Abstract
Cellular CCAAT/enhancer binding protein α (C/EBPα) promotes cellular differentiation and has antimitotic activities involving cell cycle arrest at G1/S through stabilization of p21CIP-1/WAF1 and through transcriptional activation of the p21 promoter. The Epstein-Barr virus lytic-cycle transactivator protein ZTA is known to arrest the host cell cycle at G1/S via a p53-independent p21 pathway, but the detailed molecular mechanisms involved have not been defined. To further evaluate the role of ZTA in cell cycle arrest, we constructed a recombinant adenovirus vector expressing ZTA (Ad-ZTA), whose level of expression at a low multiplicity of infection in normal human diploid fibroblast (HF) cells was lower than or equal to the physiological level seen in Akata cells lytically induced by EBV (EBV-Akata cells). Fluorescence-activated cell sorting analysis of HF cells infected with Ad-ZTA confirmed that G1/S cell cycle arrest occurred in the majority of ZTA-positive cells, but not with an adenovirus vector expressing green fluorescent protein. Double-label immunofluorescence assays (IFA) performed with Ad-ZTA-infected HF cells revealed that only ZTA-positive cells induced the expression of both endogenous C/EBPα and p21 and blocked the progression into S phase, as detected by a lack of incorporation of bromodeoxyuridine. The stimulation of endogenous ZTA protein expression either through treatment with tetradecanoyl phorbol acetate in D98/HR1 cells or through B-cell receptor cross-linking with anti-immunoglobulin G antibody in EBV-Akata cells also coincided with the induction of both C/EBPα and p21 and their mRNAs, as assayed by Northern blot, Western blot, and IFA experiments. Mechanistically, the ZTA protein proved to directly interact with C/EBPα by coimmunoprecipitation in EBV-Akata cells and with DNA-bound C/EBPα in electrophoretic mobility shift assay experiments, and the in vitro interaction domain encompassed the basic leucine zipper domain of ZTA. ZTA also specifically protected C/EBPα from degradation in a protein stability assay with a non-EBV-induced Akata cell proteasome extract. Furthermore, both C/EBPα and ZTA were found to specifically associate with the C/EBPα promoter in chromatin immunoprecipitation assays, but the interaction with ZTA appeared to be mediated by C/EBPα because it was abolished by clearing with anti-C/EBPα antibody. ZTA did not bind to or activate the C/EBPα promoter directly but cooperatively enhanced the positive autoregulation of the C/EBPα promoter by cotransfected C/EBPα in transient luciferase reporter gene assays with Vero and HeLa cells as well as with DG75 B lymphocytes. Similarly, ZTA alone had little effect on the p21 promoter in transient reporter gene assays, but in the presence of cotransfected C/EBPα, ZTA enhanced the level of C/EBPα activation. This effect proved to require a previously unrecognized region in the proximal p21 promoter that contains three high-affinity C/EBPα binding sites. Finally, in C/EBPα-deficient mouse embryonic fibroblasts (MEF), Ad-ZTA was unable to induce either p21 or G1 arrest, whereas it was able to induce both in wild-type MEF. Overall, we conclude that C/EBPα is essential for at least one pathway of ZTA-induced G1 arrest during EBV lytic-cycle DNA replication and that this process involves a physical piggyback interaction between ZTA and C/EBPα leading to greatly enhanced C/EBPα and p21 levels through both transcriptional and posttranslational mechanisms.
In cytomegalovirus, herpes simplex virus, Epstein-Barr virus (EBV), and Kaposi's sarcoma-associated herpesvirus (KSHV) infections, viral DNA replication appears to take place only in G1-arrested host cells (21, 53). The cell cycle arrest may be imposed by herpesviruses during viral lytic-cycle DNA replication to prevent competition with host cell DNA synthesis for limited free nucleotides and to provide nuclear space for progeny viral DNA storage. Herpesviruses in general encode many of their own viral DNA replication proteins and nucleotide synthesis enzymes, thus partially obviating the need for cellular S-phase-associated replication machinery (54, 59). These virus-encoded proteins always include six core DNA replication proteins (DNA polymerase, polymerase processivity factor, helicase, primase, primase-associated factor, and single-stranded DNA binding protein), together with an origin binding initiator protein, and may also include enzymes such as thymidylate synthase, thymidine kinase, ribonucleotide reductase, uracil DNA glycosylase, and dihydrofolate reductase.
EBV is a human gamma-1 herpesvirus that is associated with several B-cell and epithelial cell malignancies, including the endemic form of Burkitt's lymphoma, posttransplant lymphoproliferative diseases, AIDS-associated lymphomas, Hodgkin's disease, and nasopharyngeal carcinoma (29, 39, 55). During the EBV lytic cycle in vivo, viral DNA replication preferentially takes place in the growth-arrested and more differentiated layers of the oral epithelium (1, 13, 56). The introduction of the EBV lytic-cycle transactivator protein ZTA (also known as BZLF1 or ZEBRA) is known to induce G0/G1 cell cycle arrest in EBV-negative mammalian cell lines (5, 21). EBV ZTA has been extensively studied as a DNA binding basic leucine zipper (bZIP) family transcription factor (6, 15, 20, 33, 34) that triggers the EBV lytic cycle at both the transcriptional and the DNA replication levels (8, 10, 15, 17-19, 26, 28, 33, 34, 57).
Although some reports have suggested that the cell cycle arrest observed during induction of the EBV lytic cycle is the result of lytic-cycle-inducing agents (27, 42), Cayrol and Flemington (5) showed that ZTA protein stably expressed in tetracycline-inducible HeLa cells in the absence of any lytic-cycle-inducing agents still induces G1 arrest. Recently, Mauser et al. (38) found that ZTA alone expressed from a defective adenovirus induces G1 arrest, although at a high multiplicity of infection (MOI) it also appears to induce G2/M arrest (38). Studies to date on the mechanism of ZTA-mediated G1 arrest have suggested that ZTA may interact at multiple distinct points with the cell cycle regulatory machinery and is associated with higher levels of p53, p27, and p21CIP-1/WAF1 expression during the lytic cycle (38, 41). ZTA also inhibits the expression of c-MYC, an important factor in EBV latency and oncogenesis (42). However, Cayrol and Flemington (5) found little change in p53 and p27 mRNA expression, implying that a posttranscriptional mechanism may be used by ZTA to increase the levels of these proteins. In contrast, ZTA expression results in the accumulation of both p21 protein and p21 mRNA, suggesting that ZTA mediates p21 accumulation through an unknown transcriptional mechanism (5). Mutagenesis of ZTA showed that the C-terminal basic DNA binding domain and bZIP dimerization domain are essential for ZTA induction of p21, whereas the N-terminal region of ZTA is needed in addition for p53 and p27 stabilization (4, 41). Furthermore, ZTA can readily induce the expression of p21 and growth arrest in p53-deficient SAOS-2 cells, implying that p53 is not necessary as an upstream regulator of p21 expression in ZTA-mediated growth arrest (41), especially since the p53 gene is often mutated in a majority of Burkitt's lymphoma cells (52). Therefore, the precise pathway that links ZTA and p21 transcriptional induction has remained elusive.
Recently, Wu et al. found that a cellular protein, CCAAT/enhancer binding protein α (C/EBPα), binds to the KSHV-encoded replication-associated protein (RAP, or K8), a distant evolutionary homologue of ZTA, and mediates RAP-induced cell cycle arrest in both cells lytically induced by KSHV and adenovirus RAP-infected cells (53). C/EBPα also has been found to physically interact with EBV ZTA, and in the present study, we investigated the possible role of C/EBPα in ZTA-induced growth arrest.
C/EBPα belongs to a family of cellular bZIP DNA binding nuclear transcription factors that includes c-JUN, c-FOS, and CREB (30) and that is known to be associated with growth arrest during adipocyte, granulocyte, and neutrophil differentiation (11, 31, 51, 58). The C/EBPα gene encodes two predominant isoforms, a 42-kDa full-length form and a 30-kDa truncated form (2, 35, 40). C/EBPα positively autoregulates its own gene promoter (9, 45) and is thought to control differentiation and G1 cell cycle arrest through three reported mechanisms: (i) up-regulation of the expression of p21CIP-1, an inhibitor of the cell cycle kinases Cdk2, Cdk4, and Cdk6 (46, 47); (ii) inhibition of E2F transcription (44); and (iii) inhibition of Cdk2 and Cdk4 (23, 48). We show here that EBV ZTA enhances C/EBPα-mediated up-regulation of both the C/EBPα and the p21 gene promoters and physically interacts with C/EBPα in vitro via the bZIP region. Furthermore, C/EBPα expression is greatly increased in ZTA-expressing cells during the EBV lytic cycle, and ZTA-induced p21 induction and cell cycle arrest were found to be mediated by and dependent on C/EBPα. Finally, we identified a new region on the p21 promoter that binds to C/EBPα and mediates induction.
MATERIALS AND METHODS
Cell lines and plasmids.
Human EBV-positive Burkitt's lymphoma cells (Akata cells), herpesvirus-negative DG75 B cells, and human primary effusion lymphoma (PEL) cells (BCBL-1 and JSC-1) (3), were grown in RPMI 1640 medium containing 5% fetal bovine serum in a humidified 5% CO2 incubator at 37oC. Human primary foreskin fibroblast (HF) cells, EBV-positive epithelial cell-B-cell hybrid cell line D98/HR1, Vero cells, HeLa cells, C/EBPα-deficient (C/EBPα−/−) mouse embryonic fibroblast (MEF) cells, and C/EBPα-expressing (C/EBPα+/+) MEF cells (46) were grown in Dulbecco's modified Eagle's medium (DMEM) containing 10% fetal bovine serum. Fusion proteins used included wild-type GST-ZTA(1-245) (a fusion of glutathione S-transferase [GST] with ZTA residues 1 to 245 [ZTA(1-245)] (in plasmid pDH237) and deletion mutants GST-Z(Δ24-86) (pGL14), GST-Z(Δ94-140) (pGL15), GST-Z(Δ4-140) (pGL16), GST-Z(1-133) (pDH245), and GST-Z(27-133) (pDH279) (32).
Plasmid pRTS21 carries EBV ZTA genomic DNA encoding simian virus 2 (SV2) ZTA in a pSG5 background (Stratagene) (43). Plasmid pFYW03 carries ZTA cDNA in the pSG5 background (54). The construction of plasmid pSG5-Z-DM (carrying Zdbm1; a gift from Erik K. Flemington, Tulane University) is described elsewhere (20a). Plasmid pcDNA-C/EBPα (pSEW-CFL) is a cDNA expression vector for C/EBPα in a pcDNA background (50), and plasmid pC/EBPα-LUC (pSEW-CP1) contains the 500-bp proximal C/EBPα promoter region driving a luciferase reporter gene (C/EBPα-LUC) in a pGL3-Basic (Promega) background. Plasmid pYNC100 is a black beetle virus leader region (BBV)-enhanced T7 in vitro transcription-translation vector encoding ZTA cDNA (6). Plasmid pYNC172a (6) is a BBV-enhanced T7 in vitro transcription-translation vector encoding full-length C/EBPα. Plasmid pHC85 is a BBV-enhanced T7 in vitro transcription-translation vector encoding full-length CHOP-10 protein. Plasmid pWWP (a gift from B. Vogelstein, School of Medicine, The Johns Hopkins University) is a luciferase reporter containing 2.4 kb of the p21CIP-1/WAF1 promoter fragment [p21(WT)-LUC]. Deletion derivatives containing positions −994 to +1, −779 to +1, and −224 to +1 of the p21CIP-1/WAF1 promoter and referred to as p21-M1-LUC (pFYW35), p21-M2-LUC (pFYW36), and p21-M3-LUC (pFYW37), respectively, were generated by PCR amplification with primers LGH4280 (5′-GATCCGGGGTACCGCAAAAGTCCTGTGTTCCAA-3′) and LGH4281 (5′-TCGACTCCACACTCGTCGACGAGCTCGCCCTAG-3′) for p21-M1-LUC; LGH4240 (5′-GATCCGGGGTACCGTAGGGAGATTGGTTCAATG-3′) and LGH4281 for p21-M2-LUC; and LGH4282 (5′-GATCCGGGGTACCAAGTGCCCTCCTGCAGCAC-3′) and LGH4281 for p21-M3-LUC.
Adenovirus preparation and infection.
A BamHI fragment encompassing the full-length cDNA encoding the EBV ZTA protein (amino acids [aa] 1 to 245) from pFYW02 (54) was subcloned into the BamHI site of the recombinant transfer vector pAdLOX to make plasmid pFYW31. Vector pAdLOX, Ψ5 virus, and CRE8 cells (22) were generous gifts from S. Hardy (Somatix Therapy Corp.). Recombinant adenovirus was made by cotransfecting Ψ5 virus DNA with pFYW31 into CRE8 cells. Virus was passed sequentially through CRE8 cells four times. Viral lysates were harvested by freezing-thawing three times and cleared by centrifugation to generate the recombinant replication-defective (ΔE1A/E1B) adenovirus Ad-ZTA.
HF, C/EBPα−/− MEF, and C/EBPα+/+ MEF cells were seeded in two-well slide chambers for immunofluorescence assays (IFA) or in a T175 flask for flow cytometric analysis. The cells were washed with phosphate-buffered saline (PBS) and incubated for 1 h in serum-free DMEM containing adenovirus vectors at an MOI of 0.5. The medium was changed to DMEM containing 10% fetal bovine serum after 1 h, and viral expression was assayed after 24 to 48 h.
Indirect IFA.
Procedures for bromodeoxyuridine (BrdU) labeling, indirect IFA, and fluorescence microscopy were performed as described elsewhere (54). For IFA of adenovirus vector infections, BrdU pulse-labeling of HF and MEF cells was performed for 45 min at 30 h. Sheep anti-BrdU primary antibody (20-BS17; Fitzgerald Co., Concord, Mass.) was used to detect BrdU incorporation, and donkey or goat fluorescein isothiocyanate (FITC)- or rhodamine-conjugated immunoglobulin G (IgG) (Jackson Pharmaceuticals, West Grove, Pa.) secondary antibody was used to detect the primary antibody. Mounting solution with 4′,6′-diamidino-2-phenylindole (DAPI) (Vector Shield) was used to visualize cellular DNA. Primary antibodies included rabbit anti-ZTA antiserum (1:800) (18), mouse anti-ZTA monoclonal antibody (MAb) (Argene), mouse anti-human p21 MAb (sc-817; 1:200; Santa Cruz Biotechnology), mouse anti-mouse p21 MAb (sc-6246; 1:200; Santa Cruz Biotechnology), rabbit anti-C/EBPα and -C/EBPβ polyclonal antibodies (PAbs) (1:800) (gifts from M. D. Lane, School of Medicine, The Johns Hopkins University) (45).
