Abstract
Recurrent respiratory papillomatosis (RRP) is characterised by multiple laryngeal papillomas. Left untreated, the lesions enlarge, spread, and endanger the airway. Medical treatments are unsatisfactory, and repeated surgery remains the mainstay of therapy. RRP is caused by human papillomavirus (HPV) infection. However, since oral HPV infection is common and RRP is rare, other host and/or viral factors may contribute to pathogenesis. In an attempt to identify such factors, we have investigated 60 patients. The patients were HLA class I, II, and tumor necrosis factor TNF typed by sequence-specific primer PCR, and the results compared to those for 554 healthy controls by using Fisher's exact test. Peripheral blood mononuclear cell proliferative responses of 25 controls and 10 patients to HPV-11 L1 virus-like particles (VLP) were compared. Short-term VLP-specific T-cell lines were established, and recognition of L1 was analyzed. Finally, the L1 open reading frames of HPV isolates from four patients were sequenced. Susceptibility to RRP was associated with HLA DRB1*0301 (33 of 60 patients versus 136 of 554 controls, P < 0.0001). The three most severely affected patients were homozygous for this allele. A range of T-cell proliferative responses to HPV-11 VLP were observed in DRB1*0301-positive healthy donors which were comparable to those in DRB1*0301-negative controls. Individuals with juvenile-onset RRP also mounted a range of VLP responses, and their magnitude was negatively correlated with the clinical staging score (P = 0.012 by the Spearman rank correlation). DRB1*0301-positive patients who responded to L1 recognized the same epitope as did matched controls and produced similar cytokines. Sequencing of clinical isolates excluded the possibility that nonresponsiveness was the result of mutation(s) in L1.
Recurrent respiratory papillomatosis (RRP) is a life-threatening disease characterised by the growth of multiple benign tumours in the larynx and other sites within the upper aerodigestive tract. RRP has a bimodal age distribution, with one peak in early infancy and early childhood and the second in young adults (15). Although a variety of medical treatments have been investigated, repeated surgical ablation remains the mainstay of therapy (15). Despite its rarity (the incidence in the United States is approximately 1 in 100,000 [5]), the impact of the disease on patients, their families, and health care systems is immense. It is not unusual for patients to require more than 100 surgical procedures, and in the United States the average lifetime costs per juvenile-onset patient exceed $200,000 (8).
RRP is caused by human papillomavirus (HPV) infection, usually by HPV-6 and -11 (34), which are more commonly associated with benign genital warts (21). This led to the hypothesis that juvenile RRP is transmitted from mother to child during delivery while adult RRP is transmitted sexually (18). However, in contrast to the low incidence of RRP, genital HPV infection is common; indeed, it has been estimated that visible warts are present in 1% of American women of childbearing age and that another 15% have evidence of subclinical infection (21). Furthermore the carriage rate of HPV in the oropharynx is at least 10% in both children (12, 29) and adults (23), indicating that other factors must contribute to the pathogenesis of RRP.
One such factor might be a host immune deficit, although, since patients do not appear to be more susceptible to other infectious agents and no consistent immune deficit has previously been demonstrated (15), any such deficit is likely to be either subtle or HPV specific. Susceptibility to a number of infectious diseases has been associated with polymorphisms in immune response genes (reviewed by Hill [19]). Of particular note are reports that susceptibility to cervical cancer, which is also caused by HPV infection (37), is associated with several HLA class II alleles (38-40). The role of HLA class II polymorphisms in RRP pathogenesis have not been analyzed in detail, although there is an unconfirmed report from a small study of an association between HLA-DQ3 and disease (see Discussion) (9).
In this study, we have examined a cohort of 60 patients with RRP for defects in immune responsiveness. We report a novel HLA class II disease association, and have gone on to investigate T-cell proliferative responses to HPV-11 virus-like particles (VLP) in patients and matched controls.
MATERIALS AND METHODS
Patients and controls.
Sixty patients with RRP were recruited from Ear Nose and Throat clinics in the United Kingdom. Written informed consent was obtained from patients, and ethical approval was obtained from the Multi Centre Research Ethics Committee for Wales, the Bro Taf Local Research Ethics Committee, and other appropriate Local Research Ethics Committees. All work was conducted in accordance with the Helsinki Declaration of 1975 as revised in 1983. Patients were staged using the system of Derkey et al. by an experienced Ear Nose and Throat surgeon who was blinded to the scientific data (16). In brief, disease activity was scored as 0 (none), 1 (surface lesion), 2 (raised lesion), and 3 (bulky lesion) at 25 sites (13 laryngeal, 6 tracheal, and 6 other).
The controls for the HLA typing were 554 cadaveric organ donors in the United Kingdom. The representative nature of these controls for the British population has been established (10). Initially all patients were HLA class II typed by low-resolution sequence-specific primer PCR (SSP-PCR) (11) Subsequently they were HLA class I and tumor necrosis factor (TNF) typed using SSP-PCR (11, 17) For cellular studies, venous blood was obtained from 25 healthy, unrelated, adult donors.
HPV typing and HPV L1 sequencing.
HPV typing was performed using PCR with enzyme immunoassay as previously described (20). Sequencing of the L1 gene from HPV-11 was performed on a CEQ 2000 automated sequencer (Coulter-Beckman, Fullerton, Calif.) using reagents and protocols specified by the supplier. All samples were sequenced three times in both directions (primer details available on request).
Antigens.
HPV-11 VLP, composed of the surface coat protein L1, were the gift Merck Sharp & Dohme (West Point, Pa.) (14). A panel of 95 synthetic 15-mer peptides spanning the entire sequence of the HPV-11 L1 (accession number AAC53712) was synthesized by Eurogentec (Seraing, Belguim). Peptides overlapped by a minimum of 9 residues. Peptide pools contained five peptides and were named by the N-terminal residue of their first peptide and the C-terminal residue of their final peptide. Individual peptide sequences and details of peptide pool composition have been previously described (41).
The sources of the four control antigens were as follows: influenza A virus/Beijing/32/92 (H3N2) hemagglutinin was the generous gift of Ruud Brands (Solvay Duphar B.V., Weesp, The Netherlands); tuberculin purified protein derivative (PPD) and absorbed tetanus vaccine were obtained from Evans Medical Ltd., Leatherhead, United Kingdom; and keyhole limpet hemocyanin (KLH) was obtained from Calbiochem-Behring, La Jolla, Calif.
PBMC responses to HPV L1 and control antigens.
Peripheral blood mononuclear cells (PBMC) were examined as previously described (41). In brief, PBMC were incubated at 0.3 × 106 cells per 1-ml well in 48-well flat-bottom tissue culture plates (Greiner) for 2 h at 37°C under 5.8% CO2 in complete medium (minimal Eagle medium [Sigma, Poole, United Kingdom] with 5% heat-inactivated pooled AB serum, 100 IU of penicillin-streptomycin (Gibco Life Technologies, Paisley, United Kingdom), and 2 mM l-glutamine [Gibco]) with either HPV-11 VLP (1, 5, or 10 μg/ml) or positive control antigen (15 μg of tetanus toxoid per ml, 5 μg of influenza virus hemagglutinin per ml, 200 U of PPD per ml, or 10 μg of KLH per ml) or complete medium alone (negative control). The cells were washed, and 0.7 × 106 autologous PBMC were added to each well. On days 6, 7, and 8 three 80-μl aliquots were taken from each well, pulsed with 0.5 μCi of [3H]thymidine (Amersham UK Ltd., Little Chalfont, United Kingdom) per well, and harvested 16 h later. Proliferation was measured by liquid scintillation spectroscopy. Experiments were conducted in triplicate, and responses of five times the geometric mean of controls (T cells, antigen-presenting cells [APC], and media) were taken to be significant. Results are presented as stimulation indices (SI = cpm of PBMC response to antigen/cpm of PBMC response to control).
Establishment of CD4+ T-cell lines.
