Abstract
Failure to detoxify the intermediary metabolite glyoxylate in human hepatocytes underlies the metabolic pathology of two potentially lethal hereditary calcium oxalate kidney stone diseases, PH (primary hyperoxaluria) types 1 and 2. In order to define more clearly the roles of enzymes involved in the metabolism of glyoxylate, we have established singly, doubly and triply transformed CHO (Chinese-hamster ovary) cell lines, expressing all combinations of normal human AGT (alanine:glyoxylate aminotransferase; the enzyme deficient in PH1), GR/HPR (glyoxylate/hydroxypyruvate reductase; the enzyme deficient in PH2), and GO (glycolate oxidase). We have embarked on the preliminary metabolic analysis of these transformants by studying the indirect toxicity of glycolate as a simple measure of the net intracellular production of glyoxylate. Our results show that glycolate is toxic only to those cells expressing GO and that this toxicity is diminished when AGT and/or GR/HPR are expressed in addition to GO. This finding indicates that we have been able to reconstruct the glycolate→glyoxylate, glyoxylate→glycine, and glyoxylate→glycolate metabolic pathways, catalysed by GO, AGT, and GR/HPR respectively, in cells that do not normally express them. These results are compatible with the findings in PH1 and PH2, in which AGT and GR/HPR deficiencies lead to increased oxalate synthesis, due to the failure to detoxify its immediate precursor glyoxylate. These CHO cell transformants have a potential use as a cell-based bioassay for screening small molecules that stabilize AGT or GR/HPR and might have use in the treatment of PH1 or PH2.
Keywords: alanine:glyoxylate aminotransferase, glycolate oxidase, glyoxylate reductase, hydroxypyruvate reductase, kidney stone, oxalate metabolism
Abbreviations: AGT, alanine:glyoxylate aminotransferase; CHO, Chinese-hamster ovary; GO, glycolate oxidase; GR/HPR, glyoxylate/hydroxypyruvate reductase; LDH, L-lactate dehydrogenase; PH, primary hyperoxaluria; PTS1, peroxisomal targeting sequences type 1
INTRODUCTION
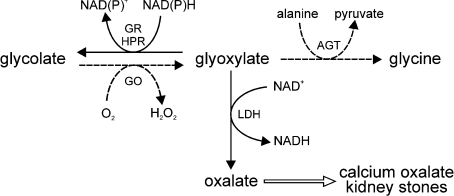
In mammals, oxalate is a metabolic end-product of no known use. In fact, too much oxalate is detrimental because of the low solubility of its calcium salt, which can readily crystallize out as calculi in the kidney and urinary tract [1]. The most important precursor of oxalate in humans is the intermediary metabolite glyoxylate, which is itself synthesized from glycolate catalysed by the liver-specific [2] peroxisomal [3] enzyme GO (glycolate oxidase; EC 1.1.3.15) (Scheme 1). The source of glycolate is unclear, but it most probably arises directly from the diet or from carbohydrate dietary precursors [4]. Glyoxylate is normally metabolized to glycine, a process catalysed by another liver-specific [5] peroxisomal [6] enzyme, AGT (alanine:glyoxylate aminotransferase; EC 2.6.1.44), or back to glycolate catalysed by GR/HPR (glyoxylate/hydroxypyruvate reductase; EC 1.1.1.26/79) (Scheme 1). Although GR/HPR is more widely distributed than GO or AGT, it is found in the liver at much higher levels than in other organs/tissues [7,8]. In human liver, GR/HPR is found in both cytosol and mitochondria [9]. Any glyoxylate that is not reduced back to glycolate or transaminated to glycine will be oxidized to oxalate, a reaction catalysed by LDH (L-lactate dehydrogenase; EC 1.1.1.27) and occuring mainly in the cytosol (Scheme 1). The best evidence for these pathways, as well as for the pivotal role of glyoxylate, comes from two rare autosomal recessive kidney stone diseases, PH (primary hyperoxaluria) types 1 (MIM 259900) and 2 (MIM 260000), which are caused by deficiencies of AGT [10] and GR/HPR [11] respectively.
Scheme 1. Enzymes involved in glyoxylate metabolism in human hepatocytes.
Solid arrows indicate reactions occurring mainly in the cytosol and dashed arrows indicate those in the peroxisomes of hepatocytes. GR/HPR is also partly mitochondrial.
Although the metabolic pathways leading to oxalate have been investigated for many years, their relative quantitative importance in normal humans and patients with PH has been difficult to ascertain directly due to the absence of suitable model systems. Numerous studies have indicated that most of the oxalate synthesized in the body is made in the liver [12–14]. Certainly, most of the enzymes thought to be involved are either liver (hepatocyte)-specific or are present in high concentrations in the liver (see above). Hepatocyte metabolic pathways are especially difficult to investigate in normal and pathological states, because of the lack of availability of normal and diseased human hepatocytes suitable for study, the difficulty of culturing mammalian hepatocytes in primary culture without loss of hepatocyte-specific functions, significant differences between the properties of primary hepatocytes and transformed hepatocyte-derived cell lines, and numerous stress-related problems associated with mammalian liver organ cultures. Although whole organism metabolic studies are frequently dominated by reactions occurring in the liver, it is often impossible to exclude, or even allow for, the contributions from other organs and tissues.
GO, AGT and GR/HPR are all known to be important determinants of glyoxylate, and hence oxalate, synthesis, but their relative importance, and how this shifts in disease states, is unclear. In an attempt to rectify this, we have set up a model cellular system in which the roles of each enzyme can be determined, not only in isolation, but also in relationship to the others. The system we have developed is based on CHO (Chinese-hamster ovary) cells, which do not normally express GO, AGT or GR/HPR, that have been singly, doubly and triply stably transformed with all possible combinations of these enzymes. Preliminary characterization of these cell lines indicates that we have been able to reconstruct successfully the glycolate→glyoxylate, glyoxylate→glycine, and glyoxylate→glycolate metabolic pathways in cells which would not normally contain them.
EXPERIMENTAL
Expression constructs
Full-length cDNAs of the three genes encoding human AGT, GR/HPR and GO were subcloned into the mammalian expression vectors pcDNA3(+)neo, pcDNA3.1(−)hygro and pcDNA3.1(−)-zeo respectively (Invitrogen). Expression of these plasmids confers resistance to the antibiotics G-418, hygromycin B and zeocin respectively. The construction of the AGT expression construct has been described previously [15]. For the GR/HPR construct, a 1214 bp XhoI/HindIII fragment of GR/HPR cDNA [16] in pTrcHisB (Invitrogen) was directionally ligated into pcDNA3.1-(−)hygro. A similar strategy was employed to sub-clone a 1180 bp BamHI/HindIII fragment of GO cDNA [17] into BamHI/HindIII digested pcDNA3.1(−)zeo.
