Kinetoplastid protozoa include the parasites Trypanosoma brucei, Trypanosoma cruzi, and various species of Leishmania that are responsible for African trypanosomiasis, Chagas' disease, and leishmaniasis, devastating diseases that afflict millions of victims largely in the tropical regions of the globe. In addition to eliciting long-standing interest from medical microbiologists, these intriguing protists have become more recently the objects of intense scrutiny by cellular and molecular biologists. Because these single-cell eukaryotes occupy a very ancient position on the evolutionary tree (1), they exhibit many unusual biological characteristics that distinguish them from their mammalian hosts and from many other organisms used as model systems by biologists. The increasing number of molecular, genetic, and cellular studies on these unusual protozoa have thus allowed a window into various biological phenomena that have often been obscured in the more routinely studied organisms. In some cases, the apparent peculiarities of these protists have served simply to highlight phenomena that are widespread within the biological universe. Accordingly, glycosylphosphatidyl inositol (GPI) membrane anchors were first discovered in T. brucei (2, 3); their discovery was facilitated by the abundance of the variant surface glycoproteins that cover the surface of this parasite, and which are involved in antigenic variation and tethered into the extracellular face of the plasma membrane by GPI moieties. Subsequent studies revealed that many eukaryotes, including mammalian cells, also express a plethora of GPI-linked membrane proteins. In other cases, the idiosyncrasies have proven to be more esoteric, one example being the astonishing RNA editing that extensively alters the sequence of many mitochondrial transcripts (4) and which has correlates among other protozoa, though not, apparently, among higher eukaryotes. An article by Das and Bellofatto (11) in this issue of PNAS now advances our rather rudimentary knowledge of transcription by RNA polymerase II (Pol II) in the Kinetoplastida.
The spliced leader gene is important because transcription of mRNA encoding genes occurs by a flamboyantly idiosyncratic mode.
One remarkable phenomenon in gene expression among the Kinetoplastida is the existence of a 39-nt spliced leader (SL) sequence that is placed onto the 5′ ends of all mRNAs by a trans-splicing reaction (5). Although such trans-splicing has been observed subsequently for a subpopulation of mRNAs in other eukaryotes such as Caenorhabditis elegans, the distinctive feature of the kinetoplastid protozoa is the universal requirement for SL addition onto every mRNA yet analyzed. The SL sequence is derived from a larger transcript (ranging in size from 96- to 141-nt, depending upon the species of parasite) that is transcribed from arrays of tandemly repeated genes. This SL RNA is thus an abundant small nuclear RNA (snRNA). Transcription of SL RNA, which initiates from a separate promoter upstream of each gene, had been ascribed previously to RNA Pol II, RNA Pol III, or an RNA polymerase of intermediate character (6). More recent studies by the Bellofatto group (7) have confirmed that Pol II is indeed the RNA polymerase responsible for transcription of SL RNA. These experiments were done in the trypanosomatid Leptomonas seymouri, a parasite of insects that has the biochemical virtue that it can be grown to very high cell densities and is thus a useful organism for isolating amounts of proteins required for in vitro characterization and functional analysis of individual polypeptides. The approach involved the development of an in vitro transcription system for the L. seymouri SL gene, followed by the demonstration that immunodepletion of the Pol II largest subunit specifically inhibited transcription of this template.
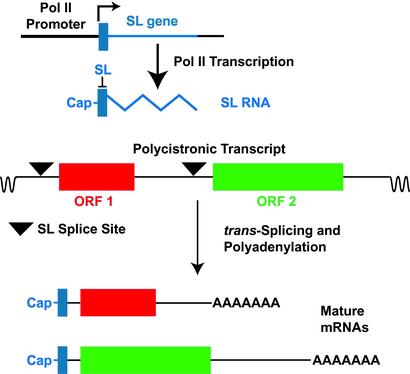
These studies have now placed the SL gene at center stage for understanding Pol II transcription in kinetoplastid protozoa. The principal reason why the SL gene is so important is that transcription of mRNA encoding genes occurs by a flamboyantly idiosyncratic mode in these protists. Genes that encode mRNAs are typically arranged in long arrays encompassing scores of coding regions that are separated by relatively short intergenic spacers. In Leishmania major, the sequence of the smallest chromosome revealed two apparently divergent transcriptional units, with all of the genes in each unit oriented in the same direction (8), and assembly of sequence from additional chromosomes has shown similar if somewhat more complex transcriptional patterns (9). Each unit is thought to be transcribed as a very long polycistronic primary transcript that is cotranscriptionally processed into the individual mRNAs (see Fig. 1). Notably, this processing is achieved by the trans-splicing of the SL sequence onto the 5′ end of the mRNA and by polyadenylation of the 3′ end of the mature message. This unusual mode of resolving individual mRNAs explains the universal requirement for the SL sequence on each mRNA. In addition, because the SL transcript is cotranscriptionally capped (10), the SL also provides the 5′ cap that is required, among other things, for efficient translation of protein-coding RNAs. However, this remarkable scenario for gene expression has made it exceedingly difficult to identify Pol II promoters among the Kinetoplastida. The reason is because transcription does not initiate shortly upstream of the 5′ end of the mature mRNA but rather at an indeterminate location that may be many hundreds of kilobases away from the body of the gene, thus frustrating the standard methods used for promoter mapping in other eukaryotes. At present, it is not clear whether the relatively limited number of apparent transcriptional start sites (e.g., segments between two divergent transcription units) encompass “promoters” as such, or whether transcription even begins at a discrete location. In short, the SL genes contain the only clearly defined Pol II promoters in any of these organisms.