HPV-11 L1-specific T-cell lines were established as previously described (41), by culturing PBMC at 106 cells per 2-ml well in flat-bottom 24-well tissue culture plates (Greiner) with 10 μg of HPV-11 VLP/ml in complete medium at 37°C under 5.8% CO2 for 2 h. The cells were washed, and 4 × 106 autologous PBMC were added to each well. On day 7, the emerging line was recovered and restimulated with autologous irradiated (3,000 rads) PBMC prepulsed with HPV-11 VLP (1 μg/ml in complete medium). Restimulated lines were supplemented with 10% (vol/vol) Lymphocult-LT (Biotest Folex, Frankfurt, Germany) on days 1 and 3. Every 7 days, lines were recovered and restimulated as above.
Definition of L1 epitopes.
On days 21, 28, and 35 of in vitro culture, the proliferative responses of emerging lines to HPV-11 VLP and HPV-11 L1 peptide pools were examined. Lines were cultured at 4 × 104 cells/150 ml in compete medium in 96-well U-bottom tissue culture plates with 104 autologous irradiated (3,000 rads) PBMC prepulsed with medium (control), HPV-11 L1 VLP (1, 5, or 10 μg/ml), or L1 peptide pools (19 peptide pools each containing five peptides). At 48 h, the cells were pulsed with 0.5 μCi of tritiated thymidine (Amersham Pharmacia Biotech) per well, harvested, and analyzed as above. Responses of three times the geometric mean of background (T cells, APC, and medium) were taken to be significant. Responses to individual peptides (5 μg/ml) within dominant pools were then examined as detailed above.
Peptide binding competition assay.
Binding assays were carried out using the principle of competition with biotinylated CLIP peptide as previously described (26).
Analysis of cytokine production by L1-specific T-cell lines.
The production of gamma interferon (IFN-γ), interleukin-4 (IL-4), or IL-10 by L1-specific T-cell lines was examined using ELISpot kits (Mabtech AB, Nacka, Sweden). Experiments were performed in triplicate as specified by the manufacturer. After overnight drying at room temperature, the number of colored spots was determined under a dissecting microscope.
Statistical analysis.
Fisher's exact test, odds ratio analyses, the Mann-Whitney test, the Kruskal-Wallis test, and the Spearman rank correlation were carried out using InStat3 software version 3.0a for Macintosh OS X (Intuitive Software for Science, San Diego, Calif.). All tests were two sided, and a P value of <0.05 with a 95% confidence interval was considered significant.
RESULTS
Patient demographics and immune screen.
Clinical and virological details for the patients are summarized in Table 1. As previously reported, the juvenile-onset patients as a group had undergone more surgical procedures than had the adult-onset patients (28). The first eight patients were screened for a gross immunological deficit. Studies included enumeration of B cells (CD19+), immunoglobulin subsets, secretory immunoglobulin A levels, complement hemolytic activity, T-lymphocyte subsets (CD3+ CD4+ and CD3+ CD8+), and specific antibodies against childhood vaccines. No significant abnormalities were detected.
TABLE 1.
Clinical and virological details of the patients
| Patient typea | No. of patients | Age (yrs)
|
% Male | No. of procedures
|
% with >10 procedures in total | No. of procedures/patient/yr
|
|||
|---|---|---|---|---|---|---|---|---|---|
| Mean | Range | Mean | Range | Mean | Range | ||||
| All | 50 | 14.9 | 0.25-62 | 52.0 | 24.6 | 2-200 | 55 | 4.7 | 0.14-20.0 |
| JORRP | 36 | 4.3 | 0.25-13 | 38.0 | 31.4b | 2-200 | 73 | 5.1c | 0.67-20.0 |
| AORRP | 24 | 38.7 | 17-69 | 70.0 | 9.9b | 2-33 | 26 | 1.4c | 0.14-3.15 |
| HPV-6 positive | 20 | 12.3 | 0.25-52 | 60.0 | 10.6d | 2-38 | 46 | 2.3 | 0.14-6.80 |
| HPV-11 positive | 13 | 6.5 | 3.0-35 | 46.0 | 34.6d | 2-104 | 72 | 5.3 | 2.0-15.8 |
JORRP, juvenile-onset RRP; AORRP, adult-onset RRP.
The difference in total number of surgical procedures between patients with JORRP and AORRP is significant (P = 0.01, Mann-Whitney test).
The difference between patients with JORRP and AORRP in the number of procedures per year is significant (P < 0.0001 Mann-Whitney test).
The difference in the total number of procedures between patients with HPV-11 infection and those with HPV-6 infection is significant (p = 0.03, Mann-Whitney test), although the apparent difference between these groups in the number of procedures per year is not significant (P = 0.07 Mann-Whitney test).
HLA class II typing.
Susceptibility to HPV-associated cervical cancer has been associated with specific HLA class II alleles (3, 38-40). To investigate if this was also true of RRP, all patients were genotyped for HLA class II (HLA-DR, HLA-DRw, and HLA-DQ) using low-resolution SSP-PCR (Table 2). The most striking finding was an increase in the number of HLA-DRB1*03-positive individuals. This association remained statistically significant after correction for multiple comparisons (corrected P < 0.0024). DRB1*03 subtyping revealed that all patients were DRB1*0301 positive. Interestingly, the three most severely affected patients were homozygous for DRB1*0301; one has parenchymal lung involvement, one has severe tracheal and esophageal disease, and one has severe tracheal disease. Weaker positive associations were also seen with HLA-DRB1*14 and HLA-DQB1*02 (the latter being in linkage disequilibrium with HLA-DRB1*03), and a negative association was seen with HLA-DQB1*03 (previously reported to be positively associated with RRP [9] [see Discussion]).
TABLE 2.
HLA class II types
| HLA alleled | No. (%) of patients with allele
|
P
|
Odds ratio | 95% confidence interval | ||
|---|---|---|---|---|---|---|
| RRP (n = 60) | Control (n = 554) | Fisher's exact test | Corrected (×24) | |||
| DRB1*01 | 9 (0.15) | 124 (0.22) | NSa | |||
| DRB1*03 | 33 (0.55) | 136 (0.25) | <0.0001 | <0.0024 | 3.76 | 2.18-6.47 |
| DRB1*04 | 19 (0.32) | 206 (0.37) | NS | |||
| DRB1*07 | 13 (0.22) | 147 (0.27) | NS | |||
| DRB1*08 | 3 (0.05) | 28 (0.05) | NS | |||
| DRB1*09 | 1 (0.02) | 21 (0.04) | NS | |||
| DRB1*10 | 1 (0.02) | 10 (0.02) | NS | |||
| DRB1*11 | 6 (0.10) | 79 (0.14) | NS | |||
| DRB1*12 | 0 | 16 (0.03) | NS | |||
| DRB1*13 | 6 (0.10) | 102 (0.18) | NS | |||
| DRB1*14 | 8 (0.13) | 25 (0.05) | 0.01 | NS | 3.26 | 1.40-7.58 |
| DRB1*15 | 10 (0.17) | 8 (0.20) | NS | |||
| DRB1*16 | 1 (0.02) | 6 (0.01) | NS | |||
| DRB3*0x | 41 (0.68) | 319 (0.58) | NS | |||
| DRB4*0101-5 | 28 (0.47) | 301 (0.54) | NS | |||
| DRB5*01/02/03 | 11 (0.18) | 164 (0.30) | NS | |||
| DQB1*02 | 36 (0.60) | 232 (0.42) | 0.009 | NS | 2.08 | 1.21-3.59 |
| DQB1*03 | 29 (0.48) | 364 (0.66) | 0.01 | NS | 0.46 | 0.27-0.78 |
| DQB1*0301/4 | 19 (0.32) | 71 (0.38)b | NS | |||
| DQB1*0302/7/8 | 4 (0.07) | 36 (0.19)b | NS | |||
| DQB1*0303 | 5 (0.08) | 19 (0.10)b | NS | |||
| DQB1*04 | 3 (0.05) | 21 (0.04) | NS | |||
| DQB1*05 | 20 (0.33) | 88 (0.16)c | NS | |||
| DQB1*06 | 15 (0.25) | 108 (0.19)c | NS | |||
NS, not significant.