Cells, cell culture and cell transformation
CHO cells (A.T.C.C. number CCl-61) were used in these studies, because they are a well established tissue culture cell line that were considered unlikely to express liver-specific metabolic enzymes (confirmed by later studies) such as AGT, GO and GR/HPR. CHO cells were cultured at 37 °C under O2/CO2 (19:1) in Ham's F12 medium (Invitrogen) supplemented with fetal calf serum (10%, v/v), L-glutamine (2 mM), penicillin (100 units/ml) and streptomycin (100 μg/ml). When the cells were 70–80% confluent they were trypsinized, counted and re-seeded into 24-well plates at a density of 30000 cells/well. After 24 h incubation at 37 °C, the cells were transfected with one of the three gene constructs described above (i.e. AGT, GO or GR/HPR), using Superfect transfection reagent according to the manufacturer's instructions (Qiagen). After 24 h, the appropriate antibiotic (see Table 1) was added and the cultures were left for 3–7 days to grow. To generate pure (i.e. clonal) stably transformed cell lines, the antibiotic-resistant transformants were trypsinized and seeded at a very low density in 10 cm culture dishes containing complete medium supplemented with the appropriate antibiotic at the selection concentrations (see Table 1) and incubated until isolated single colonies started growing. Cells were washed twice with PBS, and then diluted trypsin-EDTA in PBS was added briefly to loosen the colonies. Single colonies were picked using a sterile micropipette tip and transferred to a fresh culture flask for expansion. If the transformants appeared not to be ‘pure’ following immunofluorescence analysis, the procedure was repeated. At least 12 pure, singly-transformed clones were grown for each construct. Clones were designated as follows: CHO-A (cell lines expressing AGT), CHO-G (cell lines expressing GO), and CHO-H (cell lines expressing GR/HPR). After preliminary characterization, one of each (i.e. CHO-A, CHO-G6, and CHO-H9) was used for further study. The double transformants were made by the same procedure as above, except that the single transformants CHO-A and CHO-G6 were used as the parent cell lines rather than wild-type CHO cells, as follows. CHO-AH and CHO-AG cells were made by transfecting CHO-A cells with GR/HPR and GO respectively, and CHO-GH cells by transfecting CHO-G6 cells with GR/HPR. Selection was as described above, but using the appropriate two antibiotics instead of just one. At least twelve pure doublytransformed clones were grown for each combination of constructs (CHO-AG, CHO-AH, and CHO-GH) and, after preliminary characterization, one of each (i.e. CHO-AG2, CHO-AH8, and CHO-G6H1) was used for further study. The triple transformants (CHO-AGH) were made by the same procedure as above, except that the double transformant CHO-AH8 was used as the parent cell line. CHO-AH8 was transfected with GO, and pure clones were isolated as above, except that all three antibiotics were used. At least twelve pure, triply-transformed clones were isolated, one of which (CHO-AH8G1) was used for further study. For simplicity, the clones selected for further study are referred to as CHO-A, CHO-G, CHO-H, CHO-AG, CHO-AH, CHO-GH, and CHO-AGH throughout the rest of the present study.
Table 1. Characteristics of transformed CHO cells.
G-418 was used at a concentration of 1000 μg/ml for selection and 800 μg/ml for maintenance. Zeocin was used at a concentration of 600 μg/ml for selection and 400 μg/ml for maintenance. Hygromycin B was used at a concentration of 1000 μg/ml for selection and 800 μg/ml for maintenance. The same number of cells for each transformed line was plated out into multiwell plates and cultured for 3 days in medium containing the appropriate antibiotics at their maintenance concentrations (see Experimental section for details). The cells were then harvested and the viable (Trypan Blue excluding) cells counted. The values are expressed as means±S.D. (n=6) and are percentages of control value (i.e. CHO-O cells cultured without additional antibiotics), which was normalized to 100%. The probabilities of significance of the differences between the means of the transformed lines and the control were determined by the Student-Newman-Keul's multiple comparison t test. NS, not significant.
| Cell line | Enzymes expressed by stable transformants | Antibiotics used for selection and maintenance (μg/ml) | Effect on cell division (%) |
|---|---|---|---|
| CHO-O | None (non-transformed cells) | None | 100±13.7 |
| CHO-A | AGT | G-418 (1000/800) | 107.8±5.4 (NS) |
| CHO-G | GO | Zeocin (600/400) | 105.1±5.3 (NS) |
| CHO-H | GR/HPR | Hygromycin B (1000/800) | 67.4±4.7 (P<0.001) |
| CHO-AG | AGT and GO | G-418 and zeocin | 64.3±2.4 (P<0.001) |
| CHO-AH | AGT and GR/HPR | G-418 and hygromycin B | 76.9±1.9 (P<0.005) |
| CHO-GH | GO and GR/HPR | Zeocin and hygromycin B | 75.2±8.9 (P<0.005) |
| CHO-AGH | AGT, GO and GR/HPR | G-418, zeocin and hygromycin B | 48.5±5.6 (P<0.001) |
Protein purification
N-terminally His-tagged AGT [18], GR/HPR [16] and GO [17] were expressed in Escherichia coli and purified using Qiagen Ni-NTA (Ni2+–nitrilotriacetate) spin columns with imidazole according to the manufacturer's recommended procedure (QIAexpressionist™; Qiagen). These constructs were used as positive controls and molecular mass markers for the SDS/PAGE analysis of the transformants (see below).
Immunoblotting
Cells were vortexed in lysis buffer [1% (v/v) Triton X-100, 3 mM EDTA and protease inhibitor cocktail tablets Complete mini; catalogue number 1 836 153; Boehringer Mannheim] before ten cycles of sonication on ice in a Branson Sonifier 450 (VWR Scientific) with the output control set to 7, snap-freezing on dry ice and re-thawing at 37 °C. Human liver sonicates and His-tagged purified proteins were used as positive controls and the Medium or Low-range Rainbow™ markers (Amersham Biosciences) as molecular mass markers. The total protein concentrations of the sonicates were measured using the Bradford reagent kit (Sigma–Aldrich) and aliquots of each sonicate, containing 10 μg of protein, were loaded per lane on SDS/12% PAGE (Protogel; National Diagnostics). Following transfer to Hybond C Extra nylon membranes (Amersham Biosciences), three identical filters were incubated separately with polyclonal rabbit primary antisera, or purified IgG, against human AGT, GO (both at a dilution of 1:20000) and GR/HPR (dilution 1:10000). The filters were washed in PBST [PBS containing 1% (v/v) Tween 20] four times for 5 min each, with a final wash for 15 min at 20 °C, before the addition of peroxidase-conjugated goat-anti rabbit IgG (dilution 1:200000). The signal was detected using an ECL® Advance kit (Amersham Biosciences), autoradiographed and scanned for documentation and band sizing using a Bio-Rad GS-710 calibrated imaging densitometer and Quantity One package (version 4.2.1).