EBV lytic-cycle induction, coimmunoprecipitation, and Western immunoblot analysis.
Induction of the EBV lytic cycle in D98/HR1 cells was done by incubating 105 cells with 20 ng of tetradecanoyl phorbol acetate (TPA)/ml for 48 h prior to fixation and IFA analyses. Induction of the EBV lytic phase in Akata cells was done with 500 ml of Akata cells (5 × 105 cells/ml). IgG (Cappel) was added to the cell culture to reach a final concentration of 50 μg/ml. For coimmunoprecipitation analysis, after 48 h of induction, the cells were pelleted at 800 rpm for 5 min and washed once with 10 ml of PBS at room temperature. The cells were pelleted again at 3,000 × g for 3 min and resuspended gently in 10 ml of hypotonic buffer A (10 mM HEPES [pH 7.9], 10 mM KCl, 0.1 mM EDTA, 0.1 mM EGTA, 1 mM dithiothreitol [DTT], 1 mM phenylmethylsulfonyl fluoride [PMSF]). The mixture was placed on ice for 15 min to swell cells. Then, NP-40 was added to reach a final concentration of 0.62%, and the mixture was vortexed vigorously for 10 s and pelleted at 3,500 rpm for 2 min at 4°C to obtain the nuclear pellet. The nuclear pellet was resuspended in 3 ml of cold immunoprecipitation buffer (50 mM Tris-HCl [pH 7.9], 50 mM NaCl, 0.1 mM EDTA, 1% glycerol, 0.2% NP-40, 1 μg of ethidium bromide/ml, 0.2 U of DNase I/μl, 0.2 μg of RNase A/μl, 1 mM DTT, 1 mM PMSF) and sonicated for 5 s. The mixture was pelleted at 13,000 rpm for 1 min at 4°C, and the clarified supernatant was collected as the nuclear extract. The nuclear extract was divided into three 1-ml aliquots and stored at −80°C. For coimmunoprecipitation, each nuclear extract aliquot was precleared with 5 μl of rabbit or goat preimmune serum and 100 μl of 50% protein A- and 50% protein G-Sepharose beads for 1 h. After removal of the protein A- and G-Sepharose beads from the extract, 3 μg of rabbit anti-ZTA PAb, 3 μg of goat anti-C/EBPα PAb, or 3 μg of goat preimmune serum was added to each nuclear extract aliquot and incubated at 4°C with gentle rocking for 2 h. Protein A- and G-Sepharose beads (100 μl) were added, and the mixture was incubated for 1 h at 4°C. The beads were washed five times at 3-min intervals with cold immunoprecipitation buffer and resuspended in 20 μl of sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) loading buffer. The mixture was boiled for 3 min, and the supernatant was fractionated by SDS-PAGE. A 10-μl aliquot from the original nuclear extract was also separated by SDS-PAGE.
For timed Akata cell lytic-cycle induction and protein detection by Western immunoblot analysis, 5 × 106 Akata cells were removed from a 50-ml culture containing IgG (50 μg/ml) at 0, 24, 48, and 72 h after IgG treatment. The proteasome inhibitor MG132 was added to Akata cell culture aliquots at a final concentration of 50 μM for 5 to 10 min prior to lysis. For detection of Ad-ZTA expression, 106 HF cells were seeded in a 10-mm dish and infected with Ad-ZTA (MOI, 0.5), and cell lysates were collected 30 h after infection. The total protein concentrations in the different cell lysates were measured with a bicinchoninic acid protein assay kit (Pierce Inc., Rockford, Ill.) according to the bicinchoninic acid protocol and normalized to equal concentrations with PBS. Cell lysates for Western immunoblot analyses were prepared as described elsewhere (60). Primary antibodies used included anti-ZTA MAb (1:1,000), anti-C/EBPα PAb (1:1,000), anti-p21 MAb (sc-817; 1:100), anti-p27 MAb (Ab-2; Oncogene) (1:500), and β-actin MAb (1:5,000; Sigma).
GST affinity assay.
GST and GST fusion proteins were purified from DNA plasmid-transformed Escherichia coli strain BL21 as described elsewhere (32). Samples of 30 μl of 50% Sepharose beads with attached GST fusion proteins were washed twice with EBC buffer (140 mM NaCl, 0.2% NP-40, 100 mM NaF, 200 μM NaO3VO4, 50 mM Tris-HCl [pH 8], 1 μg of ethidium bromide/ml, 0.2 μg of RNase A/ml, 0.2 U of DNase I/μl), resuspended in 400 μl of fresh EBC buffer containing 50 μg of bovine serum albumin (BSA)/μl (5% BSA), and gently rocked at 4°C for 1 h. Then, 10 μl of in vitro-translated [35S]methionine (Amersham)-labeled protein made with a TNT quick coupled transcription-translation system (Promega) was added to each sample, and the samples were gently rocked at 4°C for 3 h. The samples were subsequently centrifuged to pellet the Sepharose beads, and after removal of the supernatant, the beads were washed with 1 ml of cold NETN buffer (100 mM NaCl, 1 mM EDTA, 0.2% NP-40, 20 mM Tris-HCl [pH 8]) five times at 15-min intervals. After the final wash, the beads were resuspended in 20 μl of SDS loading buffer and boiled for 5 min. The mixture was separated by SDS-PAGE. After drying of the SDS-polyacrylamide gel, the [35S]methionine-labeled proteins were visualized by autoradiography with Kodak film.
Protein stability assay with proteasome extract.
The detailed protocol for the protein stability assay was modified from a previously published version (46). For the preparation of a cellular proteasome extract, 200 ml of Akata cells latently infected with EBV (5 × 105 cells/ml) was pelleted at 3,000 rpm for 5 min at room temperature. After the cells were washed with PBS, the cell pellet was resuspended in 1.5 ml of cold proteasome buffer (10 mM ATP, 20 mM Tris-HCl [pH 8], 5 mM DTT, 1 mM MgCl2, 0.1 mM EDTA, 1% glycerol) and incubated on ice for 45 min. The cells were lysed with a Dounce homogenizer and centrifuged at 2,000 × g for 10 min at 4°C. The supernatant containing cellular proteasomes was collected, brought to 150 mM NaCl, and stored in aliquots at −80°C for 2 weeks. For the protein stability assay, 50 μl of 35S-labeled protein was synthesized from the appropriate template with a TNT coupled in vitro transcription-translation kit (Promega); then, 1 μl of the labeled protein was diluted with 25 μl of proteasome extract, and 10 μl of the diluted protein was added to a 150-μl proteasome extract aliquot. The mixture was placed in a 37°C water bath, and 20-μl aliquots were removed after 0, 0.5, 1, 3, 6, or 24 h of incubation and then boiled for 5 min with 2× protein loading dye. The samples were separated by SDS-PAGE. To reduce the 35S background, the gel was treated with fixing solution (50% methanol, 40% distilled water, 10% acetic acid) for 15 min and rehydrated with hydration solution (5% methanol, 1% acetic acid, 94% distilled water) for 20 min. After drying of the SDS-polyacrylamide gel, the [35S]methionine-labeled proteins were visualized by autoradiography with Kodak film.
DNA transfection and luciferase assays.
Lipofectamine (Invitrogen) DNA transfection was performed according to Invitrogen protocols. Vero or HeLa cells were split at 5 × 105 cells/ml, transfected with a total of 2.5 μg of DNA (adjusted with vector plasmid pSG5 as a carrier) per well in a six-well plate (0.2 μg of luciferase reporter gene, 2 μg of C/EBPα, and/or 0.1 μg of ZTA), and harvested 48 h posttransfection. DG75 cells were transfected by using the electroporation method as described elsewhere (49). A total of 107 cells were mixed with 5 to 10 μg of total plasmid DNA (2 μg of reporter gene, 10 μg of C/EBPα, and/or 1 μg of ZTA) in 0.5 ml of RPMI 1640 medium and electroporated at 300 V and 950 μF by using a GenePulser (Bio-Rad, Hercules, Calif.). A luciferase assay system (Promega) was used for the luciferase assay, and all procedures were done according to the Promega protocol. Luciferase activity was measured for 10 s with a Lumat LB9501 luminometer (Berthold Systems, Inc.) after injection of 100 μl of 1 mM luciferin (Invitrogen). For Western immunoblot detection of endogenous C/EBPα in Vero cells, 0, 0.1, or 1.0 μg of ZTA expression plasmid (adjusted with pSG5 vector DNA as a carrier) was transfected into Vero cells, and cells were harvested 48 h posttransfection.
Chromatin immunoprecipitation (ChIP) assays.
EBV-positive Akata cells (5 × 108 cells) were induced by treatment with anti-IgG antiserum for 40 h. Harvesting and chromatin extraction were performed as described elsewhere (50). For immunoprecipitation, 1 μg of anti-CHOP-10 MAb (Santa Cruz), C/EBPα PAb, or ZTA MAb was used with 500 μl of extract. Depletion of endogenous proteins with specific antibodies in the extracts was done by incubating the appropriate antibodies attached to Sepharose beads with the extracts for 24 h at 4°C and subsequently collecting the antibody-cleared supernatant for a new round of PCR assays. For detection of the immunoprecipitated C/EBPα promoter region, two primers, LGH4273 (5′-ACTTCGG TACCGCTACCGACCACGTGGGCG-3′) and LGH4275 (5′-GTGAACTCGAGCACCTCCGGGTCGCGAATGG-3′), specific for a 280-bp region in the C/EBPα promoter that encompasses the C/EBP binding site, were used for PCR amplification. For detection of the C/EBPα coding region, two primers, LGH4268 (5′-ATTCACTCGAGGATCCCCATGAGCGCGCTGAAGGGGCTG-3′) and LGH4270 (5′-GTGAACTCGAGGTACCTCACGCGCAGTTGCCCAT-3′), specific for the C/EBPα coding region (aa 251 to 358), were used as negative controls to detect a nonpromoter region. The PCR products were analyzed on a 2.5% agarose gel, and quantitation of PCR products was conducted by using a MultiImage light cabinet (Alpha-Innotech Corp.) with the accompanying FluorChem (version 1.02) software.
Extraction of total RNA and Northern blot assay.
Akata cells were induced by the addition of anti-IgG antiserum, and total RNA was isolated at different times by using Trizol reagent (GIBCO BRL) according to the manufacturer's protocol. RNA samples (50 μg) were size fractionated on a 1.2% formaldehyde-agarose gel in morpholinopropanesulfonic acid (MOPS) running buffer and transferred to a Nytran nylon membrane (Schleicher & Schuell). For the generation of Northern blot probes, a DNA product of the C/EBPα bZIP domain (aa 251 to 358) was obtained by PCR amplification with primers LGH4268 and LGH4270, an 800-bp ZTA DNA fragment was obtained by BamHI cleavage of plasmid pFYW02, a full-length p21 cDNA fragment was obtained by BamHI/EcoRI cleavage of plasmid pFYW45, and a fragment containing a 286-bp glyceraldehyde-3-phosphate dehydrogenase (GAPDH) cDNA region was used as a control. The probes were labeled by using a Prime-a-Gene labeling system (Promega) and a 50-μl mixture containing labeling buffer, 60 μM deoxynucleoside triphosphates (dATP, dTTP, and dGTP), 25 ng of denatured DNA template, 20 μg of BSA, 333 nM [α-32P]dCTP (3,000 Ci/mmol), and 5 U of Klenow DNA polymerase. After incubation at room temperature for 60 min, the probes were purified with Sephadex G-50, denatured, and added to the membrane in a hybridization mixture containing 50% formamide, 5× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate), Denhardt's solution, 20 mM NaPO4 (pH 6.5), 10% dextran sulfate, and 100 μg of calf thymus DNA/ml. Hybridization was carried out overnight at 42°C, and the membrane was washed twice in 2× SSC-0.1% SDS at room temperature and twice in 0.5× SSC-0.1% SDS at 50°C before autoradiography. As an RNA loading control, the membrane was reprobed with GAPDH cDNA.
Flow cytometric analysis.
Primary HF cells were seeded in T175 flasks at 2 × 106 cells/ml. Adenovirus infection was carried out at 40% confluence and an MOI of 0.5, and the cells were harvested 24 to 48 h postinfection. After trypsinization for 2 min, the cells were washed once with PBS and resuspended in 2% paraformaldehyde for 20 min at room temperature. Cells were gently washed once in 3% BSA-PBS for 3 min and resuspended in permeabilization solution (0.5% saponin, 1% BSA, PBS) at room temperature for 30 min. After one wash with 1% BSA-PBS, the cells were resuspended in 5% goat serum and blocked for 30 min at room temperature. Cells were resuspended in 1% BSA-PBS containing mouse anti-ZTA MAb and incubated for 1 h at 37°C. After two washes with 1% BSA-PBS for 3 min each, the cells were resuspended in 1% BSA-PBS containing secondary anti-mouse IgG conjugated to FITC (Jackson Pharmaceuticals) and were incubated for 30 min at 37°C. After two washes with 1% BSA-PBS, the cells were fixed again in 2% paraformaldehyde for 20 min at room temperature. After one wash with 1% BSA-PBS, the cells were completely resuspended in Hoechst solution (10 ng of Hoechst reagent/ml, 0.5% NP-40, 3.5% formaldehyde, PBS). FITC channel-gated flow cytometric analysis was performed with a Becton Dickinson BDLSR FACScan. Each analysis was concluded upon the gated acquisition of 10,000 FITC-positive events per sample, and the results were analyzed with the Cell Quest program.
EMSAs.