DQ3 subtypes had 169 controls.
DQ5 and DQ6 subtypes had 169 controls.
HLA DR and DQ alleles do not add up to twice number of donors because of homozygosity.
HLA class I and TNF typing.
HLA-DRB1*0301 is an important component of the 8.1 extended ancestral HLA haplotype (HLA A-1, B-8, Cw7, DR3, DQ2, TNF-2, termed A1-B8-DR3). To determine whether the association was with HLA-DRB1*0301 itself or with other components of this haplotype, HLA class I and TNF typing was undertaken (Table 3). A significant association with the A1-B8-DR3 haplotype was found in the whole population and in the juvenile-onset population. The TNF haplotype G−308G−238G+488 (frequently known as TNF-1) was significantly decreased, and this was paralleled by a rise in the TNF haplotype A−308G−238G+488 (TNF-2), a component of the A1-B8-DR3 ancestral haplotype.
TABLE 3.
HLA class I and TNF types
| HLA allele | No. (%) of patients with allele
|
P (Fisher's exact test) | Odds ratio | 95% confidence interval | |
|---|---|---|---|---|---|
| RRP (n = 60) | Control (n = 554) | ||||
| A*01 | 27 (0.45) | 172 (0.31) | 0.04 | 1.82 | 1.06-3.12 |
| B*08 | 21 (0.35) | 133 (0.24) | NSb (0.08) | 1.70 | 0.97-3.00 |
| B*27 | 8 (0.13) | 40 (0.07) | NS (0.12) | 1.98 | 0.88-4.50 |
| C*w7 | 35 (0.58) | 248 (0.47) | NS (0.06) | 1.73 | 1.01-2.96 |
| TNF-GGG* | 45 (0.76) | 103a (0.93) | 0.004 | 0.25 | 0.10-0.64 |
| TNF-AGG* | 32 (0.54) | 35a (0.32) | 0.005 | 2.57 | 1.34-4.93 |
| A1B8DR3DQ2 | 17 (0.28) | 61 (0.11) | 0.0007 | 3.20 | 1.72-5.95 |
TNF comparisons used 111 controls and 59 patients since insufficient DNA was obtained from one patient with juvenile-onset RRP.
NS, not significant.
HLA analysis as two cohorts of 30 patients.
To confirm the validity of the HLA disease associations described above, we reexamined our data by treating the first 30 patients recruited as one cohort and the second group of patients as a second cohort (Table 4). Both the DRB1*0301 and the A1-B8-DR3-DQ2 disease associations remained significant in this analysis.
TABLE 4.
Data analyzed as two independent cohorts
| HLA allele | No. (%) of patients with allele
|
P (Fisher's exact test) | Odds ratio | 95% confidence interval | |
|---|---|---|---|---|---|
| RRP | Control | ||||
| First 30 patients (control n = 554) | |||||
| A*01 | 14 (0.47) | 172 (0.31) | NSb | ||
| B*08 | 10 (0.33) | 133 (0.24) | NS | ||
| DRB1*03 | 15 (0.50) | 136 (0.25) | 0.004 | 3.07 | 1.46-6.45 |
| DRB1*14 | 3 (0.10) | 25 (0.05) | NS | ||
| DQB1*02 | 17 (0.57) | 232 (0.42) | NS | ||
| TNF-GGGa | 21 (0.70) | 103 (0.93) | 0.002 | 0.18 | 0.06-0.52 |
| TNF-AGGa | 15 (0.50) | 35 (0.32) | NS | ||
| A1B8DR3DQ2 | 8 (0.27) | 61 (0.11) | 0.02 | 2.94 | 1.25-6.89 |
| Second 30 patients (control n = 554) | |||||
| A*01 | 13 (0.43) | 172 (0.31) | NS | ||
| B*08 | 11 (0.37) | 133 (0.24) | NS | ||
| DRB1*03 | 18 (0.60) | 136 (0.25) | <0.0001 | 4.61 | 2.17-9.82 |
| DRB1*14 | 5 (0.17) | 25 (0.05) | 0.01 | 4.23 | 1.50-11.98 |
| DQB1*02 | 19 (0.63) | 232 (0.42) | 0.02 | 2.40 | 1.12-5.14 |
| TNF-GGGa | 24 (0.80) | 103 (0.93) | NS | ||
| TNF-AGGa | 17 (0.57) | 35 (0.32) | 0.01 | 3.08 | 1.33-7.13 |
| A1B8DR3DQ2 | 9 (0.30) | 61 (0.11) | 0.006 | 3.46 | 1.52-7.91 |
TNF comparisons used 111 controls in both cohorts, and 29 patients in the second cohort.
NS, not significant.
HLA subgroup analysis by age of onset.
We were interested in determining whether the HLA associations observed in the whole population remained significant when the juvenile- and adult-onset cohorts were analyzed separately (Table 5). While the HLA-DRB1*0301 association described above held true in both subgroups, the HLA-DRB1*14 association was confined to the juvenile patients and an additional association with HLA-B*27 was observed in the adult-onset patients.
TABLE 5.
Comparison of juvenile- and adult-onset subgroups of patients with RRP
| HLA alleleb | No. (%) of patients with allele
|
P (Fisher's exact test) | Odds ratio | 95% confidence interval | |
|---|---|---|---|---|---|
| RRP | Control | ||||
| JORRP (n = 36, control n = 554) | |||||
| A*01 | 18 (0.50) | 172 (0.31) | 0.03 | 2.22 | 1.13-4.37 |
| B*08 | 13 (0.36) | 133 (0.24) | NSc (0.11) | 1.79 | 0.88-3.63 |
| B*27 | 2 (0.06) | 40 (0.07) | NS (1.0) | 0.76 | 0.18-3.26 |
| C*w7 | 22 (0.61) | 248 (0.47) | NS (0.06) | 1.94 | 0.97-3.87 |
| DRB1*03 | 21 (0.56) | 136 (0.25) | <0.0001 | 4.30 | 2.16-8.58 |
| DR1*14 | 6 (0.16) | 25 (0.05) | 0.008 | 4.23 | 1.61-11.10 |
| DQB1*02 | 23 (0.62) | 232 (0.42) | 0.01 | 2.46 | 1.22-4.95 |
| DQB1*03 | 18 (0.5) | 364 (0.66) | NS (0.07) | 0.52 | 0.27-1.03 |
| TNF-GGGa | 27 (0.77) | 103a (0.93) | 0.03 | 0.26 | 0.09-0.76 |
| TNF-AGGa | 20 (0.57) | 35a (0.32) | 0.009 | 2.9 | 1.33-6.32 |
| A1B8DR3DQ2 | 12 (0.33) | 61 (0.11) | 0.0006 | 4.04 | 1.92-8.49 |
| AORRP (n = 24, control n = 554) | |||||
| A*01 | 9 (0.38) | 172 (0.31) | NS (0.51) | 1.83 | 0.57-3.11 |
| B*08 | 8 (0.33) | 133 (0.24) | NS (0.33) | 1.58 | 0.66-3.78 |
| B*27 | 6 (0.25) | 40 (0.07) | 0.008 | 4.28 | 1.61-11.40 |
| C*w7 | 13 (0.54) | 248 (0.47) | NS (0.41) | 1.46 | 0.64-3.31 |
| DRB1*03 | 12 (0.44) | 136 (0.25) | 0.008 | 3.07 | 1.35-7.00 |
| DRB1*14 | 2 (0.06) | 25 (0.05) | NS (0.31) | 1.92 | 0.43-8.64 |
| DQB1*02 | 13 (0.50) | 232 (0.42) | NS (0.29) | 1.64 | 0.72-3.73 |
| DQB1*03 | 10 (0.44) | 364 (0.66) | 0.03 | 0.37 | 0.16-0.86 |
| TNF-GGGa | 18 (0.75) | 103a (0.93) | 0.02 | 0.23 | 0.07-0.75 |
| TNF-AGGa | 12 (0.50) | 35a (0.32) | NS (0.10) | 2.17 | 0.89-5.31 |
| A1B8DR3DQ2 | 5 (0.21) | 61 (0.11) | NS (0.18) | 2.13 | 0.77-5.90 |
TNF comparisons used 111 controls and 35 juvenile-onset patients.
