Abstract
aaRSs (aminoacyl-tRNA synthetases) are responsible for the covalent linking of amino acids to their cognate tRNAs via the aminoacylation reaction and play a vital role in maintaining the fidelity of protein synthesis. LeuRS (leucyl-tRNA synthetase) can link not only the cognate leucine but also the nearly cognate residues Ile and Met to tRNALeu. The editing domain of LeuRS deacylates the mischarged Ile–tRNALeu and Met–tRNALeu. We report here the crystal structures of ecLeuRS-ED (the editing domain of Escherichia coli LeuRS) in both the apo form and in complexes with Met and Ile at 2.0 Å, 2.4 Å, and 3.2 Å resolution respectively. The editing active site consists of a number of conserved amino acids, which are involved in the precise recognition and binding of the noncognate amino acids. The substrate-binding pocket has a rigid structure which has an optimal stereochemical fit for Ile and Met, but has steric hindrance for leucine. Based on our structural results and previously available biochemical data, we propose that ecLeuRS-ED uses a lock-and-key mechanism to recognize and discriminate between the amino acids. Structural comparison also reveals that all subclass Ia aaRSs share a conserved structure core consisting of the editing domain and conserved residues at the editing active site, suggesting that these enzymes may use a common mechanism for the editing function.
Keywords: aminoacyl-tRNA synthetase, aminoacylation, editing domain, lock-and-key mechanism, recognition
Abbreviations: aaRS, aminoacyl-tRNA synthetase; CP1, connective polypeptide 1; ecLeuRS, Escherichia coli LeuRS; ED, editing domain; IleRS, isoleucyl-tRNA synthetase; LeuRS, leucyl-tRNA synthetase; Nva2AA, 2′-(L-norvalyl)amino-2′-deoxyadenosine; phLeuRS, Pyrococcus horikoshii LeuRS; RF, Rossmann fold; saIleRS, Staphylococcus aureus IleRS; ttLeuRS, Thermus thermophilus LeuRS; ValRS, valyl-tRNA synthetase
INTRODUCTION
aaRSs (aminoacyl-tRNA synthetases) are a family of enzymes that catalyse the esterification of an amino acid to its cognate tRNA (for a review see [1]). The aminoacylation reaction usually takes place in two steps: the activation of the amino acid by ATP to form an aminoacyl-AMP and the transfer of the aminoacyl-AMP to the cognate tRNA to form an aminoacyl-tRNA. The selectivity and specificity of the recognition of both the amino acid and tRNA by aaRSs plays a vital role in maintaining the fidelity of the translation of the genetic code during protein synthesis. The fidelity of the aminoacylation reaction is controlled by regulatory determinants in both tRNA and aaRSs, which permit the correct recognition and productive binding of cognate substrates (both amino acid and tRNA) and discrimination against non-productive binding of non-cognate analogues. To ensure that the correct amino acids are selected, aaRSs have either evolved highly specific structural motifs at the catalytic active site that can discriminate between amino acids and/or acquired an extra editing domain that has the ability to remove the misactivated amino acids [2–4].
Subclass Ia aaRSs contain three closely related enzymes, LeuRS, IleRS and ValRS (leucyl-, isoleucyl-, and valyl-tRNA synthetase respectively). All of them are large monomers (approx. 100 kDa) and have an unusually large insertion, CP1 (connective polypeptide 1), in the aminoacylation catalytic domain which adopts a typical RF (Rossmann fold) [5–8]. Subclass Ia aaRSs can aminoacylate other structurally similar, nearly cognate amino acids, in addition to their cognate amino acids, which poses a fundamental challenge to the molecular recognition mechanism of these enzymes. Based on biochemical data, Fersht [9] proposed a ‘double-sieve’ (two-step substrate selection) model as the mechanism for amino acid selection and discrimination by IleRS. In this model, amino acids larger than L-Ile are excluded by the aminoacylation site, serving as the ‘coarse sieve’, and smaller ones, such as L-Val, are eliminated by the ‘fine sieve’ at the putative editing site. This model was first visualized in the crystal structure of IleRS [5,6]. The large CP1 insertion was found to be responsible for the editing function and, therefore, is also called the ED (editing domain) [5,10]. The editing active site hydrolyses the misactivated aminoacyl-adenylate (pre-transfer editing) or the mischarged tRNA (post-transfer editing). The two different sieves enable subclass Ia aaRSs to achieve a high specificity in the recognition and selection of the amino acids.
LeuRS can recognize and misactivate a number of nearly cognate amino acids, such as Ile, Met, and norvaline, and transfer all of them to tRNALeu. The mischarged Met-tRNALeu and Ile-tRNALeu are hydrolytically cleaved into Met or Ile and tRNALeu by the ED of LeuRS through the post-transfer editing pathway [11–13]. A pre-transfer editing pathway also exists in which the misactivated aminoacyl-AMP is directly hydrolysed to amino acid and AMP at the editing active site in the presence of tRNALeu by LeuRS [11]. So far, crystal structures of LeuRS from two bacterial species (Thermus thermophilus and Pyrococcus horikoshii) have been reported [7,14]. In addition, the crystal structures of ttLeuRS (T. thermophilus LeuRS) in complex with the pre- and post-transfer editing substrate analogues have been reported, showing that both analogues are bound at the same pocket of the ED and preserve the same mode of adenine recognition [15]. However, no structure of LeuRS in complex with the editing product is available and the mechanism by which LeuRS-ED selectively recognizes and binds Met and Ile remains unknown.
In the present study, we report the crystal structures of ecLeuRS-ED (the ED of Escherichia coli LeuRS) in both apo form and complexes with Met and Ile at 2.0 Å, 2.4 Å, and 3.2 Å resolution respectively. These structures provide new insight into the molecular basis of the editing function of ecLeuRS-ED. Analyses of these structures revealed the precise binding and recognition mode of Met and Ile at the editing active site. Structural comparison also revealed important structural differences between ecLeuRS-ED and ttLeuRS-ED which are implicated in tRNA binding. Based on our structural and previously available biochemical data, we propose that ecLeuRS-ED uses a lock-and-key mechanism to recognize and discriminate the amino acids.