Immunofluorescence microscopy
Approx. 105 cells were seeded into each well of an 8-well chamber glass slide (Lab-Tel®; Nalgene) and grown overnight in complete Ham's F12 medium (500 μl), supplemented with the appropriate antibiotic(s) (see Table 1) at 37 °C under O2/CO2 (19:1). In those experiments in which mitochondrial labelling was required, mitochondria were vitally stained prior to fixation by incubating the cells for 15 min with MitoTracker (Red CMXRos version; Molecular Probes, Invitrogen) in complete Ham's F12 medium following the manufacturer's instructions. Cells were fixed in freshly prepared paraformaldehyde (4%, w/v) followed by permeabilization with 1% Triton X-100 for 15 min at 20 °C. The cells were then processed for double label indirect immunofluorescence microscopy, using various combinations of guinea pig polyclonal anti-human catalase antibodies, rabbit or guinea pig polyclonal anti-human AGT antibodies, rabbit or sheep anti-human GR/HPR antbodies and rabbit anti-human GO antibodies. Secondary antibodies were FITC-conjugated goat anti-rabbit IgG (Sigma–Aldrich), FITC-conjugated donkey anti-rabbit IgG (Vector Laboratories), biotinylated goat anti-rabbit IgG, biotinylated swine anti-rabbit IgG (Dako) and biotinylated donkey anti-sheep IgG (Jackson ImmunoResearch Laboratories) followed by Texas Red–avidin DSC (Vector Laboratories). All incubations were performed at 20 °C for 15 min in PBS containing 3% (w/v) BSA, except when Texas Red–avidin DSC was used, when the incubation buffer was 3% (w/v) BSA in 0.1 M sodium bicarbonate buffer (pH 8.5)/0.15 M NaCl. Glass coverslips (22 mm×60 mm) were mounted over each slide in Vectashield® mounting medium (Vector Laboratories). The fluorescence images were captured at a magnification of 40× or 100× and a resolution of 1024×1024 pixels, using a confocal laser-scanning fluorescence microscope (either the MRC 1024 or the Radiance 2100 Upright Multiphoton, Bio-Rad Laboratories). FITC was visualized using excitation and emission wavelengths of 488 nm and 522 nm respectively. For Texas Red and MitoTracker the wavelengths were 568 nm and 605 nm respectively. The captured images were processed using Adobe Photoshop software.
Catalytic activity
AGT [19], GR/HPR [7] and GO [20] catalytic activities were measured spectrophotometrically as previously described.
Indirect glycolate toxicity
In order to test the effect of glycolate on cell viability, aliquots of 5×104 cells from all eight cell lines (one control and seven stable transformants) were seeded into 24-well culture plates (six replicates per treatment) in complete Ham's F12 medium supplemented with the appropriate antibiotic(s) and cultured at 37 °C for 24 h. Various concentrations of glycolate (glycolic acid in Hepes buffer, pH 7.0) were added to the medium and the cells cultured for a further 48 h. After washing twice with PBS, the cells were trypsinized and the viable cell count obtained using Trypan Blue exclusion.
In order to determine the short-term effect of glycolate on GO expression, 105 CHO-G cells were plated on to 8-chamber slides in complete Ham's F12 medium supplemented with zeocin (400 μg/ml) and cultured at 37 °C for 24 h. Various concentrations of glycolate were added to the medium and the cells cultured for a further 48 h. The cells were then fixed, permeabilized, and immunolabelled for GO, as described above.
In order to determine the long-term effect of glycolate on GO expression, 106 CHO-G cells were plated out into each of four 75 cm2 culture flasks and cultured in complete Ham's F12 medium supplemented with zeocin (400 μg/ml) and containing various concentrations of glycolate. Upon reaching 70–80% confluence (on average every 3 days), the cells were trypsinized and re-seeded at one tenth concentration and cultured in fresh complete Ham's F12 medium supplemented with zeocin (400 μg/ml) and the same concentrations of glycolate as used previously. This procedure was repeated until 14 days had elapsed, after which the cells were trypsinized, and 105 cells were plated out into an eight-well chamber slide and cultured overnight in the complete glycolate-containing medium. The cells were then fixed, permeabilized, and immunolabelled for GO, as described above. In a parallel experiment, the cells were harvested by trypsinization and assayed for GO catalytic activity (see above).
RESULTS
In order to check the characteristics of enzyme expression in the CHO cell transformants, one clone from each cell line was picked and assayed for AGT, GR/HPR and GO catalytic activity, immunoreactivity and subcellular distribution (as described in the Experimental section).
GO, AGT and GR/HPR are expressed at comparable levels in single, double and triple transformants
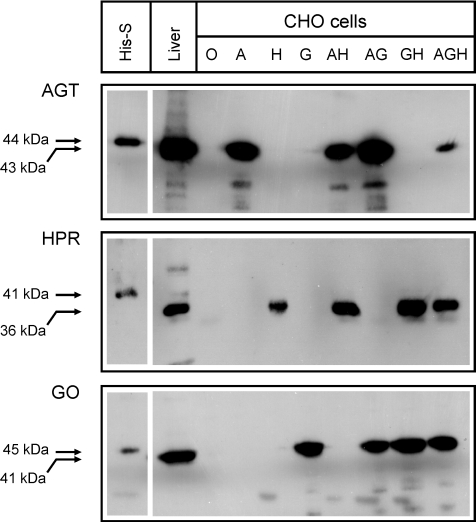
The total levels of expression of immunoreactive GO, AGT and GR/HPR were determined by SDS/PAGE and immunoblotting (Figure 1). The anti-AGT antibodies recognized a single major band of approx. 43 kDa in the CHO-A, CHO-AG, CHO-AH, and CHO-AGH cell lines. The anti-GO antibodies recognized a single major band of approx. 41 kDa in the CHO-G, CHO-AG, CHO-GH, and CHO-AGH cell lines. The anti-GR/HPR antibodies recognized a single major band of approx. 36 kDa in the CHO-H, CHO-AH, CHO-GH, and CHO-AGH cell lines. The sizes of the bands were very similar to those detected in a normal human liver sample electrophoresed at the same time (Figure 1). The estimated molecular masses are very close to the values predicted from the cDNAs (i.e. 43.0 kDa for AGT, 40.9 kDa for GO, and 36.5 kDa for GR/HPR), implying the absence of any significant post-translational modification. Immunoreactive AGT, GO and GR/HPR were not detectable in the untransformed cells (CHO-O; Figure 1). With the exception of AGT expression in the CHO-AGH triple transformant, the overall levels of expression of the enzymes in the transformed cells were similar to those in normal human liver (Figure 1).
Figure 1. SDS/PAGE immunoblot analysis of transformed CHO cells.
Samples (10 μg of protein) of transformed CHO cells (O, CHO-O; A, CHO-A; H, CHO-H; G, CHO-G; AH, CHO-AH; AG, CHO-AG; GH, CHO-GH; AGH, CHO-AGH), normal human liver (10 μg of protein), and purified His-tagged enzymes (His-S, approx. 50 ng of protein) were electrophoresed through an SDS/PAGE gel and immunoblotted against anti-AGT (top panel), anti-GR/HPR (middle panel) or anti-GO (bottom panel) antibodies (see Experimental section for details). The estimated molecular masses of the His-tagged standards (higher values) are indicated, as are the estimated values for liver and transformed CHO cells (lower values).