The in vitro-translated protein used in these studies was made with a TNT quick coupled transcription-translation system by adding 2.0 μg of pBBV-C/EBPα or pBBV-ZTA plasmid to a mixture containing 40 μl of TNT quick master mix, 1 μl of RNase inhibitor, and 2 μl of cold methionine. The mixture was incubated at 30°C for 90 min and then stored at −80°C. For verification of protein expression, 2 μl of [35S]methionione (Amersham) was added in place of nonradioactive methionine, and the reaction mixture was separated by SDS-PAGE and detected by autoradiography. For electrophoretic mobility shift assays (EMSAs), 2 μl of in vitro-translated protein was used for each reaction and added to a 19-μl reaction mixture containing binding buffer (10 mM HEPES [pH 7.5], 50 mM KCl, 1 mM EDTA, 1 mM DTT, 1 mM PMSF, 1% Triton X-100, 5% glycerol) and 2 μg of poly-dI/dC (Sigma). After incubation for 5 min at room temperature, 1 μl of 32P-labeled dCTP oligonucleotides (50,000 cpm) was added to the mixture and incubated for 1 h. For supershift experiments, the reaction mixture was incubated at room temperature for 30 min, and 0.5 μl of anti-C/EBPα PAb or 1 μl of anti-ZTA MAb was added to the mixture and incubated for 30 min.
Oligonucleotides were synthesized commercially (Invitrogen). LGH3973 (5-′-GATCGGTTGATTGTGACTATTTGTGAAACAATAATGA-3′) and LGH3974 (5′-GATCTCATTATTGTTTCACAAATAGTCACAATCAACC-3′) were annealed to form the control KSHV RAP promoter RTA response element (RRE) probe, which contained a known C/EBP binding site (50). LGH4228 (5′-GATCGTAGGGAGATTGGTTCAATGTCCAATTCTTCTG-3′) and LGH4229 (5′-GATCCAGAAGAATTGGACATTGAACCAATCTCCCTAC-3′) were annealed to form the p21P-1 probe. LGH4230 (5′-GATCGCCCTTTTTTGGTAGTCTCTCCAATTCCCTCCT-3′) and LGH4231 (5′-GATCAGGAGGGAATTGGAGAGACTACCAAAAAAGGGC-3′) were annealed to form the p21P-2 probe. LGH4232 (5′-GATCGAAGCATGTGACAATCAACAACTTTGTATACTT-3′) and LGH4233 (5′-GATCAAGTATACAAAGTTGTTGATTGTCACATGCTTC-3′) were annealed to form the p21P-3 probe. LGH4234 (5′-GATCAAAAATCATTCTGGCCTCAAGATGCTTTGTTGG-3′) and LGH4235 (5′-GATCCCAACAAAGCATCTTGAGGCCAGAATGATTTTT-3′) were annealed to form the p21P-4 probe. LGH4236 (5′-GATCGCTGCATTGGGTAAATCCTTGCCTGCCAGAGTG-3′) and LGH4237 (5′-GATCCACTCTGGCAGGCAAGGATTTACCCAATGCAGC-3′) were annealed to form the p21P-5 probe. LGH4238 (5′-GATCCCTGGAGAGTGCCAACTCATTCTCCAAGTAAAA-3′) and LGH4239 (5′-GATCTTTTACTTGGAGAATGAGTTGGCACTCTCCAGG-3′) were annealed to form the p21P-6 probe. LGH4242 (5′-GATCCAATCCATCCTCTGCAATTTTTTAAAAGCAAAA-3′) and LGH4243 (5′-GATCTTTTGCTTTTAAAAAATTGCAGAGGATGGATTG-3′) were annealed to form the p21P-GRE probe. LGH4276 (5′-GATCGAGGCGGTGGGCGTTGCGCCGCGGCCTGCCTGG-3′) and LGH4277 (5′-GATCCCAGGCAGGCCGCGGCGCAACGCCCACCGCCTC-3′ were annealed to form the C/EBPα-P probe. The preparation of 32P-labeled oligonucleotides and other procedures involved in the preparation of the electrophoretic gel for EMSAs were done according to protocols described elsewhere (7). Binding efficiency was quantitated by using an Instant Imager (Packard Instrument Company) and its accompanying computer software.
RESULTS
Adenovirus vector-expressed ZTA inhibits G1-to-S cell cycle progression in infected HF cells.
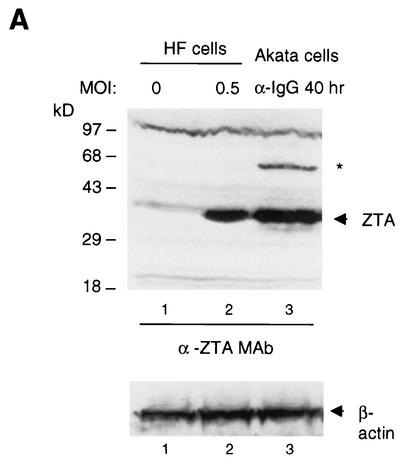
ZTA was previously reported to induce G1 cell cycle arrest (5). To examine the mechanisms underlying this effect, we constructed a recombinant ΔE1A/E1B adenovirus vector expressing either the ZTA protein (Ad-ZTA) or GFP as a negative control (Ad-GFP). To avoid complications seen in previous studies with abnormal tumor cells, p53 mutations, p53 destabilization by human papillomavirus genes, or high adenovirus MOIs, we used HF cells. These cells were infected with the adenovirus vectors at a low MOI (0.5) to prevent cytotoxicity and to permit the simultaneous correlation of ZTA-positive and ZTA-negative cells in the same culture. Prior to fluorescence-activated cell sorting (FACS) analysis, we compared the protein expression level of Ad-ZTA with the physiological ZTA level obtained in a lytically induced Akata cell culture with almost the same percentage of ZTA-positive cells. After normalizing the total protein concentrations, we collected an HF cell lysate 30 h after Ad-ZTA infection (25% ZTA-positive cells) and compared it with an Akata cell lysate collected 48 h after anti-IgG antibody treatment (15% ZTA-positive cells). Under these conditions, the Ad-ZTA protein was not overexpressed; rather, the level was lower than or equal to the ZTA expression level observed in Akata cells, as assayed by both Western immunoblotting (Fig. 1A) and determination of the relative IFA signal intensities (data not shown).
FIG. 1.
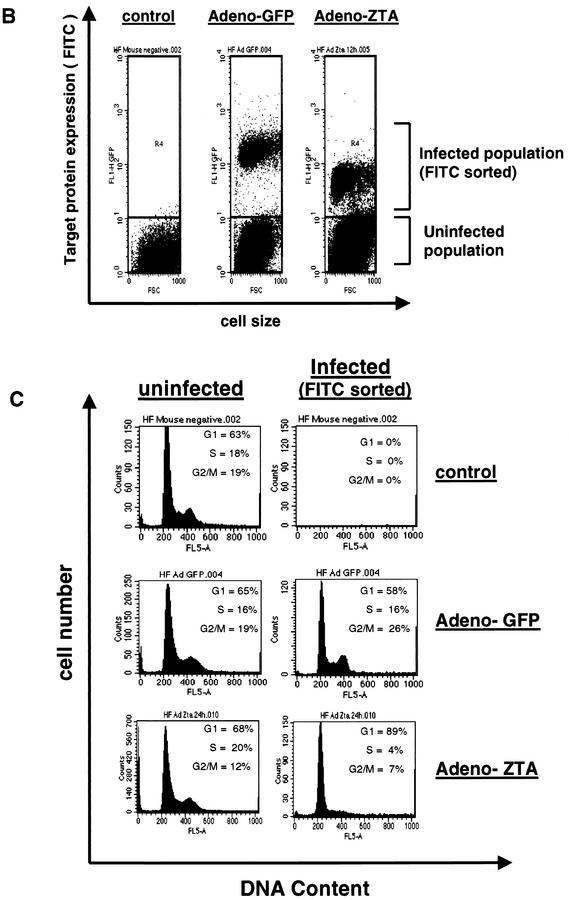
Analytical FACS demonstrating that EBV ZTA causes G1/S cell cycle arrest in diploid HF cells. (A) Western immunoblot detection of ZTA protein in Ad-ZTA (MOI, 0.5)-infected HF cells and Akata cells treated with anti-IgG (α-IgG) antiserum for 40 h showed that ZTA expression mediated by Ad-ZTA is similar to physiological levels. Detection of β-actin showed equal loading of samples. The asterisk indicates an unidentified band representing either dimeric or SUMO-modified ZTA. (B) Fluorescence-positive cells in an Ad-ZTA-infected HF cell population after labeling with anti-ZTA (α-ZTA) antibody or a control Ad-GFP-infected cell population were sorted from uninfected populations through an FITC-gated channel based on the difference in their fluorescence signals. (C) Flow cytometric analysis showed that EBV ZTA-expressing cells (FITC channel gated) are predominantly arrested in G1 in comparison to either uninfected cells from the same population or GFP-positive cells from the Ad-GFP-infected population.
Subsequently, we performed cell cycle analysis of Ad-ZTA-infected HF cells under the same low-MOI conditions by analytical FACS with anti-ZTA MAb to determine whether HF cells expressing ZTA were indeed cell cycle arrested. For the gated fluorescence-positive cell population expressing wild-type ZTA, dramatic G1/S cell cycle arrest (89% G1, 4% S, 7% G2/M) was observed, in contrast to the normal cell cycle profile of the nonexpressing cell population (68% G1, 20% S, 12% G2/M) from the same culture (Fig. 1C). Furthermore, in contrast, control cells expressing Ad-GFP exhibited a normal cell cycle profile, with distinct G1 (58%), S (16%), and G2/M (26%) peaks (Fig. 1C). These results showed that the adenovirus vector did not significantly affect the cell cycle and further confirmed that ZTA alone can inhibit G1/S cell cycle progression in infected (not just transfected) normal diploid human cells.
Expression of C/EBPα and its interaction with ZTA.
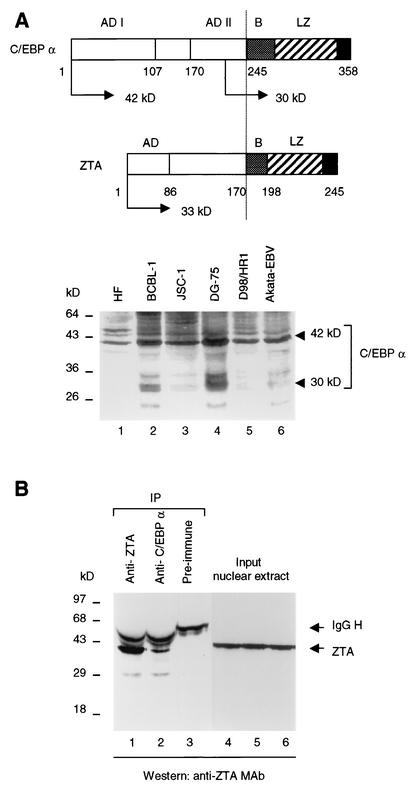
Like ZTA, C/EBPα is a member of the bZIP family of DNA binding transcriptional factors. It is expressed either as a full-length 42-kDa form that has antimitotic functions or as a nonfunctional N-terminally truncated 30-kDa form (Fig. 2A, upper panel). To determine whether the functional form of C/EBPα is present in B-lymphoblastoid and other relevant cell lines, endogenous C/EBPα expression was examined in two Burkitt's lymphoma cell lines, DG75 (EBV negative) and Akata (EBV positive), in the PEL cell lines BCBL-1 (KSHV positive) and JSC-1 (KSHV and EBV positive), and in the EBV-positive epithelial cell-B-cell hybrid cell line D98/HR1. After gel electrophoresis of total protein extracts, Western immunoblotting with anti-C/EBPα PAb revealed the presence of low levels of the 42-kDa active antimitotic form of C/EBPα as well as less of the 30-kDa N-terminally truncated form in each of these cell lines (Fig. 2A, lower panel).
FIG. 2.
Expression of C/EBPα in human lymphoma cell lines and evidence obtained by immunoprecipitation for interactions between C/EBPα and EBV ZTA. (A) (Upper panel) Diagrams of C/EBPα and ZTA. AD, activation domain; B, basic region; LZ, leucine zipper region. (Lower panel) Western immunoblot analysis showing the levels of endogenous C/EBPα in primary HF cells, in different lymphoma cell lines (BCBL-1, JSC-1, DG75, and Akata), and in the EBV-positive epithelial cell-B-cell hybrid cell line D98/HR1. (B) Coimmunoprecipitation of EBV ZTA with C/EBPα in EBV-positive Akata cells undergoing EBV lytic-cycle progression after IgG cross-linking. Lanes 1 to 3, recovery of endogenous 35-kDa ZTA protein (detected by Western immunoblotting with anti-ZTA PAb) from immunoprecipitates (IP) with either anti-ZTA MAb or anti-C/EBPα PAb but not with preimmune serum. Lanes 4 to 6, Western blot showing equal amounts of ZTA protein in input nuclear extracts. H, heavy chain.
Because C/EBPα was recently found to interact with KSHV RAP (K8) (Wu et al., unpublished), we examined whether C/EBPα might also interact with ZTA. We initially examined this interaction by performing a coimmunoprecipitation experiment with EBV-positive Akata cells after anti-IgG cross-linking to induce the lytic cycle. A nuclear extract of the Akata cells was prepared 48 h after induction, and immunoprecipitation was performed with goat anti-C/EBPα PAb. The immunoprecipitate was analyzed by SDS-PAGE and Western blotting, which detected coprecipitating ZTA protein (Fig. 2B, lane 2). A negative control performed with preimmune goat serum yielded no ZTA signal (Fig. 2B, lane 3). ZTA protein was confirmed to be present at equal levels in all of the input nuclear extracts (Fig. 2B, lanes 4 to 6). Because both C/EBPα and ZTA can bind to DNA, we also included a DNase I incubation and added ethidium bromide to the immunoprecipitation reaction to destroy double-stranded DNA and prevent nonspecific protein-protein linking via DNA binding. Therefore, ZTA binds strongly to C/EBPα and can be immunoprecipitated with endogenous C/EBPα from lytically infected Akata cells.
Ad-ZTA causes G1 arrest through the induction of both C/EBPα and p21.