MATERIALS AND METHODS
Protein expression and purification
The gene fragment encoding the ED of ecLeuRS (residues 228–413) was subcloned into the pET3E-His expression plasmid (Novagen). The plasmid was transformed into the E. coli strain BL-21(DE3) and the bacterial cells were grown in Luria–Bertani medium at 37 °C until an attenuance at 600 nm of 0.8 was reached. Protein expression was induced with 0.4 mM isopropylβ-D-thiogalactopyranoside at 30 °C for 4 h. The cells were harvested by centrifugation at 5000 g for 15 min at 4 °C, resuspended in a lysis buffer (50 mM K2HPO4, pH 8.0), and then lysed by sonication on ice. The insoluble cell debris were removed by centrifugation at 15000 g for 30 min at 4 °C. The supernatant was loaded onto a Ni-NTA (Ni2+-nitrilotriacetate) column (Amersham Biosciences) which was first washed with buffer A (lysis buffer supplemented with 25 mM imidazole) and then eluted with buffer B (lysis buffer supplemented with 200 mM imidazole). The elution fractions were pooled together and further purified by gel filtration in the lysis buffer. Finally, the protein sample was dialysed against a buffer containing 5 mM K2HPO4 (pH 9.0) and 5 mM dithiothreitol, and concentrated to approx. 30 mg/ml by centrifugation at 5000 g for approx. 4 h at 4 °C for further structural studies. The purity of the protein was analysed by SDS/14% PAGE, which showed a single band with a molecular mass of 22 kDa. The purity and homogeneity of the protein were also confirmed by the dynamic light scattering results.
Crystallization, X-ray diffraction data collection and structure determination
Crystallization of ecLeuRS-ED was performed using the hanging drop vapour diffusion method, by mixing 2 μl of protein solution (30 mg/ml) and 2 μl of crystallization solution [2.0 M (NH4)2SO4] equilibrated against 500 μl of the crystallization solution at 20 °C. Single crystals of the apo form ecLeuRS-ED were grown to a maximum size of 0.2 mm×0.2 mm×1.0 mm in 3 days and could diffract X-ray to approx. 2.0 Å resolution. Crystals of the Met and Ile complexes were prepared by either soaking the apo form crystals of ecLeuRS-ED in the crystallization solution containing the amino acid (100 mM L-Met or L-Ile) for 3–7 days or by co-crystallization in the presence of 20 mM substrate in the protein solution. Attempts to obtain crystals of ecLeuRS-ED in complexes with L-Leu, L-Thr, L-Val, and L-Ala by both soaking and co-crystallization were unsuccessful, resulting in no electron density for the amino acid at the substrate-binding pocket. Diffraction data were collected from flash-cooled crystals cryoprotected with the crystallization solution containing 25% (v/v) glycerol at −150 °C on an in-house R-Axis IV++ image-plate detector equipped with a Rigaku rotating-anode generator. Data processing and scaling were performed using the Crystal-Clear suite [16]. Crystals of ecLeuRS-ED in both apo form and substrate-bound complexes are of space group P6322 containing two molecules per asymmetric unit with a solvent content of approx. 55%. A summary of the diffraction data used for structure determination is given in Table 1.
Table 1. Summary of diffraction data and structure refinement statistics.
Numbers in parentheses refer to the highest resolution shell. Rmerge=||I0| − 〈I〉|/〈I〉. R factor=||Fo| − |Fc||/|Fo|. RMS, root mean square.
| Apo | L-Met | L-Ile | |
|---|---|---|---|
| Statistics of diffraction data | |||
| Wavelength (Å) | 1.5418 | 1.5418 | 1.5418 |
| Space group | P6322 | P6322 | P6322 |
| Cell parameters | |||
| a=b (Å) | 111.5 | 111.5 | 112.5 |
| c (Å) | 136.8 | 134.9 | 135.0 |
| Resolution range (Å) | 50.0–2.0 (2.07–2.00) | 50.0–2.4 (2.49–2.40) | 50.0–3.2 (3.37–3.20) |
| No. of observed reflections | 180755 | 123565 | 33885 |
| No. of unique reflections (I/σ>0) | 33890 | 19784 | 7726 |
| Mosaicity | 0.49 | 0.60 | 0.96 |
| Average redundancy | 5.3 (5.5) | 6.2 (6.0) | 4.4 (4.4) |
| I/σ | 7.8 (1.7) | 4.5 (1.4) | 8.7 (2.1) |
| Completeness (%) | 98.6 (98.3) | 98.9 (99.7) | 89.4 (89.4) |
| Rmerge (%) | 7.2 (40.6) | 12.1 (45.2) | 9.1 (41.0) |
| Statistics of refinement and model | |||
| Resolution range (Å) | 20.0–2.0 | 20.0–2.4 | 20.0–3.2 |
| No. of reflections (Fo≥0σ(Fo)) | 33867 | 19758 | 7692 |
| Working set | 32157 | 18751 | 7329 |
| Test set | 1710 | 1007 | 363 |
| R factor (%) | 21.6 (29.3) | 24.1 (30.4) | 23.2 (28.2) |
| Free R factor (%) | 24.9 (27.8) | 28.4 (31.8) | 28.4 (40.1) |
| No. of residues | 371 | 371 | 371 |
| No. of protein atoms | 2732 | 2712 | 2706 |
| No. of water molecules | 214 | 78 | 0 |
| Average B factor of all atoms (Å2) | 41.0 | 52.6 | 55.1 |
| Protein main-chain atoms | 39.4 | 52.1 | 55.0 |
| Protein side-chain atoms | 42.0 | 53.3 | 55.4 |
| Ligand atoms | 45.6 | 40.2 | |
| RMS bond lengths (Å) | 0.009 | 0.014 | 0.021 |
| RMS bond angles (°) | 1.4 | 1.6 | 1.5 |
| RMS dihedral angles (°) | 26.9 | 26.9 | 27.1 |
| RMS improper angles (°) | 1.57 | 1.51 | 1.29 |
| Luzzati atomic positional error (Å) | 0.26 | 0.48 | 0.40 |
| Ramachandran plot (%) | |||
| Most favoured regions | 92.1 | 90.9 | 91.2 |
| Allowed regions | 7.9 | 9.1 | 8.8 |
The structure of the apo form of ecLeuRS-ED was solved using the molecular replacement method implemented in the program CNS (crystallography & NMR system) [17], using the coordinates of the editing domain of ttLeuRS (Protein Data Bank accession number 1H3N) as the search model [7]. The structures of ecLeuRS in complexes with L-Met and L-Ile were solved by the molecular replacement method using the apo form ecLeuRS-ED structure as the starting model. Structure refinement was performed with CNS using standard protocols (energy minimization, simulated annealing, and B factor refinement) and the model building was carried out using program O [18]. The final structure refinement was performed using the maximum likelihood algorithm implemented in the program REFMAC5 [19]. A bulk solvent correction and a free R factor monitor (calculated with 5% of randomly chosen reflections) were applied throughout the refinement. The stereochemical quality of the structure models during the course of refinement and model building was evaluated with the program PROCHECK [20]. A summary of the structure refinement statistics is given in Table 1.
