Abstract
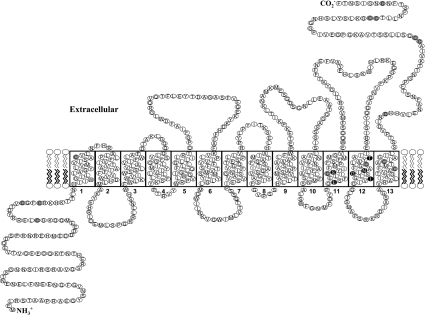
hCNT3 (human concentrative nucleoside transporter 3) is a nucleoside–sodium symporter that transports a broad range of naturally occurring purine and pyrimidine nucleosides as well as anticancer nucleoside drugs. To understand its uridine binding and translocation mechanisms, a cysteine-less version of hCNT3 was constructed and used for cysteine-accessibility and permeant-protection assays. Cysteine-less hCNT3, with 14 endogenous cysteine residues changed to serine, displayed wild-type properties in a yeast expression system, indicating that endogenous cysteine residues are not essential for hCNT3-mediated nucleoside transport. A series of cysteine-substitution mutants spanning predicted TMs (transmembrane domains) 11–13 was constructed and tested for accessibility to thiol-specific reagents. Mutants M496C, G498C, F563C, A594C, G598C and A606C had no detectable transport activity, indicating that a cysteine substitution at each of these positions was not tolerated. Two functional mutants in putative TM 11 (L480C and S487C) and four in putative TM 12 (N565C, T557C, G567C and I571C) were partially inhibited by MTS (methanethiosulphonate) reagent and high concentrations of uridine protected against inhibition, indicating that TMs 11 and 12 may form part of the nucleoside translocation pathway. The lack of accessibility of MTS reagents to TM 13 mutants suggests that TM 13 is not exposed to the nucleoside translocation pathway. Furthermore, G567C, N565C and I571C mutants were only sensitive to MTSEA (MTS-ethylammonium), a membranepermeant thiol reagent, indicating that these residues may be accessible from the cytoplasmic side of the membrane, providing evidence in support of the predicted orientation of TM 12 in the current putative topology model of hCNT3.
Keywords: cysteine-scanning mutagenesis, human concentrative nucleoside transporter, methanethiosulphonate modification, permeant translocation pathway, structure–function relationship, transmembrane domain
Abbreviations: CMM, complete minimal medium; CNT, concentrative nucleoside transporter; ENT, equilibrative nucleoside transporter; GFP, green fluorescent protein; hCNT, human CNT; hENT, human ENT; MTS, methanethiosulphonate; MTSEA, MTS-ethylammonium; MTSES, MTS-ethylsulphonate; MTSET, MTS-ethyltrimethylammonium; SCAM, substituted-cysteine-accessibility method; TM, transmembrane domain
INTRODUCTION
Nucleosides are central metabolites in all life forms and, as precursors of nucleotides, play an essential role in intermediary metabolism, biosynthesis of macromolecules and cell signalling through interaction with purinergic receptors. Since most nucleosides are hydrophilic molecules and do not cross cell membranes readily by diffusion, their cellular uptake is dependent on the activity of specialized membrane transporter proteins. Two distinct nucleoside transporter families, the ENTs (equilibrative nucleoside transporters) and CNTs (concentrative nucleoside transporters), have been identified by molecular cloning and functional expression of cDNAs encoding transporter proteins from a variety of species, including mammals, protozoan parasites and bacteria. In humans (h), hENT1 and hENT2 mediate facilitated diffusion of nucleosides down their concentration gradients, whereas hCNT1 (human CNT1), hCNT2 and hCNT3 couple uphill nucleoside transport with downhill sodium transport and, in the case of hCNT3, also with downhill proton transport. hENT1 and hENT2 are functionally distinguished by different sensitivities (hENT1≫hENT2) to NBMPR {6-[(4-nitrobenzyl)thio]-9-β-D-ribofuranosylpurine; nitrobenzylthioinosine} and have therefore been assigned the functional designations equilibrative sensitive (es) and equilibrative insensitive (ei) respectively [1,2]. hENT3, which is proton-dependent and, like hENT1, broadly selective, is believed to be a transporter of intracellular membranes [3–5], and hENT4, which mediates equilibrative transport of adenosine [3,6], also transport monoamine neurotransmitters [7]. Three major CNT subtypes differ functionally with respect to their permeant selectivities. In humans as in other mammals, hCNT1 prefers pyrimidine nucleosides but also transports adenosine, whereas hCNT2, which is 72% identical with hCNT1, prefers purine nucleosides but also transports uridine [8,9]. hCNT3, which is 48% identical with either hCNT1 or hCNT2, transports both pyrimidine and purine nucleosides [10]. The ENTs appear to be expressed ubiquitously in human cells. In contrast, the CNTs are found primarily in specialized cell types [9–13], suggesting an important role in absorption, secretion, distribution and elimination of nucleoside and nucleoside analogue drugs.
Interest in nucleoside transporters has been increased because of their potential and proven therapeutic applications in cancer and stroke as well as cardiovascular, parasitic and viral diseases. Nucleoside transporter proteins are critical in controlling extracellular concentrations of adenosine, a physiological ligand for purinergic receptors that is involved in coronary vasodilation, neuromodulation and platelet aggregation [14,15]. The presence of nucleoside transporters in plasma membranes is required for effective cellular uptake of many anticancer nucleoside drugs (e.g. gemcitabine, capecitabine and fludarabine) and is linked mechanistically with drug sensitivities and toxicities [16–18]. Results from an oligonucleotide array study revealed that expression of mRNA encoding hENTs and hCNTs positively correlated with chemosensitivity to nucleoside anticancer drugs [19]. For example, a positive correlation existed between the expression level of hCNT3 mRNA and cytotoxicity of cytorabine and gemcitabine.
While considerable progress has been made in elucidating the structural basis of ENT proteins [20–24], studies on the structurally and functionally important residues of the CNT family are still at an early stage. Mammalian CNT proteins (∼650 amino acid residues) of human, mouse, rat, rabbit and pig share high amino acid sequence identities (>50%). The presence of 13 TMs (transmembrane domains) for mammalian CNTs was predicted from sequence alignment, which has been experimentally verified using glycosylation-scanning mutagenesis [25]. However, the three-dimensional packing of the TMs is still largely unknown, as are the molecular mechanisms by which CNTs bind and translocate their permeants. The first three TMs of mammalian CNTs, which are absent from prokaryote CNTs, are not essential for transport, since both rat CNT1 and hCNT1 that lacked TMs 1–3 retained wild-type transporter properties [25]. TMs 7–9 are thought to form part of the substrate translocation pore for CNT proteins, and four critical residues (Ser319, Glu320, Ser353 and Leu354) in this region of hCNT1 determine permeant selectivities [26,27]. Furthermore, conserved residues Phe316 (putative TM 7) and Gly476 (putative TM 11) were identified as determinants for guanosine sensitivity and membrane expression of hCNT1 respectively [28]. Chimaeric proteins comprising hCNT3 (TMs 1–6) and hCNT1 (TMs 7–13) and hfCNT (hagfish CNT; TMs 1–6) and hCNT1 (TMs 7–13) produced in Xenopus laevis oocytes exhibited hCNT1-like cation interactions as well as hCNT1-like permeant selectivities [29,30], establishing that the structural determinants of cation stoichiometry and binding affinity are located within the C-terminal half of the protein. The loss of proton dependence of the hCNT3–hCNT1 chimaera indicated that the proton-binding site resides in the C-terminal half of the protein [29]. Studies of transporters and their interactions with uridine analogues showed that the three hCNTs exhibit distinct permeant selectivities and nucleoside-binding motifs [31,32].