GO, AGT and GR/HPR are all catalytically active in single, double and triple transformants
In order to check that the liver-specific enzymes were actually working in the CHO cell transformants, their catalytic activities were measured and compared to those found in normal human livers (Table 2). In all the transformants expressing GO and or GR/HPR, the enzyme activities were within or above the reference ranges for normal human liver. However in the transformants expressing AGT, the AGT enzyme activity was within, or nearly within, the normal range only for the single and double transformants (i.e. CHO-A, CHO-AG and CHO-AH). In the triple transformant CHO-AGH, AGT catalytic activity was approx. 10-fold lower.
Table 2. Enzyme specific activities in transformed CHO cells.
Enzyme activities are expressed as means±S.D. (n=5). Units are as follows: AGT, μmol of pyruvate utilized per h per mg of cell protein; GO, nmol of glycolate utilized per min per mg of cell protein; GR/HPR, μmol of NADPH oxidized per min per mg of cell protein. n/d, not detectable.
| Enzyme activity | |||
|---|---|---|---|
| Cell lines | AGT | GO | GR |
| CHO-O | n/d | n/d | n/d |
| CHO-A | 26.8 (±2.0) | n/d | n/d |
| CHO-G | n/d | 154.1 (±8.6) | n/d |
| CHO-H | n/d | n/d | 52.1 (±1.4) |
| CHO-AG | 22.5 (±1.4) | 122.2 (±9.7) | n/d |
| CHO-AH | 18.6 (±2.8) | n/d | 280.4 (±9.0) |
| CHO-GH | n/d | 232.1 (±4.4) | 115.6 (±6.0) |
| CHO-AGH | 2.3 (±0.2) | 110 (±14.6) | 265.1 (±43.0) |
| Reference range for normal human liver | 19.1–47.9 | 13.2–101.6 | 49–213 |
| Limit of detection | 0.24 | 0.19 | 5.5 |
GO, AGT and GR/HPR are correctly localized in single, double and triple transformants
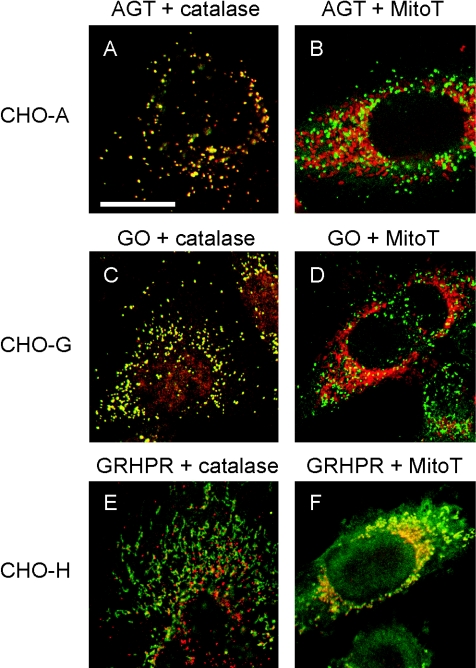
In order to check that the enzymes were compartmentalized within CHO cells and hepatocytes in a similar fashion, their subcellular localization was determined by confocal immunofluorescence microscopy. There was considerable heterogeneity in levels of enzyme expression in different individual cells and this had a knock-on effect on their subcellular localization. For example, although both AGT and GO were clearly peroxisomal in most cells, variable amounts were also found in the cytosol (Figures 2–4). Generally, but not exclusively, the more highly-expressing cells tended to have a greater proportion of their enzyme immunoreactivity in the cytosol than did cells expressing lower levels. The reasons for this are unclear, but may be the result of exceeding the peroxisomal import capacity in the high expressing cells. Similar results were found with a variety of transiently transfected tissue culture cell lines, including CHO, COS-1 and U2OS cells (results not shown). Numerous studies have shown that AGT is peroxisomal, not only in human liver [6], but also when expressed in a variety of tissue culture cell lines [21]. Although much less work has been done on GO, it is also generally considered to be mainly peroxisomal [22].
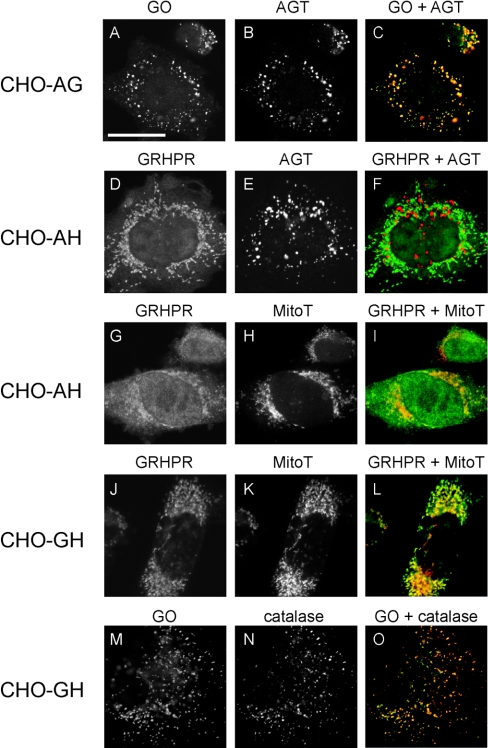
Figure 2. Analysis of the intracellular distribution of AGT, GO and GR/HPR in singly transformed CHO cells by immunofluorescence microscopy.
CHO-A cells were double labelled for AGT and the peroxisomal marker catalase (A), or AGT and the mitochondrial marker MitoTracker (MitoT) (B). CHO-G cells were double labelled for GO and catalase (C), or GO and MitoTracker (D). CHO-H cells were double labelled for GR/HPR and catalase (E), or GR/HPR and MitoTracker (F). Catalase and MitoTracker are shown in red, whereas AGT, GO and GR/HPR are shown in green. Bar=20 μm, with all images being at the same magnification. AGT and GO are mainly peroxisomal (i.e. they co-localize with catalase, but not MitoTracker). GR/HPR is both cytosolic and mitochondrial (i.e. it is both diffuse and co-localized with MitoTracker, but not catalase).