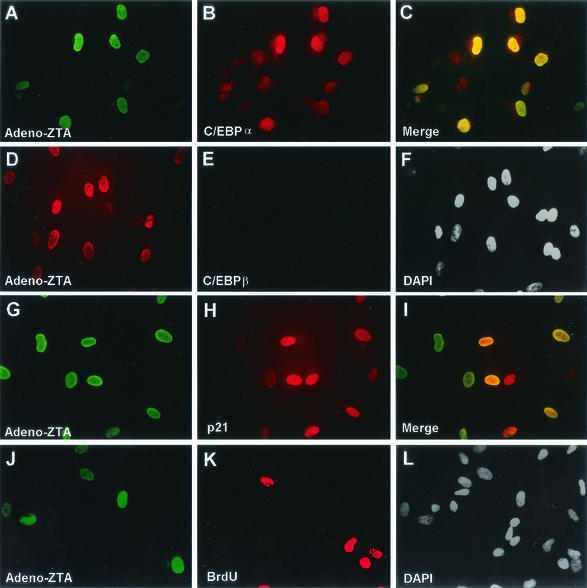
C/EBPα is known to cause G1 cell cycle arrest by one or more mechanisms involving stabilization and up-regulation of p21, inhibition of E2F DNA binding, and direct inhibition of Cdk2 and Cdk4 (44, 46-48). To attempt to establish whether ZTA-induced G1 cell cycle arrest is mediated by C/EBPα, we performed IFA with adenovirus vector-infected HF cell populations and also measured S phase by detection of pulse-labeled BrdU incorporated into newly synthesized DNA in the same cells. Less than 1% of uninfected HF cells were found positive by IFA for either C/EBPα or p21, as detected with a specific MAb, and no expression of ZTA was detectable in uninfected cells. However, after infection with wild-type Ad-ZTA at a low MOI, many cells expressed highly induced levels of C/EBPα and p21. Among the HF cells positive for wild-type ZTA, 78% were also positive for endogenous C/EBPα (Fig. 3A to C). In contrast, endogenous C/EBPβ expression was not induced by Ad-ZTA (Fig. 3D to F). Among the ZTA-positive HF cells, 86% were also positive for highly induced levels of p21 (Fig. 3G to I). In addition, after BrdU pulse-labeling of the Ad-ZTA-infected cell culture, 24% of the cells expressed ZTA and 27% of the cells were positive for BrdU (S phase), but <1% of the cells were positive for both ZTA and BrdU; these results suggested that S-phase progression was blocked only in cells expressing ZTA (Fig. 3J to L). Overall, these results implied that ZTA inhibits S-phase cell cycle progression directly though the induction of either or both C/EBPα and p21.
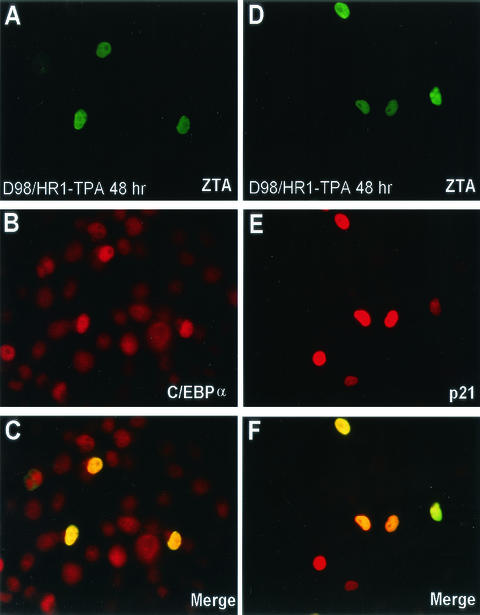
FIG. 3.
A recombinant adenovirus vector expressing ZTA (Ad-ZTA) induces both C/EBPα expression and p21 expression in infected HF cells. (A to C) Double-label IFA showing expression of the ZTA protein (A; FITC, green) and enhanced expression of C/EBPα (B; rhodamine, red) in the same cell population. The merge of the two frames (C) shows the up-regulation of C/EBPα only in ZTA-expressing cells. (D to F) Double-label IFA showing ZTA protein expression (D; red) and the absence of enhanced C/EBPβ expression (E; green) in the same cell population. The merge of the two frames (F) shows that ZTA did not induce C/EBPβ expression. (G to I) Double-label IFA showing expression of the ZTA protein (G; green) and p21 induction (H; red) in the same infected cell population. The merge of the two frames (I) shows the induction of p21 protein only in ZTA-positive cells. (J to L) Double-label IFA showing expression of the ZTA protein (J; green) and BrdU incorporation (K; red) in the same Ad-ZTA-infected cell population. DAPI nuclear staining of the total cell population is shown in panel L. ZTA-positive cells and BrdU-positive cells are mutually exclusive, indicating that ZTA-positive cells do not undergo S phase.
Expression of endogenous p21 and C/EBPα is increased after lytic-cycle induction in both D98/HR1 and EBV-positive Akata cells.
To examine the situation in EBV-infected cells, we used the D98/HR1 cell line, in which adherent cells can be induced by TPA to undergo productive EBV lytic-cycle expression. The ZTA protein was rarely detected in untreated D98/HR1 cells by IFA (0.7%), but its expression was induced in 11% of the cells by 48 h after TPA treatment (Fig. 4). Furthermore, the fraction of D98/HR1 cells expressing a high level of nuclear C/EBPα increased from 2% before lytic-cycle induction to 12% after TPA treatment (Fig. 4B), coinciding with the dramatic increase in the level of ZTA-positive cells (Fig. 4A and D). Importantly, 96% of the ZTA-positive D98/HR-1 cells proved to also express C/EBPα (Fig. 4B), confirming that most lytically infected cells displayed increased C/EBPα expression. Similarly, 71% of the C/EBPα-positive cells were also ZTA positive (Fig. 4B).
FIG. 4.
Induction of C/EBPα and p21 in TPA-induced EBV-positive D98/HR1 cells. (A to C) Double-label IFA showing expression of the ZTA protein (A; green) in cells undergoing the lytic cycle and induction of C/EBPα (B; red) in the same cells. The merge of the two frames (C) shows that enhanced ZTA expression can induce C/EBPα expression. (D to F) Double-label IFA showing the expression of ZTA (D; green) and the expression of p21 (E; red) in the same cells. The merge of the two frames (F) shows that ZTA expression coincides with p21 expression in the same cells.
Increased expression of both ZTA and C/EBPα also coincided with increased overall expression of p21 in TPA-induced D98/HR1 cells (Fig. 4E). By IFA, less than 1.0% of D98/HR1 cells were found to express detectable p21 protein prior to lytic-cycle induction, but after induction of the lytic cycle, 13% of the cells expressed p21 protein in the nucleus (Fig. 4E). Again, double-label IFA revealed that 97% of the ZTA-positive D98/HR1 cells also expressed p21 and, conversely, that 73% of the p21-positive cells were also ZTA positive (Fig. 4E).
We also investigated whether ZTA-positive B lymphoblasts induce C/EBPα by using lytically induced Akata cells. After inducing the EBV lytic cycle in Akata cells with anti-IgG antiserum for 40 h, we conducted double-label IFA with anti-C/EBPα PAb and anti-ZTA MAb. The ZTA protein was rarely detected in Akata cells not treated with anti-IgG antiserum by IFA (<0.05% of the cells) (Fig. 5B) but was found to be induced at 40 h after anti-IgG antiserum treatment (>10%) (Fig. 5E). Furthermore, the fraction of Akata cells expressing a high level of nuclear C/EBPα increased from 1.0% before lytic-cycle induction (Fig. 5A) to 11% after anti-IgG antiserum treatment (Fig. 5D), again coinciding with the dramatic increase in the fraction of ZTA-positive cells. Similarly, 75% of the ZTA-positive Akata cells proved to also express C/EBPα (Fig. 5F), and 68% of the C/EBPα-positive cells were also ZTA positive, confirming that a majority of the ZTA-expressing cells displayed increased C/EBPα expression (Fig. 5F). Importantly, very few Akata cells that failed to express ZTA were positive for C/EBPα (3.5%).
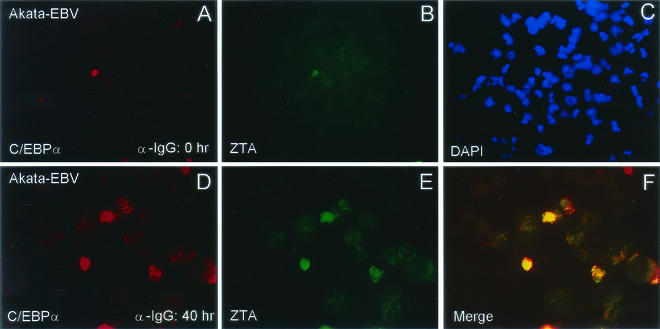
FIG. 5.
Coinduction of C/EBPα with ZTA in lytically induced Akata cells. (A to C) Double-label IFA showing low basal-level expression of both C/EBPα (A; rhodamine, red) and ZTA (B; FITC, green) in the same field of uninduced Akata cells (B; red). DAPI nuclear staining (C) shows the whole cell population. (D to F) Double-label IFA showing high-level expression of induced C/EBPα (D; red) and ZTA (E; green) in lytically induced Akata cells. The merge of the two frames (F) confirms that ZTA expression coincides with C/EBPα expression in the same cells.
FIG. 11.
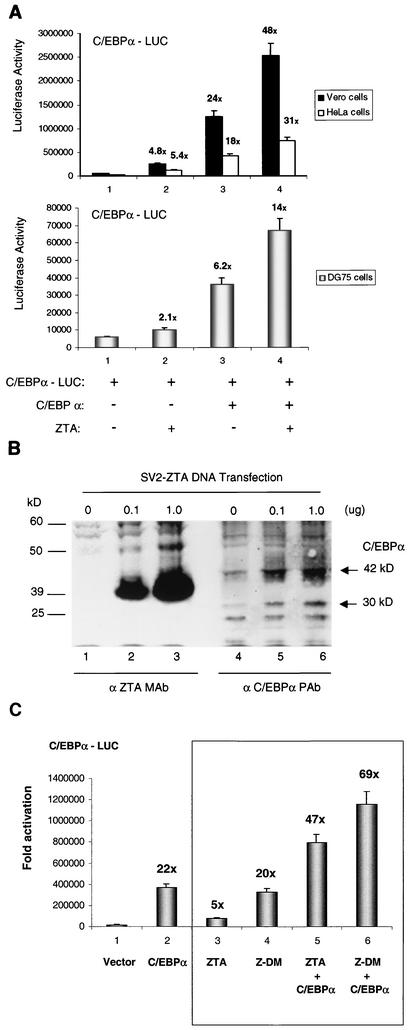
EBV ZTA enhances the transcriptional autoregulation of the C/EBPα gene promoter by C/EBPα. (A) Luciferase assays comparing the levels of transactivation of a target C/EBPα-LUC reporter gene after transfection into Vero, HeLa, or DG75 cells, either alone orin the presence of mammalian expression plasmids encoding C/EBPα or ZTA. (B) Western immunoblot showing the dose-responsive induction of endogenous C/EBPα by the transfected ZTA expression plasmid in Vero cells. The specificity of both the C/EBPα PAb (α C/EBPα PAb) and the ZTA MAb (α ZTA MAb) was also directly confirmed in this experiment, which shows that the C/EBPα PAb is not reactive against the ZTA protein and vice versa. (C) Luciferase assays comparing the effects of cotransfected wild-type ZTA and a DNA binding-negative form of ZTA (Z-DM, or Zdbm1) on the C/EBPα-LUC reporter gene in Vero cells. The results show that the ability of ZTA to bind to standard ZRE or AP1 motifs is not required for either direct activation or enhancement of C/EBPα autoregulation.
Similar results were obtained for EBV-positive Akata cells by Western immunoblot analysis. The 35-kDa ZTA protein was almost undetectable by Western blot analysis of Akata cells during EBV latency but was significantly induced at 24 h after the induction of the lytic cycle by IgG treatment, and its expression then persisted throughout the lytic cycle (Fig. 6A). Similarly, the overall levels of 42-kDa C/EBPα and two more slowly migrating, presumably phosphorylated or sumo)-modified C/EBPα bands detected by Western immunoblotting increased steadily from 24 to 72 h after IgG treatment of Akata cells (Fig. 6B). Note that the absence of any cross-reactions between C/EBPα and ZTA and their respective antibodies is unambiguously demonstrated in another experiment (see Fig. 11B), where both antibodies are compared for the same samples containing both proteins. Increased expression of both ZTA and C/EBPα also proved to correlate with the strongly increased expression of p21 detected by Western immunoblotting from 24 to 72 h in IgG-treated Akata cells (Fig. 6C). Importantly, the level of p27 protein, another Cdk inhibitor associated with cell cycle arrest, was found to be relatively unchanged throughout lytic-cycle induction in IgG-treated Akata cells by Western blotting (Fig. 6C).
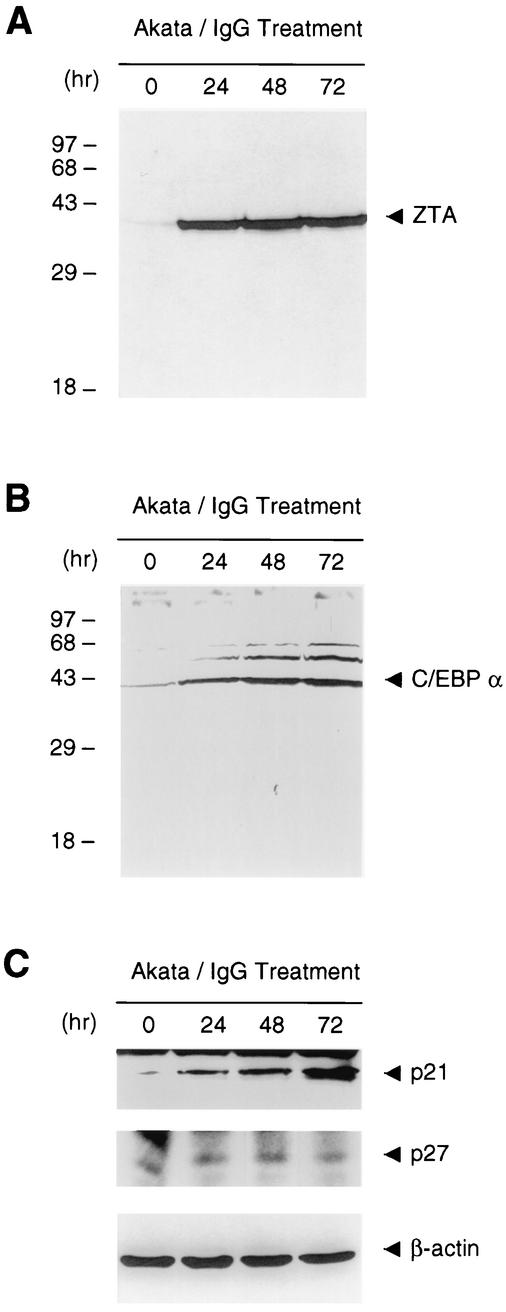
FIG. 6.