Several of the 13 putative TMs possess the potential to form amphipathic α-helices, which led to the hypothesis that these helices cluster together in the membrane to form the walls of a water-filled tunnel through which nucleosides translocate the membrane. It was further suggested that the hydroxyl- and amide-containing amino acid side chains within these helices form the nucleoside-binding pocket of hCNT3 via the formation of hydrogen bonds with the hydroxy groups of nucleosides [31]. In the present study, we used SCAM (substituted-cysteine-accessibility method) in conjunction with three thiol-specific MTS (methanethiosulphonate) reagents to systematically address the roles of TMs 11–13 in the formation of the nucleoside translocation pathway. A fully functional cysteine-less hCNT3 mutant was constructed by substitution of the endogenous cysteine residues with serine. The single-cysteine mutants were constructed using cysteine-less hCNT3 as the starting point, and their expression patterns, transport activities and sensitivities to MTS reagents were determined in Saccharomyces cerevisiae. Our results suggest that portions of TMs 11 and 12 face the water-accessible uridine permeation pathway.
EXPERIMENTAL
Strains and media
fui1::TRP1 (MATα, gal, ura3-52, trp1, lys2, ade2, hisd2000 and Δfui1::TRP1), which contains a disruption in the gene encoding the endogenous uridine permease (FUI1) [33], was the parental yeast strain used to produce the recombinant human nucleoside transporters [31,34]. Other strains were generated by transformation of the yeast Escherichia coli shuttle vector pYPGE15 [containing the constitutive PGK (phosphoglycerate kinase) promoter] [35] into fui1::TRP1 with a standard lithium acetate method [36]. Yeast strains were maintained in CMM (complete minimal medium) containing 0.67% yeast nitrogen base (Difco, Detroit, MI, U.S.A.), amino acids (as required to maintain auxotrophic selection) and 2% (w/v) glucose (CMM/GLU). Agar plates contained CMM with various supplements and 2% (w/v) agar (Difco). Plasmids were propagated in E. coli strain DH5α (Invitrogen, Carlsbad, CA, U.S.A.) and maintained in Luria–Bertani broth with 100 μg/ml ampicillin.
Construction of cysteine-scanning mutants
The hCNT3 open reading frame (GenBank® accession number AF305210) was subcloned into the yeast expression vector pYPGE15 to generate pYPhCNT3 as described previously [31]. pYPhCNT3 served as the template to generate plasmid containing cDNAs encoding cysteine-less hCNT3 (pYPhCNT3-cysteine-less) and the pYPhCNT3-cysteine-less served as the template to generate single-cysteine mutants using the QuikChange site-directed mutagenesis kit (Stratagene, La Jolla, CA, U.S.A.) according to the manufacturer's instructions. The sequences of all the constructs were confirmed by DNA sequencing using an ABI PRISM 310 sequence detection system (PerkinElmer Life and Analytical Sciences, Boston, MA, U.S.A.).
Immunofluorescence and confocal microscopy of yeast
Exponentially growing yeast (10 absorbance units, A600 0.7–1.0) were fixed using 3.7% (w/v) formaldehyde for 30 min with occasional mixing, after which cells were centrifuged (5 min, 3000 g) and washed with 4 ml of doubly distilled water. The resulting pellets were resuspended in 1 ml of 700 μg/ml Zymolyase-100 T (MP Biomedicals, Irvine, CA, U.S.A.) in solution B (1.2 M sorbitol and 100 mM potassium phosphate, pH 7.5) for 30–40 min at 30 °C. The yeast suspensions (300 μl) were applied on to poly(L-lysine)-coated coverslips, permeabilized using chilled acetone/methanol (1:1, v/v) and incubated first with blocking buffer [2% (v/v) goat serum in PBS (pH 7.2)] for 30 min and then with anti-hCNT3 monoclonal antibodies in PBST (PBS with 1% Triton X-100) for 30 min. An immunogenic epitope (residues 45–69 of hCNT3, REHTNTKQDEEQVTVEQDSPRNREH) was used to generate the monoclonal antibodies against hCNT3 [31,37]. After extensive washing with PBST, the yeast were stained with the secondary antibodies [Alexa Fluor 488 goat-anti mouse IgM (1:250 dilution in PBS; Molecular Probes)] for 30 min, followed by extensive washing with PBS. The coverslips were mounted and dried overnight. Confocal images were collected using a Zeiss LSM510 confocal laser scanning microscope with a 60×1.4 objective (F-Fluar) using a frame size of 512×512 pixels with a pixel resolution of 0.08 μm and a pixel depth of 8 bits.
Transport assay in S. cerevisiae
Yeast cells producing recombinant hCNT3 mutant proteins were grown in CMM/GLU medium to an A600 (absorbance) of 0.7–1.2, washed twice in sodium-containing transport buffer (5 mM D-glucose, 20 mM Tris/HCl, 3 mM K2HPO4, 1 mM MgCl2, 2 mM CaCl2 and 130 mM NaCl, pH 7.4) and resuspended to A600 4.0 in sodium-containing transport buffer (pH 7.4). All transport assays were performed at room temperature (24 °C) and pH 7.4 using a cell-harvester-based method as described previously [31,38]. Transport reactions were initiated by rapid mixing of 50 μl of yeast suspension with 50 μl of sodium-containing transport buffer (pH 7.4) with [3H]nucleoside at twice the final concentration in each of the individual wells of 96-well microtitre plates. Nucleoside uptake into yeast cells producing recombinant hCNT3 mutants was linear for approx. 10–15 min, and kinetic studies were therefore performed using rates obtained from 5 min exposures to 3H-labelled nucleosides. At the end of 5 min incubations, the yeast cells were collected on glass-fibre filtermats (Skatron Instruments, Lier, Norway) using the semi-automated cell harvester (Micro96 Harvester; Skatron Instruments) with continuous washing with demineralized water. The individual filter portions, which corresponded to individual wells of the microtitre plates, were excised and transferred to scintillation vials for liquid-scintillation counting. Uptake rates are presented as pmol/mg of yeast protein. The quantification of yeast protein was performed using a Bio-Rad protein assay kit (Bio-Rad, Hercules, CA, U.S.A.). Km (permeant concentration at half-maximal unidirectional flux) and Vmax (maximum transport rate) values for transport of nucleosides by yeast producing cysteine-less hCNT3 or single-cysteine mutant were calculated from rate-versus-concentration plots using PRISM GraphPad version 3.0 software (GraphPad Software, San Diego, CA, U.S.A.). Statistical significance of the reported data sets was evaluated using t tests.