Docking model of the ecLeuRS-ED–tRNA complex
The docking model of the ecLeuRS-ED–tRNA complex was constructed in three steps, based on the structure of the ttValRS–tRNA complex in which the 3′ end of the tRNA is bound at the editing active site [8] and on the structure of ttLeuRS [7]. ttValRS shares a similar overall structure with that of ttLeuRS in both the RF catalytic domain and the ED. However, the ED is inserted at a different location of the RF domain and is positioned with a different orientation relative to the RF domain in these two enzymes. The ttValRS–tRNA complex was first superimposed onto ttLeuRS, based on the conserved structural elements of the RF domain, to construct a tRNA model covering nt 1–72. To build nt 75 and 76 of the tRNA model at the editing active site, the ttValRS–tRNA complex was superimposed onto the editing domain of ttLeuRS. The nt 73–74 linker region was generated by energy minimization, after fixing the positions and conformations of nt 1–72 and 75–76 of the tRNA model. The assembled tRNA model was further docked to ecLeuRS-ED, based on the super-position of ecLeuRS-ED and ttLeuRS-ED. The docked tRNA model has no steric conflict with any amino acid residue of the protein. The 3′ terminal CCA of the tRNA acceptor arm is well positioned in the editing pocket, without obvious steric conflict with the surrounding residues of the protein and nt A76 is located in a similar position as that of the post-transfer analogue in the ttLeuRS complex [15]. This ecLeuRS-ED–tRNA complex model provides a basis on which to interpret the biochemical data reasonably well.
RESULTS AND DISCUSSION
Structure of ecLeuRS-ED
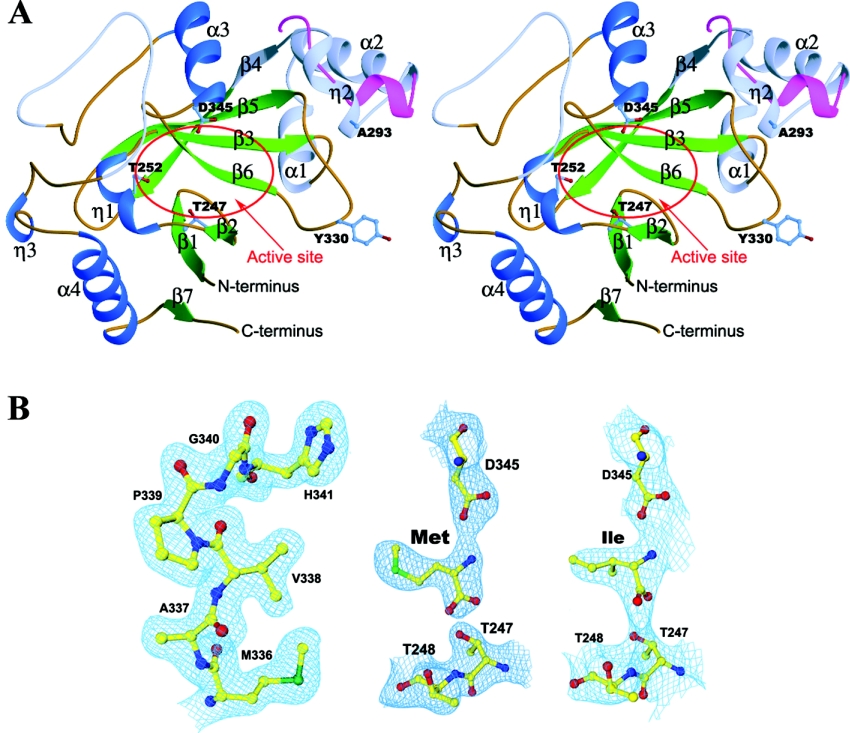
The crystal structure of ecLeuRS-ED has been determined at 2.0 Å resolution using the molecular replacement method and refined to an R factor of 0.216 and a free R factor of 0.249. The crystal structures of ecLeuRS-ED in complexes with L-Met and L-Ile have been solved at 2.4 Å and 3.2 Å resolution and refined to an R factor (a free R factor) of 0.241 (0.284) and 0.232 (0.284) respectively. The overall structure of ecLeuRS-ED is illustrated in Figure 1(A). Figure 1(B) shows the representative SIGMAA-weighted (2Fo−Fc) composite omit maps at the editing active site. The statistics of the structure refinement and the quality of the structure models are listed in Table 1.
Figure 1. Structure of ecLeuRS-ED.
(A) Stereoview of the overall structure of ecLeuRS-ED and its comparison with ttLeuRS-ED. The α-helices, β-strands, and coils are coloured in blue, green and brown respectively. The non-conserved regions are coloured in light blue. The three strictly conserved residues at the editing active site (Thr247, Thr252, and Asp345) are shown with side chains. The major structural difference between ecLeuRS-ED and ttLeuRS-ED (in red) occurs in the Ala293-containing region. (B) Representative SIGMAA-weighted 2Fo−Fc composite omit maps (1.0 σ contour level) at the editing active site. Left panel: apo form ecLeuRS-ED structure at 2.0 Å resolution; middle panel: the Met-bound complex at 2.4 Å resolution; and right panel: the Ile-bound complex at 3.2 Å resolution. The final coordinates of the structures are shown as ball-and-stick models.
ecLeuRS-ED is composed of a globular β-barrel surrounded by four α-helices. The editing active site is located at the edge of the core β-sheet and is exposed to solvent and accessible to substrates (Figure 1A). Although ecLeuRS-ED has a moderate sequence similarity with ttLeuRS-ED (42% identity), they share a very similar overall structure with an R.M.S.D. (root mean square deviation) of 1.25 Å for 174 Cα atoms. The Met- and Ile-bound ecLeuRS-ED complexes represent the product states of the editing reaction. The noncognate amino acids Met and Ile are bound at the same location of the editing active site. Comparisons of the apo structure with the Met- and Ile-bound structures indicate that the binding of Met or Ile did not induce obvious conformational changes in the overall structure and the structure of the editing active site.