Quantitation and time course of enhanced ZTA, C/EBPα, and p21 expression after lytic-cycle induction in EBV-positive Akata cells. Western immunoblot analyses illustrate the overall levels of increases in the expression of total ZTA, C/EBPα, and p21 (but not p27) in Akata cell cultures at 0, 24, 48, and 72 h after lytic-cycle induction by IgG treatment. β-Actin levels are shown as a loading control.
Similar experiments were performed with DG75 B cells as EBV-negative controls, but no induction of either C/EBPα or p21 was observed after either TPA treatment or cross-linking of the B-cell receptor with anti-IgG antibodies, suggesting that these treatments do not nonspecifically up-regulate these two cellular proteins in the absence of EBV lytic-cycle induction and ZTA.
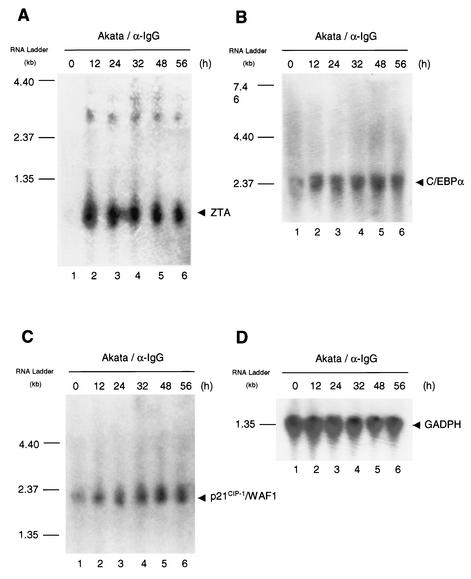
Induction of C/EBPα and p21 mRNAs during the EBV lytic cycle in Akata cells.
To evaluate whether ZTA induction of C/EBPα and p21 might involve a transcriptional mechanism, we performed Northern blot hybridization with total RNA samples extracted from Akata cells treated with anti-IgG antiserum for 0, 12, 24, 32, 48, or 56 h. Three specific 32P-labeled DNA probes were used to detect the 2.5-kb C/EBPα mRNA, 1.0-kb ZTA mRNA, and 2.1-kb p21 mRNA species. The level of C/EBPα mRNA proved to be limited in uninduced Akata cells (0 h) but became elevated within 12 h after anti-IgG antiserum treatment, suggesting that some induction of C/EBPα occurs at the transcriptional level (Fig. 7B). The kinetics of the increase in the level of C/EBPα mRNA coincided with the sharp increase in the level of 1.0-kb ZTA mRNA at 12 h (Fig. 7A).
FIG. 7.
Induction of C/EBPα and p21CIP-1 mRNAs in lytically induced Akata cells. Northern blot hybridization was carried out by using total RNA extracted from Akata cells at different times after anti-IgG (α-IgG) antiserum treatment (0, 12, 24, 32, 48, and 56 h). For the detection of specific mRNA, the membrane was sequentially incubated, stripped, and reprobed with denatured 32P-labeled DNA probes presenting fragments of the ZTA (A), C/EBPα (B), or p21 (C) cDNA coding regions. Detection of GAPDH mRNA (D) was used as a loading control for the samples in different lanes. Positions of size markers in an RNA ladder are indicated to the left of each panel.
Cayrol and Flemington (4, 5) previously reported that the induction of p21 protein is caused by an increase in the p21 mRNA level in stably transfected HeLa cells expressing the tetracycline-inducible ZTA protein. In our experiments, the low level of uninduced p21 mRNA also increased detectably after lytic-cycle induction in Akata cells (Fig. 7C), suggesting that transactivation of the p21 promoter contributed to the large accumulation of p21 protein in ZTA-positive EBV-infected cells. Detection of GAPDH mRNA was performed to confirm equal loading of total RNA in the samples (Fig. 7D). Although the increased C/EBPα and p21 levels in this experiment are rather modest, it is important to remember that only 10 to 15% of the cells in the Akata cell cultures were ZTA positive and showed enhanced C/EBPα and p21 levels.
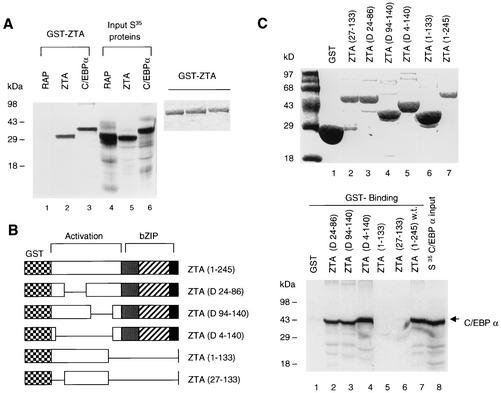
In vitro mapping of the specificity and bZIP domain requirement for the ZTA-C/EBPα interaction.
To further confirm the physical interaction of ZTA with C/EBPα and to map the domain of ZTA that interacts with C/EBPα, we performed GST affinity assays with GST-ZTA and five GST-ZTA fusion proteins with deletions (Fig. 8). We found that 35S-labeled in vitro-translated C/EBPα was able to interact very strongly with GST-ZTA (Fig. 8A), and by quantitative Instant Imager analysis, we measured the C/EBPα-ZTA interaction to be twofold stronger than the ZTA-ZTA self-interaction. C/EBPα failed to interact with the GST control protein, and we also detected no interaction between GST-ZTA and KSHV RAP (Fig. 8A), demonstrating that the C/EBPα-ZTA interaction in the GST affinity assays was highly specific. Mapping of the interaction domain with deletion mutants (Fig. 8B) revealed that C/EBPα interacts specifically with the domain of ZTA encompassing the bZIP region. Two truncated GST-ZTA mutants that lacked the domain encompassing the bZIP region (aa 134 to 245) failed to bind to C/EBPα, whereas a deletion across the N-terminal region from aa 4 to 140 did not interfere with binding (Fig. 8C). These observations correlate with previous evidence that the ability of ZTA to induce p21 and G1 cell cycle arrest rests in its bZIP domain (4). Therefore, ZTA binds strongly and specifically to C/EBPα in vitro, and the interaction maps to the C-terminal segment of ZTA encompassing the bZIP domain.
FIG. 8.
C/EBPα interacts with the C-terminal region of the ZTA protein (aa 140 to 245) that encompasses the bZIP domain. (A) GST affinity assay showing that GST-ZTA interacts specifically with itself and with C/EBPα but does not interact with KSHV RAP. (B) Diagram of the wild-type GST-ZTA fusion protein and GST-ZTA deletion mutants. D, deletion. (C) (Upper panel) Relative sizes of the purified GST products from plasmid-transformed E. coli strain BL21. (Lower panel) In vitro GST affinity binding assay providing evidence that in vitro-translated 35S-labeled C/EBPα is able to interact only with GST-ZTA fusion proteins that contain the bZIP domain. w.t., wild type.
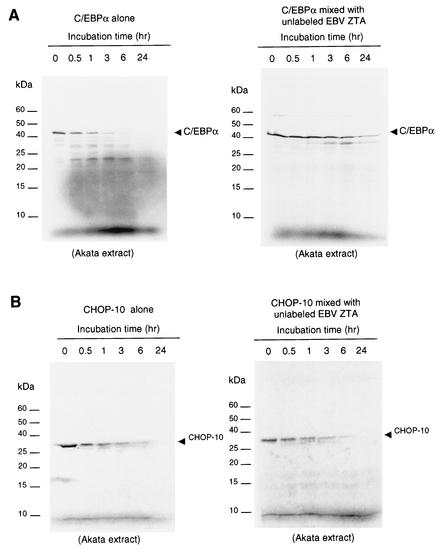
ZTA stabilizes C/EBPα in an in vitro proteasome-dependent stability assay.
Because ZTA interacts strongly with C/EBPα, we examined whether ZTA might affect the normally relatively short half-life of C/EBPα (46). A protein stability assay carried out by incubation of 35S-labeled in vitro-translated C/EBPα with an uninduced Akata cell proteasome extract showed that it had an in vitro half-life of 30 min and was completely degraded after 6 h (Fig. 9A). However, when we preincubated 35S-labeled C/EBPα with unlabeled in vitro-translated ZTA protein for 30 min before adding a proteasome extract, the stability assay showed that the half-life of C/EBPα was increased from 30 min to 3 h (Fig. 9A). As a negative control, we used the CHOP-10 protein, which is a member of the C/EBP family but which does not interact with ZTA in vitro (unpublished data). A parallel protein stability assay showed that the 35S-labeled CHOP-10 protein had an in vitro half-life similar to that of C/EBPα (30 min) and was completely degraded after 6 h of incubation (Fig. 9B). However, preincubation of the 35S-labeled CHOP-10 protein with the same ZTA protein sample did not affect the half-life of the CHOP-10 protein at all, showing that the protective effect of ZTA was specifically targeted to C/EBPα (Fig. 9B). Therefore, ZTA-dependent stabilization of C/EBPα against degradation may partially explain the increased level of C/EBPα and is probably mediated by the C/EBPα-ZTA interaction.
FIG. 9.
ZTA stabilizes C/EBPα in a protein stability assay. (A) (Left panel) Incubation time course showing that the half-life of 35S-labeled C/EBPα in an Akata cell proteasome extract is 30 min. (Right panel) Parallel time course after preincubation of 35S-labeled C/EBPα with unlabeled ZTA protein showing that ZTA increased the half-life of C/EBPα from 30 min to 3 h in the same Akata cell proteasome extract. (B) (Left panel) Incubation time course showing that the half-life of 35S-labeled CHOP-10 protein in an Akata cell proteasome extract is 30 min. (Right panel) Parallel time course after preincubation of 35S-labeled CHOP-10 protein with unlabeled ZTA protein showing that ZTA did not affect the half-life of the CHOP-10 protein.
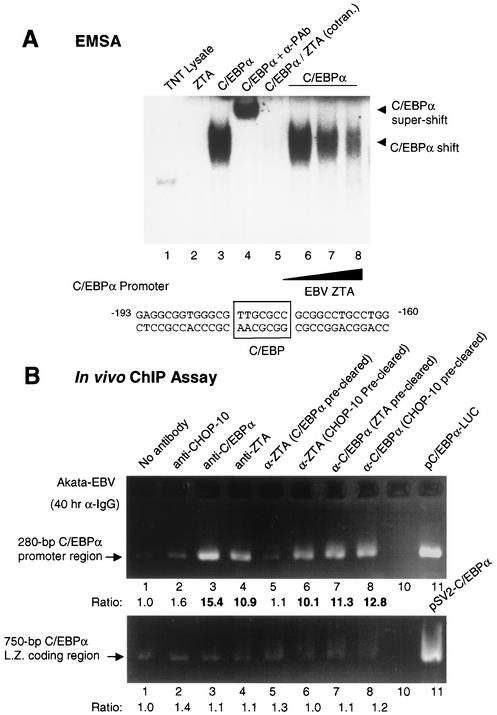
ZTA affects C/EBPα binding to DNA in EMSAs.
C/EBPα positively autoregulates its own gene promoter through a defined C/EBPα binding motif at positions −180 to −173 (9, 45). We were unable to recognize any potential ZRE or AP1 sites that might bind to ZTA within the C/EBPα promoter DNA sequence and also found that ZTA only weakly transactivated a C/EBPα-LUC reporter gene (see below). Furthermore, in vitro-translated ZTA protein failed to bind to our 40-bp C/EBP motif DNA probe containing the known C/EBP binding site from the C/EBPα promoter in an EMSA experiment (Fig. 10A, lane 2). As expected, the same sample of ZTA protein bound efficiently to a ZRE DNA probe in an EMSA experiment (data not shown). In contrast, in vitro-translated C/EBPα bound strongly to the same probe and was supershifted by anti-C/EBPα antibody (Fig. 10A, lanes 3 and 4). When both C/EBPα and ZTA were added together to the binding mixture containing the labeled C/EBP probe, C/EBPα binding to the C/EBP probe was eliminated, although no new supershifted complexes were observed (Fig. 10A, lane 5). The inhibitory effect was dose responsive and specific for C/EBPα. This result implies that ZTA also interacts physically with DNA-bound C/EBPα, although we cannot yet distinguish between the alternatives that the ZTA interaction inhibits C/EBPα binding to the C/EBP motif or that ZTA-C/EBPα complexes retain the ability to bind to DNA but either are too large to enter the gel or are not stably maintained in the in vitro system, leading to the failure to observe a supershifted band.
FIG. 10.
Both C/EBPα and ZTA associate with the C/EBPα promoter in ChIP assays, but C/EBPα is required for ZTA to associate efficiently. (A) EMSA confirming that ZTA interacts with DNA-bound C/EBPα. A 40-bp 32P-labeled C/EBP motif probe from positions −193 to −160 in the C/EBPα promoter was used. Lane 1, a control sample of unprogrammed reticulocyte lysate alone (2 μl) does not bind to the C/EBP motif. Lane 2, in vitro-translated ZTA protein fails to bind to the C/EBP motif. Lane 3, in vitro-translated C/EBPα binds efficiently to the C/EBP motif. Lane 4, supershifting of the DNA-bound C/EBPα band with anti-C/EBPα PAb (α-PAb). Lane 5, C/EBPα fails to bind to the C/EBP probe when cotranslated with ZTA. Lanes 6 to 8, loss of the C/EBPα complex bound to the C/EBP probe occurs in a dose-responsive manner after the addition of increasing amounts of in vitro-translated ZTA protein. (B) ChIP assay results obtained with anti-IgG antiserum (α-IgG)-treated Akata cell nuclear extracts showing that both C/EBPα and ZTA indeed associate with C/EBPα promoter DNA in vivo. (Upper panel) C/EBPα promoter DNA recovered with PCR primers LGH4273 and LGH4275. Lanes 1 to 4, C/EBPα promoter DNA can be recovered from immunoprecipitates obtained with either C/EBPα (α-C/EBPα) or ZTA (α-ZTA) antibody but not from immunoprecipitates obtained with CHOP-10 antibody or with a no-antibody control. Lanes 5 and 6, removal of C/EBPα from the lysate (preclearing) with C/EBPα antibody abolishes the association of the ZTA protein with the C/EBPα promoter, whereas preclearing with CHOP-10 antibody does not. Lanes 7 and 8, the association of C/EBPα with C/EBPα promoter DNA is not affected by preclearing with either ZTA or CHOP-10 antibody. Lane 11, size control showing the PCR product amplified directly from C/EBPα promoter template DNA (plasmid pSEW-CP1) with the same primers. (Lower panel) Negative-control ChIP assay results confirming that no nonspecific C/EBPα coding region DNA was recovered from immunoprecipitates obtained with PCR primers LGH4268 and LGH4270. Lanes 1 to 8, same as described above. Bold ratio numbers indicate significant fold increases over basal level.