MTS modification experiments
Yeast containing pYPhCNT3-cysteine-less or one of the single-cysteine mutant plasmids were grown in CMM to an A600 of 0.7–1.2, washed twice in sodium-containing transport buffer (pH 7.4) and resuspended to an A600 of 2.2. All MTS reagents were dissolved in ice-cold doubly distilled water and kept on ice until use. Portions (0.9 ml) of the yeast suspension were distributed to microcentrifuge tubes to which had been added 100 μl of 25 mM MTSEA (MTS-ethylammonium), 100 mM MTSES (MTS-ethylsulphonate) or 10 mM MTSET (MTS-ethyltrimethylammonium; Toronto Research Chemicals, Toronto, ON, Canada) alone or together with 10 mM uridine as a protection permeant. After a 5 min incubation period, the cells were centrifuged and washed three times with ice-cold PBS to remove unchanged MTS reagent and uridine. The cells were resuspended to an A600 of 4.0 in sodium-containing transport buffer (pH 7.4) and distributed to 96-well microtitre plates for uridine transport assays. For each individual mutant, the accessibility to MTS reagents was normalized by computing the percentages of uridine uptake in the presence of the MTS reagent relative to that in its absence.
RESULTS
Generation and characterization of cysteine-less hCNT3
Most endogenous cysteine residues are clustered in the C-terminal part of hCNT3 (Figure 1). To test the role of endogenous cysteine residue in hCNT3, 14 mutants in which a single cysteine residue was changed to serine were constructed and the kinetic properties of uridine and adenosine transport were compared with those of wild-type hCNT3 (results not shown). No obvious differences were observed between the mutants and wild-type hCNT3, indicating that none of the individual endogenous cysteine residues was important for the function of activity of hCNT3. Therefore a cysteine-less hCNT3 mutant, in which all 14 endogenous cysteine residues, were replaced with serine residues was generated by site-directed mutagenesis. The resulting cysteine-less hCNT3 protein was produced in yeast and characterized for its ability to transport several naturally occurring nucleosides (uridine, cytidine, adenosine and inosine) and nucleoside analogues (gemcitabine and clofarabine). Compared with wild-type hCNT3, the cysteine-less transporter exhibited similar affinities and a 4-fold increase in transport activities (Table 1). The global increase in Vmax values for various nucleosides may have been due to an increased production of cysteine-less hCNT3 in yeast cells.
Figure 1. Predicated topology model of hCNT3.
Residues shaded in grey are endogenous cysteine residues and in black are MTS-sensitive residues.
Table 1. Kinetic properties of wild-type and cysteine-less hCNT3 produced in S. cerevisiae.
The Km and Vmax values shown are the means±S.E.M. for three or four separate experiments.
| hCNT3 | Cysteine-less hCNT3 | |||
|---|---|---|---|---|
| Substrate | Km (μM) | Vmax (pmol·min−1·mg−1) | Km (μM) | Vmax (pmol·min−1·mg−1) |
| Uridine | 1.7±0.3 | 1402±286 | 2.5±0.4 | 5800±142 |
| Cytidine | 3.6±1.3 | 1310±113 | 2.3±0.4 | 5448±112 |
| Adenosine | 2.2±0.7 | 1020±44 | 2.1±0.5 | 5020±110 |
| Inosine | 2.1±0.6 | 1740±114 | 3.1±0.5 | 6420±180 |
| Gemcitabine | 3.8±1.0 | 390±50 | 4.1±1.1 | 1600±50 |
| Clofarabine | 4.1±1.3 | 410±40 | 4.6±1.5 | 1800±120 |
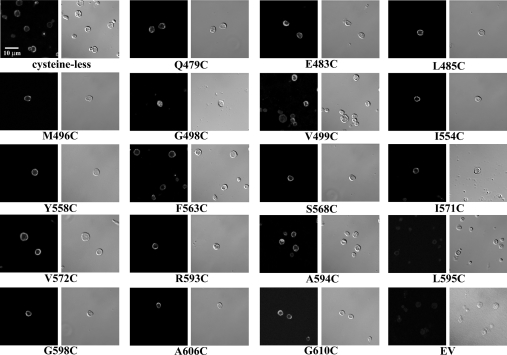
Indirect immunostaining was used to compare the localization of hCNT3 and cysteine-less hCNT3 proteins in yeast. As shown in Figure 2, yeast producing recombinant cysteine-less hCNT3 exhibited staining patterns similar to those of wild-type hCNT3, in which staining was predominantly localized to the plasma membrane. No significant fluorescence was observed for yeast transformed with the insert-free vector (pYPGE15) or for yeast producing recombinant hCNT3 or cysteine-less hCNT3 that were treated with either IgM isotype antibodies or secondary antibodies only (results not shown).
Figure 2. Immunostaining of yeast producing cysteine-less hCNT3 or some single-cysteine hCNT3 mutants.
Yeast cells producing wild-type hCNT3, cysteine-less hCNT3 or one of the 63 single-cysteine hCNT3 mutants were treated with anti-hCNT3 IgM as primary antibodies followed by Alexa Fluor 488 goat anti-mouse IgM as secondary antibodies. The images were captured by confocal microscopy as described in the Materials and methods section using a ×60 objective; and the scale bar represents a distance of 10 μm. Negative control cells containing empty vector (EV; pYPGE15) with no insert were stained with both the anti-hCNT3 IgM and Alexa Fluor 488 IgG antibodies. Other controls included yeast producing cysteine-less hCNT3 stained with IgM isotype primary antibodies in place of the anti-hCNT3 IgM or only with the secondary antibodies. No significant fluorescence signals were detected under these control conditions (results not shown). Images presented are representative of 10–20 similar images recorded for cysteine-less hCNT3 and some of the single-cysteine hCNT3 mutants.
Recombinant hCNT3 produced in X. laevis oocytes exhibits a sodium/nucleoside coupling ratio of 2:1 [10]. Using a radio-isotope assay, the sodium dependence of uridine influx mediated by recombinant cysteine-less hCNT3 produced in yeast was analysed. When uridine uptake was measured as a function of sodium concentration, a K50 (sodium concentration at half-maximal activation rate) value for sodium activation of 4.5±0.6 mM was observed for cysteine-less hCNT3. Fitting the data to the Hill equation, v=Vmax·[Na+]/(K50n + [Na+]n), gave Hill coefficients (n) of 2.1±0.2 (wild-type hCNT3) and 2.0±0.1 (cysteine-less hCNT3), indicating that a sodium/nucleoside coupling ratio of 2:1 remained unchanged for cysteine-less hCNT3. These results demonstrated that the cysteine-less transporter, which maintained wild-type properties, is an appropriate substituent of hCNT3 for SCAM studies.