JORRP, juvenile-onset RRP; AORRP, adult-onset RRP.
NS, not significant.
HLA subgroup analysis by HPV type.
HLA associations in cervical cancer appear to be HPV type specific (3). We were therefore interested in discovering whether HLA associations in RRP were type specific. Papilloma biopsy samples were available from 33 patients; HPV-6 was detected in 20 patients, and HPV-11 was detected in 13 patients. No examples of multiple HPV infection was seen. Patients with HPV-11 infection had more aggressive disease than did those infected with HPV-6 (Table 1). Of the 13 HPV-11-positive patients, 11 were HLA DRB1*0301 positive, while of the 20 HPV-6-positive patients, 9 were DRB1*0301 positive, although this apparent difference did not attain statistical significance.
Cellular responses to HPV-11 VLP in healthy controls.
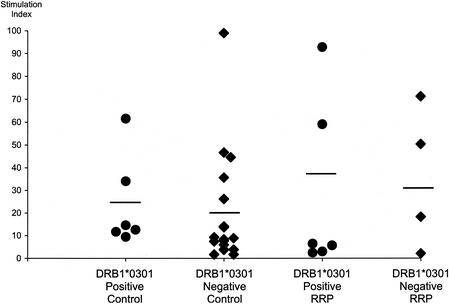
The genetic analysis detailed above suggested an HLA class II disease association, particularly with HLA-DRB1*0301, and raised the possibility that aberrant CD4+ T-cell responses to HPV-6 and HPV-11 might underlie the pathogenesis of RRP. Since HPV-11 does not grow in tissue culture, it is difficult to examine T-cell responses to the intact virus directly. We therefore elected to investigate T-cell proliferative responses to HPV-11 VLP (see Discussion) and four control antigens (tetanus toxoid, influenza virus hemagglutinin, PPD, and KLH). When 25 normal controls were examined, all responded to at least two control antigens (SI >5 [data not shown]) and 20, including all 6 who were DRB1*0301 positive, demonstrated significant proliferative responses to the HPV-11 VLP (Fig. 1). No significant differences in the magnitude of the response were seen between DRB1*0301-negative and DRB1*0301-positive donors (For DRB1*0301-negative controls, mean SI = 19.0; for DRB1*0301-positive controls, mean S.I. = 24.0 [P = 0.14 by the Mann-Whitney test]).
FIG. 1.
Proliferative responses to HPV-11 L1 VLP. Proliferative responses of PBMC to HPV-11 VLP from DRB1*0301-positive and -negative controls and DRB1*0301-positive and -negative patients with RRP, as assessed by [3H]thymidine incorporation. PBMC were incubated with either HPV-11 L1 VLP (1, 5, or 10 μg/ml) or complete medium (control). On days 6, 7, and 8, three 80-μl aliquots were taken from each emerging line, pulsed with 0.5 μCi of tritiated thymidine per well, and harvested 16 h later. Proliferation was measured by liquid scintillation spectroscopy. Experiments were conducted in triplicate, and a response five times the geometric mean of controls (T cells, APC, and medium) was taken to be significant. Results are presented as SI and represent the strongest response seen during the course of the experiment. No significant differences were seen among the four groups either as a whole (P = 0.43 by the Kruskal-Wallis test [nonparametric analysis of variance]) or when pairs of groups were compared using the Mann-Whitney test: DRB1*0301-positive controls versus DRB1*0301-negative controls, P = 0.14; DRB1*0301-positive patients versus DRB1*0301-positive controls, P = 0.31; DRB1*0301-negative patients versus DRB1*0301-negative controls, P = 0.30; DRB1*0301-positive versus DRB1*0361-negative patients, P = 0.91.
Cellular responses to HPV-11 VLP in patients with RRP.
The proliferative responses of 10 patients with RRP (7 juvenile onset and 3 adult onset [clinical summaries are given in Table 6 ]) to the same panel of antigens was then examined. Nine recognized at least three control antigens; the exception was patient 1, who responded only to tetanus toxoid (Fig. 2). As regards recognition of the HPV-11 VLP, a very similar range of responses was seen to that previously observed in normal controls (Fig. 1 and 2): In particular, no significant differences were seen between DRB1*0301-positive patients and positive controls, between DRB1*0301-negative patients and negative controls, between DRB1*0301-positive and negative patients.
TABLE 6.
Clinical summaries for patients—cellular experiments
| Patient no. | Juvenile or adult onset | Site of disease | Notes |
|---|---|---|---|
| 1 | Juvenile | Larynx, trachea, and lung | Recently developed squamous cell carcinoma of lung |
| 2 | Juvenile | Larynx | In remission when seen 2 mo after venesection |
| 3 | Juvenile | Single lesion at carina | Previously laryngeal, tracheal, and esphogeal disease |
| 4 | Juvenile | Larynx and trachea | Has tracheostomy, and currently no attempt is being made to clear larynx |
| 5 | Juvenile | None | In remission for 15 yr; previously 200 surgical procedures |
| 7 | Juvenile | Larynx and trachea | |
| 8 | Juvenile | Larynx and trachea | Improving, as evidenced by decreased staging score and increasing intervals between procedures |
| 14 | Adult | Larynx and subglottis | |
| 22 | Adult | Larynx | Relapsed to staging score of 14 within 3 mo of venesection |
| 23 | Adult | Larynx |
FIG. 2.
Proliferative responses of RRP patients to HPV-11 L1 VLP and four control antigens. Proliferative responses of PBMC from individuals with RRP to HPV-11 VLP (1, 5, or 10 μg/ml), tetanus toxoid (Tet) (15 μg/ml), influenza virus hemagglutinin (HA) [A/Beijing/32/92 (H3N2), 5 μg/ml], PPD (200 U/ml), and KLH (10 μg/ml) were assessed by [3H]thymidine incorporation as detailed in the legend to Fig. 1. Results are presented as SI.
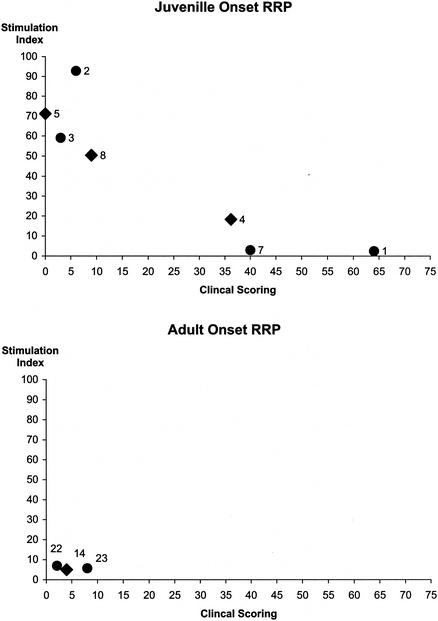
Given that the patients in this study had a wide range of clinical phenotypes, ranging from remission through parencyhmal lung disease (Table 6), we were curious to discover if there was any relationship between the magnitude of their proliferative T-cell responses to HPV-11 VLP and their clinical status when formally staged by the system of Derkay et al. (16). Because of the differences in clinical phenotype between juvenile- and adult-onset RRP (both noted above and previously described [15]), the two groups of patients were analyzed separately (Fig. 3). Interestingly, a significant negative correlation was seen between the clinical staging of patients with juvenile-onset RRP and the SI of their proliferative response to the HPV-11 VLP (P = 0.012, Spearman rank correlation). No such association was seen between clinical status and responses to any of the four control antigens (Fig. 4).
FIG. 3.
Comparison of clinical status and proliferative responses to HPV-11 L1 by patients with RRP. Proliferative responses of PBMC to HPV-11 VLP from individuals with RRP were assessed by [3H]thymidine incorporation as detailed in the legend to Fig. 1. Results are presented as SI. Clinical status was assessed by the method of Derkay et al. (16). Results for DRB1*0301-positive patients are represented by black circles, and results for DRB1*0301-negative patients are represented by black diamonds. A significant negative correlation was observed between the clinical status of juvenile-onset patients and the magnitude of their proliferative responses to HPV-11 VLP (Spearman rank correlation: r = −0.89, P = 0.012).