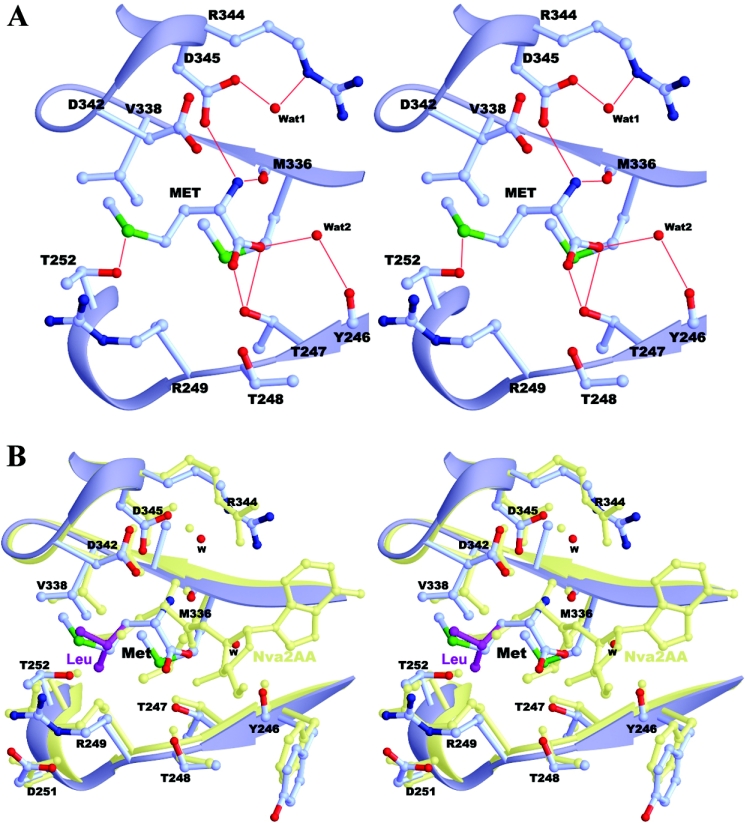
The editing active site of ecLeuRS consists of amino acids from structural elements of β2 (Thr247 and Thr248), η1 (Arg249 and Thr252), β6 (Met336 and Val338), and α3 (His341 and Asp345) (Figures 1A and 2). These residues are highly conserved in LeuRSs across different species (Figure 3A). Structural comparisons of ecLeuRS-ED with ttLeuRS complexed with the pre-transfer analogue [NvaAMS (norvalyl-adenylate sulphamoyl)] and post-transfer analogue {Nva2AA [2′-(L-norvalyl)amino-2′-deoxyadenosine]} [15] show no obvious conformational difference at the editing active site (Figure 2B). In the unliganded ecLeuRS structure, the empty substrate-binding pocket is filled with a number of water molecules which form a hydrogen-bonding network with the surrounding residues of the protein to stabilize the structure of the editing active site. In the substrate-bound complexes, the main chains of the bound Met and Ile take the same position as that of norvaline of the pre-transfer and post-transfer analogues in the ttLeuRS complexes [15]. Specifically, the main-chain amide group of Met forms two hydrogen bonds with the main-chain carbonyl group of Met336 (2.9 Å) and the side-chain Oδ2 atom of Asp345 (2.8 Å), respectively. Its carboxylate oxygen atoms form two hydrogen bonds with the side-chain Oγ1 atom of Thr247 (2.7 and 2.9 Å). The side chain of Met is surrounded by Arg249, Thr252, Val338 and His341, and forms extensive hydrophobic interactions. In particular, the side-chain Sδ atom of Met has a weak hydrogen bond with the Oγ1 atom of Thr252 (3.6 Å). In the Ile-bound complex, the Ile substrate occupies the same position and maintains similar interactions with the surrounding residues as Met. Its main-chain amide group retains the same hydrogen-bonding interactions with Met336 (3.0 Å) and Asp345 (2.8 Å); its carboxylate group is hydrogen-bonded with the side chain of Thr247 (2.8 Å) and the main-chain carbonyl group of Met336 (3.5 Å), and its side chain also has extensive hydrophobic interactions with the surrounding residues. These results suggest that LeuRSs from different species have a very similar and relatively rigid structure at the editing active site and are very likely to use a common mechanism for the editing function.
Figure 2. Structure of the editing active site.
(A) The Met-bound ecLeuRS-ED complex. The hydrogen-bonding interactions between Met and the surrounding residues are indicated with thin red lines. (B) Superposition of ecLeuRS-ED in complexes with Met (in light blue) and docked Leu (in purple) and ttLeuRS in complex with the post-transfer analogue Nva2AA (in light yellow).
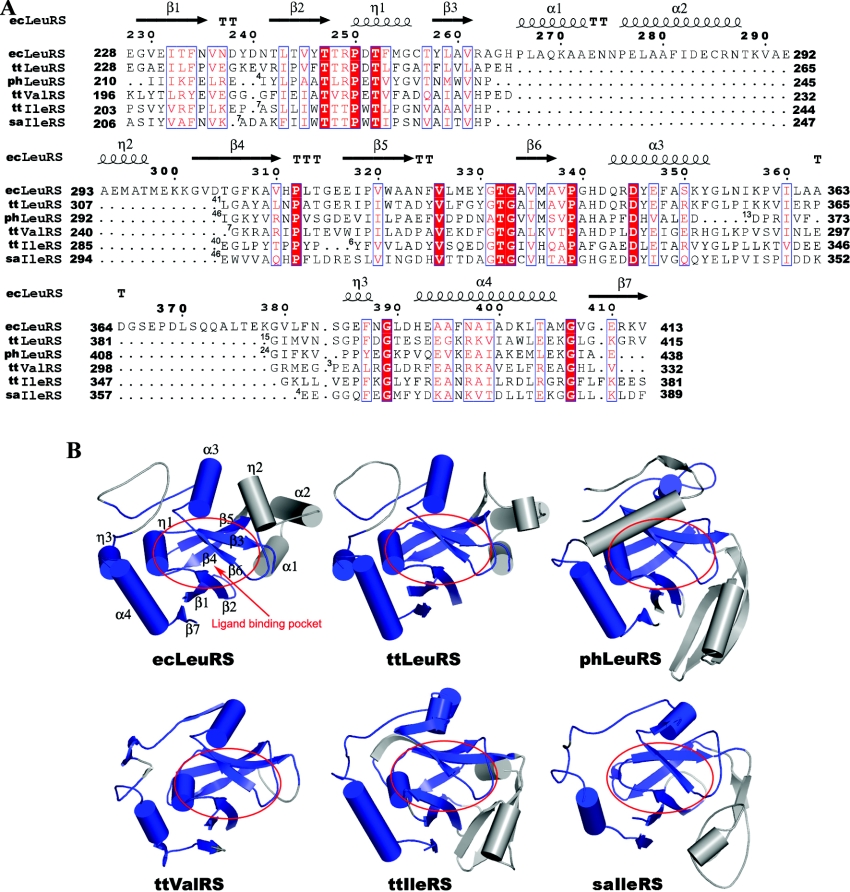
Figure 3. Comparison of the ED of ecLeuRS with other subclass Ia aaRSs with known structures.
(A) Structure-based sequence alignment of the ED between ecLeuRS and other subclass Ia aaRSs. ecLeuRS, ttLeuRS, phLeuRS, ttValRS, ttIleRS, and saIleRS refer to the editing domain of LeuRS in E. coli, T. thermophilus, and P. horikoshii, ValRS in T. thermophilus, and IleRS in T. thermophilus and S. aureus respectively. Invariant residues are highlighted in shaded red boxes and conserved residues in open red boxes. The secondary structure of ecLeuRS-ED is placed on top of the alignment. The numbers of residues omitted in the alignment are indicated by numbers at the beginning of each gap. The alignment was drawn with ESPript [32]. (B) Structural comparison of ecLeuRS-ED with the editing domain of other subclass Ia aaRSs. The conserved and non-conserved regions are coloured in blue and grey, respectively. The ligand binding pocket is indicated by red circles.