Functional expression of single-cysteine mutants in yeast
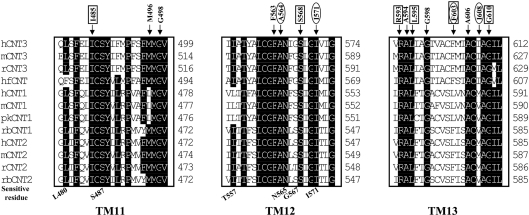
The cysteine-less hCNT3 was used as the template to construct 63 site-directed mutants in which single-cysteine mutations were systematically introduced at positions 479–499, 554–574 and 592–612, encompassing all of predicted TMs 11–13 of hCNT3. The kinetic properties of each single-cysteine mutant were assessed by measurements of the concentration dependence of transport of [3H]uridine and [3H]adenosine (see Supplementary Tables 1–3 at http://www.BiochemJ.org/bj/394/bj3940389add.htm). Although the Vmax values varied, most of the cysteine-substituted mutants were functional, exhibiting micromolar Km values for uridine and adenosine similar to those of wild-type and cysteine-less hCNT3 (Supplementary Tables 1–3; http://www.BiochemJ.org/bj/394/bj3940389add.htm). Among the functional mutants, only three, A564C, I571C and F603C, exhibited large increases in Km values for uridine and adenosine relative to those of cysteine-less hCNT3. I608C displayed higher affinity and capacity for uridine than for adenosine: Km values of 6.0±1.2 and 18±3.2 μM respectively; Vmax values of 1250±90 and 250±50 pmol·mg−1·min−1 respectively. Six mutants (I485C, S568C, R593C, L595C, F603C and G610C) showed very low transport capacity, with Vmax values ≤200 pmol·mg−1·min−1 for both uridine and adenosine. While the activities of these mutants were low, they were high enough above background to measure the effects of MTS reagents and thus were included in subsequent analyses. However, no significant transport activity was detected for the six other mutants (M496C, G498C, F563C, A594C, G598C and A606C). Multiple sequence alignments across the mammalian CNT family (Figure 3) revealed that most of the mutants that exhibited compromised transport affinity and/or capacity were highly conserved residues, indicating that these residues are structurally and/or functionally important for hCNT3 transport activity. Substitution of Met496, Gly498, Phe563 and Gly598 with an alanine residue resulted in two functional mutants. M496A exhibited Km values of 2.7±0.3 and 3.1±0.6 μM and Vmax values of 950±30 and 990±50 pmol·(mg of protein)−1·min−1 for uridine and adenosine respectively, and G598A exhibited Km values of 3.1±0.6 and 2.7±0.6 μM and Vmax values of 320±30 and 300±50 pmol·mg−1·min−1 for uridine and adenosine respectively. However, mutants G498A and F563A remained non-functional, indicating that something other than the hydrophobic property of glycine or phenylalanine may be required at these positions.
Figure 3. Protein sequence alignment of TMs 11–13 of the CNT transporter family.
Single-cysteine mutants that were inactivated by cysteine substitution are indicated. Single-cysteine mutants that showed large decreases in Vmax values or increase in Km values are boxed or circled respectively. Abbreviations: h, human; hf, hagfish; r, rat; m, mouse; pk, pig; and rb, rabbit.
Expression of the single-cysteine mutants in yeast was assessed visually by indirect immunofluorescence laser confocal microscopy using monoclonal anti-hCNT3 antibodies (Figure 2). All recombinant single-cysteine hCNT3 mutants except G498C and L595C exhibited staining patterns similar to those of wild-type and cysteine-less hCNT3, for which staining was predominantly localized to the plasma membrane (Figure 2). Stronger intracellular staining was observed in yeast producing G598C and very weak staining was observed in yeast producing L595C (for which staining could be visualized in the magnified on-screen version of the high-resolution digital image shown in Figure 2); these results were consistent with the absence of detectable transport for mutant G598C and extremely low transport activity for mutant L595C (Supplementary Tables 1 and 3; http://www.BiochemJ.org/bj/394/bj3940389add.htm). The lack of nucleoside transport activity observed in other substitution mutants was not the result of reduced protein expression, since strong plasma membrane staining was observed for yeast that produced either of these mutants. No significant fluorescent signal was observed for yeast transformed with the insert-free vector (pYPGE15) or yeast producing recombinant hCNT3 that were treated with either IgM isotype antibodies or secondary antibodies only (results not shown).
SCAM of TMs 11–13
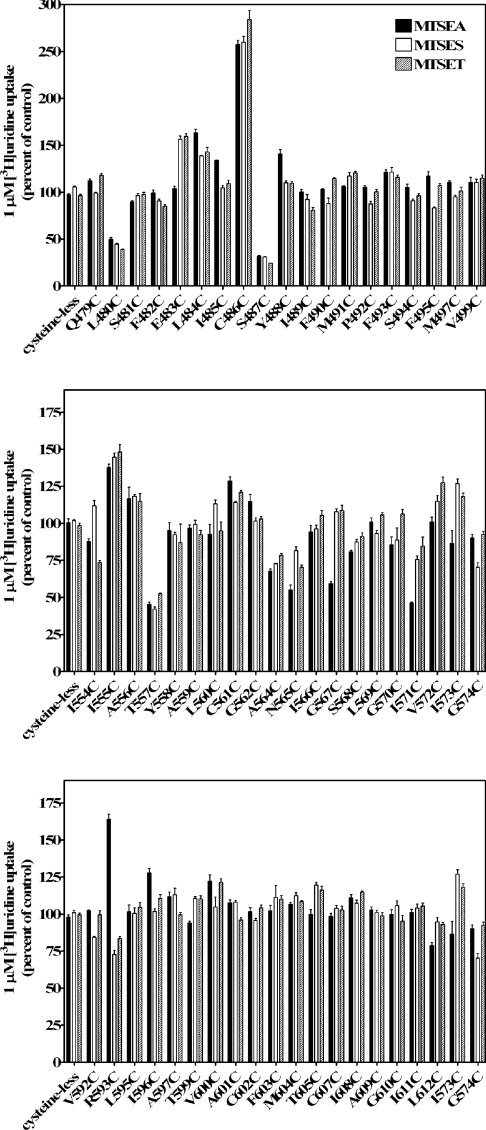
The function of cysteine-less hCNT3 was not affected by the thiol-specific reagents, p-chloromercuribenzenesulphonate (results not shown) and MTS reagents (Figure 4). The goal of SCAM was to determine if any portions of TMs 11–13 contribute to the structure of the water-filled pore through which the permeant (uridine) is translocated into the cell. Three MTS reagents (MTSEA, MTSES and MTSET) with different sizes, charges and membrane permeabilities were used. Any time that a cysteine residue has replaced an amino acid residue that is normally exposed to the aqueous translocation pore, MTSES, MTSET or MTESA should have access to the cysteine thiol group, resulting in formation of cysteine–MTS adduct, thus blocking permeant translocation.
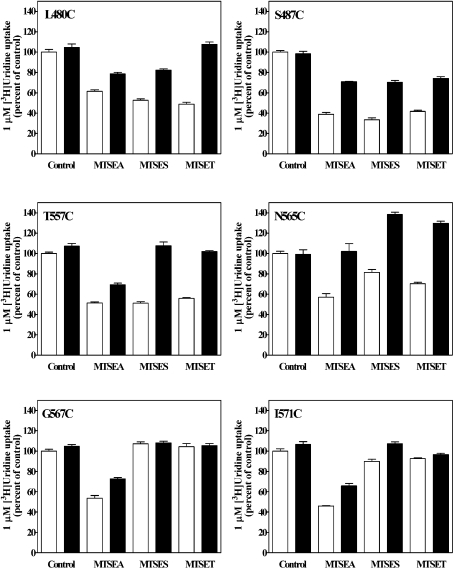
Figure 4. Reactivity of cysteine residues within TM 11 (top panel), TM 12 (middle panel) and TM 13 (bottom panel) with three MTS reagents.
Mutants are designated by the single-letter amino acid abbreviation for the targeted residue, followed by the sequence position number in hCNT3 and a second letter indicating the cysteine replacement. These three panels show the effects of MTSEA, MTSES and MTSET on the ability of single-cysteine substituted mutants to transport 1 μM [3H]uridine. Activity is expressed as the mean percentage of control value (no MTS reagent) for each individual mutant with standard deviations of quadruplicate values from a representative experiment. Black bars, 2.5 mM MTSEA; white bars, 10 mM MTSES; shaded bars, 1 mM MTSET. Three independent experiments were performed and similar results were obtained.