FIG. 4.
Comparison of clinical status and proliferative responses to four control antigens by patients with juvenile-onset RRP. Proliferative responses of PBMC from juvenille-onset RRP patients to tetanus toxoid, influenza virus hemagglutinin (A/Beijing/32/92 H3N2), PPD, and KLH were assessed by [3H]thymidine incorporation as detailed in the legend to Fig. 2. Results are presented as SI. Clinical status was assessed by the method of Derkay et al. (16). Results for DRB1*0301-positive patients are represented by black circles, and results for DRB1*0301-negative patients are represented by black diamonds. No significant correlations were observed between clinical status and the magnitude of the proliferative responses to any of the four control antigens by using Spearman rank correlation (influenza virus hemagglutinin, r = −0.61, P = 0.17; PPD, r = −0.43, P = 0.35; KLH, r = −0.46, P = 0.30; tetanus toxoid, r = −0.61, P = 0.17).
Proliferative studies were carried out on three patients with adult onset disease. While the small size of this group makes it difficult to draw firm conclusions, as a group they mounted weak responses to the VLP (Fig. 3).
Fine specificity of cellular responses to HPV-11 VLP.
To establish whether the subset of DRB1*0301 patients who responded to the HPV-11 VLP recognized the same L1 epitopes as did the DRB1*0301-positive controls, CD4+ T-cell responses to HPV-11 VLP were mapped by using short-term CD4+ T-cell lines and pools of 15-mer synthetic peptides spanning the entire sequence of L1 (Fig. 5). Peptide pool 121-153 was recognized by all six DRB1*0301-positive controls and was the sole target of control 1, who was homozygous for DRB1*0301. Responses within this pool were localized to peptide 139-153 (sequence 5′ DNRVNVGMDYKQTQL 3′). This peptide represents a region of L1 that is homologous between HPV-6 and HPV-11 but not to the region in common cutaneous and genital HPV (HPV-1, HPV-2, HPV-3, HPV-4, HPV-16 and HPV-18) (41) and that contains a binding motif consistent with HLA-DRB1*0301 restriction (P1 = V, P4 = D, P6 = K [peptide residues 6, 9, and 11] [22] and bound strongly to purified DRB1*0301 in a competition assay (data not shown). Responses to other peptide pools were heterogeneous, reflecting differences in other HLA -class II alleles.
FIG. 5.
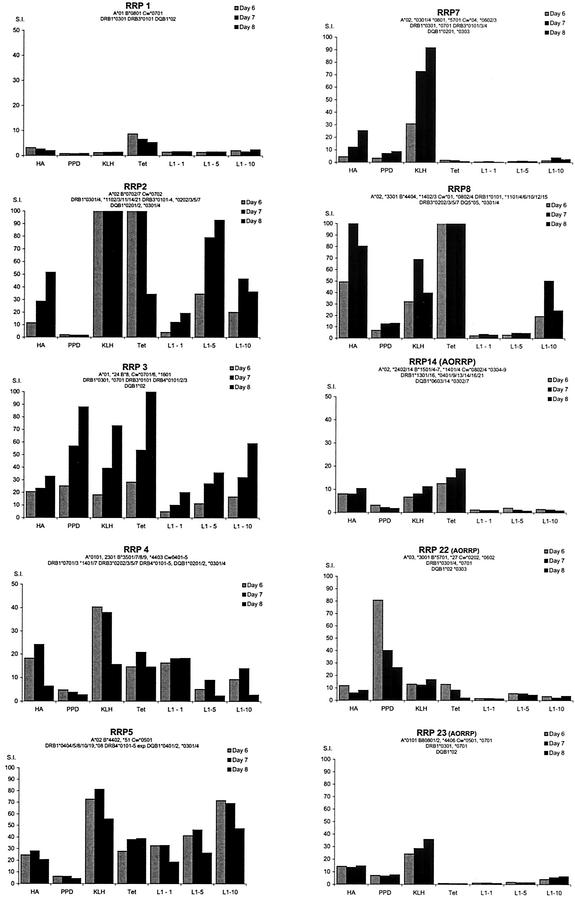
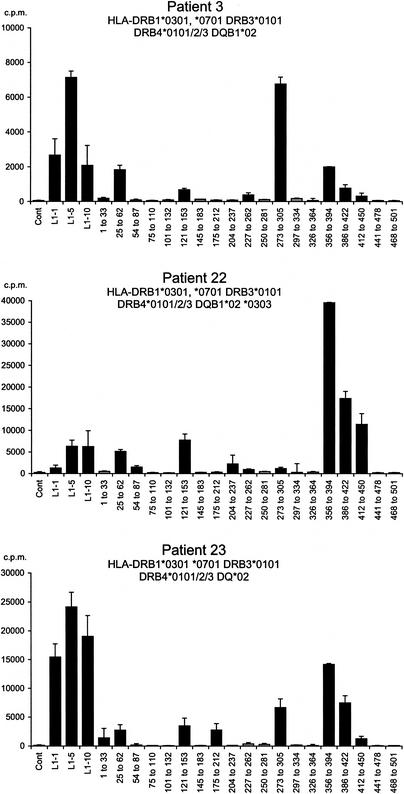
Responses of short-term CD4+ T-cell lines derived from DRB1*0301-positive control donors to HPV-11 L1 VLP. L1-specific T-cell lines were established from six DRB1*0301-positive healthy controls by culturing PBMC with HPV-11 L1 VLP. Every 7 days, the emerging line was recovered and restimulated with autologous irradiated PBMC which had been prepulsed with the VLP. On days 21, 28, and 35, the proliferative responses of the emerging lines to HPV-11 L1 and the HPV-11 L1 peptide pools was examined. The lines were washed and cultured with autologous irradiated PBMC prepulsed for 2 h with medium (control), HPV-11 L1 VLP (1, 5, and 10 μg/ml), or L1 peptide pools (19 peptide pools each containing five peptides). At 48 h, the cells were pulsed with [3H]thymidine and harvested and analyzed as described in the legend to Fig. 1. Experiments were conducted in triplicate, and the geometric mean of results is shown. Black bars represent SI of >3; grey bars represent SI of <3. Error bars represent standard errors. The HLA class I types of the controls were as follows: control 1, HLA-A*0101/2*0201-17, B*0801/2 *15, Cw*0701 *0303; control 2, HLA- A*0101/2 *0301/2, *0801 *4001, Bw6, Cw*0701 *1203; control 3, HLA-A*0101/2 *0201-17, B*0801 *4001, Bw6, Cw*0701; control 4, HLA- A*01, B*0801/2 *3501-4, Cw*0701 *0401-3; and control 5, HLA- A*0101*0201-17, B*0801 *1801/2, Bw6, Cw*1203: control 6, HLA- A*0101/2 *0201-17, B*0801/2 *3501/4, Cw*0701, *0401.
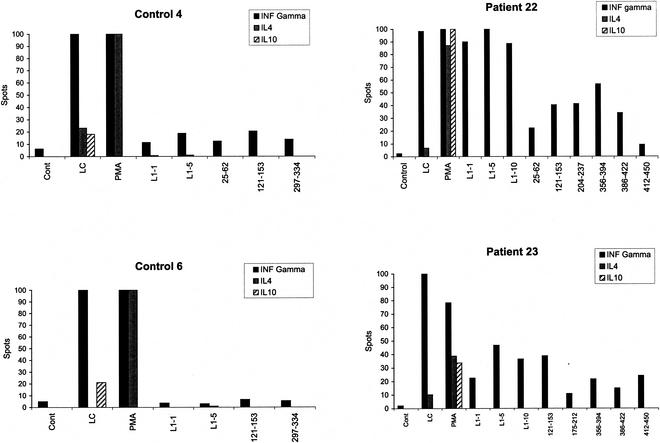
We attempted to establish short-term CD4+ T-cell lines from four DRB1*0301-positive donors old enough to donate the volume of blood required for this experiment. A line could not be established from patient 1 (three separate failures), who has severe disease with lung parenchymal involvement. Lines were established in three patients, and all three recognized peptide pool 121-153 (Fig. 6). Within the pool, all focused on p139. By chance, these patients had the same second DR allele, and their responses to the peptide pools were similar. Finally, the production of IFN-γ, IL-4, and IL-10 by T-cell lines selected from two DRB1*0301 patients and two DRB1*0301-positive controls in response to the VLP and p139 was compared by using ELISpots. The production of IFN-γ dominated responses to the VLP and pool 121-153 in both patients and controls (Fig. 7).