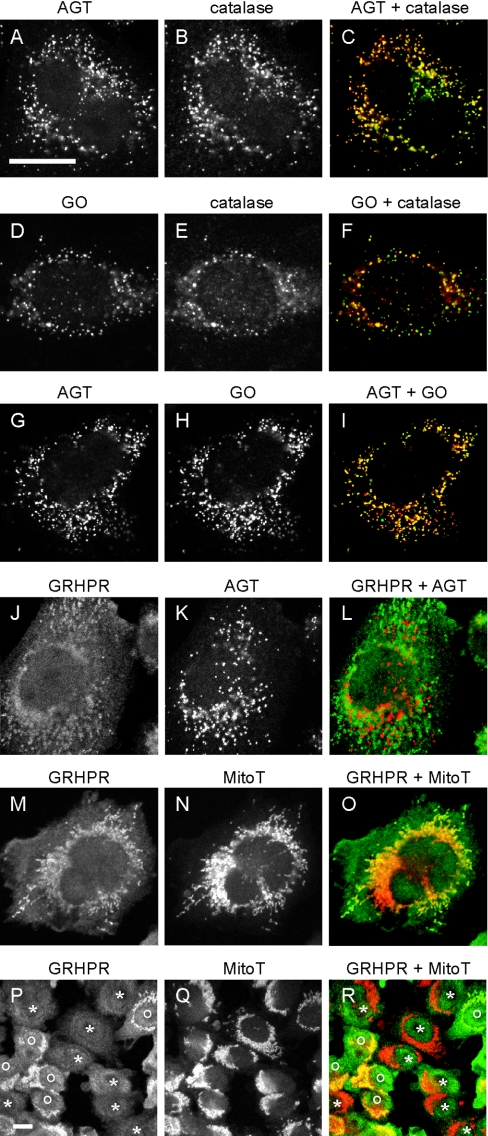
GR/HPR compartmentalization has been only poorly studied. We have previously reported that in human liver it is >90% cytosolic, with the remainder mitochondrial [9]. In the present study, the subcellular distribution of GR/HPR was much more variable than those of AGT and GO. It was found to be both cytosolic and mitochondrial in varying proportions, partly depending on the level of expression and the method of fixation and permeabilization (Figures 2–4). Mitochondrial localization of GR/HPR was much more noticeable when the cells were fixed in methanol/acetic acid rather than paraformaldehyde, or when the Triton X-100 concentration was increased from 1% to 2% (v/v) (results not shown). However, notwithstanding the effects of level of expression and fixation/permeabilization protocol, the variable distribution of GR/HPR was even present in neighbouring cells in the same culture (Figures 4P–4R). Transient expression of GR/HPR in various tissue culture cell lines, such as COS-1, CHO, and U2OS, showed a similar cytosolic and mitochondrial distribution (results not shown).
Figure 4. Analysis of the intracellular distribution of AGT, GO and GR/HPR in triply transformed CHO cells by immunofluorescence microscopy.
CHO-AGH cells were double-labelled for all combinations of test enzymes and markers, except GO+GR/HPR due to secondary antibody cross-reactivity. Cell were labelled as follows: AGT (green) and catalase (red) (A–C), GO (green) and catalase (red) (D–F), AGT (red) and GO (green) (G–I), GR/HPR (green) and AGT (red) (J–L), or GR/HPR (green) and MitoTracker (red) (M–R). Bars=20 μm. (A–O) are the same magnification, as are (P–R). AGT and GO are mainly peroxisomal, whereas GR/HPR is partly cytosolic and partly mitochondrial. In the lower magnification images (P–R): *, cells in which the GR/HPR distribution is entirely diffuse (cytosolic); o, cells in which the GR/HPR distribution is also mitochondrial.
Long-term stability of transformants
The transformants were stable when subjected to continuous culture for a minimum of 3 months, as long as the appropriate antibiotic was included in the culture medium. They were also stable following freezing at −80 °C in 50% (v/v) fetal calf serum or when frozen in liquid nitrogen in 10% (v/v) fetal calf serum.
Multiple transformations increase cell doubling times
There were significant differences in the rates of division of the various transformants. In general, the triple transformants grew less well than the double, which in turn grew less well than the single, and these grew less well than the untransformed cells (Table 1). The CHO-A and CHO-G cell lines grew as well as untransformed CHO-O cells. However, CHO-H, CHO-AG, CHO-AH and CHO-GH cells grew only 65–75% as well, and CHO-AGH cells only 50% as well, as the untransformed cells. These effects were almost certainly due to the presence of the antibiotics, rather than the expression of the various enzymes, as their temporary removal allowed the cells to rapidly recover and decrease their doubling times to that of the untransformed cells (results not shown).
Glycolate is toxic only to cells expressing GO
Unlike glyoxylate, which is highly reactive and toxic in biological systems, glycolate is much less reactive and relatively innocuous [23]. As expected, glycolate showed no signs of toxicity in untransformed CHO-O cells (Figure 5), even when incubated at a concentration of 2 mM for 2 days. On the other hand, glycolate was markedly toxic to single transformants expressing GO (CHO-G cells). Cell survival after 2 days of incubation with 2 mM glycolate was only one third of that found with untransformed cells under the same conditions (Figure 5). The extensive presence of cell debris suggested that the low cell numbers were due to cell death rather than a decrease in the rate of division. Glycolate showed no toxicity to cells not expressing GO, such as CHO-A, CHO-H, and CHO-AH cells (results not shown).
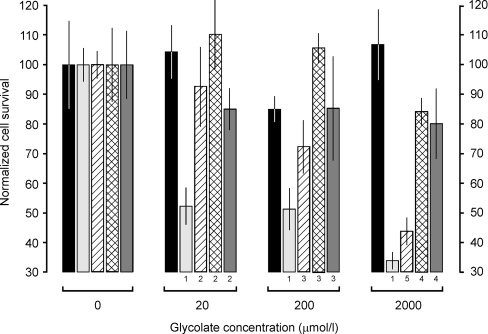
Figure 5. Indirect toxicity of glycolate in transformed CHO cells.
The normalized cell survival is the number of cells surviving after 2 days of culture in the presence of various concentrations of glycolate expressed as a percentage of those surviving in the absence of glycolate (see Experimental section for details). Normalization of the 0 μM glycolate points to 100% was necessary because of the different growth rates of the clones in the different antibiotics (see Table 1). The histograms are coded left to right as follows: black, untransformed CHO-O cells; light grey, CHO-G cells; hatch, CHO-AG cells; crosshatch, CHO-GH cells; dark grey, CHO-AGH cells. The results are expressed as means±S.D. (n=6). The numbers underneath some of the histograms indicate the probabilities of significance (P) of the differences between the means, as determined by the Student–Newman–Keul's multiple comparison t test, as follows: 1, P<0.001 compared with CHO-G (0 μM glycolate); 2, P<0.001 compared with CHO-G (20 μM glycolate); 3, P<0.001 compared with CHO-G (200 μM glycolate); 4, P<0.001 compared with CHO-G (2000 μM glycolate); 5, P<0.01 compared with CHO-G (2000 μM glycolate). The results for CHO-A, CHO-H and CHO-AH were similar to that for CHO-O (results not shown).