Figure 4 presents the normalized uridine transport activities of 57 single-cysteine mutants after treatment with 2.5 mM MTSEA, 10 mM MTSES or 1 mM MTSET (100% was set as the activity of individual mutants without being incubated with MTS reagents). In every series of SCAM experiments, the cysteine-less hCNT3 served as a negative control and displayed no sensitivity towards the MTS reagents. Most of the mutants in TMs 11 and 12 and all mutants of TM 13 were not affected by the MTS reagents, suggesting that either modification of these cysteine residues did not affect uridine translocation or, most likely, these residues were not accessible to MTS reagents. The uridine transport activities of single-cysteine mutants L480C and S487C in TM 11 and T557C in TM 12 were markedly reduced (<60% of control values) by all three reagents, indicating that these residues were accessible to hydrophilic reagents and probably faced the permeant translocation channel. Although positively charged, MTSEA is small and can cross plasma membranes by diffusion and react with cysteine side chains from both the extracellular and cytoplasmic sides of the membrane [39]. The transport activities of N565C, G567C and I571C in TM 12 were inhibited only by MTSEA (<60% of control values) but not by MTSES and MTSET, which are membrane-impermeant and have larger sizes than MTSEA. Several mutants (E483C, L484C and C486C in TM 11, I555C in TM 12 and R593C in TM 13) showed higher uridine transportability when treated with MTS reagents; the reason for the increased activities is unknown.
Uridine protection of MTS modifications
To test the hypothesis that the mutants that showed high sensitivity to modifications by the MTS reagents were exposed to the aqueous translocation pore, permeant protection analysis was undertaken. It is assumed that occupying the translocation pathway with permeant before and during exposure to MTS reagents blocks access of the reagents to the pore-lining cysteine residues, thus protecting the residues from thiol modifications. In preliminary experiments, the protection effect was shown to be concentration-dependent, with maximal protection achieved at high concentration of uridine (5–10 mM); subsequent protection experiments used 10 mM uridine. Uridine at high concentration (10 mM) protected all six MTS-sensitive mutants from inhibition by MTS modification, although the extent of protection varied among mutants and MTS reagent applied (Figure 5). These results provided evidence that Leu480, Ser487, Thr557, Asn565, Gly567 and Ile571 in TMs 11 and 12 formed part of the nucleoside permeation pathway. For MTSEA, application of 10 mM uridine completely protected N565C from inhibitor, whereas the other mutants were partially protected. For MTSES, uridine completely protected T557C but only partially protected L480C and S487C. For MTSET, L480C and T557C were completely protected by 10 mM uridine, whereas S487C was only partially protected. The experimental method used in the permeant protection assays assessed the effects of uridine on MTS modifications by co-incubation of cells with uridine and the MTS reagent, conditions that were designed to examine the ability of a reversible reaction (uridine binding and translocation) to slow an irreversible reaction (MTS modifications). With the co-incubation method, a less-than-complete protection is usually obtained (for a review, see [40]). The incomplete protection by high concentrations of nucleosides against thiol reagent modifications was also observed in studies with other nucleoside transporters [22,41,42]. A 40% stimulation of uridine uptake after co-incubating mutant N565C with 10 mM uridine and MTSES (or MTSET) was observed and the reason remains unclear. A stimulation was also observed with lower concentrations of uridine (e.g. 1 and 5 mM; results not shown). It is possible that the local steric structure of the transporter mutant was altered by MTS reagents and uridine and that this conformational change favoured uridine transport.
Figure 5. Protection from MTS reagent inhibition by uridine.
Yeast producing recombinant hCNT3 mutants were incubated with MTS reagents as indicated in the presence (black bars) or absence (control; white bars) of 10 mM uridine for 5 min, washed three times with PBS and assayed for their ability to transport 1 μM [3H]uridine. Activity is expressed as the mean percentage of control value (no MTS reagent and no 10 mM uridine) for each individual mutant, with standard deviations of quadruplicate values from a representative experiment. Three independent experiments were performed and similar results were obtained.
DISCUSSION
The role of putative TMs 11–13 in the current topology model of hCNT3 (Figure 1) was investigated using cysteine-scanning mutagenesis in conjunction with thiol-specific MTS reagents. A total of 63 single-cysteine hCNT3 mutants were created from fully functional cysteine-less hCNT3 by individually changing each residue along TMs 11–13 to a cysteine residue. Immunofluorescence staining and transport assays were used to assess the membrane abundance, kinetic properties and sensitivities of the mutant proteins to MTS modifications. Cysteine substitutions at Met496, Gly498, Phe563, Ala594, Gly598 and Ala606 abolished uridine and adenosine transport activities, and substitutions at Ile485, Ala564, Ile571, Arg593, Leu595, Phe603, Ile608 and Gly610 resulted in markedly decreased Vmax values and/or increased Km values, whereas substitutions at other positions resulted in either little or no effect on transport properties. Most of the residues, the mutation of which resulted in large functional changes, are highly conserved across the CNT family (Figure 2), consistent with their importance in transporter structure and/or function.
Immunostaining demonstrated that non-functional single-cysteine mutants, except G498C, were produced and localized to cell surfaces at levels similar to those of wild-type and cysteine-less hCNT3, indicating that the impaired transport activity of the mutants was not due to defective trafficking. Cysteine substitution might have caused changes in transporter folding that resulted in compromised transportability. Future studies using a screen for second site suppressor residue(s) could be used to further assess the potential roles of these residues in protein processing. The decreased transport capacities of mutant L595C were due to its low abundance in yeast. Mutant G498C was detected mainly in intracellular regions, consistent with the observation that the GFP (green fluorescent protein)-tagged corresponding mutants of hCNT1 (G476A–GFP and G476L–GFP) were non-functional and could not be detected on plasma membranes in undifferentiated or differentiated Madin–Darby canine kidney cells [28]. No transport activity was detected when Gly498 or Phe563 was converted into the hydrophilic small-side-chain residue alanine, suggesting that these two residues are essential and cannot be substituted.