Cotransfected ZTA enhances C/EBPα autoregulation in transient reporter gene assays.
To observe any functional effects of ZTA on C/EBPα autoregulation of its own promoter, we performed transient luciferase assays with both adherent cells (Vero and HeLa cells) and EBV-negative B lymphoblasts (DG75 cells) and a target C/EBPα promoter-driven luciferase reporter gene (C/EBPα-LUC). Cotransfection of the C/EBPα-LUC target with a mammalian expression plasmid encoding C/EBPα showed that C/EBPα can up-regulate its own promoter by 24-, 18-, or 6.2-fold in Vero, HeLa, or DG75 cells, respectively (Fig. 11A). In comparison, cotransfection of C/EBPα-LUC with a ZTA-encoding mammalian expression plasmid up-regulated the C/EBPα promoter by only 4.8-, 5.4-, or 2.1-fold in Vero, HeLa, or DG75 cells, respectively (Fig. 11A). In addition, a dose-responsive increase in the endogenous C/EBPα level in Vero cells was detected when increasing amounts of the ZTA expression plasmid were transfected into the cells (Fig. 11B). In other studies, it was found that both ZTA and KSHV RAP weakly transactivated isolated consensus C/EBP motifs added to a minimal promoter-driven luciferase reporter gene and that mutagenesis of these C/EBP motifs in their natural promoter contexts abolished ZTA- or RAP-mediated enhancement of C/EBPα transactivation (50; Wu et al., unpublished). Therefore, because ZTA cannot bind to the C/EBP motif directly, the small fold activation of C/EBPα-LUC by ZTA alone could be the result of ZTA-mediated stabilization of endogenous C/EBPα.
In addition, cotransfection of both the C/EBPα and the ZTA expression plasmids increased the activation of the C/EBPα promoter up to 48-, 31-, or 14-fold in Vero, HeLa, or DG75 cells, respectively (Fig. 11A). This result implies that ZTA can also activate the C/EBPα promoter cooperatively with C/EBPα, possibly like KSHV RAP (Wu et al., unpublished) through a piggyback mechanism by binding to and stabilizing DNA-bound C/EBPα. Although ZTA was reported to be able to interact directly with TATA binding protein on the TATA box to transactivate certain promoters, ZTA failed to transactivate C/EBPα-LUC in C/EBPα−/− MEF cells (data not shown), suggesting that C/EBPα is required for this process and that stabilization of C/EBPα is a more likely explanation for the activation observed here.
Evidence that neither the activation of the C/EBPα promoter nor the enhancement of C/EBPα autoregulation observed here involves the direct targeting of ZTA to any unknown ZRE motifs in the C/EBPα promoter was obtained by using a DNA binding-negative Z-DM mutant (Zdbm1). This protein, which contains three amino acid point mutations in the ZTA basic domain that abolish its ability to recognize and bind to standard ZRE motifs (20a), has been shown to have enhanced transcriptional activity for the EBV ZTA promoter as well as to retain the ability to induce G1 cell cycle arrest (4). Cotransfection of Z-DM (Zdbm1) alone compared to that of wild-type ZTA alone produced a 20-fold activation of the C/EBPα-LUC reporter gene in Vero cells and still enhanced C/EBPα autoregulation threefold (Fig. 11C). Because Z-DM (Zbdm1) retained its ability to interact with C/EBPα in vitro (data not shown), we conclude that both effects likely are mediated through piggyback binding to C/EBPα targeted to the C/EBP binding site. Further studies to confirm that this mechanism explains the previously enigmatic properties of Z-DM (Zbdm1) are in progress (F. Y. Wu, Q. Tang, H. Chen, S. Wang, M. Fujimuro, C. Chiou, S. D. Hayward, M. D. Lane, and G. Hayward, unpublished data).
ChIP assays show that ZTA associates with C/EBPα promoter DNA in vivo through an interaction with C/EBPα.
To attempt to confirm our model that a C/EBPα promoter-bound ZTA-C/EBPα supercomplex forms in vivo, we performed an in vivo ChIP assay with lytically induced Akata cells. Because the related CHOP-10 protein cannot interact with ZTA (Wu et al., unpublished) or activate the C/EBPα promoter, we used CHOP-10 protein as a negative control for our ChIP assay. First, we immunoprecipitated CHOP-10, C/EBPα, and ZTA from the DNA-protein cross-linked cell lysates. After removal of all proteins through phenol extraction, the recovered DNA was ethanol precipitated and amplified by PCR with primers specific for the 280-bp region of the C/EBPα promoter encompassing the C/EBP motif (LGH4273 and LGH4275). C/EBPα promoter DNA was indeed found to be associated with both C/EBPα and ZTA immunoprecipitates (Fig. 10B, lanes 3 and 4). However, the negative controls with CHOP-10 antibody or with PBS solution failed to precipitate C/EBPα promoter DNA above the basal level (Fig. 10B, lanes 1 and 2).
To address the question of whether ZTA and C/EBPα bind to the C/EBP binding site on C/EBPα promoter DNA cooperatively, we precleared C/EBPα from the lysate with anti-C/EBPα PAb overnight and then reimmunoprecipitated the ZTA protein with anti-ZTA MAb in a second round. The results showed that less than 10% of C/EBPα promoter DNA could still be recovered from the ZTA immunoprecipitate in the second round (Fig. 10B, lane 5), suggesting that in the absence of C/EBPα, no ZTA is associated with the C/EBPα promoter. A parallel control second immunoprecipitation with C/EBPα confirmed that over 95% of C/EBPα-bound DNA was removed (data not shown). To confirm the specificity of this effect, we precleared the CHOP-10 protein from the lysate with anti-CHOP-10 MAb overnight and then reimmunoprecipitated the ZTA protein with anti-ZTA MAb in a second round. The results showed that C/EBPα promoter DNA could still be recovered from the ZTA immunoprecipitate in the CHOP-10-precleared sample (Fig. 10B, lane 6). These results suggested that the removal of C/EBPα significantly impaired the ability of ZTA to bind to C/EBPα promoter DNA, whereas the removal of CHOP-10 protein did not affect this association at all. Finally, the removal of ZTA protein (or CHOP-10 protein in the negative control) by antibody preclearing did not significantly affect the association of C/EBPα with C/EBPα promoter DNA (Fig. 10B, lanes 7 and 8), suggesting that, although C/EBPα contributes to the binding of ZTA to the C/EBPα promoter, ZTA does not seem to play a major reciprocal role in mediating the binding of C/EBPα to the C/EBPα promoter. These conclusions are consistent with our earlier EMSA data, which showed that, in contrast to ZTA, in vitro-translated C/EBPα alone bound to the C/EBP motif probe very efficiently. Therefore, our in vivo ChIP assay data support the piggyback model that ZTA binding to the C/EBPα promoter requires a cooperative interaction with C/EBPα.
A negative-control PCR assay with a pair of primers that specifically detected only coding region DNA sequences from within the C/EBPα gene (LGH4268 and LGH4270) failed to yield any positive signals beyond the background level (Fig. 10B, lower panel), confirming that our ChIP assay was specific and recovered only promoter region DNA sequences specifically bound by these proteins.
C/EBPα and ZTA cooperatively up-regulate the p21 promoter through a proximal region containing three strong C/EBP binding sites.
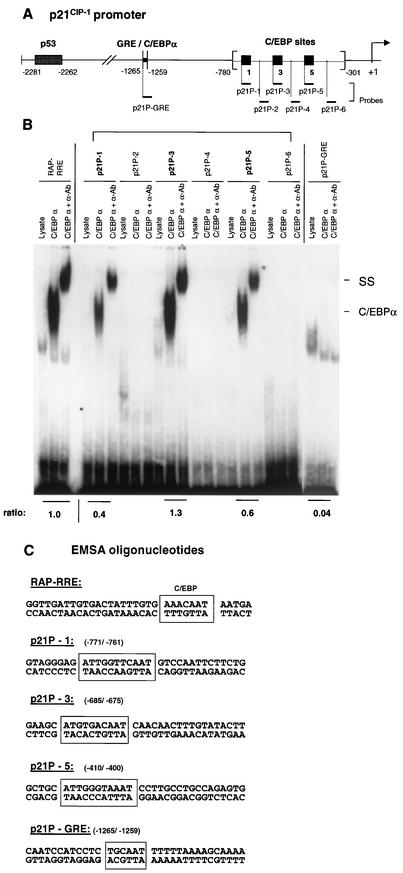
Since C/EBPα is known to stabilize the level of the p21 Cdk2 inhibitor protein as well as to up-regulate p21 gene expression (46, 47), we examined whether the interaction of ZTA with C/EBPα may affect this process in any way. A glucocorticoid response element (GRE) mapping at positions −1265 to −1259 within the p21 gene promoter has been reported to contain a putative DNA binding site for C/EBPα (16). We performed an EMSA with an oligonucleotide containing this GRE binding site and found it to be an extremely weak binding site for C/EBPα (only 4% the efficiency of a known strong C/EBPα binding site from the KSHV genome) (Fig. 12B). Therefore, we reexamined the human p21 gene promoter region and selected six putative C/EBPα binding sites based on a resemblance to known binding sites in the KSHV genome. Indeed, three of these six putative binding sites were found to bind to C/EBPα strongly (Fig. 12B and C). The newly identified binding sites all lay between −779 and −301 in the p21 gene promoter (Fig. 12A).
FIG. 12.
C/EBPα binds to a novel proximal region on the p21CIP-1/WAF1 promoter. (A) Diagram showing the locations of relevant features of the p21CIP-1/WAF1 promoter. (B) EMSA experiment showing that C/EBPα binds strongly to three sites mapped between −780 and −301 on the p21 promoter (probes in bold type) but only very weakly to the more distal site overlapping the GRE. SS, supershift. Ratios indicate the measured relative efficiencies of C/EBPα binding to various test oligonucleotide probes compared to that for a positive-control probe encompassing the RAP RRE C/EBP motif as the standard. α-Ab, antibody to C/EBPα. (C) DNA sequences of the double-stranded oligonucleotide probes used here, including the control RAP RRE C/EBP binding site, the three new p21 promoter C/EBP binding sites, and the far-upstream p21 GRE site (boxed).
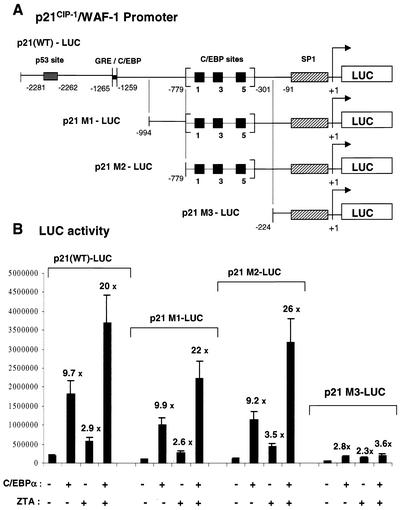
We constructed a series of deletion mutants of the 2.4-kb p21 gene promoter in luciferase reporter genes to test the role of these binding sites in mediating the up-regulation of the p21 gene promoter by C/EBPα (Fig. 13A). Plasmid p21-M1-LUC (positions −994 to +1) no longer contains either the p53 response element (14) or the proposed GRE C/EBP binding site (16), p21-M2-LUC (−779 to +1) encompasses all three C/EBP binding sites demonstrated above as well as the proximal (upstream) SP1 elements and TATA box, and p21-M3-LUC (−224 to +1) contains only the minimal upstream SP1 elements and TATA box.
FIG. 13.
ZTA enhances the transcriptional activation of the p21CIP-1/WAF1 promoter by C/EBPα in transient cotransfection assays. (A) Structures of the wild-type and deleted p21 promoter regions driving the target luciferase reporter genes used. (B) Transient reporter gene assays in which an empty vector DNA control or effector plasmids encoding SV2 C/EBPα or SV2 ZTA were cotransfected into Vero cells with target luciferase plasmids containing either the intact 2.4-kb p21CIP-1/WAF1 promoter [p21(WT)-LUC] or deletion derivatives p21-M1-LUC, p21-M2-LUC, and p21-M3-LUC.
Cotransfection of the 2.4-kb p21(WT)-LUC target with a C/EBPα expression plasmid gave 10-fold up-regulation, whereas a ZTA expression plasmid alone gave only 3-fold direct activation. However, cotransfection of both the C/EBPα and the ZTA effector plasmids together resulted in 20-fold activation of the intact p21 promoter (Fig. 13B). This observation implies that ZTA activates the p21 promoter cooperatively with C/EBPα.
Additional luciferase assays performed with our set of p21 promoter deletion mutant-luciferase reporters revealed that the absence of the p53 and GRE C/EBPα binding sites in p21-M1-LUC and p21-M2-LUC did not affect the inducibility of the p21 promoter by C/EBPα. Because the GRE C/EBP probe binds to C/EBPα only very weakly in vitro (Fig. 12B), it is not surprising that its deletion had very little effect. Furthermore, the levels of combined activation by C/EBPα and ZTA were similar for both the shorter p21-M1-LUC reporter (22-fold) and the p21-M2-LUC reporter (26-fold) (Fig. 13B). In contrast, very little induction by C/EBPα was observed with the p21-M3-LUC reporter, which no longer contains the region encompassing the three strong C/EBPα binding sites, and there was no significant enhancement of C/EBPα by ZTA (Fig. 13B). Evidently, the region from positions −779 to −301, which contains the C/EBPα binding sites, is essential for the ability of C/EBPα to activate the p21 promoter and for the C/EBPα-mediated cooperative activation of the p21 promoter by ZTA.
