann, and J. M. Vlak. 1994. Identification of genes involved in DNA replication of the Autographa californica baculovirus. Proc. Natl. Acad. Sci. USA 91:11212-11216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Korioth, F., G. G. Maul, B. Plachter, T. Stamminger, and J. Frey. 1996. The nuclear domain 10 (ND10) is disrupted by the human cytomegalovirus gene product IE1. Exp. Cell Res. 229:155-158. [DOI] [PubMed] [Google Scholar]
- 35.Laney, J. D., and M. Hochstrasser. 1999. Substrate targeting in the ubiquitin system. Cell 97:427-430. [DOI] [PubMed] [Google Scholar]
- 36.Li, X., Y. Yang, and J. D. Ashwell. 2002. TNF-RII and c-IAP1 mediate ubiquitination and degradation of TRAF2. Nature 416:345-347. [DOI] [PubMed] [Google Scholar]
- 37.Lorenzo, M. E., J. U. Jung, and H. L. Ploegh. 2002. Kaposi's sarcoma-associated herpesvirus K3 utilizes the ubiquitin-proteasome system in routing class major histocompatibility complexes to late endocytic compartments. J. Virol. 76:5522-5531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lorick, K. L., J. P. Jensen, S. Fang, A. M. Ong, S. Hatakeyama, and A. M. Weissman. 1999. RING fingers mediate ubiquitin-conjugating enzyme (E2)-dependent ubiquitination. Proc. Natl. Acad. Sci. USA 96:11364-11369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mainz, D., I. Quadt, and D. Knebel-Morsdorf. 2002. Nuclear IE2 structures are related to viral DNA replication sites during baculovirus infection. J. Virol. 76:5198-5207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Matsuda, N., T. Suzuki, K. Tanaka, and A. Nakano. 2001. Rma1, a novel type of RING finger protein conserved from Arabidopsis to human, is a membrane-bound ubiquitin ligase. J. Cell Sci. 114:1949-1957. [DOI] [PubMed] [Google Scholar]
- 41.Maul, G. G. 1998. Nuclear domain 10, the site of DNA virus transcription and replication. Bioessays 20:660-667. [DOI] [PubMed] [Google Scholar]
- 42.Maul, G. G., H. H. Guldner, and J. G. Spivack. 1993. Modification of discrete nuclear domains induced by herpes simplex virus type 1 immediate early gene 1 product (ICP0). J. Gen. Virol. 74:2679-2690. [DOI] [PubMed] [Google Scholar]
- 43.Miller, L. K. 1999. An exegesis of IAPs: salvation and surprises from BIR motifs. Trends Cell Biol. 9:323-328. [DOI] [PubMed] [Google Scholar]
- 44.Murges, D., I. Quadt, J. Schroer, and D. Knebel-Morsdorf. 2001. Dynamic nuclear localization of the baculovirus proteins IE2 and PE38 during the infection cycle: the promyelocytic leukemia protein colocalizes with IE2. Exp. Cell Res. 264:219-232. [DOI] [PubMed] [Google Scholar]
- 45.Okano, K., V. S. Mikhailov, and S. Maeda. 1999. Colocalization of baculovirus IE-1 and two DNA-binding proteins, DBP and LEF-3, to viral replication factories. J. Virol. 73:110-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ott, D. E., L. V. Coren, T. D. Copeland, B. P. Kane, D. G. Johnson, R. C. Sowder II, Y. Yoshinaka, S. Oroszlan, L. O. Arthur, and L. E. Henderson. 1998. Ubiquitin is covalently attached to the p6Gag proteins of human immunodeficiency virus type 1 and simian immunodeficiency virus and to the p12Gag protein of Moloney murine leukemia virus. J. Virol. 72:2962-2968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Parkinson, J., and R. D. Everett. 2000. Alphaherpesvirus proteins related to herpes simplex virus type 1 ICP0 affect cellular structures and proteins. J. Virol. 74:10006-10017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Parkinson, J., S. P. Lees-Miller, and R. D. Everett. 1999. Herpes simplex virus type 1 immediate-early protein vmw110 induces the proteasome-dependent degradation of the catalytic subunit of DNA-dependent protein kinase. J. Virol. 73:650-657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Passarelli, A. L., and L. K. Miller. 1993. Three baculovirus genes involved in late and very late gene expression: ie-1, ie-n, and lef-2. J. Virol. 67:2149-2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Patnaik, A., V. Chau, and J. W. Wills. 2000. Ubiquitin is part of the retrovirus budding machinery. Proc. Natl. Acad. Sci. USA 97:13069-13074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pickart, C. M. 2001. Mechanisms underlying ubiquitination. Annu. Rev. Biochem. 70:503-533. [DOI] [PubMed] [Google Scholar]