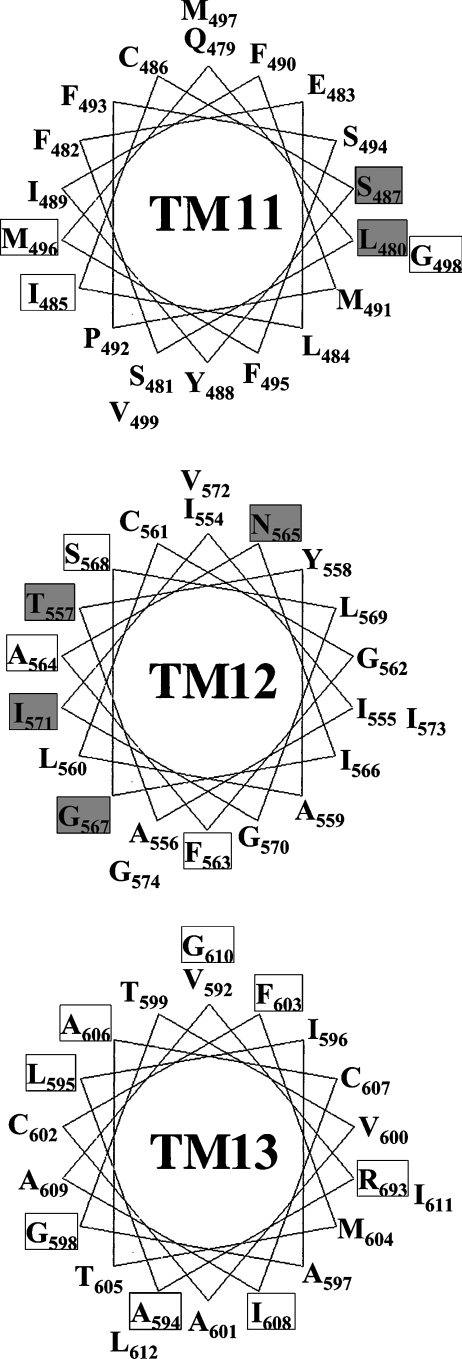
To determine the structural locations of the residues identified in the present study, helical wheel projections of TMs 11–13 were generated (Figure 6). Both TM 11 and TM 12 are moderately amphipathic, suggesting that these two domains could line the permeant translocation pore. All of the MTS-accessible residues (Leu480, Ser487, Thr557, Asn565, Gly567 and Ile571) except Gly567 are highly conserved across the CNT family. Although the extent of uridine protection varied among the mutants and with different MTS reagents, all six MTS-sensitive residues were protected by uridine, supplying further evidence that these residues face the pore-lining pathway. Generally, uridine protection was higher for modifications by MTSES and MTSET than by MTSEA, probably because MTSEA was able to diffuse across the plasma membrane, affecting uridine binding from both sides of the membrane. S487C and L480C were accessible to three MTS reagents and are located close to each other on the hydrophilic face of TM 11 (Figure 6). Residues Met496 and Ile485, which were sensitive to cysteine substitution, are buried on the hydrophilic face of TM 11, suggesting that these two residues might be involved in stabilizing the structure of the transporter. Only two residues of TM 11 were MTS-sensitive, suggesting that TM 11 is partially buried in the lipid membrane and contributes only a small portion of the nucleoside translocation pathway. TM 11 also contains a proline residue, which could induce a kink in its structure bringing residues Met496 and Ile485 close to the water-filling pore. It is possible that TM 11 has a high degree of conformational flexibility and is involved in helical movements during the transport cycle, allowing the pore-lining side of the α-helix to be solvent accessible during nucleoside flux.
Figure 6. Helix wheel projection of putative TMs 11–13 of hCNT3.
Mutants that were sensitive to MTS reagents are in shaded boxes. Mutants that showed no assayable activity or significantly changed kinetic properties are labelled in white boxes.
Four MTS-sensitive residues and most of the cysteine substitution-sensitive residues in TM 12 are clustered close to each other on the hydrophilic face of the putative TM segment (Figure 6), suggesting that TM 12 forms part of the nucleoside translocation pore. Ile571 is the second residue of a highly conserved GXXXG helix–helix interaction motif involving Gly570 and Gly574. Given the accessibility of MTSEA to I571C and the effect of I571C on permeant binding affinity changes (20-fold decrease in affinities for both uridine and adenosine compared with cysteine-less hCNT3), Ile571 might be a pore-lining residue that is also involved in permeant interactions. It could also play a role in stabilizing the helix–helix packing interface involving Gly570 and Gly574. G567C, N565C and I571C mutants were only sensitive to the membrane-permeant thiol reagent MTSEA, indicating that these residues may be accessible from the cytoplasmic side of the membrane, providing evidence in support of the predicted orientation of TM 12 in the current putative topology model of hCNT3 in which these three residues are located in the cytoplasmic side of the plasma membrane (Figure 1).
Putative TM 13 mainly contained hydrophobic residues and displayed no accessibility to any MTS reagent, suggesting that it is not in the permeabilization pathway. However, several of the cysteine substitution mutants in TM 13 displayed altered kinetic properties for nucleoside transport. Mutation of the highly conserved hydrophobic residues in TM 13 to hydrophilic cysteine residues was poorly tolerated. It is possible that these mutations created a conformational distortion that resulted in a non-functional transporter. TM 13 might be one of the outer helices that surround the inner helical bundle that comprises the aqueous permeant-binding cavity. The compromised transportability observed in nearly half of the TM 13 mutants suggests an important structural role for this TM segment, and the possibility that some of these unsubstitutable residues serve as direct determinants for permeant interactions cannot be excluded. The differential binding affinities and transport capacities that mutant I608C displayed for uridine and adenosine suggest a possible role of TM 13 in the recognition of pyrimidine and purine nucleosides.
Knowing the structure and mechanism of a transporter protein is crucial for understanding its biological function and designing drugs to interact with it. The lack of a crystal structure for any CNT family member is an impediment to interpreting the significance of the residues with functional roles within the TM regions. Although labour intensive, SCAM is a powerful tool in the elucidation and estimation of the tertiary structures of various membrane proteins. Mutagenesis approaches have identified several highly conserved residues that may be potential determinants for permeant interaction and/or structural stability and are worth further investigation. Direct evidence from SCAM studies suggested that part of the hydrophilic faces of TMs 11 and 12 contributes to the nucleoside translocation pathway, and TM 13 is not involved in permeation but may play a crucial role in maintaining proper protein functions. Extension of the present study to the remaining TM segments will provide further insights into the mechanism and structure of CNTs.
Acknowledgments
This research was funded by the National Cancer Institute of Canada (to C. E. C. and J. D. Y.) and the Alberta Cancer Foundation (to C. E. C. and J. D. Y.). C. E. C. is Canada Research Chair in Oncology and J. D. Y. is Heritage Scientist of the Alberta Heritage Foundation for Medical Research. Studentship support to J. Z. was from the Canadian Institutes of Health Research, the Alberta Heritage Foundation for Medical Research and the Department of Oncology Endowed Studentship.