Figure 1.
Transcription of the SL gene (blue) generates SL RNA and trans-splicing of the 39 nucleotide SL (solid blue rectangle) onto polycistronic primary transcripts resolves individual mRNAs. Each SL gene, arranged in a tandem repeat, is transcribed by Pol II from a promoter located immediately upstream of the gene (L-shaped arrow indicates transcriptional start site) and is cotranscriptionally capped. The resulting SL RNA is then trans-spliced onto consensus splice sites (triangles) located on the polycistronic RNAs to generate the 5′ end of each mRNA, and the 3′ end of the mRNA is polyadenylated in a step that is probably associated with trans-splicing at the downstream splice site. ORF 1 (red) and ORF 2 (green) indicate ORFs within the polycistronic RNA that will generate two distinct mRNAs. Intervening sequences that are removed by cis-splicing are exceedingly rare among the Kinetoplastida.
Two editorial notes are, however, worth keeping in mind. First, the SL promoters may not tell us a great deal about how transcription initiates at long polycistronic gene arrays such as those typically encoding proteins. Second, workers in this field have for some time been aware that much of the control of gene expression must occur at posttranscriptional levels, given the fact that many differentially regulated genes occur within the same transcriptional cluster and thus may initiate transcription from the same site. Hence, these organisms may serve as a biological cornucopia for understanding posttranscriptional gene regulation but exhibit a more limited repertoire of transcriptional control.
Das and Bellofatto (11) have now identified two components of a transcription factor among the Kinetoplastida. Their study was founded upon previous work from the same laboratory that identified three sequence elements required for transcription of the L. seymouri SL gene in vitro (12). These sequences include an initiator element near the transcription start site and two proximal elements designated PBP-1E and PBP-2E. Similar upstream elements required for transcription of SL genes in T. brucei, T. cruzi, and Leishmania amazonensis have also been reported by various groups (reviewed in ref. 6), but the details of promoter structure appear to differ somewhat from one organism to another. The Bellofatto laboratory also showed by electrophoretic mobility shift assays and DNA footprinting (13) that the PBP-1E element specifically binds a protein of ≈122 kDa, designated PBP-1. The PBP-1 protein consists of three subunits of 57, 46, and 36 kDa, and the genes encoding the two larger subunits have now been cloned and sequenced (11). The p46 protein is a unique polypeptide not even present in the genome database from the related parasite T. brucei. Distinctive features of its sequence include a plethora of di-leucine motifs that are predicted by a structural algorithm to potentially lie on a concave surface that could be involved in protein–protein interactions. In contrast, the p57 protein is related (23% identity, 43% similarity) to the snRNA-activating protein complex subunit of 50 kDa (SNAP50 protein) and to similar predicted polypeptides from T. brucei and L. major. In higher eukaryotes, SNAP50 is a subunit of the SNAPc transcription factor that is required for transcription of most, if not all, snRNAs, whether they are synthesized by Pol II or Pol III (14). Moreover, a minimal version of SNAPc that entails various deletions but is still functional in transcription consists of three subunits and has a molecular weight of ≈130 kDa, similar to the L. seymouri PBP-1 transcription factor, suggesting that the protozoan factor might be a primordial version of the mammalian protein. Significantly, both p57 and the Pol II largest subunit are present in transcriptionally competent preinitiation complexes that can be isolated from nuclear extracts by using immobilized SL gene promoter, further supporting the likely importance of p57 in promoter binding and transcription.
This study complements previous work on the SL gene of T. cruzi reported by Buck and colleagues (15). By using a proximal sequence element (PSE) that had been identified for that gene and a yeast one-hybrid system, they identified the ≈45-kDa PPB1 protein and demonstrated that it binds to the PSE in a gel shift assay and dramatically enhances the level of SL RNA when overexpressed in T. cruzi epimastigotes. The predicted sequence of PPB1 is not related to p57 or p46; rather, it contains a leucine zipper-like motif, a helix-turn-helix motif, and two helix-loop-helix motifs, leading to the suggestion that it may be a bZIP-type transcription factor.