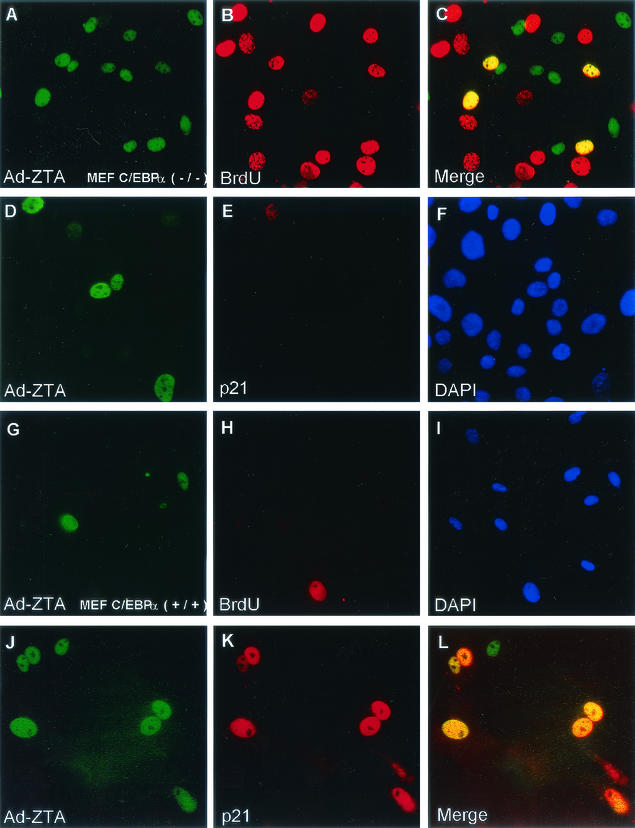
Recombinant Ad-ZTA fails to induce p21 or inhibit S-phase progression in C/EBPα-negative cells.
Experiments were carried out to address whether the EBV ZTA protein could induce p21 and cell cycle arrest in cells that lacked the C/EBPα gene. We used C/EBPα−/− and C/EBPα+/+ MEF cells (46). In Ad-ZTA-infected C/EBPα−/− cells, 23% of the cells were positive for BrdU, 24% were positive for ZTA, and 14% were positive for both ZTA and BrdU (Fig. 14A to C). However, in control (C/EBPα+/+) cells, less than 1% of ZTA-positive cells incorporated BrdU, showing that S-phase progression was arrested normally in ZTA-positive wild-type mouse cells but not in ZTA-positive C/EBPα−/− mouse cells (Fig. 14G to I). In addition, no induced expression of p21 was detected in C/EBPα−/− cells infected with Ad-ZTA, even though one-third of the total cells proved to be positive for ZTA (Fig. 14D to F). In contrast, in C/EBPα+/+ cells, over 77% of the ZTA-positive cells also expressed p21 (Fig. 14J to L). In summary, in contrast to what was seen for wild-type MEF cells, no correlation existed between ZTA expression and the inhibition of S-phase progression in C/EBPα−/− MEF cells. Therefore, these experiments provide important evidence for the essential role of C/EBPα in both ZTA-mediated induction of p21 and ZTA-induced G1 cell cycle arrest.
FIG. 14.
ZTA fails to induce G1/S cell cycle arrest or to up-regulate p21CIP-1 expression in C/EBPα−/− MEF cells. (A to C) Double-label IFA of ZTA protein expression (A; FITC, green) and BrdU incorporation (B; rhodamine, red) in Ad-ZTA-infected C/EBPα−/− MEF cells. (C) The merge of the two frames shows that S phase proceeds normally in ZTA-positive C/EBPα−/− mouse cells. (D to F) Double-label IFA showing ZTAprotein expression (D; green) but the absence of any corresponding mouse p21 protein induction (E; red) in ZTA-positive C/EBPα−/− MEF cells after Ad-ZTA infection. (F) DAPI nuclear staining (blue) of the same cell population. (G to I) Double-label IFA of ZTA protein expression (G; FITC, green) and BrdU incorporation (H; rhodamine, red) in Ad-ZTA-infected C/EBPα+/+ MEF cells. (I) The merge of the two frames shows that S-phase arrest still occurs in control ZTA-positive wild-type mouse cells. (J to L) Double-label IFA showing ZTA protein expression (J; green) and the corresponding induction of mouse p21 protein (K; red) in ZTA-positive C/EBPα+/+ MEF cells after Ad-ZTA infection. (L) The merge of the two frames shows the colocalization of ZTA and induced p21 proteins in control wild-type mouse cells.
DISCUSSION
Stabilization of the p53 tumor suppressor protein represents a classic model for the induction of cellular growth arrest that involves p21 function. However, in the absence of functional p53, as in Burkitt's lymphoma cells that carry p53 mutations (52), the p53-independent mechanism of host cell cycle arrest is not very well understood. The results of recent studies suggest that the gamma-2 herpesvirus KSHV circumvents this requirement by instead using the early lytic-cycle nuclear protein RAP (K8) to induce C/EBPα expression, which independently induces p21CIP-1 even in p53-negative HepB cells (53; Wu et al., unpublished). We have now shown that C/EBPα is also a new and important cellular factor in the gamma-1 herpesvirus EBV lytic-cycle process. The EBV-encoded lytic-cycle triggering protein ZTA is a positional homologue of RAP but lacks any significant residual amino acid identity. ZTA both binds to C/EBPα and directly induces C/EBPα expression either when introduced exogenously or when present in EBV-infected cells undergoing lytic-cycle progression. Furthermore, in the absence of C/EBPα, ZTA is unable to mediate p21 activation or cell cycle arrest.
The mechanism for the induction of C/EBPα by ZTA apparently involves ZTA-mediated protein stabilization through a direct interaction with C/EBPα. ZTA-mediated induction of p21 protein levels seems to involve the cooperative up-regulation of p21 gene transcription with stabilized C/EBPα, which binds to multiple C/EBP binding sites in the p21 promoter either directly or as a complex with ZTA. Furthermore, ZTA-mediated induction of C/EBPα levels also involves the enhancement of C/EBPα promoter autoregulation, likely through a piggyback mechanism involving C/EBPα-ZTA complexes.
Although some reports have attributed the cell cycle arrest observed during the induction of the EBV lytic cycle to chemical or biological inducing agents (27, 42), we have found that neither anti-IgG antiserum nor TPA treatment of EBV-negative DG75 Burkitt's lymphoma cells induces C/EBPα above the basal level, suggesting that the EBV genome is required for this process. Furthermore, cell cycle arrest occurs only in ZTA-positive cells in populations of anti-IgG-antiserum-treated Akata cells or TPA-treated D98/HR1 cells. Similarly, for KSHV, we have shown that the induction of C/EBPα during KSHV lytic-cycle activation in PEL and dermal microvascular endothelial cells also occurs only in cells expressing KSHV RAP (53;Wu et al., unpublished). These results are consistent with our Ad-ZTA results and with the conclusion of Cayrol and Flemington (5) that, in the absence of any lytic-cycle-inducing agents, only HeLa cells in the population that are expressing the ZTA protein are arrested in G1.
The results presented in this report lead to six principal conclusions about the mechanism of ZTA-induced cell cycle arrest. First, we confirmed that EBV ZTA alone is sufficient for the induction of the p21 Cdk inhibitor and G1 cell cycle arrest. Second, ZTA-mediated G1 cell cycle arrest correlates with the induction of both C/EBPα expression and p21 expression in all systems examined. Third, ZTA interacts with and stabilizes C/EBPα as well as enhances C/EBPα promoter autoregulation in a manner that is independent of ZRE-targeted DNA binding. Fourth, the C/EBPα responsiveness of the p21 promoter involves a previously unrecognized proximal region that contains three strong C/EBPα binding sites. Fifth, ZTA cooperatively induces the transcriptional activation of the p21 gene promoter by C/EBPα via the proximal (positions −779 to −301) response region. Sixth, the absence of C/EBPα severely compromises the ability of Ad-ZTA to induce p21 and cell cycle arrest, implying that C/EBPα plays an essential role.
The ability to block the cell cycle in G1/S appears to be an important early step in the lytic cycle for most herpesviruses. The utilization of immediate-early viral nuclear factors to elicit growth arrest may ensure that cells are arrested prior to engaging in viral DNA replication, as a way to eliminate cellular competition for resources. Other candidate herpesvirus-encoded cell cycle arrest regulatory genes include IE2 and UL69 in human cytomegalovirus and ICP0 (IE110) in herpes simplex virus (12, 21, 24, 25, 36, 37). However, the molecular targets and mechanisms of action of these proteins have not been fully elucidated.
Previous studies have demonstrated that C/EBPα both induces and stabilizes p21 expression (46, 47) and can also directly inhibit Cdk2 and Cdk4 in the presence or absence of p21 (23, 48). Therefore, the stabilization of C/EBPα by ZTA may also indirectly stabilize p21 as well. The combination of these two properties is believed to account for the growth arrest function of C/EBPα. In this study, we found that the ability of ZTA to induce p21 depends upon the presence of C/EBPα and that C/EBPα binds to a previously unrecognized region of the p21 promoter. Deletion of this newly identified 480-bp region from the wild-type p21 promoter abolished both the ability of C/EBPα to transcriptionally activate the p21 promoter and the cooperative enhancement by ZTA.
We believe that during EBV lytic infection, ZTA first stabilizes low preexisting levels of C/EBPα; the accumulation of C/EBPα then leads to the up-regulation of both the C/EBPα and the p21 promoters, which is also enhanced cooperatively by ZTA. Once sufficient levels of C/EBPα and p21 are expressed, C/EBPα can further stabilize p21 against proteasome-mediated degradation. The summation of these events probably leads to G1 cell cycle arrest in host cells.
While the induction and stabilization of the p21 protein can be attributed to both C/EBPα activity (46, 47) and ZTA- plus C/EBPα-mediated transcriptional activation of the p21 promoter (Wu et al., unpublished), it is not yet clear whether p21 is absolutely necessary for ZTA-mediated G1 cell cycle arrest, because C/EBPα itself can apparently induce cell cycle arrest directly by inhibiting Cdk function and E2F transcription in the absence of p21 (44, 48). Unfortunately, the p21-deficient MEF and HF cells that we have tested quickly go into crisis and apoptosis after two or three passages (unpublished observations), making further studies difficult to perform with such cells.
Previous studies of ZTA-mediated cell cycle arrest were performed with transformed cell lines, such as HeLa, SAOS-2, C33 carcinoma, and Akata cells. However, we believed that it was important to also evaluate cell cycle arrest in normal diploid HF cells, which are devoid of the aberrant cell cycle profiles and mutant p53 effects often associated with transformed or tumor cell lines. The strong inhibition of G1/S cell cycle progression that we observed in HF cells suggests that the interaction between C/EBPα and ZTA in up-regulating C/EBPα and p21 is a biologically relevant mechanism. Cayrol and Flemington (4) showed that G1 growth arrest is mediated by a region of ZTA encompassing the bZIP domain, which is required for both dimerization and direct DNA binding, and Rodriguez et al. (41) showed that replacement of the ZTA bZIP domain with the bZIP domain of c-Fos abolishes the ability of ZTA to arrest the cell cycle. Consistent with these results, we have shown that the region of the ZTA protein encompassing the bZIP domain is both necessary and sufficient for the interaction with C/EBPα in vitro, but it remains to be determined whether the N-terminal activation domain of ZTA that has been reported to be required for p53 and p27 stabilization plays a contributory role. Our work does not address whether or not cell cycle arrest signals may play a role in ZTA or lytic-cycle induction or whether the ZTA-mediated regulation of p27, p53, or c-MYC may also play a role in cell cycle arrest functions, but it does strongly argue that these other effects are not sufficient for cell cycle arrest in MEF cells in the absence of C/EBPα.
Acknowledgments
These studies were funded by National Cancer Institute research grants R01 CA73585 and R01 CA81400 to G.S.H. and R01 CA30356 to S.D.H. from the National Institutes of Health. F.Y.W. was a graduate student at the Viral Oncology Program, The Sidney Kimmel Comprehensive Cancer Center, School of Medicine, The Johns Hopkins University, and was partially supported by the Anti-Cancer Drug Development Training Program (grant T32 CA09243).
We thank Gretchen J. Darlington (Baylor College of Medicine) for kindly providing C/EBPα−/− MEF cells, M. Daniel Lane (Department of Biological Chemistry, School of Medicine, The Johns Hopkins University) for generously providing the rabbit anti-C/EBPα antibody, and E. K. Flemington for generously providing the Zdbm1 mutant construct.