References
- 1.Cass C. E., Young J. D., Baldwin S. A., Cabrita M. A., Graham K. A., Griffiths M., Jennings L. L., Mackey J. R., Ng A. M. L., Ritzel M. W. L., et al. Nucleoside transporters of mammalian cells. In: Amidon G. L., Sadee W., editors. Membrane Transporters as Drug Targets. Dordrecht: Kluwer Academic/Plenum Publishers; 1999. pp. 313–352. [DOI] [PubMed] [Google Scholar]
- 2.Damaraju V. L., Damaraju S., Young J. D., Baldwin S. A., Mackey J., Sawyer M. B., Cass C. E. Nucleoside anticancer drugs: the role of nucleoside transporters in resistance to cancer chemotherapy. Oncogene. 2003;22:7524–7536. doi: 10.1038/sj.onc.1206952. [DOI] [PubMed] [Google Scholar]
- 3.Acimovic Y., Coe I. R. Molecular evolution of the equilibrative nucleoside transporter family: identification of novel family members in prokaryotes and eukaryotes. Mol. Biol. Evol. 2002;19:2199–2210. doi: 10.1093/oxfordjournals.molbev.a004044. [DOI] [PubMed] [Google Scholar]
- 4.Baldwin S. A., Yao S. Y., Hyde R. J., Ng A. M., Foppolo S., Barnes K., Ritzel M. W., Cass C. E., Young J. D. Functional characterization of novel human and mouse equilibrative nucleoside transporters (hENT3 and mENT3) located in intracellular membranes. J. Biol. Chem. 2005;280:15880–15887. doi: 10.1074/jbc.M414337200. [DOI] [PubMed] [Google Scholar]
- 5.Hyde R. J., Cass C. E., Young J. D., Baldwin S. A. The ENT family of eukaryote nucleoside and nucleobase transporters: recent advances in the investigation of structure/function relationships and the identification of novel isoforms. Mol. Membr. Biol. 2001;18:53–63. [PubMed] [Google Scholar]
- 6.Baldwin S. A., Beal P. R., Yao S. Y., King A. E., Cass C. E., Young J. D. The equilibrative nucleoside transporter family, SLC29. Pflugers Arch. 2004;447:735–743. doi: 10.1007/s00424-003-1103-2. [DOI] [PubMed] [Google Scholar]
- 7.Engel K., Zhou M., Wang J. Identification and characterization of a novel monoamine transporter in the human brain. J. Biol. Chem. 2004;279:50042–50049. doi: 10.1074/jbc.M407913200. [DOI] [PubMed] [Google Scholar]
- 8.Ritzel M. W., Yao S. Y., Huang M. Y., Elliott J. F., Cass C. E., Young J. D. Molecular cloning and functional expression of cDNAs encoding a human Na+-nucleoside cotransporter (hCNT1) Am. J. Physiol. 1997;272:C707–C714. doi: 10.1152/ajpcell.1997.272.2.C707. [DOI] [PubMed] [Google Scholar]
- 9.Ritzel M. W., Yao S. Y., Ng A. M., Mackey J. R., Cass C. E., Young J. D. Molecular cloning, functional expression and chromosomal localization of a cDNA encoding a human Na+/nucleoside cotransporter (hCNT2) selective for purine nucleosides and uridine. Mol. Membr. Biol. 1998;15:203–211. doi: 10.3109/09687689709044322. [DOI] [PubMed] [Google Scholar]
- 10.Ritzel M. W. L., Ng A. M., Yao S. Y. M., Graham K., Loewen S. K., Smith K. M., Ritzel R. G., Mowles D. A., Carpenter P., Chen X., et al. Molecular identification and characterization of novel human and mouse concentrative Na+-nucleoside cotransporter proteins (hCNT3 and mCNT3) broadly selective for purine and pyrimidine nucleosides (system cib) J. Biol. Chem. 2001;276:2914–2927. doi: 10.1074/jbc.M007746200. [DOI] [PubMed] [Google Scholar]
- 11.Ngo L. Y., Patil S. D., Unadkat J. D. Ontogenic and longitudinal activity of Na+-nucleoside transporters in the human intestine. Am. J. Physiol. Gastrointest. Liver Physiol. 2001;280:G475–G481. doi: 10.1152/ajpgi.2001.280.3.G475. [DOI] [PubMed] [Google Scholar]
- 12.del Santo B., Tarafa G., Felipe A., Casado F. J., Pastor-Anglada M. Developmental regulation of the concentrative nucleoside transporters CNT1 and CNT2 in rat liver. J. Hepatol. 2001;34:873–880. doi: 10.1016/s0168-8278(01)00036-8. [DOI] [PubMed] [Google Scholar]
- 13.Li J. Y., Boado R. J., Pardridge W. M. Differential kinetics of transport of 2′,3′-dideoxyinosine and adenosine via concentrative Na+ nucleoside transporter CNT2 cloned from rat blood-brain barrier. J. Pharmacol. Exp. Ther. 2001;299:735–740. [PubMed] [Google Scholar]
- 14.Van Belle H. Nucleoside transport inhibition: a therapeutic approach to cardioprotection via adenosine? Cardiovasc. Res. 1993;27:68–76. doi: 10.1093/cvr/27.1.68. [DOI] [PubMed] [Google Scholar]
- 15.Baldwin S. A., Mackey J. R., Cass C. E., Young J. D. Nucleoside transporters: molecular biology and implications for therapeutic development. Mol. Med. Today. 1999;5:216–224. doi: 10.1016/S1357-4310(99)01459-8. [DOI] [PubMed] [Google Scholar]
- 16.Mackey J. R., Jennings L. L., Clarke M. L., Santos C. L., Dabbagh L., Vsianska M., Koski S. L., Coupland R. W., Baldwin S. A., Young J. D., et al. Immunohistochemical variation of human equilibrative nucleoside transporter 1 protein in primary breast cancers. Clin. Cancer Res. 2002;8:110–116. [PubMed] [Google Scholar]
- 17.Mackey J. R., Galmarini C. M., Graham K. A., Joy A. A., Delmer A., Dabbagh L., Glubrecht D., Jewell L. D., Lai R., Lang T., et al. Quantitative analysis of nucleoside transporter and metabolism gene expression in chronic lymphocytic leukemia (CLL): identification of fludarabine-sensitive and -insensitive populations. Blood. 2005;105:767–774. doi: 10.1182/blood-2004-03-1046. [DOI] [PubMed] [Google Scholar]
- 18.Mackey J. R., Baldwin S. A., Young J. D., Cass C. E. Nucleoside transport and its significance for anticancer drug resistance. Drug Resist. Updates. 1998;1:310–324. doi: 10.1016/s1368-7646(98)80047-2. [DOI] [PubMed] [Google Scholar]
- 19.Huang Y., Anderle P., Bussey K. J., Barbacioru C., Shankavaram U., Dai Z., Reinhold W. C., Papp A., Weinstein J. N., Sadee W. Membrane transporters and channels: role of the transportome in cancer chemosensitivity and chemoresistance. Cancer Res. 2004;64:4294–4301. doi: 10.1158/0008-5472.CAN-03-3884. [DOI] [PubMed] [Google Scholar]
- 20.Vickers M. F., Mani R. S., Sundaram M., Hogue D. L., Young J. D., Baldwin S. A., Cass C. E. Functional production and reconstitution of the human equilibrative nucleoside transporter (hENT1) in Saccharomyces cerevisiae. Interaction of inhibitors of nucleoside transport with recombinant hENT1 and a glycosylation-defective derivative (hENT1/N48Q) Biochem. J. 1999;339:21–32. [PMC free article] [PubMed] [Google Scholar]
- 21.Visser F., Vickers M. F., Ng A. M., Baldwin S. A., Young J. D., Cass C. E. Mutation of residue 33 of human equilibrative nucleoside transporters 1 and 2 alters sensitivity to inhibition of transport by dilazep and dipyridamole. J. Biol. Chem. 2002;277:395–401. doi: 10.1074/jbc.M105324200. [DOI] [PubMed] [Google Scholar]
- 22.Visser F., Zhang J., Raborn R. T., Baldwin S. A., Young J. D., Cass C. E. Residue 33 of human equilibrative nucleoside transporter 2 is a functionally important component of both the dipyridamole and nucleoside binding sites. Mol. Pharmacol. 2005;67:1291–1298. doi: 10.1124/mol.104.005884. [DOI] [PubMed] [Google Scholar]