FIG. 6.
Responses of short-term CD4+ T-cell lines derived from DRB1*0301-positive patients to HPV-11 VLP. The responses of CD4+ T-cell lines from three DRB1*0301-positive patients with RRP are shown. Experimental details are as in Fig. 2. The HLA class I types of the patients were as follows: patient 3, HLA-A*01, *24, B*8, Cw*0701/6 *1601; patient 22, HLA- A*03 *3001, B*5701 *27, Cw*0202 *0602; and patient 23, HLA- A*0101, B*0801/2 *4406, Cw*0501 *0701. Experiments were conducted in triplicate, and the geometric mean of the results is shown. Black bars represent SI of >3; grey bars represent SI of <3. Error bars represent standard errors.
FIG. 7.
Cytokine production by HPV-11 L1-specific T-cell lines assessed by ELISpot. The cytokine production profiles of two representative short-term T-cell lines derived from two HLA DRB1*0301-positive controls and two HLA-DRB1*0301-positive patients were examined after 4 weeks of in vitro culture. L1-specific T-cell lines were activated for 48 h in triplicate at a density of 4 × 104 cells/150 ml of complete minimal Eagle medium in 96-well U-bottom tissue culture plates with 104 autologous irradiated (3,000 rads) PBMC prepulsed for 2 h with either medium (control), or L1 (5 μg/ml), or relevant L1 peptide pools (5 μg/ml), as previously determined by proliferative responses. As positive controls, Lymphocult-LT (LC) or PMA plus ionomycin (PMA) was added 12 h before the cells were washed and transferred to precoated INF-γ, IL-4, or IL-10 plates. After 16 h, the cells were discarded and cytokines were detected using ELISpot kits. After overnight drying at room temperature, the colored spots were counted under a dissecting microscope. Geometric means of triplicate experiments are shown.
Sequencing of HPV-11 L1.
To determine if variation of proliferative responses to L1 in DRB1*0301-positive patients was a result of sequence variation within the immunodominant DR3-restricted epitope, the L1 ORF of clinical isolates from two HPV-11-positive donors who failed to respond to the HPV-11 VLP (patients 1 and 7) and two donors who responded strongly to the VLP (patients 2 and 3) were sequenced. No sequence variation was identified in L1 (data not shown).
Response of control donors to L1 and p139 when using PBMC from patient A as APC.
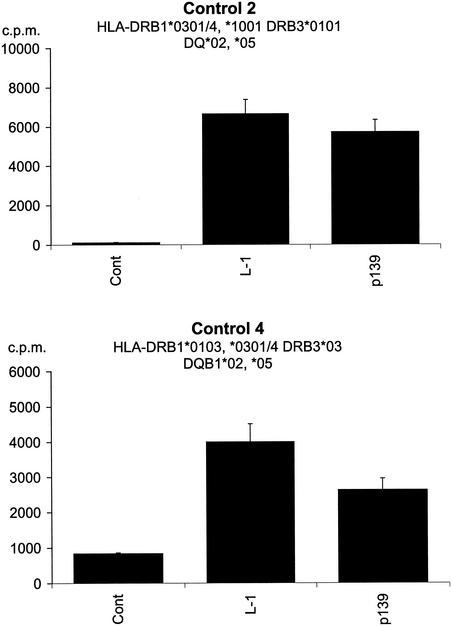
Finally, since sequence variation in L1 did not account for the failure of donors with severe RRP to respond to the VLP, we were interested is discovering if PBMC from a nonresponding DRB1*0301-positive patient were capable of processing and presenting L1 to DRB1*0301-positive control donors. Irradiated PBMC from patient 1, the most severely affected individual, were used to present L1 and p139 to 5-week-old L1-specific lines established form two DRB1*0301-positive control donors (Fig. 8). Both donors responded to the HPV-11 VLP and to p139, indicating that PBMC from the most severely affected patient in this study are capable of processing and presenting L1.
FIG. 8.
Response of two DRB1*0301-positive donors to HPV-11 L1 and p139, using PBMC from a nonresponding patient as APC. PBMC from patient 1 were irradiated (3,000 rads) and incubated for 2 h with complete medium (control), L1 (5 μg/ml), or p139 (5 μg/ml). The cells were then washed twice in complete medium and used as APC with 35-day-old CD4+ T-cell lines from controls 2 and 4 as described in the legend to Fig. 4. Experiments were conducted in triplicate, and the geometric means of the results are shown. Error bars represent standard errors.
DISCUSSION
The principal finding of this investigation is the identification of a genetic association between the presence of HLA DRB1*0301 and susceptibility to RRP. This association is novel and has not been identified in two previous studies. This may be explained by the small number of patients in the first study (n = 16, of whom only 6 were children) (9) and the fact that the second study was confined to HLA-DQB1 alleles in adult patients (1). In the present investigation, the presence of DRB1*0301 was associated with more severe disease, since the three most severely affected patients were homozygous for DRB1*0301. In addition, preliminary evidence is presented for an HLA-DRB1*14 association in juvenile-onset disease and an HLA-B*27 association in adult-onset disease. These additional findings may reflect the clinical heterogeneity of RRP (15).
The HLA DRB1*0301 allele is part of the A1-B8-DR3 ancestral haplotype (HLA DQB1*0201, HLA DRB1*0301, C4B-Sf, C4A-0, TNF-AGG, HLA-B*0801, HLA-Cw*0701, HLA-A*01), which has been linked to several autoimmune diseases (27). Analysis of linked class I and TNF alleles suggested that the association was with HLA-DRB1*0301 itself, although it is difficult to exclude other closely linked variants on the basis of genetic analysis alone (32).
The identification of an association with an HLA class II allele raised the possibility that susceptibility to RRP might be the result of aberrant CD4+ T-cell responses. Several lines of evidence indicate that cellular immunity plays an important role in controlling HPV-mediated diseases. First, there is a high incidence of HPV-associated diseases in individuals with iatrogenic (e.g., allograft recipients) or acquired cell-mediated (e.g., advanced human immunodeficiency virus infection) immune deficiencies (2, 4, 24, 36); second, infiltrates of CD4+ and CD8+ T cells and macrophages have been observed in spontaneously regressing plane warts, (30, 35) common warts, (7) genital warts, (13), and low-grade cervical intraepithelial neoplasia (33). It is difficult to examine T-cell responses to intact HPV-11 directly since the virus does not grow in tissue culture. Therefore, a pragmatic decision was made to examine responses to highly purified HPV-11 VLP composed of the surface coat protein L1, which are under investigation as potential vaccines in genital HPV infection (25) and hence are available as highly purified whole proteins, rather than to examine immune responses to early viral proteins (such as E6 and E7), which, while more directly involved in disease pathogenesis, are not readily available as intact antigens.
All six HLA-DRB1*0301-positive controls exhibited significant proliferative responses to the HPV-11 VLP which were comparable to those seen in non-DRB1*0301-positive controls, indicating that DRB1*0301 itself is not a low-responder allele. We then examined T-cell proliferative responses to HPV-11 VLP in patients with RRP. These studies were hampered by the geographical location of individual patients and by difficulties in collecting adequate amounts of blood from children; however, we were able to investigate 10 patients (7 with juvenile-onset disease). Considerable variation was observed in the magnitude of their responses, and once again this was not related to the presence or absence of the DRB1*0301 allele. Interestingly, however, the magnitude of the proliferative response to HPV-11 L1 VLP was inversely correlated with the clinical status of the juvenile-onset patients, providing support for the genetic observation that susceptibility to RRP is linked to polymorphisms in HLA class II alleles.