- 52.Prikhod'ko, E. A., and L. K. Miller. 1999. The baculovirus PE38 protein augments apoptosis induced by transactivator IE1. J. Virol. 73:6691-6699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Prikhod'ko, E. A., and L. K. Miller. 1998. Role of baculovirus IE2 and its RING finger in cell cycle arrest. J. Virol. 72:684-692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Querido, E., P. Blanchette, Q. Yan, T. Kamura, M. Morrison, D. Boivin, W. G. Kaelin, R. C. Conaway, J. W. Conaway, and P. E. Branton. 2001. Degradation of p53 by adenovirus E4orf6 and E1B55K proteins occurs via a novel mechanism involving a Cullin-containing complex. Genes Dev. 15:3104-3117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Reilly, L. M., and L. A. Guarino. 1996. The viral ubiquitin gene of Autographa californica nuclear polyhedrosis virus is not essential for viral replication. Virology 218:243-247. [DOI] [PubMed] [Google Scholar]
- 56.Salvesen, G. S., and C. S. Duckett. 2002. IAP proteins: blocking the road to death's door. Nat. Rev. Mol. Cell. Biol. 3:401-410. [DOI] [PubMed] [Google Scholar]
- 57.Saurin, A. J., K. L. Borden, M. N. Boddy, and P. S. Freemont. 1996. Does this have a familiar RING? Trends Biochem. Sci. 21:208-214. [PubMed] [Google Scholar]
- 58.Scheffner, M., J. M. Huibregtse, R. D. Vierstra, and P. M. Howley. 1993. The HPV-16 E6 and E6-AP complex functions as a ubiquitin-protein ligase in the ubiquitination of p53. Cell 75:495-505. [DOI] [PubMed] [Google Scholar]
- 59.Schubert, U., D. E. Ott, E. N. Chertova, R. Welker, U. Tessmer, M. F. Princiotta, J. R. Bennink, H. G. Krausslich, and J. W. Yewdell. 2000. Proteasome inhibition interferes with gag polyprotein processing, release, and maturation of HIV-1 and HIV-2. Proc. Natl. Acad. Sci. USA 97:13057-13062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Strack, B., A. Calistri, M. A. Accola, G. Palu, and H. G. Gottlinger. 2000. A role for ubiquitin ligase recruitment in retrovirus release. Proc. Natl. Acad. Sci. USA 97:13063-13068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Suzuki, Y., Y. Nakabayashi, and R. Takahashi. 2001. Ubiquitin-protein ligase activity of X-linked inhibitor of apoptosis protein promotes proteasomal degradation of caspase-3 and enhances its anti-apoptotic effect in Fas-induced cell death. Proc. Natl. Acad. Sci. USA 98:8662-8667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Takai, R., N. Matsuda, A. Nakano, K. Hasegawa, C. Akimoto, N. Shibuya, and E. Minami. 2002. EL5, a rice N-acetylchitooligosaccharide elicitor-responsive RING-H2 finger protein, is a ubiquitin ligase which functions in vitro in co-operation with an elicitor-responsive ubiquitin-conjugating enzyme, OsUBC5b. Plant J. 30:447-455. [DOI] [PubMed] [Google Scholar]
- 63.Theilmann, D. A., and S. Stewart. 1992. Molecular analysis of the trans-activating IE-2 gene of Orgyia pseudotsugata multicapsid nuclear polyhedrosis virus. Virology 187:84-96. [DOI] [PubMed] [Google Scholar]
- 64.Van Sant, C., R. Hagglund, P. Lopez, and B. Roizman. 2001. The infected cell protein 0 of herpes simplex virus 1 dynamically interacts with proteasomes, binds and activates the cdc34 E2 ubiquitin-conjugating enzyme, and possesses in vitro E3 ubiquitin ligase activity. Proc. Natl. Acad. Sci. USA 98:8815-8820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wilkinson, K. D., M. K. Urban, and A. L. Haas. 1980. Ubiquitin is the ATP-dependent proteolysis factor I of rabbit reticulocytes. J. Biol. Chem. 255:7529-7532. [PubMed] [Google Scholar]
- 66.Yoo, S., and L. A. Guarino. 1994. The Autographa californica nuclear polyhedrosis virus ie2 gene encodes a transcriptional regulator. Virology 202:746-753. [DOI] [PubMed] [Google Scholar]
- 67.Zheng, N., P. Wang, P. D. Jeffrey, and N. P. Pavletich. 2000. Structure of a c-Cbl-UbcH7 complex: RING domain function in ubiquitin-protein ligases. Cell 102:533-539. [DOI] [PubMed] [Google Scholar]