- 23.Damaraju S., Zhang J., Visser F., Tackaberry T., Dufour J., Smith K. M., Slugoski M., Ritzel M. W., Baldwin S. A., Young J. D., et al. Identification and functional characterization of variants in human concentrative nucleoside transporter 3, hCNT3 (SLC28A3), arising from single nucleotide polymorphisms in coding regions of the hCNT3 gene. Pharmacogenet. Genomics. 2005;15:173–182. doi: 10.1097/01213011-200503000-00006. [DOI] [PubMed] [Google Scholar]
- 24.Sundaram M., Yao S. Y. M., Ng A. M. L., Griffiths M., Cass C. E., Baldwin S. A., Young J. D. Chimeric constructs between human and rat equilibrative nucleoside transporters (hENT1 and rENT1) reveal hENT1 structural domains interacting with coronary vasoactive drugs. J. Biol. Chem. 1998;273:21519–21525. doi: 10.1074/jbc.273.34.21519. [DOI] [PubMed] [Google Scholar]
- 25.Hamilton S. R., Yao S. Y., Ingram J. C., Hadden D. A., Ritzel M. W., Gallagher M. P., Henderson P. J., Cass C. E., Young J. D., Baldwin S. A. Subcellular distribution and membrane topology of the mammalian concentrative Na+-nucleoside cotransporter rCNT1. J. Biol. Chem. 2001;276:27981–27988. doi: 10.1074/jbc.M100518200. [DOI] [PubMed] [Google Scholar]
- 26.Loewen S. K., Ng A. M., Yao S. Y., Cass C. E., Baldwin S. A., Young J. D. Identification of amino acid residues responsible for the pyrimidine and purine nucleoside specificities of human concentrative Na+ nucleoside cotransporters hCNT1 and hCNT2. J. Biol. Chem. 1999;274:24475–24484. doi: 10.1074/jbc.274.35.24475. [DOI] [PubMed] [Google Scholar]
- 27.Wang J., Giacomini K. M. Molecular determinants of substrate selectivity in Na+-dependent nucleoside transporters. J. Biol. Chem. 1997;272:28845–28848. doi: 10.1074/jbc.272.46.28845. [DOI] [PubMed] [Google Scholar]
- 28.Lai Y., Lee E. W., Ton C. C., Vijay S., Zhang H., Unadkat J. D. Conserved residues F316 and G476 in the concentrative nucleoside transporter 1 (hCNT1) affect guanosine sensitivity and membrane expression, respectively. Am. J. Physiol. Cell Physiol. 2005;288:C39–C45. doi: 10.1152/ajpcell.00192.2004. [DOI] [PubMed] [Google Scholar]
- 29.Smith K. M., Slugoski M. D., Loewen S. K., Ng A. M., Yao S. Y., Chen X. Z., Karpinski E., Cass C. E., Baldwin S. A., Young J. D. The broadly selective human Na+/nucleoside cotransporter (hCNT3) exhibits novel cation-coupled nucleoside transport characteristics. J. Biol. Chem. 2005;280:25436–25449. doi: 10.1074/jbc.M409454200. [DOI] [PubMed] [Google Scholar]
- 30.Yao S. Y., Ng A. M., Vickers M. F., Sundaram M., Cass C. E., Baldwin S. A., Young J. D. Functional and molecular characterization of nucleobase transport by recombinant human and rat equilibrative nucleoside transporters 1 and 2. Chimeric constructs reveal a role for the ENT2 helix 5-6 region in nucleobase translocation. J. Biol. Chem. 2002;277:24938–24948. doi: 10.1074/jbc.M200966200. [DOI] [PubMed] [Google Scholar]
- 31.Zhang J., Visser F., Vickers M. F., Lang T., Robins M. J., Nielsen L. P., Nowak I., Baldwin S. A., Young J. D., Cass C. E. Uridine binding motifs of human concentrative nucleoside transporters 1 and 3 produced in Saccharomyces cerevisiae. Mol. Pharmacol. 2003;64:1512–1520. doi: 10.1124/mol.64.6.1512. [DOI] [PubMed] [Google Scholar]
- 32.Zhang J., Smith K. M., Tackaberry T., Visser F., Robins M. J., Nielsen L. P., Nowak I., Baldwin S. A., Young J. D., Cass C. E. Uridine binding and transportability determinants of human concentrative nucleoside transporters. Mol. Pharmacol. 2005;68:830–839. doi: 10.1124/mol.105.012187. [DOI] [PubMed] [Google Scholar]
- 33.Vickers M. F., Yao S. Y., Baldwin S. A., Young J. D., Cass C. E. Nucleoside transporter proteins of Saccharomyces cerevisiae. Demonstration of a transporter (FUI1) with high uridine selectivity in plasma membranes and a transporter (FUN26) with broad nucleoside selectivity in intracellular membranes. J. Biol. Chem. 2000;275:25931–25938. doi: 10.1074/jbc.M000239200. [DOI] [PubMed] [Google Scholar]
- 34.Vickers M. F., Kumar R., Visser F., Zhang J., Charania J., Raborn R. T., Baldwin S. A., Young J. D., Cass C. E. Comparison of the interaction of uridine, cytidine, and other pyrimidine nucleoside analogues with recombinant human equilibrative nucleoside transporter 2 (hENT2) produced in Saccharomyces cerevisiae. Biochem. Cell Biol. 2002;80:639–644. doi: 10.1139/o02-148. [DOI] [PubMed] [Google Scholar]
- 35.Brunelli J. P., Pall M. L. A series of yeast/Escherichia coli 1 expression vectors designed for directional cloning of cDNAs and cre/lox-mediated plasmid excision. Yeast. 1993;9:1309–1318. doi: 10.1002/yea.320091204. [DOI] [PubMed] [Google Scholar]
- 36.Crawford C. R., Ng C. Y., Ullman B., Belt J. A. Identification and reconstitution of the nucleoside transporter of CEM human leukemia cells. Biochim. Biophys. Acta. 1990;1024:289–297. doi: 10.1016/0005-2736(90)90357-t. [DOI] [PubMed] [Google Scholar]
- 37.Lang T. University of Alberta; 2003. Studies of recombinant and native human concentrative nucleoside transporters and their role in nucleoside pharmacology and adenosine signaling. Ph.D. Thesis. [Google Scholar]
- 38.Vickers M. F., Zhang J., Visser F., Tackaberry T., Robins M. J., Nielsen L. P., Nowak I., Baldwin S. A., Young J. D., Cass C. E. Uridine recognition motifs of human equilibrative nucleoside transporters 1 and 2 produced in Saccharomyces cerevisiae. Nucleosides Nucleotides Nucleic Acids. 2004;23:361–373. doi: 10.1081/ncn-120028333. [DOI] [PubMed] [Google Scholar]
- 39.Frillingos S., Sahin-Toth M., Wu J., Kaback H. R. Cys-scanning mutagenesis: a novel approach to structure function relationships in polytopic membrane proteins. FASEB J. 1998;12:1281–1299. doi: 10.1096/fasebj.12.13.1281. [DOI] [PubMed] [Google Scholar]
- 40.Javitch J. A. Probing structure of neurotransmitter transporters by substituted-cysteine accessibility method. Methods Enzymol. 1998;296:331–346. doi: 10.1016/s0076-6879(98)96025-6. [DOI] [PubMed] [Google Scholar]
- 41.Valdes R., Vasudevan G., Conklin D., Landfear S. M. Transmembrane domain 5 of the LdNT1.1 nucleoside transporter is an amphipathic helix that forms part of the nucleoside translocation pathway. Biochemistry. 2004;43:6793–6802. doi: 10.1021/bi049873m. [DOI] [PubMed] [Google Scholar]
- 42.Yao S. M., Sundaram M., Chomey E. G., Cass C. E., Baldwin S. A., Young J. D. Identification of Cys140 in helix 4 as an exofacial cysteine residue within the substrate-translocation channel of rat equilibrative nitrobenzylthioinosine (NBMPR)-insensitive nucleoside transporter rENT2. Biochem. J. 2001;353:387–393. doi: 10.1042/0264-6021:3530387. [DOI] [PMC free article] [PubMed] [Google Scholar]