Previous biochemical data have shown that two highly conserved residues Thr247 and Thr248, at the editing active site of ecLeuRS, together play an important role in binding either the ribose or phosphate of the substrate, but that mutation of an individual residue has only a minor effect on the editing function [13,21]. Human mitochondrial LeuRS, which contains two alanines at the equivalent positions, has no tRNA-dependent hydrolysis editing function [22]. Indeed, Thr247 and Thr248 have hydrogen-bonding interactions with the ribose and/or phosphate groups of the substrates in the ttLeuRS complexes with the pre-transfer and post-transfer analogues [15]. In the apo form and substrate-bound ecLeuRS-ED structures, the absence of an AMP moiety has created space for a few water molecules which form a network of hydrogen bonds with Tyr246, Thr247, Thr248, and Asp345, indicating that this subsite has hydrophilic properties. Both Thr247 and Thr248 are in the proper positions to interact with the sugar and phosphate moieties of an aminoacyl-adenylate or a mischarged tRNA, suggesting that they might play a similar role in the hydrolysis by assisting a catalytic water molecule to initiate the nucleophilic attack.
Ala293- and Tyr330-containing regions are probably involved in tRNA binding during the editing reaction
Structural comparison indicates that, although most of the protein can be superimposed very well between the two ecLeuRS-ED molecules in the asymmetric unit, there are notable differences in two surface exposed regions containing residues Ala293 and Tyr330 respectively, at or near the editing active site (Figure 1A). These conformational differences exist in both the apo structure and the substrate-bound structures. Residues 292–297 form a short α-helix (η2) at the entrance to the editing active site in molecule A, but are completely disordered in molecule B. The corresponding region adopts a similar short α-helix in ttLeuRS-ED which, however, is positioned much further away from the editing active site (Figure 1A). The other marked conformational difference occurs at Tyr330 at the editing active site. In the structure of ttLeuRS complexed with the pre-transfer analogue, Tyr332 (equivalent to Tyr330 of ecLeuRS) has a disordered side chain [15]. In the structure of ttLeuRS complexed with the post-transfer analogue, Tyr332 is well ordered, its main-chain carbonyl group forms a hydrogen bond with the N6 atom of the nucleotide base of the analogue which mimics adenine 76 of the tRNA, and its side-chain hydroxyl group forms a hydrogen bond with the O5′ atom of the analogue (and presumably will interact with the phosphate of adenine 76 of the tRNA) [15]. In the ecLeuRS-ED structure, Tyr330 in molecule A assumes the same main-chain conformation as that of Tyr332 in ttLeuRS; however, the main-chain phi/psi angles of the Glu329–Tyr330 peptide is flipped by 180° in molecule B. The flexibility of both Ala293- and Tyr330-containing regions is also reflected in their relatively higher B factors and appears to be an inherited property of the ED. Because of the proximity of these regions to the editing active site and their intrinsic flexibilities, it is possible that these regions might play some roles in the editing function or aminoacylation.
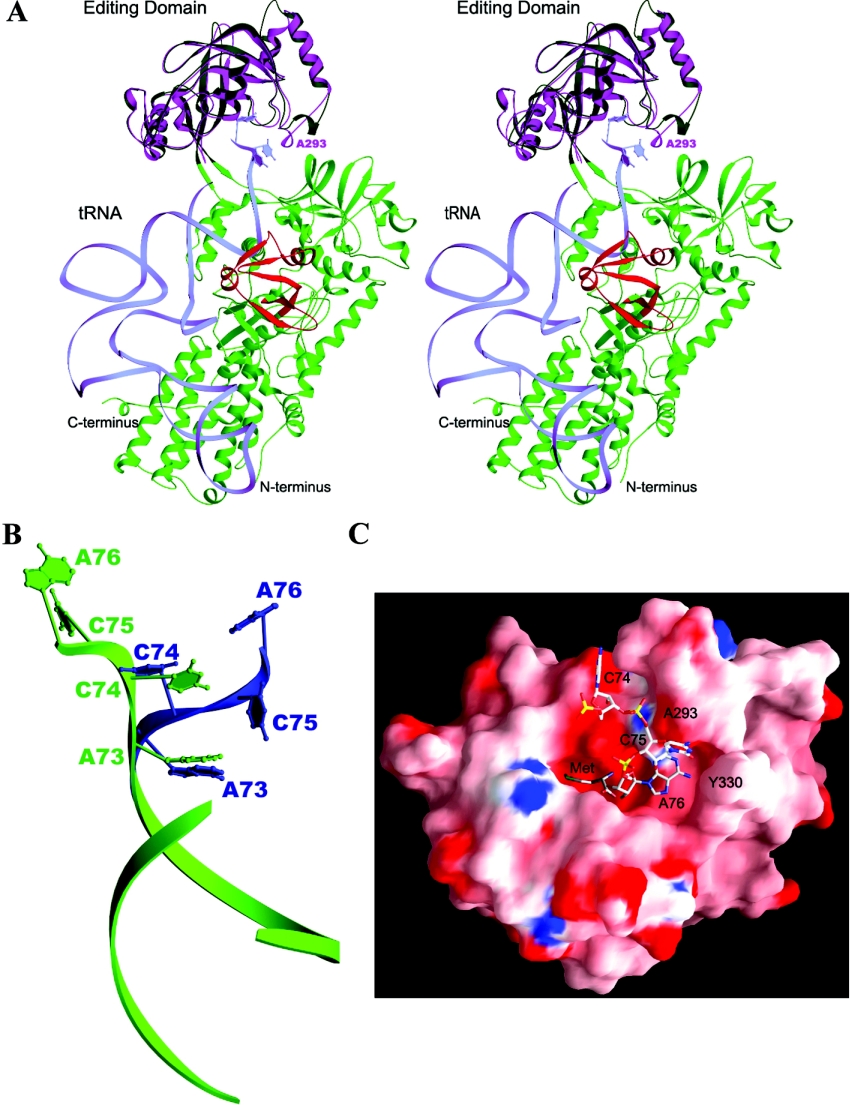
To investigate this possibility, we constructed a docking model of the ecLeuRS-ED–tRNA complex based on the ttValRS–tRNA complex structure and the ttLeuRS structure (Figure 4). In the docking model, the 3′ terminal CCA of the tRNA acceptor arm is well positioned in the editing pocket and adenine 76 takes a similar position as the nucleotide moiety of the post-transfer analogue in the ttLeuRS complex [15]. The Ala293-containing helix interacts with the tRNA, suggesting that this region may help the binding of the 3′ terminus of the tRNA. This suggestion is supported by the biochemical data showing that Glu292 and Ala293 of ecLeuRS play a functional role in the editing function and aminoacylation. ecLeuRS-ED alone can deacylate Ile-minhelix-RNA, whereas ttLeuRS-ED has no deacylation activity for Ile-minhelix-RNA [23]. Mutation of Glu292 to other amino acids (Asp, Gln, Ala, Ser, Lys, and Phe) in ecLeuRS produced mutants which can mischarge tRNA with Ile [24]. Similarly, substitution of Ala293 with Tyr, Gly, Ile, Arg, or Asp affects both the editing function and the aminoacylation activity [25]. The effects on the enzymatic activities of these mutations appear to occur through alteration of the interaction of the Ala293-containing region with the tRNA. Since ecLeuRS lacks the editing function for the pre-transfer substrate [11], the enzyme may have evolved a more effective editing function for the post-transfer mischarged tRNA than other LeuRSs. The intrinsic flexibility of the Ala293-containing region may facilitate the tRNA binding and enhance the efficiency of the editing function.