REFERENCES
- 1.Becker, J., U. Leser, M. Marshall, A. Langford, W. Jilg, H. Gelderblom, P. Reichart, and H. Wolf. 1991. Expression of protein encoded by Epstein-Barr virus transactivator genes depends on differentiation of epithelial cells in oral hairy leukoplakia. Proc. Natl. Acad. Sci. USA 88:8332-8336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Calkhoven, C. F., C. Muller, and A. Leutz. 2000. Translational control of C/EBPalpha and C/EBPbeta isoform expression. Genes Dev. 14:1920-1932. [PMC free article] [PubMed] [Google Scholar]
- 3.Cannon, J. S., D. Ciufo, A. L. Hawkins, C. A. Griffin, M. J. Borowitz, G. S. Hayward, and R. F. Ambinder. 2000. A new primary effusion lymphoma-derived cell line yields a highly infectious Kaposi's sarcoma herpesvirus-containing supernatant. J. Virol. 74:10187-10193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cayrol, C., and E. Flemington. 1996. G0/G1 growth arrest mediated by a region encompassing the basic leucine zipper (bZIP) domain of the Epstein-Barr virus transactivator Zta. J. Biol. Chem. 271:31799-31802. [DOI] [PubMed] [Google Scholar]
- 5.Cayrol, C., and E. K. Flemington. 1996. The Epstein-Barr virus bZIP transcription factor Zta causes G0/G1 cell cycle arrest through induction of cyclin-dependent kinase inhibitors. EMBO J. 15:2748-2759. [PMC free article] [PubMed] [Google Scholar]
- 6.Chang, Y. N., D. L. Dong, G. S. Hayward, and S. D. Hayward. 1990. The Epstein-Barr virus Zta transactivator: a member of the bZIP family with unique DNA-binding specificity and a dimerization domain that lacks the characteristic heptad leucine zipper motif. J. Virol. 64:3358-3369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen, H., J. Lee, Y. Wang, D. Huang, R. Ambinder, and S. Hayward. 1999. The Epstein-Barr virus latency BamHI-Q promoter is positively regulated by STATs and ZTA interference with JAK/STAT activation leads to loss of BamHI-Q promoter activity. Proc. Natl. Acad. Sci. USA 96:9339-9344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chevallier-Greco, A., E. Manet, P. Chavrier, C. Mosnier, J. Daillie, and A. Sergeant. 1986. Both Epstein-Barr virus (EBV)-encoded trans-acting factors, EB1 and EB2, are required to activate transcription from an EBV early promoter. EMBO J. 5:3243-3249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Christy, R. J., K. H. Kaestner, D. E. Geiman, and M. D. Lane. 1991. CCAAT/enhancer binding protein gene promoter: binding of nuclear factors during differentiation of 3T3-L1 preadipocytes. Proc. Natl. Acad. Sci. USA 88:2593-2597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Countryman, J., and G. Miller. 1985. Activation of expression of latent Epstein-Barr herpesvirus after gene transfer with a small cloned subfragment of heterogeneous viral DNA. Proc. Natl. Acad. Sci. USA 82:4085-4089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Darlington, G. J., S. E. Ross, and O. A. MacDougald. 1998. The role of C/EBP genes in adipocyte differentiation. J. Biol. Chem. 273:30057-30060. [DOI] [PubMed] [Google Scholar]
- 12.de Bruyn Kops, A., and D. M. Knipe. 1988. Formation of DNA replication structures in herpes virus-infected cells requires a viral DNA binding protein. Cell 55:857-868. [DOI] [PubMed] [Google Scholar]
- 13.De Souza, Y. G., D. Greenspan, J. R. Felton, G. A. Hartzog, M. Hammer, and J. S. Greenspan. 1989. Localization of Epstein-Barr virus DNA in the epithelial cells of oral hairy leukoplakia by in situ hybridization of tissue sections. N. Engl. J. Med. 320:1559-1560. [DOI] [PubMed] [Google Scholar]
- 14.el-Deiry, W. S., T. Tokino, V. E. Velculescu, D. B. Levy, R. Parsons, J. M. Trent, D. Lin, W. E. Mercer, K. W. Kinzler, and B. Vogelstein. 1993. WAF1, a potential mediator of p53 tumor suppression. Cell 75:817-825. [DOI] [PubMed] [Google Scholar]
- 15.Farrell, P. J., D. T. Rowe, C. M. Rooney, and T. Kouzarides. 1989. Epstein-Barr virus BZLF1 trans-activator specifically binds to a consensus AP-1 site and is related to c-fos. EMBO J. 8:127-132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Firestone, G., Y. Nishio, H. Cha, E. Wang, R. Ramos, and E. Cram. 1998. Role of the CCAAT/enhancer binding protein-alpha transcription factor in the glucocorticoid stimulation of p21waf1/cip1 gene promoter activity in growth-arrested rat hepatoma cells. J. Biol. Chem. 273:2008-2014. [DOI] [PubMed] [Google Scholar]
- 17.Fixman, E. D., G. S. Hayward, and S. D. Hayward. 1995. Replication of Epstein-Barr virus oriLyt: lack of a dedicated virus-encoded origin-binding protein and dependence on Zta in cotransfection assays. J. Virol. 69:2998-3006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fixman, E. D., G. S. Hayward, and S. D. Hayward. 1992. trans-Acting requirements for replication of Epstein-Barr virus ori-Lyt. J. Virol. 66:5030-5039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Flemington, E., and S. H. Speck. 1990. Autoregulation of Epstein-Barr virus putative lytic switch gene BZLF1. J. Virol. 64:1227-1232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Flemington, E., and S. H. Speck. 1990. Evidence for coiled-coil dimer formation by an Epstein-Barr virus transactivator that lacks a heptad repeat of leucine residues. Proc. Natl. Acad. Sci. USA 87:9459-9463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20a.Flemington, E., J. P. Lytle, C. Cayrol, A. M. Borras, and S. H. Speck. 1994. DNA-binding-defective mutants of the Epstein-Barr virus lytic switch activator ZTA transactivate with altered specificities. Mol. Cell. Biol. 14:3041-3052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Flemington, E. K. 2001. Herpesvirus lytic replication and the cell cycle: arresting new developments. J. Virol. 75:4475-4481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hardy, S., M. Kitamura, T. Harris-Stansil, Y. Dai, and M. L. Phipps. 1997. Construction of adenovirus vectors through Cre-lox recombination. J. Virol. 71:1842-1849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Harris, T. E., J. H. Albrecht, M. Nakanishi, and G. J. Darlington. 2001. CCAAT/enhancer-binding protein-alpha cooperates with p21 to inhibit cyclin-dependent kinase-2 activity and induces growth arrest independent of DNA binding. J. Biol. Chem. 276:29200-29209. [DOI] [PubMed] [Google Scholar]
- 24.Hayashi, M. L., C. Blankenship, and T. Shenk. 2000. Human cytomegalovirus UL69 protein is required for efficient accumulation of infected cells in the G1 phase of the cell cycle. Proc. Natl. Acad. Sci. USA 97:2692-2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hobbs, W. E., II, and N. A. DeLuca. 1999. Perturbation of cell cycle progression and cellular gene expression as a function of herpes simplex virus ICP0. J. Virol. 73:8245-8255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Holley-Guthrie, E. A., E. B. Quinlivan, E. C. Mar, and S. Kenney. 1990. The Epstein-Barr virus (EBV) BMRF1 promoter for early antigen (EA-D) is regulated by the EBV transactivators, BRLF1 and BZLF1, in a cell-specific manner. J. Virol. 64:3753-3759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Inman, G. J., U. K. Binne, G. A. Parker, P. J. Farrell, and M. J. Allday. 2001. Activators of the Epstein-Barr virus lytic program concomitantly induce apoptosis, but lytic gene expression protects from cell death. J. Virol. 75:2400-2410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kenney, S., E. Holley-Guthrie, E. C. Mar, and M. Smith. 1989. The Epstein-Barr virus BMLF1 promoter contains an enhancer element that is responsive to the BZLF1 and BRLF1 transactivators. J. Virol. 63:3878-3883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kieff, E. 1996. Epstein-Barr virus and its replication. Lippincott-Raven Press, New York, N.Y.
- 30.Landschulz, W. H., P. F. Johnson, and S. L. McKnight. 1988. The leucine zipper: a hypothetical structure common to a new class of DNA binding proteins. Science 240:1759-1764. [DOI] [PubMed] [Google Scholar]
- 31.Lane, M. D., Q. Q. Tang, and M. S. Jiang. 1999. Role of the CCAAT enhancer binding proteins (C/EBPs) in adipocyte differentiation. Biochem. Biophys. Res. Commun. 266:677-683. [DOI] [PubMed] [Google Scholar]
- 32.Liao, G., F. Y. Wu, and S. D. Hayward. 2001. Interaction with the Epstein-Barr virus helicase targets Zta to DNA replication compartments. J. Virol. 75:8792-8802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lieberman, P. M., and A. J. Berk. 1990. In vitro transcriptional activation, dimerization, and DNA-binding specificity of the Epstein-Barr virus Zta protein. J. Virol. 64:2560-2568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lieberman, P. M., J. M. Hardwick, J. Sample, G. S. Hayward, and S. D. Hayward. 1990. The Zta transactivator involved in induction of lytic cycle gene expression in Epstein-Barr virus-infected lymphocytes binds to both AP-1 and ZRE sites in target promoter and enhancer regions. J. Virol. 64:1143-1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lin, F. T., O. A. MacDougald, A. M. Diehl, and M. D. Lane. 1993. A 30-kDa alternative translation product of the CCAAT/enhancer binding protein alpha message: transcriptional activator lacking antimitotic activity. Proc. Natl. Acad. Sci. USA 90:9606-9610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lomonte, P., and R. D. Everett. 1999. Herpes simplex virus type 1 immediate-early protein Vmw110 inhibits progression of cells through mitosis and from G1 into S phase of the cell cycle. J. Virol. 73:9456-9467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lu, M., and T. Shenk. 1999. Human cytomegalovirus UL69 protein induces cells to accumulate in G1 phase of the cell cycle. J. Virol. 73:676-683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mauser, A., E. Holley-Guthrie, D. Simpson, W. Kaufmann, and S. Kenney. 2002. The Epstein-Barr virus immediate-early protein BZLF1 induces both a G2 and a mitotic block. J. Virol. 76:10030-10037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Miller, G. 1990. Epstein-Barr virus, p. 1921-1958. In B. N. Fields and D. M. Knipe (ed.), Virology. Raven Press, New York, N.Y.
- 40.Ossipow, V., P. Descombes, and U. Schibler. 1993. CCAAT/enhancer-binding protein mRNA is translated into multiple proteins with different transcription activation potentials. Proc. Natl. Acad. Sci. USA 90:8219-8223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rodriguez, A., M. Armstrong, D. Dwyer, and E. Flemington. 1999. Genetic dissection of cell growth arrest functions mediated by the Epstein-Barr virus lytic gene product, Zta. J. Virol. 73:9029-9038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rodriguez, A., E. J. Jung, Q. Yin, C. Cayrol, and E. K. Flemington. 2001. Role of c-myc regulation in Zta-mediated induction of the cyclin-dependent kinase inhibitors p21 and p27 and cell growth arrest. Virology 284:159-169. [DOI] [PubMed] [Google Scholar]
- 43.Sarisky, R. T., Z. Gao, P. M. Lieberman, E. D. Fixman, G. S. Hayward, and S. D. Hayward. 1996. A replication function associated with the activation domain of the Epstein-Barr virus Zta transactivator. J. Virol. 70:8340-8347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Slomiany, B. A., K. L. D'Arigo, M. M. Kelly, and D. T. Kurtz. 2000. C/EBPalpha inhibits cell growth via direct repression of E2F-DP-mediated transcription. Mol. Cell. Biol. 20:5986-5997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tang, Q. Q., and M. D. Lane. 1999. Activation and centromeric localization of CCAAT/enhancer-binding proteins during the mitotic clonal expansion of adipocyte differentiation. Genes Dev. 13:2231-2241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Timchenko, N. A., T. E. Harris, M. Wilde, T. A. Bilyeu, B. L. Burgess-Beusse, M. J. Finegold, and G. J. Darlington. 1997. CCAAT/enhancer binding protein alpha regulates p21 protein and hepatocyte proliferation in newborn mice. Mol. Cell. Biol. 17:7353-7361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Timchenko, N. A., M. Wilde, M. Nakanishi, J. R. Smith, and G. J. Darlington. 1996. CCAAT/enhancer-binding protein alpha (C/EBP alpha) inhibits cell proliferation through the p21 (WAF-1/CIP-1/SDI-1) protein. Genes Dev. 10:804-815. [DOI] [PubMed] [Google Scholar]
- 48.Wang, H., P. Iakova, M. Wilde, A. Welm, T. Goode, W. J. Roesler, and N. A. Timchenko. 2001. C/EBPalpha arrests cell proliferation through direct inhibition of Cdk2 and Cdk4. Mol. Cell 8:817-828. [DOI] [PubMed] [Google Scholar]
- 49.Wang, S., S. Liu, M. H. Wu, Y. Geng, and C. Wood. 2001. Identification of a cellular protein that interacts and synergizes with the RTA (ORF50) protein of Kaposi's sarcoma-associated herpesvirus in transcriptional activation. J. Virol. 75:11961-11973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang, S. E., F. Y. Wu, M. Fujimuro, J. C. Zong, S. D. Hayward, and G. S. Hayward. 2003. Role of the CCAAT/enhancer-binding protein α in activation of the Kaposi's sarcoma-associated herpesvirus (KSHV) lytic replication-associated protein (RAP) promoter in cooperation with the KSHV replication and transcription activator (RTA) and RAP. J. Virol. 77:600-623. [DOI] [PMC free article] [PubMed]
- 51.Wang, X., E. Scott, C. L. Sawyers, and A. D. Friedman. 1999. C/EBPalpha bypasses granulocyte colony-stimulating factor signals to rapidly induce PU.1 gene expression, stimulate granulocytic differentiation, and limit proliferation in 32D cl3 myeloblasts. Blood 94:560-571. [PubMed] [Google Scholar]
- 52.Wiman, K. G., K. P. Magnusson, T. Ramqvist, and G. Klein. 1991. Mutant p53 detected in a majority of Burkitt lymphoma cell lines by monoclonal antibody PAb240. Oncogene 6:1633-1639. [PubMed] [Google Scholar]
- 53.Wu, F., Q. Tang, H. Chen, C. ApRhys, C. Farrell, J. Chen, M. Fujimuro, M. Lane, and G. Hayward. 2002. Lytic replication-associated protein (RAP) encoded by Kaposi's sarcoma-associated herpesvirus causes p21CIP-1-mediated G1 cell cycle arrest through CCAAT/enhancer-binding protein-alpha. Proc. Natl. Acad. Sci. USA 99:10683-10688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wu, F. Y., J. H. Ahn, D. J. Alcendor, W. J. Jang, J. Xiao, S. D. Hayward, and G. S. Hayward. 2001. Origin-independent assembly of Kaposi's sarcoma-associated herpesvirus DNA replication compartments in transient cotransfection assays and association with the ORF-K8 protein and cellular PML. J. Virol. 75:1487-1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Young, L., and M. Rowe. 1992. Epstein-Barr virus, lymphomas and Hodgkin's disease. Semin. Cancer Biol. 3:273-284. [PubMed] [Google Scholar]
- 56.Young, L. S., M. Rowe, G. Niedobitek, G. Packham, F. Shanahan, D. T. Rowe, D. Greenspan, J. S. Greenspan, and A. B. Rickinson. 1991. Differentiation-associated expression of the Epstein-Barr virus BZLF1 transactivator protein in oral hairy leukoplakia. J. Virol. 65:2868-2874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zalani, S., E. Holley-Guthrie, and S. Kenney. 1996. Epstein-Barr viral latency is disrupted by the immediate-early BRLF1 protein through a cell-specific mechanism. Proc. Natl. Acad. Sci. USA 93:9194-9199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhang, D. E., P. Zhang, N. D. Wang, C. J. Hetherington, G. J. Darlington, and D. G. Tenen. 1997. Absence of granulocyte colony-stimulating factor signaling and neutrophil development in CCAAT enhancer binding protein alpha-deficient mice. Proc. Natl. Acad. Sci. USA 94:569-574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhong, L., and G. S. Hayward. 1997. Assembly of complete, functionally active herpes simplex virus DNA replication compartments and recruitment of associated viral and cellular proteins in transient cotransfection assays. J. Virol. 71:3146-3160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhou, S., M. Fujimuro, J. J. Hsieh, L. Chen, A. Miyamoto, G. Weinmaster, and S. D. Hayward. 2000. SKIP, a CBF1-associated protein, interacts with the ankyrin repeat domain of NotchIC to facilitate NotchIC function. Mol. Cell. Biol. 20:2400-2410. [DOI] [PMC free article] [PubMed] [Google Scholar]