Glycolate toxicity is reduced in those GO-expressing cells that co-express AGT and/or GR/HPR
AGT and GR/HPR expression appeared to be able to counter the GO-dependent toxic effects of glycolate. This was demonstrated by the fact that more CHO-AG cells survived incubation with glycolate than did CHO-G cells, especially at the lower concentrations of 20 μM and 200 μM, and more CHO-GH cells survived at all glycolate concentrations (Figure 5). In addition, glycolate did not appear to be toxic to cells that expressed both AGT and GR/HPR in addition to GO (i.e. CHO-AGH cells).
Glycolate toxicity in CHO-G cells is inversely related to the level of GO expression
Although the CHO-G cells, like all the transformants investigated in the present study, were isolated from a single clone, the level of GO expression varied significantly from cell to cell. This heterogeneity allowed a preliminary investigation into whether glycolate toxicity in CHO-G cells was related to the level of GO expression. Preliminary results showed that, when these cells were cultured either for 2 days with up to 2000 μM glycolate, or for 14 days with up to 200 μM glycolate (see the Experimental section for details), only the low GO expressing cells survived, as determined by immunofluorescence microscopy (results not shown). In addition, the specific catalytic activity of GO in CHO-G cells incubated with 20 μM, 200 μM, and 2000 μM glycolate for 14 days was reduced to 37%, 17%, and 11% respectively of the control value.
DISCUSSION
Characterization of single, double and triple CHO cell transformants
In order for a simplified cellular–metabolic model of the human hepatocyte to be useful, the reconstructed metabolic pathways, or at least the enzymes that catalyse these pathways, must have levels of activity, in both absolute and relative terms, similar to those found in human hepatocytes. With the single exception of AGT expression in the triple transformant CHO-AGH, this was indeed the case. The reason for the lower levels of AGT in CHO-AGH was not clear as this cell line was resistant to G-418, the selectable marker for AGT. Preliminary studies on a number of other CHO-AGH clones suggested that the levels of AGT expression were much more similar to those in the CHO-A, CHO-AG and CHO-AH single and double transformants (results not shown). Much of the intercellular heterogeneity within individual cell clones probably results from the fact that the enzymes are transcribed from episomally replicating plasmids rather than being integrated into the nuclear genome.
AGT, GO and GR/HPR expressed in the single, double and triple transformants described in the present study were all correctly localized – AGT and GO to the peroxisomes, and GR/HPR to the cytosol and mitochondria (but see below). In addition, all the enzymes appeared to have folded correctly to become catalytically active. These findings indicate that expression of AGT, GO and GR/HPR in cells that do not normally express them (i.e. CHO cells) has not deprived them of any folding factors (e.g. molecular chaperones), cofactors [e.g. PLP (pyridoxal 5′-phosphate) for AGT and FMN for GO] or any other cell components necessary for their correct function. To our knowledge, this is the first report in the literature of a mammalian cell line (i.e. CHO) that has been stably transformed to express three different enzymes normally located in a completely different cell type (i.e. human hepatocytes).
Intracellular compartmentalization of GR/HPR
The localization of AGT and GO to the peroxisomes in the transformed CHO cells is compatible with all previous data showing that both are imported via the Pex5p pathway due to the presence of C-terminal PTS1s (peroxisomal targeting sequences type 1), KKL in the case of human AGT [21,24] and SKI in the case of GO [22]. PTS1s are not cleaved after import, so it is not surprising that the estimated molecular masses of AGT and GO in human liver and transformed CHO cells were very close to those calculated from the full-length cDNA sequence.
The dual localization of GR/HPR to the cytosol and mitochondria also agrees with the only previous data available for the human enzyme [9], except perhaps that in the present study more appeared to be mitochondrial. However, a more extensive study in the domestic cat showed that glyoxylate reductase, hydroxypyruvate reductase and D-glycerate dehydrogenase catalytic activities, which are all the result of the same enzyme (i.e. GR/HPR), were almost entirely cytosolic [25]. The molecular basis for human GR/HPR targeting has not been studied. Various computer algorithms (PSORT II, Predator, and TargetP) predict that GR/HPR might be at least partly mitochondrial. PSORT II and PLOG also suggest a cytosolic localization. The N-terminus of human GR/HPR (i.e. MRPVRLMKVFVTRRIPAEGRVALARA) has some similarities to mitochondrial targeting sequences [26]. However, definitive data on whether this sequence is actually responsible for targeting GR/HPR to mitochondria is currently unavailable. One surprising finding in the present study is that the estimated molecular mass of GR/HPR in both transformed CHO cells and human liver suggests little or no post-translational modification. This indicates that, unlike most mitochondrial proteins, the mitochondrial targeting sequence of GR/HPR is not cleaved after import [27,28]. Although it is not known why GR/HPR is atypical in this respect, it is interesting to note that a similar finding has been made for mitochondrial LDH, a previously well-documented cytosolic enzyme [29].
Evidence for the reconstruction of glyoxylate metabolic pathways
It is one thing to demonstrate that transformed cells express enzymes that catalyse particular metabolic reactions, but it is another matter to show that the metabolic pathways themselves have been successfully reconstructed in the cells. In order to demonstrate this, we have taken advantage of the fact that glyoxylate is much more reactive and toxic to cells and biological molecules than are any of its immediate metabolites, such as glycolate or glycine (for example see [23]). The observation that glycolate is only toxic to CHO cells if they are expressing GO demonstrates that the glycolate→glyoxylate metabolic pathway has been successfully reconstructed (see Scheme 1 and Figure 5). Similarly, the observation that expression of AGT and GR/HPR decrease the indirect toxicity of glycolate in GO-expressing cells shows that the glyoxylate→glycine and glyoxylate→glycolate metabolic pathways, respectively, have also been reconstructed (Scheme 1 and Figure 5). The ‘apparent’ toxicity of glycolate is likely to be a measure of its conversion to, and the intracellular concentration of, glyoxylate. This is important in human genetic disease, because it is presumed to be the increased levels of intracellular glyoxylate, due to failure to convert it to glycine or back to glycolate, that results in its increased conversion to oxalate and hence all the pathological manifestations of PH1 and PH2. How much of the glyoxylate synthesized from glycolate is eventually oxidized to oxalate in the present system is unclear. Presumably oxidation of glyoxylate would be dependent on the activity of LDH and the concentrations of appropriate cofactors [30]. Although the cells in the present study have not been transformed with LDH, they do express it endogenously, unlike AGT, GO or GR/HPR.
Although the toxic effect of glycolate in cells expressing GO can only be due to the reconstruction of the glycolate→glyoxylate metabolic pathway, the toxic entity could be either of the two products of this reaction (i.e. glyoxylate or H2O2; see Scheme 1). Catalase is present in large quantities in most mammalian cells, including CHO cells as demonstrated by our immunofluorescence microscopy studies; therefore any H2O2 generated would be expected to be rapidly degraded. In addition, there is no reason to believe that the level of H2O2 would be diminished by the co-expression of AGT and/or GR/HPR. The rescuing effect of the latter enzymes argues strongly in favour of glyoxylate being the toxic entity rather than H2O2.