Figure 4. Docking model of the ecLeuRS-ED–tRNA complex.
(A) The docking model of ttLeuRS in complex with tRNA. The subdomains of ttLeuRS are coloured as the RF domain and the small helical domain in green, ED in dark green, and LeuRS specific domain in red. The structure of ecLeuRS-ED (in purple) is superimposed onto the editing domain of ttLeuRS. tRNA is shown as light blue ribbon and the 3′ terminal CCA are shown with nucleotide bases. (B) Comparison of the 3′ terminal ACCA of the tRNA between the ValRS–tRNA complex (in blue) and the docking model of the ecLeuRS-ED–tRNA complex (in green). (C) Molecular surface of the Met-bound ecLeuRS-ED structure viewing from the entrance to the editing active site. Both the bound Met substrate and the docked 3′ terminal CCA of the tRNA are shown as ball-and-stick models.
In the ecLeuRD-ED–tRNA docking model, the adenine 76 base has potential interactions with the main-chain carbonyl groups of Tyr330 and Leu327 in molecule A. Similar interactions are observed in the ttValRS-tRNA complex, in which the N1 and N6 atoms of the adenine 76 base have hydrogen-bonding interactions with the main-chain amide and carbonyl groups of Glu261 (equivalent to Leu327 of ecLeuRS) and the adenine 76 base has a π-π stacking interaction with the aromatic side chain of Phe264 (equivalent to Tyr330 of ecLeuRS) [8]. In molecule B, due to the flip of the main-chain conformation, the interaction of the adenine 76 base with Tyr330 is replaced by that with the carbonyl group of Glu329. It seems possible that both Glu329 and Tyr330 play a functional role in the recognition of the adenine76 of the tRNA and the two conformations of Tyr330 seen in the crystal structure might represent two intermediate states of the enzyme during the deacylation reaction. Based on these results, we propose that during the editing process the Ala293 containing region might help to induce the binding of the 3′ terminal of the tRNA at the editing active site, via its interactions with the cytosine 75 base, and the Tyr330 containing region might help to position adenine 76 precisely at the editing active site for deacylation via its interactions with the base and sugar-phosphate of adenine 76.
Lock-and-key mechanism for amino acid discrimination
LeuRS can misactivate the nearly cognate amino acids, such as Ile, Met, or norvaline, and the editing active site of LeuRS can recognize the misactivated amino acids and hydrolyse them [11,12,22]. The crystal structures of ecLeuRS-ED, in the apo form and in complexes with Met and Ile, have revealed the detailed structure of the editing active site and the conserved residues involved in interaction and recognition of the substrates. Analyses of these structures and comparisons with other structures of LeuRSs permit us to explore the mechanism to recognize and discriminate the amino acids by ecLeuRS-ED (and possibly other LeuRSs).
Although the Met- and Ile-bound ecLeuRS-ED complexes represent the editing product of the hydrolysis reaction, the amino acid substrates take the same position as the amino acid moiety of the pre-transfer and post-transfer analogues in the ttLeuRS complexes. The main chain of the substrates is recognized by the conserved residues Thr247, Met336, and Asp345. Specifically, the main-chain amide group of the amino acids forms hydrogen bonds with the main-chain carbonyl group of Met336 and the side-chain Oδ2 atom of Asp345, and their carboxylate groups with the side-chain hydroxyl group of Thr247. Similar interactions are conserved in the ttLeuRS and ttIleRS complexes [15,26]. Biochemical data have shown that Asp345 plays an important role in the binding and recognition of the substrates during the editing reaction. Mutation D345A in ecLeuRS (D419A in yeast LeuRS or D347A in ttLeuRS) abolishes the editing function of the enzyme and causes the misaminoacylation of the misactivated Ile and Met to tRNALeu [13,15,27]. Similarly, mutation of the corresponding residues (D342A) in ecIleRS also results in deficiency of the editing function in both pre-transfer and post-transfer hydrolysis of the misactivated aminoacyl-adenylate and mischarged tRNAIle, while substitution of Asp342 with Glu retains a significant level of the editing function [28]. Near to the strictly conserved Asp345, there is a conserved Asp342 which is also located in the vicinity of the bound amino acid. However, mutation of the equivalent residue in ttLeuRS (D344A) had very little effect on the editing activity [15]. In addition, Thr247, together with Thr248, was shown to play an important role in the binding and recognition of the sugar and/or phosphate groups of the adenine moiety [13,15,21]. These results suggest that the main-chain position of the amino acid substrates is fixed and is specifically recognized by Thr247, Met336, and Asp345. Since these residues are strongly conserved in LeuRSs, IleRSs, and ValRSs, they might play a similar role in the recognition and binding of the amino acid substrate during the editing function in all subclass Ia aaRSs.
In the Met- and Ile-bound ecLeuRS-ED structures, the side chain of the amino acid substrates is surrounded by Arg249, Thr252, Met336, Val338, His341, and Asp342. These residues form a narrow and deep pocket to accommodate the side chain of the mischarged amino acids. The geometry of the side-chain binding pocket is precisely configured such that it has an optimal fit for Met and Ile, less fit for Thr, Val, and Ala, but no fit for Leu. Any significant change of the shape and/or property of the pocket would result in functional impairment. In particular, at the bottom of the side chain binding pocket, there is a strictly conserved Thr252 across all LeuRSs, IleRSs, and ValRSs (Figure 3A). In the Met- and Ile-bound ecLeuRS-ED structures, the side chain of Thr252 has extensive hydrophobic interactions with the side chain of both Met and Ile. In addition, the side-chain Oγ atom of Thr252 forms a weak hydrogen bond with the Sδ atom of Met (3.6 Å) which might play some role in the specific recognition of Met. Docking experiments indicate that Leu cannot bind at the substrate-binding pocket because the side-chain γ-methyl group of Leu would sterically hinder the side chains of Arg249, Thr252, Val338, or Asp342, in agreement with the docking results with ttLeuRS [15]. However, if Thr252 is mutated to Ala or Ser, Leu would be able to fit into the pocket without any steric conflict. In contrast, norvaline can very easily be modelled into the pocket without steric conflict with the surrounding residues. Although Thr, Val, or Ala can also be docked into the substrate-binding pocket very easily, they have less interaction with the surrounding residues, consistent with the results of the crystallization experiments indicating that these small residues cannot bind stably at the editing active site. These results provide a molecular basis for the previous biochemical data. Mutation of Thr252 to Glu or Asp in ecLeuRS impairs the editing function, leading to the mischarge of Ile-tRNALeu, while mutation of Thr252 to Gly increases the editing activity but loses the selectivity for amino acids [29]. Similarly, mutant ecLeuRS containing the T252V mutation cannot hydrolyse the cognate Leu-tRNALeu because the side chain of Val still causes steric hindrance with the side chain of Leu [21]. However, mutation of Thr252 to Ser or Ala relaxes the editing specificity and enlarges the size of the binding pocket, resulting in the hydrolysis of the cognate Leu-tRNALeu [13,15,21]. In contrast, substitution of Thr252 with bulkier residues such as Leu, Phe, or Tyr reduces the size of the side chain binding pocket so that Ile and Val cannot bind at the pocket and are stably mischarged to tRNALeu without editing [30,31]. These results indicate that Thr252 plays an important role in the determination of the substrate specificity.