True to form, these protozoa appear to exhibit once again their dual character of commonality and uniqueness compared with other eukaryotes. Thus, a p57 homolog that is involved in snRNA transcription spans the evolutionary spectrum from these ancient protozoa to humans. In contrast, the p46 subunit does not even have an apparent homolog in the closely related trypanosomes (unless it is lurking somewhere in the remaining unsequenced genomic DNA from the T. brucei genome project), raising the question of what such a distinctive subunit might be doing for transcription of a major snRNA that is conserved among all Kinetoplastida. Furthermore, it is currently unclear what the relationship is between the two Leptomonas subunits and the PPB1 polypeptide from T. cruzi. Could PPB1 represent a homolog of the third subunit of the Leptomonas PBP-1, p36, whose gene has not been cloned yet, or is the T. cruzi PPB1 simply unrelated to the Leptomonas transcription factor? Is the mode of SL gene transcription similar or divergent between L. seymouri and T. cruzi? Whatever the specific functions of these recently identified proteins from these two parasites, they are welcome arrivals on the molecular parasitology stage. Although increasingly sophisticated molecular genetic techniques have been introduced to this field over the past decade, including transfection, homologous gene replacement, and RNA interference, to mention a few, there has been a notable scarcity among the Kinetoplastida of specific proteins involved in this fundamental step of gene expression that we call transcription. The identification of these and other putative transcription factors and the development of techniques for the isolation of functional transcription complexes should now open the way to rapid advances in understanding the transcriptional machinery of these ever-illuminating microbes. Vladimir and Estragon will be left waiting no longer.
Footnotes
See companion article on page 80.
References
- 1.Sogin M L, Selwood H J, Gunderson J H. Proc Natl Acad Sci USA. 1986;83:1383–1387. doi: 10.1073/pnas.83.5.1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cardosa de Almeida M L, Turner M J. Nature. 1983;302:349–352. doi: 10.1038/302349a0. [DOI] [PubMed] [Google Scholar]
- 3.McConville M J, Ferguson M A J. Biochem J. 1993;294:305–324. doi: 10.1042/bj2940305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kable M L, Heidmann S, Stuart K D. Trends Biochem Sci. 1997;22:162–168. doi: 10.1016/s0968-0004(97)01041-4. [DOI] [PubMed] [Google Scholar]
- 5.Laird P. Trends Genet. 1989;5:204–208. doi: 10.1016/0168-9525(89)90082-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Campbell D A, Sturm N R, Yu M C. Parasitol Today. 2000;16:78–82. doi: 10.1016/s0169-4758(99)01545-8. [DOI] [PubMed] [Google Scholar]
- 7.Gillinger G, Bellofatto V. Nucleic Acids Res. 2001;29:1556–1564. doi: 10.1093/nar/29.7.1556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Myler P J, Audleman L, deVos T, Hixson G, Kiser P, Lemley C, Magness C, Rickel E, Sisk E, Sunkin S, et al. Proc Natl Acad Sci USA. 1999;96:2902–2906. doi: 10.1073/pnas.96.6.2902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Myler P J, Stuart K D. Curr Opin Microbiol. 2000;3:412–416. doi: 10.1016/s1369-5274(00)00113-2. [DOI] [PubMed] [Google Scholar]
- 10.Mair C, Ullu E, Tschudi C. J Biol Chem. 2000;275:28994–28999. doi: 10.1074/jbc.M004193200. [DOI] [PubMed] [Google Scholar]
- 11.Das A, Bellofatto V. Proc Natl Acad Sci USA. 2003;100:80–85. doi: 10.1073/pnas.262609399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Luo H, Gilinger G, Mukherjee D, Bellofatto V. J Biol Chem. 1999;274:31947–31954. doi: 10.1074/jbc.274.45.31947. [DOI] [PubMed] [Google Scholar]
- 13.Luo H, Bellofatto V. J Biol Chem. 1997;272:33344–33352. doi: 10.1074/jbc.272.52.33344. [DOI] [PubMed] [Google Scholar]
- 14.Hernandez N. J Biol Chem. 2001;276:26733–26736. doi: 10.1074/jbc.R100032200. [DOI] [PubMed] [Google Scholar]
- 15.Wen L-M, Xu P, Benegal G, Carvalho R M, Buck G A. Mol Biochem Parasitol. 2000;110:207–221. doi: 10.1016/s0166-6851(00)00271-1. [DOI] [PubMed] [Google Scholar]