Possible use of multiple CHO cell transformants in the understanding of the molecular aetiology of calcium oxalate kidney stone disease
The relative importance of different metabolic pathways in the generation of toxic intermediates, such as glyoxylate, or metabolic end-products, such as oxalate, in normal and diseased states can be very difficult to determine. One potential solution to this difficulty is to reconstruct the pathways thought to be involved in well-established cell lines that do not normally express them. Thus normal and dysfunctional pathways can be reconstructed by the expression of normal and mutant enzymes respectively without the interference from other peripheral pathways. In the specific context of metabolic calcium oxalate kidney stone disease, this type of cellular reductionist approach has the potential to determine such things as the relative importance of AGT and GR/HPR in the detoxification of glyoxylate, and how this balance is shifted when the subcellular distribution of AGT is altered by a mutation in the AGXT (alanine:glyoxylate aminotransferase) gene [31], and the effect of pharmacological intervention (e.g. pyridoxine [32]) on the metabolic pathway.
The CHO cell transformants described in the present study have already provided some surprising pointers about the relative importance of AGT and GR/HPR in detoxifying glyoxylate. The observation that GR/HPR can protect GO-expressing cells from higher levels of glycolate than can AGT suggests that, in the present system at least, GR/HPR utilizes glyoxylate more efficiently than AGT. This is surprising because PH1 (AGT deficiency) is generally regarded as a more serious disorder than PH2 (GR/HPR deficiency) [1]. The explanation might lie in the availability of co-substrates (i.e. alanine in the case of AGT, or NADH/NADPH in the case of GR/HPR) in the CHO cells rather than in the enzymes themselves.
The next step is to test triple transformants in which either AGT or GR/HPR contains mutations found in PH1 or PH2 respectively. If normal AGT was replaced with AGT[P11L,G170R], which is mistargeted from the peroxisomes to the mitochondria [31,33], then both the efficiency with which it is able to detoxify glyoxylate could be directly determined and the effects of chemical chaperones known to correct its targeting, at least in vitro, could be investigated [34]. Preliminary experiments indeed show that mistargeted mitochondrial AGT is much less effective that normal peroxisomal AGT in protecting cells from the effects of indirect glycolate toxicity.
Figure 3. Analysis of the intracellular distribution of AGT, GO and GR/HPR in doubly transformed CHO cells by immunofluorescence microscopy.
CHO-AG cells were double-labelled for AGT (red) and GO (green) (A–C). CHO-AH cells were double-labelled either for AGT (red) and GR/HPR (green) (D–F), or MitoTracker (red) and GR/HPR (green) (G–I). CHO-GH cells were double-labelled either for MitoTracker (red) and GR/HPR (green) (J–L), or catalase (red) and GO (green) (M–O). Bar=20 μm, with all images being the same magnification. AGT and GO are mainly peroxisomal, co-localizing with catalase and with each other. GR/HPR is partly diffuse (cytosolic) and partly co-localized with MitoTracker (mitochondrial). AGT and GR/HPR do not co-localize.
Acknowledgments
This work was supported by a grant from the Jules Thorn Charitable Trust.
References
- 1.Danpure C. J. Primary hyperoxaluria. In: Scriver C. R., Beaudet A. L., Sly W. S., Valle D., Childs B., Kinzler K. W., Vogelstein B., editors. The Metabolic and Molecular Bases of Inherited Disease. New York: McGraw-Hill; 2001. pp. 3323–3367. [Google Scholar]
- 2.Duley J. A., Holmes R. S. L-α-hydroxyacid oxidase isozymes. Purification and properties. Eur. J. Biochem. 1976;63:163–173. doi: 10.1111/j.1432-1033.1976.tb10219.x. [DOI] [PubMed] [Google Scholar]
- 3.Angermuller S., Leupold C., Volkl A., Fahimi H. D. Electron microscopic cytochemical localization of α-hydroxyacid oxidase in rat liver. Association with the crystalline core and matrix of peroxisomes. Histochemistry. 1986;85:403–409. doi: 10.1007/BF00982670. [DOI] [PubMed] [Google Scholar]
- 4.Bais R., Nairn J. M., Rofe A. M., Conyers R. A. Enzymology of endogenous oxalate formation. Adv. Clin. Enzymol. 1987;5:43–52. [Google Scholar]
- 5.Kamoda N., Minatogawa Y., Nakamura M., Nakanishi J., Okuno E., Kido R. The organ distribution of human alanine-2-oxoglutarate aminotransferase and alanine-glyoxylate aminotransferase. Biochem. Med. 1980;23:25–34. doi: 10.1016/0006-2944(80)90051-4. [DOI] [PubMed] [Google Scholar]
- 6.Cooper P. J., Danpure C. J., Wise P. J., Guttridge K. M. Immunocytochemical localization of human hepatic alanine: glyoxylate aminotransferase in control subjects and patients with primary hyperoxaluria type 1. J. Histochem. Cytochem. 1988;36:1285–1294. doi: 10.1177/36.10.3418107. [DOI] [PubMed] [Google Scholar]
- 7.Giafi C. F., Rumsby G. Kinetic analysis and tissue distribution of human D-glycerate dehydrogenase/glyoxylare reductase and its relevance to the diagnosis of primary hyperoxaluria type 2. Ann. Clin. Biochem. 1998;35:104–109. doi: 10.1177/000456329803500114. [DOI] [PubMed] [Google Scholar]
- 8.Cregeen D. P., Williams E. L., Hulton S. A., Rumsby G. Molecular analysis of the glyoxylate reductase (GRHPR) gene and description of mutations underlying primary hyperoxaluria type 2. Hum. Mutat. 2003;22:497. doi: 10.1002/humu.9200. [DOI] [PubMed] [Google Scholar]
- 9.Mistry J., Danpure C. J., Chalmers R. A. Hepatic D-glycerate dehydrogenase and glyoxylate reductase deficiency in primary hyperoxaluria type 2. Biochem. Soc. Trans. 1988;16:626–627. [Google Scholar]
- 10.Danpure C. J., Jennings P. R. Peroxisomal alanine:glyoxylate aminotransferase deficiency in primary hyperoxaluria type I. FEBS Lett. 1986;201:20–24. doi: 10.1016/0014-5793(86)80563-4. [DOI] [PubMed] [Google Scholar]
- 11.Williams H. E., Smith L. H., Jr L-glyceric aciduria. A new genetic variant of primary hyperoxaluria. N. Engl. J. Med. 1968;278:233–238. doi: 10.1056/NEJM196802012780502. [DOI] [PubMed] [Google Scholar]