Taking these data together, we propose a lock-and-key mechanism for the recognition and discrimination of the amino acids by LeuRS-ED. Hydrolysis of the mischarged amino acids in the editing reaction will rely on the proper binding and correct recognition of both the main chain and side chain of the substrate. The substrate-binding pocket of ecLeuRS-ED is precisely configured for optimal binding of Met, Ile, or norvaline, but for rejection of the cognate Leu. In general, the editing active site of LeuRS-ED has a very rigid structure (a lock) with a defined stereochemistry and uses a number of conserved residues to precisely bind and recognize the misactivated amino acids and discriminate and reject the cognate amino acid, specifically using Met336, Asp345, and possibly Thr247 for main-chain recognition and Thr252, Met336, and Val338 for side-chain recognition. An amino acid substrate (a key) that fits the substrate-binding pocket in both geometry (size and length) and chemical properties can bind into the pocket and be properly positioned for hydrolysis. Amino acids that cannot fit into the editing site would be rejected because they are either too small to bind tightly at the active site (such as Ala) or are sterically hindered by the surrounding residues (such as Leu).
EDs of subclass Ia aaRSs share a conserved structure
Structural comparisons and structure-based sequences alignments were performed among ecLeuRS, ttLeuRS, phLeuRS (P. horikoshii LeuRS), ttIleRS, saIleRS (Staphylococcus aureus IleRS), and ttValRS whose structures are known (Figure 3). Although most of these enzymes have very low sequence similarity in both the whole protein and the ED, the core structure of the ED is conserved and can easily be aligned among all subclass Ia aaRSs (LeuRS, IleRS, and ValRS) with sequence identity ranging from 45% for ttLeuRS to 23% for saIleRS and R.M.S.D. for equivalent atoms ranging from 1.25 Å to 2.19 Å (see Table S1 at http://www.BiochemJ.org/bj/394/bj3940399add.htm). Overall, the protein fold of the ED consists of three segments, corresponding to three sequential primary sequence fragments, albeit there are insertions of varying lengths between them among different enzymes (Figure 3). The first segment comprises of structural elements β1, β2, β3, and η1, including conserved residues Thr247, Thr248, and Thr252. The second is composed of structural elements of β4, β5, β6, and α3. The third consists of structural elements η3, α4, and β7. These conserved structural elements share a similar secondary structure topology among all subclass Ia aaRSs (Figure 3). Moreover, the residues forming the editing active site or involved in the binding and recognition of the noncognate amino acids are very well conserved among these enzymes, including the strictly conserved Asp (Asp345 in ecLeuRS) and the highly conserved threonine-rich peptide (Thr247–Thr248–Thr252 in ecLeuRS). The strong conservation of the structural fold of the ED and the amino acid residues forming the editing active site among all subclass Ia aaRSs suggests that these enzymes might use a common mechanism for the editing function and that the editing function may have evolved convergently by acquisition of a common editing domain.
Online data
Acknowledgments
This work is supported by National Natural Science Foundation of China grants (30125011 and 30130080), Ministry of Science and Technology of China grants (2002BA711A13, 2004AA235091, 2004CB520801, and 2004CB720102), and Chinese Academy of Sciences grant (KSCX1-SW-17).
References
- 1.Woese C., Olsen G. J., Ibba M., Soll D. Aminoacyl-tRNA synthetases, the genetic code, and the evolutionary process. Microbiol. Mol. Biol. Rev. 2000;64:202–236. doi: 10.1128/mmbr.64.1.202-236.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Freist W., Sternbach H., Cramer F. Isoleucyl-tRNA synthetase from baker's yeast and from Escherichia coli MRE 600. Discrimination of 20 amino acids in aminoacylation of tRNA(Ile)-C-C-A. Eur. J. Biochem. 1988;173:27–34. doi: 10.1111/j.1432-1033.1988.tb13962.x. [DOI] [PubMed] [Google Scholar]
- 3.Carter C. W., Jr Cognition, mechanism, and evolutionary relationships in aminoacyl-tRNA synthetases. Annu. Rev. Biochem. 1993;62:715–748. doi: 10.1146/annurev.bi.62.070193.003435. [DOI] [PubMed] [Google Scholar]
- 4.Giege R., Sissler M., Florentz C. Universal rules and idiosyncratic features in tRNA identity. Nucleic Acids Res. 1998;26:5017–5035. doi: 10.1093/nar/26.22.5017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nureki O., Vassylyev D. G., Tateno M., Shimada A., Nakama T., Fukai S., Konno M., Hendrickson T. L., Schimmel P., Yokoyama S. Enzyme structure with two catalytic sites for double-sieve selection of substrate. Science (Washington, D.C.) 1998;280:578–582. doi: 10.1126/science.280.5363.578. [DOI] [PubMed] [Google Scholar]
- 6.Silvian L. F., Wang J., Steitz T. A. Insights into editing from an Ile-tRNA synthetase structure with tRNAIle and mupirocin. Science (Washington, D.C.) 1999;285:1074–1077. [PubMed] [Google Scholar]
- 7.Cusack S., Yaremchuk A., Tukalo M. The 2 Å crystal structure of leucyl-tRNA synthetase and its complex with a leucyl-adenylate analogue. EMBO J. 2000;19:2351–2361. doi: 10.1093/emboj/19.10.2351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fukai S., Nureki O., Sekine S., Shimada A., Tao J., Vassylyev D. G., Yokoyama S. Structural basis for double-sieve discrimination of L-Val from L-Ile and L-threonine by the complex of tRNAVal and valyl-tRNA synthetase. Cell (Cambridge, Mass.) 2000;103:793–803. doi: 10.1016/s0092-8674(00)00182-3. [DOI] [PubMed] [Google Scholar]