Detailed epitope mapping using CD4+ T-cell lines demonstrated that the subset of DRB1*0301-positive patients capable of responding to the HPV-11 VLP recognize the immunodominant DRB1*0301-restricted L1 epitope, which is located within a region with sequence identity between HPV-6 and HPV-11 (but not to HPV-1 to HPV-4, HPV-16, and HPV-18 [41]). Responding patients also had a similar cytokine profile to normal HLA-DRB1*0301-positive controls. This indicates that at least some patients with RRP have the capacity to mount an apparently normal response to HPV-11 L1. By sequencing HPV isolated from papilloma tissue, we have excluded the possibility that nonresponders are infected with a clade of HPV-11 with mutations in or around the immunodominant DRB1*0301-restricted T-cell epitope.
The questions therefore arise why some patients have not responded to HPV-11 L1 and why, although susceptibility to RRP is linked to HLA DRB1*0301, T-cell proliferative responses to HPV-11 VLP are not directly correlated to the presence of this allele. We excluded the possibility that nonresponding patients have a major deficit in HLA class II antigen processing by demonstrating that PBMC from the most severely affected patient in this study were capable of presenting both L1 and p139 to two DRB1*0301-positive control donors. The possibility that nonresponding patients have a hole in their T-cell receptor repertoire seems unlikely since we have observed vigorous responses to L1 in patients who historically have had extensive disease and are now either considerably improved or in remission. A more attractive hypothesis is that patients with severe disease have the capacity to respond to HPV-11 L1 normally but that this response has been delayed either because they have a low frequency of HPV L1-specific T cells or because the response been modulated by immunoregulatory networks. Indirect support for a delay in the immune response comes from the observations, noted above, that a range of HPV-mediated diseases undergo spontaneous regression and that during this regression there is a prominent infiltration of lymphocytes. There is also increasing evidence for the important role of regulatory T cells in other conditions including inflammatory bowel disease (6), which is also HLA DRB1*0301 associated, albeit weakly (31), and it is conceivable that the immune response in RRP has been delayed or down regulated by regulatory networks which involve HLA DRB1*0301 or a closely linked allele.
In conclusion, we have identified a genetic association between HLA DRB1*0301 and susceptibility to RRP. While the mechanism underlying this association is unknown, it will be of great interest to determine whether the presence of this allele predicts clinical severity or response to therapy in prospective studies.
Acknowledgments
This work was funded by The Wellcome Trust. C.M.G. is a Wellcome Senior Fellow in Clinical Research, O.M.W. is a Wellcome Medical Research Training Fellow, and S.E.M. is an MRC Clinician Scientist.
We are most grateful to all the patients who have helped us with this study. We are grateful to Richard Mills for staging the patients in this study, to G. S. Barr, D. Bosman, R. A. Evans, C. P. Fielder, M. V. Griffiths, Z. Hammad, D. Ingrams, M. Preece, M. Saunders, and H. Williams for help with patient recruitment and sample collection, and to Martin Rowe and Ita Askonas for support and encouragement.
REFERENCES
- 1.Aaltonen, L., J. Partanen, E. Auvinen, H. Rihkanen, and A. Vaheri. 1999. HLA-DQ alleles and human papillomavirus DNA in adult-onset laryngeal papillomatosis. J. Infect. Dis. 179:682-685. [DOI] [PubMed] [Google Scholar]
- 2.Alloub, M. I., B. B. Barr, K. M. McLaren, I. W. Smith, M. H. Bunney, and G. E. Smart. 1989. Human papillomavirus infection and cervical intraepithelial neoplasia in women with renal allografts. Br. Med. J. 298:153-156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Apple, R. J., H. A. Erlich, W. Klitz, M. M. Manos, T. M. Becker, and C. M. Wheeler. 1994. HLA DR-DQ associations with cervical carcinoma show papillomavirus-type specificity. Nat. Genet. 6:157-162. [DOI] [PubMed] [Google Scholar]
- 4.Arends, M. J., E. C. Benton, K. M. McLaren, L. A. Stark, J. A. Hunter, and C. C. Bird. 1997. Renal allograft recipients with high susceptibility to cutaneous malignancy have an increased prevalence of human papillomavirus DNA in skin tumours and a greater risk of anogenital malignancy. Br. J. Cancer 75:722-728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Armstrong, L. R., E. J. Preston, M. Reichert, D. L. Phillips, R. Nisenbaum, N. W. Todd, I. N. Jacobs, A. F. Inglis, S. C. Manning, and W. C. Reeves. 2000. Incidence and prevalence of recurrent respiratory papillomatosis among children in Atlanta and Seattle. Clin. Infect. Dis. 31:107-109. [DOI] [PubMed] [Google Scholar]
- 6.Asseman, C., S. Fowler, and F. Powrie. 2000. Control of experimental inflammatory bowel disease by regulatory T cells. Am. J. Respir. Crit. Care Med. 162:S185-S189. [DOI] [PubMed] [Google Scholar]
- 7.Bender, M. E. 1986. Concepts of wart regression. Arch. Dermatol. 122:644-647. [PubMed] [Google Scholar]
- 8.Bishai, D., H. Kashima, and K. Shah. 2000. The cost of juvenile-onset recurrent respiratory papillomatosis. Arch. Otolaryngol. Head Neck Surg. 126:935-939. [DOI] [PubMed] [Google Scholar]
- 9.Bonagura, V. R., F. P. Siegal, A. L. Abramson, F. Santiago-Schwarz, M. E. O'Reilly, K. Shah, D. Drake, and B. M. Steinberg. 1994. Enriched HLA-DQ3 phenotype and decreased class I major histocompatibility complex antigen expression in recurrent respiratory papillomatosis. Clin. Diagn. Lab. Immunol. 1:357-360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bunce, M., M. C. Barnardo, J. Procter, S. G. Marsh, C. Vilches, and K. I. Welsh. 1997. High resolution HLA-C typing by PCR-SSP: identification of allelic frequencies and linkage disequilibria in 604 unrelated random UK Caucasoids and a comparison with serology. Tissue Antigens 50:100-111. [DOI] [PubMed] [Google Scholar]
- 11.Bunce, M., C. M. O'Neill, M. C. Barnardo, P. Krausa, M. J. Browning, P. J. Morris, and K. I. Welsh. 1995. Phototyping: comprehensive DNA typing for HLA-A, B, C, DRB1, DRB3, DRB4, DRB5 and DQB1 by PCR with 144 primer mixes utilizing sequence-specific primers (PCR-SSP). Tissue Antigens 46:355-367. [DOI] [PubMed] [Google Scholar]
- 12.Cason, J., J. N. Kaye, R. J. Jewers, P. K. Kambo, J. M. Bible, B. Kell, B. Shergill, F. Pakarian, K. S. Raju, and J. M. Best. 1995. Perinatal infection and persistence of human papillomavirus types 16 and 18 in infants. J. Med. Virol. 47:209-218. [DOI] [PubMed] [Google Scholar]
- 13.Coleman, N., H. D. Birley, A. M. Renton, N. F. Hanna, B. K. Ryait, M. Byrne, D. Taylor-Robinson, and M. A. Stanley. 1994. Immunological events in regressing genital warts. Am. J. Clin. Pathol. 102:768-774. [DOI] [PubMed] [Google Scholar]
- 14.Cook, J. C., J. G. Joyce, H. A. George, L. D. Schultz, W. M. Hurni, K. U. Jansen, R. W. Hepler, C. Ip, R. S. Lowe, P. M. Keller, and E. D. Lehman. 1999. Purification of virus-like particles of recombinant human papillomavirus type 11 major capsid protein L1 from Saccharomyces cerevisiae. Protein Expression Purif. 17:477-484. [DOI] [PubMed] [Google Scholar]
- 15.Derkay, C. S. 2001. Recurrent respiratory papillomatosis. Laryngoscope 111:57-69. [DOI] [PubMed] [Google Scholar]
- 16.Derkay, C. S., D. J. Malis, G. Zalzal, B. J. Wiatrak, H. K. Kashima, and M. D. Coltrera. 1998. A staging system for assessing severity of disease and response to therapy in recurrent respiratory papillomatosis. Laryngoscope 108:935-937. [DOI] [PubMed] [Google Scholar]
- 17.Fanning, G. C., M. Bunce, C. M. Black, and K. I. Welsh. 1997. Polymerase chain reaction haplotyping using 3′ mismatches in the forward and reverse primers: application to the biallelic polymorphisms of tumor necrosis factor and lymphotoxin alpha. Tissue Antigens 50:23-31. [DOI] [PubMed] [Google Scholar]
- 18.Hallden, C., and B. Majmudar. 1986. The relationship between juvenile laryngeal papillomatosis and maternal condylomata acuminata. J. Reprod. Med. 31:804-807. [PubMed] [Google Scholar]
- 19.Hill, A. V. 1998. The immunogenetics of human infectious diseases. Annu. Rev. Immunol. 16:593-617. [DOI] [PubMed] [Google Scholar]
- 20.Jacobs, M. V., P. J. Snijders, F. J. Voorhorst, J. Dillner, O. Forslund, B. Johansson, M. von Knebel Doeberitz, C. J. Meijer, T. Meyer, I. Nindl, H. Pfister, E. Stockfleth, A. Strand, G. Wadell, and J. M. Walboomers. 1999. Reliable high risk HPV DNA testing by polymerase chain reaction: an intermethod and intramethod comparison. J. Clin. Pathol. 52:498-503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Koutsky, L. 1997. Epidemiology of genital human papillomavirus infection. Am. J. Med. 102:3-8. [DOI] [PubMed] [Google Scholar]
- 22.Marsh, S. G. E., P. Parham, and L. D. Barber. 2000. The HLA facts book. Academic Press, Ltd., London, United Kingdom.