- 12.Farinelli M. P., Richardson K. E. Oxalate synthesis from (14C1)glycollate and (14C1)glyoxylate in the hepatectomized rat. Biochim. Biophys. Acta. 1983;757:8–14. doi: 10.1016/0304-4165(83)90146-0. [DOI] [PubMed] [Google Scholar]
- 13.Liao L. L., Richardson K. E. The metabolism of oxalate precursors in isolated perfused rat livers. Arch. Biochem. Biophys. 1972;153:438–448. doi: 10.1016/0003-9861(72)90361-x. [DOI] [PubMed] [Google Scholar]
- 14.James H. M., Williams S. G., Bais R., Rofe A. M., Edwards J. B., Conyers R. A. The metabolic production of oxalate from xylitol: activities of transketolase, transaldolase, fructokinase and aldolase in liver, kidney, brain, heart and muscle in the rat, mouse, guinea pig, rabbit and human. Int. J. Vitam. Nutr. Res. Suppl. 1985;28:29–46. [PubMed] [Google Scholar]
- 15.Knott T. G., Birdsey G. M., Sinclair K. E., Gallagher I. M., Purdue P. E., Danpure C. J. The peroxisomal targeting sequence type 1 receptor, Pex5p, and the peroxisomal import efficiency of alanine:glyoxylate aminotransferase. Biochem. J. 2000;352:409–418. [PMC free article] [PubMed] [Google Scholar]
- 16.Rumsby G., Cregeen D. P. Identification and expression of a cDNA for human hydroxypyruvate/glyoxylate reductase. Biochim. Biophys. Acta. 1999;1446:383–388. doi: 10.1016/s0167-4781(99)00105-0. [DOI] [PubMed] [Google Scholar]
- 17.Williams E., Cregeen D., Rumsby G. Identification and expression of a cDNA for human glycolate oxidase. Biochim. Biophys. Acta. 2000;1493:246–248. doi: 10.1016/s0167-4781(00)00161-5. [DOI] [PubMed] [Google Scholar]
- 18.Lumb M. J., Danpure C. J. Functional synergism between the most common polymorphism in human alanine:glyoxylate aminotransferase and four of the most common disease-causing mutations. J. Biol. Chem. 2000;275:36415–36422. doi: 10.1074/jbc.M006693200. [DOI] [PubMed] [Google Scholar]
- 19.Rumsby G., Weir T., Samuell C. T. A semiautomated alanine:glyoxylate aminotransferase assay for the tissue diagnosis of primary hyperoxaluria type 1. Ann. Clin. Biochem. 1997;34:400–404. doi: 10.1177/000456329703400411. [DOI] [PubMed] [Google Scholar]
- 20.Williams E. L. University of London; 2003. Human liver glycolate oxidase – gene identification and protein studies. PhD Thesis. [Google Scholar]
- 21.Motley A., Lumb M. J., Oatey P. B., Jennings P. R., De Zoysa P. A., Wanders R. J., Tabak H. F., Danpure C. J. Mammalian alanine:glyoxylate aminotransferase 1 is imported into peroxisomes via the PTS1 translocation pathway. Increased degeneracy and context specificity of the mammalian PTS1 motif and implications for the peroxisome-to-mitochondrion mistargeting of AGT in primary hyperoxaluria type 1. J. Cell Biol. 1995;131:95–109. doi: 10.1083/jcb.131.1.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jones J. M., Morrell J. C., Gould S. J. Identification and characterization of HAOX1, HAOX2, and HAOX3, three human peroxisomal 2-hydroxy acid oxidases. J. Biol. Chem. 2000;275:12590–12597. doi: 10.1074/jbc.275.17.12590. [DOI] [PubMed] [Google Scholar]
- 23.Poldelski V., Johnson A., Wright S., Rosa V. D., Zager R. A. Ethylene glycol-mediated tubular injury: identification of critical metabolites and injury pathways. Am. J. Kidney Dis. 2001;38:339–348. doi: 10.1053/ajkd.2001.26099. [DOI] [PubMed] [Google Scholar]
- 24.Takada Y., Kaneko N., Esumi H., Purdue P. E., Danpure C. J. Human peroxisomal L-alanine: glyoxylate aminotransferase. Evolutionary loss of a mitochondrial targeting signal by point mutation of the initiation codon. Biochem. J. 1990;268:517–520. doi: 10.1042/bj2680517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Danpure C. J., Jennings P. R., Mistry J., Chalmers R. A., McKerrell R. E., Blakemore W. F., Heath M. F. Enzymological characterization of a feline analogue of primary hyperoxaluria type 2: a model for the human disease. J. Inherited Metab. Dis. 1989;12:403–414. doi: 10.1007/BF01802035. [DOI] [PubMed] [Google Scholar]
- 26.von Heijne G. Mitochondrial targeting sequences may form amphiphilic helices. EMBO J. 1986;5:1335–1342. doi: 10.1002/j.1460-2075.1986.tb04364.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ramage L., Junne T., Hahne K., Lithgow T., Schatz G. Functional cooperation of mitochondrial protein import receptors in yeast. EMBO J. 1993;12:4115–4123. doi: 10.1002/j.1460-2075.1993.tb06095.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Neupert W. Protein import into mitochondria. Annu. Rev. Biochem. 1997;66:863–917. doi: 10.1146/annurev.biochem.66.1.863. [DOI] [PubMed] [Google Scholar]
- 29.Brooks G. A., Dubouchaud H., Brown M., Sicurello J. P., Butz C. E. Role of mitochondrial lactate dehydrogenase and lactate oxidation in the intracellular lactate shuttle. Proc. Natl. Acad. Sci. U.S.A. 1999;96:1129–1134. doi: 10.1073/pnas.96.3.1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mdluli K., Booth M. P., Brady R. L., Rumsby G. A preliminary account of the properties of recombinant human glyoxylate reductase (GRHPR), LDHA and LDHB with glyoxylate, and their potential roles in its metabolism. Biochim. Biophys. Acta. 2005;1753:209–216. doi: 10.1016/j.bbapap.2005.08.004. [DOI] [PubMed] [Google Scholar]
- 31.Purdue P. E., Takada Y., Danpure C. J. Identification of mutations associated with peroxisome-to-mitochondrion mistargeting of alanine/glyoxylate aminotransferase in primary hyperoxaluria type 1. J. Cell Biol. 1990;111:2341–2351. doi: 10.1083/jcb.111.6.2341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Monico C. G., Rosetti S., Olson J. B., Milliner D. S. Pyridoxine effect in type 1 primary hyperoxaluria is associated with the commonest mutant allele. Kidney Int. 2005;67:1704–1709. doi: 10.1111/j.1523-1755.2005.00267.x. [DOI] [PubMed] [Google Scholar]
- 33.Danpure C. J., Cooper P. J., Wise P. J., Jennings P. R. An enzyme trafficking defect in two patients with primary hyperoxaluria type 1: peroxisomal alanine/glyoxylate aminotransferase rerouted to mitochondria. J. Cell Biol. 1989;108:1345–1352. doi: 10.1083/jcb.108.4.1345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lumb M. J., Birdsey G. M., Danpure C. J. Correction of an enzyme trafficking defect in hereditary kidney stone disease in vitro. Biochem. J. 2003;374:79–87. doi: 10.1042/BJ20030371. [DOI] [PMC free article] [PubMed] [Google Scholar]