- 9.Fersht A. R. Editing mechanisms in protein synthesis. Rejection of Val by the isoleucyl-tRNA synthetase. Biochemistry. 1977;16:1025–1030. doi: 10.1021/bi00624a034. [DOI] [PubMed] [Google Scholar]
- 10.Lin L., Hale S. P., Schimmel P. Aminoacylation error correction. Nature (London) 1996;384:33–34. doi: 10.1038/384033b0. [DOI] [PubMed] [Google Scholar]
- 11.Englisch S., Englisch U., von der Haar F., Cramer F. The proofreading of hydroxy analogues of leucine and Ile by leucyl-tRNA synthetases from E. coli and yeast. Nucleic Acids Res. 1986;14:7529–7539. doi: 10.1093/nar/14.19.7529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen J. F., Guo N. N., Li T., Wang E. D., Wang Y. L. CP1 domain in Escherichia coli leucyl-tRNA synthetase is crucial for its editing function. Biochemistry. 2000;39:6726–6731. doi: 10.1021/bi000108r. [DOI] [PubMed] [Google Scholar]
- 13.Mursinna R. S., Lincecum T. L., Jr, Martinis S. A. A conserved threonine within Escherichia coli leucyl-tRNA synthetase prevents hydrolytic editing of leucyl-tRNALeu. Biochemistry. 2001;40:5376–5381. doi: 10.1021/bi002915w. [DOI] [PubMed] [Google Scholar]
- 14.Fukunaga R., Yokoyama S. Crystal structure of leucyl-tRNA synthetase from the archaeon Pyrococcus horikoshii reveals a novel editing domain orientation. J. Mol. Biol. 2005;346:57–71. doi: 10.1016/j.jmb.2004.11.060. [DOI] [PubMed] [Google Scholar]
- 15.Lincecum T. L., Jr, Tukalo M., Yaremchuk A., Mursinna R. S., Williams A. M., Sproat B. S., van den Eynde W., Link A., van Calenbergh S., Grotli M., et al. Structural and mechanistic basis of pre- and posttransfer editing by leucyl-tRNA synthetase. Mol. Cell. 2003;11:951–963. doi: 10.1016/s1097-2765(03)00098-4. [DOI] [PubMed] [Google Scholar]
- 16.Pflugrath J. W. The finer things in X-ray diffraction data collection. Acta Crystallogr. Sect. D Biol. Crystallogr. 1999;55:1718–1725. doi: 10.1107/s090744499900935x. [DOI] [PubMed] [Google Scholar]
- 17.Brunger A. T., Adams P. D., Clore G. M., DeLano W. L., Gros P., Grosse-Kunstleve R. W., Jiang J. S., Kuszewski J., Nilges M., Pannu N. S., et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr. Sect. D Biol. Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 18.Jones T. A., Zou J. Y., Cowan S. W., Kjeldgaard M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. Sect. A Found. Crystallogr. 1991;47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 19.Murshudov G. N., Vagin A. A., Dodson E. J. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. Sect D Biol. Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 20.Laskowski R. A. M., Moss D. S., Thornton J. M. PROCHECK: a program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 1993;26:283–291. [Google Scholar]
- 21.Mursinna R. S., Lee K. W., Briggs J. M., Martinis S. A. Molecular dissection of a critical specificity determinant within the amino acid editing domain of leucyl-tRNA synthetase. Biochemistry. 2004;43:155–165. doi: 10.1021/bi034919h. [DOI] [PubMed] [Google Scholar]
- 22.Lue S. W., Kelley S. O. An aminoacyl-tRNA synthetase with a defunct editing site. Biochemistry. 2005;44:3010–3016. doi: 10.1021/bi047901v. [DOI] [PubMed] [Google Scholar]
- 23.Zhao M. W., Zhu B., Hao R., Xu M. G., Eriani G., Wang E. D. Leucyl-tRNA synthetase from the ancestral bacterium Aquifex aeolicus contains relics of synthetase evolution. EMBO J. 2005;24:1430–1439. doi: 10.1038/sj.emboj.7600618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Du X., Wang E. D. Discrimination of tRNALeu isoacceptors by the mutants of Escherichia coli leucyl-tRNA synthetase in editing. Biochemistry. 2002;41:10623–10628. doi: 10.1021/bi026000o. [DOI] [PubMed] [Google Scholar]
- 25.Chen J. F., Li T., Wang E. D., Wang Y. L. Effect of alanine-293 replacement on the activity, ATP binding, and editing of Escherichia coli leucyl-tRNA synthetase. Biochemistry. 2001;40:1144–1149. doi: 10.1021/bi0017226. [DOI] [PubMed] [Google Scholar]
- 26.Fukunaga R., Fukai S., Ishitani R., Nureki O., Yokoyama S. Crystal structures of the CP1 domain from Thermus thermophilus isoleucyl-tRNA synthetase and its complex with L-Val. J. Biol. Chem. 2004;279:8396–8402. doi: 10.1074/jbc.M312830200. [DOI] [PubMed] [Google Scholar]
- 27.Hendrickson T. L., Nomanbhoy T. K., Schimmel P. Errors from selective disruption of the editing center in a tRNA synthetase. Biochemistry. 2000;39:8180–8186. doi: 10.1021/bi0004798. [DOI] [PubMed] [Google Scholar]
- 28.Bishop A. C., Nomanbhoy T. K., Schimmel P. Blocking site-to-site translocation of a misactivated amino acid by mutation of a class I tRNA synthetase. Proc. Natl. Acad. Sci. U.S.A. 2002;99:585–590. doi: 10.1073/pnas.012611299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xu M. G., Li J., Du X., Wang E. D. Groups on the side chain of T252 in Escherichia coli leucyl-tRNA synthetase are important for discrimination of amino acids and cell viability. Biochem. Biophys. Res. Commun. 2004;318:11–16. doi: 10.1016/j.bbrc.2004.03.180. [DOI] [PubMed] [Google Scholar]
- 30.Mursinna R. S., Martinis S. A. Rational design to block amino acid editing of a tRNA synthetase. J. Am. Chem. Soc. 2002;124:7286–7287. doi: 10.1021/ja025879s. [DOI] [PubMed] [Google Scholar]
- 31.Tang Y., Tirrell D. A. Attenuation of the editing activity of the Escherichia coli leucyl-tRNA synthetase allows incorporation of novel amino acids into proteins in vivo. Biochemistry. 2002;41:10635–10645. doi: 10.1021/bi026130x. [DOI] [PubMed] [Google Scholar]
- 32.Gouet P., Courcelle E., Stuart D. I., Metoz F. ESPript: analysis of multiple sequence alignments in PostScript. Bioinformatics. 1999;15:305–308. doi: 10.1093/bioinformatics/15.4.305. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.