- 23.Miller, C. S., and B. M. Johnstone. 2001. Human papillomavirus as a risk factor for oral squamous cell carcinoma: a meta-analysis, 1982-1997. Oral Surg. Oral Med. Oral Pathol. Oral Radiol. Endod. 91:622-635. [DOI] [PubMed] [Google Scholar]
- 24.Palefsky, J. M. 1997. Cutaneous and genital HPV-associated lesions in HIV-infected patients. Clin. Dermatol. 15:439-447. [DOI] [PubMed] [Google Scholar]
- 25.Palker, T. J., J. M. Monteiro, M. M. Martin, C. Kakareka, J. F. Smith, J. C. Cook, J. G. Joyce, and K. U. Jansen. 2001. Antibody, cytokine and cytotoxic T lymphocyte responses in chimpanzees immunized with human papillomavirus virus-like particles. Vaccine 19:3733-3743. [DOI] [PubMed] [Google Scholar]
- 26.Plebanski, M., K. L. Flanagan, E. A. Lee, W. H. Reece, K. Hart, C. Gelder, G. Gillespie, M. Pinder, and A. V. Hill. 1999. Interleukin 10-mediated immunosuppression by a variant CD4 T cell epitope of Plasmodium falciparum. Immunity 10:651-660. [DOI] [PubMed] [Google Scholar]
- 27.Price, P., C. Witt, R. Allcock, D. Sayer, M. Garlepp, C. C. Kok, M. French, S. Mallal, and F. Christiansen. 1999. The genetic basis for the association of the 8.1 ancestral haplotype (A1, B8, DR3) with multiple immunopathological diseases. Immunol. Rev. 167:257-274. [DOI] [PubMed] [Google Scholar]
- 28.Rabah, R., W. D. Lancaster, R. Thomas, and L. Gregoire. 2001. Human papillomavirus-11-associated recurrent respiratory papillomatosis is more aggressive than human papillomavirus-6-associated disease. Pediatr. Dev. Pathol. 4:68-72. [DOI] [PubMed] [Google Scholar]
- 29.Rice, P. S., C. Mant, J. Cason, J. M. Bible, P. Muir, B. Kell, and J. M. Best. 2000. High prevalence of human papillomavirus type 16 infection among children. J. Med. Virol. 61:70-75. [DOI] [PubMed] [Google Scholar]
- 30.Rogozinski, T. T., S. Jablonska, and M. Jarzabek-Chorzelska. 1988. Role of cell-mediated immunity in spontaneous regression of plane warts. Int. J. Dermatol. 27:322-326. [DOI] [PubMed] [Google Scholar]
- 31.Satsangi, J., K. I. Welsh, M. Bunce, C. Julier, J. M. Farrant, J. I. Bell, and D. P. Jewell. 1996. Contribution of genes of the major histocompatibility complex to susceptibility and disease phenotype in inflammatory bowel disease. Lancet 347:1212-1217. [DOI] [PubMed] [Google Scholar]
- 32.Svejgaard, A., and L. P. Ryder. 1994. HLA and disease associations: detecting the strongest association. Tissue Antigens 43:18-27. [DOI] [PubMed] [Google Scholar]
- 33.Tay, S. K., D. Jenkins, P. Maddox, N. Hogg, and A. Singer. 1987. Tissue macrophage response in human papillomavirus infection and cervical intraepithelial neoplasia. Br. J. Obstet. Gynaecol. 94:1094-1097. [DOI] [PubMed] [Google Scholar]
- 34.Terry, R. M., F. A. Lewis, S. Griffiths, M. Wells, and C. C. Bird. 1987. Demonstration of human papillomavirus types 6 and 11 in juvenile laryngeal papillomatosis by in-situ DNA hybridization. J. Pathol. 153:245-248. [DOI] [PubMed] [Google Scholar]
- 35.Vardy, D. A., O. Baadsgaard, E. R. Hansen, S. Lisby, and G. L. Vejlsgaard. 1990. The cellular immune response to human papillomavirus infection. Int. J. Dermatol. 29:603-610. [DOI] [PubMed] [Google Scholar]
- 36.Vermund, S. H., K. F. Kelley, R. S. Klein, A. R. Feingold, K. Schreiber, G. Munk, and R. D. Burk. 1991. High risk of human papillomavirus infection and cervical squamous intraepithelial lesions among women with symptomatic human immunodeficiency virus infection. Am. J. Obstet. Gynecol. 165:392-400. [DOI] [PubMed] [Google Scholar]
- 37.Walboomers, J. M., M. V. Jacobs, M. M. Manos, F. X. Bosch, J. A. Kummer, K. V. Shah, P. J. Snijders, J. Peto, C. J. Meijer, and N. Munoz. 1999. Human papillomavirus is a necessary cause of invasive cervical cancer worldwide. J. Pathol. 189:12-19. [DOI] [PubMed] [Google Scholar]
- 38.Wank, R., J. T. Meulen, J. Luande, H. C. Eberhardt, and M. Pawlita. 1993. Cervical intraepithelial neoplasia, cervical carcinoma, and risk for patients with HLA-DQB1*0602, *301, *0303 alleles. Lancet 341:1215.. [DOI] [PubMed] [Google Scholar]
- 39.Wank, R., D. J. Schendel, and C. Thomssen. 1992. HLA antigens and cervical carcinoma. Nature 356:22-23. [DOI] [PubMed] [Google Scholar]
- 40.Wank, R., and C. Thomssen. 1991. High risk of squamous cell carcinoma of the cervix for women with HLA-DQw3. Nature 352:723-725. [DOI] [PubMed] [Google Scholar]
- 41.Williams, O. M., K. W. Hart, E. C. Wang, and C. M. Gelder. 2002. Analysis of CD4+ T-cell responses to human papillomavirus (HPV) type 11 L1 in healthy adults reveals a high degree of responsiveness and cross-reactivity with other HPV types. J. Virol. 76:7418-7429. [DOI] [PMC free article] [PubMed] [Google Scholar]